Abstract
DYRK1A (dual specificity tyrosine phosphorylation-regulated kinase 1A) has been shown to be involved in learning and memory impairments in Alzheimer disease and Down syndrome. As a homolog of Drosophila minibrain gene, DYRK1A also plays important roles in neurodevelopment; however, the function and regulatory mechanism of DYRK1A in neurodevelopment remain elusive. REST (RE1 silencing transcription factor) plays vital roles in neuronal differentiation. Here, we found that REST can activate DYRK1A transcription via a neuron-restrictive silencer element at bp −833 to −815 of human DYRK1A promoter. The coordinated expression of DYRK1A and REST in mouse brain further supports the cross-interaction of DYRK1A and REST during neurodevelopment. Moreover, we showed that DYRK1A dosage imbalance reduced REST protein stability and transcriptional activity through facilitating ubiquitination and subsequent degradation of REST protein. Therefore, the regulation of DYRK1A by REST in a negative feedback loop suggests that DYRK1A and REST are closely related in neurodevelopment.
Keywords: Alzheimer Disease, Dual Specificity Kinase, Neurodevelopment, Transcription Factors, Transcription Regulation, DYRK1A, NRSE, REST
Introduction
DYRK1A, an evolutionarily conserved and proline-directed protein kinase (1), has been shown to be involved in learning and memory impairments in Alzheimer disease and Down syndrome (DS)2 (2). As a member of the DYRK family, DYRK1A can catalyze tyrosine-directed autophosphorylation as well as phosphorylation of serine/threonine residues in exogenous substrates. By phosphorylating the SP motif of NFAT, DYRK1A can cooperate with RCAN1 (regulator of calcineurin 1) and counteract with calcineurin in the regulation of NFAT signaling pathways (3, 4). Substrates or interacting proteins of DYRK1A comprise more than two dozen proteins including Notch, STAT3, FHKR, Gli-1, eIF2Bϵ, dynamin, glycogen synthase, 14-3-3, CREB, cyclin L2, Arip4, Hip-1, PAHX-AP1, HPV16E7, and caspase 9 (6, 8–10). The substrate diversity indicates the pleiotropic roles of DYRK1A in multiple pathways. The homolog protein of DYRK1A in Drosophila is mnb (minibrain), mutation of which caused specific defects in neurogenesis. Transgenic mice overexpressing DYRK1A exhibited delayed craniocaudal maturation with impairments in motor skill acquisition and spatial learning (11, 12). The Dyrk1a knock-out mice are embryonically lethal, and the reduced postnatal viability, small body size, and neurobehavioral abnormalities of Dyrk1a heterozygotes indicate the vital and nonredundant role of DYRK1A in neurodevelopment (13, 14). However, the function and molecular mechanism of DYRK1A regulation in neurodevelopment remain elusive.
REST (RE1 silencing transcription factor)/neuron-restrictive silencer factor blocks transcription of its target genes by binding to a 21-bp DNA element, RE1-binding site/neuron-restrictive silencer element (RE1/NRSE), through its eight C2H2 zinc fingers in the regulatory regions of target genes (15, 16). REST modulates expression of genes encoding fundamental neuronal functions including ion channels, synaptic proteins, and neurotransmitter receptors (15, 17). Although serving as a master repressor of target genes in non-neuronal tissues (18), REST is essential for the orchestrated activation of target genes during neuronal development, acting as either a silencer or an activator (19, 20). Recent reports have shown that REST is regulated by phosphorylation and subsequent ubiquitin-mediated proteolysis in a SCFβ-TRCP (E3 ubiquitin ligase)-dependent manner (21, 22). DS is characterized by a variety of phenotypic traits including mental retardation and early onset of Alzheimer disease neuropathology in the brain. It was found that REST expression was decreased in cultured fetal DS brain cell-derived neurospheres (17) as well as in the brains of DS mouse models (23). Using partially trisomic embryonic stem cells, a recent study indicated that the dosage imbalance in the DYRK1A gene contributed to the reduction in REST expression (23). Another recent study showed that Dyrk1a can interact with a SWI/SNF complex known to interact with REST, and its dosage imbalance deregulates Rest gene expression in DS mouse models (24). These reports suggest that there is cross-talk between REST and DYRK1A. DYRK1A may fulfill its function in neurodevelopment through its regulation of REST expression.
In the present study, we found that REST can activate DYRK1A transcription via a NRSE site at bp −833 to −815 of human DYRK1A promoter, whereas DYRK1A imbalance can destabilize REST protein expression and reduce its transcriptional activity, thus forming a negative feedback loop in regulating DYRK1A expression. Our study showed that REST and DYRK1A are coordinately expressed during neurodevelopment. These results suggested that the cross-interaction between DYRK1A and REST plays a vital role in neurodevelopment.
EXPERIMENTAL PROCEDURES
Cloning of Human DYRK1A Promoter and Construction of Chimeric Luciferase Reporter Plasmids
The 5′ upstream region of human DYRK1A gene (−1079 to +48 bp, pDYluc-A) was obtained by PCR of genomic DNA isolated from human HEK293 cells using a pair of primers (5′-TTCCCCAGCCCCTACCCT-3′ and 5′-CCGCTTCCTCCGGTGAGT-3′). The transcription start site of DYRK1A gene was referred as bp +1 according to a previous study (29). Various 5′ upstream fragments of human DYRK1A gene were amplified by PCR from the 1127-bp fragment and inserted in front of the luciferase reporter gene in pGL3-Basic vector. The primers used to generate different promoter deletion plasmids were as follows: forward, 5′-GAGACTGCTGCTGTCGCT-3′ (pDYluc-B) and 5′-TGTTGGTGCCGCTGCC-3′ (pDYluc-C), and reverse: GLprimer2 (Promega). The pDYlucm was made to contain mutations of the first NRSE site (GTCAGCACGTCAGCCGGGGTTT was mutated to GTtcgtACGTCtcgtGGGGTTT). All of the constructs were verified by sequencing and restriction enzyme digestions.
Cell Culture and Luciferase Assay
HEK293 cells, a transformed human embryonic kidney cell line, C6, rat glial cell strain, and neuro-2a (N2a), a mouse neuroblastoma cell line, were cultured in high glucose Dulbecco's modified Eagle's medium (Hyclone, South Logan, UT) supplemented with 10% fetal bovine serum, 1 mm sodium pyruvate, 2 mm l-glutamine, and 100 units/ml penicillin-streptomycin (Invitrogen). PC12 cells, a cell line derived from a pheochromocytoma of the rat adrenal medulla, were cultured in the same condition except for 5% fetal bovine serum and 10% horse serum. All of the cells were maintained at 37 °C in an incubator containing 5% CO2. All of the transfections were carried out with LipofectamineTM 2000 transfection reagent (Invitrogen) according to the manufacturer's instructions. Luciferase activity was determined following a protocol supplied by the dual luciferase reporter assay kit (Promega) as described previously (25). Firefly luciferase activity was normalized to the Renilla luciferase activity and expressed as relative luciferase units. For protein half-life experiments, the cells were chased with cycloheximide (50 μg/ml) for the indicated times and lysed in radioimmune precipitation assay buffer.
mRNA Quantification
Total RNA was isolated from mouse brain or cells by TRIzol (Invitrogen). Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI) was used to synthesize the first strand of cDNA from equal amounts of RNA samples, according to the manufacturer's instructions. Here, 20–35 cycles of PCR were used to cover the linear range of the PCR amplification. The mRNA of DYRK1A expression was quantified using the ABI 7900 HT Fast real time PCR system (Applied Biosystems, Foster City, CA) using the TaqMan® gene expression assay (ABI assay ID: Hs00680802_m1). 18 S rRNA was chosen for internal control. The mRNA of REST and BDNF (brain-derived neurotrophic factor) was quantified by SYBR® Green-based gene expression analysis (Applied Biosystems). The primers for real time quantitative and semi-quantitative PCR were as follows: human DYRK1A (450 bp), forward, 5′-CGTGGTTCATTTGCTTGTGT-3′, and reverse, 5′-CGAGGAAGAAGTCGTTGAGG-3′; mouse DYRK1A (300 bp), forward, 5′-GAGACACACAGTCCCCAGGT-3′, and reverse, 5′-ACTGTGGCCAACCTCCATAG-3′; human REST (137 bp), forward, 5′-ACTCATCACGGAGAACGCCC-3′, and reverse, 5′-GAGGCCACATAACTGCACTG-3′; mouse REST (244 bp), forward, 5′-CGAGTCTCAGGAAATTGATGA-3′, and reverse, 5′-GCGATTGAGGTGTTTGCTATA-3′; human BDNF (168 bp), forward, 5′-GATGCTCAGTAGTCAAGTGCC-3′, and reverse, 5′-GCCGTTACCCACTCACTAATAC-3′; mouse BDNF (114 bp), forward, 5′-CCATAAGGACGCGGACTTGTA-3′, and reverse, 5′-TTTGCGGCATCCAGGTAATTT-3′; human and mouse β-actin (141 bp), forward, 5′-GACAGGATGCAGAAGGAGATTACT-3′, and reverse, 5′-TGATCCACATCTGCTGGAAGGT-3′; human β-actin (250 bp), forward, 5′-CATGTACGTTGCTATCCAGGC-3′, and reverse, 5′-CTCCTTAATGTCACGCACGAT-3′; and human β-actin (462 bp), forward, 5′-GGACTTGCAAGAGATGG-3′, and reverse, 5′-GAAGCATTTGCGGTGGAG-3′.
Immunoblotting
The cell lysates were resolved by 12% SDS-PAGE, and immunoblotting was performed as described previously (26). The primary antibodies used were rabbit anti-REST polyclonal antibody (07-579; Upstate, Waltham, MA), anti-FLAG M2 monoclonal antibody (F1804; Sigma-Aldrich), and β-actin mAb (AC-15; Sigma-Aldrich) according to the manufacturers' instructions. Detection and quantifications were performed with the Li-Cor Odyssey imaging system and its software.
Immunofluorescence
Brains from 6-month-old C57BL/6 mice were fixed and sectioned to 8-μm thickness with a Leica cryostat. The slices were immunostained with rabbit anti-DYRK1A (1:25; Cell Signaling Technology, Danvers, MA), rabbit anti-REST (1:500; Upstate), or mouse anti-MAP-2 (1:100; Vector Laboratories, Burlingame, CA) primary antibodies at 4 °C overnight. Fluorescein-conjugated AffiniPure donkey anti-mouse (1:100; Jackson ImmunoResearch, Luton, United Kingdom) or Cy3-conjugated AffiniPure Goat anti-rabbit secondary antibodies (1:100; Jackson ImmunoResearch) were used to visualize the signals. The slices were also stained with DAPI (1 μg/ml; Roche Applied Science) to visualize the nucleus. The results were analyzed with a fluorescence microscopy (DMI4000B; Leica).
Electrophoretic Mobility Shift Assay
Consensus oligonucleotides NRSE sense: 5′-TTCAGCACCACGGACAGCGCC-3′, and NRSE antisense: 5′-GGCGCTGTCCGTGGTGCTGAA-3′, were end-labeled with nearly infrared IRDye 700 (Bioneer) to generate a double-stranded oligonucleotide probe. Consensus NRSE, mutant consensus NRSE, and three putative NRSE sites in the promoter region of human DYRK1A gene were also synthesized to generate double-stranded competitors. The sequences of consensus NRSE and mutant NRSE oligonucleotides (sense strand) are 5′-TTCAGCACCACGGACAGCGCC-3′ and 5′-TTtcgtACCACtcgtAGCGCC-3′ (Bioneer), respectively. The sense sequence of three putative NRSE sites were NRSE-DY1: 5′-CGTCAGCACGTCAGCCGGGGTTT-3′; NRSE-DY2: 5′-GACTGCTGCTGTCGCTGCTGCTGATCGCG-3′; and NRSE-DY3: 5′-TGCTGCTGCTGTTCCTGCTGCTGCTGT-3′. Briefly, DYRK1A-NRSE infrared probes (50 nm) were incubated with or without HeLa nuclear extract (Promega) in gel shift binding buffer containing 100 mm Tris, 500 mm KCl, 10 mm DTT, pH 7.5, 25 mm DTT, 2.5% Tween® 20, poly(dI·dC), 1 μg/μl in 10 mm Tris, 1 mm EDTA at room temperature for 20 min. For the competition assay, HeLa nuclear extract was incubated with excessive competitor as indicated. The nuclear extraction from N2a cells was performed using a Chemicon nuclear extraction kit (2900; Millipore) according to the manufacturer's instructions. The samples were analyzed by 4% (upper gel) and 8% (lower gel) nondenaturing PAGE. The gel was scanned and analyzed by Li-Cor Odyssey imaging system.
Chromatin Immunoprecipitation
ChIP experiments were performed according to protocols of the EZ ChIPTM assay kit (17-371; Upstate). Briefly, 5 × 105 cells were fixed with 1% formaldehyde for 10 min at 37 °C. The cells were washed extensively with PBS, and the chromatin was sheared by sonication to 200–1000 bp fragments. The cross-linked REST-DNA complex was immunoprecipitated with anti-REST antibody (1:500; Upstate). Normal mouse IgG was used as negative controls. PCR was performed using the DNA reversed from the cross-linked complex with two pairs of primers (NRSE-DY1: 5′-AGGAGGGAACGGAATCT-3′ and 5′-ATTATTATTCGGGCGGGA-3′; and NRSE-DY3: 5′-TTCTGCTGCTGCTGTTCC-3′ and 5′-ACACTCGCACTCACACCG-3′).
Expression Vectors and siRNA Assay
The vector pBDNFluc was kindly provided by Dr. Masaaki Tsuda (27). Human REST cDNA was cloned using a pair of primers: 5′-GGAATTCATGGCCACCCAGGTAATGGG-3′ and 5′-CCCAAGCTTAGCTCCTGCCCTTGAGCTG-3′. REST-FS mutant was amplified using reverse primer 5′-CCCAAGCTTCATTTTTTCATGCAAGTATTTTG-3′ and the same forward primer as wild type. Human DYRK1A vector was cloned using primers 5′-GGGGTACCATGCATACAGGAGGAGAGAC-3 and 5′-CCGCTCGAGTCACGAGCTAGCTACAGGACTC-3′. Human REST siRNA was generated using pSUPER vector as described previously (26). The target sequence for human REST siRNA is GCTACAATACTAATCGATA. DYRK1A siRNA was generated using pGFP-V-RS vector (OriGene, Rockville, MD). The target sequence for DYRK1A siRNA is GCCACTTTATGTTTCGAAA, which is homologous in both human and mouse DYRK1A gene.
Data Analysis
All of the experiments were repeated three to five times. For immunoblotting, immunofluorescence, and quantitative real time RT-PCR, one representative picture is shown; quantifications are from three or four independent experiments. The values represent the means ± S.E. The data were evaluated for statistical significance with analysis of variance or Student's t test analysis.
RESULTS
REST Can Activate Human DYRK1A Gene Transcription
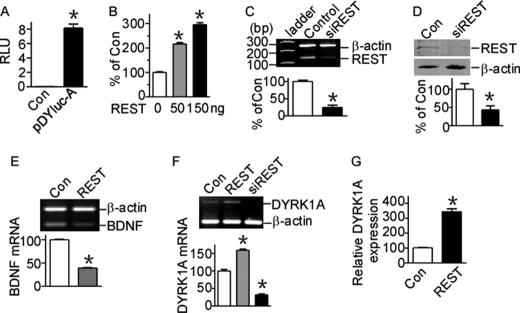
To further clarify the molecular mechanism of DYRK1A gene transcription, we cloned a 1127-bp fragment of the 5′-flanking region of the human DYRK1A gene into promoter-less plasmid pGL3-Basic. Because pGL3-Basic vector lacks eukaryotic promoter and enhancer sequences upstream of a reporter luciferase gene, the expression of luciferase activity in cells transfected with this plasmid depends on the insertion and proper orientation of a functional promoter upstream of the luciferase gene. Plasmid DNA pDYluc-A (−1079 to +48 bp) was transfected into HEK293 cells, and luciferase activity was measured by a luminometer to reflect promoter activity. pDYluc-A transfected cells had significantly higher luciferase activity compared with controls (8.18 ± 0.56 compared with 0.058 ± 0.001 relative luciferase units, p < 0.0001; Fig. 1A), indicating that the 1.1-kb fragment contains the functional promoter region of the human DYRK1A gene. A computer-based transcription factor-binding site search by the JASPAR database revealed that this 1127-bp 5′-flanking region contains three putative NRSE, which are binding sites for the zinc finger protein REST. To investigate whether DYRK1A transcription can be regulated by REST, we transfected HEK293 cells with REST expression construct and examined the promoter activity of human DYRK1A gene. Luciferase assay showed that REST overexpression increased DYRK1A promoter activity in a dosage-dependent manner (p < 0.001; Fig. 1B), indicating DYRK1A expression may be up-regulated by REST.
FIGURE 1.
REST activates human DYRK1A gene transcription. A, pDYluc-A contains the functional promoter of DYRK1A gene. The promoter construct pDYluc-A, containing the 1127-bp fragment from the human DYRK1A gene 5′-UTR, was transfected into HEK293 cells. Luciferase activity was measured 24 h after transfection by a luminometer. The values represent the means ± S.E. (n = 3). *, p < 0.001 by Student's t test. B, DYRK1A gene promoter is activated by REST. REST was co-transfected with pDYluc-A into HEK293 cells. Luciferase assay was performed 24 h after transfection. The values represent the means ± S.E. (n = 3). *, p < 0.001 by Student's t test. C, RT-PCR showed that REST knockdown construct psiREST can knock down Rest mRNA expression. β-Actin was used as the internal control. The values represent the means ± S.E. (n = 3). *, p = 0.0006 by Student's t test. D, the knockdown effect of psiREST was verified in protein level by Western blot. HEK293 cells were co-transfected with REST expression plasmid and psiREST. REST was detected with anti-REST antibody. β-Actin was used as loading control. The values represent the means ± S.E. (n = 3). *, p = 0.04 by Student's t test. E, RT-PCR showed that BDNF mRNA was repressed by REST overexpression in HEK293 cells. β-Actin was amplified as the internal control. *, p < 0.001 by Student's t test. F, RT-PCR showed that REST overexpression significantly increased DYRK1A mRNA, whereas REST knockdown significantly decreased DYRK1A mRNA levels. β-Actin was amplified as the internal control. The values represent the means ± S.E. (n = 3). *, p < 0.001 by Student's t test. G, real time PCR showed that DYRK1A expression was elevated with REST overexpression. TaqMan gene expression assay of DYRK1A was performed with an ABI 7900HT real time PCR system. 18 S rRNA was chosen for the internal control. The values represent the means ± S.E. (n = 4). *, p = 0.0032 by Student's t test. Con, control.
To further examine whether REST can regulate DYRK1A transcription, a pSuper-based REST siRNA (psiREST) was constructed to knock down REST. REST expression can be reduced by psiREST to 24.83 ± 6.52% of controls at mRNA level (p = 0.0006; Fig. 1C) and 43.83 ± 10.66% of controls at protein level (p = 0.04; Fig. 1D) in HEK293 cells. Although usually working as a repressor to down-regulate its target genes such as BDNF (39.39 ± 0.38% of controls, p < 0.0001; Fig. 1E), overexpression of REST in HEK293 cells can significantly increase DYRK1A mRNA levels to 158.8 ± 3.69% of controls (p = 0.0006; Fig. 1F, second lane). Moreover, knockdown of REST expression in cells can markedly reduce the DYRK1A mRNA levels to 31.94 ± 2.59% of controls (p = 0.0003; Fig. 1F, third lane). To further confirm this, real time PCR of DYRK1A was performed using ABI TaqMan gene expression assay. Real time PCR also confirmed that DYRK1A expression was elevated with REST overexpression to 309.4 ± 21.89% of controls (Fig. 1G). Similar DYRK1A transcriptional activation by REST was observed in N2a and PC12 cells (data not shown). Together, these results indicate that the transcription of human DYRK1A gene can be activated by REST.
Identification of the Functional NRSE Site in DYRK1A Gene Promoter
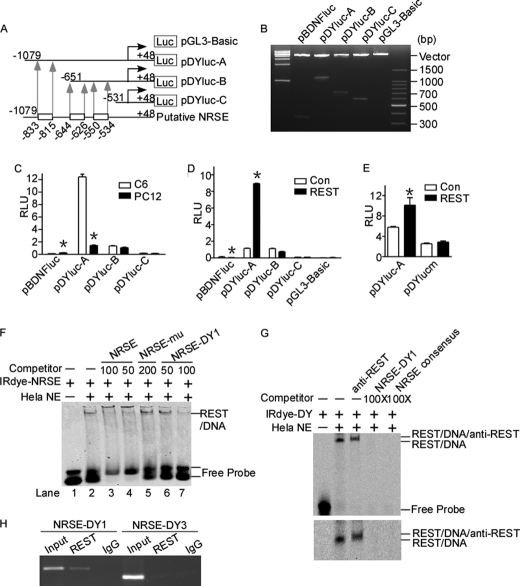
Our above results showed that DYRK1A can be regulated by REST. REST regulates the transcription of a large number of neuronal genes by binding to NRSE, a 21-bp consensus DNA sequence, which is present in the regulatory regions of neuronal genes. The sequence analysis of DYRK1A gene promoter region revealed three putative NRSE elements, located from −833 to −815 bp, −644 to −626 bp, and −550 to −534 bp, respectively. To determine whether DYRK1A gene promoter contains functional NRSE, a series of luciferase reporter gene plasmids with different upstream deletions of human DYRK1A gene promoter region were constructed (Fig. 2, A and B). Different deletion plasmids were transfected into REST-competent C6 or REST-incompetent PC12 cells (Fig. 2C). Luciferase assay showed that pBDNFluc, containing a repressing NRSE site in the rat Bdnf promoter upstream of luciferase gene, had higher promoter activity in PC12 cells (0.24 ± 0.006 relative luciferase units) compared with C6 cells (0.13 ± 0.007 relative luciferase units) (p < 0.0001; Fig. 2C, first and second bars). DYRK1A promoter constructs pDYluc-A showed higher promoter activity in C6 cells. However, further deletion of the first NRSE from −1079 bp in pDYluc-A to −651 bp in pDYluc-B abolished the differential effect in C6 and PC12 cell lines (Fig. 2C), indicating that the NRSE site was located in the region of −1079 to −651 bp. The region from −1079 to −651 bp contains the first putative NRSE site located from −833 to −815 bp (Fig. 2A). No differential effect was observed in deletion constructs pDYluc-B and pDYluc-C, indicating that the other two predicted NRSE sites did not respond to REST (Fig. 2C). To further confirm the location of NRSE, we co-transfected the DYRK1A promoter deletion constructs with REST expression vector into PC12 cells. Luciferase assay revealed that REST expression can decrease the promoter activity of Bdnf and increase the promoter activity of DYRK1A that contains the NRSE site from −833 to −815 bp (Fig. 2D). The constructs of pDYluc-B and C, without the first putative NRSE site, did not respond to REST expression in PC12 cells (Fig. 2D). Furthermore, the mutation of the NRSE site at −835 to −815 bp of DYRK1A promoter abolished the effect of REST overexpression (Fig. 2E, third and fourth bars). These data indicate that the NRSE site located from −833 to −815 bp may be responsible for the activational effect of REST in DYRK1A gene transcription.
FIGURE 2.
Identification of a functional NRSE site in human DYRK1A promoter. A, schematic diagram of the human DYRK1A promoter deletion constructs consisting of a 5′-flanking region with serial deletions cloned into pGL3-Basic in front of the luciferase gene (Luc). Arrowheads indicate the directions of transcription. The numbers represent the end points of each construct. The positions of three putative NRSE sites are shown at the bottom. B, the deletion plasmids were confirmed by sequencing and restriction enzyme digestion checking on a 1.5% agarose gel. Vector size is 4.7 kb, and the DYRK1A gene 5′-flanking fragment inserts range from 0.5 to 1.5 kb. C, the deletion plasmids were transfected into REST-competent C6 or REST-incompetent PC12 cells. Plasmid pRL-TK was used to normalize the transfection efficiency, and luciferase activity was measured at 24 h by a luminometer. The values represent the means ± S.E. (n = 5). *, p < 0.001 by Student's t test. D, PC12 cells were co-transfected with REST expression vector and various DYRK1A promoter deletion constructs. Luciferase assay was performed 24 h after transfection. The values represent the means ± S.E. (n = 5). *, p < 0.001 by Student's t test. E, pDYlucm was made to contain the mutant NRES site of DYRK1A promoter. The mutant plasmid, as well as the wild type pDYluc-A, was co-transfected with REST expression plasmid into HEK293 cells. Plasmid pRL-TK was used to normalize the transfection efficiency, and luciferase activity was measured at 24 h by a luminometer. The values represent the means ± S.E. (n = 3). *, p < 0.01 by Student's t test. F and G, EMSA was performed with IRDye 700-labeled consensus NRSE oligonucleotides (F) or NRSE site of DYRK1A promoter (G). First lane, labeled probe without nuclear extract. Second lane, incubation of infrared labeled NRSE probe with HeLa nuclear extracts retarded the migration rate of the labeled probe, which formed a new shifted DNA-protein complex band. The addition of anti-REST antibody further shifted the band to a larger mass shown by a longer running time (third lane in the lower panel of G). Competition assays were performed by further adding different concentrations of molar excess of unlabeled competitive oligonucleotides. H, anti-REST antibody was used to immunoprecipitate the cross-linked REST-DNA complex in ChIP assay in SH-SY5Y cells. A pair of primers targeting DYRK1A was used to amplify NRSE-DY1 and NRSE-DY3. Signals amplified from input were used as size markers. Mouse IgG was used as a negative control. Con, control.
To determine whether the NRSE located from −833 to −815 bp in DYRK1A promoter can bind to REST, EMSA was performed. Consensus NRSE, a 21-bp double-stranded oligonucleotide probe corresponding to NRSE canonical sequence, was synthesized and end-labeled with IRDye 700 infrared dye. Consensus NRSE, mutant consensus NRSE, and three putative NRSE sites in DYRK1A promoter were also synthesized as cold probes. A shifted DNA-protein complex band was detected after incubation of the labeled consensus NRSE probe with HeLa nuclear extract (Fig. 2F, lane 2). The binding intensity of this shifted band was significantly reduced by applying 50- or 100-fold molar excess of unlabeled consensus NRSE competitive oligonucleotides (Fig. 2F, lanes 3 and 4). The incubation of 200-fold excessive mutant NRSE oligonucleotides had no competitive effect on the shifted NRSE-REST band (Fig. 2F, lane 5). The addition of 50- and 100-fold excess of NRSE-DY1, corresponding to the NRSE site from −833 to −815 bp of the DYRK1A promoter region, reduced the NRSE-REST shifted band in a dosage-dependent manner (Fig. 2F, lanes 6 and 7). However, the NRSE-DY2 and NRSE-DY3, corresponding to −644 to −626 bp and −550 to −534 bp of DYRK1A promoter region, respectively, had no competitive effect at 50-, 100-, or 200-fold excess (data not shown). Furthermore, similar results were achieved by using the NRSE site at −835 to −815 bp of DYRK1A promoter as probe (Fig. 2G). The supershift band with the addition of anti-REST antibody further confirmed the specific binding complexes (Fig. 2G, third lane). These results demonstrate that the NRSE-DY1 site at −833 to −815 bp in human DYRK1A promoter can bind to REST protein in vitro.
To confirm that the DY-NRSE1 site verified in vitro can actually bind to REST in vivo, ChIP was employed. ChIP-PCR results showed that REST antibody effectively immunoprecipitated the NRSE-DY1 site (Fig. 2H, second lane), indicating that REST binds to the NRSE located at −833 to −815 bp. The region of NRSE-DY3 (Fig. 2H, fifth lane) cannot be amplified from immunoprecipitates, suggesting that REST did not bind to the NRSE-DY3 site in DYRK1A promoter. Together, these data indicate that it is the first NRSE site, corresponding to DYRK1A promoter −833 to −815 bp, that is actually responsible for the up-regulation of DYRK1A gene by REST.
DYRK1A and REST Are Coordinately Expressed in Mouse Brain during Neurodevelopment and in Cell Lines
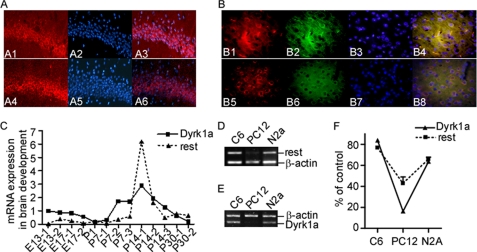
To provide further evidence that REST expression can be regulated by DYRK1A, we detected the expression of DYRK1A and REST proteins in adult mouse brains. Immunofluorescence staining of adult mice brain slices showed that both DYRK1A and REST were enriched in neurons of hippocampus (Fig. 3A) and cerebral cortex (Fig. 3B). The co-localization of DYRK1A and REST further supports the interactions of these two proteins. Both REST and DYRK1A are critical factors in neurogenesis. To investigate whether there are interactions between these two proteins during neurodevelopment, we detected expression levels of Dyrk1a and Rest from the developing mouse brains. RNA were extracted from normal mouse brain aging at embryonic days 13 and 17 and postnatal day 1 (P1), P7, P14, and P30. The real time PCR results showed that mRNA levels of Dyrk1a and Rest were coordinately expressed with significant correlation during neurodevelopment (p = 0.0028; Fig. 3C). Both Dyrk1a and Rest showed increased expression at P7–P14 and then decreased at P30. We also examined the expression of Dyrk1a and Rest in several cell lines including the C6, N2a, and PC12. Consistent with previous reports, PC12 cells had very low levels of Rest expression (28), and correlatively, Dyrk1a expression is also almost undetectable in PC12 cell line (Fig. 3, D and E). Strong Rest and Dyrk1a signals were detected coordinately in C6 and N2a cell lines (p = 0.0002; Fig. 3F).
FIGURE 3.
DYRK1A and REST coordinately expressed in mouse brain during neurodevelopment and in cell lines. A and B, immunofluorescence staining was performed on coronal slices from adult mouse brain. A Cy3-conjugated anti-rabbit secondary antibody was used to visualize DYRK1A (A1 and B1, red) and REST (A4 and B5, red). MAP2 (microtubule-associated protein 2) was stained to serve as a neuronal marker. A fluorescein-conjugated anti-mouse secondary antibody was used to visualize MAP2 (B2 and B6, green). The nuclei were counterstained with blue DAPI (A2, A5, B3, and B7). Overlay showed that both DYRK1A and REST are enriched in neurons in hippocampus (A3 and A6) and cerebral cortex (B4 and B8). The images were captured with a Leica fluorescence microscope at ×400 (A) or ×600 (B). C, quantitative RT-PCR was performed on cDNA templates prepared from normal mouse brain aging at embryonic days 13 (E13) and 17 (E17); at P1, P7, P14, and P30; and in adult mice. One to three mice were used in each time point as indicated by the numbers after the hyphens. p = 0.0028 by correlation test. Rest (D) and Dyrk1a (E) mRNA levels were determined by RT-PCR in C6, N2A, and PC12 cells. β-Actin was amplified as the internal control. F, quantification of D and E showed that Rest and Dyrk1a were coordinately expressed at low levels in PC12 cells and at high levels in N2a and C6 cells. The values represent the means ± S.E. (n = 3). *, p = 0.0002 by correlation test.
REST Protein Stability Is Disturbed by DYRK1A Dosage Imbalance
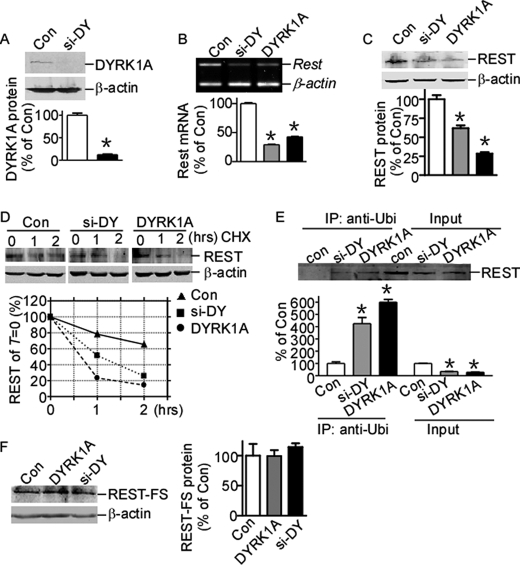
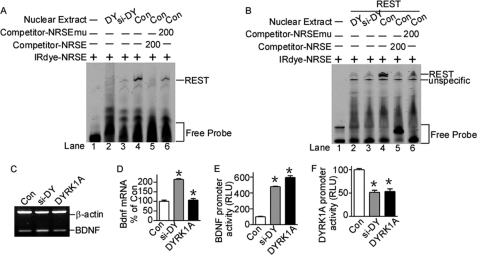
Rest mRNA has been shown to be down-regulated by DYRK1A protein imbalance in the mouse ES cell line (23); however, whether REST protein is regulated by DYRK1A is still unknown. To investigate the interaction of DYRK1A and REST proteins, we constructed DYRK1A expression vector pDYRK1A and siRNA knock-out plasmid psi-DY. The target sequence of psi-DY is homologous in human and mouse. RT-PCR showed that psi-DY can efficiently knock down DYRK1A mRNA expression in both HEK293 and murine N2a cells (data not shown). Western blot assay showed that psi-DY can significantly knock down the expression of DYRK1A in N2a cells transfected with DYRK1A expression vector (Fig. 4A). Consistent with previous reports, RT-PCR clearly showed that Rest gene expression was reduced by both knockdown and overexpression of DYRK1A in N2a cells by 29.16 ± 0.82% (p < 0.0001) and 42.62 ± 1.57% (p < 0.0001) of controls, respectively (Fig. 4B). To further examine whether REST protein level was also affected by DYRK1A expression, HEK293 cells were co-transfected with REST expression vector and psi-DY or pDYRK1A. Western blot assay showed that REST protein levels were also markedly reduced by DYRK1A knockdown or overexpression by 61.85 ± 3.70% (p = 0.0041) and 28.46 ± 1.80% (p = 0.0002) (Fig. 4C). The anti-REST antibody could not detect endogenous REST expression in our Western blot assay (data not shown), indicating that the reduction observed here is not due to the down-regulation of endogenous REST mRNA. The reduction of exogenously expressed REST implies that the down-regulation by DYRK1A imbalance may affect post-translational modifications and subsequent protein stability. To further investigate whether the REST reduction is associated with its protein stability, HEK293 cells co-transfected with REST and pDYRK1A or psi-DY were chased with cycloheximide for 1 and 2 h. Western blot clearly showed that the half-life of REST protein was shortened with DYRK1A or psi-DY transfection (Fig. 4D), indicating that DYRK1A imbalance in cells can disturb the protein stability of REST.
FIGURE 4.
REST expression is down-regulated by DYRK1A protein imbalance. A, psi-DY (siRNA against DYRK1A) was co-transfected with DYRK1A expression plasmid into N2a cells. DYRK1A was detected by anti-FLAG (M2) antibody. β-Actin was used as loading control. The values represent the means ± S.E. (n = 3). *, p < 0.0001 by Student's t test. B, N2a cells were transfected with DYRK1A expression vector pDYRK1A or knockdown vector psi-DY. Rest mRNA expression was detected by RT-PCR. β-Actin was amplified as the internal control. The values represent the means ± S.E. (n = 3). *, p < 0.0001 by Student's t test. C, N2a cells were co-transfected with REST expression vector and pDYRK1A or psi-DY. The cell lysates were separated with 12% SDS-PAGE. REST expression was detected by anti-REST antibody. β-Actin was used as loading control. The values represent the means ± S.E. (n = 3). *, p < 0.01 by Student's t test. D, HEK293 cells co-transfected with REST expression vector, pUbi-His, and pDYRK1A or psi-DY were chased with 50 μg/ml cycloheximide (CHX) for 1 and 2 h. REST expression was detected by anti-REST antibody. β-Actin was used as loading control. E, HEK293 cells co-transfected with REST expression vector and pDYRK1A or psi-DY were lysed and immunoprecipitated with anti-ubiquitin antibody (1:1000) and detected with anti-REST antibody. The values represent the means ± S.E. (n = 3). *, p < 0.01 by Student's t test. F, N2a cells were co-transfected with C terminus REST truncated mutant, FLAG-tagged REST-FS, together with pDYRK1A or psi-DY. REST-FS protein was detected by anti-FLAG (M2) antibody. β-Actin was used as loading control. The values represent the means ± S.E. (n = 3). p > 0.05 by Student's t test. Con, control; IP, immunoprecipitation.
Recent reports showed that REST can be degraded by the ubiquitin-proteasome system via phosphorylation at its C terminus (21, 22). Next we examine whether this REST protein instability caused by DYRK1A imbalance is associated with its ubiquitination. HEK293 cells co-transfected with REST, pUbi-His, and psi-DY or pDYRK1A were immunoprecipitated with ubiquitin antibody and detected with anti-REST antibody. Our results clearly demonstrated the increase of REST ubiquitination with DYRK1A imbalance (Fig. 4E, second and third lanes versus first lane), although the REST input signal were reduced with DYRK1A imbalance (Fig. 4E, fifth and sixth lanes versus fourth lane). These results clearly demonstrated that DYRK1A imbalance can reduce REST level via increasing the ubiquitination and subsequent degradation of REST.
REST-FS, a C-terminally truncated mutant of REST that was firstly discovered in a colon cancer cell line, cannot be degraded via the ubiquitin-proteasome system because it lacks the C terminus containing the phosphorylation and ubiquitination sites. To further confirm that the reduction of REST expression by DYRK1A imbalance is related to its protein degradation, REST-FS was co-transfected with psi-DY or pDYRK1A into HEK293 cells. In contrast to the reduction in wild type REST, there is no reduction in the C-terminally truncated REST-FS expression with DYRK1A overexpression or knockdown (Fig. 4F), suggesting that the REST C terminus contributes to the reductions induced by DYRK1A imbalance. These data suggest that imbalance of DYRK1A may perturb the phosphorylation and degradation pathway and subsequently reduces REST expression through its C terminus.
Transcriptional Activity of REST Is Reduced by DYRK1A Imbalance
The above results showed that REST mRNA and protein was reduced by DYRK1A imbalance at transcriptional and post-translational levels. To further confirm that REST expression was perturbed by DYRK1A imbalance, EMSA was performed to examine the binding ability of REST to its consensus NRSE oligonucleotides. EMSA results showed that nuclear extracts from N2a cells transfected with psi-DY and pDYRK1A had significantly lower binding ability to REST protein compared with controls (Fig. 5A, lanes 2 and 3 compared with lane 4). The EMSA was also validated in cells co-transfected with REST expression plasmid and DYRK1A knockdown or overexpression plasmid (Fig. 5B, lanes 2 and 3 compared with lane 4), indicating that the transcriptional activity of REST is also down-regulated by either knockdown or overexpression of DYRK1A in cells. Taken together, our results demonstrated that REST transcriptional activity was also reduced by DYRK1A dosage imbalance.
FIGURE 5.
Transcriptional activity of REST was reduced by DYRK1A protein imbalance. A and B, EMSA showed that REST activity was perturbed by DYRK1A imbalance. N2a cells were transfected with pDYRK1A and psi-DY (A) or co-transfected with REST expression vector (B). EMSA was performed with IRDye 700-labeled consensus NRSE. Nuclear extracts were obtained from N2a cells transfected with pDYRK1A (lane 2), psi-DY (lane 3), or empty vector (lane 4). The specificity of REST-NRSE binding was indicated by competition by consensus NRSE oligonucleotides (lane 5) or NRSE mutant oligonucleotides (lane 6). C and D, N2a cells were transfected with DYRK1A expression vector pDYRK1A or knockdown vector psi-DY. Bdnf mRNA levels were detected by RT-PCR. β-Actin was amplified as the internal control. The values represent the means ± S.E. (n = 3). *, p < 0.001 by Student's t test. E, the BDNF promoter construct pBDNFluc was co-transfected with pDYRK1A or psi-DY. Luciferase assay was performed 24 h after transfection. The values represent the means ± S.E. (n = 3). *, p < 0.001 by Student's t test. F, the DYRK1A promoter construct pDYluc-A was co-transfected with pDYRK1A or psi-DY into N2a cells. Luciferase activity was measured at 24 h by a luminometer. The values represent the means ± S.E. (n = 4). *, p < 0.001 by Student's t test. Con, control.
To determine whether the transcriptional activity of REST was also affected by DYRK1A, we examined the expression of REST target gene Bdnf. Because Bdnf is a known gene repressed by REST, elevation of Bdnf would indicate the reduced transcriptional activity of REST. RT-PCR results showed that Bdnf mRNA was significantly increased by DYRK1A knockdown or overexpression by 215.0 ± 5.90% (p = 0.0012) and 106.9 ± 12.54% (p = 0.0083) (Fig. 5, C and D). Consistent with this, the promoter activity of BDNF was also significantly elevated by overexpression or knockdown of DYRK1A by 596.1 ± 45.99% (p < 0.0001) or 481.0 ± 12.34% (p < 0.0001) (Fig. 5E).
Considering the reduction of REST by DYRK1A as well as activation of DYRK1A transcription by REST, a negative feedback loop was formed in the regulation of DYRK1A, in which DYRK1A transcription should be indirectly affected by its own protein expression. Consistent with this, luciferase assay showed that the promoter activity of DYRK1A was reduced by either overexpression or knockdown of DYRK1A protein by 53.43 ± 11.06% (p < 0.0001) and 51.54 ± 9.10% (p = 0.0003) of controls, respectively (Fig. 5F), which is due to the reduced REST expression by DYRK1A imbalance. The sensitivity of DYRK1A expression to its own protein levels helps in maintaining the homeostasis of DYRK1A in cells, suggesting that DYRK1A may play vital roles in cell function.
DISCUSSION
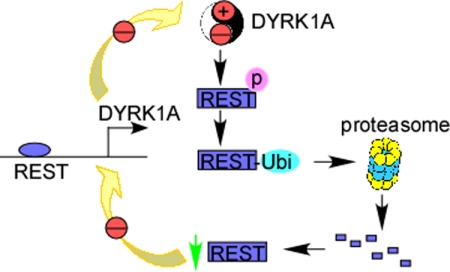
This is the first study to show that DYRK1A is activated by REST through a NRSE at −833 to −815 bp at its gene promoter. In this study, we also first showed that DYRK1A dosage imbalance can reduce REST expression through facilitating the ubiquitination and subsequent degradation of REST. Thus, a negative loop is formed in the regulation of DYRK1A by REST (Fig. 6).
FIGURE 6.
Schematic diagram of the cross-interaction of REST and DYRK1A. The imbalance of DYRK1A expression in cells increases the phosphorylation and subsequent ubiquitination of REST protein. Ubiquitinated REST can be degraded by proteasome, which results in the weaker transcription of DYRK1A via a NRSE site located at DYRK1A gene promoter. Thus, a negative feedback loop is formed in DYRK1A gene regulation.
DYRK1A is highly expressed in neuronal cells in central nervous system (39, 40). The Allen Brain Atlas showed a particularly high expression of DYRK1A in neuronal cells in hippocampus. In accordance with this, our data revealed that DYRK1A is enriched in neurons, particularly in neurons of hippocampus. The mechanism of DYRK1A gene regulation in the brain remains elusive. Here our data showed that DYRK1A is activated by REST, a key regulator of neurogenesis. The parallel expression of DYRK1A and REST during neurodevelopment further supports the regulatory role of REST in DYRK1A expression.
DYRK1A expression can be enhanced by treatment with β-amyloid or overexpression of the transcription factor E2F1 (29, 38). DYRK1A, located in the Down syndrome critical region on chromosome 21, can phosphorylate microtubule-associated protein Tau at several sites in cultured cells, DYRK1A transgenic mice, and adult DS brains (34–36). Amyloid precursor protein can also be phosphorylated by DYRK1A at Thr668 in vitro, and the Aβ production was increased in vitro and in vivo by DYRK1A overexpression (37). DYRK1A level was also shown to be significantly increased in hippocampus from Alzheimer disease patients (38). It remains to be examined whether DYRK1A will interact with REST and contribute to Alzheimer disease pathogenesis.
Overexpression of REST has been found in human medulloblastomas and neuroblastomas (16), in which it is thought to maintain the stem character of neural cells. Expression of a C terminus truncated form of REST has been shown to be associated with neuronal tumors and small cell lung carcinomas (30, 31). REST-FS, a frameshift mutant truncated at C terminus, was found to have oncogenic properties (32). REST level declines with neuronal differentiation from embryonic stem cells to neural stem and progenitor cells (19), indicating that REST is a key transcription factor in neurogenesis. These studies suggest that regulation of REST has important physiological and pathological consequences. Our studies here showed that DYRK1A can regulate REST expression from transcriptional as well as post-translational levels. The sensitivity of REST expression to either overexpression or knockdown of DYRK1A implies that DYRK1A is a vital factor in regulation of REST level. Consistent with this, recent reports showed that DYRK1A is elevated in DS patients because of a gene dosage effect, which induced a 30–60% reduction of REST in DS brains and subsequently disturbed the development of all embryonic lineages (23, 33). Neurobehavioral abnormalities observed in both transgenic and heterozygotes of DYRK1A indicate that the balance of DYRK1A is crucial to neurogenesis. Our study here showed that the reduction of REST may be the underlying mechanism through which neurobehavioral changes developed in DYRK1A transgenic or heterozygotes.
The activation of DYRK1A by REST and reduction of REST by DYRK1A together form a negative feedback loop in regulation of DYRK1A (Fig. 6). Because both transgenic and heterozygotes of DYRK1A showed neurobehavioral abnormalities, the negative feedback loop in the regulation of DYRK1A by REST is very important to the balance of DYRK1A expression as well as its function. This negative feedback loop also plays important roles in maintaining the homoeostasis of REST, supporting the vital roles of REST and DYRK1A in the central nervous system.
Acknowledgments
We thank Dr. Masaaki Tsuda for providing the pBDNFluc plasmid. We also thank Dr. Fan Yi for helpful discussions.
This work was supported by the National Natural Science Foundation of China.
- DS
- Down syndrome
- NRSE
- neuron-restrictive silencer element
- Pn
- postnatal day n.
REFERENCES
- 1. Tejedor F., Zhu X. R., Kaltenbach E., Ackermann A., Baumann A., Canal I., Heisenberg M., Fischbach K. F., Pongs O. (1995) Neuron 14, 287–301 [DOI] [PubMed] [Google Scholar]
- 2. Smith D. J., Stevens M. E., Sudanagunta S. P., Bronson R. T., Makhinson M., Watabe A. M., O'Dell T. J., Fung J., Weier H. U., Cheng J. F., Rubin E. M. (1997) Nat. Genet. 16, 28–36 [DOI] [PubMed] [Google Scholar]
- 3. Arron J. R., Winslow M. M., Polleri A., Chang C. P., Wu H., Gao X., Neilson J. R., Chen L., Heit J. J., Kim S. K., Yamasaki N., Miyakawa T., Francke U., Graef I. A., Crabtree G. R. (2006) Nature 441, 595–600 [DOI] [PubMed] [Google Scholar]
- 4. Gwack Y., Sharma S., Nardone J., Tanasa B., Iuga A., Srikanth S., Okamura H., Bolton D., Feske S., Hogan P. G., Rao A. (2006) Nature 441, 646–650 [DOI] [PubMed] [Google Scholar]
- 5. Deleted in proof.
- 6. Hämmerle B., Carnicero A., Elizalde C., Ceron J., Martínez S., Tejedor F. J. (2003) Eur. J. Neurosci. 17, 2277–2286 [DOI] [PubMed] [Google Scholar]
- 7. Deleted in proof.
- 8. Laguna A., Aranda S., Barallobre M. J., Barhoum R., Fernández E., Fotaki V., Delabar J. M., de la Luna S., de la Villa P., Arbonés M. L. (2008) Dev. Cell 15, 841–853 [DOI] [PubMed] [Google Scholar]
- 9. Seifert A., Allan L. A., Clarke P. R. (2008) FEBS J. 275, 6268–6280 [DOI] [PubMed] [Google Scholar]
- 10. Fernandez-Martinez J., Vela E. M., Tora-Ponsioen M., Ocaña O. H., Nieto M. A., Galceran J. (2009) J. Cell Sci. 122, 1574–1583 [DOI] [PubMed] [Google Scholar]
- 11. Altafaj X., Dierssen M., Baamonde C., Martí E., Visa J., Guimerà J., Oset M., González J. R., Flórez J., Fillat C., Estivill X. (2001) Hum. Mol. Genet. 10, 1915–1923 [DOI] [PubMed] [Google Scholar]
- 12. Kim M. Y., Jeong B. C., Lee J. H., Kee H. J., Kook H., Kim N. S., Kim Y. H., Kim J. K., Ahn K. Y., Kim K. K. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13074–13079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fotaki V., Dierssen M., Alcántara S., Martínez S., Martí E., Casas C., Visa J., Soriano E., Estivill X., Arbonés M. L. (2002) Mol. Cell. Biol. 22, 6636–6647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fotaki V., Martínez De Lagrán M., Estivill X., Arbonés M., Dierssen M. (2004) Behav. Neurosci. 118, 815–821 [DOI] [PubMed] [Google Scholar]
- 15. Chong J. A., Tapia-Ramírez J., Kim S., Toledo-Aral J. J., Zheng Y., Boutros M. C., Altshuller Y. M., Frohman M. A., Kraner S. D., Mandel G. (1995) Cell 80, 949–957 [DOI] [PubMed] [Google Scholar]
- 16. Lawinger P., Venugopal R., Guo Z. S., Immaneni A., Sengupta D., Lu W., Rastelli L., Marin Dias Carneiro A., Levin V., Fuller G. N., Echelard Y., Majumder S. (2000) Nat. Med. 6, 826–831 [DOI] [PubMed] [Google Scholar]
- 17. Bahn S., Mimmack M., Ryan M., Caldwell M. A., Jauniaux E., Starkey M., Svendsen C. N., Emson P. (2002) Lancet 359, 310–315 [DOI] [PubMed] [Google Scholar]
- 18. Schoenherr C. J., Anderson D. J. (1995) Science 267, 1360–1363 [DOI] [PubMed] [Google Scholar]
- 19. Ballas N., Grunseich C., Lu D. D., Speh J. C., Mandel G. (2005) Cell 121, 645–657 [DOI] [PubMed] [Google Scholar]
- 20. Sun Y. M., Greenway D. J., Johnson R., Street M., Belyaev N. D., Deuchars J., Bee T., Wilde S., Buckley N. J. (2005) Mol. Biol. Cell 16, 5630–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guardavaccaro D., Frescas D., Dorrello N. V., Peschiaroli A., Multani A. S., Cardozo T., Lasorella A., Iavarone A., Chang S., Hernando E., Pagano M. (2008) Nature 452, 365–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Westbrook T. F., Hu G., Ang X. L., Mulligan P., Pavlova N. N., Liang A., Leng Y., Maehr R., Shi Y., Harper J. W., Elledge S. J. (2008) Nature 452, 370–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Canzonetta C., Mulligan C., Deutsch S., Ruf S., O'Doherty A., Lyle R., Borel C., Lin-Marq N., Delom F., Groet J., Schnappauf F., De Vita S., Averill S., Priestley J. V., Martin J. E., Shipley J., Denyer G., Epstein C. J., Fillat C., Estivill X., Tybulewicz V. L., Fisher E. M., Antonarakis S. E., Nizetic D. (2008) Am. J. Hum. Genet. 83, 388–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lepagnol-Bestel A. M., Zvara A., Maussion G., Quignon F., Ngimbous B., Ramoz N., Imbeaud S., Loe-Mie Y., Benihoud K., Agier N., Salin P. A., Cardona A., Khung-Savatovsky S., Kallunki P., Delabar J. M., Puskas L. G., Delacroix H., Aggerbeck L., Delezoide A. L., Delattre O., Gorwood P., Moalic J. M., Simonneau M. (2009) Hum. Mol. Genet. 18, 1405–1414 [DOI] [PubMed] [Google Scholar]
- 25. Sun X., Wang Y., Qing H., Christensen M. A., Liu Y., Zhou W., Tong Y., Xiao C., Huang Y., Zhang S., Liu X., Song W. (2005) FASEB J. 19, 739–749 [DOI] [PubMed] [Google Scholar]
- 26. Liu H., Wang P., Song W., Sun X. (2009) FASEB J. 23, 3383–3392 [DOI] [PubMed] [Google Scholar]
- 27. Hara D., Fukuchi M., Miyashita T., Tabuchi A., Takasaki I., Naruse Y., Mori N., Kondo T., Tsuda M. (2009) Biochem. Biophys. Res. Commun. 384, 506–511 [DOI] [PubMed] [Google Scholar]
- 28. Ballas N., Battaglioli E., Atouf F., Andres M. E., Chenoweth J., Anderson M. E., Burger C., Moniwa M., Davie J. R., Bowers W. J., Federoff H. J., Rose D. W., Rosenfeld M. G., Brehm P., Mandel G. (2001) Neuron 31, 353–365 [DOI] [PubMed] [Google Scholar]
- 29. Maenz B., Hekerman P., Vela E. M., Galceran J., Becker W. (2008) BMC Mol. Biol. 9, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Coulson J. M., Edgson J. L., Woll P. J., Quinn J. P. (2000) Cancer Res. 60, 1840–1844 [PubMed] [Google Scholar]
- 31. Gurrola-Diaz C., Lacroix J., Dihlmann S., Becker C. M., von Knebel Doeberitz M. (2003) Oncogene 22, 5636–5645 [DOI] [PubMed] [Google Scholar]
- 32. Westbrook T. F., Martin E. S., Schlabach M. R., Leng Y., Liang A. C., Feng B., Zhao J. J., Roberts T. M., Mandel G., Hannon G. J., Depinho R. A., Chin L., Elledge S. J. (2005) Cell 121, 837–848 [DOI] [PubMed] [Google Scholar]
- 33. Dowjat W. K., Adayev T., Kuchna I., Nowicki K., Palminiello S., Hwang Y. W., Wegiel J. (2007) Neurosci. Lett. 413, 77–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Azorsa D. O., Robeson R. H., Frost D., Meec hoovet B., Brautigam G. R., Dickey C., Beaudry C., Basu G. D., Holz D. R., Hernandez J. A., Bisanz K. M., Gwinn L., Grover A., Rogers J., Reiman E. M., Hutton M., Stephan D. A., Mousses S., Dunckley T. (2010) BMC Genomics 11, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu F., Liang Z., Wegiel J., Hwang Y. W., Iqbal K., Grundke-Iqbal I., Ramakrishna N., Gong C. X. (2008) FASEB J. 22, 3224–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ryoo S. R., Jeong H. K., Radnaabazar C., Yoo J. J., Cho H. J., Lee H. W., Kim I. S., Cheon Y. H., Ahn Y. S., Chung S. H., Song W. J. (2007) J. Biol. Chem. 282, 34850–34857 [DOI] [PubMed] [Google Scholar]
- 37. Ryoo S. R., Cho H. J., Lee H. W., Jeong H. K., Radnaabazar C., Kim Y. S., Kim M. J., Son M. Y., Seo H., Chung S. H., Song W. J. (2008) J. Neurochem. 104, 1333–1344 [DOI] [PubMed] [Google Scholar]
- 38. Kimura R., Kamino K., Yamamoto M., Nuripa A., Kida T., Kazui H., Hashimoto R., Tanaka T., Kudo T., Yamagata H., Tabara Y., Miki T., Akatsu H., Kosaka K., Funakoshi E., Nishitomi K., Sakaguchi G., Kato A., Hattori H., Uema T., Takeda M. (2007) Hum. Mol. Genet. 16, 15–23 [DOI] [PubMed] [Google Scholar]
- 39. Martí E., Altafaj X., Dierssen M., de la Luna S., Fotaki V., Alvarez M., Pérez-Riba M., Ferrer I., Estivill X. (2003) Brain Res. 964, 250–263 [DOI] [PubMed] [Google Scholar]
- 40. Okui M., Ide T., Morita K., Funakoshi E., Ito F., Ogita K., Yoneda Y., Kudoh J., Shimizu N. (1999) Genomics 62, 165–171 [DOI] [PubMed] [Google Scholar]