Abstract
Germ cells possess the unique ability to acquire totipotency during development in vivo as well as give rise to pluripotent stem cells under the appropriate conditions in vitro. Recent studies in which somatic cells were experimentally converted into pluripotent stem cells revealed that genes expressed in primordial germ cells (PGCs), such as Oct3/4, Sox2, and Lin28, are involved in this reprogramming. These findings suggest that PGCs may be useful for identifying factors that successfully and efficiently reprogram somatic cells into toti- and/or pluripotent stem cells. Here, we show that Blimp-1, Prdm14, and Prmt5, each of which is crucial for PGC development, have the potential to reprogram somatic cells into pluripotent stem cells. Among them, Prmt5 exhibited remarkable reprogramming of mouse embryonic fibroblasts into which Prmt5, Klf4, and Oct3/4 were introduced. The resulting cells exhibited pluripotent gene expression, teratoma formation, and germline transmission in chimeric mice, all of which were indistinguishable from those induced with embryonic stem cells. These data indicate that some of the factors that play essential roles in germ cell development are also active in somatic cell reprogramming.
Keywords: Development, Embryo, Embryonic Stem Cell, Epigenetics, Induced Pluripotent Stem (iPS) Cell, Primordial Germ Cell
Introduction
Yamanaka and colleagues (1) showed that somatic cells could be reprogrammed by specific factors. To achieve this, the authors induced somatic cells to adopt an embryonic stem (ES)4 cell-like state by introducing Oct3/4, Sox2, Klf4, and c-Myc into mouse fibroblasts. This technique for producing induced pluripotent stem (iPS) cells has also been applied to human somatic cells (2, 3). Because the generation of iPS cells is not plagued by either ethical considerations or immune rejection, the potential of this technology is of great importance for the field of regenerative medicine. Apart from the potential clinical applications, iPS cell generation also provides an excellent model system for investigating the mechanisms underlying cellular reprogramming in vitro through the identification of alternative or additional factors involved in this process. So far, a number of reprogramming factors, including Oct3/4, Sox2, Klf2/4/5, c-Myc, and Lin28, as well as reprogramming-inhibitory factors such as p53 and let7, have been identified in this way (3–5). Of note, each factor is not only active during reprogramming but it also has biological significance in development and homeostasis in the cells and tissues. Frequency of the conversion into iPS cells by transfection with these four reprogramming factors is still very low, implying that other, unidentified reprogramming factors may exist.
Before the discovery of iPS cells, pluripotent cells could only be established from germ cells in post-implantation embryos. Primordial germ cells (PGCs) and spermatogonia are known to de-differentiate into pluripotent stem cells, embryonic germ (EG) cells and multipotent germline stem cells, respectively (6, 7). Consistent with their ability to de-differentiate, cells from the germ cell lineage express key factors that maintain pluripotency, such as Oct3/4 and Sox2 (8). In addition, the efficiency of deriving of EG cells from PGCs is much higher than the efficiency of generating iPS cells from mouse embryonic fibroblasts (MEFs), suggesting that germ cells already possess critical reprogramming factors. Recent studies have identified several factors that are crucial for germ cell specification and development (9). Of particular interest, a pair of PR domain-containing factors, Blimp-1/Prdm1 and Prdm14, is essential for early PGC development. Blimp-1 is detectable at the earliest stage of PGC specification and is essential for repressing somatic programs such as Hox gene expression. Prdm14 expression follows Blimp-1 expression in PGCs and also plays a pivotal role in germ cell specification (10, 11). Both mutants of these genes fail to allocate the PGC population, which has been attributed to the impaired repression of somatic programs followed by genome-wide reprogramming. Although it is unclear whether PR domains are essential for reprogramming, Blimp-1 is known to interact with many epigenetic factors, including the arginine-specific histone methyltransferase, Prmt5. Prmt5 is thought to play a role in epigenetic reprogramming in germ cells (12, 13). During germ cell development, epigenetic reprogramming occurs via massive DNA demethylation and altered histone modification that normally occurs in the embryonic gonad (14), suggesting that the combined expression of Blimp-1, Prmd14, and Prmt5 evokes proper epigenetic reprogramming in PGCs. Likewise, during iPS cell induction, somatic cell gene expression is repressed and histone modification patterns are markedly altered (15). These findings suggest that there is a functional relationship between germ cell development and somatic cell reprogramming.
Here, we investigated whether Blimp-1, Prdm14, and Prmt5, which are important for reprogramming in PGCs, are also active in somatic cell reprogramming. We screened cells expressing combinations of these transcription factors and found that all of these factors had reprogramming activity. Somatic cell reprogramming of MEFs was especially effective when Prmt5 was introduced in combination with Klf4 and Oct3/4 (PKO cells). PKO cells were indistinguishable from ES cells in terms of their gene expression profile, DNA methylation status, capacity to differentiate into the three germ layers, and germline transmission. Here we show that factors with essential roles in germ cell development are also active in somatic cell reprogramming.
EXPERIMENTAL PROCEDURES
Mice
A Nanog-GFP-IRES-puro transgenic mouse (RBRC02290) was provided by RIKEN BRC, which is participating in the National Bio-Resource Project of the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan (16). C57BL/6 and Balb/c nude mice were purchased from Japan SLC (Shizuoka, Japan). Animal care was in accordance with the guidelines of Keio University for animal and recombinant DNA experiments. MEFs were prepared from E13.5 embryos obtained by crossing Nanog-GFP mice with C57BL/6.
Cell Culture
PKO cells were cultured on mitomycin C-treated STO cells with knock-out DMEM containing 15% FCS, LIF, penicillin/streptomycin, l-glutamine, β-mercaptoethanol, and non-essential amino acids. The PKO cells were depleted of feeder cells by being incubated twice on a 2% gelatin-coated dish. Collected cells were used for DNA and RNA analysis.
Feeder Cell Preparation
STO cells were treated with 12 μg/ml of mitomycin C for 2 h and plated at a density of 1 × 106 cells/55 cm2.
Plasmids
Retroviral plasmids for iPS cell induction were provided by Addgene as follows: pMXs-Sox2 (Addgene plasmid 13367), pMXs-Oct3/4 (Addgene plasmid 13366), pMXs-Klf4 (Addgene plasmid 13370), and pMXs-c-Myc (Addgene plasmid 13375). The Blimp-1-IRES-GFP vector was a gift from Dr. Kiyoshi Takatsu and the IRES-GFP region was excised (17). Prdm14 and Prmt5 were cloned by PCR, inserted into the pGEM-T-easy plasmid (Promega), and converted to pMXs via the BamHI and XhoI sites. The PCR primers used were: Prdm14, forward, 5′-CCGCGGTCCGCAAAACTCAGGCCACCATGG-3′, and Prdm14, reverse, 5′-CTCGAGAACCATGCCCACGCGACACAGACA-3′; Prmt5, forward, 5′-GGATCCGGCCGCGCGAGGGCCACCATGGCG-3′, and Prmt5, reverse, 5′-CTCGAGAACCAGCAGATGTTCTACACCTTC-3′.
Reprogramming of MEFs by Blimp-1, Prdm14, and Prmt5
PKO cells were generated by the iPS cell induction method as described previously (16), except for cell density at the time of re-seeding. MEFs transfected with Prmt5, Klf4, and Oct3/4 were reseeded on mitomycin C-treated STO cells at 5 days after infection, at the cell densities indicated under Table 2. Primary ES cell-like colonies of MEFs transfected with Blimp-1, Prdm14, and c-Myc were reseeded onto a 0.1% gelatin-coated dish without STO feeder cells at a density of 580,000 cells/35-mm dish.
TABLE 2.
PKO cell colony numbers
| Well No. | Number of cells at reseeding | Puromycin selection | Numbers of Nanog-GFP positive colonies |
Total | ||
|---|---|---|---|---|---|---|
| Day 23 | Day 26 | Day 29 | ||||
| ×105/35 mm | days after infection | |||||
| 1 | 8.75 | 16 | 2 | 0 | 0 | 2 |
| 2 | 5.8 | 16 | 3 | 0 | 0 | 3 |
| 3 | 0.58 | 16 | 1 | 0 | 0 | 1 |
| 4 | 8.75 | 23 | 3 | 5 | 2 | 10 |
| 5 | 5.8 | 23 | 0 | 9 | 3 | 12 |
| 6 | 0.58 | 23 | 0 | 2 | 0 | 2 |
| Yamanaka's method | 0.58 | 14 | ||||
Gene Expression Analysis
Primers and probes for Oct3/4, Sox2, Klf4, and c-Myc were designed to distinguish between endogenous and viral transcripts (see supplemental Table S1). Transcript levels were determined using the 7500 Fast Real Time PCR system (Applied Biosystems).
Bisulfate Sequencing
Bisulfate reactions were performed with the EpiTecht Bisulfite Kit (Qiagen) according to the manufacturer's instructions. Primers used for the PCR have been described previously (7, 16). PCR products were cloned into the pGEM-T Easy plasmid (Promega) and sequenced by conventional methods.
Teratoma Formation
To produce teratomas, 1.0 × 106 cells were suspended in BD Matrigel (BD Biosciences) and injected into nude mice. Three to 4 weeks later, tumors were fixed with 4% paraformaldehyde in PBS, embedded in paraffin, sectioned, and stained with hematoxylin and eosin.
Creation of Chimeric Mice
To analyze the germ cell contribution of PKO cells, PKO cells were injected into blastocysts from ICR mice. Gonads were isolated at E12.5 and examined for PKO cell-derived Nanog-GFP fluorescence. At the same time, the contribution of PKO cells to the eyes was also analyzed by the examination of pigment cells. To generate adult chimeric mice from PKO cells, blastocysts with a B6 background were used. Functional germ cell development was analyzed by the fluorescence of Nanog-GFP in the F1 morula.
Microarray Data Analysis
Expression profiles were analyzed using the three-dimensional Gene Mouse Oligo chip 24K (Toray Industries, Tokyo, Japan). The fluorescence intensities were detected using the Scan-Array Life Scanner (PerkinElmer Life Sciences). PMT levels were adjusted to achieve 0.1–0.5% pixel saturation. Each TIFF image was analyzed using GenePix Pro version 6.0 software (Molecular Devices, Sunnyvale, CA). The data were filtered to remove low confidence measurements and globally normalized per array, such that the median signal intensity was adjusted to 50 after normalization (accession number GSE18813).
Detection of Introduced Alleles
Genomic integration of introduced factors were analyzed with the following primers: Virus Klf4, forward, 5′-CATCCTCTAGACTGCCGGATCT-3′ and Virus Klf4, reverse, 5′-GATTCAATATAAACCGGCGATGTC-3′; Virus Sox2, forward, 5′-TTAAGGATCCCAGTGTGGTGGTA-3′, and Virus Sox2, reverse, 5′-TTCAGCTCCGTCTCCATCATG-3′; Virus Oct3/4, forward, 5′-CATCCTCTAGACTGCCGGATCT-3′, and Virus Oct3/4, reverse, 5′-TGCTTCAGCAGCTTGGCAAACTGT-3′; Virus c-Myc, forward, 5′-CATCCTCTAGACTGCCGGATCT-3′, and Virus c-Myc, reverse, 5′-AGGTCATAGTTCCTGTTGGTGAAGT-3′; Virus Prmt5, forward, 5′-CATCCTCTAGACTGCCGGATCT-3′ and Virus Prmt5, reverse, 5′-AATTCAGGTCCCTCCCGCTGGACA-3′; control (IL-2), forward, 5′-CTAGGCCACAGAATTGAAAGATCT-3′, and control (IL-2), reverse, 5′-GTAGGTGGAAATTCTAGCATCATCC-3′.
Knockdown of Prmt5
Previously reported sequences were used for the knockdown of Prmt5 (18). The sequences were introduced with a retroviral expression vector (19). Virus production and infection during the reprogramming process were performed as described as above (16). The viruses for knockdown were added at a 3 times higher concentration than the reprogramming factors.
Purification of MEFs after Three- or Four-factor Infection
MEFs infected with three or four factors were harvested at days 3 and 12. The cells were incubated with anti-FcγR antibody (2.4G2) (eBioscience) at 4 °C for 30 min. Then, cells were continuously incubated with allophycocyanin-conjugated anti-Thy-1 monoclonal antibody (53-2.1) (BD Biosciences) and PE-conjugated anti-SSEA-1 monoclonal antibody (MC-480) (BD Biosciences) for 30 min at 4 °C. After washing, samples were sorted by FACS AriaII (BD Biosciences).
RESULTS
Screen for the Reprogramming Activity of Blimp-1, Prdm14, and Prmt5
The unique relationship between PGCs and pluripotency led to an investigation of the reprogramming activity of factors involved in PGC development, EG cell derivation, or both. Factors that met the following criteria were selected: (i) preferential expression in early PGCs during reprogramming, (ii) a role in early PGC development, and (iii) involvement in pluripotent EG cell derivation. This screen yielded three candidate genes: Blimp-1 (also known as Prdm1), Prdm14, and Prmt5.
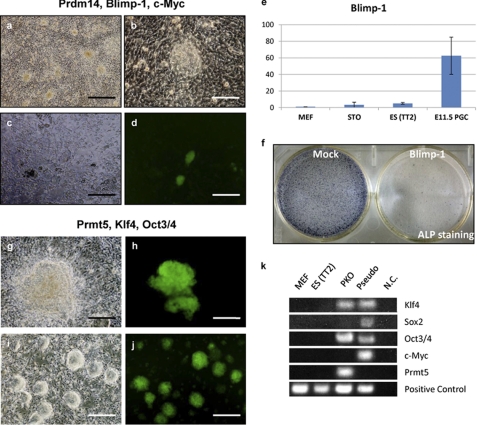
Subsequently, the reprogramming activity of Blimp-1, Prdm14, and Prmt5 was examined in somatic cells. Various combinations of the three factors were introduced, along with Oct3/4, Sox2, Klf4, or c-Myc, into MEFs carrying the Nanog-GFP reporter gene (Table 1). Although no Nanog-positive colonies grew after the expression of each of the three factors alone, Nanog-GFP-positive colonies were detected in two conditions when the cells were co-infected with the three known factors. In the first instance, MEFs co-infected with Blimp-1, Prdm14, and c-Myc (Table 1, case 4) produced primary colonies with an ES cell-like morphology (Fig. 1). A few colonies occasionally expressed Nanog-GFP but most did not (Fig. 1), and the Nanog-positive colonies exhibited severely inhibited growth. Moreover, cell growth was arrested when the Nanog-GFP-positive colonies were isolated and re-plated. Thus, despite the emergence of primary Nanog-GFP-positive colonies after infection with Blimp-1, Prdm14, and c-Myc, the MEFs did not become completely reprogrammed pluripotent stem cells. Because Blimp-1 is not expressed in ES cells (Fig. 1), it is possible that this factor acts only during the induction phase. Indeed, overexpression of Blimp-1 in ES cells resulted in cell growth defects (Fig. 1). This may explain why the appearance of Nanog-GFP colonies was very rare and why a cell line could not be established by infection with Blimp-1, Prdm14, and c-Myc. Complete silencing of Blimp-1 may be necessary to establish a cell line during reprogramming.
TABLE 1.
Screened combination of introduced genes
| Case | Name | Induced factors |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| GFPa | Klf4 | Sox2 | Oct3/4 | c-Myc | Blimp-1 | Prdm14 | Prmt5 | ||
| 1 | K,1,14 | + | + | + | |||||
| 2 | S,1,14 | + | + | + | |||||
| 3 | O,1,14 | + | + | + | |||||
| 4 | M,1,14 | + | + | + | + | ||||
| 5 | K,1,5 | + | + | + | |||||
| 6 | S,1,5 | + | + | + | |||||
| 7 | O,1,5 | + | + | + | |||||
| 8 | M,1,5 | + | + | + | |||||
| 9 | K,14,5 | + | + | + | |||||
| 10 | S,14,5 | + | + | + | |||||
| 11 | O,14,5 | + | + | + | |||||
| 12 | M,14,5 | + | + | + | |||||
| 13 | K,S,1 | + | + | + | |||||
| 14 | K,S,14 | + | + | + | |||||
| 15 | K,S,5 | + | + | + | |||||
| 16 | K,O,1 | + | + | + | |||||
| 17 | K,O,14 | + | + | + | |||||
| 18 | K,O,5 | + | + | + | + | ||||
| 19 | S,O,1 | + | + | + | |||||
| 20 | S,O,14 | + | + | + | |||||
| 21 | S,O,5 | + | + | + | |||||
| 22 | K,S,1,14 | + | + | + | + | ||||
| 23 | K,O,1,14 | + | + | + | + | ||||
| 24 | S,O,1,14 | + | + | + | + | ||||
| 25 | K,S,1,5 | + | + | + | + | ||||
| 26 | K,O,1,5 | + | + | + | + | ||||
| 27 | S,O,1,5 | + | + | + | + | ||||
| 28 | K,S,14,5 | + | + | + | + | ||||
| 29 | K,O,14,5 | + | + | + | + | ||||
| 30 | S,O,14,5 | + | + | + | + | ||||
| 31 | 1,14 | + | + | ||||||
| 32 | 1,5 | + | + | ||||||
| 33 | 14,5 | + | + | ||||||
| 34 | 1 | + | |||||||
| 35 | 14 | + | |||||||
| 36 | 5 | + | |||||||
| 37 | K,1 | + | + | ||||||
| 38 | S,1 | + | + | ||||||
| 39 | O,1 | + | + | ||||||
| 40 | M,1 | + | + | ||||||
| 41 | K,14 | + | + | ||||||
| 42 | S,14 | + | + | ||||||
| 43 | O,14 | + | + | ||||||
| 44 | M,14 | + | + | ||||||
| 45 | K,5 | + | + | ||||||
| 46 | S,5 | + | + | ||||||
| 47 | O,5 | + | + | ||||||
| 48 | M,5 | + | + | ||||||
| 49 | 1,14,5 | + | + | + | |||||
| 50 | K,S,O | + | + | + | + | ||||
| 51 | K,S,O,M | + | + | + | + | + | |||
a Nanog-GFP positive colonies.
FIGURE 1.
Reprogramming of MEFs by Blimp-1, Prdm14, and Prmt5. a–d, the morphology of MEFs transfected with Prdm14, Blimp-1, and c-Myc. ES cell-like colonies are shown at 16 days after induction (a and b). Panel b shows a higher magnification of a. Nanog-GFP-positive colonies are shown at 14 days after induction (c and d). Representative phase-contrast (c) and Nanog-GFP fluorescence (d) images are shown. e and f, endogenous expression and exogenous overexpression of Blimp-1 in ES cells. The relative expression profiles of endogenous Blimp-1 in ES cells compared with those in MEFs, STO cells, and E11.5 PGCs (e). ES cells transfected with a CAG-Blimp-1 vector contained the Blastsidin resistance gene. Two days after transfection, cells were re-plated and 10 μg/ml of Blastsidin was added. After 4 days (6 days post-transfection), cells were fixed and stained for alkaline phosphatase (ALP) activity (f). g-j, morphologies of MEFs transfected with Prmt5, Klf4, and Oct3/4. Nanog-GFP-positive colonies at 28 days after induction (g and h) and subsequently established PKO cells (i and j) are depicted. Scale bars = 500 μm (a, c, d, i, and j), 100 (b), and 200 μm (g and h). k, integration of retroviral genes. Genomic DNA was purified from the indicated cells, and PCR with primers specific for Klf4, Sox2, Oct3/4, c-Myc, Prmt5, and the IL-2 locus (control) was performed. Pseudo-colony cells were generated by infection with the four Yamanaka factors, but these cells were morphologically dissimilar to ES-like cells and negative for Nanog-GFP.
Successful Generation of a Stable Line of Nanog-GFP-positive Cells by the Introduction of Prmt5, Klf4, and Oct3/4
The second instance in which Nanog-GFP-positive primary colonies were detected occurred following the co-infection of cells with Prmt5, Klf4, and Oct3/4 (Table 1, case 18). Under these conditions, we observed ES cell-like primary colonies, many of which were positive for Nanog-GFP (Fig. 1). In contrast to the primary colonies derived from Blimp-1/Prdm14/c-Myc co-infection, Nanog-GFP-positive colonies from Prmt5/Klf4/Oct3/4 co-infection grew and were successfully maintained as stable lines (Fig. 1). These cells were subsequently designated PKO cells (Prmt5, Klf4, and Oct3/4). Genomic PCR analysis showed that PKO cells actually integrated exogenous Prmt5, Klf4, and Oct3/4, but not Sox2 or c-Myc (Fig. 1). PKO cells were successfully re-plated and their growth rate was comparable with the growth rate of ES cells derived from the inner cell mass of the blastocyst. Even with continuous passage, the level of Nanog-GFP expression in PKO cells remained as high as during the early passage. Upon induction of differentiation, Nanog-GFP expression was down-regulated, supporting that PKO cells are reprogrammed pluripotent stem cells with ES cell characteristics.
Gene Expression and DNA Methylation Patterns in PKO Cells
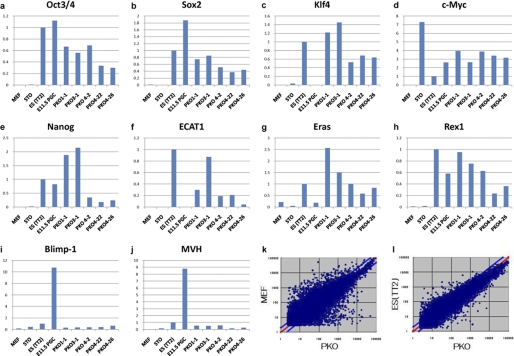
The gene expression patterns in PKO cells were analyzed. PKO cells exhibited up-regulated endogenous expression of many known reprogramming factors, including Oct3/4, Sox2, Klf4, and c-Myc (Fig. 2). In addition, PKO cells expressed other ES cell-specific genes such as Nanog, ECAT-1, and Eras, whereas parental MEFs did not (Fig. 2) (20, 21). Although PKO cells were induced by Prmt5, which is critical for germ cell specificity, the gene expression patterns of PKO cells were distinct from those of germ cells, as shown by the expression of MVH (mouse vasa homolog) and Blimp-1 (Fig. 2) (10, 22). Furthermore, microarray analysis of PKO cells showed that the gene expression patterns of PKO cells became ES cell-like (Fig. 2).
FIGURE 2.
Gene expression profiles of PKO cells. a–j, the relative gene expression profiles of PKO cells were compared with those of ES cells, MEFs, STOs, and PGCs. The endogenous expression profiles of the four iPS cell-inducible factors (a–d), ES cell-specific markers (e–h), and germ cell markers (i and j) for each cell type are shown. k and l, microarray analysis of PKO cells. PKO cells were compared with MEFs (k) or ES cells (l) by microarray.
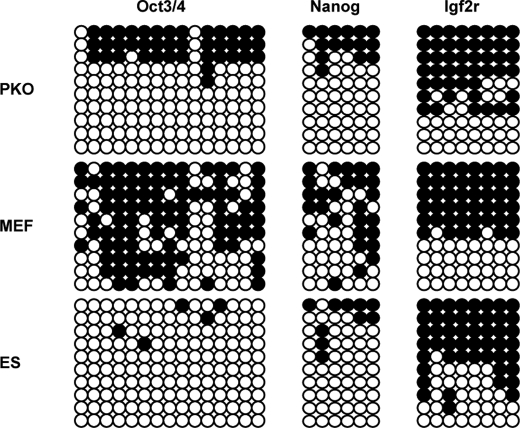
Next, the DNA methylation states of the Oct3/4- and Nanog-regulatory elements were examined (16). In contrast to the promoter regions of these two genes in MEFs, the promoter regions of these two genes were demethylated in PKO cells (Fig. 3). Compared with ES cells, the Oct3/4 locus of PKO cells was partially methylated, indicating that the PKO was composed of heterogeneous cells, some of which could have been incompletely reprogrammed. To distinguish whether PKO cells were derived from the direct reprogramming of MEFs or by mediating MEF-derived PGCs from Prmt5/Klf4/Oct3/4 co-infection, the DNA methylation pattern of the imprinted loci, Igf2r, was examined. Because germ cells lose the parental methylation pattern, even the early stage PGCs, the methylation status would be altered if the PKO cells were derived from PGCs. However, the Igf2r locus was not altered after reprogramming (Fig. 3). Thus, PKO cells did not progress through the germ cell phase during reprogramming, indicating that they are similar to ES cells but not EG cells.
FIGURE 3.
DNA methylation of ES cell-specific genes and imprinted genes in PKO cells. The bisulfite sequence at the promoter regions of Oct3/4 and Nanog in PKO cells and the differentially methylated region of Igf2r are shown. White circles indicate unmethylated CpG dinucleotides, whereas black circles indicate methylated CpG dinucleotides.
In Vivo Differentiation Capacity of PKO Cells
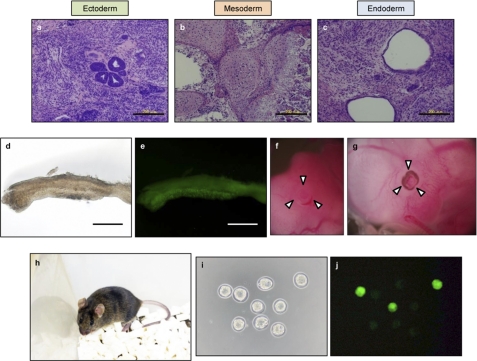
Finally, we tested the pluripotency of PKO cells. When PKO cells were injected into nude mice, PKO cells produced teratomas that contained tissues derived from all three germ layers (Fig. 4). Although some PKO cells contained a methylated Oct3/4-regulatory element, PKO cells were able to differentiate into germ cells through the formation of a chimeric embryo (Fig. 4). PKO cells were also detected in the eyes of chimeric embryos (Fig. 4). In addition to their presence in the embryo, adult chimeras were also produced from PKO cells and some of these demonstrated the incorporation of PKO cells by their coat color (Fig. 4). Furthermore, chimeric mice produced PKO cell-derived F1 progeny, confirmed by Nanog-GFP fluorescence of the morula (Fig. 4). These results demonstrated that somatic cells were reprogrammed to a pluripotent status by expression of reprogramming factors Prmt5, Klf4, and Oct3/4.
FIGURE 4.
In vivo differentiation capacity of PKO cells. a–c, teratomas formed from PKO cells contained ectoderm (a), mesoderm (b), and endoderm (c). Scale bars = 200 μm. d and e, the genital ridge from an E12.5 PKO chimeric embryo. Phase-contrast image (d) and PKO cell-derived Nanog-GFP fluorescence (e). Scale bars = 500 μm. f and g, contribution of PKO cells in a chimeric embryo. PKO cell contribution to the eyes of a chimera (g) compared with a non-chimeric embryo (f). h, adult chimera of PKO cells. Chimerism was roughly estimated by aguti coat color. i and j, morula of F1 progeny. Phase-contrast image (i) and PKO cell-derived Nanog-GFP fluorescence (j) are shown.
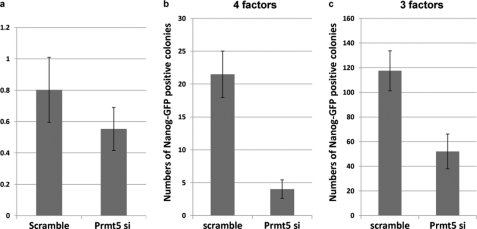
Knockdown of Prmt5 Decreases the Formation of Nanog-GFP Colonies in 4- and 3-Factor-mediated Reprogramming
To investigate whether endogenous Prmt5 also contributed to reprogramming by Yamanaka factors, we knocked down the expression of Prmt5 in combination with the exogenous expression of four or three known reprogramming factors (Oct3/4, Klf4, and Sox2 with or without c-Myc). The knockdown efficiency was estimated in ES cells by RT-PCR (Fig. 5). The number of Nanog-GFP positive colonies was counted at day 14 for four factors and at day 18 for three factors, respectively (Fig. 5). In both cases, Nanog-GFP colonies were decreased by Prmt5 knockdown, especially in the case of four factors. These data indicate that Prmt5 also participates in the reprogramming process mediated by Yamanaka factors.
FIGURE 5.
Knockdown of Prmt5 in the reprogramming process. a, vectors that contained the sequence for Prmt5 knockdown or a scrambled sequence were electroporated into ES cells. Two days after electroporation, bulk cells were collected and analyzed for expression of Prmt5. Relative expression compared with non-electroporated ES cells is shown. b and c, MEFs that contain the Nanog-GFP-IRES-puro allele were infected with a retrovirus containing the Prmt5 knockdown sequence or a scrambled sequence and a combination of three or four factors (Oct3/4, Klf4, and Sox2 with or without c-Myc). The numbers of puromycin-resistant colonies were counted at day 14 for four factors (b) or day 18 for three factors (c) post-infection, respectively.
DISCUSSION
Because germ cells are closely related to pluripotent stem cells, certain endogenous factors in germ cells are predicted to have reprogramming activity. In this study, we demonstrated that the induction of Blimp-1, Prdm14, and c-Myc resulted in the partial reprogramming of somatic cells, whereas the induction of Prmt5, Klf4, and Oct3/4 conferred pluripotency to somatic cells, making them comparable with ES cells with respect to gene expression, epigenetic modification of the promoter regions of Nanog and Oct3/4, and the potential to differentiate into all three germ layers. Blimp-1 is an essential factor in germ cell specification, demonstrated by the failure of Blimp-1-deficient PGCs to suppress Hox gene expression (10). Prdm14 is also essential for establishing PGCs, and Prdm14 deficiency leads to the failure of Sox2 reactivation in PGCs. More importantly, Prdm14-deficient PGCs fail to give rise to pluripotent EG cells (11). In addition, in human ES cells, PRDM14 is important for maintaining pluripotency (23). The third factor, Prmt5, forms a complex with Blimp-1 in PGCs and is thought to be important for PGC development (13). Furthermore, Prmt5 is important for PGC conversion into EG cells (24). Therefore, each of the three factors is essential factors for germ cell specificities.
iPS cell generation was first characterized by exogenous expression of transcription factors Oct4, Sox2, Klf4, and c-Myc (9). Thereafter, it became evident that the control of epigenetic regulators, such as histone deacetylase and G9a (histone H3K9-specific methyltransferase), could improve the efficiency of somatic cell reprogramming (25, 26). It is noteworthy that both histone deacetylase and G9a play inhibitory roles in reprogramming. Until now, no epigenetic factor was known to act as a positive regulator of somatic cell reprogramming. The work described in this study demonstrates that Prmt5 could be one such factor, as it promoted somatic cell reprogramming when it was overexpressed in MEFs together with Klf4 and Oct3/4. Furthermore, the knockdown of Prmt5 showed that it also works in the reprogramming process mediated by Yamanaka factors. Taken together, it is important to consider the role of Prmt5 from both genetic and epigenetic aspects of the reprogramming process. The reprogramming activity of Prmt5 could depend on its ability to symmetrically methylate arginine R3 of histone H2/4 and arginine R8 of H3 (27), a process that has been linked to gene silencing (18). Alternatively, arginine methylation of proteins other than histones could be crucial for somatic cell reprogramming. Indeed, it has been reported that Prmt5-mediated arginine methylation of p53 weakens its target specificity and function (28). Because p53 inhibits the generation of iPS cells (5), it is also feasible that functional inhibition of p53 by Prmt5 contributes to the generation of PKO cells. To evaluate these possibilities, it will be important to identify the target of Prmt5 methylation and the effect of p53 activity during PKO cell generation. Furthermore, to address the function of Prmt5 during PKO cell generation, the subcellular localization of Prmt5 must also be evaluated because Prmt5 localization changes during the conversion of unipotent PGCs to pluripotent EG cells (24). Although the significance of the location and activation of Prmt5 is not yet clear, it is likely to be important for future identification of the molecular targets of Prmt5. Dign et al. (29) demonstrated that a combination with Oct3/4 introduction, inhibitor for Prmt, and TGF-βR could induce iPS cells. A Prmt inhibitor, AMI-5, which they used inhibits the activities of Prmt1/3/4/6. It suggests unique reprogramming activity of Prmt5.
Although Prmt5, Klf4, and Oct3/4 induced pluripotency in somatic cells, the process was largely inefficient and the timing of reprogramming was relatively late compared with the established method (Table 2). This indicates that reprogramming in these cells occurred in a novel manner. Supporting this idea, the expression of Prmt5 was not up-regulated after infection of three or four factors (supplemental Fig. S1). The repression of somatic cell genes is an early event in reprogramming (30), and c-Myc overexpression has a unique effect on repression compared with the other three reprogramming factors (Oct3/4, Klf4, and Sox2) (31). Interestingly, our preliminary data showed that the activity of Prmt5 in repressing the somatic cell gene, Thy-1, is similar to the activity of c-Myc. Because germ cell development is accompanied by the repression of somatic lineage differentiation, it is likely that Prmt5 may repress Thy-1 expression.
PGCs arise directly from the epiblast (9). During the process of cell commitment, PGCs have a unique gene expression profile that includes Sox2 reactivation and Hox gene repression (9). Blimp-1 and Prdm14 play central roles in establishing these gene expression patterns. Thereafter, PGCs change their genome-wide chromatin structure during development. As shown here, factors that are important in early PGCs also have reprogramming activity, we show that several genes expressed in PGCs could be involved in the reprogramming of somatic cells. During development, genome-wide reprogramming that occurs in PGCs is the sole process by which differentiating epiblast cells are efficiently reprogrammed (32). As such, there are a number of similarities between the reprogramming in PGCs and generation of iPS cells, such as the expression of Sox2 and Nanog, a functional requirement for Lin28, and repression of G9a activity (3, 26, 32, 33). These data, together with the results of the present study, strongly indicate that other reprogramming factors exist that are involved in both PGC development and iPS cell derivation. A more thorough understanding of how germ cells undergo genome-wide cellular reprogramming should provide useful clues about the identities of these factors and increase our knowledge of somatic cell reprogramming.
Acknowledgments
We thank Dr. N. Takeda (Kumamoto University) for the PKO cell microinjection used to analyze the germ cell contribution of PKO cells in chimeric embryos. We also thank Dr. T. Kitamura (Tokyo University) for the Plat-E cells and Dr. K. Hayashi (Kyoto University) for critical reading of this manuscript.
This work was supported by PREST of the Japan Science and Technology Agency and Grant-in-Aid for Young Scientists (B) and a Grant-in-aid for Scientific Research (KAKENHI) on Innovative Areas, “Regulatory Mechanism of Gamate Stem Cells.” This work was also supported in part by a grant from the Project for Realization of Regenerative Medicine, and support for the core institutes for iPS cell research was provided by MEXT, a Grant-in-aid for the Global Century COE Program from MEXT to Keio University, and the Keio University Medical Science Fund.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Fig. S1.
- ES
- embryonic stem
- iPS
- induced pluripotent stem
- PGC
- primordial germ cell
- PKO cells
- Prmt5, Klf4, and Oct3/4-induced pluripotent cells
- MEF
- mouse embryonic fibroblast
- EG
- embryonic germ.
REFERENCES
- 1. Takahashi K., Yamanaka S. (2006) Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 2. Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. (2007) Cell 131, 861–872 [DOI] [PubMed] [Google Scholar]
- 3. Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R., Slukvin I. I., Thomson J. A. (2007) Science 318, 1917–1920 [DOI] [PubMed] [Google Scholar]
- 4. Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., Aoi T., Okita K., Mochiduki Y., Takizawa N., Yamanaka S. (2008) Nat. Biotechnol. 26, 101–106 [DOI] [PubMed] [Google Scholar]
- 5. Zhao Y., Yin X., Qin H., Zhu F., Liu H., Yang W., Zhang Q., Xiang C., Hou P., Song Z., Liu Y., Yong J., Zhang P., Cai J., Liu M., Li H., Li Y., Qu X., Cui K., Zhang W., Xiang T., Wu Y., Liu C., Yu C., Yuan K., Lou J., Ding M., Deng H. (2008) Cell Stem Cell 3, 475–479 [DOI] [PubMed] [Google Scholar]
- 6. Matsui Y., Zsebo K., Hogan B. L. (1992) Cell 70, 841–847 [DOI] [PubMed] [Google Scholar]
- 7. Kanatsu-Shinohara M., Inoue K., Lee J., Yoshimoto M., Ogonuki N., Miki H., Baba S., Kato T., Kazuki Y., Toyokuni S., Toyoshima M., Niwa O., Oshimura M., Heike T., Nakahata T., Ishino F., Ogura A., Shinohara T. (2004) Cell 119, 1001–1012 [DOI] [PubMed] [Google Scholar]
- 8. Mise N., Fuchikami T., Sugimoto M., Kobayakawa S., Ike F., Ogawa T., Tada T., Kanaya S., Noce T., Abe K. (2008) Genes Cells 13, 863–877 [DOI] [PubMed] [Google Scholar]
- 9. Saitou M. (2009) Curr. Opin. Genet. Dev. 19, 386–395 [DOI] [PubMed] [Google Scholar]
- 10. Ohinata Y., Payer B., O'Carroll D., Ancelin K., Ono Y., Sano M., Barton S. C., Obukhanych T., Nussenzweig M., Tarakhovsky A., Saitou M., Surani M. A. (2005) Nature 436, 207–213 [DOI] [PubMed] [Google Scholar]
- 11. Yamaji M., Seki Y., Kurimoto K., Yabuta Y., Yuasa M., Shigeta M., Yamanaka K., Ohinata Y., Saitou M. (2008) Nat. Genet. 40, 1016–1022 [DOI] [PubMed] [Google Scholar]
- 12. Pal S., Sif S. (2007) J. Cell Physiol. 213, 306–315 [DOI] [PubMed] [Google Scholar]
- 13. Ancelin K., Lange U. C., Hajkova P., Schneider R., Bannister A. J., Kouzarides T., Surani M. A. (2006) Nat. Cell Biol. 8, 623–630 [DOI] [PubMed] [Google Scholar]
- 14. Hajkova P., Ancelin K., Waldmann T., Lacoste N., Lange U. C., Cesari F., Lee C., Almouzni G., Schneider R., Surani M. A. (2008) Nature 452, 877–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mikkelsen T. S., Hanna J., Zhang X., Ku M., Wernig M., Schorderet P., Bernstein B. E., Jaenisch R., Lander E. S., Meissner A. (2008) Nature 454, 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Okita K., Ichisaka T., Yamanaka S. (2007) Nature 448, 313–317 [DOI] [PubMed] [Google Scholar]
- 17. Horikawa K., Takatsu K. (2006) Immunology 118, 497–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao Q., Rank G., Tan Y. T., Li H., Moritz R. L., Simpson R. J., Cerruti L., Curtis D. J., Patel D. J., Allis C. D., Cunningham J. M., Jane S. M. (2009) Nat. Struct. Mol. Biol. 16, 304–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fujino R. S., Tanaka K., Morimatsu M., Tamura K., Kogo H., Hara T. (2006) Mol. Endocrinol. 20, 904–915 [DOI] [PubMed] [Google Scholar]
- 20. Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S. (2003) Cell 113, 631–642 [DOI] [PubMed] [Google Scholar]
- 21. Takahashi K., Mitsui K., Yamanaka S. (2003) Nature 423, 541–545 [DOI] [PubMed] [Google Scholar]
- 22. Tanaka S. S., Toyooka Y., Akasu R., Katoh-Fukui Y., Nakahara Y., Suzuki R., Yokoyama M., Noce T. (2000) Genes Dev. 14, 841–853 [PMC free article] [PubMed] [Google Scholar]
- 23. Tsuneyoshi N., Sumi T., Onda H., Nojima H., Nakatsuji N., Suemori H. (2008) Biochem. Biophys. Res. Commun. 367, 899–905 [DOI] [PubMed] [Google Scholar]
- 24. Durcova-Hills G., Tang F., Doody G., Tooze R., Surani M. A. (2008) PLoS ONE 3, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huangfu D., Maehr R., Guo W., Eijkelenboom A., Snitow M., Chen A. E., Melton D. A. (2008) Nat. Biotechnol. 26, 795–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Toyooka Y., Shimosato D., Murakami K., Takahashi K., Niwa H. (2008) Development 135, 909–918 [DOI] [PubMed] [Google Scholar]
- 27. Lacroix M., El Messaoudi S., Rodier G., Le Cam A., Sardet C., Fabbrizio E. (2008) EMBO Rep. 9, 452–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jansson M., Durant S. T., Cho E. C., Sheahan S., Edelmann M., Kessler B., La Thangue N. B. (2008) Nat. Cell Biol. 10, 1431–1439 [DOI] [PubMed] [Google Scholar]
- 29. Yuan X., Wan H., Zhao X., Zhu S., Zhou Q., Ding S. (2011) Stem Cells, in press [DOI] [PubMed] [Google Scholar]
- 30. Stadtfeld M., Maherali N., Breault D. T., Hochedlinger K. (2008) Cell Stem Cell 2, 230–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sridharan R., Tchieu J., Mason M. J., Yachechko R., Kuoy E., Horvath S., Zhou Q., Plath K. (2009) Cell 136, 364–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Seki Y., Yamaji M., Yabuta Y., Sano M., Shigeta M., Matsui Y., Saga Y., Tachibana M., Shinkai Y., Saitou M. (2007) Development 134, 2627–2638 [DOI] [PubMed] [Google Scholar]
- 33. West J. A., Viswanathan S. R., Yabuuchi A., Cunniff K., Takeuchi A., Park I. H., Sero J. E., Zhu H., Perez-Atayde A., Frazier A. L., Surani M. A., Daley G. Q. (2009) Nature 460, 909–913 [DOI] [PMC free article] [PubMed] [Google Scholar]