Abstract
The membrane-integrated transcriptional regulator CadC of Escherichia coli activates expression of the cadBA operon at low external pH with concomitantly available lysine, providing adaptation to mild acidic stress. CadC is a representative of the ToxR-like proteins that combine sensory, signal transduction, and DNA-binding activities within a single polypeptide. Although several ToxR-like regulators such as CadC, as well as the main regulator of Vibrio cholerae virulence, ToxR itself, which activate gene expression at acidic pH, have been intensively investigated, their molecular activation mechanism is still unclear. In this study, a structure-guided mutational analysis was performed to elucidate the mechanism by which CadC detects acidification of the external milieu. Thus, a cluster of negatively charged amino acids (Asp-198, Asp-200, Glu-461, Glu-468, and Asp-471) was found to be crucial for pH detection. These amino acids form a negatively charged patch on the surface of the periplasmic domain of CadC that stretches across its two subdomains. The results of different combinations of amino acid replacements within this patch indicated that the N-terminal subdomain integrates and transduces the signals coming from both subdomains to the transmembrane domain. Alterations in the phospholipid composition did not influence pH-dependent cadBA expression, and therefore, interplay of the acidic surface patch with the negatively charged headgroups is unlikely. Models are discussed according to which protonation of these acidic amino acid side chains reduces repulsive forces between the two subdomains and/or between two monomers within a CadC dimer and thereby enables receptor activation upon lowering of the environmental pH.
Keywords: Amino Acid, Bacterial Signal Transduction, Cell pH, Protein Domains, Protein Structure, Dimer, ToxR-like Proteins, Acid Stress Response, pH Sensor
Introduction
Bacteria that prefer the human intestine as a natural habitat encounter a broad range of environmental stresses, including acidic pH (1). To survive a decrease in the external pH, neutrophilic bacteria such as Escherichia coli, Salmonella typhimurium, and Vibrio cholerae possess different stress response systems to maintain the cytoplasmic pH in the physiological range. One system that is important for the response to mild acidic stress in E. coli is the Cad system (1). The Cad system of E. coli consists of the cytoplasmic protein CadA and the transmembrane proteins CadB and CadC. The lysine decarboxylase CadA converts lysine upon consumption of a cytoplasmic proton to cadaverine and carbon dioxide. Subsequently, CadB transports the basic reaction product cadaverine in exchange for lysine out of the cell (2, 3). Transcription of the cadBA operon in E. coli is activated when the external pH drops below pH 6.6 if external lysine is available (4, 5). The main activator of the Cad system is the inner membrane protein CadC (6).
CadC is a member of the family of ToxR-like transcriptional activators that comprise several low pH-regulated activators, including the main regulator for virulence of V. cholerae, ToxR. ToxR-like regulators are characterized by a conserved modular composition. They exhibit a cytoplasmic DNA-binding domain with a winged helix-turn-helix motif, a single transmembrane domain, and a periplasmic sensor domain. In contrast to histidine kinase/response regulator systems, in which signal transduction is accompanied by phosphorylation reactions, signal transduction in ToxR-like regulators is mediated without chemical modification (7). Thus, ToxR-like regulators are one-component systems that exhibit sensory, signal transduction, and effector function within one polypeptide (8). Although transcriptional activation by ToxR as well as CadC has been intensively investigated using genetic and biochemical methods, the precise molecular mechanism is hardly understood. Previously, we showed that CadC is not the direct sensor for lysine but senses lysine via interplay with the lysine-permease LysP as co-sensor (9). Using random mutagenesis, Dell et al. (10) identified several CadC variants with mutations in the periplasmic domain that were impaired in the pH-dependent regulation of the Cad system. Therefore, the pH sensory property was assigned to the periplasmic domain of CadC. Recently, the three-dimensional structure of the periplasmic domain of E. coli CadC was solved (41). In this study, we employed a structure-guided mutational analysis that led to the identification of a negatively charged patch comprising acidic amino acids on the CadC periplasmic domain surface as crucial for sensing low pH.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
E. coli JM109 or DH5α (11, 12) was used as a carrier for the plasmids described. All plasmids used in this study are listed in supplemental Table 1. E. coli BL21(DE3)pLysS (13) was used for expression of cadC and its variants from the T7 promoter. E. coli EP314 (14), which carries a cadA-lacZ fusion gene and a Tn10 transposon in cadC, was complemented with plasmids (pET16b-based) encoding cadC and its variants and used for cadBA transcriptional analysis. For strain maintenance, plasmid preparation, and protein overproduction, strains were grown in LB medium (15). To monitor signal transduction in vivo, E. coli EP314 transformed with the indicated plasmids was grown in minimal medium (16). The pH was adjusted to either pH 5.8 or pH 7.6 using the corresponding phosphate-buffered medium. E. coli strains with modified phospholipid content (E. coli AD93 with or without plasmid pDD72 (17) and HDL1001 (18)) were grown in LB medium that was pH-adjusted using 100 mm MOPS (pH 7.6 or 7.0) or MES (pH 5.8). To enable growth of AD93, 50 mm MgCl2 was added to the medium (17). Where indicated, lysine was added to a final concentration of 10 mm. Antibiotics were added for selection purpose at concentrations of 100 μg/ml (ampicillin), 50 μg/ml (kanamycin), and 34 μg/ml (chloramphenicol).
Construction of cadC Variants
All cadC variants were constructed by either one- or two-step PCR using mismatched primers (19). To facilitate construction, four unique restriction sites (XhoI, 1338T→C and 1341A→G; XmaI, 1443A→C; SacI, 1002T→A, 1005G→T, and 1006T→C; and SacII, 588A→G and 591C→G; numbers indicate the nucleotide positions in the cadC sequence) were introduced into plasmid pET16b-cadC2 (9) by silent mutation, resulting in plasmids pET16b-cadC3 (XhoI), pET16b-cadC4 (XhoI and XmaI), pET16b-cadC5 (XhoI and SacI), pET16b-cadC6 (XhoI, XmaI, and SacI), and pET16b-cadC7 (XhoI, XmaI, SacI, and SacII) (supplemental Table 1).
Detection of CadC in the Membrane Fraction
Verification of production and membrane integration was carried out for all CadC variants as described previously (9).
Measurement of CadC Signal Transduction Activity in Vivo
Cultivation of cells and measurements of the signal transduction activity of different CadC variants by β-galactosidase assays were performed as described previously (9). To analyze the influence of changes in the phospholipid composition, cells of an overnight culture (grown in LB medium at pH 7.6) were inoculated into fresh LB medium (pH 7.6) with the A600 adjusted to 0.05. Cells were grown under aerobic conditions at 37 °C to mid-logarithmic growth phase (A600 ∼ 0.7) and harvested by centrifugation (37 °C). Pellets were resuspended and transferred to prewarmed LB medium (pH 5.8–7.6) by adjusting the A600 to 0.7. After growth for another 1.5 h, cells were harvested by centrifugation. To test CadA activity, a modified spectrophotometric assay for lysine decarboxylase as described by Lemonnier and Lane (20) was used. For this purpose, the cell density was adjusted to A600 = 1 with 20 mm potassium Pi buffer (pH 5.8). Subsequently, 200 μl of this cell suspension was mixed with 20 μl of chloroform to disrupt cells. After settling of the chloroform, 10 μl of the chloroform-free supernatant was added to 120 μl of either a lysine-free (control) or a lysine-containing reaction mixture consisting of 16 mm potassium Pi buffer (pH 5.8) and 0.1 mm pyridoxal phosphate (Sigma) with and without 5 mm lysine (Sigma) 37 °C. The enzymatic reaction was performed at 37 °C for 15 min and thereafter stopped by the addition of 120 μl of 1 m Na2CO3. Subsequently, lysine and cadaverine were derivatized by adding 120 μl of 10 mm picrylsulfonic acid (2,4,6-trinitrobenzenesulfonic acid; Sigma), followed by incubation at 40 °C for 4 min. To each sample toluene (1 ml) was added, followed by vigorously mixing for 20 s. The toluene and aqueous layers were separated by centrifugation. N,N′-Bistrinitrophenyllysine is soluble in water but not in toluene, whereas the reverse is true for N,N′-bistrinitrophenyl cadaverine (21). The absorption of N,N′-bistrinitrophenyl cadaverine (toluene phase) was determined at 340 nm, and the specific activity of CadA (μmol/min*mg) was calculated.
Three-dimensional Structure Analysis
Analysis of the three-dimensional structure of the periplasmic domain (Protein Data Bank code 3LY7) was carried out using the PyMOL molecular viewer, version 0.99rc6 (22). Calculation of electrostatic surface was carried out with the program GRASP (23).
RESULTS
Histidine Residues Are Not Involved in pH Detection by CadC
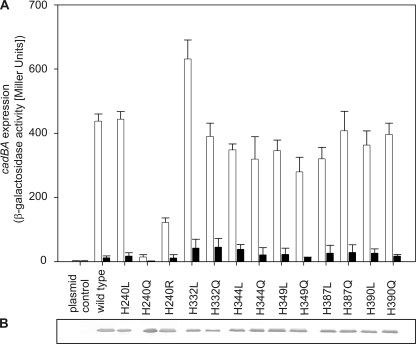
Moderate acidification (pH 6.6) of the surrounding medium in the presence of lysine is sufficient to activate transcription of the cadBA operon in E. coli. Full induction of this operon is accomplished at or below pH 5.8 (4). These values are close to the pH value at which histidine side chains become protonated (pKa around 6). In addition, histidines have already been shown to mediate the pH response in several proteins such as the sensor kinase PhoQ of Salmonella typhimurium (24) and the histidine kinase ArsS of Helicobacter pylori (25). The periplasmic domain of CadC contains six histidine residues (His-240, His-332, His-345, His-349, His-387, and His-390), which were all replaced with either leucine or glutamine to substitute the imidazole ring with different side chains of similar size. Plasmids encoding the corresponding CadC variants were introduced into the reporter strain E. coli EP314, which lacks a functional chromosomal cadC gene and carries a cadA-lacZ fusion gene (9, 14). With the exception of CadC-H240Q, all variants of CadC with histidine replacements induced cadBA expression at low pH but not at pH 7.6, similar to wild-type CadC (Fig. 1A). Because all CadC variants were inserted into the membrane (Fig. 1B), the lack of response of CadC-H240Q seemed to be related to defects in sensing and/or signal transduction (off-state). To test whether the off-state of CadC-H240Q was due to the loss of the positive charge of the protonated imidazole ring or other structural effects, arginine and leucine were introduced at this position, respectively. Indeed, replacement of His-240 with leucine and also with arginine resulted in pH-responsive CadC variants, albeit the signaling capacity of CadC-H240R was somewhat reduced (Fig. 1A). These results indicate that the imidazole side chain of His-240 is not important for pH sensing but that this amino acid seems to play a more intricate role in signaling.
FIGURE 1.
A, influence of histidine replacements in the periplasmic domain of CadC on cadBA expression. Reporter gene assays were performed with E. coli EP314 (cadC1::Tn10, cadA′::lacZ) that was complemented with plasmid-encoded cadC or the mutant indicated. Cells were cultivated under microaerobic conditions in minimal medium in the presence of 10 mm lysine at pH 7.6 (black bars) or at pH 5.8 (white bars). The activity of the reporter enzyme β-galactosidase was determined according to Miller (40) and served as a measure for cadBA expression. The experiment was performed in triplicate, and error bars indicate S.D. B, verification of production and integration of CadC variants into the cytoplasmic membrane of E. coli. E. coli BL21(DE3)pLysS was transformed with plasmids encoding either wild-type or mutant cadC. Each lane contained 25 μg of membrane protein. CadC was detected by a monoclonal mouse antibody against the His tag and an alkaline phosphatase-coupled secondary antibody.
Glu-461, Glu-468, and Asp-471 Are Involved in pH Sensing
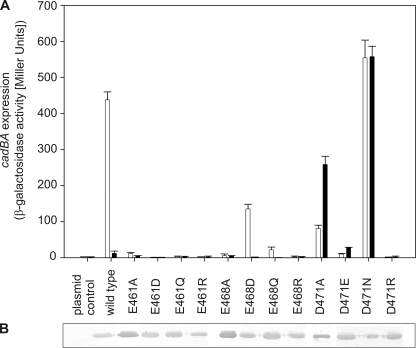
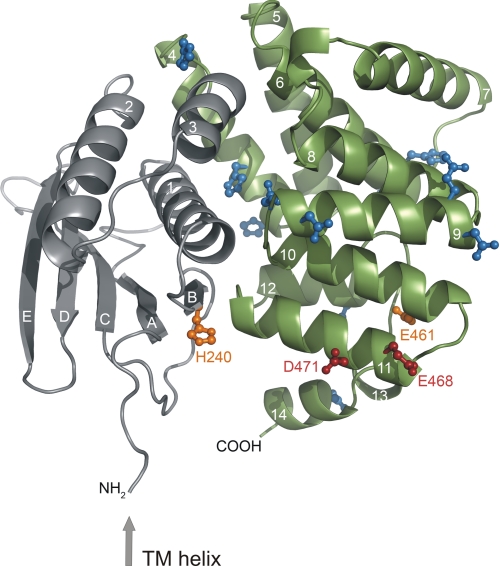
Full induction of cadBA transcription occurs at pH 5.8, a pH value at which initial protonation of aspartate (side chain pKa of 3.9) or glutamate (side chain pKa of ∼4.3 (26)) residues, depending on their electrostatic environment, could be responsible for pH sensing. The periplasmic domain contains 19 aspartates and 15 glutamates. Because almost all pH-sensitive amino acids previously identified by Dell et al. (10) were located close to the C terminus, we initially focused on 10 C-terminal aspartate and glutamate residues (Asp-434, Glu-435, Asp-445, Glu-447, Glu-461, Glu-468, Asp-471, Glu-490, Asp-506, and Glu-512). Each amino acid was substituted with the related carboxamide amino acid devoid of charge (i.e. asparagine instead of aspartate and glutamine instead of glutamate) to mimic the uncharged protonated state and also with arginine to further introduce a positive net charge. The effect of each amino acid replacement was tested as described for the histidine variants. Most of the resulting CadC variants activated cadBA expression like wild-type CadC (Fig. 2 and Table 1). However, replacement of Glu-461, Glu-468, and Asp-471 caused either nonresponsive CadC variants (off-state) or pH-insensitive, i.e. constitutively activated, CadC variants (on-state) (Fig. 2A). Therefore, the role of these residues was further analyzed by additional amino acid replacements. For Glu-461, shortening the side chain (CadC-E461A), removing the negative charge (CadC-E461Q), and introducing a positive charge (CadC-E461R) resulted in nonresponsive proteins. Even the introduction of aspartate at this position (CadC-E461D) abolished the response to the external low pH signal (off-state). Hence, any alteration of Glu-461 led to CadC variants that were arrested in the off-state. More or less the same results were observed when Glu-468 was replaced. In this case, only the conservative replacement with aspartate (CadC-E468D) resulted in a pH-responsive sensor, albeit with lower activity (Fig. 2A). Notably, the replacement of aspartate at position 471 evoked different effects. Small uncharged amino acids such as alanine and glycine (CadC-D471A/G) (Fig. 2A) (10), as well as the larger uncharged asparagine (CadC-D471N) converted CadC to the on-state. On the other hand, the charged amino acids glutamate (CadC-D471E) and arginine (CadC-D471R) at this position led to nonresponsive CadC proteins, which no longer activated cadBA expression when exposed to low pH (Fig. 2A). As the three-dimensional structure of the periplasmic domain of CadC was recently solved (41), the amino acids involved in pH sensing identified thus far could be located in the tertiary structure. The CadC periplasmic domain consists of two subdomains (Fig. 3). Only His-240 is located in the N-terminal subdomain, whereas Glu-461, Glu-468, and Asp-471 are located in the C-terminal subdomain, which forms a bundle of α-helices (Fig. 3). Strikingly, Glu-468 and Asp-471 are located within the same α-helix (helix 11), like Leu-479 and Thr-475, which were previously identified by Dell et al. (10). Glu-461 is located in helix 10, and its side chain is oriented toward helices 11 and 12. Interestingly, His-240 is found in close proximity to α-helices 10 and 11 and displayed on the same protein face as Glu-468 and Asp-471.
FIGURE 2.
A, influence of replacements of acidic amino acids in the C-terminal region of the CadC periplasmic domain on cadBA expression. Reporter gene assays were performed with E. coli EP314 (cadC1::Tn10, cadA′::lacZ fusion) that was complemented with plasmid-encoded cadC or the indicated cadC mutant. Cells were cultivated under microaerobic conditions in minimal medium with 10 mm lysine at pH 7.6 (black bars) or at pH 5.8 (white bars). The reporter enzyme β-galactosidase was determined as described in the legend to Fig. 1. B, verification of production and integration of CadC variants into the cytoplasmic membrane of E. coli (see Fig. 1).
TABLE 1.
Influence of replacements of aspartate and glutamate residues in the C-terminal subdomain of the periplasmic domain of CadC on cadBA expression
Reporter gene assays were performed with E. coli EP314 (cadC1::Tn10, cadA′::lacZ) that was complemented with plasmid-encoded cadC or the indicated cadC mutant. See also Fig. 2.
| CadC variant |
cadBA expression (β-galactosidase activity) |
|
|---|---|---|
| pH 5.8 + lysine | pH 7.6 + lysine | |
| Miller units | ||
| Plasmid control | 1.9 | 2.0 |
| Wild-type CadC | 437.7 | 11.8 |
| D434N | 417.8 | 25.1 |
| D434R | 250.4 | 12.5 |
| E435Q | 465.2 | 12.7 |
| E435R | 270.4 | 10.3 |
| D445N | 384.3 | 10.2 |
| D445R | 263.6 | 9.3 |
| E447Q | 789.4 | 31.5 |
| E447R | 699.6 | 8.8 |
| E490Q | 281.0 | 53.8 |
| E490R | 338.7 | 14.5 |
| D506N | 358.8 | 29.6 |
| D506R | 243.9 | 6.6 |
| E512Q | 359.7 | 35.5 |
| E512R | 272.7 | 35.4 |
FIGURE 3.
Localization of histidine, glutamate, and aspartate residues investigated in the first mutagenesis screen within the three-dimensional structure of the periplasmic domain of CadC (41). The preceding transmembrane (TM) helix is indicated by an arrow. The N-terminal subdomain of the periplasmic domain is colored gray, and the C-terminal subdomain is colored green. The side chains of residues that were mutated but did not alter the pH response of the corresponding CadC variants are colored blue. Amino acids whose mutagenesis suggested a role in pH sensing are indicated in orange (off-state) or red (on-state). Secondary structure elements are labeled with white letters (41).
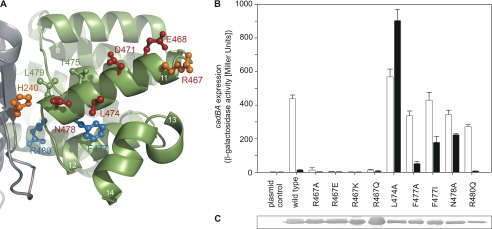
Helix 11 Harbors pH-sensing Residues
Beside Glu-468 and Asp-471 within helix 11, two CadC variants with altered pH responses were already known from the random mutagenesis experiments of Dell et al. (10), namely CadC-T475A and CadC-L479S. To further clarify the role of helix 11 in pH sensing, we systematically exchanged all remaining surface-exposed residues (Arg-467, Leu-474, and Asn-478) (Fig. 4). In addition, we mutated Arg-480 and Phe-477 to analyze the influence of the charged residue (Arg-480) and the hydrophobic side chain (Phe-477) (Fig. 4). Beyond these residues, helix 11 contains four alanines. Because these alanines are part of the contact area with the neighboring helices 10 and 14, they were not replaced. Substitution of arginine at position 467 (CadC-R467A/Q/E/K) clearly abolished the pH response (off-state) of the corresponding CadC variants (Fig. 4B), although the mutations did not affect biosynthesis or membrane integration (Fig. 4C). Substitution of the other surface-exposed residues (CadC-L474A and CadC-N478A) led to CadC variants that remained in the on-state (Fig. 4B). Because changes of Arg-467, Leu-474, and Asn-478 (all unprotonable or positively charged amino acids) caused drastic effects on the pH-dependent cadBA expression, it is conceivable that these amino acids, together with Thr-475 and Leu-479, are responsible either for the correct positioning of the protonable residues within this helix, namely Glu-468 and Asp-471, or for their electrostatic force field. Exchange of Phe-477 with different residues resulted in opposing phenotypes: isoleucine at this position led to an almost pH-insensitive CadC variant (on-state), whereas the smaller alanine (CadC-F477A) did not alter pH sensing (Fig. 4B). Because removal of the aromatic side chain (CadC-F447A) did not affect pH-dependent activation of CadC, it is likely that the effect seen for CadC-F477I was due to steric alterations evoked through insertion of the β-branched isoleucine, which is less well accommodated in α-helices. Finally, exchange of the second inward-facing residue, Arg-480 (CadC-R480Q), did not significantly affect the pH-sensing capability of CadC.
FIGURE 4.
Helix 11 of the periplasmic domain of CadC. A, amino acids of helix 11 that were investigated are colored blue (wild type-like phenotype), orange (off-state), or red (on-state). Side chains that are depicted in green were previously identified by random mutagenesis (Thr-475 and Leu-479) (10). The color code of the subdomains and the labeling of secondary structure elements is the same as described in the legend to Fig. 3. B, influence of amino acid replacements in helix 11 in the periplasmic domain of CadC on cadBA expression. Reporter gene assays were performed with E. coli EP314 (cadC1::Tn10, cadA′::lacZ fusion) that was complemented with plasmid-encoded cadC or the indicated cadC mutant as described in the legend to Fig. 1. C, verification of production and integration of CadC variants into the cytoplasmic membrane of E. coli BL21(DE3)pLysS.
Helix 10 Exerts a Stabilizing Effect
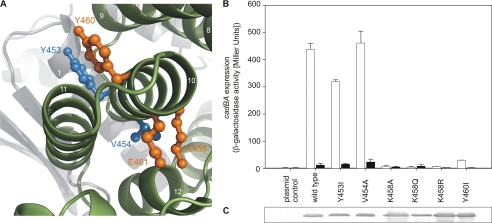
As Glu-461 is located in helix 10, the role of this helix in detecting low external pH was investigated in greater detail. Tyr-453, Val-454, Lys-458, and Tyr-460, together with Glu-461, decorate this helix on opposite sides (Fig. 5A). When Tyr-453 and Tyr-460 were replaced with isoleucine, the resulting CadC variants exhibited different phenotypes. CadC-Y453I regulated cadBA expression similarly to wild-type CadC, whereas CadC-Y460I blocked cadBA transcription (off-state) (Fig. 5B). Tyr-453 and Val-454, which were also inert to replacement (Fig. 5B), are located more at the N-terminal end of helix 10, whereas Tyr-460, as well as Lys-458 and Glu-461, i.e. sensitive residues, are situated at the C-terminal end. As in the case of Glu-461, each amino acid replacement of Lys-458 (CadC-K458A/Q/R) led to a pH-nonresponsive protein in the off-state (Fig. 5B), although these variants were synthesized and integrated into the membrane like wild-type CadC (Fig. 5C). The helix 10 residues analyzed are oriented toward helix 11 and seem to embrace it. For this reason and also because they are barely exposed to the surface of the protein, it is assumed that the nontitratable residues provide a kind of structural scaffold for the correct positioning of the pH-responsive residues Glu-468 and Asp-471 in helix 11.
FIGURE 5.
Helix 10 of the periplasmic domain of CadC. A, amino acids of helix 10 of the periplasmic domain of CadC embrace helix 11. Amino acids that were investigated are colored blue (wild type-like phenotype) or orange (off-state). The color code of the subdomains and the labeling of secondary structure elements is the same as described in the legend to Fig. 3. B, influence of amino acid replacements in helix 10 in the periplasmic domain of CadC on cadBA expression. Reporter gene assays were performed with E. coli EP314 (cadC1::Tn10, cadA′::lacZ fusion) that was complemented with plasmid-encoded cadC or the indicated cadC mutant as described in the legend to Fig. 1. Black bars, pH 7.6; white bars, pH 5.8. C, verification of production and integration of CadC variants into the cytoplasmic membrane of E. coli (see Fig. 1).
Identification of a Negatively Charged Patch That Extends across Both Subdomains
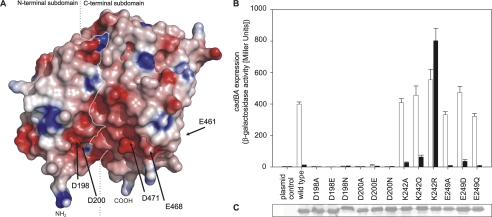
Comparison of the location of amino acids involved in pH detection and of the electrostatic charge at the protein surface (Fig. 6A) revealed that the residues in helix 11 are part of a broader negatively charged patch. Importantly, one-half of this patch is formed by the N-terminal subdomain, whereas the second half is located on the C-terminal subdomain (encompassing residues of helix 11) of the periplasmic domain of CadC. Based on this notion, all other charged amino acids contributing to this patch were analyzed by mutagenesis: Asp-198, Asp-200, Glu-249, and also the positively charged Lys-242. The role of the aspartates and the glutamate was probed by removal of the negative charge (substitution with asparagine or glutamine, respectively) or by shortening the side chain (alanine). Furthermore, CadC variants that have a conservative exchange at these positions (mutual exchange between glutamate and aspartate, respectively) were tested. Mutations of Asp-198 and Asp-200 indicated the necessity of the aspartate side chain at these two positions. Each replacement (D198A/E/N and D200A/E/N) transformed the resulting CadC variant to a pH-nonresponsive off-state (Fig. 6B), although these proteins were properly integrated into the membrane (Fig. 6C). In contrast, substitution of Glu-249 did not significantly influence pH detection (Fig. 6B); all corresponding CadC variants (CadC-E249A/D/Q) activated cadBA expression in a wild type-like manner. In addition, we analyzed Lys-242 to explore the role of this positively charged amino acid within the cluster. Unexpectedly, only the mutation to arginine (CadC-K242R), which retained the positive charge, altered the CadC-mediated pH-dependent activation, whereas CadC-K242A and CadC-K242Q showed a response like the wild-type protein (Fig. 6B). At first glance, this result would suggest that the Lys side chain naturally occurs in CadC in the unprotonated state such that its replacement with the more basic arginine would lead to an electrostatic effect. However, the salt bridge between the Lys Nϵ group and the negatively charged carboxylate side chain of Asp-200 provides a more plausible explanation, as this electrostatic interaction is probably different and possibly even stronger with the shorter arginine side chain. Notably, Asp-200 also plays a crucial role in this context, as described above.
FIGURE 6.
Acidic residues from both subdomains form a contiguous negatively charged surface patch. A, electrostatic surface coloring of the periplasmic domain of CadC. Negatively charged surfaces are shown in red, whereas positively charged areas are shown in blue. The positions of Asp-198, Asp-200, Glu-461, Glu-468, and Asp-471 at the protein surface are indicated by arrows. The boundary between the two subdomains is indicated by a white dashed line. The N (NH2) and C (COOH) termini are indicated. B, influence of replacements of amino acids in the N-terminal subdomain on cadBA expression. Reporter gene assays were performed as described in the legend to Fig. 1. Black bars, pH 7.6; white bars, pH 5.8. C, verification of production and integration of CadC variants into the cytoplasmic membrane of E. coli.
Role of Phospholipids in Sensing
Ca2+ sensing mediated by the histidine kinase PhoQ of S. typhimurium, for example, supposedly involves metal ion bridging between negatively charged side chains of the protein and the phospholipid headgroups of the cytoplasmic membrane (27). To test whether CadC might sense a higher H+ concentration via changes in a similar kind of interaction, cadBA expression was analyzed in different E. coli phospholipid mutant strains. These experiments were based on the hypothesis that, at neutral pH, the repulsive force between CadC and the phospholipids is stronger than after protonation of residues in the negatively charged surface patch. Hence, the cadBA expression profile was determined in two phospholipid mutants, whereby major changes in the phospholipid composition are expected to result in significant effects on CadC-mediated output. E. coli AD93 (17) has an inactivated allele of the gene encoding the phosphatidylserine synthase (pss93::kan), which catalyzes the committed step in the synthesis of phosphatidylethanolamine. Due to this defect, the mutant has a much higher content of negatively charged phospholipids (phosphatidylglycerol and cardiolipin). This property can be reverted by expressing pss from a plasmid (pDD72) (17). The second mutant, E. coli HDL1001 (18), is genetically modified such that expression of pgsA, which is crucial for the production of phosphatidylglycerol and cardiolipin, is under the control of the lac promoter. This mutant is characterized by an extremely low content of these two phospholipids (<2%). Both mutants were grown in medium at different pH values (pH 7.6, 7.0, and 5.8), and cadBA expression was recorded by measuring CadA activity (supplemental Fig. S1). In both mutants, cadBA expression was induced in a pH-dependent manner, and no shift either in the onset of expression or in the induction level was observed in comparison with the corresponding control strains (supplemental Fig. S1). Thus, alteration of the phospholipid composition was without effect on cadBA expression, and it can be concluded that the activation mechanism of CadC does not involve specific interactions with the phospholipid headgroups.
The N-terminal Subdomain Transduces Signals to the Transmembrane Domain
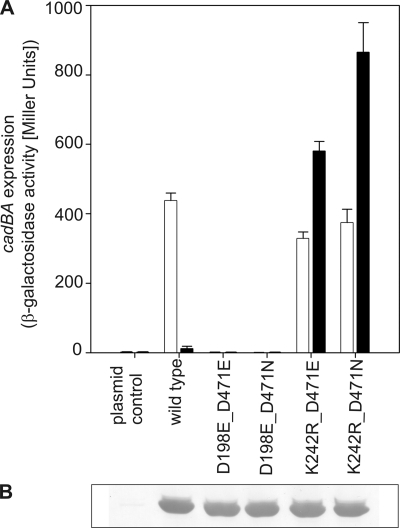
To gain insight into the signaling mechanism between the two subdomains within the periplasmic region of CadC, different amino acid replacements leading to the on- or off-state were combined within one polypeptide. In this way, a putative intramolecular complementation of different phenotypes was tested. For these experiments, the amino acid replacements D198E (off-phenotype) and K242R (on-phenotype) in the N-terminal subdomain and the replacements D471E (off-phenotype) and D471N (on-phenotype) in the C-terminal subdomain were assembled in all pairwise combinations. Interestingly, an intramolecular complementation was not achieved with any combination (Fig. 7). However, amino acid exchanges in the N-terminal subdomain were always dominant over exchanges in the C-terminal subdomain. Thus, the replacements D198E and K242R transformed CadC into either the off- or on-state, respectively, regardless of the second site mutation. These results suggest that the N-terminal subdomain integrates the signals coming from both subdomains and transduces them to the transmembrane and thus to the cytoplasmic domain (see Fig. 3).
FIGURE 7.
Intramolecular complementation of signaling off- and on-state mutants. A, influence of different combinations of amino acid replacements in the two subdomains on cadBA expression. Reporter gene assays were performed with E. coli EP314 (cadC1::Tn10, cadA′::lacZ fusion) as described in the legend to Fig. 1. Black bars, pH 7.6; white bars, pH 5.8. B, verification of production and integration of CadC variants into the cytoplasmic membrane of E. coli.
DISCUSSION
The capability to adapt to changes in the external pH is vital throughout all three domains of life. Changes in the intracellular pH can have severe effects such as induction of apoptosis in human tissues (28–30) and cell death in bacteria. To cope with an acidification of the environment, different mechanisms have evolved. Besides complete adaptation of the mode of life (i.e. for acidophilic bacteria), cells often respond by changes in gene expression. Therefore, pH sensors are crucial to monitor the pH of the environment. Different strategies to detect acidic stress are conceivable, e.g. pH-dependent protein folding and unfolding (31), but direct measurement of protons would provide the most straightforward and rapid response.
The ToxR-like regulator CadC of E. coli senses acidification of the environment through its periplasmic domain, thereby enabling the bacterium to respond to unfavorable conditions before experiencing challenges to the cellular interior. Until now, the molecular mechanism by which CadC senses an increase in the external proton concentration was not clear.
Histidine residues often mediate pH detection through protonation as shown for the pH-dependent folding switch of the Pseudomonas syringae effector protein AvrPto (32), for interaction of the diphtheria toxin T domain (33), and the pH-dependent dimer dissociation of the dynein light chain LC8 (34). Therefore and due to the fact that the activation range of cadBA expression (4) essentially matches the pKa value of the free histidine side chain, we analyzed the role of all histidines present in the periplasmic domain of CadC by site-directed mutagenesis. Our mutational analyses revealed, however, that protonation of histidines is not essentially involved in sensing low pH.
Further analyses of protonable residues in the periplasmic domain of CadC led to the identification of a negatively charged patch at the protein surface (Fig. 6). This patch is formed by Asp-198, Asp-200, Glu-461, Glu-468, and Asp-471 and extends across both subdomains. Strikingly, almost all residues are located on the same side of the protein surface, with Asp-198 and Asp-200 in the N-terminal subdomain and Glu-468 and Asp-471 in the C-terminal subdomain (Fig. 6). Glu-461 is almost buried within the protein but close to Glu-468. Substitution of the titratable residues within this patch had drastic effects (on-state, CadC-D471A/N; and off-state, CadC-D198A/E/N, CadC-D200A/E/N, CadC-E461A/D/Q, CadC-E468A/D/Q, and CadC-D471E/R). This might be due to a concerted effect of the entire group of negatively charged amino acids, also considering that the pKa of titratable side chains depends on the electrostatic environment and thus on the protonation state of groups in its surroundings. One could speculate that sensing the external H+ concentration with such an elaborate network could protect the cell from investing in an energy-consuming process (protein expression) under the wrong conditions and allows a graduated response of CadC to alterations of the external pH as it was already demonstrated (4).
Interestingly, a similar network of acidic residues was found to be involved in the pH-dependent gating of the acid-sensing ion channel 1a (35). In contrast, neither the crystal structure nor the site-directed mutagenesis studies presented here provided any evidence for the formation of specific salt bridges at neutral pH that may be broken upon proton binding within the CadC monomer, as was discussed for the pH-sensing mechanism of the bacterial potassium channel KcsA (36). Instead, it seems likely that the overall electrostatic potential at the protein surface (and associated variations in the Coulomb force field) plays a role in pH-dependent conformational changes.
Perturbation of the negatively charged patch, even via replacement of unprotonable or positively charged residues, led to CadC variants with either pH-insensitive (on-state, CadC-L479A and CadC-N478A) or nonresponsive (off-state, CadC-H240Q, CadC-L479A, CadC-N478A, CadC-T475A, CadC-L474A, CadC-R467A/E/K/Q, and CadC-K458A/Q/R) phenotypes. Because of the severe effects caused by these amino acid exchanges, one might speculate that these residues are vital for the precise structural or dynamic integrity of the protein, thus indirectly affecting pH sensing.
For the membrane-integrated kinase PhoQ, it was proposed that the positively charged calcium ions form bridges between the carboxylate side chains of the protein and the negatively charged headgroups of phospholipids (27). A similar bridging mechanism was described for the C2 membrane-binding domain of protein kinase Cα and its interaction with phosphatidylserine (37). Alternatively, protonation of the negatively charged surface patch of CadC might reduce repulsive forces between CadC and phospholipid headgroups. Thus, protonation instead of metal ion binding might lead to alterations in the protein orientation with respect to the lipid bilayer and hence affect activity. However, based on our results obtained with two different phospholipid mutants of E. coli, the protonation-mediated involvement of phospholipids seems unlikely.
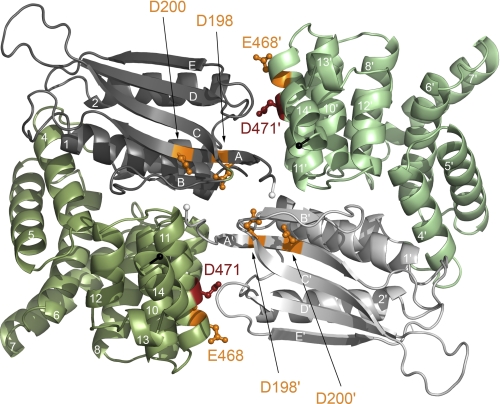
Three putative mechanisms by which protonation of the negatively charged residues within the surface patch affects the conformation or dynamics of CadC are conceivable: (1) reduction of intramolecular Coulomb repulsion, (2) reduction of intermolecular Coulomb repulsion, and (3) repositioning of monomer subunits within a functional dimer relative to each other. Mechanism 1 is based on the assumption that repelling surfaces exist between the two subdomains within one CadC monomer, more precisely between the two halves of the negatively charged patch. The two subdomains could alter their mutual arrangement via hinge movement at the connecting loop (Leu-330–Arg-333). Structural changes could be propagated to the transmembrane helix, resulting in binding of the cytoplasmic domain to the DNA and/or RNA polymerase. According to Mechanism 2, protonation would weaken the electrostatic repulsion between two CadC monomers, leading to pH-dependent oligomerization and corresponding signal transduction across the cytoplasmic membrane. Indeed, two CadC-binding sites have been identified in the Pcad promoter (38), such that a CadC dimer would be needed for efficient cadBA activation. This hypothesis is supported by recent observations from our structural analysis according to which the periplasmic domain of CadC forms a dimer (41). It is important to note that this protein-protein interaction is based mainly on polar interactions. Notably, the pH-responsive residues identified in this study are located mostly at the dimer interface (Fig. 8). Thus, it is conceivable that protonation of the corresponding residues would reduce intermolecular repulsion and allow approximation or, at least, spatial rearrangement (corresponding to Mechanism 3) of the two monomers. Unfortunately, such a signal-dependent change in the oligomerization state is difficult to prove experimentally due to the naturally extremely low copy number of CadC. Notably, none of the crystal structures solved so far indicates major pH-dependent structural changes. The crystal structure of the periplasmic wild-type domain, as well as the domain of the off-state variant CadC-D471E and the on-state variant CadC-D471N, showed nearly the same structure (41). This could be related to the fact that only a truncated version was crystallized so far that lacks the ability to undergo the changes of the full-length protein. Mechanism 3 also takes into account that a CadC dimer is needed to activate cadBA expression. In this case, the dimer is persistent, and the monomers would change their position relative to each other only as a consequence of a change in pH. Such a movement could be achieved, for example, by a rotation similar to that described for signaling in HAMP domains (39).
FIGURE 8.
Residues involved in pH sensing are located at the CadC periplasmic domain dimer interface. To distinguish the monomers, they are colored in dark and pale colors, respectively. Residues that are crucial for pH sensing are indicated in orange (off-state) or red (on-state). The N-terminal subdomains of the periplasmic domain are colored gray, and the C-terminal subdomains are colored green. The N termini, which are preceded by the transmembrane helices, are indicated by white spheres, whereas the C termini are indicated by black spheres.
Regardless of the precise mechanism, the pH signal arising from protonation of different side chains within the acidic patch seems to be integrated and further transduced by the N-terminal subdomain. This notion is based on our study of combined amino acid replacements within the N- and C-terminal subdomains. All tested amino acid replacements in the N-terminal subdomain were found to dominate the phenotype of the resulting CadC variants (CadC-D198E, off-state; and CadC-K242R, on-state), regardless of the amino acid replacement in the N-terminal subdomain.
In conclusion, it is proposed that CadC senses a decrease in the external pH through direct binding of protons and associated conformational and/or oligomerization effects. Because of the dose-dependent activation of cadBA expression between pH 6.6 and 5.8 (4), it is conceivable that multiple residues are involved in pH detection, giving rise to a network of pH-responsive side chains that may sense the external proton concentration in a cooperative manner. Depending on the external pH, the charge of the identified surface patch varies due to different protonation propensities of each titratable residue within the network. Once a certain degree of charge neutralization is achieved, cooperative conformational changes and/or oligomerization occurs, resulting in the activation of cadBA expression in the cytoplasm.
Acknowledgments
We thank Sophie Buchner and Ulla Kyrylenko for help with construction and analysis of some CadC variants.
Footnotes
This work was supported by Deutsche Forschungsgemeinschaft Grants JU270/5-1 and Exc114/1 and by an Elitenetzwerk Bayern research scholarship (BayEFG) (to I. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Table 1.
The atomic coordinates and structure factors (code 3LY7) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
REFERENCES
- 1. Bearson S., Bearson B., Foster J. W. (1997) FEMS Microbiol. Lett. 147, 173–180 [DOI] [PubMed] [Google Scholar]
- 2. Auger E. A., Redding K. E., Plumb T., Childs L. C., Meng S. Y., Bennett G. N. (1989) Mol. Microbiol. 3, 609–620 [DOI] [PubMed] [Google Scholar]
- 3. Soksawatmaekhin W., Kuraishi A., Sakata K., Kashiwagi K., Igarashi K. (2004) Mol. Microbiol. 51, 1401–1412 [DOI] [PubMed] [Google Scholar]
- 4. Fritz G., Koller C., Burdack K., Tetsch L., Haneburger I., Jung K., Gerland U. (2009) J. Mol. Biol. 393, 272–286 [DOI] [PubMed] [Google Scholar]
- 5. Meng S. Y., Bennett G. N. (1992) J. Bacteriol. 174, 2670–2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Watson N., Dunyak D. S., Rosey E. L., Slonczewski J. L., Olson E. R. (1992) J. Bacteriol. 174, 530–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Miller V. L., Taylor R. K., Mekalanos J. J. (1987) Cell 48, 271–279 [DOI] [PubMed] [Google Scholar]
- 8. Ulrich L. E., Koonin E. V., Zhulin I. B. (2005) Trends Microbiol. 13, 52–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tetsch L., Koller C., Haneburger I., Jung K. (2008) Mol. Microbiol. 67, 570–583 [DOI] [PubMed] [Google Scholar]
- 10. Dell C. L., Neely M. N., Olson E. R. (1994) Mol. Microbiol. 14, 7–16 [DOI] [PubMed] [Google Scholar]
- 11. Taylor R. G., Walker D. C., McInnes R. R. (1993) Nucleic Acids Res. 21, 1677–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yanisch-Perron C., Vieira J., Messing J. (1985) Gene 33, 103–119 [DOI] [PubMed] [Google Scholar]
- 13. Studier F. W., Moffatt B. A. (1986) J. Mol. Biol. 189, 113–130 [DOI] [PubMed] [Google Scholar]
- 14. Neely M. N., Dell C. L., Olson E. R. (1994) J. Bacteriol. 176, 3278–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 16. Epstein W., Kim B. S. (1971) J. Bacteriol. 108, 639–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. DeChavigny A., Heacock P. N., Dowhan W. (1991) J. Biol. Chem. 266, 5323–5332 [PubMed] [Google Scholar]
- 18. Heacock P. N., Dowhan W. (1989) J. Biol. Chem. 264, 14972–14977 [PubMed] [Google Scholar]
- 19. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 20. Lemonnier M., Lane D. (1998) Microbiology 144, 751–760 [DOI] [PubMed] [Google Scholar]
- 21. Phan A. P., Ngo T. T., Lenhoff H. M. (1982) Anal. Biochem. 120, 193–197 [DOI] [PubMed] [Google Scholar]
- 22. DeLano W. L. (2008) The PyMOL Molecular Graphics System, DeLano Scientific, Palo Alto, CA [Google Scholar]
- 23. Nicholls A., Sharp K. A., Honig B. (1991) Proteins 11, 281–296 [DOI] [PubMed] [Google Scholar]
- 24. Prost L. R., Daley M. E., Le Sage V., Bader M. W., Le Moual H., Klevit R. E., Miller S. I. (2007) Mol. Cell 26, 165–174 [DOI] [PubMed] [Google Scholar]
- 25. Müller S., Götz M., Beier D. (2009) PLoS One 4, e6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nozaki Y., Tanford C. (1967) J. Biol. Chem. 242, 4731–4735 [PubMed] [Google Scholar]
- 27. Cho U. S., Bader M. W., Amaya M. F., Daley M. E., Klevit R. E., Miller S. I., Xu W. (2006) J. Mol. Biol. 356, 1193–1206 [DOI] [PubMed] [Google Scholar]
- 28. Lagadic-Gossmann D., Huc L., Lecureur V. (2004) Cell Death Differ. 11, 953–961 [DOI] [PubMed] [Google Scholar]
- 29. Matsuyama S., Llopis J., Deveraux Q. L., Tsien R. Y., Reed J. C. (2000) Nat. Cell Biol. 2, 318–325 [DOI] [PubMed] [Google Scholar]
- 30. Yu J., Tian S., Metheny-Barlow L., Chew L. J., Hayes A. J., Pan H., Yu G. L., Li L. Y. (2001) Circ. Res. 89, 1161–1167 [DOI] [PubMed] [Google Scholar]
- 31. Damaghi M., Bippes C., Köster S., Yildiz O., Mari S. A., Kühlbrandt W., Muller D. J. (2010) J. Mol. Biol. 397, 878–882 [DOI] [PubMed] [Google Scholar]
- 32. Dawson J. E., Seckute J., De S., Schueler S. A., Oswald A. B., Nicholson L. K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8543–8548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Perier A., Chassaing A., Raffestin S., Pichard S., Masella M., Ménez A., Forge V., Chenal A., Gillet D. (2007) J. Biol. Chem. 282, 24239–24245 [DOI] [PubMed] [Google Scholar]
- 34. Nyarko A., Cochrun L., Norwood S., Pursifull N., Voth A., Barbar E. (2005) Biochemistry 44, 14248–14255 [DOI] [PubMed] [Google Scholar]
- 35. Liechti L. A., Bernèche S., Bargeton B., Iwaszkiewicz J., Roy S., Michielin O., Kellenberger S. (2010) J. Biol. Chem. 285, 16315–16329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thompson A. N., Posson D. J., Parsa P. V., Nimigean C. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6900–6905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Verdaguer N., Corbalan-Garcia S., Ochoa W. F., Fita I., Gómez-Fernández J. C. (1999) EMBO J. 18, 6329–6338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kuper C., Jung K. (2005) J. Mol. Microbiol. Biotechnol. 10, 26–39 [DOI] [PubMed] [Google Scholar]
- 39. Hulko M., Berndt F., Gruber M., Linder J. U., Truffault V., Schultz A., Martin J., Schultz J. E., Lupas A. N., Coles M. (2006) Cell 126, 929–940 [DOI] [PubMed] [Google Scholar]
- 40. Miller J. H. (1992) Experiments in Molecular Genetics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 41. Eichinger A., Haneburger I., Koller C., Jung K., Skerra A. (2011) Protein Sci., in press [DOI] [PMC free article] [PubMed] [Google Scholar]