Abstract
Discovery of proteins expressed in the central nervous system sharing the three-finger structure with snake α-neurotoxins provoked much interest to their role in brain functions. Prototoxin LYNX1, having homology both to Ly6 proteins and three-finger neurotoxins, is the first identified member of this family membrane-tethered by a GPI anchor, which considerably complicates in vitro studies. We report for the first time the NMR spatial structure for the water-soluble domain of human LYNX1 lacking a GPI anchor (ws-LYNX1) and its concentration-dependent activity on nicotinic acetylcholine receptors (nAChRs). At 5–30 μm, ws-LYNX1 competed with 125I-α-bungarotoxin for binding to the acetylcholine-binding proteins (AChBPs) and to Torpedo nAChR. Exposure of Xenopus oocytes expressing α7 nAChRs to 1 μm ws-LYNX1 enhanced the response to acetylcholine, but no effect was detected on α4β2 and α3β2 nAChRs. Increasing ws-LYNX1 concentration to 10 μm caused a modest inhibition of these three nAChR subtypes. A common feature for ws-LYNX1 and LYNX1 is a decrease of nAChR sensitivity to high concentrations of acetylcholine. NMR and functional analysis both demonstrate that ws-LYNX1 is an appropriate model to shed light on the mechanism of LYNX1 action. Computer modeling, based on ws-LYNX1 NMR structure and AChBP x-ray structure, revealed a possible mode of ws-LYNX1 binding.
Keywords: Neurobiology, Neuroscience, Neurotoxin, Nicotinic Acetylcholine Receptors, NMR, Ly6 Proteins, LYNX, Bacterial Expression
Introduction
Endogenous “prototoxins” like LYNX1, LYNX2, SLURP-1, and SLURP-2, belonging to the Ly6 protein family, modulate nicotinic acetylcholine receptors (nAChRs)3 (1–8). In the central nervous system, LYNX1 and LYNX2 regulate nAChR activity, preventing excessive excitation (3, 4). Gene deletion of LYNX1 or LYNX2 indicates that these modulators are critical for nAChR function in the brain. LYNX1 knock-out mice demonstrated enhanced performance in specific tests of learning ability and memory, whereas loss of LYNX2 results in increased anxiety-related behaviors (3, 4). Prototoxins have also been shown to affect cell growth in lung carcinoma (9), are involved in skin diseases (6, 7), and are related to prostate stem cell antigen (10).
LYNX1 and LYNX2 are tethered to the membrane by a GPI anchor, which considerably complicates in vitro studies. LYNX1 is co-localized in the brain with α4β2 and α7 nAChRs (1–3), and its modulatory activity on α4β2 nAChR was shown in experiments on Xenopus oocytes (1, 3). It was reported that soluble form of LYNX1 (not containing a GPI anchor) potentiates α4β2 receptor (1), but the concentration at which it acts remains unknown. A secreted water-soluble protein SLURP-1 expressed in palmoplantar skin acts on α7 nAChR and regulates keratinocyte proliferation (5).
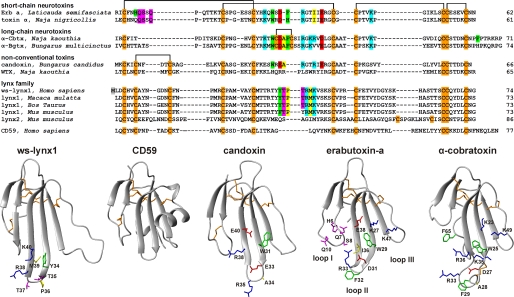
It was predicted that the prototoxins should have a spatial structure similar to that of snake venom α-neurotoxins, effective competitive inhibitors of nAChR (1). α-Neurotoxins are characterized by a three-finger fold formed by three adjacent loops arising from a small globular hydrophobic core, cross-linked by four conserved disulfide bonds (11–13). Nicotinic acetylcholine receptors are targeted by short-chain α-neurotoxins, by long-chain α-neurotoxins with additional fifth disulfide in the central loop II and an extended C-terminal tail, and by structurally similar κ-bungarotoxins, as well as by some so-called nonconventional (or weak) neurotoxins. The latter, similarly to Ly6 proteins, have the additional fifth disulfide bond in the N-terminal loop I (see Fig. 1).
FIGURE 1.
Amino acid sequence alignment of ws-LYNX1, other members of LYNX family (shown without GPI consensus sequence at the C terminus), CD59, and three-finger α-neurotoxins from snake venoms (upper panel) and comparison of spatial structures of ws-LYNX1, CD59, candoxin, erabutoxin-a, and α-cobratoxin (lower panel). The sequence data were obtained from Swiss-Prot Protein Database. The artificially introduced Met1 residue in the ws-LYNX1 sequence is highlighted gray. Cysteine residues are colored in orange, and the disulfide linkages are shown. The atomic coordinates for α-Cbtx, erabutoxin-a, candoxin, and CD59 were taken from the Protein Data Bank (Protein Data Bank codes 2CTX, 1QKD, 1JGK, and 2J8B, respectively). Residues of α-Cbtx and erabutoxin-a important for binding to nAChRs according to mutagenesis data (14, 15), homologous residues in candoxin (16), and corresponding residues in loop II of ws-LYNX1 are shown. Aromatic, hydrophobic, positively charged, negatively charged, and polar residues are colored in green, yellow, blue, red, and magenta, respectively.
Elucidating molecular mechanisms of the prototoxins interaction with nAChRs requires the knowledge of their three-dimensional structures. Because the production of GPI-linked LYNX1 as an individual protein represents an unfeasible task, a water-soluble LYNX1 appears to be a good substitute for structural analysis and may provide an adequate model for functional studies. Here, we present the expression in Escherichia coli of a water-soluble LYNX1 lacking a GPI anchor (ws-LYNX1) and its high resolution NMR structure. It was found that the protein has classical a three-finger fold formed by two β-sheets composed of six antiparallel strands. A high degree of structural homology between ws-LYNX1 and other members of the Ly6/neurotoxin family was observed. Furthermore, we demonstrated the interaction of ws-LYNX1 with acetylcholine-binding proteins (AChBPs) and several nAChR subtypes. The observed competition with 125I-α-bungarotoxin (α-Bgtx) for binding to AChBPs and Torpedo nAChR revealed partial overlap in binding sites for ws-LYNX1 and α-neurotoxins on the receptor surface. The concentration-dependent activation/deactivation effects of ws-LYNX1 on α7 nAChR were observed in electrophysiological experiments. This is of special interest because for LYNX1 itself, no concentration dependences were analyzed earlier.
EXPERIMENTAL PROCEDURES
Cloning and Bacterial Expression of ws-LYNX1
The ws-lynx1 gene encoding 73 amino acids of water-soluble fragment of a human LYNX1 (UniProt database accession no. Q9BZG9) was constructed from six overlapping synthetic oligonucleotides (supplemental Table S1) using a three-stage PCR. The ws-lynx1 gene was cloned into the expression vector pET-22b(+) (Novagen) on the NdeI and BamHI restriction sites. E. coli BL21(DE3) cells transformed with pET-22b(+)/ws-lynx1 vector were grown at 37 °C on Terrific Broth medium using a fermenter (Bioflow 3000, New Brunswick Scientific) under automatic maintenance of oxygen content in the system at a level of 30%. Gene expression was induced by addition of isopropyl 1-thio-β-d-galactopyranoside to a final concentration of 0.025 mm at A600 0.6, and cells were grown additionally for 18 h. Ws-LYNX1 was purified and refolded as was described for nonconventional neurotoxin WTX from Naja kaouthia venom (17). Briefly, ws-LYNX1 was extracted from inclusion bodies after incubation with 50 mm NaPi, 8 m urea, 1 mm tris(2-carboxyethyl)phosphine, 5 mm DTT, pH 7.4. Next, reduced ws-LYNX1 was purified on a SP Sepharose resin (GE Healthcare) equilibrated in 50 mm NaPi, 8 m urea, 5 mm DTT, pH 5.0. The protein was eluted by a gradient of NaCl. DTT and NaCl were removed by gel filtration on a NAP-25 column (GE Healthcare) equilibrated in 50 mm Tris/HCl, 8 m urea, pH 9.5. Refolding of ws-LYNX1 was induced by dissolving of reduced protein in a renaturation buffer (50 mm Tris/HCl, 1.5 m urea, 0.5 m l-Arg, 3 mm GSH, and 0.3 mm GSSG, pH 9.5) to final concentration of 0.1 mg/ml. Renaturation was performed during 3 days at 4 °C. The refolded ws-LYNX1 was analyzed and purified on a reverse-phase C4 HPLC column (4.6 × 250 mm, A300, Jupiter, Phenomenex). For production of 15N-labeled ws-LYNX1, transformed cells were grown on Terrific Broth medium until A600 of 0.6. The cells were harvested (2000 × g for 20 min) and resuspended in an equal volume of minimal medium M9 containing 15NH4Cl as a nitrogen source, and afterward, gene expression was induced.
NMR Experiments and Spatial Structure Calculation
The NMR investigation was done using 0.5 mm samples of 15N-labeled or unlabeled ws-LYNX1 in 5% D2O or 100% D2O at pH 5.3 and 25 °C. NMR spectra were acquired on a Bruker Avance 800 spectrometer equipped with a cryoprobe. 1H and 15N resonance assignment was obtained by a standard procedure using combination of two- and three-dimensional total correlation spectroscopy (TOCSY) and NOESY spectra (18). The 3JHNHα and 3JNHβ coupling constants were determined using three-dimensional HNHA and HNHB experiments (19). The 3JHαHβ and 3JHβHγ coupling constants were measured using the ACME program (20) in the COSY spectrum in 100% D2O solution. Spatial structure calculation was performed in the CYANA program (21). Upper interproton distance constraints were derived from NOESY (τm = 80 ms) cross-peaks via a “1/r6” calibration. Torsion angle restraints and stereospecific assignments were obtained from J coupling constants and NOE intensities. Hydrogen bonds were introduced basing on temperature coefficients and deuterium exchange rates of HN protons (supplemental Fig. S2). The disulfide bond connectivity pattern was established on the basis of observed NOE contacts (22) and verified during preliminary stages of spatial structure calculation. In the final rounds of structure calculation, lower distance constraints (3.0 Å), based on the expected cross-peaks but not present in the NOESY spectra, were introduced. The NMR-derived data (atomic coordinates, chemical shifts, and restraints) have been deposited in the Protein Data Bank under accession code 2L03.
Binding of ws-LYNX1 to nAChRs and AChBPs
The binding of ws-LYNX1 to GH4C1 cells expressing human α7 nAChR, nAChR-enriched Torpedo californica membranes, Aplysia californica AChBP (Ac-AChBP), and Lymnaea stagnalis AChBP (Ls-AChBP) was carried out in competitive experiments with 125I-α-Bgtx as described in Ref. 23. The tested amounts of ws-LYNX1 (from 1 to 30 μm) were preincubated 3 h with the GH4Cl cells expressing human α7 nAChR (final concentration of toxin-binding sites, 0.4 nm), or nAChR-enriched Torpedo membranes (final concentration of toxin-binding sites, 1.25 nm), or the Ls-AChBP, or Ac-AChBP (final concentration of toxin-binding sites 2.4 and 150 nm, respectively). Binding with the GH4C1 cells and Torpedo membranes was carried out in 50 μl of 20 mm Tris/HCl buffer (containing 1 mg/ml of BSA, pH 8.0) and with the AChBPs, in 50 μl of binding buffer (PBS, containing 0.7 mg/ml of BSA and 0.05% Tween 20, pH 7.5), at 25 °C. Next, 125I-α-Bgtx (∼2000 Ci/mmol) was added to a final concentration of 0.15–0.35 nm for 5 min followed by filtration of reaction mixture on GF/C filters (Whatman, Maidstone) presoaked in 0.25% polyethylenimine (for cells and membranes) or on double DE-81 filters (Whatman) presoaked in binding buffer (for AChBPs). Unbound radioactivity was removed from the filters by washes (3 × 3 ml) with the respective incubation buffers. Nonspecific binding was determined in the presence of 10 μm α-cobratoxin (α-Cbtx; 3-h preincubation).
The binding of ws-LYNX1 to SH-EP1 epithelial cells expressing human α4β2 nAChR was carried out in competitive experiments with [3H]epibatidine (49 Ci/mmol, Amersham Biosciences) as follows. The tested amounts of ws-LYNX1 (from 1 to 30 μm) were incubated 4 h with SH-EP1 cells (final concentration of epibatidine-binding sites, 0.2 nm) and 2 nm [3H]epibatidine. Filtration was performed on GF/C filters as mentioned above, and nonspecific binding was determined in the presence of 4 mm nicotine. Competition data were fit using ORIGIN (version 7.5; OriginLab Corp.) to a one-site dose-response curve using the Hill equation.
Binding of ws-LYNX1 to Muscarinic Acetylcholine Receptors
Membranes from CHO cells expressing individual subtypes of muscarinic receptors (for M1 and M3, 5 μg of membrane protein; for M2, M4, and M5, 20 μg of membrane protein) were incubated for 60 min at 30 °C in the presence of 150 pm [3H]NMS without (control) or with 10 μm ws-LYNX1. Incubation was terminated by fast filtration through GF/B filters (Whatman) essentially as described previously (24). Nonspecific binding was determined in the presence of 10 μm atropine.
Electrophysiology
Two-electrode voltage clamp experiments were performed on Xenopus laevis oocytes. Oocytes were prepared and injected as described in Ref. 25. Briefly, oocytes were injected with 2 ng of cDNA encoding for human α4β2 (ratio 1:1), α3β2 (ratio 1:1), or α7 nAChRs and utilized 2–3 days later. All recordings were performed with an automated two-electrode voltage clamp robot. Oocytes were clamped at −100 mV and perfused with OR2 (oocyte ringer) containing 82.5 mm NaCl, 2.5 mm KCl, 2.5 mm Ca2Cl, 1 mm MgCl2, 5 mm HEPES, and 20 μg/ml BSA. OR2 was adjusted to pH 7.4. Atropine (0.5 μm) was added to all solutions to block activity of endogenous muscarinic receptors. Acetylcholine and ws-LYNX1 were dissolved in OR2 just before use. Data were digitized and analyzed off-line using MATLAB (Mathworks, Natick, MA).
Computer Modeling of ws-LYNX1·Ls-AChBP Complex
The x-ray structures of Ls-AChBP with nicotine, carbamylcholine, and α-Cbtx (Protein Data Bank codes 1UW6, 1UV6, and 1YI5, respectively) were used for modeling of the ws-LYNX1·Ls-AChBP complex. All 20 NMR structures of ws-LYNX1 were docked independently using both AutoDock (version 4) and HEX (version 4.5) programs. The solutions of the complexes were relaxed and underwent molecular dynamics simulation with GROMACS for 20-ns trajectories as described in Ref. 26 (the OPLS-AA force field was used). Ws-LYNX1 structure was also analyzed by molecular dynamics for 50 ns. The root mean square deviation to starting coordinates did not exceed 2.5 Å.
RESULTS
Expression of ws-LYNX1 in E. coli
The structural and functional studies require the milligram quantities of a protein. For obtaining the water-soluble domain of human LYNX1 (amino acid residues 1–73), the production in bacterial E. coli cells was chosen, as previous studies reported a successful application of bacterial systems for expression of α-neurotoxins (27). Several approaches, including the expression of thioredoxin-fused protein, secretion of ws-LYNX1 fused with the signal peptide of E. coli heat-stable enterotoxin II (STII) (NCBI accession number M35729), and the production in the form of E. coli inclusion bodies with subsequent refolding, were tested. Despite the fact that secretion system was the most efficient for production of short-chain and long-chain α-neurotoxins (28), its application for ws-LYNX1 as well as production in the form of thioredoxin-fused protein (29) were unsuccessful. However, the direct expression into inclusion bodies followed by the refolding (see “Experimental Procedures”) resulted in the milligram scaled production of ws-LYNX1. Interestingly, a similar protocol was previously successfully applied for bacterial production of nonconventional (weak) toxin WTX from N. kaouthia venom, which, similarly to LYNX1, contains a fifth disulfide bond in the N-terminal loop (Fig. 1) (17). The yield of refolded ws-LYNX1 and of its 15N-labeled analog was 1.8 and 0.5 mg/l of bacterial culture, respectively.
The homogeneity of the refolded ws-LYNX1 was confirmed by SDS-PAGE, analytical HPLC, and mass spectrometry (supplemental Fig. S1). The measured molecular mass of the recombinant protein (8400 Da) within experimental error coincides with the calculated mass of water-soluble domain of human LYNX1 (amino acids 1–73) with an additional Met residue at the N terminus and five closed disulfide bridges (8399.6 Da, supplemental Fig. S1C). Formation of disulfide bonds for the refolded ws-LYNX1 was additionally confirmed using Ellman's reagent. CD spectroscopy of the refolded ws-LYNX1 revealed a preferentially β-structural organization (supplemental Fig. S1D).
Spatial Structure of ws-LYNX1 in Comparison with Structures of Other Three-finger Proteins
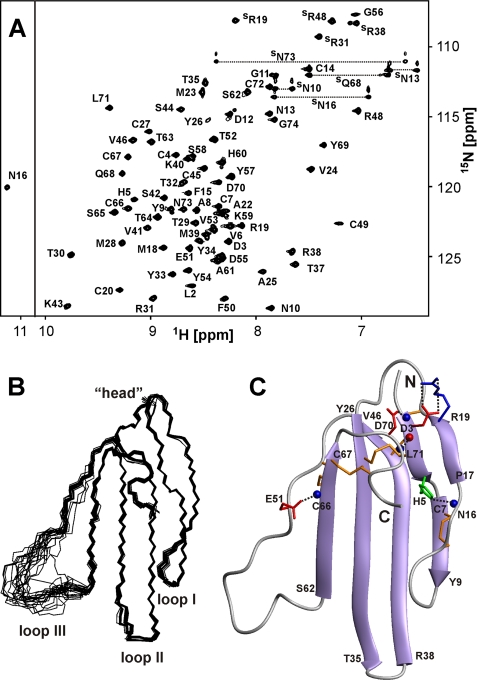
The spatial structure of ws-LYNX1 was studied by 1H-15N NMR spectroscopy in aqueous solution at pH 5.3 (supplemental Fig. S2 and Table S2). The calculated set of 20 ws-LYNX1 structures and the representative structure are shown on Fig. 2. The protein is composed by three prolonged loops (I, II, and III) protruding from the “head” region. The secondary structure of ws-LYNX1 represents two antiparallel β-sheets, one consisting of two strands (Asp3–His5 and Pro17–Arg19 (loop I)) and another of four strands (Cys7–Tyr9 (loop I), Tyr26–Thr35 (loop II), Arg38–Val46 (loop II), and Ser62–Cys67 (loop III)). Almost the whole structure is well defined, except for a part of the third loop (residues Thr52–Ala61) (Fig. 2B). The residues in this region demonstrate close to random coil values of 1H chemical shifts and averaged 3JHNHα coupling constants with magnitudes ∼7 Hz. Moreover, no long and medium range NOE contacts were detected for these residues indicating enhanced mobility of this fragment.
FIGURE 2.
NMR data define the spatial structure of ws-LYNX1 in aqueous solution. A, two-dimensional 1H,15N-HSQC spectrum of ws-LYNX1 (0.5 mm, pH 5.3, 25 °C). The obtained resonance assignments are shown. The resonances of side chain groups are marked with an S. The resonances of Asn and Gln NH2 groups are connected by dotted lines. B, the set of the best 20 ws-LYNX1 structures superimposed over the backbone atoms in regions with well defined structure. The three loops and head of the protein are labeled. C, ribbon representation of ws-LYNX1 spatial structure. The electrostatic and hydrogen bonding interactions that stabilize the protein fold are shown. The Arg, Asp/Glu, and His side chains participating in salt bridges and hydrogen bonds with backbone amides are in blue, red, and green, respectively. The hydrogen bond HN Leu71–CO Asp3 is also shown. Backbone amide and carbonyl groups are shown by blue and red spheres, respectively. The disulfide bonds are shown in orange.
The ws-LYNX1 structure is stabilized by four disulfide bonds in the head (Cys4–Cys27, Cys20–Cys45, Cys49–Cys66, and Cys67–Cys72) and one in the first loop (Cys7–Cys14). These results confirm the disposition of disulfide bonds earlier proposed for LYNX1 (1). Apart from the backbone-backbone hydrogen bonds associated with canonical elements of secondary structure, the head and loops of the protein are stabilized by additional H-bonding and electrostatic interactions (Fig. 2C). For instance, two hydrogen bonds (HN Leu71–CO Asp3 and HN Asp3–Oδ1 Asp70) control the relative spatial arrangement of N and C termini of the protein. The N-H–N hydrogen bond (HN Asn16–Nδ1 His5) and salt bridge (Asp3–Arg19) stabilize the first loop. The hydrogen bond HN Cys66–Oϵ1 Glu51 stabilizes the third loop. In addition, several tight β- and γ-turns present in the tips of the loops I and II and in the head region (supplemental Fig. S2). The comparison of NMR spectra of the ws-LYNX1 measured at pH 5.3 and 7.0 (supplemental Fig. S3) indicates that the protein has similar spatial structure at neutral and moderately acidic conditions.
Because LYNX1 targets nAChRs, it is interesting to compare its three-dimensional structure with those of snake venom α-neurotoxins and some nonconventional toxins acting on nAChRs (16, 30, 31). CD59, the complement regulatory protein of the Ly6 family, was included for comparison because the spatial structure is known for its GPI-lacking form (32, 33). Fig. 1 shows a similarity in the overall fold for all these proteins. The fifth disulfide bond in the loop I is common for ws-LYNX1, nonconventional toxins, and CD59. However, a large and mostly disordered loop III is a unique feature of ws-LYNX1; CD59 protein also contains quite long insertion in the loop III, but this element is well structured and adopts an α-helical conformation in crystal and solution (32, 33).
ws-LYNX1 Binding to AChBPs, nAChRs, and Muscarinic AChRs
Previous studies demonstrated LYNX1 effects on the ACh-induced currents in Xenopus oocytes and cell lines expressing α4β2 nAChR (1–3). However, it was not possible to quantify the observed activity and measure Kd or EC50 for LYNX1 effects. Here, we measured the binding parameters for ws-LYNX1 interaction with nAChRs as well as with Ac-AChBP and Ls-AChBP, excellent structural models for nAChR ligand-binding domains (34). Because homo-oligomeric AChBPs are most similar to α7 homo-oligomeric nAChR, and as Ls-AChBP binds α-Bgtx with nanomolar affinity, competition with radioactive α-Bgtx allows the detection of compounds that should bind to α7 nAChR (34, 35). Moreover, compounds that target other nAChR subtypes may also interact with AChBPs, although with a lower affinity (36).
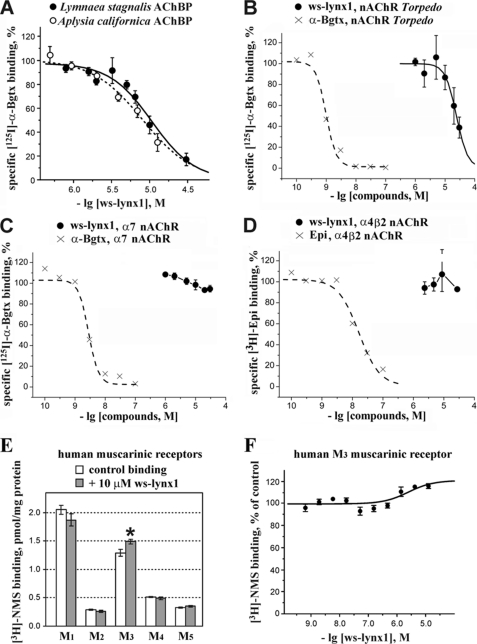
The competition experiments with 125I-α-Bgtx yielded an IC50 ∼ 10 μm for ws-LYNX1 binding to AChBPs (Fig. 3A). Although the affinity is not high, the attempts to crystallize the ws-LYNX1·AChBP complexes were undertaken but still did not yield positive results.4 The binding of ws-LYNX1 to AChBPs suggests that it may also bind to nAChRs. Indeed, the presence of tight complexes of LYNX1 and α7 or α4β2 nAChRs was earlier shown by co-immunoprecipitation (1–3). However, in the case of human α7 nAChRs overexpressed in the GH4C1 cell line, no competition with α-Bgtx was detected at 30 μm ws-LYNX1 (Fig. 3C). Under the conditions necessary to analyze competition with [3H]epibatidine for binding to α4β2 nAChR in the SH-EP1 epithelial cell line, we achieved the same 30 μm concentration of ws-LYNX1, which did not show any inhibition (Fig. 3D). For comparison, the displacements of these radioactive ligands by α-bungarotoxin and epibatidine, respectively, are shown in these figures.
FIGURE 3.
Binding of ws-LYNX1 to AChBPs and nicotinic and muscarinic acetylcholine receptors. Competition of ws-LYNX1 with 125I-α-Bgtx for binding to Ls-AChBP and Ac-AChBP (A), to membrane-bound nAChR from T. californica (B), and to α7 nAChR in the GH4C1 cell line (C). Competition of ws-LYNX1 with [3H]epibatidine for binding to α4β2 nAChR in the SH-EP1 cell line is shown in D. For comparison, the displacements of 125I-α-Bgtx and [3H]epibatidine by unlabeled Bgtx and epibatidine are also shown in these panels. Each point is mean ± S.E. of three independent experiments. The Hill equation (y = 100/(1 + ([toxin]/IC50)nH)) was fitted to normalized data (% of control binding). The calculated parameters IC50 and nH were 10.7 μm and 1.5 for Ls-AChBP, 9.4 μm and 1.2 for Ac-AChBP, and 24 μm and 2.3 for muscle-type nAChR. E, effects of 10 μm ws-LYNX1 on [3H]NMS binding at M1–M5 human muscarinic receptors expressed in membranes of CHO cells. The asterisk indicates that binding of [3H]NMS to the M3 receptor in the presence of ws-LYNX1 was significantly different from control (p < 0.05, according to t test). Each point is mean ± S.E. of quadruplicates. F, interaction of ws-LYNX1 with [3H]NMS binding at muscarinic M3 receptor. Membranes expressing M3 muscarinic receptor (5 μg of protein) were incubated in the presence of indicated concentrations of ws-LYNX1 and 108 pm [3H]NMS. [3H]NMS binding is expressed in percent of control binding in the absence of ws-LYNX1. Each point is mean ± S.E. of quadruplicates. The equation (y = 100 × ([NMS] + Kd)/([NMS] + Kd × (Ka + 10[ws-LYNX1])/(Ka + 10[ws-LYNX1]/α)) was fitted to normalized data (% of control binding). Kd of [3H]NMS binding (207 pm) was determined in parallel saturation experiment. Estimated parameters are Ka (equilibrium dissociation constant of ws-LYNX1) 3.0 μm and a factor of cooperativity (α) of 0.8. The correlation coefficient of the fit is 0.8.
Because LYNX2 was co-immunoprecipitated with muscle nAChR (4), we tested ws-LYNX1 binding to the muscle-type nAChR from T. californica and found IC50 to be ∼30 μm (Fig. 3B). Thus, the observed competitions pointed to a possible overlap in ws-LYNX1 and α-neurotoxin binding sites on the surfaces of AChBPs and muscle type nAChR.
The additional disulfide bond in the loop I and micromolar affinity to the muscle type and α7 nAChRs are characteristic for some nonconventional neurotoxins, e.g. WTX (37). Because WTX binding to muscarinic acetylcholine receptors has been reported recently (38), we tested the interaction of ws-LYNX1 with M1–M5 muscarinic receptors. Fig. 3E shows that 10 μm ws-LYNX1 had no effect on all tested subtypes, with the exception of the M3 receptor, for which a statistically significant 15% increase in [3H]N-methylscopolamine (NMS) binding in the presence of ws-LYNX1 was observed. A weak positive allosteric interaction (factor of cooperativity α ∼ 0.8) between ws-LYNX1 and [3H]NMS was confirmed in a pseudocompetition experiment at the M3 receptor (Fig. 3F). The affinity of ws-LYNX1 was relatively low (Ka ∼ 3 μm) as compared with the reported affinity of WTX (Ka ∼ 0.3 μm). Nevertheless, the obtained preliminary data point to a possible existence of non-nAChR molecular targets for LYNX1.
Electrophysiology Studies and Effect of ws-LYNX1 on Human nAChRs
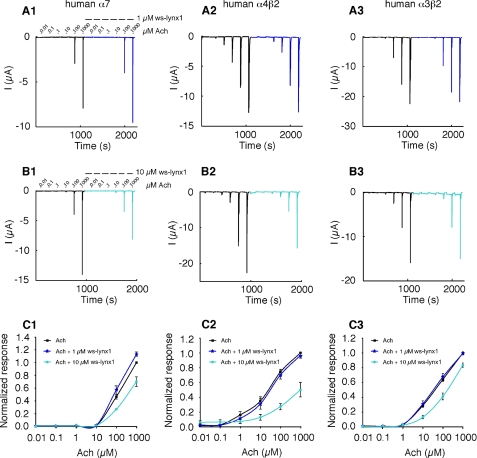
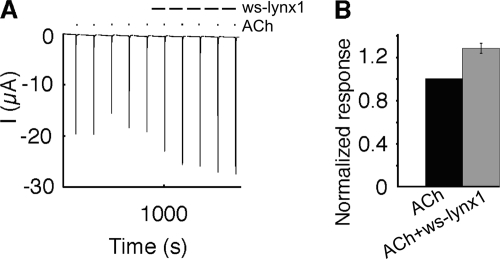
Electrophysiological recordings were performed on Xenopus oocytes expressing α4β2, α3β2, and α7 nAChRs in the absence and presence of 1 μm or 10 μm ws-LYNX1. Currents were measured at −100 mV and elicited by different ACh concentrations ranging from 10 nm to 1 mm. Application of 1 μm ws-LYNX1 showed no effect on α4β2 and α3β2; however, a slight enhancement of the ACh-induced currents was observed for α7 nAChR (Fig. 4, A and C). This effect was confirmed in another batch of cells using a single concentration of 1 mm ACh with or without exposure to 1 μm ws-LYNX1 (n = 6) (Fig. 5). Contrary to that, the application of 10 μm ws-LYNX1 resulted in inhibition of the ACh-induced currents for all three receptors, but the effect was more profound on α4β2 nAChRs (Fig. 4, B and C). Interestingly, the blocking effect of ws-LYNX is not suppressed by increasing ACh concentrations but becomes even more visible, especially for α4β2 nAChRs as was earlier observed for the membrane-tethered LYNX1 (2). This suggests that inhibition caused by ws-LYNX1 is probably not competitive. For naturally occurring LYNX1 (although at unknown concentrations), an increase in desensitization rate has been described for α4β2 nAChR (2), but our data did not reveal significant modification of the response time course in the presence of 1 μm or 10 μm ws-LYNX1 for any of receptors tested.
FIGURE 4.
Effect of ws-LYNX1 on nAChRs expressed in Xenopus oocytes. A1–A3, concentration-response curve for ACh on human α7 (A1), human α4β2 (A2), and human α3β2 nAChRs (A3) in the absence and presence of 1 μm ws-LYNX1. B1–B3, ACh-induced responses on human α7 (B1), human α4β2 (B2), and human α3β2 nAChRs (B3) in the absence and presence of 10 μm ws-LYNX1. C1–C3, ACh-evoked activation curves of the fitted data for human α7 (C1), human α4β2 (C2), and human α3β2 nAChRs (C3) in the absence and presence of 1 and 10 μm ws-LYNX1 (n = 3–11). The highest ws-LYNX1 concentration (10 μm) affects the higher ACh-induced responses. At 1 μm ws-LYNX1 an increased response was observed for human α7 nAChR at 1000 μm ACh.
FIGURE 5.
Effect of 1 μm ws-LYNX1 on human α7 nAChR expressed in Xenopus oocytes. A, five repetitive ACh (1000 μm) responses in the absence and presence of 1 μm ws-LYNX1. B, bar diagram of the data (n = 6), 1 μm ws-LYNX1 increases the ACh response.
Computer Modeling of ws-LYNX1 Interaction with AChBP
The established NMR structure of ws-LYNX1 made possible the docking of ws-LYNX1 to Ls-AChBP and the comparison with binding modes for three-finger α-neurotoxins. The Ls-AChBP structures, with either loop C in an outward position (as in complex with competitive antagonist α-Cbtx) or shifted to the central axis (as in complexes with agonists carbamylcholine and nicotine) (39, 40), were used. The docking and subsequent molecular dynamics studies revealed that the ws-LYNX1 affinity to Ls-AChBP in the latter case was considerably higher (estimated energies of interaction are −239 and −342 kJ/mol/binding site, respectively). Thus, ws-LYNX1 appears to stabilize loop C in the position characteristic for Ls-AChBP complexes with agonists.
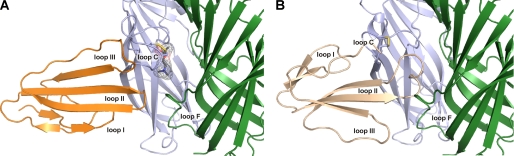
The resulting model for complex ws-LYNX1·Ls-AChBP is shown in Fig. 6A, and the structure of the α-Cbtx·Ls-AChBP complex (39) is given for comparison (Fig. 6B). Similarly to α-neurotoxins, the central loop II of ws-LYNX1 interacts with loop C of the AChBP protomer. At the same time, the model revealed some differences in the binding modes of ws-LYNX1 and α-Cbtx with Ls-AChBP. These include the disposition of the loop C and opposite orientations of ligands; the loop I of ws-LYNX1 is below loop II, and the loop I of α-Cbtx is above.
FIGURE 6.
Model of ws-LYNX1·Ls-AChBP complex (A) in comparison with the x-ray structure of α-Cbtx·Ls-AChBP complex (Protein Data Bank code 1YI5) (B). One subunit of AChBP (principal side of the interface) is in blue; the other (complementary side) is in green. The vicinal disulfide bond in C-loop of AChBP is marked by yellow. Ws-LYNX1 and α-Cbtx are in orange and beige, respectively. An ACh molecule is shown in A using a space-filled representation.
DISCUSSION
Membrane-tethered proteins LYNX1 and LYNX2 are co-localized with some nAChR subtypes and modulate their activity. The structure-function investigations of these natural GPI-tagged proteins would meet considerable problems, including their production in sufficient amounts, solubilization, and estimation of the effective concentrations. Thus, we focused our study on a recombinant water-soluble analog of LYNX1 that can permit detailed structure-function characterization. Due to expected spatial similarity of LYNX1 with secreted three-finger neurotoxins, we supposed that their action on nAChRs should have some common features and, at least in part, does not require the GPI anchor. Indeed, our NMR study revealed a high degree of structural homology between ws-LYNX1 expressed in E. coli and snake venom α-neurotoxins (Fig. 1). Moreover, the comparison of obtained electrophysiological data on ws-LYNX1 with previous investigations of natural membrane-tethered LYNX1 (2) revealed some similarities in their action on nAChRs (see below).
The presented data reveal new facets in the molecular mechanism of LYNX1 action on the nicotinic receptors. One such finding is the observation of competition between ws-LYNX1 and long-chain α-neurotoxin (α-Bgtx) for binding to Torpedo nAChR and to AChBPs. The measured binding parameters (Fig. 3, A and B) supplement the earlier co-immunoprecipitation data on binding of LYNX1 or LYNX2 to α4β2, α7, and muscle-type nAChRs (2–4). The observed profound inhibition of ACh-evoked currents by 10 μm ws-LYNX1 in α4β2 nAChR (Fig. 4) agrees well with the previous proposal that this receptor subtype is possibly the major target for GPI-anchored prototoxins LYNX1 and LYNX2 (2–4). Fig. 4 shows that 10 μm ws-LYNX1 also inhibited (reduced the current amplitude) α7 and α3β2 nAChRs. No substantial effects were observed for α7 nAChR at 10 μm ACh, but a clear reduction of the current was registered at 100 and 1000 μm ACh. Interestingly, ws-LYNX1 at 1 μm had no effect on currents induced in α4β2 and α3β2 nAChRs but significantly increased the current of α7 nAChRs (Fig. 5). These data imply the presence of concentration-dependent effects of ws-LYNX1 on α7 nAChRs resulting in potentiation or inhibition of the current amplitude. It should be noted that attempts to elucidate the concentration dependences were made previously only for the insect analog of LYNX1 by varying the amount of its cRNA introduced into oocytes (41). Moreover, LYNX1 action on α3β2 nAChRs was not reported previously.
Because in binding studies we saw neither inhibition of [3H]epibatidine binding to α4β2 nAChR nor of 125I-α-Bgtx association with α7 nAChR at 30 μm ws-LYNX1, the effects observed in electrophysiological experiments with 1 and 10 μm ws-LYNX1 indicate that, at least in part, its activity could arise from binding outside the classical nAChR binding site for agonists/competitive antagonists. This assumption is further supported by the fact that in electrophysiological experiments, the inhibitory effect of ws-LYNX1 becomes even more pronounced at high concentrations of ACh (Fig. 4, C2).
The observed competition between ws-LYNX1 and α-Bgtx for binding to muscle type nAChR and AChBPs suggests that the ws-LYNX1 binding site at least partially overlaps with the α-neurotoxin binding sites on these targets. The differences in the ws-LYNX1 interactions with distinct nAChR subtypes can be compared with those of the three-finger neurotoxins and α-conotoxins: α-Bgtx, a long-chain neurotoxin, binds to the α7 and muscle-type nAChRs, whereas short-chain neurotoxins target only the latter type. The most relevant to our case is the example of α-conotoxin ImII; it has the same potency as α-conotoxin ImI in blocking α7 nAChR but displaces 125I-α-Bgtx much more weakly (42). Moreover, it has been recently shown that binding of α-conotoxin ImII occurs at the classical site for agonists and competitive antagonists on AChBPs and α7 nAChR, whereas at Torpedo nAChR, this conotoxin binds mainly beyond this site (23).
The different action modes of ws-LYNX1 and snake neurotoxins can be attributed to considerable differences in the nature of their functional groups brought to the binding sites at the receptor. Fig. 1 depicts α-Cbtx side chains important for interaction with α7 nAChRs (14), whereas those necessary for binding of short-chain α-neurotoxins to muscle-type receptors (15) are shown on erabutoxin-a. The short- and long-chain neurotoxins, together with those nonconventional neurotoxins that block nAChRs with high affinity (e.g. candoxin (16)), have in loop II a combination of aromatic, positively and negatively charged residues (Fig. 1), which possibly determine the targeting of the toxin to the ligand binding pocket on the receptor surface (39, 43). A positively charged residue located in the tip of loop II (Arg33 in α-Cbtx and erabutoxin-a or Arg35 in candoxin) imitates a positive charge of ACh, thus preventing the simultaneous binding of neurotoxin and the agonist. Ws-LYNX1 does not have such a set of residues and in principle can be targeted to another site on the receptor surface, leaving a possibility for simultaneous ACh binding.
There is another group of three-finger proteins, so-called muscarinic toxins, mostly found in black mamba venoms, which block muscarinic acetylcholine receptors (44). The dual action of a three-finger protein on the nicotinic and muscarinic AChRs was described for WTX, a nonconventional weak neurotoxin from the N. kaouthia venom (38). In the present report, we showed the preliminary data about the ws-LYNX1 action on the M3 subtype of human muscarinic AChR (Fig. 3, E and F), which, in view of the described similarities in the activity of ws-LYNX1 and LYNX1 on the nAChRs, suggest the necessity to check the action of GPI-attached LYNX1 on muscarinic AChRs and other putative targets.
Attaching α-Bgtx, a three-finger neurotoxin, to the membrane via GPI anchor preserved its inhibition of α7 nAChR (45). Our work, on the other hand, shows that the detachment of LYNX1, also a three-finger protein, from the membrane does not destroy its activity. The release of water-soluble forms from GPI-containing proteins due to effects of proteases and phospholipases is well known (46); such forms were detected for CD59 (33) and for uPAR proteins (composed of the prototypical three-finger domains), especially in cancer cells (47, 48). Thus, our results suggest that if membrane-detached LYNX1 arises, some in vivo activities similar to those of ws-LYNX1 may be anticipated.
The determined high resolution spatial structure of ws-lynx1 makes it possible to analyze how this molecule, and probably its progenitor LYNX1, recognizes the target. As the highest affinity for ws-LYNX1 was observed in binding to AChBPs, and the x-ray structures for these proteins are known, at the first stage, we confined this analysis to computer modeling of the ws-LYNX1 complex with the L. stagnalis AChBP. We have chosen this particular protein because the x-ray structure of its complex with α-Cbtx, a three-finger neurotoxin, is known (39).
The results of docking and molecular dynamics illustrate a general similarity between binding of ws-LYNX1 and α-Cbtx (Fig. 6). However, marked dissimilarities were also identified; α-Cbtx approaches the target with one side of its relatively flat molecule, whereas ws-LYNX1 approaches with the opposite side. In addition, the binding site for ws-LYNX1 overlaps only partially with that for snake neurotoxin. Contrary to α-Cbtx (Fig. 6), ws-LYNX1 appears to stabilize the loop C of AChBP in position closer to the central axis in a conformation that was observed previously in complexes of AChBPs with agonists (40). In principle, this would even allow a simultaneous binding of ws-LYNX1 and acetylcholine depicted in the model. However, further studies are needed to verify how this model is close to reality and to what extent it is applicable to different nAChR subtypes.
In summary, the results presented here demonstrate that the water-soluble domain of human LYNX1 (ws-LYNX1), obtained by expression in E. coli, is an individual protein with the correct primary structure and well defined three-dimensional structure. A high resolution NMR study of ws-LYNX1 demonstrated a three-finger fold well characterized earlier for snake venom neurotoxins. However, the marked differences between ws-LYNX1 and neurotoxins in the conformation of individual loops were revealed. In general, it was shown that ws-LYNX1 provides a good model to elucidate at the molecular level the modulatory effects of LYNX1 on nAChRs. Indeed, we characterized the interaction of ws-LYNX1 with several nAChR subtypes and AChBPs. Our results for the first time demonstrated a concentration dependence of ws-LYNX1 modulatory effects and indicated that the activity of the naturally occurring LYNX1 and its congeners is most probably not restricted to α4β2 nAChR. Determination of the high resolution NMR structure of ws-LYNX1, one of the initial stages of our research in this field, made docking and molecular dynamics analysis of ws-LYNX1 interaction with its targets possible. We hope that future mutagenesis studies, based on the solved spatial structure of ws-LYNX1 and computational models, will shed light on the detailed molecular mechanisms of LYNX1 modulating effects.
Acknowledgments
We are grateful to T. Sixma for the samples of acetylcholine binding proteins, F. Hucho for the membrane-bound nicotinic receptor from T. californica, Eli Lilly for the GH4C1 line with the overexpressed α7 nicotinic receptor, R. Lukas for the line with α4β2 receptor, and R. V. Tikhonov for assistance in recombinant protein production. Fruitful discussions with I. Ibanez-Tallon are highly appreciated. The computations were performed at the Research Computing Center SKIF-MSU Chebyshev.
This work was supported by grants from the Russian Academy of Sciences “Molecular and Cellular Biology,” Russian Foundation for Basic Research (RFBR) Grants 08-04-01603 and 09-04-01567, RFBR-NWO (Netherlands Organization for Scientific Research) Grant 06-04-89400, and FP7 Grant “Neurocypres.”

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Tables S1 and S2, Figs. S1–S3, and additional references.
The atomic coordinates and structure factors (code 2L03) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
P. Rucktooa and T. Sixma, private communication.
- nAChR
- nicotinic acetylcholine receptor
- ws-LYNX1
- water-soluble LYNX1
- ACh
- acetylcholine
- AChBP
- acetylcholine-binding protein
- GPI
- glycosylphosphatidylinositol
- Ac-AChBP
- Aplysia californica AChBP
- Ls-AChBP
- Lymnaea stagnalis AChBP
- mAChR
- muscarinic acetylcholine receptor
- α-Cbtx
- α-cobratoxin
- WTX
- weak toxin
- α-Bgtx
- α-bungarotoxin
- NMS
- N-methylscopolamine
- Epi
- epibatidine.
REFERENCES
- 1. Miwa J. M., Ibanez-Tallon I., Crabtree G. W., Sánchez R., Sali A., Role L. W., Heintz N. (1999) Neuron 23, 105–114 [DOI] [PubMed] [Google Scholar]
- 2. Ibañez-Tallon I., Miwa J. M., Wang H. L., Adams N. C., Crabtree G. W., Sine S. M., Heintz N. (2002) Neuron 33, 893–903 [DOI] [PubMed] [Google Scholar]
- 3. Miwa J. M., Stevens T. R., King S. L., Caldarone B. J., Ibanez-Tallon I., Xiao C., Fitzsimonds R. M., Pavlides C., Lester H. A., Picciotto M. R., Heintz N. (2006) Neuron 51, 587–600 [DOI] [PubMed] [Google Scholar]
- 4. Tekinay A. B., Nong Y., Miwa J. M., Lieberam I., Ibanez-Tallon I., Greengard P., Heintz N. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4477–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chimienti F., Hogg R. C., Plantard L., Lehmann C., Brakch N., Fischer J., Huber M., Bertrand D., Hohl D. (2003) Hum. Mol. Genet. 12, 3017–3024 [DOI] [PubMed] [Google Scholar]
- 6. Tsuji H., Okamoto K., Matsuzaka Y., Iizuka H., Tamiya G., Inoko H. (2003) Genomics 81, 26–33 [DOI] [PubMed] [Google Scholar]
- 7. Moriwaki Y., Watanabe Y., Shinagawa T., Kai M., Miyazawa M., Okuda T., Kawashima K., Yabashi A., Waguri S., Misawa H. (2009) Neurosci. Res. 64, 403–412 [DOI] [PubMed] [Google Scholar]
- 8. Arredondo J., Chernyavsky A. I., Jolkovsky D. L., Webber R. J., Grando S. A. (2006) J. Cell. Physiol. 208, 238–245 [DOI] [PubMed] [Google Scholar]
- 9. Song P., Sekhon H. S., Fu X. W., Maier M., Jia Y., Duan J., Proskosil B. J., Gravett C., Lindstrom J., Mark G. P., Saha S., Spindel E. R. (2008) Cancer Res. 68, 4693–4700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hruska M., Keefe J., Wert D., Tekinay A. B., Hulce J. J., Ibañez-Tallon I., Nishi R. (2009) J. Neurosci. 29, 14847–14854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tsetlin V. (1999) Eur. J. Biochem. 264, 281–286 [DOI] [PubMed] [Google Scholar]
- 12. Nirthanan S., Gwee M. C. (2004) J. Pharmacol. Sci. 94, 1–17 [DOI] [PubMed] [Google Scholar]
- 13. Tsetlin V. I., Hucho F. (2004) FEBS Lett. 557, 9–13 [DOI] [PubMed] [Google Scholar]
- 14. Antil-Delbeke S., Gaillard C., Tamiya T., Corringer P. J., Changeux J. P., Servent D., Ménez A. (2000) J. Biol. Chem. 275, 29594–29601 [DOI] [PubMed] [Google Scholar]
- 15. Pillet L., Trémeau O., Ducancel F., Drevet P., Zinn-Justin S., Pinkasfeld S., Boulain J. C., Ménez A. (1993) J. Biol. Chem. 268, 909–916 [PubMed] [Google Scholar]
- 16. Nirthanan S., Charpantier E., Gopalakrishnakone P., Gwee M. C., Khoo H. E., Cheah L. S., Bertrand D., Kini R. M. (2002) J. Biol. Chem. 277, 17811–17820 [DOI] [PubMed] [Google Scholar]
- 17. Lyukmanova E. N., Shulepko M. A., Tikhonov R. V., Shenkarev Z. O., Paramonov A. S., Wulfson A. N., Kasheverov I. E., Ustich T. L., Utkin Y. N., Arseniev A. S., Tsetlin V. I., Dolgikh D. A., Kirpichnikov M. P. (2009) Biochemistry 74, 1142–1149 [DOI] [PubMed] [Google Scholar]
- 18. Wuthrich K. (1986) NMR of Proteins and Nucleic Acids, John Wiley and Sons, New York [Google Scholar]
- 19. Bax A., Vuister G. W., Grzesiek S., Delaglio F., Wang A. C., Tschudin R., Zhu G. (1994) Methods Enzymol. 239, 79–105 [DOI] [PubMed] [Google Scholar]
- 20. Delaglio F., Wu Z., Bax A. (2001) J. Magn. Reson. 149, 276–281 [DOI] [PubMed] [Google Scholar]
- 21. Güntert P. (2004) Methods Mol. Biol. 278, 353–378 [DOI] [PubMed] [Google Scholar]
- 22. Arseniev A. S., Kondakov V. I., Maiorov V. N., Bystrov V. F. (1984) FEBS Lett. 165, 57–62 [Google Scholar]
- 23. Kasheverov I. E., Zhmak M. N., Fish A., Rucktooa P., Khruschov A. Y., Osipov A. V., Ziganshin R. H., D'hoedt D., Bertrand D., Sixma T. K., Smit A. B., Tsetlin V. I. (2009) J. Neurochem. 111, 934–944 [DOI] [PubMed] [Google Scholar]
- 24. Jakubík J., El-Fakahany E. E., Dolezal V. (2006) Mol. Pharmacol. 70, 656–666 [DOI] [PubMed] [Google Scholar]
- 25. Hogg R. C., Bandelier F., Benoit A., Dosch R., Bertrand D. (2008) J. Neurosci. Methods 169, 65–75 [DOI] [PubMed] [Google Scholar]
- 26. Mordvitsev D. Y., Polyak Y. L., Kuzmin D. A., Levtsova O. V., Tourleigh Y. V., Utkin Y. N., Shaitan K. V., Tsetlin V. I. (2007) Comput. Biol. Chem. 31, 72–81 [DOI] [PubMed] [Google Scholar]
- 27. Lyukmanova E. N., Shulepko M. A., Shenkarev Z. O., Dolgikh D. A., Kirpichnikov M. P. (2010) Russian J. Bioorg. Chem. 36, 137–145 [DOI] [PubMed] [Google Scholar]
- 28. Lyukmanova E. N., Shenkarev Z. O., Schulga A. A., Ermolyuk Y. S., Mordvintsev D. Y., Utkin Y. N., Shoulepko M. A., Hogg R. C., Bertrand D., Dolgikh D. A., Tsetlin V. I., Kirpichnikov M. P. (2007) J. Biol. Chem. 282, 24784–24791 [DOI] [PubMed] [Google Scholar]
- 29. Lyukmanova E. N., Shulga A. A., Arsenieva D. A., Pluzhnikov K. A., Dolgikh D. A., Arseniev A. S., Kirpichnikov M. P. (2004) Russian J. Bioorg. Chem. 30, 25–34 [Google Scholar]
- 30. Betzel C., Lange G., Pal G. P., Wilson K. S., Maelicke A., Saenger W. (1991) J. Biol. Chem. 266, 21530–21536 [DOI] [PubMed] [Google Scholar]
- 31. Nastopoulos V., Kanellopoulos P. N., Tsernoglou D. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 964–974 [DOI] [PubMed] [Google Scholar]
- 32. Leath K. J., Johnson S., Roversi P., Hughes T. R., Smith R. A., Mackenzie L., Morgan B. P., Lea S. M. (2007) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63, 648–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fletcher C. M., Harrison R. A., Lachmann P. J., Neuhaus D. (1994) Structure 2, 185–199 [DOI] [PubMed] [Google Scholar]
- 34. Rucktooa P., Smit A. B., Sixma T. K. (2009) Biochem. Pharmacol. 78, 777–787 [DOI] [PubMed] [Google Scholar]
- 35. Celie P. H., Kasheverov I. E., Mordvintsev D. Y., Hogg R. C., van Nierop P., van Elk R., van Rossum-Fikkert S. E., Zhmak M. N., Bertrand D., Tsetlin V., Sixma T. K., Smit A. B. (2005) Nat. Struct. Mol. Biol. 12, 582–588 [DOI] [PubMed] [Google Scholar]
- 36. Dutertre S., Ulens C., Büttner R., Fish A., van Elk R., Kendel Y., Hopping G., Alewood P. F., Schroeder C., Nicke A., Smit A. B., Sixma T. K., Lewis R. J. (2007) EMBO J. 26, 3858–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Utkin Y. N., Kukhtina V. V., Kryukova E. V., Chiodini F., Bertrand D., Methfessel C., Tsetlin V. I. (2001) J. Biol. Chem. 276, 15810–15815 [DOI] [PubMed] [Google Scholar]
- 38. Mordvintsev D. Y., Polyak Y. L., Rodionov D. I., Jakubik J., Dolezal V., Karlsson E., Tsetlin V. I., Utkin Y. N. (2009) FEBS J. 276, 5065–5075 [DOI] [PubMed] [Google Scholar]
- 39. Bourne Y., Talley T. T., Hansen S. B., Taylor P., Marchot P. (2005) EMBO J. 24, 1512–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Celie P. H., van Rossum-Fikkert S. E., van Dijk W. J., Brejc K., Smit A. B., Sixma T. K. (2004) Neuron 41, 907–914 [DOI] [PubMed] [Google Scholar]
- 41. Liu Z., Cao G., Li J., Bao H., Zhang Y. (2009) J. Neurochem. 110, 1707–1714 [DOI] [PubMed] [Google Scholar]
- 42. Ellison M., McIntosh J. M., Olivera B. M. (2003) J. Biol. Chem. 278, 757–764 [DOI] [PubMed] [Google Scholar]
- 43. Dellisanti C. D., Yao Y., Stroud J. C., Wang Z. Z., Chen L. (2007) Nat. Neurosci. 10, 953–962 [DOI] [PubMed] [Google Scholar]
- 44. Servent D., Fruchart-Gaillard C. (2009) J. Neurochem. 109, 1193–1202 [DOI] [PubMed] [Google Scholar]
- 45. Ibañez-Tallon I., Wen H., Miwa J. M., Xing J., Tekinay A. B., Ono F., Brehm P., Heintz N. (2004) Neuron 43, 305–311 [DOI] [PubMed] [Google Scholar]
- 46. Montuori N., Ragno P. (2009) Front. Biosci. 14, 2494–2503 [DOI] [PubMed] [Google Scholar]
- 47. Lomholt A. F., Høyer-Hansen G., Nielsen H. J., Christensen I. J. (2009) Br. J. Cancer 101, 992–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rasch M. G., Lund I. K., Almasi C. E., Hoyer-Hansen G. (2008) Front. Biosci. 13, 6752–6762 [DOI] [PubMed] [Google Scholar]