Abstract
Poly(ADP-ribose) polymerase-1 (PARP-1) has two homologous zinc finger domains, Zn1 and Zn2, that bind to a variety of DNA structures to stimulate poly(ADP-ribose) synthesis activity and to mediate PARP-1 interaction with chromatin. The structural basis for interaction with DNA is unknown, which limits our understanding of PARP-1 regulation and involvement in DNA repair and transcription. Here, we have determined crystal structures for the individual Zn1 and Zn2 domains in complex with a DNA double strand break, providing the first views of PARP-1 zinc fingers bound to DNA. The Zn1-DNA and Zn2-DNA structures establish a novel, bipartite mode of sequence-independent DNA interaction that engages a continuous region of the phosphodiester backbone and the hydrophobic faces of exposed nucleotide bases. Biochemical and cell biological analysis indicate that the Zn1 and Zn2 domains perform distinct functions. The Zn2 domain exhibits high binding affinity to DNA compared with the Zn1 domain. However, the Zn1 domain is essential for DNA-dependent PARP-1 activity in vitro and in vivo, whereas the Zn2 domain is not strictly required. Structural differences between the Zn1-DNA and Zn2-DNA complexes, combined with mutational and structural analysis, indicate that a specialized region of the Zn1 domain is re-configured through the hydrophobic interaction with exposed nucleotide bases to initiate PARP-1 activation.
Keywords: ADP-ribosylation, DNA Damage, Protein-DNA Interaction, Protein Structure, X-ray Crystallography, Zinc Finger, Poly(ADP-ribose) Polymerase
Introduction
Poly(ADP-ribose) (or PAR),2 is a unique post-translational modification synthesized by poly(ADP-ribose) polymerases (PARPs) using NAD+ as a precursor (1, 2). PARP-1 is an abundant nuclear enzyme and a prolific source of PAR production, particularly in stressed cells (3). PARP-1 is a multifunction enzyme with roles in maintaining genome integrity, regulation of transcription and chromatin architecture, and cell death signaling pathways (1, 2, 4). The catalytic activity of PARP-1 is robustly stimulated through interaction with DNA, in particular damaged DNA structures such as single and double strand breaks (3). PARP-1 rapidly localizes to sites of DNA damage and appears to regulate access to DNA damage and facilitate assembly of repair factors (5). PAR synthesized by PARP-1 can serve as a ligand for PAR-binding factors (6–10), and it can act as a signaling molecule that initiates programmed cell death (11, 12).
PARP-1 has a six-domain modular construction (Fig. 1A). Two homologous zinc finger domains, Zn1 and Zn2, are located at the extreme N terminus of PARP-1. The Zn1 and Zn2 domains are members of the PARP-like zinc finger family, which includes zinc fingers found in mammalian DNA ligase III and plant DNA 3′-phosphatases (13–16). The PARP-like zinc fingers are specialized zinc fingers that do not bind to specific DNA sequences, but rather appear to recognize DNA structure (17–19). Mutational and deletion analysis of the Zn1 and Zn2 domains have indicated a pivotal role for the Zn1 domain in PARP-1 DNA-dependent activity in vitro, whereas the Zn2 domain is not essential (20, 21), but perhaps plays a role in PARP-1 binding to particular damaged DNA structures (20, 22). A combination of mutations that simultaneously disrupts both the Zn1 and Zn2 domains interferes with PARP-1 interaction with chromatin in the absence of DNA damage (23). This further highlights that PARP-1 interaction with DNA through the Zn1 and Zn2 domains can adapt to a range of DNA structures, including nucleosome-bound conformations of continuous, undamaged DNA. The mode of interaction with DNA for the Zn1 and Zn2 domains has been a notable deficiency in our understanding of PARP-1, and the mechanism by which the Zn1 domain specifically contributes to DNA-dependent activation of PARP-1 is unknown. Furthermore, the individual contributions of the Zn1 and Zn2 domains to PARP-1 DNA binding affinity have not been directly assessed.
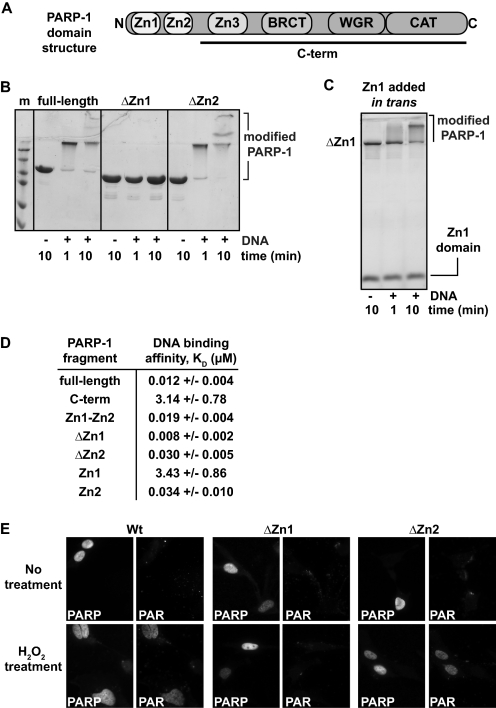
FIGURE 1.
The Zn1 and Zn2 domains of PARP-1 have distinct biochemical activities. A, schematic representation of PARP-1 domain structure. B, DNA-dependent automodification assay of full-length PARP-1, ΔZn1, and ΔZn2 (1 μm) in the presence of 1 μm DNA and 5 mm NAD+. Time points were analyzed by 12% SDS-PAGE. C, DNA-dependent automodification assay of ΔZn1 PARP-1 in the presence/absence of the isolated Zn1 domain added in trans. Time points were analyzed by 15% SDS-PAGE. D, DNA binding affinity constants (KD) derived from fluorescence polarization experiments using an 18-bp DNA. The values represent the average and mean ± S.D. of three or more independent experiments. E, PARP-1−/− mouse embryonic fibroblasts were transiently transfected with an expression vector coding for PARP-1 full-length, ΔZn1 or ΔZn2. Fixed cells were stained with antibodies against PAR or PARP-1 as indicated. Top panels, no treatment; bottom panels, treatment with H2O2 for 10 min.
In addition to the Zn1 domain, the Zn3 domain and the WGR domain of PARP-1 are also required to support DNA-dependent PAR synthesis activity of the catalytic domain (21, 24, 25). The Zn3 domain is a unique zinc finger domain that is distinct in structure and function from the Zn1 and Zn2 domains (24, 25). Biochemical studies indicate that the zinc ribbon motif of the Zn3 domain contributes key residues that mediate an interdomain contact essential for the efficient assembly of PARP-1 domains (26). The function of the WGR domain is unknown. The BRCT domain of PARP-1 is not essential for DNA-dependent PAR synthesis activity (21, 23), but might play a role in orchestrating PARP-1 interaction with partner proteins in DNA repair pathways (27, 28).
NMR structural analyses of the isolated Zn1 and Zn2 domains have established the overall fold of these domains and their solution conformations in the absence of DNA (PDB accession codes: Zn1, 2dmj; Zn2, 2cs2; no publication record). An NMR study of the PARP-like zinc finger domain found in human DNA ligase III demonstrated a similar overall structure (29). DNA titration NMR experiments with human DNA ligase III indicated multiple chemical shift perturbations upon interaction with DNA (29), but a structure for this complex has not been determined. Thus, the mode of interaction with DNA and the DNA features recognized by PARP-1 zinc fingers are still unknown.
Here, we have determined crystal structures for the individual human PARP-1 zinc fingers in complex with blunt-ended duplex DNA, a potent stimulator of PARP-1 activity (17, 21, 24, 26, 30) and a model for a double-stranded break in DNA. The Zn1-DNA and Zn2-DNA structures demonstrate a consistent mode of interaction with DNA that is distinct from other known DNA binding factors. The Zn1 and Zn2 domains bind to an uninterrupted segment of the phosphate backbone using a region that we have termed the phosphate backbone grip, and they engage the exposed nucleotide bases of DNA through a second region termed the base stacking loop. Biochemical analysis demonstrates that the Zn1 domain has relatively weak DNA binding affinity, but this activity is required for activation of PARP-1. In contrast, the Zn2 domain binds to DNA with much higher affinity yet is not essential for DNA-dependent PARP-1 activation in vitro or in vivo. The Zn1-DNA complex structure combined with mutational and structural analysis indicate that a specialized loop region of the Zn1 domain is repositioned upon binding to DNA, and this situates key residues that contribute to the activation of PAR synthesis in a DNA-dependent manner.
EXPERIMENTAL PROCEDURES
Gene Cloning and Mutagenesis
The following PARP-1 constructs were cloned into the NdeI/XhoI sites of the pET28 expression vector (Novagen): full-length wild-type (WT) PARP-1 (residues 1–1014) and mutants, WT Zn1 domain (residues 1–96) and mutants, Zn1–Zn2 fragment (residues 1–215), C-terminal fragment (residues 216–1014; Zn3-BRCT-WGR-CAT domains), ΔZn1 (deletion of residues 1–96), and ΔZn2 (deletion of residues 97–206). The WT Zn2 domain (residues 105–206) and mutants were cloned into the pET24 expression vector (NdeI/XhoI). All mutations and deletions were performed using the QuikChange protocol (Stratagene), and verified by automated DNA sequencing. The following PARP-1 constructs were cloned into the NheI/XhoI sites of the pCDNA 3.1/V5-HisA vector (Invitrogen) for mammalian cell transfection: full-length, ΔZn1, and ΔZn2.
Protein Expression and Purification
Full-length PARP-1 WT and mutants were expressed and purified as described previously (24, 26) using three chromatographic steps: Ni2+ affinity, heparin-sepharose, and gel filtration. Selenomethionine-containing Zn1 was expressed in Escherichia coli grown in defined medium (31) and purified as WT Zn1. Specific details for the purification of the different PARP-1 domains are presented under supplemental “Experimental Procedures” and supplemental Table 1.
DNA-dependent Automodification Assay
The DNA-dependent automodification assay was performed essentially as described (24, 26). PARP-1 full-length WT and mutants, ΔZn1 or ΔZn2 (1 μm), were preincubated with 1 μm of an 18-bp DNA duplex for 10 min at room temperature (22 °C). In the complementation experiments, a mixture of domains (1 μm each) was preincubated for 10 min at RT with DNA (1 μm). 5 mm NAD+ was then added to the reaction, and the mixture was incubated for various times. For each experiment, reactions were stopped by the addition of SDS loading buffer containing 0.1 m EDTA. The samples were resolved on SDS-PAGE and stained with Imperial Protein Stain (Pierce).
Fluorescence Polarization DNA Binding Assay
Fluorescence polarization DNA binding experiments were performed as described previously (26) in 20 mm Hepes, pH 8.0, 8 mm MgCl2, 60 mm KCl, 0.12 mm EDTA, 5.5 μm β-mercaptoethanol, 50 μg/ml of bovine serum albumin (BSA), and 4% glycerol, and using a DNA probe at 5 nm. The probe was either an 18-base pair (bp) duplex DNA (5′-GGGTTGCGGCCGCTTGGG-3′ plus complement) or a 10-bp duplex DNA (5′-GCCGCTTGGG-3′ plus complement) carrying a fluorescein derivative (6-carboxyfluorescein) on the 5′ terminus of one strand. The DNA binding experiments in lower ionic strength buffer for Zn1 WT and mutants were performed in 20 mm Hepes, pH 8.0, 30 mm KCl, 0.12 mm EDTA, 5.5 μm β-mercaptoethanol, 50 μg/ml of BSA, and 4% glycerol. The observed binding constants were obtained from a non-linear least squares fit to the data using a two state binding model (see supplemental Fig. S6).
Transient Transfection and Immunofluorescence of Mouse Embryonic Fibroblasts
Embryonic fibroblasts derived from a PARP-1 knock-out mouse (32) (PARP-1−/− mouse embryonic fibroblasts) were grown in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin until ∼80–90% confluent. Cells in 6-well plates (2 × 105 cells/well) with sterilized glass coverslips were grown 24 h before transfection using 4 μg of DNA and 10 μl of Lipofectamine 2000 (Invitrogen) following the recommended protocol. 24 h post-transfection, fresh medium supplemented with 1 mm H2O2 was added for 10 min at 37 °C. Cells were washed with cold PBS, fixed with 2 ml of ice-cold methanol for 2 min at RT, washed with PBS, and blocked for 20 min at RT (blocking buffer: 25 mm Tris, pH 7.5, 150 mm NaCl, 1% Triton X-100, 2.5% dry nonfat milk). Cells were incubated 16 h at 4 °C with 100 μl of 1:200 dilution of primary antibody (mouse mAb (10H) to PAR (Enzo Lifesciences), rabbit PARP-1/2 (H250) (Santa Cruz)), washed with blocking buffer, incubated 30 min at RT with 50 μl of 1:100 dilution of secondary antibody (goat α-mouse Alexa Fluor 594 (Invitrogen); goat α-rabbit Alexa Fluor 488 (Invitrogen)), washed with TBS, 1% Triton X-100, rinsed with ddH2O, and air dried. Coverslips were mounted onto glass slides with 10 μl of ProLong Gold Antifade Reagent with DAPI (Invitrogen), dried 16 h in the dark, and imaged with an Olympus BX-61 upright microscope with ORCA-ER (Hamamatsu, Bridgewater NJ).
Crystallization of Zn1-DNA and Zn2-DNA Complexes
Native and selenomethionine Zn1-DNA complexes were formed by incubating protein (5 mg/ml) with a 10-bp DNA duplex (210 μm) in gel filtration buffer for 45 min at RT. Two DNA duplexes were assembled: (i) DNA sequence B, 5′-GCCGCTTGGG-3′ plus complementary strand, and (ii) DNA sequence G, 5′-GCCTGCAGGC-3′ (palindromic). Crystals were grown in sitting drops at RT by mixing the Zn1-DNA complex with an equal amount of 28–30% PEG 3350, 0.1 m sodium acetate, 0.1 m Tris, pH 8.5, 0.1 mm Tris(2-carboxyethyl)phosphine, and 15–20% ethylene glycol. Crystals were flash-cooled in liquid nitrogen prior to data collection. Zn2-DNA complexes were formed by incubating protein (5 mg/ml) with an 8-bp DNA duplex (216 μm) in gel filtration buffer for 45 min at RT. Two DNA duplexes were used: (i) DNA sequence A, 5′-CGCTTGGG-3′ plus complementary strand, and (ii) DNA sequence H, 5′-CGTCTGGG-3′ plus complementary strand. Crystals were grown in sitting drops at RT by mixing the Zn2-DNA complex with an equal amount of 1.36–1.42 m sodium citrate, pH 6.5. Prior to flash-cooling with liquid nitrogen, crystals were transferred to a cryosolution of 1.38–1.42 m sodium citrate, pH 6.5, and 10–14% glycerol. Diffraction data were collected at beamlines X29 and X12C at the National Synchrotron Light Source (NSLS, Brookhaven National Laboratory) and the SIBYLS beamline at the Advanced Light Source (ALS, Berkeley Laboratory), and were processed using HKL2000 (33) and the CCP4 suite of programs (34) (Table 1).
TABLE 1.
Crystallographic data and refinement statistics
| Zn1-DNA | Zn2-DNA | ||||
|---|---|---|---|---|---|
| Data collectiona | |||||
| Space group | P21 | P312 | |||
| Unit cell dimensions | a = 62.8 Å, b = 107.3 Å, c = 87.0 Å | a = b = 63.7 Å, c = 192.4 Å | |||
| α = γ = 90°, β = 100.6° | α = β = 90°, γ = 120° | ||||
| Crystal | Selenomethionine, DNA seq. B | Native, DNA seq. G | Native, DNA seq. A | Native, DNA seq. H | Native, DNA seq. A |
| Wavelength (Å) | 0.98 | 0.99 | 1.28 | 0.99 | 0.99 |
| Resolution range (Å) | 50–2.4 (2.44–2.4) | 50–2.65 (2.7–2.65) | 50–4.0 (4.14–4.0) | 50–2.8 (2.85–2.8) | 50–2.95 (3.0–2.95) |
| Completeness (%) | 100 (100) | 99.9 (99.9) | 98.9 (91.8) | 100 (99.9) | 98.6 (96.0) |
| Average redundancy | 8.3 (8.4) | 4.2 (4.3) | 3.6 (3.2) | 8.1 (8.2) | 7.7 (7.5) |
| Mean I/σ(I) | 14.2 (3.0) | 15.0 (2.5) | 23.9 (13.7) | 23.9 (3.0) | 15.2 (3.1) |
| Rmerge (%)b | 13.8 (70.6) | 9.0 (50.9) | 5.3 (8.8) | 9.2 (71.7) | 10.3 (86.9) |
| Phasing FOMc | 0.40/0.72 | 0.29/0.77 | |||
| Model refinementa | |||||
| Resolution range (Å) | 50–2.4 (2.48–2.4) | 50–2.65 (2.74–2.65) | 50–2.8 (3.08–2.8) | 50–2.95 (3.38–2.95) | |
| Number of reflections | 44,511 (4,340) | 33,211 (3,224) | 11,798 (2,760) | 9,907 (3,037) | |
| Rcrystd | 0.191 (0.234) | 0.196 (0.256) | 0.193 (0.286) | 0.199 (0.246) | |
| Rfreed | 0.239 (0.311) | 0.246 (0.327) | 0.240 (0.347) | 0.244 (0.314) | |
| Number of atoms/average B-factor (Å2) | 9,313/32.7 | 7,584/35.0 | 2,205/48.4 | 2,135/58.2 | |
| Protein | 5,731/30.8 | 5,727/34.0 | 1,520/49.6 | 1,470/62.0 | |
| Zinc | 8/19.6 | 8/21.9 | 2/42.9 | 2/51.1 | |
| Solvent | 342/30.6 | 233/29.0 | 39/43.0 | 19/41.0 | |
| DNA | 3,232/36.3 | 1,616/39.6 | 644/45.9 | 644/50.0 | |
| φ/ψ, most favored (%) | 96.4 | 95.5 | 96.3 | 91.7 | |
| Root mean square deviation bond angles (°) | 1.354 | 1.288 | 1.305 | 1.47 | |
| Root mean square deviation bond lengths (Å) | 0.009 | 0.011 | 0.010 | 0.011 | |
a Values in parentheses refer to data in the highest resolution shell.
b Rmerge = ΣhklΣj|Ij − 〈I〉|/ΣhklΣjIj. 〈I〉 is the mean intensity of j observations of reflection hkl and its symmetry equivalents.
c Figure of merit (FOM) before and after density modification as reported in SOLVE/RESOLVE or PHENIX (35).
d Rcryst = Σhkl|Fobs − kFcalc|/Σhkl|Fobs|. Rfree = Rcryst for 5% of reflections excluded from crystallographic refinement.
Zn1-DNA and Zn2-DNA Structure Determination
The initial electron density map of the Zn1-DNA complex structure was calculated using PHENIX (35) provided with SAD data collected at the selenium edge (Table 1 and supplemental Fig. S2). The initial map allowed manual positioning of four copies of an ideal B-form DNA duplex and eight copies of an NMR-derived model of the Zn1 domain (PDB accession code 2dmj). An anomalous Fourier difference map indicated the locations of the selenium atoms in selenomethionine residues, and thus guided the manual positioning of the Zn1 domains (supplemental Fig. S2). Residues 45 to 56 were removed from the model at this point due to a poor fit with the experimental electron density. These residues were added to the model during iterative rounds of model building in COOT (36) and refinement in PHENIX (35). The non-palindromic DNA sequence B was positioned randomly in two possible overlapping orientations (see supplemental Figs. S2 and S3); therefore the final model of Zn1 in complex with DNA sequence B contains both orientations, with 0.5 occupancy for each.
The initial electron density map of the Zn2-DNA complex structure was calculated using PHENIX (35) provided with SAD data collected at the Zn edge (Table 1 and supplemental Fig. S2). The initial map allowed manual positioning of two copies of an ideal B-form DNA duplex and two copies of an NMR-derived model of the Zn2 domain (PDB accession code 2cs2). An anomalous Fourier difference map indicated the locations of zinc atoms and guided the manual positioning of the Zn2 domain (supplemental Fig. S2). The model was improved through iterative rounds of model building in COOT (36) and refinement in PHENIX (35).
Circular Dichroism (CD) Spectroscopy
CD experiments were performed for WT Zn1 and Zn2 domains and mutants using 10 μm of protein, essentially as described (24).
Illustrations
Structure images were created using PyMOL Molecular Graphics System (Schrödinger, LLC), Illustrator and Photoshop (Adobe Systems).
Structural Data
Atomic coordinates and structure factor amplitudes have been deposited in the Protein Data Bank for the two Zn1-DNA complexes (accession numbers 3OD8 and 3ODA) and the two Zn2-DNA complexes (accession numbers 3ODC and 3ODE).
RESULTS
The Homologous PARP-1 Zinc Fingers Zn1 and Zn2 Have Distinct Biochemical Activities
To understand the respective roles of the Zn1 and Zn2 domains in DNA-dependent PARP-1 activation, the Zn1 and Zn2 domains were individually deleted from full-length PARP-1 (ΔZn1 and ΔZn2 constructs). A PARP-1 automodification assay was used to compare the activity of full-length PARP-1, ΔZn1, and ΔZn2. PARP-1 automodification was monitored on SDS-PAGE by a decrease in electrophoretic mobility due to the addition of PAR to PARP-1 itself (Fig. 1B). The ΔZn1 construct showed no signs of DNA-dependent PARP-1 automodification (Fig. 1B). In contrast, the ΔZn2 construct showed robust DNA-dependent automodification activity at a level similar to full-length PARP-1 (Fig. 1B). The same result was obtained using a quantitative assay of PAR production comparing full-length and ΔZn2 DNA-dependent activity (not shown). The results of our deletion mutagenesis are consistent with the results of a recent study (21). Interestingly, DNA-dependent activity of the ΔZn1 construct is restored when the isolated Zn1 domain is added in trans (Fig. 1C), demonstrating that the ΔZn1 construct is folded properly and that the deficiency is strictly related to the deletion of the Zn1 domain. Thus, the Zn1 domain fulfills a special role in DNA-dependent PARP-1 activation that cannot be carried out by the Zn2 domain.
We also assessed the individual contributions of the Zn1 and Zn2 domains to the overall DNA binding affinity of PARP-1. DNA binding affinities were measured by monitoring the polarization of light emitted from a fluorescently labeled DNA duplex during protein titration experiments (Fig. 1D and supplemental Fig. S1). Full-length PARP-1 binds robustly to duplex DNA with a KD of 12 nm in this assay (Fig. 1D). Deletion of both the Zn1 and Zn2 domains, resulting in a C-terminal fragment of PARP-1 (referred to herein as C-term), reduces the DNA binding affinity to 3.1 μm (over 250-fold). The Zn1–Zn2 fragment of PARP-1 binds with a KD of 19 nm, only slightly less than full-length PARP-1. These data indicate that the Zn1 and Zn2 zinc fingers are the primary contributors to PARP-1 DNA binding affinity, consistent with other studies (18, 22, 37). However, C-term does make a minor contribution to the overall binding affinity. Deletion of the Zn1 domain had no significant affect on the measured DNA binding affinity, with a KD of 8 nm. Deletion of the Zn2 domain reduced the DNA binding affinity to 30 nm (Fig. 1D), roughly a 3-fold reduction in affinity compared with full-length PARP-1. These data suggest that the Zn2 domain makes a more substantial contribution than the Zn1 domain does to the overall DNA binding affinity of PARP-1.
The isolated Zn1 and Zn2 domains were cloned and purified to directly analyze their individual DNA binding affinities. Interestingly, the Zn2 domain alone binds to DNA with a KD of 34 nm, whereas the isolated Zn1 domain binds to DNA with a KD of 3.4 μm. Thus, in contrast with the fact that the Zn1 domain is essential for DNA-dependent activity, the Zn1 domain binds to DNA with ∼100-fold less affinity than the Zn2 domain. The marked difference in Zn1 and Zn2 binding affinities was also observed with other DNA structures (duplex DNA containing an overhang or a nick; not shown); therefore the higher binding affinity appears to be a general feature of the Zn2 domain. Interestingly, even though the isolated Zn2 domain binds to DNA robustly on its own, deletion of the Zn2 domain does not have a dramatic impact on the overall binding affinity of PARP-1. This suggests that the relatively weak binding affinity of the Zn1 domain (3.4 μm) and the C-term fragment (3.1 μm) collectively establish a robust DNA binding affinity (30 nm).
Although our biochemical analysis and a recent study (21) have indicated that the Zn2 domain is not essential for DNA-dependent PARP-1 activity in vitro, we wanted to exclude the possibility that the Zn2 domain, with a significantly higher DNA binding affinity, plays a more critical role in a cellular context. Embryonic fibroblasts derived from a PARP-1−/− knock-out mouse (32) were transiently transfected with mammalian expression vectors coding for full-length, ΔZn1, or ΔZn2 human PARP-1. Twenty-four hours after transfection, cells were treated with hydrogen peroxide (H2O2) for 10 min at 37 °C to generate DNA damage. PARP-1 and PAR were detected in fixed cells using PARP-1 and PAR antibodies, respectively (Fig. 1E). PARP-1−/− cells transfected with a control empty vector do not support PAR production in the presence or absence of H2O2 (not shown). PAR is produced in cells transfected with full-length PARP-1 after treatment with H2O2 (Fig. 1E). ΔZn2 PARP-1 also produces PAR in transfected cells, indicating that the Zn2 domain does not play a critical role in DNA damage-dependent PARP-1 activation under the conditions tested. In contrast, cells transfected with ΔZn1 PARP-1 show no signs of PAR staining, indicating that Zn1 is essential for PARP-1 catalytic activity in vivo, consistent with in vitro analysis (Fig. 1B).
Crystallization of the Individual PARP-1 Zinc Fingers in Complex with DNA
We endeavored to determine the structure of PARP-1 zinc fingers bound to DNA to define their mode of interaction with DNA, to identify the structural basis for their distinct biochemical activities, and to gain insights into the mechanism of DNA-dependent PARP-1 activation. Attempts were made to crystallize the individual Zn1 and Zn2 domains, and the Zn1-Zn2 fragment of PARP-1, in complex with a variety of DNA structures. Crystallization trials were successful for the individual Zn1 and Zn2 domains using duplex DNA with blunt ends. Duplex DNA containing blunt ends is a model for DNA double strand break damage and is a potent stimulator of PARP-1 activity in vitro (17, 21, 24, 26, 30). Furthermore, PARP-1 shows a high binding affinity for blunt-ended DNA (Fig. 1B) (17, 24, 38). The presented crystal structures of PARP-1 zinc fingers provide the first views of this unique type of zinc finger bound to DNA.
A 10-bp DNA duplex was compatible with crystallization of a Zn1-DNA complex. The structure was determined using single-wavelength anomalous dispersion (SAD) phasing methods (Table 1 and supplemental Fig. S2 and “Experimental Procedures”). Structures were determined for the Zn1 domain in complex with two different 10-bp DNA sequences, and the two Zn1-DNA models were refined to a resolution of 2.4 Å and 2.65 Å with a crystallographic R/Rfree of 0.19/0.24 and 0.20/0.25, respectively (Table 1). An 8-bp DNA duplex was compatible with crystallization of a Zn2-DNA complex, and the structure was determined using SAD phasing methods (Table 1 and supplemental Fig. S2 and “Experimental Procedures”). Structures were determined for the Zn2 domain in complex with two different 8-bp DNA sequences, and the two Zn2-DNA models were refined to resolutions of 2.8 and 2.95 Å with crystallographic R/Rfree of 0.19/0.24 and 0.20/0.24, respectively (Table 1).
The asymmetric unit of Zn1-DNA crystals contains eight Zn1 molecules and four DNA duplexes; a Zn1 molecule binds at each end of the duplex DNA (supplemental Fig. S3). The asymmetric unit of the Zn2-DNA crystal contains two Zn2 molecules and two DNA duplexes (supplemental Fig. S4). The 8-bp duplex in the Zn2-DNA structures can accommodate only a single Zn2 molecule. Crystal contacts introduce some differences in the eight copies of the Zn1 domain, and the two copies of the Zn2 domain (see supplemental Figs. S3 and S4 for details); however, the following sections discuss features of the Zn1-DNA and Zn2-DNA structures that are consistent between all Zn1 and Zn2 molecules contained in the crystal structures.
Zn1-DNA and Zn2-DNA Complex Crystal Structures Reveal the Molecular Basis for PARP-1 Interaction with DNA
The crystal structures of the Zn1-DNA and Zn2-DNA complexes reveal that both PARP-1 zinc fingers bind in a similar manner to the end of duplex DNA, and recognize structural features that are independent of DNA sequence (Fig. 2, A and B). The Zn1 and Zn2 domains do not influence the overall shape of the bound DNA; each of the DNA duplexes exhibits a B-form conformation. The Zn1 and Zn2 domains each form a continuous interaction surface with DNA that can be described in two regions based on the type of contacts made with the DNA, which we have termed (i) the phosphate backbone grip and (ii) the base stacking loop (Fig. 2, A and B).
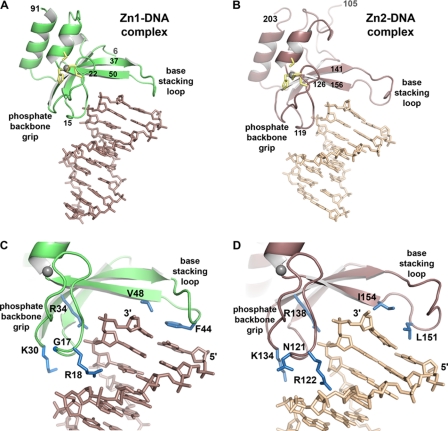
FIGURE 2.
Crystal structures of PARP-1 zinc fingers in complex with DNA demonstrate the molecular basis for structure-specific DNA binding. A, x-ray structure of the Zn1 domain bound to a 10-bp DNA duplex (DNA sequence G is shown). For clarity, the Zn1 molecule bound to the other end of the DNA is not shown (see supplemental Fig. S3). Zinc-coordinating residues are drawn as yellow sticks; the zinc atom is drawn as a gray sphere. B, x-ray structure of the Zn2 domain bound to an 8-bp DNA duplex (DNA sequence B is shown). Zinc-coordinating residues are drawn as yellow sticks; the zinc atom is drawn as a gray sphere. C and D, a more detailed view of the backbone grip and base stacking loop of the Zn1 and Zn2 domains, respectively. Residues mentioned in the text are drawn as blue sticks.
The phosphate backbone grip covers roughly 3 nucleotides of DNA toward the 3′ end of one DNA strand (Fig. 2, C and D); however, the 3′ terminus of the DNA strand remains solvent accessible and is not involved in contacts with the protein. Thus, the phosphate backbone grip does not contact the DNA strand break (i.e. the 3′ end), but rather contacts a continuous region of the DNA phosphate backbone. Residues 15 to 22 of the Zn1 domain and residues 119 to 126 of the Zn2 domain form the majority of the contacts with the DNA phosphodiester backbone (Fig. 2, A and B). An additional contact is formed between the phosphate group of the penultimate nucleotide and the guanidinium group of an arginine side chain that extends out from the interior of the zinc finger-fold (Arg-34 in Zn1 and Arg-138 in Zn2) (Fig. 2, C and D). Although the phosphate backbone grip primarily engages phosphate groups, an arginine side chain inserts into the minor groove of the duplex (Arg-18 in Zn1 and Arg-122 in Zn2) (Fig. 2, C and D). Positioning of the minor groove Arg is dictated by bound solvent molecules and varies in the Zn1 and Zn2 structures with different DNA sequences; therefore it does not make base-specific contacts in the minor groove.
The phosphate backbone grip of the Zn2 domain makes additional contacts with the phosphate backbone of the complementary DNA strand; thus the Zn2 domain spans the DNA minor groove (Fig. 2D). In particular, Zn2 residues Asn-121 and Lys-134 are positioned close to the phosphate backbone of the complementary strand. The Zn1 domain does not span the minor groove and therefore lacks these additional contacts (Fig. 2C). The additional Zn2 contacts with DNA could contribute to the observed higher affinity for DNA compared with the Zn1 domain.
The base stacking loop extends from the phosphate backbone grip at roughly a 90° angle, approaching the major groove of the DNA and capping the terminal base pair at the end of the DNA duplex (Fig. 2). Residues 37 to 50 of the Zn1 domain, and residues 141 to 156 of the Zn2 domain, compose the base stacking loop (Fig. 2, A and B). Unlike the phosphate backbone grip, the Zn1-DNA and Zn2-DNA structures vary in the number of residues within the base stacking loop, and in the overall conformation of the loop. As discussed in a later section, these structural differences account for Zn1 ability to stimulate PARP-1 activity, whereas the Zn2 domain is not able to perform this function. Despite these important differences, there are consistent features between the base stacking loops of the Zn1 and Zn2 domains that define the interaction with the exposed nucleotide bases of DNA. Hydrophobic residues Phe-44 and Val-48 of Zn1, and Leu-151 and Ile-154 of Zn2, stack against the hydrophobic face of the paired nucleotide bases at the end of the duplex (Fig. 2, C and D). Phe-44, in particular, is a striking mimic of an additional nucleotide base stacking on the DNA end. Similar to the phosphate backbone grip, the base stacking loop does not contact the 3′ or 5′ ends of the DNA strands.
Overall, the Zn1-DNA and Zn2-DNA crystal structures indicate that PARP-1 binds to DNA by recognizing an uninterrupted phosphate backbone through the phosphate backbone grip, and by contacting exposed bases via a hydrophobic interaction with the base stacking loop. These DNA features are present in the various structures bound by PARP-1, including single strand breaks in DNA as well as undamaged, continuous DNA structures. Therefore, the crystal structures of the Zn1-DNA and Zn2-DNA on duplex DNA provide insights into how PARP-1 will bind to different types of DNA structures (see “Discussion”).
Structure-based Mutagenesis Identifies Critical Zn1 and Zn2 DNA Binding Residues
The Zn1-DNA and Zn2-DNA crystal structures indicate the residues of the phosphate backbone grip and the base stacking loop that are directly involved in DNA binding. Several of these residues were targeted by site-directed mutagenesis to confirm their expected contribution to DNA binding affinity. We also used these mutagenesis experiments to assess the relative contribution of the phosphate backbone grip versus the base stacking loop to the overall DNA binding affinity of the Zn1 and Zn2 domains. DNA binding affinities of the mutants were determined using the fluorescence polarization DNA binding assay. The mutations were studied in the context of the individual zinc finger domains to directly measure the affect on the DNA binding ability of the isolated Zn1 and Zn2 domains. Circular dichroism (CD) spectroscopy analysis indicated that there were no gross overall structural perturbations in the Zn1 and Zn2 domain mutants compared with the WT proteins (supplemental Fig. S5).
Two mutations in the Zn1 domain phosphate backbone grip (R18A and R34A), and two in the base stacking loop (F44A and F44A/V48A), effectively abolished Zn1 DNA binding affinity (Table 2). This is consistent with an important role for these residues in DNA binding activity. We estimated the KD values for these mutants to be greater than 30 μm (the highest protein concentration tested) compared with 2 μm for WT Zn1. The base stacking loop mutant V48A was less affected with a KD of 10 μm (Table 2).
TABLE 2.
DNA binding affinities for WT Zn1 and Zn2 domains and mutants
| Zn1 domain | KD | KDa | Zn2 domain | KD |
|---|---|---|---|---|
| μm | μm | μm | ||
| WT | 1.96 ± 0.54 | 0.19 ± 0.03 | WT | 0.032 ± 0.004 |
| Phosphate backbone grip mutants | ||||
| R18A | >30 | >30 | R122A | 2.58 ± 1.15 |
| R34A | >30 | >30 | R138A | >8 |
| Base stacking loop mutants | ||||
| F44A | >30 | 1.04 ± 0.33 | L151A | 0.115 ± 0.035 |
| V48A | 10.2 ± 1.3 | 2.05 ± 1.16 | I154A | 0.829 ± 0.117 |
| F44A/V48A | >30 | 5.16 ± 0.72 | L151A/I154A | >8 |
| Q40A | 3.93 ± 0.86 | 0.24 ± 0.03 | ||
| D45A | 2.08 ± 0.54 | 0.10 ± 0.01 | ||
a Experiment performed in lower ionic strength buffer. See text and “Experimental Procedures” for details.
The Zn2 phosphate backbone grip mutant R138A showed little evidence of interaction with DNA, with a KD value greater than 8 μm (the highest protein concentration tested). The Zn2 phosphate backbone grip mutant R122A binds to DNA with a KD of 2.6 μm, roughly 80-fold higher than that of WT Zn2 domain with a KD of 0.03 μm (Table 2). The Zn2 base stacking loop mutants L151A and I154A bind with a KD of 0.12 and 0.83 μm, respectively. The combined mutation of these residues, L151A/I154A, effectively abolished DNA binding activity (KD > 8 μm).
The results for the Zn2 domain mutagenesis suggested that the phosphate backbone grip makes a larger contribution to the overall binding affinity of the zinc finger, because point mutation of residues in this region abolished DNA binding, whereas point mutation of base stacking loop residues tended to only weaken the DNA binding activity (Table 2). To gain this type of insight into the Zn1 interaction with DNA, it was necessary to perform another set of DNA binding experiments with a lower ionic strength buffer that increases the overall Zn1 DNA binding affinity. These experiments allowed us to obtain informative measurements of the relative contribution of the phosphate backbone grip and the base stacking loop. Under the lower ionic strength conditions, the WT Zn1 domain binds to DNA duplex with a KD of 0.19 μm (Table 2). The Zn1 phosphate backbone grip mutants, R18A and R34A, show very little evidence of DNA binding (KD values greater than 30 μm). The Zn1 base stacking loop mutants, F44A, V48A, and double mutant, F44A/V48A, exhibit weakened DNA binding affinity, with KD values of 1.0, 2.1, and 5.2 μm, respectively. Thus, the phosphate backbone grip appears to make a larger contribution than the base stacking loop to the overall DNA binding affinity of the Zn1 domain, consistent with the mutagenesis results obtained for the Zn2 domain.
Collectively, the DNA binding affinity measurements confirm the importance of both the phosphate backbone grip and the base stacking loop to DNA binding affinity as suggested by the crystal structures. Furthermore, the results indicate that the phosphate backbone grip makes a larger contribution to Zn1 and Zn2 interaction with DNA, although the hydrophobic residues of the base stacking loop also make contributions to the overall binding affinity.
Mutagenesis Screen Identifies Key Residues That Underlie Zn1 Specificity Toward DNA-dependent Activation of PARP-1
We were interested in understanding the structural basis for the specificity of the Zn1 domain in regulating PARP-1 catalytic activity (Fig. 1B). Therefore, we aligned the Zn1-DNA and Zn2-DNA structures (Fig. 3A) and generated a structure-based sequence alignment (Fig. 3B). The alignment revealed prominent structural differences that might underlie the specificity of Zn1 function. Two variable regions emerged from this analysis. The first variable region corresponds to the base stacking loop. Despite having a conserved mode of hydrophobic interaction with exposed DNA bases, the base stacking loops of Zn1 and Zn2 have distinct overall conformations and different amino acid compositions (Fig. 3, A and B, variable region 1). In particular, the Zn2 base stacking loop contains a 3-residue insertion relative to the Zn1 base stacking loop.
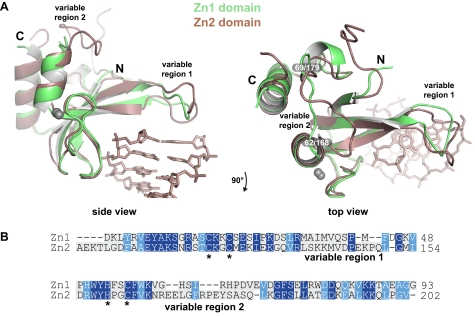
FIGURE 3.
Alignment of the Zn1 and Zn2 domain structures in complex with DNA. A, two views of an alignment of the Zn1 and Zn2 domains, highlighting two structurally distinct regions, labeled variable region 1 (base stacking loop) and variable region 2. Only the DNA duplex from the Zn1-DNA structure is shown for clarity. B, structure-based amino acid sequence alignment of human PARP-1 Zn1 and Zn2 domains. Conserved residues are shaded blue; zinc-coordinating residues are marked with stars.
The second variable region spans residues 62 to 69 of the Zn1 domain and residues 168 to 179 of the Zn2 domain, and exhibits a substantially different conformation in the Zn1-DNA and Zn2-DNA structures (Fig. 3, A and B, variable region 2). The residues in the second variable region are not located near the DNA binding interface and are therefore unlikely to make direct contributions to DNA binding activity. Variable region 2 is poorly conserved among Zn1 domains in PARP-1 proteins from multiple species, but is well conserved among Zn2 domains (not shown). Therefore, specific Zn2 functions might be mediated through this specialized region of the structure. We hypothesized that variable region 1, or the base stacking loop, is the source of Zn1 domain specificity toward DNA-dependent PARP-1 activation due to its distinct structure compared with the Zn2 domain, its high degree of conservation, and its location at an interface with the DNA.
To test whether the Zn1 base stacking loop is involved in performing the Zn1-specific function of DNA-dependent PARP-1 activation, we measured the DNA-dependent automodification activity of a series of Zn1 point mutants that targeted this region. The isolated Zn1 domain can function to stimulate the DNA-dependent activity of the ΔZn1 fragment of PARP-1 (Fig. 1C); therefore the Zn1 mutations were first generated in the isolated Zn1 domain so that we could rapidly assay for functional defects in this complementation assay. Importantly, creating the mutants in the isolated Zn1 domain also allowed us to directly measure the DNA binding activity of deficient mutants, and to assess the overall structure of the mutants using CD analysis. Interesting mutations identified in this initial screen were further tested in the context of full-length PARP-1.
Eight mutations affecting seven amino acid positions were introduced in the base stacking loop of the Zn1 domain and tested for their ability to stimulate DNA-dependent PARP-1 activity (Fig. 4, A and B). Mutations S41A, P42G, and M43A had no apparent deficiencies in their ability to stimulate PARP-1 automodification, and were therefore not investigated further (Fig. 4B). In contrast, mutations D45A, F44A, V48A, and F44A/V48A each exhibited significant deficiency in the ability to support DNA-dependent PARP-1 automodification. The mutation Q40A was also deficient in stimulating PARP-1 automodification, although to a lesser extent, mostly observed at the 5-min time point of the reaction (Fig. 4B).
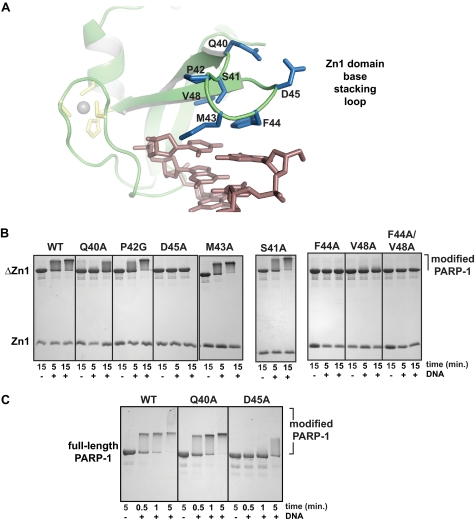
FIGURE 4.
Specific residues of the Zn1 base stacking loop are required for DNA-dependent PARP-1 activation. A, detailed view of the Zn1 base stacking loop. B, DNA-dependent automodification assay using a combination of WT or mutant Zn1 domain, the ΔZn1 fragment of PARP-1, 5 mm NAD+, and the absence or presence of 1 μm DNA. Time points were analyzed on 15% SDS-PAGE with Coomassie staining. C, DNA-dependent automodification activity of full-length PARP-1 bearing mutations in the Zn1 domain. Time points were analyzed on 12% SDS-PAGE with Coomassie staining.
We envisioned that there could be two types of base stacking loop mutations that would affect Zn1 function: one that affects the DNA binding ability and prevents/perturbs association with DNA, and one that does not affect DNA binding but rather affects the ability of Zn1 to function with the other essential domains of PARP-1. As shown previously, Zn1 mutants F44A, V48A, and F44A/V48A were deficient in DNA binding activity (Table 2); therefore the inability to support DNA-dependent PARP-1 activation could be a consequence of reduced interaction with DNA. In contrast to these mutations, the Q40A and D45A mutations were not deficient in DNA binding activity (Table 2). Indeed, these residues do not directly face the DNA duplex, and were therefore not expected to make a substantial contribution to DNA binding affinity. Thus, we anticipated that the deficiency of the Q40A and D45A mutants in stimulating PARP-1 activity was related to their inability to function with the other PARP-1 domains that are necessary for DNA-dependent activity. Importantly, these residues are well conserved in PARP-1 proteins across multiple species.
We therefore further evaluated the DNA-dependent automodification activity of the Q40A and D45A mutants in the context of full-length PARP-1 (Fig. 4C). The full-length D45A mutant was deficient in automodification compared with WT PARP-1, with only a small amount of activity visible after 5 min of incubation. This result is consistent with the deficiency observed in the in trans complementation assay. The full-length Q40A mutant had only a minor affect on DNA-dependent automodification activity (Fig. 4C), with the 5-min time point showing the only notable deficiency in production of PAR-shifted PARP-1. The Q40A mutation had a more potent affect on PARP-1 activity in the complementation assay. This is likely due to the fact that the complementation assay is a stringent test of Zn1 function, in that it requires the Zn1 domain to bind to DNA independently, and to interact with other PARP-1 domains, without the benefit of being connected to the rest of the PARP-1 polypeptide. Separating the Zn1 domain from the PARP-1 polypeptide in the complementation assay weakens Zn1 domain contacts with other PARP-1 domains. Thus, the subtle affect of the Q40A in the context of full-length PARP-1 activity is amplified in the more stringent context of the complementation assay.
Both Asp-45 and Gln-40 are directed away from the base stacking loop interface with DNA, and thus are accessible for potential interaction with other essential domains of PARP-1. The D45A mutation is clearly deficient in both the complementation assay and the context of full-length PARP-1; therefore we expect that residue Asp-45 will bear a more substantial role in functioning with other domains of PARP-1. Notably, Zn1 residues Gln-40 and Asp-45 are well conserved and are located in the region of the base stacking loop that is structurally distinct from the Zn2 domain (see Fig. 3A), providing a molecular basis for the specificity of the Zn1 domain in regulating PARP-1 activity.
Structural Transitions Occur in the Base Stacking Loop Upon Binding to DNA
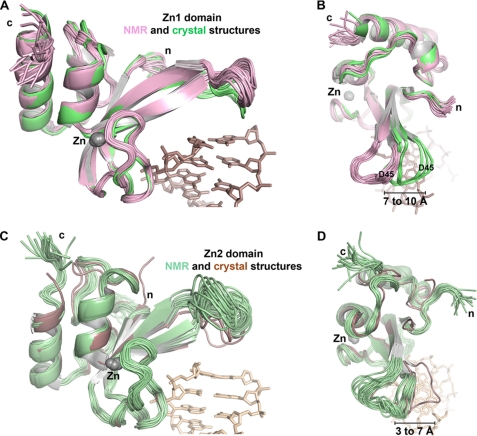
We expected that the Zn1 interaction with DNA might induce a structural change that would thereby lead to PARP-1 activation. Therefore, we compared the structures of the Zn1 domain in complex with DNA to the Zn1 domain solution structure determined by NMR in the absence of DNA (there is no publication describing this structure; it is deposited in the Protein Data Bank with accession code 2dmj). Although the overall fold of the Zn1 domain is similar in the absence or presence of DNA, there is a prominent shift in the position of the base stacking loop that is manifest when the NMR and x-ray structures are aligned (Fig. 5, A and B). There does not appear to be a substantial rearrangement of the conformation of the base stacking loop upon binding DNA. Rather, re-positioning of the base stacking loop is an overall shift of the loop region. Notably, the position of residue Asp-45 of the Zn1 domain moves 7 to 10 Å upon binding to DNA, when each of the deposited NMR models and the eight independent Zn1 domain structures are considered (Fig. 5, A and B). A similar analysis of the Zn2 domain considering all deposited NMR models (PDB accession code 2cs2) and the two independent Zn2 molecules demonstrates a 3 to 7 Å shift in the base stacking loop (Fig. 5, C and D), indicating that re-positioning of the zinc finger structure on DNA is a feature that is conserved between the two zinc finger domains. However, the Zn2 domain does not stimulate DNA-dependent PARP-1 activity because the overall structure of its base stacking loop is quite different from that of the Zn1 domain, and it therefore lacks specific features present in the Zn1 base stacking loop, for example, residue Asp-45. Consistent with the observations of this structural analysis, a DNA titration NMR experiment using the PARP-like zinc finger of human DNA ligase III showed substantial chemical shift perturbations for residues located in the region analogous to the base stacking loop (29).
FIGURE 5.
The zinc finger base stacking loop is repositioned upon binding to DNA. A, the 20 deposited NMR models of the Zn1 domain in the absence of DNA (PDB code 2dmj; pink) aligned with the 8 Zn1 domains present in the Zn1-DNA complex x-ray structure (green). B, down view of the alignment in panel A. C, the 20 deposited NMR models of the Zn2 domain in the absence of DNA (PDB 2cs2; pale green) aligned with the 2 Zn2 domains in the Zn2-DNA complex x-ray structure (brown). D, down view of the alignment in panel C.
Collectively, our biochemical assays and structural analysis support a model in which re-positioning of the base stacking loop of the Zn1 domain, through hydrophobic interaction with exposed nucleotide bases, places specific residues (i.e. Asp-45) in the optimal location to initiate DNA-dependent activation of PARP-1, most likely by forming important contacts with the other essential domains of PARP-1.
DISCUSSION
PARP-like zinc fingers are distinct from other DNA binding zinc fingers in that they recognize DNA structure rather than a specific DNA sequence (3). The Zn1-DNA and Zn2-DNA crystal structures provide the first views of PARP-like zinc fingers bound to DNA, revealing a bipartite mode of DNA interaction that contacts sequence-independent features of the DNA structure: the sugar-phosphate backbone and exposed nucleotide bases. PARP-1 binds to DNA structures containing damage such as single and double strand breaks (17, 18); yet, it is interesting to note that the Zn1 and Zn2 domains do not contact the DNA at 3′ or 5′ terminus (i.e. the breaks in the DNA strands). Rather, the Zn1 and Zn2 domains bind to exposed nucleotide bases that are indeed present in DNA structures containing breaks, but would also be present in the undamaged, abnormal DNA structures that PARP-1 binds, such as hairpin and cruciform DNA (19). Thus, the Zn1-DNA and Zn2-DNA structures provide insights into how PARP-1 zinc fingers could interact with a variety of DNA structures. We envision the phosphate backbone grip as a rigid component of the DNA interaction that will engage uninterrupted 3-nucleotide segments of DNA backbone in a consistent manner when engaging all types of DNA structure. The base stacking loop will likely serve as a flexible component of the DNA interaction that will allow the zinc fingers to adapt to variability in the DNA structures, with a common element being the interaction between hydrophobic protein side chains and exposed DNA bases. For example, we anticipate that PARP-1 zinc fingers will bind to a DNA hairpin using the backbone grip to engage the duplex region of the hairpin, and the base stacking loop will adapt to the arrangement of the DNA bases that are exposed in the hairpin portion of the DNA structure.
Collectively, our biochemical data and structural analyses have identified the base stacking loop of the Zn1 domain as a critical and specific factor that regulates DNA-dependent PARP-1 automodification activity. We propose that the base stacking loop interaction with DNA positions specific Zn1 residues (e.g. Asp-45 and Gln-40) that will form key contacts with other essential domains of PARP-1, and these DNA-induced contacts contribute to PARP-1 DNA-dependent activity. Importantly, the Zn1 residues that are important for mediating PARP-1 activation are not conserved in the Zn2 domain, providing a molecular basis for Zn1 specificity in regulating PARP-1 activity. Consistent with our model, previous biochemical analysis of PARP-1 DNA-dependent activity has demonstrated that the composition of duplex DNA (e.g. blunt ends versus 3′ or 5′ overhangs) influences the level of PARP-1 activation (17, 30, 37). We expect that these variations in DNA structure influence the positioning of the base stacking loop on DNA, and thus affect Zn1 ability to efficiently form interdomain contacts that support DNA-dependent PARP-1 activity. We envision that interdomain contacts will ultimately impose structural changes in the PARP-1 catalytic domain that increase enzymatic activity, or will promote an arrangement of PARP-1 domain architecture that increases access to substrates.
Our study indicates that the Zn1 and Zn2 domains have distinct biochemical properties, and they can work independently of each other to carry out at least some of PARP-1 functions. The Zn1 domain is essential for PARP-1 DNA-dependent activity, whereas the Zn2 domain is dispensable both in vitro and in a cellular context with regards to DNA damage-dependent activation of PARP-1 (Fig. 1, B and E). Our DNA binding experiments indicate that the Zn2 domain alone has substantial DNA binding affinity on its own, 100-fold higher than that of the Zn1 domain alone. The robust DNA binding affinity of the Zn2 domain could be important for the rapid localization of PARP-1 to sites of DNA damage, or the persistence of PARP-1 at damage sites (5), even though the Zn2 domain is not strictly required for PAR synthesis activity at sites of damage.
The relatively weak DNA binding affinity of the Zn1 domain could be an important feature for regulation of PARP-1 DNA-dependent activity. A proteomic analysis of PARP-1 phosphorylation sites identified Ser-41 of the Zn1 domain as a site of modification, and the phosphomimic S41E decreased PARP-1 recruitment and persistence at a microirradiated region of the nucleus containing DNA damage (39), presumably by disrupting Zn1 interaction with DNA. Ser-41 is located on the base stacking loop adjacent to the exposed DNA bases, and therefore should not be readily accessible to kinase activity when the Zn1 domain is bound to DNA. The weak DNA binding affinity of the Zn1 domain would allow this surface to be accessible for phosphorylation a portion of the time, supporting modification at this site as a mode of regulating PARP-1 function (39).
Our analysis of Zn1 and Zn2 specific functions indicates that the Zn2 domain does not play a pivotal role in DNA-dependent activation of PARP-1. PARP-1 has important functions outside of the DNA damage response, contributing to transcription as both a general and specific regulator of gene expression (40). DNA-dependent PAR synthesis activity is not required for all of PARP-1 transcriptional activities (40). We speculate that the Zn2 domain could serve specific roles in regards to PARP-1 function(s) in transcription. In this regard, the higher DNA binding affinity of the Zn2 domain might play a more important role. There is a precedent for distinct functions of PARP-like zinc fingers in the Drosophila melanogaster homolog of PARP-1 (dmPARP-1) (41). A mutant of dmPARP-1 that carries only a single zinc finger is capable of producing PAR in vivo and is therefore catalytically active. However, this mutant of dmPARP-1 is excluded from regions of heterochromatin, in contrast to full-length dmPARP-1 that contains two zinc fingers and is distributed to heterochromatic regions of the nucleus (41). Individual mutation of the Zn1 and Zn2 domains of human PARP-1 might reveal specific functions for the zinc fingers in regulating PARP-1 transcriptional activities or interaction with chromatin.
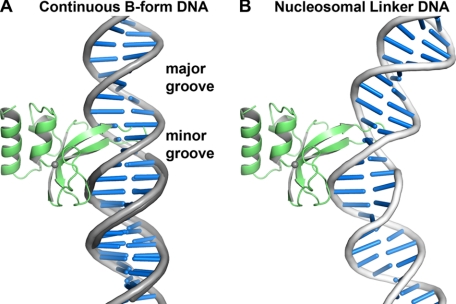
PARP-1 binds to chromatin and influences chromatin structure (23, 42, 43), and thus functions as a DNA architectural protein. Interestingly, the manner in which PARP-1 zinc fingers engage DNA bases using hydrophobic protein side chains is reminiscent of other DNA architectural proteins that bind to distorted DNA structures, or induce DNA distortions upon binding, such as high-mobility group protein HMG1 and TATA-binding protein (44). Although the Zn1 and Zn2 domains are structurally distinct compared with both of these DNA architectural proteins, there are likely to be common elements to how the PARP-1 zinc fingers will engage exposed DNA bases in distorted DNA structures. A critical distinction between the Zn1 and Zn2 domains and DNA architectural proteins with known structures is that the hydrophobic residues of the Zn1 and Zn2 domains insert into the major groove of the DNA, rather than the minor groove. This is best visualized by aligning a continuous B-form DNA helix to the duplex DNA contained in the Zn1-DNA complex (Fig. 6A). Due to the positioning of the base stacking loop in the major groove of the DNA, we expect that both the Zn1 and Zn2 domains will bind to DNA distortions that expose nucleotide bases in the major groove, and therefore bend DNA toward the minor groove. Interestingly, the structure of a tetramer of nucleosomes demonstrates this type of major groove distortion in the linker DNA connecting nucleosomes (45), where PARP-1 is known to bind (43). Rigid body positioning of a PARP-1 zinc finger on the nucleosomal linker DNA suggests that this type of major groove distortion can better accommodate the base stacking loop in the major groove (Fig. 6B). This hypothetical model provides insight into how PARP-1 zinc fingers are capable of interacting with continuous, undamaged DNA structures.
FIGURE 6.
The zinc finger base stacking loop inserts into the major groove of continuous DNA structures. A, continuous B-form DNA (light gray) aligned to the DNA duplex in the Zn1-DNA crystal structure (dark gray) highlights that the base stacking loop inserts into the DNA major groove. DNA distortions will be required to fully accommodate the base stacking loop. B, the Zn1 domain modeled on a segment of bent nucleosomal linker DNA illustrates the type of DNA distortion that is anticipated to interact with PARP-1 zinc fingers. DNA bending toward the minor groove better accommodates the base stacking loop compared with B-form DNA (DNA extracted from PDB coordinate 1zbb (45)).
In summary, the crystal structures and functional analysis of PARP-1 zinc fingers have defined the mode of binding to DNA, and thereby advanced our understanding of a principal PARP-1 regulation mechanism: interaction with DNA damage. Furthermore, our analysis has provided new insights into the activities of PARP-like zinc fingers, and their specific roles in regulating PARP-1 functions.
Acknowledgments
We thank S. Classen at the ALS SIBYLS beamline and NSLS beamline staff for assistance with data collection, and Y.-M. Hou and J. L. Benovic for insightful comments on the manuscript. Preliminary x-ray diffraction and CD analyses were performed in the Kimmel Cancer Center X-ray Crystallography and Molecular Characterization Shared Resource at Thomas Jefferson University.
This work was supported by American Cancer Society Grants RSG0918301DMC and IRG0806001, The Emerald Foundation, Inc., and The Ladies of Port Richmond.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Figs. S1–S6, and Table 1.
The atomic coordinates and structure factors (codes 3OD8, 3ODC, 3ODA, and 3ODE) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- PAR
- poly(ADP-ribose)
- PARP
- poly(ADP-ribose) polymerase
- SAD
- single-wavelength anomalous dispersion.
REFERENCES
- 1. Hassa P. O., Hottiger M. O. (2008) Front. Biosci. 13, 3046–3082 [DOI] [PubMed] [Google Scholar]
- 2. Krishnakumar R., Kraus W. L. (2010) Mol. Cell 39, 8–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. D'Amours D., Desnoyers S., D'Silva I., Poirier G. G. (1999) Biochem. J. 342, 249–268 [PMC free article] [PubMed] [Google Scholar]
- 4. Rouleau M., Patel A., Hendzel M. J., Kaufmann S. H., Poirier G. G. (2010) Nat. Rev. Cancer 10, 293–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haince J. F., McDonald D., Rodrigue A., Déry U., Masson J. Y., Hendzel M. J., Poirier G. G. (2008) J. Biol. Chem. 283, 1197–1208 [DOI] [PubMed] [Google Scholar]
- 6. Gagné J. P., Isabelle M., Lo K. S., Bourassa S., Hendzel M. J., Dawson V. L., Dawson T. M., Poirier G. G. (2008) Nucleic Acids Res. 36, 6959–6976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gottschalk A. J., Timinszky G., Kong S. E., Jin J., Cai Y., Swanson S. K., Washburn M. P., Florens L., Ladurner A. G., Conaway J. W., Conaway R. C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 13770–13774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Timinszky G., Till S., Hassa P. O., Hothorn M., Kustatscher G., Nijmeijer B., Colombelli J., Altmeyer M., Stelzer E. H., Scheffzek K., Hottiger M. O., Ladurner A. G. (2009) Nat. Struct. Mol. Biol. 16, 923–929 [DOI] [PubMed] [Google Scholar]
- 9. Ahel D., Horejsí Z., Wiechens N., Polo S. E., Garcia-Wilson E., Ahel I., Flynn H., Skehel M., West S. C., Jackson S. P., Owen-Hughes T., Boulton S. J. (2009) Science 325, 1240–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kraus W. L. (2009) Nat. Struct. Mol. Biol. 16, 904–907 [DOI] [PubMed] [Google Scholar]
- 11. Andrabi S. A., Kim N. S., Yu S. W., Wang H., Koh D. W., Sasaki M., Klaus J. A., Otsuka T., Zhang Z., Koehler R. C., Hurn P. D., Poirier G. G., Dawson V. L., Dawson T. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18308–18313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. David K. K., Andrabi S. A., Dawson T. M., Dawson V. L. (2009) Front. Biosci. 14, 1116–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mackey Z. B., Niedergang C., Murcia J. M., Leppard J., Au K., Chen J., de Murcia G., Tomkinson A. E. (1999) J. Biol. Chem. 274, 21679–21687 [DOI] [PubMed] [Google Scholar]
- 14. Taylor R. M., Whitehouse C. J., Caldecott K. W. (2000) Nucleic Acids Res. 28, 3558–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Petrucco S., Volpi G., Bolchi A., Rivetti C., Ottonello S. (2002) J. Biol. Chem. 277, 23675–23683 [DOI] [PubMed] [Google Scholar]
- 16. Petrucco S. (2003) Nucleic Acids Res. 31, 6689–6699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. D'Silva I., Pelletier J. D., Lagueux J., D'Amours D., Chaudhry M. A., Weinfeld M., Lees-Miller S. P., Poirier G. G. (1999) Biochim. Biophys. Acta 1430, 119–126 [DOI] [PubMed] [Google Scholar]
- 18. Pion E., Bombarda E., Stiegler P., Ullmann G. M., Mély Y., de Murcia G., Gérard D. (2003) Biochemistry 42, 12409–12417 [DOI] [PubMed] [Google Scholar]
- 19. Lonskaya I., Potaman V. N., Shlyakhtenko L. S., Oussatcheva E. A., Lyubchenko Y. L., Soldatenkov V. A. (2005) J. Biol. Chem. 280, 17076–17083 [DOI] [PubMed] [Google Scholar]
- 20. Ikejima M., Noguchi S., Yamashita R., Ogura T., Sugimura T., Gill D. M., Miwa M. (1990) J. Biol. Chem. 265, 21907–21913 [PubMed] [Google Scholar]
- 21. Altmeyer M., Messner S., Hassa P. O., Fey M., Hottiger M. O. (2009) Nucleic Acids Res. 37, 3723–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gradwohl G., Ménissier de Murcia J. M., Molinete M., Simonin F., Koken M., Hoeijmakers J. H., de Murcia G. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 2990–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wacker D. A., Ruhl D. D., Balagamwala E. H., Hope K. M., Zhang T., Kraus W. L. (2007) Mol. Cell. Biol. 27, 7475–7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Langelier M. F., Servent K. M., Rogers E. E., Pascal J. M. (2008) J. Biol. Chem. 283, 4105–4114 [DOI] [PubMed] [Google Scholar]
- 25. Tao Z., Gao P., Hoffman D. W., Liu H. W. (2008) Biochemistry 47, 5804–5813 [DOI] [PubMed] [Google Scholar]
- 26. Langelier M. F., Ruhl D. D., Planck J. L., Kraus W. L., Pascal J. M. (2010) J. Biol. Chem. 285, 18877–18887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. El-Khamisy S. F., Masutani M., Suzuki H., Caldecott K. W. (2003) Nucleic Acids Res. 31, 5526–5533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Masson M., Niedergang C., Schreiber V., Muller S., Menissier-de Murcia J., de Murcia G. (1998) Mol. Cell. Biol. 18, 3563–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kulczyk A. W., Yang J. C., Neuhaus D. (2004) J. Mol. Biol. 341, 723–738 [DOI] [PubMed] [Google Scholar]
- 30. Benjamin R. C., Gill D. M. (1980) J. Biol. Chem. 255, 10493–10501 [PubMed] [Google Scholar]
- 31. Van Duyne G. D., Standaert R. F., Karplus P. A., Schreiber S. L., Clardy J. (1993) J. Mol. Biol. 229, 105–124 [DOI] [PubMed] [Google Scholar]
- 32. Wang Z. Q., Auer B., Stingl L., Berghammer H., Haidacher D., Schweiger M., Wagner E. F. (1995) Genes Dev. 9, 509–520 [DOI] [PubMed] [Google Scholar]
- 33. Otwinowski Z., Minor W. (1997) in Methods in Enzymology (Carter C. W., Jr, Sweet R. M. eds) pp. 307–326, Academic Press, New York: [DOI] [PubMed] [Google Scholar]
- 34. Collaborative Computational Project, N. (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 35. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 37. Pion E., Ullmann G. M., Amé J. C., Gérard D., de Murcia G., Bombarda E. (2005) Biochemistry 44, 14670–14681 [DOI] [PubMed] [Google Scholar]
- 38. Lilyestrom W., van der Woerd M. J., Clark N., Luger K. (2010) J. Mol. Biol. 395, 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gagné J. P., Moreel X., Gagné P., Labelle Y., Droit A., Chevalier-Paré M., Bourassa S., McDonald D., Hendzel M. J., Prigent C., Poirier G. G. (2009) J. Proteome Res. 8, 1014–1029 [DOI] [PubMed] [Google Scholar]
- 40. Kraus W. L. (2008) Curr. Opin. Cell Biol. 20, 294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kotova E., Jarnik M., Tulin A. V. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 6406–6411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Poirier G. G., de Murcia G., Jongstra-Bilen J., Niedergang C., Mandel P. (1982) Proc. Natl. Acad. Sci. U.S.A. 79, 3423–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim M. Y., Mauro S., Gévry N., Lis J. T., Kraus W. L. (2004) Cell 119, 803–814 [DOI] [PubMed] [Google Scholar]
- 44. Bewley C. A., Gronenborn A. M., Clore G. M. (1998) Annu. Rev. Biophys. Biomol. Struct. 27, 105–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schalch T., Duda S., Sargent D. F., Richmond T. J. (2005) Nature 436, 138–141 [DOI] [PubMed] [Google Scholar]