Abstract
Free fatty acid receptor 2 (FFA2; GPR43) is a G protein-coupled seven-transmembrane receptor for short-chain fatty acids (SCFAs) that is implicated in inflammatory and metabolic disorders. The SCFA propionate has close to optimal ligand efficiency for FFA2 and can hence be considered as highly potent given its size. Propionate, however, does not discriminate between FFA2 and the closely related receptor FFA3 (GPR41). To identify FFA2-selective ligands and understand the molecular basis for FFA2 selectivity, a targeted library of small carboxylic acids was examined using holistic, label-free dynamic mass redistribution technology for primary screening and the receptor-proximal G protein [35S]guanosine 5′-(3-O-thio)triphosphate activation, inositol phosphate, and cAMP accumulation assays for hit confirmation. Structure-activity relationship analysis allowed formulation of a general rule to predict selectivity for small carboxylic acids at the orthosteric binding site where ligands with substituted sp3-hybridized α-carbons preferentially activate FFA3, whereas ligands with sp2- or sp-hybridized α-carbons prefer FFA2. The orthosteric binding mode was verified by site-directed mutagenesis: replacement of orthosteric site arginine residues by alanine in FFA2 prevented ligand binding, and molecular modeling predicted the detailed mode of binding. Based on this, selective mutation of three residues to their non-conserved counterparts in FFA3 was sufficient to transfer FFA3 selectivity to FFA2. Thus, selective activation of FFA2 via the orthosteric site is achievable with rather small ligands, a finding with significant implications for the rational design of therapeutic compounds selectively targeting the SCFA receptors.
Keywords: Drug Design, Fatty Acid, G Protein-coupled Receptors (GPCR), 7-Helix Receptor, Signal Transduction, FFA2, FFA3, FFAR2, GPR41, GPR43
Introduction
The seven-transmembrane G protein-coupled receptor FFA2,6 previously named GPR43, is expressed primarily on neutrophils, eosinophils, and other immune cells and responds to short-chain fatty acids (SCFAs) (1), which are produced in high concentrations by microbial fermentation in the colon (2). The receptor plays a critical role in neutrophil recruitment during intestinal inflammation (3), and FFA2-deficient mice show exacerbated or unresolved inflammation in colitis, arthritis, and asthma models, indicating that FFA2 can provide a molecular link between fermentable dietary fiber and its beneficial effects on colitis and inflammation (4). These studies suggest that FFA2 is important in the regulation of intestinal inflammation and that the receptor might represent a new drug target for the treatment of inflammatory bowel disease. The receptor is also believed to play a role in energy homeostasis and appetite regulation (5). Further studies, however, are necessary to firmly establish these novel links, and such studies will depend critically on the availability of selective agonists and antagonists for the receptor.
FFA2 and the closely related receptor FFA3 (GPR41) respond to SCFAs at high micromolar and millimolar concentrations with propionate being the most potent agonist for both receptors (6–8). Together with the medium- and long-chain free fatty acid receptor FFA1 (GPR40), the receptors form a subfamily capable of sensing free fatty acids in concentrations corresponding to elevated physiological levels (1, 9). Although the molar potency of the SCFAs on FFA2 and FFA3 must be regarded as low, the compounds are also very small. Ligand efficiency (LE) is a recently introduced concept that assists lead selection by calculating binding energy per non-hydrogen atom as smaller, potent leads increase the likelihood of generating drug candidates with appropriate pharmacokinetic characteristics (10, 11). LE has become popular in evaluation of early leads and is especially widespread in fragment-based drug discovery where such small ligands are grown or assembled to larger and more potent leads (12, 13). Analysis of successful drugs has suggested that an LE of Δg < −0.3 kcal mol−1 per non-hydrogen atom is desirable, and empirical analysis has concluded that the maximal free energy contribution per heavy atom for non-metal ligands is around −1.5 kcal mol−1 per non-hydrogen atom (14). Notably, acetate (C2) and propionate (C3) already have ligand efficiencies close to this value at FFA2 and FFA3 and can therefore be regarded as highly potent for their size (Table 1). Thus, it is unrealistic to expect that the potency of the SCFAs can be increased without at the same time considerably increasing the size of the compounds. It is unclear, however, whether the orthosteric binding site can accommodate significantly larger ligands. A detailed understanding of the interactions of the SCFAs in the orthosteric binding site is expected to indicate the prospects for this strategy and to assist the rational design of larger, more potent and selective modulators of FFA2 and FFA3.
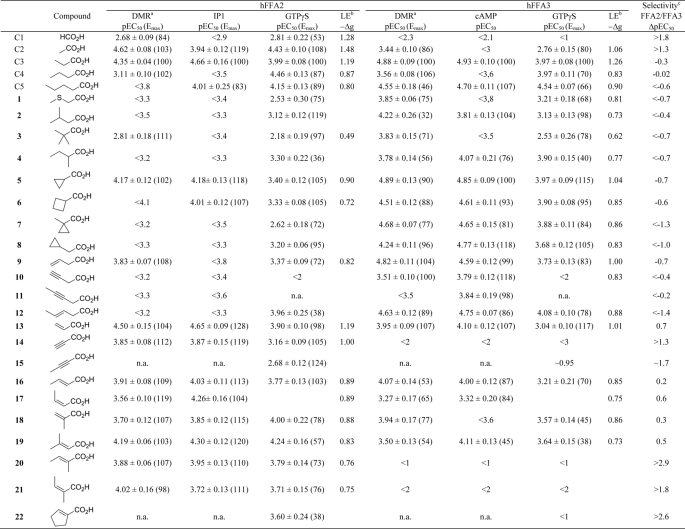
TABLE 1.
Agonist activities of small carboxylic acids on hFFA2 and hFFA3, calculated ligand efficiencies, and average selectivities
Data are mean ± S.E., n ≤ 3.
a DMR assay using the Corning Epic biosensor.
b LE is calculated by the formula −Δg = −ΔG/Nnon-hydrogen atoms where −ΔG = RTln(KD) presuming that KD ≈ EC50. The average of pEC50 values from at least two different assays was used in the calculation. Values are given as kcal mol−1 atom−1.
c Selectivity (calculated by ΔpEC50 = pEC50,FFA2 − pEC50,FFA3) is based on the average pEC50 values of the available assays.
d n.a., not assessed.
Previous structure-activity relationship (SAR) studies have found FFA2 to exhibit a preference for smaller SCFAs than does FFA3 with a rank order of potency of acetate (C2) ∼ propionate (C3) > butyrate (C4) > valerate (C5) ∼ formate (C1) for FFA2 and C3 ∼ C4 ∼ C5 > C2 > caproate (C6) for FFA3 (15). A series of FFA2-selective allosteric agonists that exhibit positive cooperativity with SCFAs has also been disclosed (16, 17). Although these compounds certainly represent useful tools for further pharmacological studies of FFA2, because they bind to a site or sites distinct from the orthosteric binding pocket, it is possible that they may generate signals that are not identical to those triggered by SCFAs. The moderate solubility and metabolic liability of these allosteric compounds furthermore represent a limitation to their use (17). In the present study, we explored the potential of small carboxylic acids (SCAs) as pharmacological tools and further investigated their interaction with the orthosteric binding site. To date, only a small number of saturated straight and branched SCAs have been investigated as ligands at these receptors. Here we report the results from structure-activity investigations of an expanded set of SCAs, including carboxylic acids with additional branched, unsaturated, and cyclic tails. The studies led to the identification of selective orthosteric ligands for both FFA2 and FFA3 and to the elucidation of a general rule to predict the FFA2/FFA3 selectivity of SCAs. Furthermore, this rule was validated by molecular modeling and site-directed mutagenesis at FFA2, resulting in the reversal of selectivity between FFA2 and FFA3 ligands at the mutated receptor.
EXPERIMENTAL PROCEDURES
Materials
Materials for cell culture were purchased from Invitrogen. Cell culture-compatible Epic® biosensor microplates and compound source plates were from Corning Inc. Restriction endonucleases and modifying enzymes were from New England Biolabs, and all other laboratory reagents were obtained from Sigma-Aldrich unless otherwise specified. The radiochemical [35S]GTPγS was from PerkinElmer Life Sciences.
Formic acid (C1), acetic acid (C2), propionic acid (C3), butyric acid (C4), valeric acid (C5), methylthioacetic acid (1), 3-methylbutyric acid (2), pivalic acid (3), 2-methylbutyric acid (4), cyclopropylcarboxylic acid (5), cyclobutylcarboxylic acid (6), 1-methylcyclopropanecarboxylic acid (7), vinylacetic acid (9), 3-pentenoic acid (12), acrylic acid (13), propiolic acid (14), 2-butynoic acid (15), trans-crotonic acid (16), 2-methylacrylic acid (18), 3-methylcrotonic acid (19), trans-2-methylcrotonic acid (20), cyclopent-1-enecarboxylic acid (22), trifluoroacetic acid (23), 3-bromopropionic acid (24), (±)-2-methylcyclopropanecarboxylic acid (25; 1:4 mixture of diastereomers), 1-cyanocyclopropanecarboxylic acid (26), pyruvic acid (27), and (±)-2-phenylpropionic acid (28) were purchased from Sigma-Aldrich. Angelic acid (21) was purchased from ABCR (Karlsruhe, Germany). Cyclopropylacetic acid (8) was purchased from Alfa Aesar. 3-Butynoic acid (10), 3-pentynoic acid (11), and cis-crotonic acid (17) were synthesized as described in the supplemental methods. The identity and purity of all compounds were confirmed by 1H and 13C NMR.
Plasmid Construction
The Flp recombinase-mediated homologous recombination system (Flp-InTM T-RExTM, Invitrogen) was used to generate cell lines stably expressing human FFA2 and FFA3 in a doxycycline-dependent manner. The coding sequences of FFA2 and FFA3 were subcloned from pcDNA3.1 (Invitrogen) into the inducible expression vector pcDNA5/FRT/TO (Invitrogen) via 5′ HindIII and 3′ XhoI. Veracity of the constructs was confirmed by restriction endonuclease digestion. Cloning and generation of enhanced yellow fluorescent protein-tagged versions of human FFA2 and FFA3 (FFA2-eYFP and FFA3-eYFP) and various binding mutants have been described elsewhere (18).
Cell Culture and Generation of Stable Flp-In T-REx HEK293 Cells
Flp-In T-REx cells were maintained in high glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal bovine serum, 1% penicillin/streptomycin mixture, and 15 μg/ml blasticidin at 37 °C and 5% CO2 in a humidified atmosphere.
To generate Flp-In T-REx 293 cells able to inducibly express a receptor of interest, the cells were transfected with a 1:9 mixture of the desired receptor cDNA in pcDNA5/FRT/TO vector and the pOG44 vector (Invitrogen's expression vector for Flp recombinase) using a calcium phosphate DNA precipitation method according to the manufacturer's instructions. After 48 h, the medium was changed to medium supplemented with 100 μg/ml hygromycin B (InvivoGen, Toulouse, France) to initiate selection of stably transfected cells. Expression of the appropriate construct from the Flp-In locus was induced by treatment with 1 μg/ml doxycycline (Sigma) for 16 h.
Dynamic Mass Redistribution (DMR) Assays (Corning Epic Biosensor)
DMR assays were performed on a beta version of the Corning Epic biosensor. A description and validation of this method is detailed in Schröder et al. (19). Briefly, cells were grown in Epic microplates, which are equipped with a resonant wave guide grating biosensor. GPCR signaling-induced mass redistribution, due to relocation of cellular constituents, leads to a change of the optical density in cells. This phenomenon can be detected by passing polarized broadband light through the bottom portion of cells and measuring changes in wavelength of the outgoing light. The wavelength shift (in picometers) is directly proportional to the amount of DMR.
For Epic assays, HEK Flp-In T-REx cells were seeded onto fibronectin-coated 384-well Epic sensor microplates at a density of 15,000 cells/well to obtain confluent monolayers. After cultivation for 20–24 h (at 37 °C and 5% CO2) cells were washed with Hanks' balanced salt solution containing 20 mm HEPES and kept for 1 h at 28 °C in the Epic reader to allow for temperature equilibration. The sensor plate was then scanned to obtain a base-line optical signature. Finally, compound solutions were transferred into the sensor plate with an integrated liquid handling device, and cell responses were recorded continuously for at least 3600 s. All data were normalized to the responses induced by 300 μm propionic acid, which were set to 100%.
[35S]GTPγS G Protein Activation Assays
Membranes expressing wild type or mutant versions of human FFA2-eYFP or FFA3-eYFP were prepared from stable cell lines using 0.5 μg/ml doxycycline (24 h) as described elsewhere (20). [35S]GTPγS binding assays were performed essentially as described (20). Briefly, 5 μg of cell membranes were preincubated for 15 min at 25 °C in assay buffer (50 mm Tris-HCl, pH 7.4, 10 mm MgCl2, 100 mm NaCl, 1 mm EDTA, 1 μm GDP, 0.5% fatty acid-free BSA) containing the indicated concentrations of ligands. Binding was initiated by addition of 50 nCi of [35S]GTPγS to each tube, and the reaction was terminated after 1-h incubation by rapid filtration through GF/C glass filters using a 24-well Brandel cell harvester (Alpha Biotech, Glasgow, UK). Unbound radioligand was washed from filters by three washes with ice-cold wash buffer (50 mm Tris-HCl, pH 7.4, 10 mm MgCl2), and [35S]GTPγS binding was determined by liquid scintillation spectrometry. Membrane expression of each receptor was assessed by measurement of enhanced yellow fluorescent protein (eYFP) using a PHERAstar FS microplate reader (excitation, 485 nm; emission, 520 nm; BMG Labtech, Offenburg, Germany).
cAMP Accumulation Assays
Inhibition of forskolin-stimulated cAMP accumulation in FFA3 cells was monitored with the MithrasLB 940 multimode reader (Berthold Technologies, Bad Wildbad, Germany) using the HTRF®-cAMP dynamic kit (CIS Bio International, Gif-sur-Yvette Cedex, France) according to the manufacturer's instructions. For the assay, cells were resuspended in assay buffer (Hanks' balanced salt solution, 20 mm HEPES, 1 mm 3-isobutyl-1-methylxanthine) and transferred to 384-well small volume microplates (Greiner Bio-One, Frickenhausen, Germany) at a density of 50,000 cells/well. Plates were incubated for 30 min at 37 °C before compounds were added in the presence of 5 μm forskolin. After further incubation for 30 min at 37 °C, the reactions were stopped by adding 1.25% Triton X-100 containing HTRF reagents. Plates were then incubated for 60 min at room temperature, and time-resolved FRET signals were measured after excitation at 320 nm. Both the emission signal from the europium cryptate-labeled anti-cAMP antibody (620 nm) and the FRET signal resulting from the labeled cAMP-d2 (665 nm) were recorded. Results were calculated from the 665/620 nm ratio and expressed as ΔF (ΔF % = ((Standard or sample ratio − Rationeg)/Rationeg) × 100). All data were normalized to the functional response obtained with 300 μm propionic acid.
Inositol Monophosphate (IP1) Accumulation Assays
The HTRF-IP One kit (CIS Bio International) was used for measuring IP1 production in cells expressing FFA2. In a 384-well format, the cell suspension was dispensed at 100,000 cells/7 μl/well. After 20-min incubation at 37 °C, 7 μl of stimulation buffer (Hanks' balanced salt solution, 10 mm HEPES, 50 mm LiCl) containing various concentrations of ligand were added. After further incubation at 37 °C for 30 min, 3 μl of IP1-d2 conjugate followed by 3 μl of europium cryptate-labeled anti-IP1 antibody were added. Time-resolved fluorescence at 620 and 665 nm was measured with the Mithras LB 940 multimode reader after incubation at room temperature for 60 min, and the ratios of the signals expressed as ΔF were calculated as described above. Data were normalized to the IP1 response obtained with 300 μm propionic acid.
Molecular Modeling and Ligand Docking
Homology modeling of hFFA2 and hFFA3 receptors using the β2-adrenergic receptor (Protein Data Bank code 2RH1) as a template was conducted using MOE software (Molecular Operating Environment, 2009, Chemical Computing Group, Inc., Montreal, Quebec, Canada) with a default homology modeling protocol. Selection of homology models was based on the location of extracellular loop 2 (ECL2). Only models where ECL2 was located relatively similarly to the available crystal structures of the ligand-bound GPCRs were considered for docking. Thus, ECL2 conformations that prevented entrance to the putative binding cavity were excluded for the next steps. The available model of FFA1 (21), with the side-chain conformers optimized according to mutagenesis data for HisVI:16/4.56, ArgV:05/5.39, HisVI:20/6.55, and ArgVII:02/7.35, was used to optimize the rough homology models of hFFA2 and hFFA3 obtained. Docking of SCAs into these models and those containing proposed mutants was performed using the Glide module of Schrödinger software (22). The Glide docking box included the cavity between transmembrane helices 3, 5, and 6 involving residues at positions V:05/5.39, VII:02/7.35, and VI:20/6.55, which we have shown to be crucial for anchoring the carboxyl group of fatty acids (18). The Glide default settings with the extraprecision scoring option were used for docking. Images were prepared using the Maestro 8.5 interface (23).
Data Analysis
All data were quantified, grouped, and analyzed using GraphPad Prism 5.02 and are expressed as mean ± S.E. Data were fit to both three-parameter (fixed Hill slope) and four-parameter non-linear regression isotherms. The three-parameter curve fit was statistically appropriate in all cases.
RESULTS
Characterization of Assays for Primary and Secondary Screening
FFA2 and FFA3 clearly mediate distinct physiological and pathological outcomes (1), yet the absence of selective ligands for these receptors has hampered further pharmacological studies of their individual functions. Both receptors couple to the Pertussis toxin-sensitive Gαi/o family of G proteins, whereas FFA2 additionally couples to the Gαq/11 family (6–8, 18). To avoid any potential bias in our assay results due to functional selectivity of tested ligands, we used the holistic, label-free Epic DMR assay, which monitors integrated traffic in the cell in real time without the need for epitope tagging or specific receptor probes (19, 24). Previously, we have demonstrated that DMR is a powerful assay platform for discerning the individual pathways activated by different GPCRs from all four G protein classes (Gαi/o, Gαs, Gαq/11, and Gα12/13) and is therefore unbiased yet also pathway-sensitive (19). Thus, we initially tested the standard straight-chain orthosteric FFA2/FFA3 agonists, formate (C1), acetate (C2), propionate (C3), butyrate (C4), and valerate (C5), using the Epic DMR assay and compared these results with those obtained with the traditional [35S]GTPγS binding assay, which measures predominantly Gαi/o G protein activation (and hence is appropriate for both FFA2 and FFA3 signaling). In addition, second messenger accumulation in whole cells was examined using IP1 assays for FFA2 and cAMP accumulation assays for FFA3.
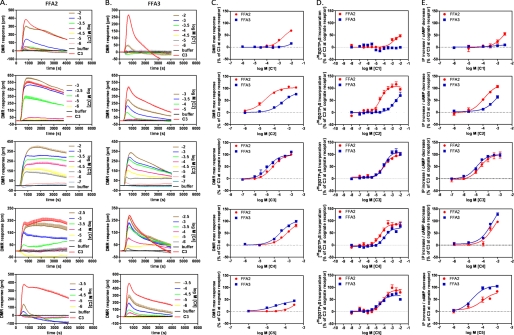
Stimulation of Flp-In T-REx 293 cells stably transfected to express human FFA2 on demand resulted in positive deflections in DMR in a concentration-dependent manner (Fig. 1A), the peaks of which can be converted into log concentration-response curves to determine potency and efficacy for each agonist (Fig. 1C, red symbols, and Table 1). Notably, the DMR traces obtained for the same ligands at FFA2 (Fig. 1A) or FFA3 (Fig. 1B) were qualitatively different, indicating the involvement of non-identical signaling pathways and corroborating previous studies using traditional second messenger assays (6–8, 18, 20). Millimolar activity of formate (C1) on FFA2 was confirmed (7), but no activity was observed on FFA3 at up to 10 mm. In accordance with previous results, acetate (C2) was more than an order of magnitude more potent on FFA2, whereas propionate (C3) was the most potent SCFA and equipotent at each receptor (Fig. 1C and Table 1). Butyrate (C4) and valerate (C5) exhibited modest selectivity for FFA3 and in the case of valerate with lower efficacy than C3 also in agreement with other reports (6, 7).
FIGURE 1.
Short-chain fatty acid responses at FFA2 and FFA3 receptors. Signaling in response to the SCFAs formate (C1), acetate (C2), propionate (C3), butyrate (C4), and valerate (C5) in Flp-In T-REx 293 cells stably expressing either human FFA2 or FFA3 was assessed by measuring DMR in a Corning Epic biosensor. A and B, wavelength shift in picometers (pm) over time (seconds) was assessed upon stimulation with increasing concentrations of ligand as indicated at FFA2 (A) and FFA3 (B). Shown are representative traces from a single experiment representative of three separate experiments. C, concentration-response curves were constructed from the maximum (max) wavelength shift per concentration normalized to C3 at the corresponding receptor, thereby allowing comparison of ligand selectivity at FFA2 (red circles) versus FFA3 (blue squares). Data are mean ± S.E., n = 3. D, concentration-response curves were also generated using the [35S]GTPγS binding assay to enable comparison of receptor selectivity using DMR with a more traditional GPCR readout. Data are mean ± S.E., n = 3. E, comparison of concentration-response curves from cAMP (FFA3) and IP1 (FFA2) second messenger assays. Data are mean ± S.E., n = 3.
Examination of each of C1–C5 in traditional assays of receptor activation resulted in a similar pattern of selectivity (Fig. 1, D and E, and Table 1), although as expected for signals with different levels of amplification and for comparisons between intact cells and cell membrane assays, absolute values of potency varied somewhat. Despite this, C1 and C2 had marked selectivity for FFA2 over FFA3 (ΔpEC50 of >1.8 and >1.3 for C1 and C2, respectively, where positive ΔpEC50 indicates selectivity for FFA2 and a negative number represents FFA3 selectivity). LEs were calculated using an average of the experimentally determined pEC50 values and indicate that C2 and C3 do indeed have LE values close to the maximal value (Table 1).
Selection and Evaluation of Compounds
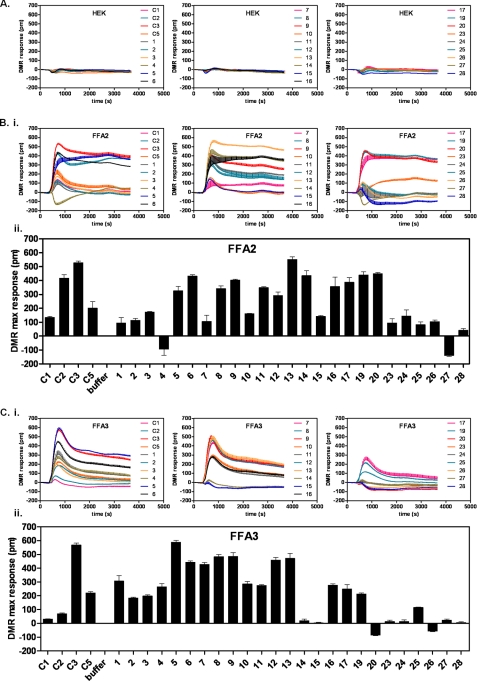
SCAs with lipophilic hydrocarbon tails consisting of four or fewer carbon atoms, including cyclic and unsaturated compounds with well defined three-dimensional structure, were selected to thoroughly explore the binding site of FFA2. We screened these SCAs at a single concentration of 300 μm using the Epic DMR (Fig. 2). None of the noted chemicals produced significant DMR responses in native HEK 293 cells lacking expression of either FFA2 or FFA3 (Fig. 2A). A large proportion of the SCAs tested displayed activity, however, at either or both FFA2 (Fig. 2B) and FFA3 (Fig. 2C). SCAs with polar substituents, like 26 and 27, or larger structures, like 28, were close to inactive on both receptors. Ligands that generated negative DMR signals were not pursued further, whereas the majority of the active compounds were then further explored with both full concentration-response curves in the DMR assay and at [35S]GTPγS binding and appropriate second messenger pathways (Table 1 and below) after being subdivided into groups based upon their structure to enable SAR investigation.
FIGURE 2.
Screening of small carboxylic acids using DMR. Cells either lacking both FFA2 and FFA3 (HEK293 cells; A) or expressing FFA2 (B) or FFA3 (C) were stimulated with a 300 μm concentration of individual SCAs or positive controls (SCFAs C1, C2, C3, and C5) and DMR-monitored over a 60-min period. B, panel i, DMR traces for SCAs over time at FFA2. B, panel ii, maximum (max) wavelength shift (pm) achieved at FFA2 during the length of the assay for each SCA in triplicate. C, DMR traces (panel i) and maximum wavelength shift (panel ii) for the FFA3 counterscreen. Data are mean ± S.E., n = 3.
Branched and Cyclic Compounds
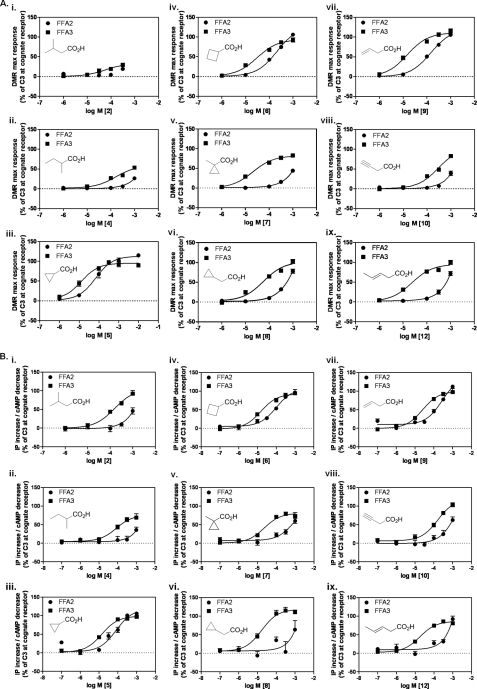
Receptor selectivity and SAR were examined with branched and cyclic SCAs using the Epic DMR assay (Fig. 3A) and conventional signaling assays (Table 1 and Fig. 3B). Isobutyrate, isovalerate (2), and pivalate (3), representing bulkier methyl-substituted analogs of propionate and butyrate, have previously been reported to be at least an order of magnitude more selective for FFA3 over FFA2 (7). We also found these ligands to be selective for FFA3 over FFA2 (ΔpEC50 <−0.4 for 2 and <−0.7 for 3; Fig. 3, A and B, and Table 1), and this relationship extended to sec-valerate (4; ΔpEC50 <−0.7; Fig. 3, A and B). Interestingly, although selective for FFA3, both 2 and 4 were poorly efficacious at this receptor in the DMR assay, an observation that was consistent at the other signaling assays for 4 but not 2. Cyclopropylcarboxylate (5), a somewhat bulkier analog of C3, exhibited moderate selectivity for FFA3, which was maintained upon replacing the cyclopropyl with cyclobutyl (6). The bulky cyclopropyl analog (7) was equipotent with C3 on FFA3 and was ∼30-fold selective for FFA3 over FFA2. A similar selectivity was observed for the cyclopropyl analog 8. In each instance these ligands displayed close to full agonism at FFA3 (Fig. 3, A and B, and Table 1). Based upon these findings, branched and cyclic SCAs appear to be selective for FFA3 over FFA2 at a variety of signaling readouts.
FIGURE 3.
Structure-activity relationships of branched, cyclic, and non-conjugated unsaturated SCAs at FFA2 and FFA3. A, DMR maximum (max) response (pm) concentration-response curves to branched/cyclic (panels i–vi) and non-conjugated unsaturated (panels vii–ix) SCAs (chemical structures are inset) were generated in cells stably expressing either FFA2 (circles) or FFA3 (squares). Data are mean ± S.E., n = 3. B, concentration-response curves of the same chemical series in IP1 (FFA2) and cAMP (FFA3) assays. Data are mean ± S.E., n = 3.
Non-conjugated Unsaturated Compounds
Introduction of a terminal double bond in C4 (9) resulted in preserved FFA3 activity and slightly increased selectivity over FFA2 compared with C4 (ΔpEC50 −0.7; Fig. 3, A and B, and Table 1), whereas introduction of a terminal alkyne (10), which produces a constrained and distinctly shaped C4 analog, resulted in an order of magnitude lower activity on both receptors while maintaining selectivity for FFA3 (ΔpEC50 < −0.4; Fig. 3, A and B). The poor potency of 10 resulted in loss of signal in the [35S]GTPγS binding assay (Table 1). Greatest selectivity for FFA3 in the Epic DMR and second messenger assays was achieved with the rigidified unsaturated analog 12 (ΔpEC50 = −1.4; Fig. 3, A and B). Interestingly, 12 displayed higher efficacy on FFA3 in the receptor-proximal [35S]GTPγS assay, yet the potency of 12 at each receptor was equivalent (Table 1). Regardless, compound 12 is also instructive because it demonstrates that a rigidly extended C4 tail can be contained within the binding sites of both receptors.
Conjugated Unsaturated Compounds
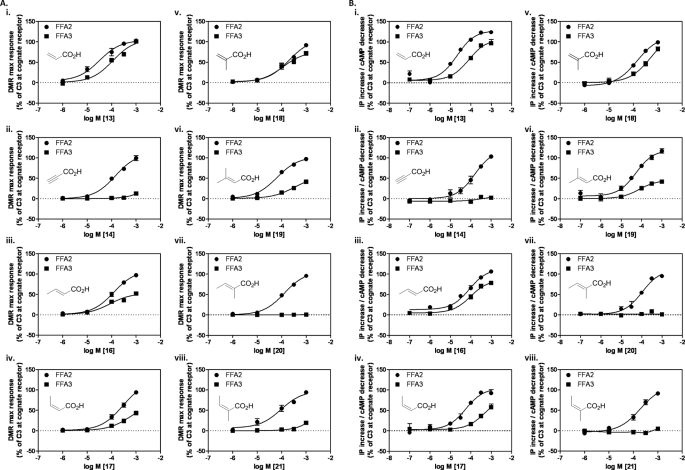
Acrylate (13) represents a narrower analog of propionate and exists preferentially in a flat conformation because of the conjugation between the alkene and the carboxylic acid. This compound was a full agonist at both FFA2 and FFA3 but was moderately selective for FFA2 (ΔpEC50 = 0.7; Fig. 4, A and B). Introduction of a triple bond with propargylate (14) resulted in a ligand markedly more selective for FFA2 (ΔpEC50 > 1.3; Fig. 4, A and B, and Table 1) with little to no activity observed in either the Epic DMR assay or second messenger assays for FFA3. For FFA2, methyl substitutions were accommodated at all positions on acrylate (16-20) (Fig. 4, A and B, and Table 1). In contrast, FFA3 was far less tolerant to conjugated unsaturated compounds. Of the ligands tested, trans-3-methyl (16), cis-3-methyl (17), and 2-methyl (18) substitutions were moderately active with reduced efficacy and, for 17, reduced potency in both the DMR and signaling assays (ΔpEC50 = 0.6). The terminally dimethyl-substituted 19 maintained the selectivity of 17, whereas 2,3-cis-dimethylacrylate (20) and 2,3-trans-dimethylacrylate (21) both exhibited ≫100-fold selectivity for FFA2 (Fig. 4, A and B, and Table 1). The conjugated cyclopentene analog 22 exhibited activity with low efficacy on FFA2 but again ≫100-fold selectivity over FFA3 (Table 1). Thus, the series of conjugated unsaturated SCAs preferentially activated the FFA2 receptor, and this was achieved without substantial loss of LE.
FIGURE 4.
Structure-activity relationships of conjugated unsaturated SCAs at FFA2 and FFA3. A, panels i–viii, DMR maximum response (pm) concentration-response curves to conjugated unsaturated SCAs were generated for either FFA2 (circles) or FFA3 (squares). Data are mean ± S.E., n = 3. B, panels i–viii, concentration-response curves of the same chemical series in IP1 (FFA2) and cAMP (FFA3) assays. Data are mean ± S.E., n = 3.
General Rule for SCA Selectivity at FFA2 and FFA3 and Molecular Modeling of Binding Pocket
Close inspection of the SAR data described above led us to hypothesize a general rule governing SCA selectivity at FFA2 and FFA3. Overall, we found that C1, C2, and all conjugated unsaturated carboxylic acids exhibited substantial selectivity for FFA2, whereas the remaining compounds exhibited selectivity for FFA3. Thus, we propose that ligands with sp2- or sp-hybridized α-carbon preferentially activate FFA2, whereas FFA3-selective ligands contain a substituted sp3-hybridized α-carbon. Furthermore, this relationship indicates that the binding pockets of FFA2 and FFA3 must be subtly different despite both receptors requiring the same four basic amino acids for SCFA binding (18).
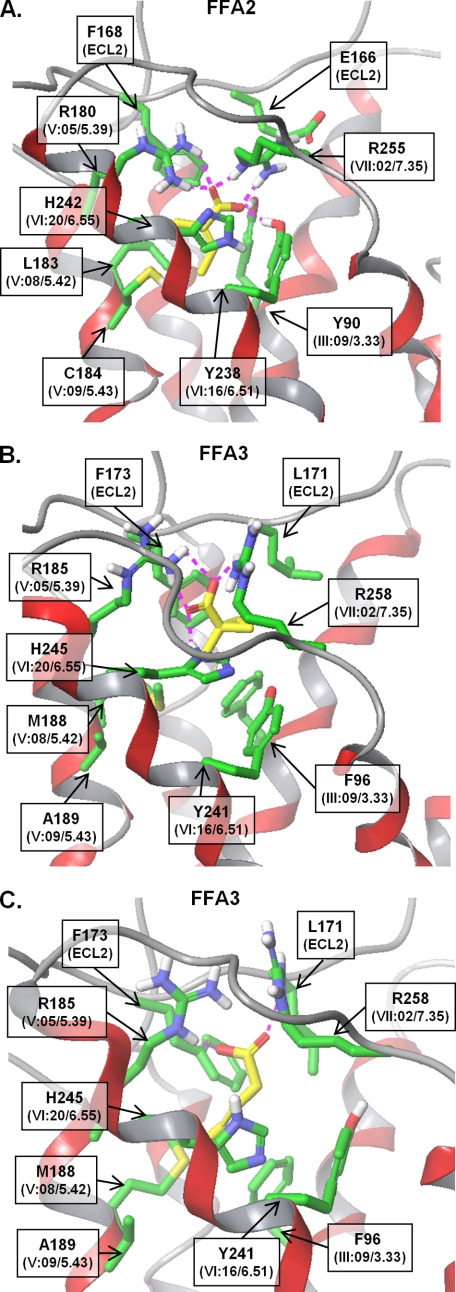
To examine the molecular determinants of SCA ligand selectivity, we docked the FFA2-selective compound 20 (Fig. 5A) and FFA3-selective compound 7 (Fig. 5B) into our molecular models of the receptors. Because 12 also showed receptor-specific effects, albeit on efficacy rather than potency in the [35S]GTPγS assay, we also modeled 12 at FFA3 (Fig. 5C). We have previously demonstrated that SCFA binding requires two conserved arginine residues in both FFA2 and FFA3, ArgV:05/5.39 and ArgVII:08/7.35 (numbered according to the systems of Schwartz and Baldwin (25, 41, 42) and Ballesteros and Weinstein (26)), which are presumed to act as anchoring residues for the carboxylic acid on SCFAs (18). As expected, the carboxyl group of each of the ligands is coordinating these arginines in the models, and the ligands are placed in the cavity comprising transmembrane helices 3, 5, and 6 (Fig. 5, A and B). In particular, the SCA is surrounded by Tyr90(III:09/3.33), Glu166(ECL2), Phe168(ECL2), Leu183(V:08/5.42), Cys184(V:09/5.43), Tyr238(VI:16/6.51), and His242(VI:20/6.55) in hFFA2 and by Phe96(III:09/3.33), Leu171(ECL2), Phe173(ECL2), Met188(V:08/5.42), Ala189(V:09/5.43), Tyr241(VI:16/6.51), and His245(VI:20/6.55) in hFFA3. We noted that the non-conserved residues closest to the aliphatic moiety are residues at positions III:09/3.33, V:08/5.42, and V:09/5.43 and Glu166/Leu171 in ECL2. The aromatic residues at position III:09/3.33 form hydrophobic interactions with the ligand; the tyrosine OH group at this position in FFA2 is predicted to interact with the carboxyl group of 20. It is unlikely that this hydrogen-bond is crucial for binding, however, because the conserved positively charged arginines already anchor this group; thus, this residue is not thought to contribute to ligand selectivity. Of the remaining non-conserved residues within the predicted binding pocket, amino acids at positions 5.42 and 5.43 are likely to provide hydrophobic interactions with the aliphatic moiety of the SCAs. Glu166(ECL2) of FFA2 forms a salt bridge with Arg255(VII:08/7.35), thereby coordinating the position of Arg255, whereas Leu171(ECL2) of FFA3 has hydrophobic interactions with ligands 7 and 12 (Fig. 5C). Because our SAR data indicated that FFA3 preferred larger saturated or alicyclic moieties within the SCAs compared with FFA2, which preferred flat unsaturated moieties, we calculated the volume of the orthosteric binding sites. In accordance with our SAR studies, we found that the modeled binding site volume of FFA3 (105 Å3) is more than double the volume of FFA2 (41 Å3). This difference may be due to the presence of the non-conserved residues discussed earlier as well as different residue packing caused by overall sequence differences between the receptors. To verify that our models and, therefore, our binding pocket hypotheses were predictive, we next examined signaling at a series of orthosteric binding site mutants.
FIGURE 5.
Molecular models of selective SCAs in orthosteric binding sites of FFA2 and FFA3. A, molecular model of compound 20 in the putative FFA2 orthosteric binding site. B, compound 7 in FFA3 binding site. C, compound 12 in FFA3 binding site. Ligand is represented by yellow sticks, side chains of the residues forming the binding site only are shown in green, and the backbone of the receptor is in ribbon style. The GPCR residue notations of Schwartz and Baldwin (25, 41, 42) and Ballesteros and Weinstein (26) are shown in parentheses.
Small Carboxylic Acids Bind to Orthosteric Binding Sites of FFA2 and FFA3
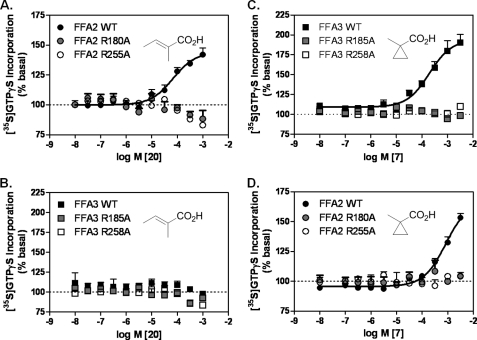
We have previously demonstrated that SCFA binding requires the two conserved arginine residues described above (18). To establish whether the selective SCAs are also accommodated within the orthosteric sites of FFA2 and FFA3, respectively, and to confirm positioning of ligands 20, 7, and 12 in our molecular models, we examined the effect of these ligands in [35S]GTPγS binding assays following mutation of either ArgV:05/5.39 or ArgVII:08/7.35 in FFA2 and FFA3. Membranes were isolated from Flp-In T-REx 293 cells stably expressing hFFA2 R180A(V:05/5.39)-eYFP, hFFA2 R255A(VII:08/7.35)-eYFP, hFFA3 R185A(V:05/5.39)-eYFP, or hFFA3 R258A(VII:08/7.35)-eYFP in an inducible manner (18). For 20, stimulation of wild type FFA2-eYFP but not hFFA2 R180A(V:05/5.39)-eYFP or hFFA2 R255A(VII:08/7.35)eYFP membranes resulted in [35S]GTPγS incorporation (pEC50 = 4.17 ± 0.09; Fig. 6A), whereas no activity was observed at any of the forms of FFA3 (Fig. 6B). Both arginine residues were also required for activity of ligand 7 as mutation of either ArgV:05/5.39 or ArgVII:08/7.35 prevented signal transduction at both FFA3 (pEC50 = 3.74 ± 0.08 for wild type; Fig. 6C) and also FFA2 where it is a weak partial agonist (pEC50 = 3.05 ± 0.09 for wild type; Fig. 6D). Thus, it is clear that the carboxylic acid head groups of the SCAs described here are also coordinating with the positively charged arginines in the orthosteric binding site.
FIGURE 6.
Activity of selective SCAs at FFA2 and FFA3 orthosteric binding mutants. A and B, the FFA2-selective SCA 20 was assessed in the [35S]GTPγS binding assay at wild type (WT) FFA2-eYFP (A) or WT FFA3-eYFP (B) and the corresponding orthosteric binding site mutants R(V:05/5.39)A and R(VII:08/7.35)A (amino acid and sequence number for each receptor construct is indicated in the key). C and D, the same orthosteric binding site mutants were assayed in response to the FFA3-selective SCA, 7. Data are mean ± S.E., n = 3.
Examination of SCA Binding Pocket at FFA2
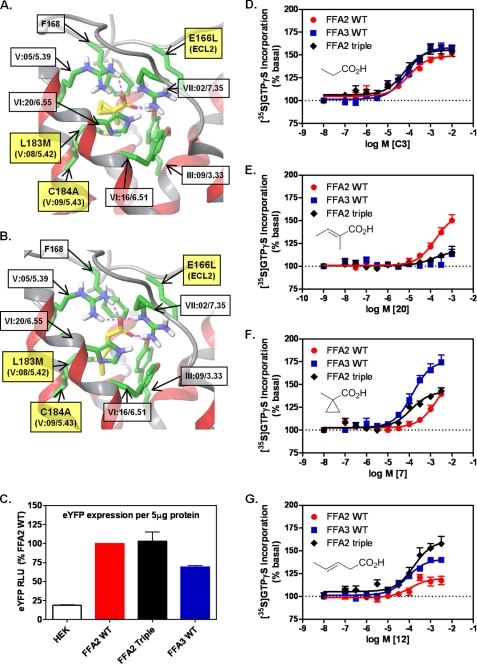
To interrogate the putative mode of binding of FFA2-selective ligands, we replaced three non-conserved residues lining the FFA2 binding site with the corresponding residues in FFA3. By making such a triple mutant receptor, we aimed to change the architecture of the binding pocket to more closely resemble that of FFA3 and, if our hypotheses were correct, change the selectivity of ligands acting at this receptor. Thus, the FFA2 residues Glu166(ECL2), Leu183(V:08/5.42), and Cys184(V:09/5.43) were substituted by the corresponding FFA3 residues Leu, Met, and Ala, respectively (hereafter referred to as “FFA2 triple”) within the context of hFFA2-eYFP (Fig. 7, A and B). Compounds 7 and 12 were subsequently docked in the model of FFA2 triple, indicating that mutation of these residues was consistent with facilitating FFA3 ligand binding. We again calculated the volume of the ligand binding site (this time at FFA2 triple) and found that exchange of the FFA3 residues within FFA2 increased the volume of the orthosteric site (50 Å3), favoring accommodation of the bulkier 7 (Fig. 7A) and 12 (Fig. 7B), which are now located deeper within the pocket. Thus, to examine whether the triple mutant could lead to reversal of ligand selectivity at FFA2, a stable inducible Flp-In T-REx 293 cell line was established. FFA2 triple expression was assessed using eYFP fluorescence as a surrogate parameter and found to be at a level similar to wild type hFFA2-eYFP and not significantly different from wild type hFFA3-eYFP (Fig. 7C).
FIGURE 7.
Triple mutation within FFA2 orthosteric binding site confers FFA3 ligand selectivity to resulting receptor chimera. A and B, molecular model of the hFFA2 E166L(ECL2)/L183M(V:08/5.42)/C184A(V:09/5.43)-eYFP chimeric receptor, FFA2 triple, showing docking of 7 (A) and 12 (B). C, expression of each receptor was established by eYFP fluorescence where HEK293 autofluorescence was used as a negative control. RLU, relative light units. D–G, [35S]GTPγS binding was assessed for non-selective (C3; D), FFA2-selective (20; E), and FFA3-selective (7 and 12; F and G, respectively) SCAs at FFA2 triple. Responses were compared with WT FFA2 and FFA3 in each experiment. Data are mean ± S.E., n = 3.
As noted earlier, propionate is equipotent at FFA2 and FFA3 in the [35S]GTPγS assay (Fig. 7D). Propionate potency at FFA2 triple was identical to wild type FFA2 and FFA3 (Fig. 7D), indicating that these modifications to the orthosteric binding site of FFA2 did not cause gross changes in receptor conformation. Consistent with the Epic DMR assay data in Fig. 4A, 20 was selective for FFA2 because it was inactive at FFA3 (Fig. 7E). Critically, however, activity of 20 at FFA2 triple was markedly impaired, suggesting that the binding pocket could no longer effectively accommodate or be activated by this sp2 ligand (Fig. 7E). Because of their differing effects on selectivity and efficacy in the [35S]GTPγS assay, we also examined both 7 and 12 at the FFA2 triple chimera. The highly selective and bulky compound 7 displayed >100-fold selectivity for FFA3 over FFA2 (pEC50 = 3.76 ± 0.26 at FFA3 and pEC50 = 1.65 ± 0.17 at FFA2; Fig. 7F). Mutation within the FFA2 orthosteric binding pocket was able to rescue 7 function at FFA2 as this ligand displayed equivalent potency at FFA2 triple and the FFA3 receptor (pEC50 = 3.80 ± 0.12) (Fig. 7F), although full agonism was not achieved. Conversely, although ligand 12 was highly selective for FFA3 in the DMR and second messenger assays (Fig. 3 and Table 1), in the [35S]GTPγS assay, it was equipotent at the wild type receptors but differed in the magnitude of signal effect. Interestingly, this poor efficacy at FFA2 wild type was completely reversed at FFA2 triple with maximum activation even exceeding that of wild type FFA3 (Fig. 7G). Thus, guided by developing a chemical rule for predicting specific activation of SCFA receptors and in conjunction with molecular modeling and site-directed mutagenesis, we were able to transfer FFA3 selectivity to FFA2.
DISCUSSION
SCFAs are the endogenous ligands for both of the closely related receptors FFA2 and FFA3, acting with similar potency for all but the very shortest chain length SCFAs, C1 and C2 (1). The physiological roles of the two receptors have largely been based on studies with knock-out mice. The expression of FFA2, however, was recently found to be reduced in FFA3 knock-out mice, making it difficult to draw clear conclusions from these studies (27). Limited endogenous ligand selectivity, the low potency of these ligands, and the absence of selective orthosteric compounds are therefore currently major obstacles to the exploration of the physiological and pathological roles of these receptors both in vitro and in vivo (15). In the present study, we sought to address this issue by examining the molecular determinants of ligand selectivity at FFA2. By screening a targeted library of SCAs at FFA2 and counterscreening at FFA3 using the holistic Epic DMR biosensor assay, we discovered a series of ligands with selectivity for either receptor. SAR analysis of the screening hits in combination with molecular modeling and site-directed mutagenesis enabled formulation of a general chemical rule to predict receptor preference of SCAs. Three amino acids were identified as critical for selective orthosteric activation of FFA2, and transfer of these residues to the binding pocket of FFA3 was sufficient to confer FFA3 ligand selectivity to the FFA2 receptor. Our findings therefore provide the first description of the determinants of selective orthosteric activation for FFA2.
Propionate is the most potent SCFA at both FFA2 and FFA3 and displays close to maximal LE for both these receptors; i.e. it is highly potent based on its size. Although increased potency requires larger ligands, here we demonstrate that development of small but selective ligands is indeed feasible. In fact, 20 and 21 represent two notable examples as they are both selective and potent given their size. These two ligands are expected to be instrumental for the design of novel selective orthosteric FFA2 activators. A multifaceted program combining the validated molecular model of FFA2 described herein with fragment-based drug discovery may facilitate the identification of ligands with enhanced potency either by “growing” the ligands stepwise or by linking the SCAs to fragments docked at accessible positions within the receptor (13). In recent times, a series of receptors have been shown to be activated by endogenous small molecule ligands that are more traditionally considered as products of intermediary metabolism (28–31). All of these ligands have modest potency at their target receptors, and the concepts elucidated and discussed here may have general value in the identification of selective and more potent orthosteric agonists. This is particularly relevant as the potential therapeutic implications of many of these receptors, e.g. GPR109A, clearly define a need to develop potent and selective agonists rather than antagonists (32, 33).
A well defined shape was a criterion for selection of screening compounds to facilitate SAR analysis and modeling. Apart from C5, they only have zero to two rotatable bonds (defined as the number of non-cyclic single bonds where rotation leads to a change in the position of non-hydrogen atoms relative to the remaining molecule; e.g. 14 has zero rotatable bonds). These ligands adopt a very limited number of possible conformations and are therefore ideally suited for modeling and docking studies at the FFA2 binding site. Indeed, evidence of the accuracy of our FFA2 model and SCA docking is provided by the fact that we were able to identify the anchoring arginines for the carboxylic acid group in transmembrane domain V and transmembrane domain VII and transfer ligand selectivity between FFA2 and FFA3 by exchanging just three key amino acids. Very few examples of such a transfer of ligand selectivity between GPCR binding sites have been described. A notable example is the transfer of the binding of AMD3100 from CXCR4 to CXCR3 achieved by a double mutation and the presence of two essential aspartates conserved in both receptors (34).
It is well known from a number of studies that receptor pharmacology may vary depending on the cellular context and the signaling end point under investigation (35–38). The SCFA receptors examined herein are known to engage with Gαi/o family proteins (FFA3) or with both Gαi/o and Gαq/11 (FFA2). This implies that, at least for FFA2, ligands may be identified with preference for individual signaling pathways. To rule out the possibility that so-called biased ligands may escape detection when using a single functional end point assay, one can either gather biological information by testing compounds in several parallel assays or rather use one of the novel, label-free assay technologies that capture whole cell responses in a manner comparable with tissue bioassays (24, 39, 40). The latter may also have the advantage that ligand activity will reflect an integrated response of a living cell rather than a readout determined in isolation and underappreciative of intertwined signaling networks that overlap in time and space. In our study, we took advantage of the Corning Epic biosensor, which translates receptor activation into real time optical traces in a pathway-unbiased yet pathway-sensitive manner (19) and chose this method for primary screening. Hit confirmation was performed with classical biochemical ([35S]GTPγS) or second messenger (cAMP and IP1) assays as validation. The majority of hits identified using the holistic approach were also confirmed in the classical assays. However, we also noted the existence of compounds such as 10 with activity in the Epic but complete inactivity in [35S]GTPγS binding assays. Lack of [35S]GTPγS incorporation induced by 10 at FFA2 might be explained by its inability to engage Gαi/o signaling; however, inactivity at FFA3 of 10 is hard to rationalize given that activity was also detected in parallel cAMP inhibition assays. Hence, it is possible that the receptor-proximal [35S]GTPγS binding assay is simply not sensitive enough to identify activators with poor potency and efficacy because this is likely to be linked closely to receptor occupancy, whereas more distal signals, such as those measured in the DMR assay, often provide substantial amplification and receptor reserve (39). In contrast, 2 was hardly active in DMR assays on either receptor but appeared to be a high efficacy agonist in GTPγS binding. As DMR is an integrated signal, the absence of positive DMR for 2 may reflect additional but opposing cellular events triggered by this compound. Overall, these data demonstrate the strength of combining traditional end point with novel holistic assays to unravel mechanistic differences of compounds that would remain unexplored when using either method in isolation.
It is interesting that introduction of an alkene or alkyne conjugated with the carboxylic acid in the SCAs consistently reversed FFA receptor selectivity and that FFA2 was able to accommodate and respond to larger compounds provided that they contained this conjugated unsaturation. Acrylate (13) is the parent compound of most of these and has agonist activity comparable with C3 yet preferentially activates FFA2 over FFA3. All substituted acrylates (16-22) were FFA2 agonists and showed preference or in most cases significant selectivity for FFA2 over FFA3. These results indicate that the binding pocket of FFA2 has a narrow shape, and steric restrictions apply in the area around the α-carbon atoms of the SCFAs. The preferred planar structure around the carboxylic acid due to conjugation is likely to be a significant factor in FFA2 selectivity. These observations have led to the identification of a general rule to predict FFA2 versus FFA3 selectivity of small (<6 carbon atoms) carboxylic acids with lipophilic tails: compounds carrying an sp2- or sp-hybridized α-carbon will be FFA2-selective, whereas compounds carrying a substituted sp3-hybridized α-carbon will be FFA3-selective. The rule has proven highly predictive, and we have yet to observe any exceptions.
In summary, the first series of molecular tools selectively activating FFA2 via the orthosteric binding site was provided, and many of these ligands showed close to optimal ligand efficiencies. Hence, it is possible to achieve selective activation of FFA2 versus FFA3 with rather small ligands. Such ligands will undoubtedly be useful to further explore the mechanism of activation of FFA2 in more detail or its signaling characteristics in vitro even in cells coexpressing the related FFA3. Future studies are needed to unravel whether selectivity can be maintained within molecules that are gradually increased in size to also achieve a gain of potency that will be a prerequisite to explore the role of FFA2 in vivo. Nevertheless, our results provide, for the first time, a molecular explanation for selective orthosteric activation of FFA2 by highlighting previously unrecognized determinants of selectivity within the orthosteric binding site.
Acknowledgment
We thank Ulrike Rick for expert technical assistance.
This work was supported in part by Danish Council for Independent Research Technology and Production Grant 09-070364, the Danish Medical Research Council, and Wellcome Trust Grant 089600/Z/09/Z.

The on-line version of this article (available at http://www.jbc.org) contains supplemental methods.
- FFA2
- free fatty acid receptor 2
- FFA3
- free fatty acid receptor 3
- FFA1
- free fatty acid receptor 1
- SCFA
- short-chain fatty acid
- SCA
- small carboxylic acid
- GTPγS
- guanosine 5′-(3-O-thio)triphosphate
- LE
- ligand efficiency
- SAR
- structure-activity relationship
- eYFP
- enhanced yellow fluorescent protein
- DMR
- dynamic mass redistribution
- GPCR
- G protein-coupled receptor
- IP1
- inositol monophosphate
- hFFA2
- human FFA2
- hFFA3
- human FFA3
- ECL2
- extracellular loop 2.
REFERENCES
- 1. Stoddart L. A., Smith N. J., Milligan G. (2008) Pharmacol. Rev. 60, 405–417 [DOI] [PubMed] [Google Scholar]
- 2. Topping D. L., Clifton P. M. (2001) Physiol. Rev. 81, 1031–1064 [DOI] [PubMed] [Google Scholar]
- 3. Sina C., Gavrilova O., Förster M., Till A., Derer S., Hildebrand F., Raabe B., Chalaris A., Scheller J., Rehmann A., Franke A., Ott S., Häsler R., Nikolaus S., Fölsch U. R., Rose-John S., Jiang H. P., Li J., Schreiber S., Rosenstiel P. (2009) J. Immunol. 183, 7514–7522 [DOI] [PubMed] [Google Scholar]
- 4. Maslowski K. M., Vieira A. T., Ng A., Kranich J., Sierro F., Yu D., Schilter H. C., Rolph M. S., Mackay F., Artis D., Xavier R. J., Teixeira M. M., Mackay C. R. (2009) Nature 461, 1282–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sleeth M. L., Thompson E. L., Ford H. E., Zac-Varghese S. E., Frost G. (2010) Nutr. Res. Rev. 23, 135–145 [DOI] [PubMed] [Google Scholar]
- 6. Brown A. J., Goldsworthy S. M., Barnes A. A., Eilert M. M., Tcheang L., Daniels D., Muir A. I., Wigglesworth M. J., Kinghorn I., Fraser N. J., Pike N. B., Strum J. C., Steplewski K. M., Murdock P. R., Holder J. C., Marshall F. H., Szekeres P. G., Wilson S., Ignar D. M., Foord S. M., Wise A., Dowell S. J. (2003) J. Biol. Chem. 278, 11312–11319 [DOI] [PubMed] [Google Scholar]
- 7. Le Poul E., Loison C., Struyf S., Springael J. Y., Lannoy V., Decobecq M. E., Brezillon S., Dupriez V., Vassart G., Van Damme J., Parmentier M., Detheux M. (2003) J. Biol. Chem. 278, 25481–25489 [DOI] [PubMed] [Google Scholar]
- 8. Nilsson N. E., Kotarsky K., Owman C., Olde B. (2003) Biochem. Biophys. Res. Commun. 303, 1047–1052 [DOI] [PubMed] [Google Scholar]
- 9. Wellendorph P., Johansen L. D., Bräuner-Osborne H. (2009) Mol. Pharmacol. 76, 453–465 [DOI] [PubMed] [Google Scholar]
- 10. Abad-Zapatero C., Metz J. T. (2005) Drug. Discov. Today 10, 464–469 [DOI] [PubMed] [Google Scholar]
- 11. Hopkins A. L., Groom C. R., Alex A. (2004) Drug. Discov. Today 9, 430–431 [DOI] [PubMed] [Google Scholar]
- 12. Coyne A. G., Scott D. E., Abell C. (2010) Curr. Opin. Chem. Biol. 14, 299–307 [DOI] [PubMed] [Google Scholar]
- 13. de Kloe G. E., Bailey D., Leurs R., de Esch I. J. (2009) Drug. Discov. Today 14, 630–646 [DOI] [PubMed] [Google Scholar]
- 14. Kuntz I. D., Chen K., Sharp K. A., Kollman P. A. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 9997–10002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Milligan G., Stoddart L. A., Smith N. J. (2009) Br. J. Pharmacol. 158, 146–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee T., Schwandner R., Swaminath G., Weiszmann J., Cardozo M., Greenberg J., Jaeckel P., Ge H., Wang Y., Jiao X., Liu J., Kayser F., Tian H., Li Y. (2008) Mol. Pharmacol. 74, 1599–1609 [DOI] [PubMed] [Google Scholar]
- 17. Wang Y., Jiao X., Kayser F., Liu J., Wang Z., Wanska M., Greenberg J., Weiszmann J., Ge H., Tian H., Wong S., Schwandner R., Lee T., Li Y. (2010) Bioorg. Med. Chem. Lett. 20, 493–498 [DOI] [PubMed] [Google Scholar]
- 18. Stoddart L. A., Smith N. J., Jenkins L., Brown A. J., Milligan G. (2008) J. Biol. Chem. 283, 32913–32924 [DOI] [PubMed] [Google Scholar]
- 19. Schröder R., Janssen N., Schmidt J., Kebig A., Merten N., Hennen S., Müller A., Blättermann S., Mohr-Andrä M., Zahn S., Wenzel J., Smith N. J., Gomeza J., Drewke C., Milligan G., Mohr K., Kostenis E. (2010) Nat. Biotechnol. 28, 943–949 [DOI] [PubMed] [Google Scholar]
- 20. Smith N. J., Stoddart L. A., Devine N. M., Jenkins L., Milligan G. (2009) J. Biol. Chem. 284, 17527–17539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sum C. S., Tikhonova I. G., Costanzi S., Gershengorn M. C. (2009) J. Biol. Chem. 284, 3529–3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schrödinger, LLC (2009) Glide, Version 5, Schrödinger, LLC, New York [Google Scholar]
- 23. Schrödinger, LLC (2009) Maestro 8.5, Version 5.5, Schrödinger, LLC, New York [Google Scholar]
- 24. Kenakin T. (2010) Nat. Biotechnol. 28, 928–929 [DOI] [PubMed] [Google Scholar]
- 25. Rosenkilde M. M., Benned-Jensen T., Frimurer T. M., Schwartz T. W. (2010) Trends Pharmacol. Sci. 31, 567–574 [DOI] [PubMed] [Google Scholar]
- 26. Ballesteros J. A., Weinstein H. (1995) Methods Neurosci. 25, 366–428 [Google Scholar]
- 27. Zaibi M. S., Stocker C. J., O'Dowd J., Davies A., Bellahcene M., Cawthorne M. A., Brown A. J., Smith D. M., Arch J. R. (2010) FEBS Lett. 584, 2381–2386 [DOI] [PubMed] [Google Scholar]
- 28. Tunaru S., Kero J., Schaub A., Wufka C., Blaukat A., Pfeffer K., Offermanns S. (2003) Nat. Med. 9, 352–355 [DOI] [PubMed] [Google Scholar]
- 29. Wise A., Foord S. M., Fraser N. J., Barnes A. A., Elshourbagy N., Eilert M., Ignar D. M., Murdock P. R., Steplewski K., Green A., Brown A. J., Dowell S. J., Szekeres P. G., Hassall D. G., Marshall F. H., Wilson S., Pike N. B. (2003) J. Biol. Chem. 278, 9869–9874 [DOI] [PubMed] [Google Scholar]
- 30. He W., Miao F. J., Lin D. C., Schwandner R. T., Wang Z., Gao J., Chen J. L., Tian H., Ling L. (2004) Nature 429, 188–193 [DOI] [PubMed] [Google Scholar]
- 31. Ahmed K., Tunaru S., Langhans C. D., Hanson J., Michalski C. W., Kölker S., Jones P. M., Okun J. G., Offermanns S. (2009) J. Biol. Chem. 284, 21928–21933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ahmed K., Tunaru S., Offermanns S. (2009) Trends Pharmacol. Sci. 30, 557–562 [DOI] [PubMed] [Google Scholar]
- 33. Offermanns S. (2006) Trends Pharmacol. Sci. 27, 384–390 [DOI] [PubMed] [Google Scholar]
- 34. Rosenkilde M. M., Gerlach L. O., Jakobsen J. S., Skerlj R. T., Bridger G. J., Schwartz T. W. (2004) J. Biol. Chem. 279, 3033–3041 [DOI] [PubMed] [Google Scholar]
- 35. Galandrin S., Oligny-Longpré G., Bouvier M. (2007) Trends Pharmacol. Sci. 28, 423–430 [DOI] [PubMed] [Google Scholar]
- 36. Kenakin T. (2007) Trends Pharmacol. Sci. 28, 407–415 [DOI] [PubMed] [Google Scholar]
- 37. Kenakin T. (2008) Comb. Chem. High Throughput Screen. 11, 337–343 [DOI] [PubMed] [Google Scholar]
- 38. Smith N. J., Bennett K. A., Milligan G. (2011) Mol. Cell. Endocrinol. 331, 241–247 [DOI] [PubMed] [Google Scholar]
- 39. Kenakin T. P. (2009) Nat. Rev. Drug Discov. 8, 617–626 [DOI] [PubMed] [Google Scholar]
- 40. Rocheville M., Jerman J. C. (2009) Curr. Opin. Pharmacol. 9, 643–649 [DOI] [PubMed] [Google Scholar]
- 41. Baldwin J. M. (1993) EMBO J. 12, 1693–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schwartz T. W. (1994) Curr. Opin. Biotechnol. 5, 434–444 [DOI] [PubMed] [Google Scholar]