Abstract
Retinoic acid-inducible gene I (RIG-I) recognizes RNA virus-derived nucleic acids, which leads to the production of type I interferon (IFN) in most cell types. Tight regulation of RIG-I activity is important to prevent ultra-immune responses. In this study, we identified an ARF-like (ARL) family member, ARL16, as a protein that interacts with RIG-I. Overexpression of ARL16, but not its homologous proteins ARL1 and ARF1, inhibited RIG-I-mediated downstream signaling and antiviral activity. Knockdown of endogenous ARL16 by RNAi potentiated Sendai virus-induced IFN-β expression and vesicular stomatitis virus replication. ARL16 interacted with the C-terminal domain (CTD) of RIG-I to suppress the association between RIG-I and RNA. ARL16 (T37N) and ARL16Δ45–54, which were restricted to the GTP-disassociated form, did not interact with RIG-I and also lost the inhibitory function. Furthermore, we suggest that endogenous ARL16 changes to GTP binding status upon viral infection and binds with the RIG-I CTD to negatively control its signaling activity. These findings suggested a novel innate immune function for an ARL family member, and a GTP-dependent model in which RIG-I is regulated.
Keywords: G Proteins, Innate Immunity, Interferon, Signal transduction, Viral Immunology, ARL16, GTP, IFN-β, RIG-I, Antivirus
Introduction
The first line of defense against infections is mediated by innate pattern recognition receptors, which include Toll-like receptors, RIG-I-like receptors (RLRs), NOD-like receptors, and C-type lectin receptors (1). RIG-I (retinoic acid-inducible gene I)2 is the prototype of the RLR family that also includes MDA5 (melanoma differentiation-associated gene 5) and LGP2 (laboratory of genetics and physiology 2). In the cytoplasm, RIG-I and MDA5 detect different species of viral RNAs in various cells, leading to activation of the transcription factors IRF3 and NF-κB, which collaborate to induce type I interferons (IFNs) and inflammatory cytokines to trigger the host antiviral program, whereas LGP2 plays a regulatory role in the signaling pathway of RIG-I and MDA5 (2). RIG-I responds to in vitro-transcribed dsRNA and RNA viruses, including vesicular stomatitis virus (VSV), Newcastle disease virus (NDV), Sendai virus (SV), Japanese encephalitis virus, and hepatitis C virus. It has been shown that RIG-I preferentially recognizes the 5′-triphosphate moiety of viral RNA (3, 4).
RIG-I belongs to the DExD/H box RNA helicase family, and contains two CARD modules at the N terminus, a DexD/H-box helicase domain in the middle, and a C-terminal domain (CTD). The CTD was recently identified as the RNA recognition domain of RIG-I (5, 6). The recognition of ligand RNA by the CTD induces a conformational change and dimerization of RIG-I, and allows the N-terminal CARD to interact with a mitochondrial adaptor, VISA/MAVS/IPS-1/Cardif, which in turn activates TAK1 and TBK1 kinases, leading to activation of NF-κB and IRF-3 and induction of type I IFNs (7, 8, 9–13, 14). Recently the adaptor protein MITA/STING was identified to links virus-sensing receptors to IRF3 activation (15, 16).
To prevent ultra-immune responses, the activity of RIG-I is tightly regulated by several mechanisms (17). The 5′-triphosphate binding C-terminal region of RIG-I was initially referred to as a repressor domain (RD), through which RIG-I controls its basal activity by auto-inhibition (13). RNA ligand binding induces RIG-I structural alteration to expose the CARD to a downstream signaling adaptor (13, 14). Key regulators of the autophagy process, Atg5 and Atg12, associate directly with the CARD domain of RIG-I and this leads to an inactive status (18). Protein modifications are also implied in the regulation of RIG-I activity. E3 ubiquitin ligase TRIM25 and Riplet/REUL conjugate Lys-63-linked ubiquitins independently and positively regulate RIG-I (19–21). It was recently reported that RIG-I binds specifically to unanchored K63-polyubiquitin chains through its tandem CARD domains (22); E3-ubiquitin ligase RNF125 specifies proteosomal degradation of RIG-I (23); and the deubiquitinating enzymes CYLD and USP17 play negative and positive roles respectively in regulation of RIG-I-mediated signaling (24, 25). Other post-translational modifications, such as phosphorylation, ISG15 conjugation and SUMOylation are also reported to associated with the regulation of RIG-I activity (26–31).
In the present study, we identified an ARF-like (ARL) family member, ARL16, as a novel RIG-I regulator. ADP-ribosylation factor (ARF) GTP-binding proteins, which have well-characterized roles in membrane trafficking and cytoskeletal reorganization, are members of the Ras superfamily of GTPases. Data base mining and phylogenetic analysis of the ARF family revealed the presence of three different groups of proteins: ARFs, ARF-like (ARLs), and Secretion associated and Ras-related(SARs), which share the characteristic structural features of the family (32, 33). ARLs were first identified as ARF-related GTP-binding proteins in Drosophila. More than ten genes encoding ARL proteins have been identified in the human genome. Many ARL proteins are highly conserved throughout eukaryotic evolution, indicating that they have important roles; however, the roles of most ARLs, including ARL16, are completely unknown (32).
Our data demonstrate that overexpression of ARL16, but not its homologous proteins ARL1 and ARF1, inhibited RIG-I-mediated downstream signaling and antiviral activity. Knockdown of endogenous ARL16 by RNAi potentiated SV-induced IFN-β expression and VSV replication. ARL16 interacted with the CTD of RIG-I and suppressed the association between RIG-I and RNA. ARL16 (T37N) and ARL16Δ45–54, which were restricted to the GTP-disassociated form, did not interact with RIG-I and also lost the inhibitory function. Furthermore, we suggested that endogenous ARL16 changes to GTP-binding status upon SV infection and binds with the RIG-I CTD to negatively control its signaling activity. These findings suggest that ARL16 functions as an inhibitor of RIG-I in a GTP-dependent manner.
EXPERIMENTAL PROCEDURES
Reagents and Cell Lines
Mouse or rabbit antibodies against Flag and HA epitopes (Sigma-Aldrich), mouse IgG (Sigma-Aldrich), poly(I:C) (Amersham Biosciences), mouse monoclonal anti-GAPDH (California Bioscience), IRDye800-conjugated anti-mouse and anti-rabbit IgG (Rockland Immunochemicals), SV (Hong-Bing Shu, Wuhan University, Wuhan, China), NDV-eGFP (Cheng Wang, Institute of Biochemistry and Cell Biology, Shanghai, China), VSV (Hong-Kui Deng, Peking University, Beijing, China), EMCV (Xin Guo, China Agriculture University, Beijing, China) were obtained from the indicated sources. Mouse anti-ARL16 antibody was prepared with recombinant protein by the Experimental Animal Center, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences. 293T, HeLa, HUVEC, HT1080, and HuH-7 cells were grown in DMEM supplemented with 10% fetal bovine serum. U937 and K562 cells were grown in RPMI 1640 supplemented with 10% fetal bovine serum.
Yeast Two-hybrid Screens
The mixtures of human fetal kidney cDNA and leukocyte cDNA libraries (Clontech) were screened with full-length RIG-I as bait, following protocols recommended by the manufacturer.
Constructs
Mammalian expression plasmids for Flag- or HA-tagged ARL16, ARL1, ARF1, RIG-I, and their mutants were constructed by standard molecular biology techniques. Mammalian expression plasmids for Flag-VISA, -MDA5, -TBK1, -TRIF, -TLR3, and ISRE, NF-κB, and IFN-β promoter luciferase reporter plasmids were provided by Dr. Hong-Bing Shu (Wuhan University, Wuhan, China).
Transfection and Luciferase Assays
293T cells were seeded on 24-well dishes and transfected the next day by standard calcium phosphate precipitation. In the same experiment, we added empty control plasmid to ensure that each transfection received the same amount of total DNA. To normalize for transfection efficiency, 0.05 μg of pRL-TK (Renilla luciferase) reporter plasmid was added to each transfection. Luciferase assays were performed using a dual-specific luciferase assay kit (Promega). Firefly luciferase activity was normalized based on Renilla luciferase activity. All reporter assays were repeated at least three times. Data shown are average values ± S.D. from one representative experiment.
Immunofluorescent Staining
Cells were fixed in ice-cold methanol for 10 min at −20 °C, rehydrated three times with PBS, and blocked in 5% bovine serum albumin/PBS for 10 min. The cells were stained with primary antibody in blocking buffer for 1 h at 37 °C, rinsed with PBS, and stained again with FITC-labeled Affinipure rabbit anti-mouse IgG or Texas Red-labeled Affinipure goat anti-rabbit IgG (Kirkegaard & Perry Laboratories) for 1 h at 37 °C. The cells were then rinsed with PBS containing DAPI and mounted. The cells were observed under an Olympus BX51 immunofluorescence microscope using a ×100 plan objective.
RT-PCR
Total RNA was isolated from 293T cells using TRIzol reagent (Tianwei Co., Beijing, China) and subjected to RT-PCR analysis to measure the expression of IFN-β, RANTES and β-actin. The gene-specific primer sequences were: IFN-β, sense: 5′-CCAACAAGTGTCTCCTCCAA-3′, antisense: 5′-ATAGTCTCATTCCAGCCAGT-3′; RANTES, sense: 5′-CCTCGCTGTCATCCTCATTG-3′, antisense: 5′-TACTCCCGAACCCATTTCTT-3′; β-actin, sense: 5′-ACGTGGACATCCGCAAAGAC-3′, antisense: 5′-CAAGAAAGGGTGTAACGCAACTA-3′.
RNAi Experiments
Double-stranded oligonucleotides corresponding to the target sequences were cloned into the pSuper.retro RNAi plasmid (Oligoengine). In this study, the target sequences for human ARL16 cDNA were 1: 5′-AACAACTTGCAGAAGCATCGG-3′; 2: 5′-ACGGAGGAGATGAAGTCATTA-3′; 3: 5′-AACATCACCACGGCAGAAATC-3′; positive control VISA RNAi target sequence 5′-GTATATCTGCCGCAATTTC-3′.
VSV Plaque Assay
293T cells (∼1 × 105) were transfected with the indicated plasmids for 24 h prior to VSV infection. At 1 h post-infection, cells were washed with PBS, and then fresh medium was added. The supernatant was harvested at the indicated times and used to infect confluent BHK21 cells cultured on 24-well plates. At 1 h post-infection, supernatant was removed and culture medium containing 2% methylcellulose was overlaid. At 60 h post-infection, the overlaid medium was removed; cells were fixed in 0.5% glutaraldehyde for 30 min and stained with 1% crystal violet dissolved in 70% ethanol. Plaques were counted, averaged, and multiplied by the dilution factor to determine viral titer as Pfu/ml. The experiments were repeated three times, and each experiment was performed in duplicate. Data shown are average values ± S.D. from one representative experiment.
Coimmunoprecipitation and Western Blot Analysis
293T cells (1 × 106) were transfected with the indicated plasmids for 20 h. The transfected cells were lysed in 0.5 ml of lysis buffer (20 mm Tris, pH 7.5, 150 mm NaCl, 1% Triton X-100, 1 mm EDTA, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mm phenylmethylsulfonyl fluoride). For each immunoprecipitation, a 0.4-ml aliquot of lysate was incubated with 0.5 μg of the indicated antibody and 25 μl of a 1:1 slurry of protein A-Sepharose (GE Healthcare) for 2 h at 4 °C. The Sepharose beads were washed three times with 1 ml of lysis buffer. The precipitates were analyzed by Western blot with the indicated antibodies and visualized by incubation with IRDye800-conjugated secondary antibodies (diluted 1:10,000) using an Odyssey infrared imaging system (LICOR Inc.).
RNA Pull-down Experiments
Template ssDNA was generated by PCR using plasmid pPOLi-NA(-)-RT (provided by Ying-Fang Liu, Institute of Biological Physics, Chinese Academy of Sciences) as template and T7 primers. Biotinylated 5′-triphosphate RNA was transcribed from the template ssDNA described above using the Riboprobe System-T7 Kit (Promega) and biotin-11-cytosine-5′-triphosphate (Roche, Germany), following protocols recommended by the manufacturer. The DNA template was then removed by DNase I treatment, and RNA was purified using NucleoSpin RNA Clean-up Columns (Macherey-Nagel, Germany). RNA pull-down was performed as described (13). Cytoplasmic extracts were prepared from 3 × 107 HEK293T cells transfected with Flag-RIG-I, Flag-RIG-I-CTD, or Flag-RIG-I-ΔCTD plasmids. The extracts were incubated with biotinylated 5′-triphosphate RNA and subjected to pull-down with streptavidin-agarose beads (Sigma-Aldrich), followed by SDS-PAGE analysis and immunoblotting with anti-Flag antibody.
GTP Overlay Experiments
Escherichia coli cells were transformed with the particular constructs pET30c-ARL16, -ARL16 mutations, -ARL1, or -ARF1, and then incubated with 1 mm isopropyl-β-d-thiogalactopyranoside (IPTG). After 4 h of induction cells were harvested and lysed in Laemmli sample buffer (4% SDS, 20% glycerol, 10% β-mercaptoethanol, 0.004% bromphenol blue, 0.125 m Tris HCl, pH 6.8). The relative amounts of recombinant proteins were estimated by Western blot analysis. The GTP overlay experiment was performed as described (34). Protein lysates containing equivalent amounts of recombinant proteins were separated by 12% SDS-PAGE. The gel was then soaked in 50 mm Tris-HCl (pH 7.5, 20% glycerol) for 30 min and transferred onto nitrocellulose filters using transfer buffer (10 mm NaHCO3, 3 mm Na2CO3, pH 9.8). After transfer, the filters were rinsed twice for 10 min each in binding buffer (50 mm NaH2PO4, pH 7.5, 10 μm MgCl2, 2 mm dithiothreitol, 0.3% Tween 20, 4 μm ATP) and then incubated for 2 h in binding buffer containing 1 μCi/ml [α-32P]GTP (Beijing Furui Bioengineering). The filters were subsequently washed twice for 3 min in binding buffer and exposed to x-ray film (Kodak Rochester).
RESULTS
Identification of ARL16
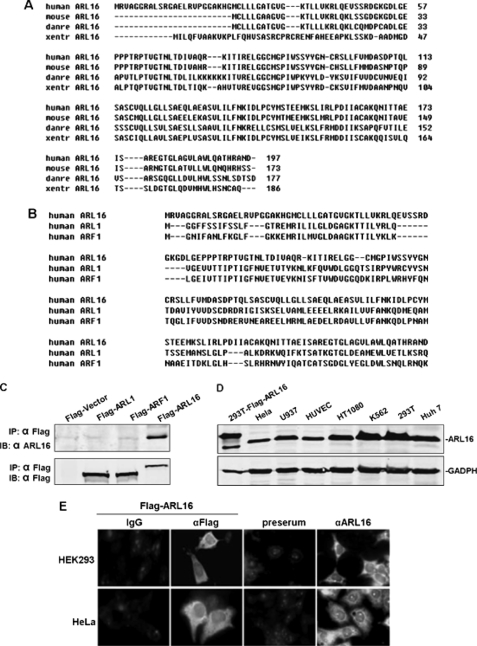
It is well established that RIG-I is an essential cytosolic sensor for the innate immune response to RNA viruses. However, the regulatory mechanisms of RIG-I-mediated signaling have not been adequately characterized. To identify proteins that potentially interact with RIG-I and regulate its signaling, we performed yeast two-hybrid screens of a mixture of human fetal kidney and leukocyte cDNA libraries using full-length RIG-I as bait. These efforts led to the identification of REUL (also known as Riplet/RNF135) as an E3 ligase and stimulator of RIG-I, which we reported in 2009 (21). We also identified a protein of unknown function, ARL family member ARL16 (NM_001177.3). From 22 independent positive clones, 3 clones encoded almost the full-length (Aa 5–197) of ARL16. We amplified full-length ARL16 cDNA from the human fetal kidney cDNA library using PCR. Human ARL16 contains 197 amino acid residues and shares 86.5% sequence identity at the amino acid level with its mouse ortholog, and 61.1% with its zebrafish ortholog (Fig. 1A). This gene belongs to the ARL group of the ARF family. Structural analysis with several programs showed that human ARL16 shares 25% identity and 43.4% similarity with ARF1, and 28.2% identity and 48.1% similarity with ARL1 (Fig. 1B).
FIGURE 1.
Sequence, expression, and location of ARL16. A, alignment of human, mouse, Xenopus, and Drosophila ARL16 amino acid sequences. B, alignment of human ARL16 with ARF1 and ARL1 amino acid sequences. C, mouse anti-ARL16 serum specifically recognizes Flag-ARL16 and endogenous ARL16, but not Flag-tagged ARF1 and ARL1. D, expression of ARL16 in mammalian cells. Lysates of the indicated cells were analyzed by Western blot with anti-ARL16 serum. E, ARL16 localization in HEK293 and HeLa cells. Upper panels: HEK293 cells transfected with an expression plasmid for Flag-tagged ARL16; immunofluorescent staining was performed with anti-Flag (αFlag) or mouse IgG; untransfected HEK293 cells stained with preimmune serum (preserum) or anti-ARL16 serum (αARL16); lower panels: transfected and untransfected HeLa cells stained as in the upper panels. The experiments were repeated three times, and similar results were obtained.
To identify ARL16 expression in mammalian cells at the protein level, we raised mouse antiserum against human ARL16 and tested its specificity (Fig. 1C). Western blot analysis showed that ARL16 was expressed as a 30 kDa protein in all human cell lines examined: HeLa, U937, HUVEC, HT1080, K562, 293T, and Huh7 cells. The endogenous ARL16 protein was slightly smaller than the Flag-tagged ARL16 overexpressed in 293T cells (Fig. 1D). These data suggest that ARL16 is expressed at the protein level.
We next identified the cellular localization of ARL16. ARL family proteins occur in different locations, such as intracellular membranes, cytosol, nucleus, mitochondria, lysosomes, and cytoskeleton (32, 35). Using immunofluorescence, we showed that overexpressed and endogenous ARL16 were localized in the cytoplasm of 293T and HeLa cells (Fig. 1E). It was reported that at position +2 of most ARF family GTPases is a myristoylated glycine that helps to interact with membrane lipids and anchor the GTPase to organelle membranes. However, the sequence of ARL16 does not contain a myristoylation site. These findings suggested that the intracellular targeting of ARL16 differs from those of the myristoylated ARF proteins like ARF1 and ARL1 (32).
ARL16 Inhibits RIG-I Signaling
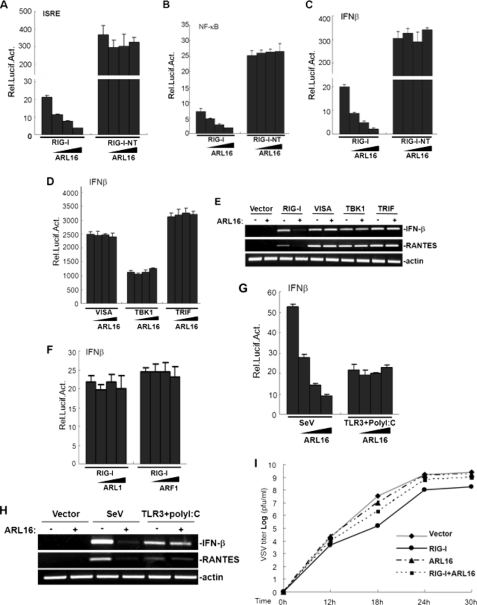
We next determined whether ARL16 is involved in the regulation of RIG-I mediated signaling. In reporter assays, overexpression of ARL16 alone had no apparent effects on the activation of ISRE, NF-κB, and IFN-β promoter, while in cotransfection experiments, RIG-I-mediated induction of ISRE, NF-κB, and IFN-β promoter activity was considerably inhibited by ARL16 expression in a dose-dependent manner (Fig. 2, A–C). Overexpression of the N-terminal CARD domain of RIG-I (RIG-I NT) alone potently activates downstream signaling. However, in these assays, ARL16 did not inhibit the overexpression of RIG-I NT-induced activation of ISRE, NF-κB, and IFN-β promoter (Fig. 3, A–C). We further tested whether ARL16 targets the downstream components in the RIG-I signaling pathway. We found that ARL16 had no effect on VISA and the TBK1-induced IFN-β promoter (Fig. 2D), suggesting that ARL16 specifically inhibits RIG-I, but not downstream effectors induced IFN-β activation.
FIGURE 2.
ARL16 negatively regulates RIG-I signaling. A–C, ARL16 inhibits RIG-I, but not RIG-I-N-terminal CARD- (RIG-I-NT)-mediated activation of ISRE (A), NF-κB (B), and IFN-β (C) promoter in a dose-dependent manner. D, ARL16 has no effect on VISA-, TBK1-, and TRIF-mediated activation of IFN-β promoter. 293T cells (2 × 105) were transfected with the indicated reporter plasmid (50 ng), pRL-TK Renilla luciferase plasmid (50 ng), expression plasmid for RIG-I, RIG-I-NT, VISA, TBK1, or TRIF (100 ng each) together with an empty vector and ARL16 construct (5, 10, and 50 ng). Luciferase assays were performed 24 h after transfection. E, ARL16 inhibits RIG-I-, but not VISA-, TBK1-, and TRIF-mediated gene expression. 293T cells (2 × 105) were transfected with indicated plasmids. Twenty-four hours after transfection, total RNA was isolated and RT-PCR was performed using indicated primers. F, ARL1 and ARF1 do not inhibit RIG-I-mediated activation of IFN-β. Transfection and luciferase assay were performed as in A–D. G, ARL16 suppresses SV-induced, but not TLR3-mediated activation of IFN-β promoter in a dose-dependent manner. Transfection and luciferase assay were performed as in A–D. For poly I:C treatment, cells were also transfected with expression plasmids for TLR3 (100 ng). At 20 h after transfection, cells were infected with SV, treated with polyI:C (2 μg/ml) or left untreated for 12 h before reporter assay. H, ARL16 inhibits SV-induced, but not TLR3-mediated gene expression. The transfection and treatment were as in G, total RNA was isolated and RT-PCR was performed using indicated primers. I, ARL16 inhibits the RIG-I-mediated anti-VSV response. 293T cells (2 × 105) were transfected with indicated plasmids (0.5 μg each). At 30 h after transfection, cells were infected with VSV (MOI = 0.01), and supernatants were harvested at 12, 18, 24, and 30 h post-infection. Supernatants were analyzed for VSV production using standard plaque assays. Plaques were counted and titers calculated as plaque-forming units (pfu/ml).
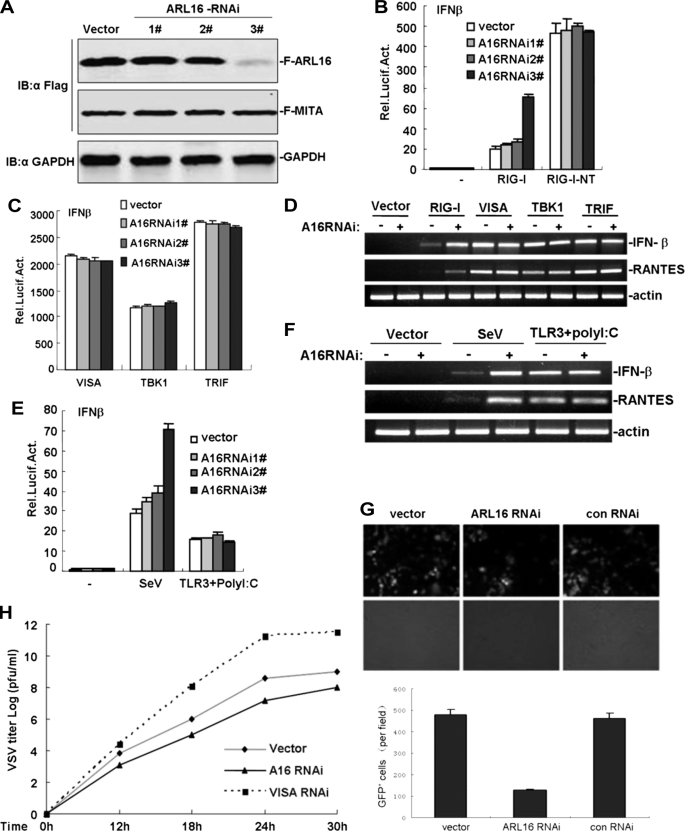
FIGURE 3.
ARL16 siRNA results in an enhanced antiviral response. A, effects of ARL16 RNAi plasmids on the expression of transfected ARL16. 293T cells (2 × 105) were transfected with expression plasmids for Flag-ARL16, with Flag-MITA as control (0.5 μg each), and the indicated RNAi plasmids (1 μg). At 48 h after transfection, cell lysates were analyzed by Western blot with anti-Flag and anti-GAPDH antibodies. B and C, ARL16 RNAi plasmid potentiates RIG-I, but not RIG-I-NT and VISA-, TBK1-, and TRIF-induced activation of IFN-β promoter. 293T cells (2 × 105) were transfected with the indicated reporter plasmid (50 ng), pRL-TK Renilla luciferase plasmid (50 ng), and the indicated ARL16 RNAi (1 μg). At 40 h after transfection, luciferase assays were performed. D, ARL16 RNAi enhances RIG-I, but not and VISA-, TBK1-, and TRIF-mediated gene expression. 293T cells (2 × 105) were transfected with indicated plasmids. Twenty-four hours after transfection, total RNA was isolated and RT-PCR was performed using indicated primers. E, ARL16 RNAi plasmid potentiates SV-induced, but not TLR3-mediated activation of IFN-β promoter. Experiments were carried out as in A and B. For polyI:C treatment, cells were also transfected with expression plasmids for TLR3 (100 ng). At 40 h after transfection, cells were infected with SV, treated with polyI:C (2 μg/ml) or left untreated for 12 h before reporter assay. F, ARL16 RNAi enhances SV-induced, but not TLR3-mediated gene expression. The transfection and treatment were carried out as in E, total RNA was isolated, and RT-PCR was performed using indicated primers. G, ARL16 RNAi enhances NDV-eGFP replication. 293T cells (2 × 105) were transfected with the indicated plasmids. At 20 h after transfection, cells were infected with NDV-eGFP at MOI 0.001. At 40 h after infection, virus replication was determined by GFP expression visualized by fluorescence microscopy. The GFP-positive cells were counted, and the percentage was calculated. H, knockdown of ARL16 potentiates the RIG-I-mediated anti-VSV response. 293T cells (2 × 105) were transfected with indicated plasmids (0.5 μg each). At 30 h after transfection, cells were infected with VSV (MOI = 0.01), and supernatants were harvested at 12, 18, 24, and 30 h post-infection. Supernatants were analyzed for VSV production using standard plaque assays. Plaques were counted and titers calculated as plaque-forming units (pfu/ml).
In RT-PCR experiments, cotransfection of ARL16 apparently inhibited RIG-I, but not VISA- and TBK1-induced expression of the endogenous antiviral and pro-inflammatory genes IFN-β and RANTES (Fig. 2E), suggesting that ARL16 inhibits RIG-I-mediated gene expression.
We also investigated whether proteins homologous to ARL16, ARF1, and ARL1, inhibited RIG-I signaling. Reporter gene assays indicated that ARF1 and ARL1 had no effects on RIG-I-induced IFN-β promoter (Fig. 2F), indicating that RIG-I activity is specifically inhibited by ARL16.
Previously, it was demonstrated that SV/VSV/NDV infections trigger IFN-β production in a RIG-I dependent manner. We further determined whether ARL16 played roles in the cellular antiviral responses mediated by RIG-I. In reporter gene assays, overexpression of ARL16 markedly inhibited SV infection-induced activation of IFN-β promoter in a dose-dependent manner (Fig. 2G). As a control, ARL16 had no effect on TLR3-mediated activation of IFN-β promoter. Consistent with this, ARL16 did not inhibit IFN-β promoter activation induced by TRIF, the key adaptor of the TLR3 signaling pathway (Fig. 2D). We also found that transfection with ARL16 markedly inhibited SV-induced, but not TLR3-mediated expression of endogenous IFN-β and RANTES in RT-PCR experiments (Fig. 2H).
Using plaque assays, we found that cotransfection with ARL16 efficiently suppressed the inhibitory effect on VSV replication mediated by overexpression of RIG-I (Fig. 2I), showing that ARL16 plays an inhibitory role in the cellular antiviral response mediated by RIG-I. These data suggest that ARL16 inhibits RIG-I-mediated signaling and antiviral activity.
Knockdown of ARL16 Potentiates RIG-I Signaling
We next determined whether endogenous ARL16 is involved in RIG-I-mediated signaling. We used three ARL16-RNAi plasmids targeting different sites of human ARL16 mRNA. Transient transfection and Western blot analysis showed that one of these RNAi plasmids (#3) markedly inhibited the expression of transfected ARL16 in 293T cells, whereas the #1 and #2 RNAi plasmids had little effect on ARL16 expression (Fig. 3A). In reporter gene assays, knockdown of ARL16 by #3 potentiated RIG-I- but not RIG-I NT-, VISA-, TBK1-, and TRIF-induced activation of IFN-β promoter, whereas the #1 and #2 RNAi plasmids had little effect (Fig. 3, B and C). Consistent with this, we found that transfection with #3 evidently potentiated RIG-I, but not VISA- and TBK1-induced expression of endogenous IFN-β and RANTES in RT-PCR experiments (Fig. 3D).
We also found that overexpression of ARL16 RNAi plasmid #3 markedly potentiated SV-induced activation of IFN-β promoter, but had no effect on TLR3-mediated IFN-β promoter activation (Fig. 3E). Consistent with this, transfection with ARL16 enhanced SV-induced, but not TLR3-mediated expression of endogenous IFN-β and RANTES in RT-PCR experiments (Fig. 3F).
We further determined whether endogenous ARL16 plays an inhibitory role in the cellular antiviral response. On infection with NDV-eGFP (Newcastle disease virus-enhanced green fluorescent protein), we found that knockdown of endogenous ARL16 in 293T rendered cells remarkably resistant to viral infection and reduced the levels of NDV-eGFP-positive cells (Fig. 3G). Using plaque assays, we found that knockdown of VISA markedly improved VSV replication, whereas ARL16 RNAi had the opposite effect, inhibiting it (Fig. 3H). These results indicated that antiviral activity was stronger when endogenous ARL16 was decreased. We concluded that ARL16 knockdown greatly increases innate cytokine production and antiviral immunity against NDV and VSV infection. Collectively, these data suggest that ARL16 plays an inhibitory role in the efficient cellular antiviral response mediated by RIG-I.
ARL16 Associates with the CTD of RIG-I
Because ARL16 interacted with RIG-I in the yeast two-hybrid system, we further determined whether this interaction occurs in mammalian cells. Coimmunoprecipitation experiments in 293T cells indicated that HA-tagged RIG-I associated with Flag-tagged ARL16 in co-transfection experiments (Fig. 4A). The interaction between ARL16 and RIG-I was specific because ARL1 and ARF1 did not interact with RIG-I under the same conditions (Fig. 4A).
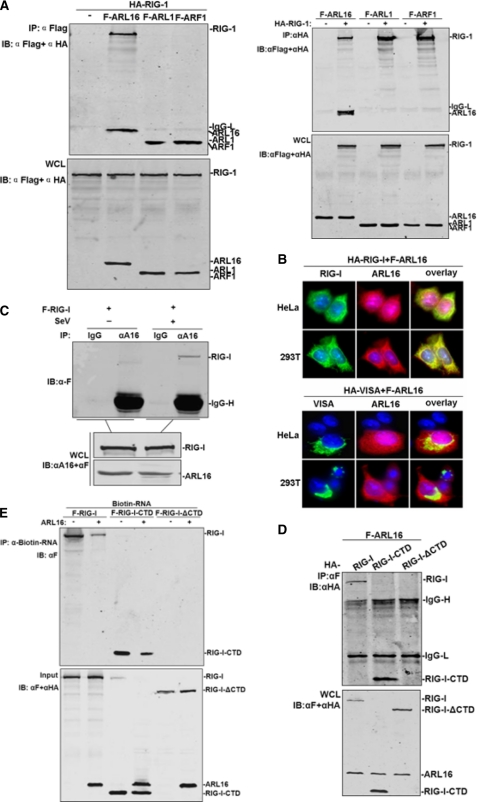
FIGURE 4.
ARL16 associates with the CTD of RIG-I. A, ARL16 but not ARF1 and ARL1 interacts with RIG-I. 293T cells (1 × 106) were transfected with indicated plasmid (5 μg each). Cell lysates were immunoprecipitated (IP) with anti-Flag (αF) or anti-HA (αHA). The immunoprecipitates and whole-cell lysates (WCL) were analyzed by Western blot (IB) with anti-HA and anti-Flag antibody. B, ARL16 colocalizes with RIG-I but not VISA in 293T and HeLa cells. Upper panels: HeLa cells were transfected with Flag-ARL16, HA-RIG-I, or HA-VISA. Immunofluorescent staining was performed with anti-HA (green) and anti-Flag (red); lower panels: 293T cells were transfected and stained as in the upper panels. The experiments were repeated twice, and similar results were obtained. C, endogenous ARL16 interacts with RIG-I upon SV infection. 293T (3 × 106) cells were treated with SV or left untreated for 12 h before lysis. Cell lysates were immunoprecipitated for Western blot by indicated antibodies. D, identification of domains of RIG-I mediating the interaction with ARL16. 293T cells (1 × 106) were transfected with expression plasmids for Flag-ARL16, together with HA-RIG-I or its mutants (5 μg each). Cell lysates were immunoprecipitated with indicated antibodies. The immunoprecipitates were analyzed by Western blots with anti-HA or anti-Flag antibody. E, ARL16 decreases the binding of RIG-I or CTD with RNA. The lysates of cells transfected with Flag-RIG-I, RIG-I-CTD, or RIG-I-ΔCTD were incubated with biotinylated 5′pppRNA transcripts. RNA-protein complexes were pulled down using streptavidin affinity beads. Input and pull-down samples were analyzed by SDS-PAGE and immunoblotting using anti-Flag antibody (one representative of three experiments is shown).
ARL16 was subsequently shown to colocalize with RIG-I in co-transfected 293T and HeLa cells (Fig. 4B). Double immunofluorescent staining showed that overexpressed ARL16 had a similar distribution pattern and overlapped with RIG-I. As a control, there was no overlap between ARL16 and VISA. These observations substantiated the yeast 2-hybrid results and established a direct interaction between RIG-I and ARL16.
To further define this interaction under physiological conditions, and the effects of viral infection on the interaction, we performed immunoprecipitation with anti-ARL16 serum with or without SV infection. The results indicated that overexpressed RIG-I interacted weakly with endogenous ARL16 without SV stimulation; however the association was markedly improved upon SV infection (Fig. 4C). These results suggest that ARL16 associates with RIG-I in a viral infection-inducible manner.
The CTD is the RNA sensor region of RIG-I and is essential for RIG-I activity (6, 27, 44). The crystal structure of the CTD reveals a zinc-binding domain that is structurally related to GDP/GTP exchange factors of Rab-like GTPases (14), indicating that CTD could bind with GTPase family proteins. We found that ARL16 interacted with the CTD-(792–925) independently, as with the full-length RIG-I, whereas ARL16 did not interact with the CTD deletion mutant ΔCTD-(1–792) (Fig. 4D). These data suggested that the CTD of RIG-I is required for interaction with ARL16, consistent with the findings that ARL16 inhibited IFN-β activation induced by full-length RIG-I but not by RIG-I NT.
We next investigated whether the interaction of ARL16 with the CTD interfered with RIG-I sensing and binding to viral RNA. Using the biotin-labeled virus RNA transcript, we did RNA-binding protein pull-down experiments to detect interactions between full-length RIG-I, the CTD and 5′-triphosphate RNA with or without ARL16 expression. We found that co-expression of ARL16 decreased the association between full-length RIG-I, the CTD and RNA transcript; as control, no RNA was detectably associated with ΔCTD (Fig. 4E). These results suggested that ARL16 interacts with the CTD of RIG-I and suppresses the RNA sensing activity of RIG-I.
ARL16 Functions in a GTP-dependent Manner
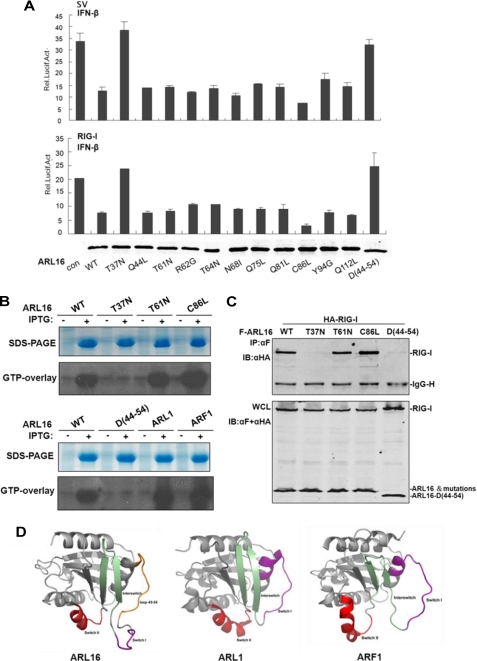
ARF proteins contain highly conserved guanine nucleotide-binding domains involved in GDP/GTP exchange. Much has been learned about how nucleotide-dependent conformational changes in ARFs modulate these cellular activities (36). Because ARL16 is an ARF family member, we tested whether GTP binding affected ARL16 functions. Study of other ARF proteins has identified amino acid sites that are involved in GTP binding (34). We made the analogous point mutations in ARL16 and assessed their functions. Reporter assay results indicated that T37N lost its inhibitory activity on RIG-I-mediated and SV-induced IFN-β promoter activation. As control, the other point mutations Q44L, T61N, R62G, T64N, N68I, Q75L, R81G, C86L, Y94G, and Q112L had inhibitory activity similar to the wild type ARL16 (Fig. 5A). These results suggested that Thr-37 is the key site for ARL16 function.
FIGURE 5.
ARL16 functions in a GTP-dependent manner. A, Thr-37 is the essential site for ARL16 inhibitory function. 293T cells (2 × 105) were transfected with the IFNβ reporter plasmid (50 ng) and pRL-TK Renilla luciferase plasmid (50 ng), together with an empty vector and ARL16 wild type and mutations (100 ng each). Cells were infected with SV for 12 h or cotransfected with plasmid for RIG-I (100 ng). Lower panel: expression of RIG-I wild type (WT) and mutations. B, T37N lost GTP binding activity. Bacterial cells were transformed with indicated expression constructs and induced with IPTG or left untreated. Total cellar proteins were separated by SDS-PAGE, transferred to nitrocellulose filters and after renaturation allowed to bind [α-32P]GTP. Bound [α-32P]GTP was visualized by autoradiography. Upper panels: SDS-PAGE stained with Coomassie brilliant blue to assess protein concentration. C, T37N and deletion-45–54 mutants do not interact with RIG-I. 293T cells (1 × 106) were transfected with expression plasmids for Flag-ARL16 wild-type or mutations with HA-RIG-I (5 μg each). Immunoprecipitation and Western blot were performed as in Fig. 4A. D, three-dimensional models of ARL16 (left), ARL1 (middle), and ARF1 (right) shown as cartoons by the program Pymol. Switch I, Interswitch, and Switch II are indicated in purple, green, and red, respectively. The excess loop 45–54 in the three-dimensional model of ARL16 is colored orange.
Sequence analysis showed that Thr37 is a conserved site in ARF family members. The T31N mutation of ARF1/ARL1 and the T37N mutation of ARL18 lose their GTP binding activity and constitutively bind to GDP (34, 38, 41). We set out to address whether the same occurs with ARL16. We expressed ARL16 in vitro and did GTP overlay experiments. The results showed that, like ARF1 and ARL1, wild type of ARL16, T61N, and C81L expressed in vitro bound to P32-labeled GTP, but T37N did not (Fig. 5B), suggesting that T37N lost its GTP binding activity.
We further determined whether GTP binding played a role in the interaction between ARL16 and RIG-I. Co-immunoprecipitation results showed that in contrast to wild-type ARL16 and mutations of T61N and C81L, T37N did not interact with RIG-I (Fig. 5C), indicating again that Thr37 is the key site for GTP binding of ARL16, and ARL16 associates with and inhibits RIG-I in a GTP-dependent manner.
We also investigated why ARL1 and ARF1 did not interact with and inhibit RIG-I. We constructed a three-dimensional model of ARL16 in the SWISS-MODEL server (42–44) using the crystal structure of ARL1 as a template (PDB ID 1UPT) (45). According to the model, the main structural feature of ARL16 shares a common subunit fold within the ARF family (Fig. 5D). However, we found that ARL16 departs from other ARFs by a unique structural device. ARL16 has an excess 10 amino acid loop region (Aa 45–54) that is close to the functional switch I. Without this loop, ARL16 is very similar in structure to ARF1 (PDB ID 1R8Q) and ARL1 (Fig. 5D). To test the function of the excess loop region, we constructed the deletion mutant ARL16Δ45–54. We found that, in vitro expressed ARL16Δ45–54 lost GTP binding activity. Like ARF1 and ARL1, ARL16Δ45–54 did not interact with RIG-I (Fig. 5C), and also did not inhibit RIG-I-mediated IFN-β promoter activation (Fig. 5A). These results indicated that Aa45–54 is a critical region for ARL16 function.
Functional Model of ARL16 during Infection
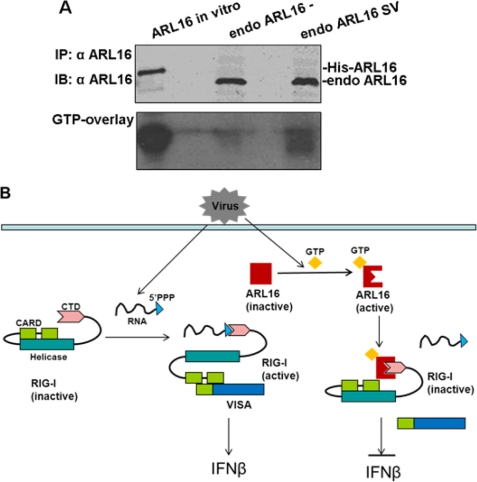
Because ARL16 inhibited RIG-I activity in a GTP-dependent manner, we further assessed the GTP binding status of endogenous ARL16 during infection. GTP overlay results showed that without stimulation, endogenous ARL16 hardly bound to GTP, however, its GTP binding status changed over a 12-h period of SV infection, while the expression of ARL16 was not induced by SV infection (Fig. 6A). These results are consistent with the hypothesis that viral infection induces ARL16 binding with RIG-I (Fig. 4C). Based on the available evidence, we proposed a functional model of ARL16 (Fig. 6B). Normally, ARL16 is localized to the cytosol in its GTP-disassociated and inactive state. Upon viral stimulation, it changes into a GTP binding and active state and interacts with the CTD of RIG-I, sequesters RIG-I to sense viral RNA, and finally inhibits RIG-I-mediated downstream signaling events.
FIGURE 6.
ARL16 working model. A, endogenous ARL16 binds with GTP during infection. 293T cells (1 × 106) were infected with SV or left untreated for 12 h before lysis. Cell lysates were immunoprecipitated with ARL16 antiserum. The immunoprecipitates were separated by SDS-PAGE, transferred to nitrocellulose filters and after renaturation allowed to bind [α-32P]GTP as in Fig. 5C. B, in the absence of viral infection, ARL16 is in its GTP-disassociated and inactive state. With virus stimulation, it changes into the GTP binding active state and sequesters the CTD of RIG-I, finally inhibiting RIG-I-mediated downstream signaling events.
DISCUSSION
As with other cytokine systems, production of type I IFN is a transient process, and can be hazardous to the host if unregulated, resulting in inflammatory and autoimmune diseases. Various molecules, including LGP2, Atg5-Atg12, A20, DUBA, CYLD, RNF125, RBCK1, NLRX1, SIKE, DAK, RNF5, ISG56, OTUB1/OTUB2, TRIAD3A, and Optineurin casein kinase II etc. have been shown to negatively regulate induction of type I IFNs by targeting distinct components of the virus-triggered signaling pathways (18, 23, 25, 29, 46–57). In this study, we identified an ARF-like family member, ARL16, as a novel negative regulator of RIG-I. Overexpression of ARL16 inhibited RIG-I-mediated downstream signaling and antiviral activity. Knockdown of endogenous ARL16 by RNAi potentiated SV-induced IFN-β expression and VSV replication. These results indicated that ARL16 is a physiological suppressor of RIG-I.
The 5′-triphosphate-binding CTD of RIG-I has been shown to interact with other domains of RIG-I, leading to RIG-I auto-inhibition in the absence of viral RNAs. Once the RIG-I CTD senses viral RNAs, conformational changes are induced, leading to RIG-I oligomerization and recruitment of the adaptor protein VISA to activate downstream signaling. The crystal structure of the CTD reveals the possibility of binding to GTPase family members (14). Our finding first identified a “CTD-specific” inhibitor which suppressed its function of sensing viral RNAs. We suggest that in the absence of viral infection, ARL16 exists in a GTP-disassociated and inactive form, and so cannot interact with RIG-I; upon viral infection, ARL16 changes into a GTP-binding and active conformation, that associates with the CTD of RIG-I and sequesters its function. As RIG-I expression is apparently up-regulated upon viral infection (58, 59), our results offer an important mechanism that efficiently prevents overactivation of RIG-I. We also showed that ARL16 expression is not induced by viral infection, so compared with traditional feedback inhibition, ARL16 represents a different type of RIG-I regulation.
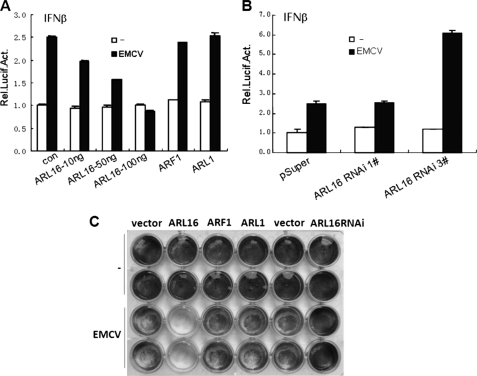
Another cytosolic RNA sensor and an RLR family member is MDA5 which is essential for the antiviral response to the encephalomyocarditis picornavirus (EMCV) (60, 61). Both RIG-I and MDA5 belong to the DEXD/H box RNA helicase family, containing similar molecular motifs. The CTD motif of RIG-I is also conserved in MDA5, suggesting a related CTD domain structure in MDA5 (14). We investigated whether ARL16 has effects on MDA5-mediated antivirus responce. We found that ARL16 inhibited EMCV-induced activation of IFN-β promoter, whereas ARF1 and ARL1 had no effects (Fig. 7A). Knockdown of endogenous ARL16 by RNAi potentiated EMCV-induced activation of IFN-β promoter (Fig. 7B). Meanwhile we found that, overexpression of ARL16 apparently decreased the number of adherent cell after EMCV infection, whereas knockdown of ARL16 by RNAi had opposite role. These results indicated that ARL16 inhibited the cellular anti-EMCV response (Fig. 7C). We also found ARL16 interacted with full-length MDA5 and its CTD (data not shown). Our results indicated that ARL16 is an inhibitor of MDA5. It would be interesting to determine whether ARL16 suppresses the interaction between MDA5 and viral RNA.
FIGURE 7.
Function of ARL16 in EMCV infection. A, ARL16, but not ARL1 and ARF1 suppresses EMCV-induced activation of IFN-β promoter. Transfection and luciferase assay were performed as in Fig. 2. At 18 h after transfection, cells were infected with EMCV (MOI = 10) or left untreated for 24 h before reporter assay. B, ARL16 RNAi plasmid potentiates EMCV-induced activation of IFN-β promoter. Experiments were carried out as in Fig. 3. At 24 h after transfection, cells were infected with EMCV (MOI = 10) or left untreated for 24 h before reporter assay. C, function of ARL16 in EMCV infection. 293T cells (2 × 105) were transfected with indicated plasmids (0.5 μg each). At 18 h after transfection, cells were infected with EMCV (MOI = 10) or left uninfected. At 36 h post-infection, the overlaid medium was removed; cells were fixed in 0.5% glutaraldehyde for 30 min and stained with 1% crystal violet dissolved in 70% ethanol. Experiments were repeated three times, one representative is shown.
ARF GTP-binding proteins are among the best-characterized members of the Ras superfamily of GTPases, with well-established functions in membrane-trafficking pathways. ARF-related protein ARLs, including ARL1 and ARL2, are also involved in membrane trafficking or organizing the cytoskeleton (38, 62–64). ARL4 may function in the nucleus (65); ARL8 promotes axonal transport (28); while ARL13 and ARL3 coordinate intraflagellar transport and ciliogenesis (40). However, most ARL proteins are so far relatively poorly characterized. In this study, we are the first to demonstrate that ARL16 is a negative regulator of the innate immune response. Our study provides the basis for expanding the functions of the ARF family into exciting new areas.
Structures of several ARF family indicated switch I' switch II segments and the interswitch segment that connects them participate in nucleotide binding (39). From our study, ARL16 contained a 10-residue loop (Aa 45–54) not found in the homologous proteins ARL1 and ARF1. This additional region adjacent to switch I seems to be important to the inhibitory function of ARL16, and may explain the difference in functions between ARL16 and other ARFs, like ARL1 and ARF1. This region is predicted to form an extensive loop that may influence switch I and switch II movement. Structural study of ARL16, both alone and in complex with the RIG-I CTD, will give insights into the process of RIG-I suppression by ARL16.
Like all small GTP-binding proteins of the Ras superfamily, ARF proteins cycle between inactive GDP-bound and active GTP-bound forms that bind selectively to effectors (37). We found that ARL16 (T37N) was restricted to the GTP-disassociated form, that it did not associate with RIG-I and also had no inhibitory activity. Furthermore, we determined that, upon SV infection, endogenous ARL16 changed to GTP-binding status and associated with RIG-I. These findings suggest that ARL16 functions as an inhibitor of RIG-I in a GTP-dependent manner. As the ARF family requires a guanine-nucleotide-exchange factor (GEF) to facilitate the conformational changes, it remains to be tested which functions as a GEF for ARL16 (perhaps RIG-I), and how regulatory molecules convert ARL16 between the GTP-associated and disassociated forms. The answers to these questions will help to further identify ARL16 functions and the regulatory mechanism of the innate immune response.
This work was supported by the National Natural Science Foundation of China (30421004 and 30871288), the China 973 Program (2010CB911801), the Foundation for Authors of National Excellent Doctoral Dissertations of PR China, and the National Key Technology R&D Program (2008BAI64B01).
- RIG-I
- retinoic acid-inducible gene I
- ARL
- ARF-like
- CTD
- C-terminal domain
- ARF
- ADP-ribosylation factor.
REFERENCES
- 1. O'Neill L. A., Bowie A. G. (2010) Curr. Biol. 20, R328–R333 [DOI] [PubMed] [Google Scholar]
- 2. Yoneyama M., Kikuchi M., Matsumoto K., Imaizumi T., Miyagishi M., Taira K., Foy E., Loo Y. M., Gale M., Jr., Akira S., Yonehara S., Kato A., Fujita T. (2005) J. Immunol. 175, 2851–2858 [DOI] [PubMed] [Google Scholar]
- 3. Hornung V., Ellegast J., Kim S., Brzózka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K. K., Schlee M., Endres S., Hartmann G. (2006) Science 314, 994–997 [DOI] [PubMed] [Google Scholar]
- 4. Pichlmair A., Schulz O., Tan C. P., Näslund T. I., Liljeström P., Weber F., Reis e Sousa C. (2006) Science 314, 997–1001 [DOI] [PubMed] [Google Scholar]
- 5. Lu C., Xu H., Ranjith-Kumar C. T., Brooks M. T., Hou T. Y., Hu F., Herr A. B., Strong R. K., Kao C. C., Li P. (2010) Structure 18, 1032–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang Y., Ludwig J., Schuberth C., Goldeck M., Schlee M., Li H., Juranek S., Sheng G., Micura R., Tuschl T., Hartmann G., Patel D. J. (2010) Nat. Struct. Mol. Biol. 17, 781–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fitzgerald K. A., McWhirter S. M., Faia K. L., Rowe D. C., Latz E., Golenbock D. T., Coyle A. J., Liao S. M., Maniatis T. (2003) Nat. Immunol. 4, 491–496 [DOI] [PubMed] [Google Scholar]
- 8. Xu L. G., Wang Y. Y., Han K. J., Li L. Y., Zhai Z., Shu H. B. (2005) Mol. Cell 19, 727–740 [DOI] [PubMed] [Google Scholar]
- 9. Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., Tschopp J. (2005) Nature 437, 1167–1172 [DOI] [PubMed] [Google Scholar]
- 10. Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K. J., Takeuchi O., Akira S. (2005) Nat. Immunol. 6, 981–988 [DOI] [PubMed] [Google Scholar]
- 11. Sharma S., tenOever B. R., Grandvaux N., Zhou G. P., Lin R., Hiscott J. (2003) Science 300, 1148–1151 [DOI] [PubMed] [Google Scholar]
- 12. Seth R. B., Sun L., Ea C. K., Chen Z. J. (2005) Cell 122, 669–682 [DOI] [PubMed] [Google Scholar]
- 13. Saito T., Hirai R., Loo Y. M., Owen D., Johnson C. L., Sinha S. C., Akira S., Fujita T., Gale M., Jr. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 582–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cui S., Eisenächer K., Kirchhofer A., Brzózka K., Lammens A., Lammens K., Fujita T., Conzelmann K. K., Krug A., Hopfner K. P. (2008) Mol. Cell 29, 169–179 [DOI] [PubMed] [Google Scholar]
- 15. Zhong B., Yang Y., Li S., Wang Y. Y., Li Y., Diao F., Lei C., He X., Zhang L., Tien P., Shu H. B. (2008) Immunity. 29, 538–550 [DOI] [PubMed] [Google Scholar]
- 16. Ishikawa H., Barber G. N. (2008) Nature 455, 674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yoneyama M., Fujita T. (2010) Rev. Med. Virol. 20, 4–22 [DOI] [PubMed] [Google Scholar]
- 18. Jounai N., Takeshita F., Kobiyama K., Sawano A., Miyawaki A., Xin K. Q., Ishii K. J., Kawai T., Akira S., Suzuki K., Okuda K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14050–14055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gack M. U., Shin Y. C., Joo C. H., Urano T., Liang C., Sun L., Takeuchi O., Akira S., Chen Z., Inoue S., Jung J. U. (2007) Nature 446, 916–920 [DOI] [PubMed] [Google Scholar]
- 20. Oshiumi H., Matsumoto M., Hatakeyama S., Seya T. (2009) J. Biol. Chem. 284, 807–817 [DOI] [PubMed] [Google Scholar]
- 21. Gao D., Yang Y. K., Wang R. P., Zhou X., Diao F. C., Li M. D., Zhai Z. H., Jiang Z. F., Chen D. Y. (2009) PLoS. One. 4, e5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zeng W., Sun L., Jiang X., Chen X., Hou F., Adhikari A., Xu M., Chen Z. J. (2010) Cell 141, 315–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Arimoto K., Takahashi H., Hishiki T., Konishi H., Fujita T., Shimotohno K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7500–7505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen R., Zhang L., Zhong B., Tan B., Liu Y., Shu H. B. (2010) Cell Res. 20, 802–811 [DOI] [PubMed] [Google Scholar]
- 25. Friedman C. S., O'Donnell M. A., Legarda-Addison D., Ng A., Cárdenas W. B., Yount J. S., Moran T. M., Basler C. F., Komuro A., Horvath C. M., Xavier R., Ting A. T. (2008) EMBO Rep. 9, 930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nistal-Villán E., Gack M. U., Martínez-Delgado G., Maharaj N. P., Inn K. S., Yang H., Wang R., Aggarwal A. K., Jung J. U., García-Sastre A. (2010) J. Biol. Chem. 285, 20252–20261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gack M. U., Nistal-Villán E., Inn K. S., García-Sastre A., Jung J. U. (2010) J. Virol. 84, 3220–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Klassen M. P., Wu Y. E., Maeder C. I., Nakae I., Cueva J. G., Lehrman E. K., Tada M., Gengyo-Ando K., Wang G. J., Goodman M., Mitani S., Kontani K., Katada T., Shen K. (2010) Neuron 66, 710–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sun Z., Ren H., Liu Y., Teeling J. L., Gu J. (2010) J. Virol. 85, 1036–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim M. J., Hwang S. Y., Imaizumi T., Yoo J. Y. (2008) J. Virol. 82, 1474–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mi Z., Fu J., Xiong Y., Tang H. (2010) Protein Cell 1, 275–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Burd C. G., Strochlic T. I., Gangi Setty, SR. (2004) Trends Cell Biol. 14, 687–694 [DOI] [PubMed] [Google Scholar]
- 33. Kahn R. A., Cherfils J., Elias M., Lovering R. C., Munro S., Schurmann A. (2006) J. Cell Biol. 172, 645–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dascher C., Balch W. E. (1994) J. Biol. Chem. 269, 1437–1448 [PubMed] [Google Scholar]
- 35. Hofmann I., Munro S. (2006) J. Cell Sci. 119, 1494–1503 [DOI] [PubMed] [Google Scholar]
- 36. Kahn R. A., Volpicelli-Daley L., Bowzard B., Shrivastava-Ranjan P., Li Y., Zhou C., Cunningham L. (2005) Biochem. Soc. Trans. 33, 1269–1272 [DOI] [PubMed] [Google Scholar]
- 37. Vetter I. R., Wittinghofer A. (2001) Science 294, 1299–1304 [DOI] [PubMed] [Google Scholar]
- 38. Lu L., Horstmann H., Ng C., Hong W. (2001) J. Cell Sci. 114, 4543–4555 [DOI] [PubMed] [Google Scholar]
- 39. Pasqualato S., Renault L., Cherfils J. (2002) EMBO Rep. 3, 1035–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li Y., Wei Q., Zhang Y., Ling K., Hu J. (2010) J. Cell Biol. 189, 1039–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Munro S. (2005) Biochem. Soc. Trans. 33, 601–605 [DOI] [PubMed] [Google Scholar]
- 42. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) Bioinformatics. 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 43. Guex N., Peitsch M. C. (1997) Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 44. Schwede T., Kopp J., Guex N., Peitsch M. C. (2003) Nucleic Acids Res. 31, 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Panic B., Perisic O., Veprintsev D. B., Williams R. L., Munro S. (2003) Mol. Cell 12, 863–874 [DOI] [PubMed] [Google Scholar]
- 46. Li Y., Li C., Xue P., Zhong B., Mao A. P., Ran Y., Chen H., Wang Y. Y., Yang F., Shu H. B. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7945–7950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nakhaei P., Mesplede T., Solis M., Sun Q., Zhao T., Yang L., Chuang T. H., Ware C. F., Lin R., Hiscott J. (2009) PLoS. Pathog. 5, e1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rothenfusser S., Goutagny N., DiPerna G., Gong M., Monks B. G., Schoenemeyer A., Yamamoto M., Akira S., Fitzgerald K. A. (2005) J. Immunol. 175, 5260–5268 [DOI] [PubMed] [Google Scholar]
- 49. Mankouri J., Fragkoudis R., Richards K. H., Wetherill L. F., Harris M., Kohl A., Elliott R. M., Macdonald A. (2010) PLoS. Pathog. 6, e1000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang Y. Y., Li L., Han K. J., Zhai Z., Shu H. B. (2004) FEBS Lett. 576, 86–90 [DOI] [PubMed] [Google Scholar]
- 51. Li S., Zheng H., Mao A. P., Zhong B., Li Y., Liu Y., Gao Y., Ran Y., Tien P., Shu H. B. (2010) J. Biol. Chem. 285, 4291–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhong B., Zhang Y., Tan B., Liu T. T., Wang Y. Y., Shu H. B. (2010) J. Immunol. 184, 6249–6255 [DOI] [PubMed] [Google Scholar]
- 53. Diao F., Li S., Tian Y., Zhang M., Xu L. G., Zhang Y., Wang R. P., Chen D., Zhai Z., Zhong B., Tien P., Shu H. B. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11706–11711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kayagaki N., Phung Q., Chan S., Chaudhari R., Quan C., O'Rourke K. M., Eby M., Pietras E., Cheng G., Bazan J. F., Zhang Z., Arnott D., Dixit V. M. (2007) Science 318, 1628–1632 [DOI] [PubMed] [Google Scholar]
- 55. Zhang M., Tian Y., Wang R. P., Gao D., Zhang Y., Diao F. C., Chen D. Y., Zhai Z. H., Shu H. B. (2008) Cell Res. 18, 1096–1104 [DOI] [PubMed] [Google Scholar]
- 56. Moore C. B., Bergstralh D. T., Duncan J. A., Lei Y., Morrison T. E., Zimmermann A. G., Accavitti-Loper M. A., Madden V. J., Sun L., Ye Z., Lich J. D., Heise M. T., Chen Z., Ting J. P. (2008) Nature 451, 573–577 [DOI] [PubMed] [Google Scholar]
- 57. Huang J., Liu T., Xu L. G., Chen D., Zhai Z., Shu H. B. (2005) EMBO J. 24, 4018–4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang X., Wang C., Schook L. B., Hawken R. J., Rutherford M. S. (2000) Microb. Pathog. 28, 267–278 [DOI] [PubMed] [Google Scholar]
- 59. Kato H., Sato S., Yoneyama M., Yamamoto M., Uematsu S., Matsui K., Tsujimura T., Takeda K., Fujita T., Takeuchi O., Akira S. (2005) Immunity. 23, 19–28 [DOI] [PubMed] [Google Scholar]
- 60. Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K. J., Yamaguchi O., Otsu K., Tsujimura T., Koh C. S., Reis e Sousa C., Matsuura Y., Fujita T., Akira S. (2006) Nature 441, 101–105 [DOI] [PubMed] [Google Scholar]
- 61. Gitlin L., Barchet W., Gilfillan S., Cella M., Beutler B., Flavell R. A., Diamond M. S., Colonna M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8459–8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gillingham A. K., Munro S. (2007) Annu. Rev. Cell Dev. Biol. 23, 579–611 [DOI] [PubMed] [Google Scholar]
- 63. Bhamidipati A., Lewis S. A., Cowan N. J. (2000) J. Cell Biol. 149, 1087–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Steinborn K., Maulbetsch C., Priester B., Trautmann S., Pacher T., Geiges B., Küttner F., Lepiniec L., Stierhof Y. D., Schwarz H., Jürgens G., Mayer U. (2002) Genes Dev. 16, 959–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lin C. Y., Huang P. H., Liao W. L., Cheng H. J., Huang C. F., Kuo J. C., Patton W. A., Massenburg D., Moss J., Lee F. J. (2000) J. Biol. Chem. 275, 37815–37823 [DOI] [PubMed] [Google Scholar]