Abstract
The utilization of phenylacetic acid (PA) in Escherichia coli occurs through a hybrid pathway that shows features of both aerobic and anaerobic metabolism. Oxygenation of the aromatic ring is performed by a multisubunit phenylacetyl-coenzyme A oxygenase complex that shares remote homology of two subunits to well studied bacterial multicomponent monooxygenases and was postulated to form a new bacterial multicomponent monooxygenase subfamily. We expressed the subunits PaaA, B, C, D, and E of the PA-CoA oxygenase and showed that PaaABC, PaaAC, and PaaBC form stable subcomplexes that can be purified. In vitro reconstitution of the oxygenase subunits showed that each of the PaaA, B, C, and E subunits are necessary for catalysis, whereas PaaD is not essential. We have determined the crystal structure of the PaaAC complex in a ligand-free form and with several CoA derivatives. We conclude that PaaAC forms a catalytic core with a monooxygenase fold with PaaA being the catalytic α subunit and PaaC, the structural β subunit. PaaAC forms heterotetramers that are organized very differently from other known multisubunit monooxygenases and lacks their conservative network of hydrogen bonds between the di-iron center and protein surface, suggesting different association with the reductase and different mechanisms of electron transport. The PaaA structure shows adaptation of the common access route to the active site for binding a CoA-bound substrate. The enzyme-substrate complex shows the orientation of the aromatic ring, which is poised for oxygenation at the ortho-position, in accordance with the expected chemistry. The PA-CoA oxygenase complex serves as a paradigm for the new subfamily multicomponent monooxygenases comprising several hundred homologs.
Keywords: Bacterial Metabolism, Crystal Structure, Enzyme Catalysis, Iron, Oxidation-Reduction, Protein Assembly, Degradation of Aromatic Compounds, Monooxygenase, Protein-Protein Interactions
Introduction
Aromatic organic compounds represent a major class of environmental pollutants (1). Many microbes can grow using these molecules as an abundant source of nutrients and a carbon source. Under anaerobic conditions, the metabolism of aromatic compounds proceeds through initial conversion to a CoA derivative, which is then reduced in an ATP-dependent reaction (2). In the presence of oxygen, bacteria utilize a wide variety of oxygenases to activate the inert aromatic ring (3). In aerobic pathways that utilize multisubunit dioxygenases or non-heme monooxygenases, both the hydroxylation of the aromatic ring and its cleavage are oxygen-dependent (1). Dioxygenases incorporate two adjacent hydroxyl groups into the aromatic ring (4). For metabolism that relies on monooxygenases, the consecutive action of two such enzymes is required, with insertion of one hydroxyl group at each step, as is the case in Pseudomonas stutzeri OX1 with toluene/o-xylene monooxygenase and phenol hydroxylase performing these reactions (5).
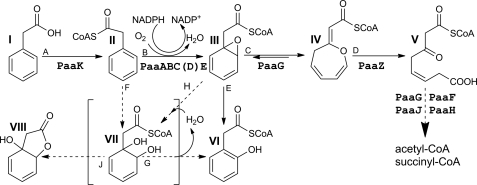
An aerobic hybrid pathway combines the features of classic aerobic and anaerobic strategies (6). As in anaerobic metabolism, the aromatic compound is attached to CoA, but the substrate is oxygenated, whereas the ring opening proceeds without oxygen. This pathway is present in many bacteria and frequently serves as a core component of an extensive metabolic network involved in degradation of 2-phenylethylamine, styrene, phenylacetaldehyde, phenylacetyl amides, n-phenylalkanoic acids with even numbers of carbon atoms, and phenylacetyl esters (7). Escherichia coli has only a subset of these enzymes that are required for the utilization of phenylacetic acid (PA)2 and of 2-phenylethylamine, encoded by the paa operon (6) (see Fig. 1). The enzymatic steps along the PA degradation pathway and enzymes associated with each step have been unequivocally established only recently (8).
FIGURE 1.
Aerobic hybrid phenylacetate catabolic pathway as updated recently (8). The product of the PA-CoA monooxygenase is the 1,2-epoxide derivative of PA-CoA (compound III). The previously proposed dihydrodiol derivative of PA-CoA (compound VII) (35) is shown in parentheses.
The key enzyme of the PA-CoA metabolic pathway is a ring oxygenase that binds the aromatic CoA thioester and performs a strictly aerobic oxygenation reaction. Phylogenetic analysis suggests that the enzyme belongs to the family of bacterial multicomponent monooxygenases (BMMs) and forms the prototype of a new branch (9). BMMs were previously divided into six major groups (10) and include well studied enzymes such as methane monooxygenase (MMO) (11, 12), phenol hydroxylase (5, 13), toluene/o-xylene monooxygenase (ToMO) (14, 15), and toluene 4-monooxygenase (16, 17).
The PA-CoA oxygenase complex was proposed to contain five subunits, PaaA, B, C, D, and E (9). The PaaA subunit is distantly related to the catalytic (α) subunit of other well characterized BMMs, sharing with them only 10–15% sequence identity, with the iron-binding motifs being the easiest to recognize. Of the other subunits, PaaC shows sequence similarity to PaaA, being analogous to the structural (β) subunit of BMMs. PaaB and PaaD have no detectable sequence similarity to subunits of other BMMs. The PaaACD complex was proposed to be analogous to αβγ hydroxylase complex of BMMs, whereas PaaB was suggested to be a regulatory subunit controlling hydroxylase-reductase interactions (9). Based on amino acid sequence, PaaE was postulated to be a class IA reductase (classification of reductases according to Ref. 18). Interestingly, this is the first example of a monooxygenase complex containing a class IA reductase. Usually, the monooxygenase-associated reductases belong to either class IB (e.g. MMO) or class III (e.g. ToMO). Reductases class IA and IB differ in domain order and in cofactor specificity.
We have investigated protein-protein interactions between the putative components of the E. coli PA-CoA monooxygenase complex and identified several stable subcomplexes in vitro. By reconstituting the monooxygenase from its components, we show that PaaA, B, C, and E are necessary for activity, whereas PaaD, which was deemed to be important for in vivo function (9), had no effect on in vitro activity. We have determined the crystal structure of the PaaAC subcomplex with and without bound PA-CoA substrate. This structure shows that PaaA is the catalytic subunit, whereas PaaC is the structural subunit. The PaaAC complex serves as the prototype for a new class of BMMs and provides the first structural example of a BMM oxygenase-substrate complex. Finally, we showed experimentally that PaaE is a reductase.
EXPERIMENTAL PROCEDURES
The detailed procedures of cloning and heterologous expression of genes, protein purification and characterization, reaction measurement, crystallization, and structure determination are described in the supplemental materials. Here we provide an abbreviated description.
Cloning, Expression, and Purification
Individual genes encoding PaaA, B, C, D, and E from E. coli K-12 were cloned into a modified pGEX-4T1 (N-GST tag) (GE Healthcare), pET15b (N-His tag), pRDSFDuet-1 MCS2 (no tag), and pCDFDuet-1 MCS2 (no tag) vectors (Novagen). The paaE gene was also cloned into pMAL-c2x (N-MBP tag) vector (New England Biolabs). Additionally the paaA-E segment of the entire paa operon was cloned into the modified pET15b. E. coli expression strains BL21(DE3) or BL21 (Novagen) were used for protein production.
The purification of individual proteins and the coexpressed complexes was done as described previously (19). Briefly, the cells were lysed by sonication, cell debris was removed by centrifugation, and the supernatant was loaded on Ni-NTA (Qiagen). The column was washed with buffer containing 50 mm HEPES, pH 7.5, 1 m NaCl, and 40 mm imidazole. The final purification step consisted of gel filtration chromatography on Superose 12 column (GE Healthcare). The purification of PaaB-His6C was accomplished following the same protocol, but with the omission of high salt wash. The growth of cells expressing PaaA-His6C in iron-enriched medium (20) or the addition of 0.1 mm ammonium ferrous sulfate to purified PaaA-His6C resulted in aggregation of the protein.
GST-PaaB and GST-PaaD were purified by affinity chromatography using glutathione-Sepharose 4B (GE Healthcare) with the subsequent cleavage from the beads by TEV protease, followed by size exclusion chromatography on a Superose 12. The expression of MBP-PaaE was done in iron-enriched medium. The protein was purified using amylose resin (New England Biolabs) and used without further purification.
Protein-Protein Interaction Assays
Protein-protein interactions were evaluated using coexpression of different combinations of subunits with only one of the subunits having a His tag. The extracted proteins were loaded on a Ni-NTA-agarose column, and the proteins retained on the column after washing were analyzed by SDS-PAGE (Table 1). The composition of buffers was varied from pH 6.5 to 8.5 with and without 0.4 m NaCl to study the effects of pH and ionic strength on complex formation. In addition, interactions of purified GST-PaaD with PaaABC as well as Paa-His6ABC with PaaD and MBP-PaaE with PaaABC and PaaD were investigated by pull-down experiments using the appropriate columns (Table 2).
TABLE 1.
Identified complexes between the Paa subunits by copurification
NA, not applicable.
| Coexpression combinations | High salt purification scheme | Low salt wide pH range purification scheme |
|---|---|---|
| His6PaaC + PaaA | His6PaaC + PaaA | NA |
| His6PaaC + PaaB | His6PaaC | His6PaaC + PaaB |
| His6PaaC + PaaD | His6PaaC | His6PaaC |
| His6PaaC + PaaA + PaaB | His6PaaC + PaaA + PaaB | NA |
| His6PaaC + PaaA + PaaD | His6PaaC + PaaA | His6PaaC + PaaA |
| His6PaaC + PaaA + PaaE | His6PaaC + PaaA | His6PaaC + PaaA |
| His6PaaE + PaaB + PaaD | Absence of specific protein | NA |
| His6PaaA + PaaB | Absence of specific protein | NA |
| His6PaaA + PaaD | Absence of specific protein | NA |
| His6PaaA + PaaB + PaaC | His6PaaA + PaaB + PaaC | NA |
| His6PaaA + PaaB + PaaC + PaaD | His6PaaA + PaaB + PaaC | His6PaaA + PaaB + PaaC |
| His6PaaA + PaaB + PaaC + PaaD + PaaE | His6PaaA + PaaB + PaaC | His6PaaA + PaaB + PaaC |
TABLE 2.
Pulldown assays to identify interactions of PaaD and PaaE with other subunits
| Affinity resin | Bait | Prey | Interact? |
|---|---|---|---|
| Glutathione-Sepharose | GST-PaaD | PaaC | No |
| Glutathione-Sepharose | GST-PaaD | PaaAC | No |
| Glutathione-Sepharose | GST-PaaD | PaaABC | No |
| Ni-NTA | PaaA-His6C | PaaD | No |
| Ni-NTA | Paa-His6ABC | PaaD | No |
| Ni-NTA | His6PaaB | PaaD | No |
| Ni-NTA | His6PaaC | PaaD | No |
| Amylose resin | MBP-PaaE | PaaABC + PaaD | No |
Enzyme Activity Assays
Mixtures of different Paa subunits, each at a final concentration of 2 μm, were added to reaction buffer containing 25 mm Tris, pH 7.5, 500 μm NADPH, 10 μm Fe(NH4)2(SO4)2, 10 μm FAD, and were incubated for 15 min at 37 °C. The reaction was initiated by the addition of phenylacetyl-CoA to the final concentration of 300 μm. After 20 h, the reaction was stopped by the addition of HCl. The appearance of 2-hydroxy-phenylacetate (2-OH-PA) was quantitatively monitored by LC-MS/MS on an Agilent 1200 HPLC system coupled to a Agilent QQQ6410 mass spectrometer (Agilent Technologies, Inc., Palo Alto, CA). Separations were performed on a SymmetryShield RP18 column (Waters).
Crystallization
Crystallization of PaaA-His6PaaC was described previously (19). All of the crystals were obtained using hanging drop vapor diffusion with protein concentration of 8 mg/ml. Two conditions were optimized for complexes of PaaAC with CoA, 3-hydroxybutyril-CoA, benzoyl-CoA, and phenylacetyl-CoA. For the first condition, the reservoir solution contained 100 mm Pipes, pH 6.5, 15% (v/v) PEG 400MME (Sigma), whereas the second condition was 100 mm Pipes, pH 6.5, 5% (v/v) PEG 400MME, 5% (v/v) isopropanol. PaaAC with acetyl-CoA was crystallized in 0.1 m sodium citrate buffer, pH 5.5, and 15% (w/v) PEG 6000 (Fluka). Crystals of ligand-free PaaAC were obtained in 100 mm ADA, pH 5.5.
X-ray Data Collection, Structure Solution, and Refinement
The crystals were cryoprotected in 25% (v/v) glycerol or 25% (v/v) 2-methyl-2,4-pentanediol. The diffraction data were collected with a Mar300CCD detector at the CMCF-1 beamline at the Canadian Light Source. Data integration and scaling was performed with HKL2000 (21). The structure of PaaA-His6C was solved by molecular replacement using the programs PHASER (22) and MOLREP (23) with the PaaC structure (Protein Data Bank code 1OTK)3 as a search model. Subsequent model building and refinement was performed using COOT (24) and REFMAC5 (25). Pertinent details are presented in Table 3. Analysis of structure using MolProbity (26) showed models of good quality.
TABLE 3.
X-ray data collection and refinement statistics
| Data set | PaaAC + CoA | PaaAC + 3-hydroxybutyryl CoA | PaaAC + benzoyl-CoA | PaaAC + phenylacetyl-CoA | PaaAC + acetyl-CoA | PaaAC | PaaAC with Fe2+ |
|---|---|---|---|---|---|---|---|
| Space group | P41212 | P41212 | P41212 | P41212 | P3221 | P212121 | P41212 |
| a, b, c (Å) | 77.5, 77.5, 300.1 | 77.4, 77.4, 301.4 | 77.6, 77.6, 300.7 | 77.6, 77.6, 300.4 | 122.8, 122.8, 153.8 | 110.3, 109.1, 305.9 | 78.4, 78.4, 309.9 |
| Wavelength (Å) | 0.97949 | 0.97949 | 0.97949 | 0.97949 | 0.97949 | 0.97949 | 0.97949 |
| Resolution (Å) | 50.0-2.15 | 50.0-2.03 | 50.0-2.06 | 50.0-2.25 | 50.0-2.97 | 50.0-2.64 | 50.0-5.8 |
| Last shell of resolution (Å) | 2.23-2.15 | 2.10-2.03 | 2.13-2.06 | 2.33-2.25 | 3.02-2.97 | 2.73-2.64 | 5.9-5.8 |
| Observed hkl | 268595 | 599496 | 339272 | 300130 | 175298 | 707383 | 19671 |
| Unique hkl | 45467 | 60180 | 58037 | 44543 | 27899 | 108855 | 2987 |
| Completeness (%)a | 88.7 (92.6) | 99.3 (94.8) | 99.5 (96.6) | 99.1 (94.4) | 99.0 (99.6) | 99.9 (99.9) | 98.4 (99.3) |
| Redundancya | 5.9 (6.0) | 10.0 (4.1) | 5.8 (4.2) | 6.7 (5.9) | 6.3 (6.3) | 6.5 (4.0) | 6.6 (7.0) |
| Rsyma | 0.055 (0.22) | 0.063 (0.382) | 0.078 (0.389) | 0.098 (0.459) | 0.15 (0.681) | 0.08 (0.547) | 0.044 (0.422) |
| I/(σI) a | 10.8 (3.8) | 11.8 (2.1) | 8.6 (2.1) | 6.5 (2.0) | 11.2 (2.0) | 10.0 (2.0) | 39.6 (3.49) |
| Rwork | 0.195 | 0.195 | 0.188 | 0.186 | 0.238 | 0.234 | |
| Rfree (% hkl) | 0.232 | 0.230 | 0.225 | 0.233 | 0.264 | 0.267 | |
| Wilson B (Å2) | 28.2 | 34.7 | 29.4 | 32.1 | 59.8 | 65.2 | |
| B-factor Å2 (# atoms) | |||||||
| Protein | 51.4 (6278) | 60.9 (6269) | 51.0 (6268) | 50.0 (6269) | 40.57 (8658) | 54.9 (24587) | |
| Solvent | 54.5 (333) | 69.5 (378) | 56.4 (457) | 49.2 (348) | NA | 43.3 (217) | |
| Ligand | 50.4 (48) | 57.8 (54) | 46.6 (56) | 39.8 (57) | 33.7 (102) | NA | |
| Ramachandran | |||||||
| Allowed (%) | 97.96 | 97.57 | 97.83 | 97.45 | 96.42 | 97.19 | |
| Generous (%) | 1.91 | 2.17 | 1.66 | 2.30 | 3.22 | 2.36 | |
| Disallowed (%) | 0.13 | 0.26 | 0.51 | 0.26 | 0.37 | 0.45 | |
| Root mean square deviation | |||||||
| Bonds (Å) | 0.009 | 0.010 | 0.01 | 0.011 | 0.011 | 0.009 | |
| Angles (°) | 1.08 | 1.15 | 1.12 | 1.20 | 1.26 | 1.06 | |
| Protein Data Bank code | 3PVY | 3PVT | 3PVR | 3PW1 | 3PW8 | 3PWQ | |
a The information for the last shell of resolution is given in parentheses.
RESULTS
Interactions between Subunits of the PA-CoA Oxygenase Complex
The five proteins encoded by paaABCDE were postulated to form a multicomponent oxygenase involved in the hydroxylation of phenylacetyl-CoA. To analyze protein-protein interactions among these five subunits, we have coexpressed and purified them in various combinations (Table 1). We were able to overexpress each subunit individually but only PaaB, C, and D were soluble, whereas PaaA and E were insoluble. However, when PaaA and PaaC were coexpressed, they formed a well behaved soluble complex. When the entire paaABCDE operon was expressed, only PaaA, B, and C but not PaaD or PaaE could be detected. Other coexpression strategies are summarized in Table 1. An attempt to improve PaaE solubility by coexpression with various combinations of other subunits was not successful. Finally, the MBP-PaaE fusion resulted in a soluble protein.
Interactions between the subunits were established by affinity purification of various combinations of coexpressed PaaA, B, C, D, and E where only one subunit carried a His6 tag. Protein association was tested at pH 6.5, 7.5, and 8.4, at low and high ionic strength (0–1 m NaCl). Under all of the tested conditions, we observed PaaA-His6C, PaaAB-His6C, and Paa-His6ABC complexes, whereas in the absence of salt, we also detected the His6PaaC-PaaB complex (Table 1). No evidence for the formation of a stable complex between PaaD or PaaE and other proteins (or their combinations) was found. Pull-down experiments using GST-PaaD and MBP-PaaE as baits and purified PaaABC or individual subunits were also negative (Table 2).
The apparent molecular mass of the PaaA-His6C complex (calculated molecular mass, 64.8 kDa) was determined to be ∼180 kDa by size exclusion chromatography (SEC) and ∼160 kDa by dynamic light scattering, consistent with either heterotetramers or heterohexamers. The molecular mass of PaaABC (molecular mass, 75.5 kDa) was ∼90–120 kDa (SEC) and 160–200 kDa (dynamic light scattering), making the prediction of stoichiometry uncertain. Finally, the molecular mass of PaaB-His6C (molecular mass, 40.0 kDa) was ∼40 kDa (SEC) and 38–43 kDa (dynamic light scattering), indicating heterodimer. Similar SEC analysis of PaaB (molecular mass, 11.0 kDa) showed that it elutes as a peak with an apparent molecular mass of 33 kDa, corresponding to trimers in solution. The PaaD (molecular mass, 18.5 kDa) elutes from the SEC column with an apparent molecular mass of 21 kDa, indicating a monomer.
Paa Components Necessary for Enzymatic Activity
Because there is a discrepancy between the in vivo (9) and in vitro (8) data as to which subunits are necessary for activity, we first reconstituted all five components, PaaABCDE, in the presence of the required cofactors and 10 μm Fe2+, and after the addition of PA-CoA, we followed the reaction by LC-MS (supplemental Fig. S1). The reaction product (III in Fig. 1) is expected to rapidly decompose to 2-OH-PA (VI in Fig. 1) (9). We observed the disappearance of the substrate and concomitant appearance of VI. We also detected PA (compound I), a byproduct of substrate hydrolysis (supplemental Fig. S1). The measured activity of the reconstituted PA oxygenase was ∼5 nmol·min−1·mg−1 (supplemental Table S1). The rather low activity of the reconstituted complex may be due to partially active PaaE, which could only be expressed in a soluble form as a MBP-tagged protein.
To determine whether all five proteins are indispensable for the oxygenation of phenylacetyl-CoA, we analyzed the reaction mixture with different subsets of PaaABCDE proteins. The maximum reaction rate was observed when PaaA, B, C, and E subunits were present (Table 4). The presence or absence of PaaD had no effect on the reaction outcome, suggesting that this component is not essential for the reaction in vitro. Absence of PaaB leads to a more than 100-fold drop in the product concentration. PaaAC and PaaABC were able to generate small amounts of the product, suggesting a single turnover utilizing the reduced Fe2+ present in the reaction mixtures.
TABLE 4.
Activity of reconstituted Paa subunits
| Paa subunits | 2-OH-PA | PA |
|---|---|---|
| μm | μm | |
| ABCDE | 86.3 | 22.7 |
| ABC[5D]Ea | 87.9 | 40.15 |
| ABCE | 80.1 | 15.1 |
| ACE | 0.34 | 112.8 |
| ACB | 1.1 | 101.9 |
| AC | 0.97 | 96.9 |
| BE | 0.13b | 56.6 |
a [5D] indicates 5-fold higher concentration of PaaD than the concentration of all other compounds.
b The value is at the edge of the detection limit, which is 0.1 μm.
To establish whether the oxygen for the hydroxylation reaction is obtained from water or from gaseous O2, we analyzed the reaction products in the presence of H2O18. No incorporation of O18 at position 2-OH of 2-OH-PA was observed by mass spectrometry, indicating that the oxygen is derived from O2 (supplemental Table S2). In the same experiment, we observed, as expected, the incorporation of O18 into the carboxyl of the 2-OH-PA and PA.
The Reductase PaaE
To identify the type of iron-sulfur cluster in PaaE, we have recorded the absorption spectrum of MBP-PaaE (supplemental Fig. S2). The spectrum with maxima at 325, 415, and 460 nm closely resembles that of spinach [2Fe-2S] ferredoxin (27).
We next investigated the cofactor preference of the PaaE, which requires nicotinamide adenine dinucleotide, NAD(P), and a riboflavin, FAD or FMN, cofactors and is expected to transfer electrons from NAD(P)H to the di-iron center of PaaA through the riboflavin cofactor and the iron-sulfur center. The measured activity was 8.5 times higher in the presence of NADPH than in the presence of NADH and 3 times higher in the presence of FAD than in the presence of FMN (supplemental Table S1). PaaE showed the same cofactor preference when cytochrome c was used as an artificial electron acceptor instead of PaaABC (supplemental Table S1).
Crystal Structure of PaaAC
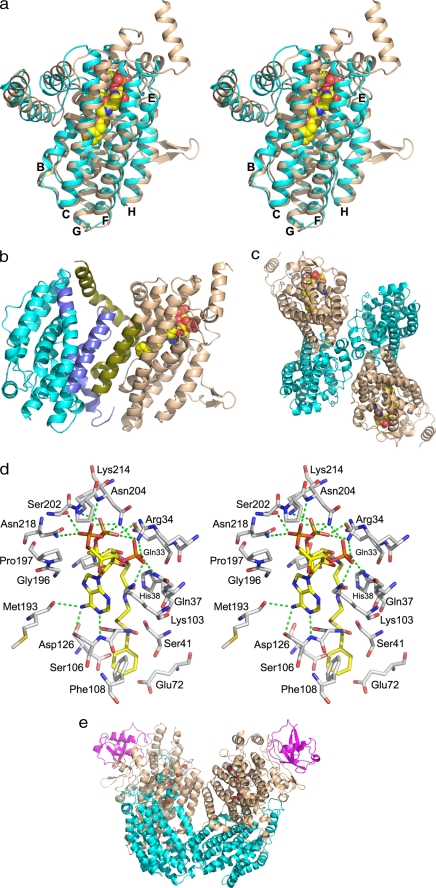
Of the three stable complexes, PaaAC, PaaABC, and PaaBC, we obtained crystals only of the PaaA-His6PaaC subcomplex. PaaA and PaaC share only 17% sequence identity, yet they can be superimposed with a root mean square deviation of 1.7 Å for 191 Cα atoms (of 302 for PaaA and 248 for PaaC). Their overall fold is characteristic of the hydroxylase components of monooxygenases (28). The core of the molecules is composed of six long α-helices: B, C, E, F, G, and H (named according to Ref. 11; Fig. 2a and supplemental Table S3) and is similar to other monooxygenases. In PaaA the α-helices B, C, E, and F form a four-helix bundle, adopting a ferritin-like fold and encompassing a long tunnel. The di-iron-binding site of PaaA is unoccupied. PaaC lacks the N- and C-terminal α-helical extensions observed in PaaA (Fig. 2a) and differs in conformation of segments between α-helices G, G′, H, and J. The space between the helices of the ferritin bundle in PaaC is filled with side chains eliminating the tunnel (supplemental Fig. S3).
FIGURE 2.
Structure of PaaAC. a, the stereo view of the superimposed PaaA (wheat) and PaaC (cyan). Core α-helices B, C, E, F, G, and H are marked. The PA-CoA substrate molecule is shown in sphere mode. b, the PaaAC heterodimer with α-helices participating in heterodimerization are shown in darker tones. c, the PaaAC heterotetramer showing head-to-tail association; d, stereo view of the PaaA substrate-binding site with bound PA-CoA. e, the ToMOH (αβγ)2 homodimer associated side-by-side. Subunits are colored the same as their homologs in PaaAC. The Fe2+ ions and toluene molecules are shown as spheres.
The PaaAC complex crystallized in three crystal forms. The stoichiometry of PaaA:PaaC was 1:2 in two forms and 1:1 in the third form. Comparison of molecular arrangements in different crystal packing environments allowed us to identify the (PaaAC)2 heterotetramer (dimer of heterodimers) as the common structural unit. PaaAC heterodimer interface is formed by helices B and C from each molecule, together forming an antiparallel four-helix bundle, with additional contacts between helix K of PaaA and the C terminus of PaaC (Fig. 2b). The interface has a significantly hydrophobic character, with a particularly large hydrophobic surface on PaaA (supplemental Fig. S4). The presence of such an extensive hydrophobic patch on the surface of PaaA is likely responsible for its high propensity for aggregation when expressed alone. Three salt bridges anchor the PaaA and PaaC subunits at the edges of the interface (Glu-33PaaC…Arg-90PaaA, Asp-35PaaC…Lys-282PaaA, and terminal COOHPaaC…Arg-61PaaA). The two PaaAC heterodimers are arranged in an antiparallel fashion to form a heterotetramer (Fig. 2c). This interface has a predominantly hydrophilic character. The interactions of unpaired PaaC subunit (“free PaaC”) are described in the supplemental materials.
Substrate Binding
To date no hydroxylase component of a BMM oxygenase was crystallized with its substrate in the active site. We have determined the structure of apo PaaAC as well as its complexes with CoA, acetyl-CoA, 3-hydroxybutytyl-CoA, benzoyl-CoA, and the substrate phenylacetyl-CoA. The ligands bind only to the PaaA subunit, accompanied by ordering of the loop between α-helices G and G′ (residues 199–205) that now forms part of the ligand-binding site. The difference map unequivocally shows the substrate position in the active site (supplemental Fig. S5). The ligand/substrate binds within the tunnel formed by helices B, C, E, and F and the tips of helices G and I. This tunnel extends ∼20 Å from the protein surface toward the center of PaaA and ends near the di-iron center (Fig. 2d and supplemental Fig. S3a). The CoA moiety of the substrate forms interactions with ∼30 residues (Fig. 2d). The ligands are accommodated in the tunnel with the adenine in an anti-conformation, folded over the diphosphate moiety at the top of the tunnel and the pantothenate moiety extending toward the bottom of the tunnel (Fig. 2d). The adenine base is anchored by hydrogen bonds between the N6 amine group and OGSer-106 and COMet-193 and by van der Waals contacts with Ile-268 and Phe-264. The phosphate groups form multiple hydrogen bonds. The 3′-phosphate hydrogen-bonds with Arg-33, Gln-37, and Lys-103; the α-phosphate bonds with Ser-202, Asn-204, and Asn-218; whereas the β-phosphate bonds with Arg-33 and Lys-214. Pantothenate and β-mercaptoethylamine moieties make numerous van der Waals contacts along the tunnel with Gln-34, His-38, Ser-41, Ser-105, Asn-132, Gln-133, Leu-136, Tyr-144, Met-194, and Phe-195. The bottom of the binding site is formed by Phe-108 and three acidic residues (Glu-49, Glu-76, and Asp126) hydrogen-bonded to the NZLys68 (Figs. 2d and 3a). Phe-108 forms an “edge-to-face” contact with the aromatic ring. Only two other hydrophobic side chains, Ile-121 and Val-125, are in the proximity of the aromatic ring of the substrate. The relatively few contacts made by the phenylacetate are compensated by the many interactions with CoA, assuring adequate binding of the substrate.
FIGURE 3.
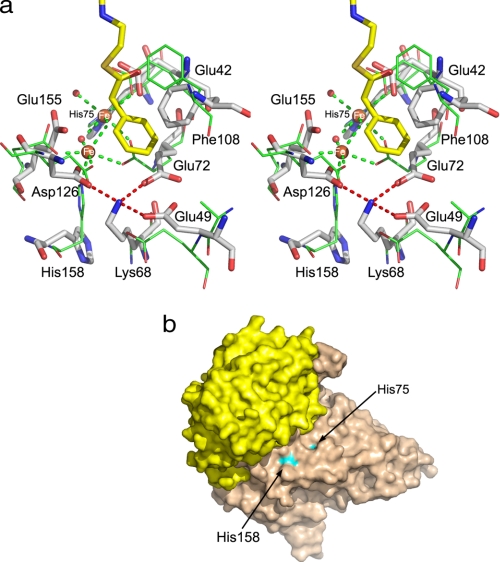
Comparison of substrate-binding sites in MMBs. a, superposition of iron binding centers in PaaA (shown as sticks and labeled) and ToMOH (lines). Glu-72, Glu-155, and His-158 in PaaA in the absence of Fe2+ are rotated away. The “lysine bridge,” Lys-68 salt-bridged to Glu-49, Glu-72, and Asp-126, is also shown. b, surface representation of PaaAC showing that His-75 and His-158 near the di-iron center are partially solvent-exposed.
To obtain iron-loaded PaaAC, we supplemented the media with iron, resulting in protein aggregation. The presence of as little as a 1:1 molar ratio of Fe(NH4)2SO4 or CoCl2 to PaaAC arrested crystal growth, whereas with a 4-fold lower concentration of Fe(NH4)2SO4 (25 μm), no iron was found in the structure. We therefore resorted to soaking PaaAC crystals with iron. Even very brief soaks led to drastically reduced diffraction resolution. Nevertheless, we were able to collect diffraction data to 5.8 Å resolution from these crystals. The anomalous map showed a large peak at the expected position of the iron center in PaaA. At this resolution, the peak could represent the expected di-iron center.
DISCUSSION
Role of Individual Components of the PA-CoA Oxygenase Complex
PaaAC displays very low sequence identity to the hydroxylase subunits of other BMMs (BMMH), and its structure shows several features unique to this hydroxylase (see below), thus supporting the phylogenetic assignment of PA-CoA oxygenase as a prototype of a new subfamily of BMMs. Indeed, BLAST search identified several hundred bacterial homologs of PaaA and PaaC with sequence identity exceeding 30%. We hypothesize that like PaaC, the β subunits in other BMMs may be essential for structural integrity of the α subunit. The PaaE subunit is a reductase with a preference for NADPH and FAD, capable of reducing cytochrome c.
We identified stable subcomplexes formed by PaaABC, PaaAC, and PaaBC. Furthermore, our in vitro reconstitution experiments showed that only PaaA, B, C, and E subunits of the E. coli monooxygenase are necessary for in vitro enzymatic activity and that the presence of PaaD is inconsequential. This supports a similar observation reported recently by investigation of PaaABCDE proteins from Pseudomanas putida (8). In this light, the previous conclusion from in vivo studies of E. coli knock-out mutants for each of the paaABCDE genes, which indicated that PaaD is essential for the appearance of the product (9), may indicate that PaaD function is related to the maturation of the monooxygenase complex, rather than direct involvement in catalysis. Indeed, PaaD shows low sequence similarity to SufT from the suf operon, which may be involved in iron-sulfur cluster assembly. We hypothesize that in vivo, PaaD could assist either in maturation of the PaaE reductase (insertion of an iron-sulfur cluster) or PaaA (insertion of di-iron center).
The function of the essential, small PaaB subunit is not yet clear. Although it has no sequence similarity to the γ or regulatory subunits of BMMs, it may share their functions. PaaB forms a heterodimeric complex with PaaC that is sensitive to ionic strength, suggesting a predominantly hydrophilic interaction. A larger complex that includes PaaA (PaaACB) is more stable, suggesting possible additional interactions with PaaA. We hypothesize that the PaaCB heterodimer binds PaaA predominantly through PaaC and that PaaB plays a regulatory rather than a structural role.
Comparison of PaaAC with Other BMMHs
Comparison of PaaA with other catalytic subunits of BMMHs shows that the α-helical core is structurally most conserved (see the multiple structure alignment in supplemental Fig. S6). Four of these helices form the ferritin fold enclosing the active center. The structures of the BMMHs show that the substrate-binding site, located in cavity 1 of the α-subunit, is connected to the surface through two consecutive cavities (cavities 2 and 3) formed within the helical core, as e.g. in MMOH (11) or through a continuous channel leading from the surface to the active center, as e.g. in ToMOH (14) or PaaA.
PaaAC provides the first structural evidence for the mode of aromatic substrate binding in the reaction center of BMM hydroxylases and shows how the common monooxygenase fold has adapted to binding CoA derivatives. The elongated PA-CoA substrate molecule uses the same access route to the active center as in other BMMHs, extending along the channel and placing the PA moiety in the active center. The acyl moiety of the substrate is positioned in the region corresponding to cavity 1. The β-mercaptoethylamine, pantothenate, and adenine moieties occupy the region equivalent to cavity 2, which in PaaA is substantially larger than in other BMMHs. The ribose and phosphates reside in cavity 3. Cavity 1 in BMMHs is usually lined with many hydrophobic residues (11, 14). Extensive mutagenesis studies as well as molecular modeling proved that residues lining cavity 1 are important for regiospecificity, for enantioselectivity, and for determining substrate specificity (29–32). This cavity in PaaA is even more hydrophilic than in other BMMHs (supplemental Table S4), with the only strictly conserved residue being Phe-108, whose side chain makes “edge-to-face” interaction with the aromatic ring of the substrate (Figs. 2d and 3a). Phe-108 corresponds to Phe-188 in MMOH, which occludes the entrance to the reaction cavity and together with Leu-110 forms the so-called “leucine gate.”
An important difference between PaaAC and other BMMHs is in their oligomerization. Although PaaAC heterodimers interact in an antiparallel fashion to form heterotetramers (Fig. 2c), the BMMH αβγ heterotrimers pack parallel to each other (Fig. 2e) (14), with different parts of the surfaces involved in these interactions.
The structure of PaaA with PA-CoA provides new insight into the precise organization of the Michaelis complex. The interaction with Phe-108 helps to orient the PA substrate such that the C1–C2 bond of the aromatic ring is located exactly above the putative di-iron center, and the acetyl group is pointing toward the entrance of the active site. This orientation is compatible with hydroxylation at the ortho-position. Our crystal structure provides a template for substrate binding in other BMMHs and will aid in protein engineering attempts that were previously made without the knowing the substrate positioning (29, 31, 32). For example, a recent attempt at modeling the toluene substrate in the active site of ToMOH (30) placed the aromatic ring in a different orientation than in our structure.
Differences in the Iron Center
The anomalous difference map based on crystals soaked with iron indicated that the iron binds at the expected site within PaaA. Aggregation of PaaAC in the presence of iron and significant reduction of resolution upon soaking crystals with iron suggest either a low affinity of PaaA for iron, possibly associated with a conformational rearrangement, or a more complex mechanism of iron insertion that requires accessory protein(s). The positions of the di-iron ligands in PaaA are similar to those in other BMMHs, with the exception of one of the four iron-binding glutamates, which is replaced here by an aspartate (Fig. 3a). However, a local adjustment of the backbone brings the carboxylic group close to its position in the glutamate of other BMMHs. Therefore, we expect a similar di-iron center in the PaaA subunit, with one Fe2+ ion coordinated by Glu-42, Glu-72, and His-75 and the second Fe2+ ion coordinated by Asp-126, Glu-155, and His-158 (Fig. 3a). In the absence of iron, the side chains of Glu-72, Glu-155, and His-158 assume different conformations than their counterparts in MMOH and ToMOH.
PaaA contains a unique structural feature in close proximity to the iron-binding center, which we designate the “lysine bridge” (Fig. 3a). At its center is Lys-68, which forms salt bridges with three acidic residues, Glu-49, Glu-72, and Asp-126; the latter two are expected to coordinate Fe2+ ions. The residues of the lysine bridge belong to three α-helices of the ferritin-like bundle (B, C, and E). The lysine and the acidic residues are together conserved in all of the BMMHs homologous to PaaA with >30% sequence identity. This lysine bridge may account for stabilization of the helices that form the ferritin core, which in PaaA are well ordered and have similar B-factors to other parts of the structure. MMOH, not having this feature, shows an increased flexibility of two α-helices of the ferritin-like four-helix bundle (E and F) in the absence of iron (33). Another possibility follows from the urease, where a lysine serves as an iron-binding ligand after carbamoylation (34).
PaaA also lacks the hydrogen bond network from the surface to the active center highly conserved in other BMMHs and formed by residues belonging to α-helices A (missing in PaaA), C, and F and two histidines from the metal-binding center (11, 14). This network was proposed to provide electron transport path from the reductase to the iron-binding center of the hydroxylase. Additionally, two residues, Thr-213 and Asn-214 (MMOH numbers) from the helix E, that are conserved in BMMs and affect catalysis (10) are replaced in PaaA by Ala-129 and Ile-130, suggesting some differences in the reaction cycle. These residues are located next to an enlarged turn (π-turn) of the α-helix E, the feature retained in PaaA.
Interactions with the Reductase
The hydroxylase component of BMMs are usually hetero-oligomers of the type (αβγ)2. However, the (PaaAC)2 complex is organized differently from other BMMHs (Fig. 2, c and e). The α-helix A from the α-subunit and the homologous α-helix from the β-subunit, which play pivotal roles in αβγ dimerization, are absent in PaaA and PaaC.
This different oligomerization of (PaaAC)2 brings the two histidines of the iron-binding center very close to the protein surface (Fig. 3b). His-158 is separated from solvent by only one layer of side chains, whereas the side chain of His-75 is partially solvent-exposed in the absence of iron. Thus the di-iron center is accessible for direct electron transfer from the PaaE reductase, suggesting interaction of PaaE with helices C and F of PaaA. The absence of the hydrogen bond network conserved in other BMMs and implicated in electron transfer supports this hypothesis. We hypothesize that the PaaB subunit, found to be essential for catalysis, may be directly involved in electron transport.
PA-CoA Oxygenase Is a Monooxygenase
Until recently, the product of the PA-CoA oxygenase complex was thought to be a nonaromatic dihydrodiol (VII; Fig. 1), suggesting that the PA-CoA oxygenase might be a dioxygenase (35). However, the PaaAC complex shows a typical monooxygenase structure. The formation of the dihydrodiol VII could, however, also be explained by a monooxygenase mechanism. Such a mechanism for PaaAC should account for the formation of two hydroxyl groups and the reduction of one of the double bonds of the benzene ring analogous to the known mechanism of complex of hydroxylase of toluene 4-monooxygenase (17). Reiner and Hegeman (36) considered this mechanism in investigation of the metabolism of benzoic acid by Alcaligenes eutrophus. According to this mechanism, the PA-CoA oxygenase would operate through the 1,2-epoxide intermediate (Fig. 1, III), which hydrolysis would give the previously expected product VII (Fig. 1, reaction H). If this was indeed the case, then VII would convert to the observed VI with the 2-OH group derived from water. We performed the reaction in H2O18 and unequivocally showed that this oxygen does not come from water but must come from O2, ruling out this proposed mechanism (III → VII → VI). That would suggest that observed product VI occurs by rearrangement of III. Indeed, the presence of III was recently observed directly as the product of the PA-CoA oxygenase (8), requiring only a monooxygenase activity. This is consistent with our data and reconciles the expected chemistry with the structure of PaaA.
Acknowledgment
We thank Allan Matte for helpful suggestions and comments on the manuscript.
This work was supported in part by the National Sciences and Engineering Research Council of Canada, the National Research Council, the Canadian Institutes of Health Research, and the University of Saskatchewan. This work was supported by Grant GSP-48370 from Canadian Institutes of Health Research (to M. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Tables S1–S4, and Figs. S1–S8.
R. Zhang, unpublished observations.
- PA
- phenylacetic acid (phenylacetate)
- BMM
- bacterial multicomponent monooxygenases
- BMMH
- hydroxylase component of BMM
- MBP
- maltose-binding protein
- MMO
- methane monooxygenase complex
- MMOH
- hydroxylase of MMO
- 2-OH-PA
- 2-hydroxy-phenylacetate
- ToMO
- toluene/o-xylene monooxygenase complex
- ToMOH
- hydroxylase of ToMO
- PaaAC
- PaaA–PaaC
- Pipes
- 1,4-piperazinediethanesulfonic acid
- ADA
- [(carbamoylmethyl)imino]diacetic acid
- SEC
- size exclusion chromatography
- Ni-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1. Cao B., Nagarajan K., Loh K. C. (2009) Appl. Microbiol. Biotechnol. 85, 207–228 [DOI] [PubMed] [Google Scholar]
- 2. Fuchs G. (2008) Ann. N.Y. Acad. Sci. 1125, 82–99 [DOI] [PubMed] [Google Scholar]
- 3. Ullrich R., Hofrichter M. (2007) Cell Mol. Life Sci. 64, 271–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gibson D. T., Parales R. E. (2000) Curr. Opin. Biotechnol. 11, 236–243 [DOI] [PubMed] [Google Scholar]
- 5. Cafaro V., Izzo V., Scognamiglio R., Notomista E., Capasso P., Casbarra A., Pucci P., Di Donato A. (2004) Appl. Environ. Microbiol. 70, 2211–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ferrández A., Miñambres B., García B., Olivera E. R., Luengo J. M., García J. L., Díaz E. (1998) J. Biol. Chem. 273, 25974–25986 [DOI] [PubMed] [Google Scholar]
- 7. Luengo J. M., García J. L., Olivera E. R. (2001) Mol. Microbiol. 39, 1434–1442 [DOI] [PubMed] [Google Scholar]
- 8. Teufel R., Mascaraque V., Ismail W., Voss M., Perera J., Eisenreich W., Haehnel W., Fuchs G. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 14390–14395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fernández C., Ferrández A., Miñambres B., Díaz E., García J. L. (2006) Appl. Environ. Microbiol. 72, 7422–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Notomista E., Lahm A., Di Donato A., Tramontano A. (2003) J. Mol. Evol. 56, 435–445 [DOI] [PubMed] [Google Scholar]
- 11. Rosenzweig A. C., Frederick C. A., Lippard S. J., Nordlund P. (1993) Nature 366, 537–543 [DOI] [PubMed] [Google Scholar]
- 12. Rosenzweig A. C., Brandstetter H., Whittington D. A., Nordlund P., Lippard S. J., Frederick C. A. (1997) Proteins 29, 141–152 [PubMed] [Google Scholar]
- 13. Sazinsky M. H., Dunten P. W., McCormick M. S., DiDonato A., Lippard S. J. (2006) Biochemistry 45, 15392–15404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sazinsky M. H., Bard J., Di Donato A., Lippard S. J. (2004) J. Biol. Chem. 279, 30600–30610 [DOI] [PubMed] [Google Scholar]
- 15. Cafaro V., Scognamiglio R., Viggiani A., Izzo V., Passaro I., Notomista E., Piaz F. D., Amoresano A., Casbarra A., Pucci P., Di Donato A. (2002) Eur. J. Biochem. 269, 5689–5699 [DOI] [PubMed] [Google Scholar]
- 16. Bailey L. J., McCoy J. G., Phillips G. N., Jr., Fox B. G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 19194–19198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mitchell K. H., Rogge C. E., Gierahn T., Fox B. G. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3784–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Batie C. J., Ballou D. P., Correll C. C. (1992) in Chemistry and Biochemistry of Flavoenzymes (Muller F. ed) Vol. 3, pp. 543–556, CRC Press, Boca Raton, FL [Google Scholar]
- 19. Grishin A. M., Ajamian E., Zhang L., Cygler M. (2010) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 1045–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jaganaman S., Pinto A., Tarasev M., Ballou D. P. (2007) Protein Expression Purif. 52, 273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 22. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 24. Emsley P., Cowtan K. (2004) Acta Crystallogr. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 25. Murshudov G. N., Vagin A. A., Lebedev A., Wilson K. S., Dodson E. J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 247–255 [DOI] [PubMed] [Google Scholar]
- 26. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) Acta Crystallogr. D 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tagawa K., Arnon D. I. (1962) Nature 195, 537–543 [DOI] [PubMed] [Google Scholar]
- 28. Sazinsky M. H., Lippard S. J. (2006) Acc. Chem. Res. 39, 558–566 [DOI] [PubMed] [Google Scholar]
- 29. Borodina E., Nichol T., Dumont M. G., Smith T. J., Murrell J. C. (2007) Appl. Environ. Microbiol. 73, 6460–6467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Notomista E., Cafaro V., Bozza G., Di Donato A. (2009) Appl. Environ. Microbiol. 75, 823–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pikus J. D., Studts J. M., McClay K., Steffan R. J., Fox B. G. (1997) Biochemistry 36, 9283–9289 [DOI] [PubMed] [Google Scholar]
- 32. Leungsakul T., Johnson G. R., Wood T. K. (2006) Appl. Environ. Microbiol. 72, 3933–3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sazinsky M. H., Merkx M., Cadieux E., Tang S., Lippard S. J. (2004) Biochemistry 43, 16263–16276 [DOI] [PubMed] [Google Scholar]
- 34. Jabri E., Carr M. B., Hausinger R. P., Karplus P. A. (1995) Science 268, 998–1004 [PubMed] [Google Scholar]
- 35. Ismail W., El-Said Mohamed M., Wanner B. L., Datsenko K. A., Eisenreich W., Rohdich F., Bacher A., Fuchs G. (2003) Eur. J. Biochem. 270, 3047–3054 [DOI] [PubMed] [Google Scholar]
- 36. Reiner A. M., Hegeman G. D. (1971) Biochemistry 10, 2530–2536 [DOI] [PubMed] [Google Scholar]