Abstract
GPR30, also known as GPER, has been suggested to mediate rapid effects induced by estrogens in diverse normal and cancer tissues. Hypoxia is a common feature of solid tumors involved in apoptosis, cell survival, and proliferation. The response to low oxygen environment is mainly mediated by the hypoxia-inducible factor named HIF-1α, which activates signaling pathways leading to adaptive mechanisms in tumor cells. Here, we demonstrate that the hypoxia induces HIF-1α expression, which in turn mediates the up-regulation of GPER and its downstream target CTGF in estrogen receptor-negative SkBr3 breast cancer cells and in HL-1 cardiomyocytes. Moreover, we show that HIF-1α-responsive elements located within the promoter region of GPER are involved in hypoxia-dependent transcription of GPER, which requires the ROS-induced activation of EGFR/ERK signaling in both SkBr3 and HL-1 and cells. Interestingly, the apoptotic response to hypoxia was prevented by estrogens through GPER in SkBr3 cells. Taken together, our data suggest that the hypoxia-induced expression of GPER may be included among the mechanisms involved in the anti-apoptotic effects elicited by estrogens, particularly in a low oxygen microenvironment.
Keywords: Breast Cancer, Estrogen, Gene Regulation, Hypoxia, Receptors, Signal Transduction
Introduction
Estrogens are a group of pleiotropic steroid hormones that regulate multiple physiological activities in different tissues (1). A large body of data has suggested that estrogens play a pivotal role also in the development of breast cancer through a complex signaling network (2). In the cardiovascular system, it has been reported that estrogens may exert a protective action against ischemia (3, 4) and related diseases that occur in premenopausal women (3) (reviewed in 5). In this context, oxidative stress and the subsequent generation of reactive oxygen species (ROS)3 are thought to trigger cardiomyocyte apoptosis (6), whereas the ability of 17β-estradiol (E2) to counter the redox intermediates has been considered as a principal factor of overall cardioprotection (reviewed in Ref. 5).
The multiple effects exerted by estrogens are mainly mediated through two well characterized members of the nuclear receptor superfamily, the estrogen receptor α (ERα) and ERβ. However, the membrane-associated G protein-coupled receptor, GPR30/GPER, has recently been suggested to mediate estrogen effects through the activation of the EGFR/MAPK signaling cascade (7–10). Not only does GPER trigger rapid events following estrogen stimulation, such as calcium mobilization and kinase activation, but accumulating evidence also indicates that GPER signaling promotes the transcription of numerous target genes (9). In this regard, we have recently characterized the gene expression profile mediated by GPER activation in breast cancer cells, determining that one of the most induced genes is the connective tissue growth factor (CTGF) (11), which has been previously implicated in cell proliferation and migration (12). Regarding the transcriptional regulation of GPER expression, our recent studies have demonstrated that EGF and TGFα are able to transactivate the promoter of GPER and up-regulate its mRNA and protein levels in cancer cells (13, 14).
Hypoxia is a common feature of solid tumors, where it promotes relevant biological adaptive responses, such as angiogenesis, anaerobic glycolysis, and the reduction of macromolecule synthesis (15–17). Mechanisms of sensing and adaptation to low oxygen tension have been extensively studied in the mammalian system. Not only does hypoxia represent a fundamental physiologic response, but it is also critical to the pathogenesis of the major causes of human mortality, such as heart disease and stroke. A great deal of progress has been made in the understanding of oxygen-regulated gene expression. In this regard, the hypoxia-inducible factor HIF-1 has been identified as the principal mediator of the cellular oxygen response that activates signaling pathways involved in the adaptation to hypoxic conditions (18–20). Indeed, HIF-1 is a highly conserved transcription factor that plays an important role in normal growth and development as well as in the pathophysiology of inflammation, ischemia, and cancer (17). HIF-1 is a heterodimeric factor composed of hypoxia-inducible HIF-1α and the constitutively expressed HIF-1β. Under hypoxic conditions, the two subunits dimerize, inducing the nuclear translocation of the HIF-1 complex, which binds to the hypoxia-responsive elements (HREs) located within the promoter region of target genes (17). Given that cells and organs need to adapt to changes in oxygen supply, it would not be surprising that a significant variety of HIF-1α target genes is regulated in a tissue-specific manner. To date, over 100 HIF-1α downstream genes, including the above mentioned CTGF (21), have been identified and attributed with varying functions, such as angiogenesis, matrix metabolism, cell proliferation/survival, and apoptosis (reviewed in Refs. 22 and 23). Moreover, the activation of HIF-1α has been closely associated with a variety of tumors and oncogenic pathways (reviewed in Refs. 22 and 24). Overexpression of HIF-1α has been found in various cancers probably as a consequence of intratumoral hypoxia or genetic alterations (24–27). The interior of the tumor mass becomes progressively hypoxic as its size increases and adequate neovascularization is obtained by tumors. Hypoxic conditions within tumors can result in increased HIF-1α expression, stability, and activity (26, 28). A correlation between HIF-1α overexpression and patient mortality, poor prognosis, or treatment resistance has been noted in many investigations (reviewed in Ref. 17).
In the present study, we demonstrate that the hypoxia-induced HIF-1α-mediated pathway up-regulates GPER expression in breast cancer cells and in cardiomyocytes. Estrogenic GPER signaling may therefore play a role in the adaptive response to hypoxia in cell contexts characterized by a low oxygen environment.
EXPERIMENTAL PROCEDURES
Materials
Cobalt Chloride (CoCl2), E2, and the ROS scavenger N-acetyl-l-cysteine (NAC) were purchased from Sigma-Aldrich Srl. (Milan, Italy). Tyrphostin AG1478 was bought from Biomol Research Laboratories, Inc. (Milan, Italy). PD98059 was obtained from Calbiochem (Milan, Italy). All compounds were dissolved in DMSO, except for E2, which was dissolved in ethanol, and NAC, which was solubilized in water.
Cell Cultures
The SkBr3 breast cancer cells were maintained in RPMI 1640 (Invitrogen) without phenol red, supplemented with 10% fetal bovine serum (FBS) and 100 μg/ml penicillin/streptomycin. The murine cardiomyocyte-like cell line HL-1 was kindly provided by Dr. William C. Claycomb (Louisiana State University Medical Center, New Orleans, LA). HL-1 cells were cultured according to the published protocol (29) in Claycomb medium (JRH Biosciences, Sigma-Aldrich) supplemented with 10% fetal bovine serum (JRH Bioscience, Sigma-Aldrich), 100 μg/ml penicillin/streptomycin, 0.1 mm norepinephrine (Sigma-Aldrich), and 2 mm l-glutamine (Invitrogen). All cell lines were grown in a 37 °C incubator with 5% CO2. For hypoxic stimulation, cells were treated with CoCl2 or cultured in the presence of low oxygen tension (2% O2) in a HeraCell incubator (ThermoScientific-Heraeus, Milan, Italy).
Gene Reporter Assays
To generate the luciferase expression vector for the GPER 5′-flanking region (pGPER 2.9 kb), a 3000-bp fragment next to the 5′-flanking region of the GPER gene was amplified by PCR using the following primer pairs: forward (MluI), 5′- CGACGCGTGCCCCAGTCACTCTCACCAACC-3′; reverse (HindIII), 5′- GCAAGCTTCTTGAAGTGAGCCTGGCATTTGTC-3′. Genomic DNA was extracted from SkBr3 cells by TRIzol reagent, as suggested by the manufacturer (Invitrogen). PCR primer pairs were selected analyzing the 5′-flanking region of the GPER gene in chromosome 7, location 7p22.3. The PCR amplification was performed using 1.25 units of GoTaq DNA polymerase according to the manufacturer's instructions (Promega, Milan, Italy). PCR conditions were as follows: 5 min at 95 °C followed by 1 min at 94 °C, 1 min at 58 °C, and 1 min at 72 °C for 30 cycles. The fragment was then inserted in the pCR 2.1 plasmid using the TA cloning kit (Invitrogen), sequenced, and cut with HindIII and XhoI. The insert was cloned in the pGL3 basic vector (Promega). Analyses of the 3000-bp GPER 5′-flanking region revealed three hypoxia-responsive elements between −1828 and −2817 bp from the GPER gene transcription start site.
The GPER luciferase expression vector (pGPER 641 bp) was described previously (13). The CTGF luciferase reporter plasmid pCTGF (−1999/+36)-luc (30), which is based on the backbone of vector pGL3-basic (Promega), was a gift from Dr. B. Chaqour.
SkBr3 cells (1 × 105) were plated into 24-well dishes with 500 μl/well RPMI1640 containing 10% FBS and transfected for 24 h. Transfections were performed using FuGENE 6 reagent as recommended by the manufacturer (Roche Applied Science) with a mixture containing 0.5 μg of reporter plasmid and 10 ng of pRL-TK. After 24 h, the medium was replaced with serum-free RPMI1640 lacking phenol red, and cells were treated with CoCl2 as indicated; luciferase activity was measured with the Dual Luciferase Kit (Promega) normalized to the internal transfection control provided by Renilla luciferase activity. The normalized relative light unit values obtained from cells treated with vehicle were set as 1-fold induction, from which the activity induced by treatments was calculated.
Gene Expression Studies
Total RNA was extracted from cell cultures using the TRIzol commercial kit (Invitrogen) according to the manufacturer's protocol. RNA was quantified spectrophotometrically, and quality was checked by electrophoresis through agarose gels stained with ethidium bromide. Only samples that were not degraded and showed clear 18 and 28 S bands under UV light were used for RT-PCR. Total cDNA was synthesized from the RNA by reverse transcription using the murine leukemia virus reverse transcriptase (Invitrogen) following the protocol provided by the manufacturer. The expression of selected genes was quantified by real-time PCR using the Step OneTM sequence detection system (Applied Biosystems Inc., Milan, Italy), following the manufacturer's instructions. Gene-specific primers were designed using Primer Express version 2.0 software (Applied Biosystems Inc.) and are as follows: HIF-1α, GPER, CTGF, and the ribosomal protein 18 S, which was used as a control gene to obtain normalized values. Primer sequences are as follows: HIF1-α (human, mouse) forward (5′-TGCATCTCCATCTTCTACCCAAGT-3′) and reverse (5′-CCGACTGTGAGTGCCACTGT-3′); GPER (human) forward (5′-ACACACCTGGGTGGACACAA-3′) and reverse (5′-GGAGCCAGAAGCCACATCTG-3′); GPER (mouse) forward (5′-CCTGGACGAGCAGTATTACGATATC-3′) and reverse (5′-TGCTGTACATGTTGATCTG-3′); CTGF (human) forward (5′-ACCTGTGGGATGGGCATCT-3′) and reverse (5′-CAGGCGGCTCTGCTTCTCTA-3′); CTGF (mouse) forward (5′-CATTAAGAAGGGCAAAAAGTGCAT-3′) and reverse (5′-TGCAGCCAGAAAGCTCAAACT-3′); 18 S (human, mouse) forward (5′-GGCGTCCCCCAACTTCTTA-3′) and reverse (5′-GGGCATCACAGACCTGTTATT-3′).
Assays were performed in triplicate and the results were normalized for 18 S expression and then calculated as -fold induction of RNA expression. For all experiments, cells were switched to medium without serum 24 h before treatments.
Western Blot Analysis
To prepare lysates, HL-1 and SkBr3 cells were washed in PBS and solubilized with 50 mm Hepes solution, pH 7.4, containing 1% (v/v) Triton X-100, 4 mm EDTA, 1 mm sodium fluoride, 0.1 mm sodium orthovanadate, 2 mm PMSF, 10 μg/ml leupeptin, and 10 μg/ml aprotinin. Protein concentrations in the supernatant were determined according to the Bradford method. Cell lysates (10–50 μg of protein) were electrophoresed through a reducing SDS/10% (w/v) polyacrylamide gel and electroblotted onto a nitrocellulose membrane. After the transfer, the membranes were stained with Red Ponceau to confirm equal loading and transfer. Membranes were blocked and incubated with primary polyclonal IgG antibody for HIF-1α (R&D Systems, Inc., Celbio, Milan, Italy), human GPER (LS-A4271) (MBL-Eppendorf, Milan, Italy), mouse GPER (N-15), CTGF (L-20), phosphorylated ERK1/2 (E-4), ERK2 (C-14), phosphorylated EGFR (Tyr1173), EGFR (1005), β-tubulin and β-actin (C2), and appropriate secondary HRP-conjugated antibodies, all purchased from Santa Cruz Biotechnology, Inc. (DBA, Milan, Italy). The levels of proteins and phosphoproteins were detected with horseradish peroxidase-linked secondary antibodies and revealed using the ECL® system (GE Healthcare, Milan, Italy).
Gene Silencing Experiments
Cells were plated onto 10-cm dishes and maintained in serum-free medium for 24 h. Then cells were transfected for 24 h before treatments with a control vector or an independent shRNA sequence for each target gene using Fugene6 (Roche Applied Science). The shRNA plasmid for EGFR and the shRNA plasmid for HIF-1α and the respective control plasmids were purchased from SABioscience Corp. (Frederick, MD). The silencing of GPER expression was obtained by the construct that we have previously described and used (13).
Chromatin Immunoprecipitation (ChIP) Assay
SkBr3 cells were grown in 10-cm dishes to 70–80% confluence, shifted to serum-free medium for 24 h, and then treated with vehicle or 100 μm CoCl2 for 2 h. Thereafter, cells were cross-linked with 1% formaldehyde and sonicated. Supernatants were immunocleared with salmon DNA/protein A-agarose (Upstate Biotechnology, Inc., Lake Placid, NY) and immunoprecipitated with anti-HIF1-α antibody (R&D Systems, Inc.) or nonspecific IgG (Santa Cruz Biotechnology, Inc.). Pellets were washed, eluted with a buffer consisting of 1% SDS and 0.1 mol/liter NaHCO3, and digested with proteinase K. DNA was obtained by phenol/chloroform extractions and precipitated with ethanol. A 4-μl volume of each sample was used as template to amplify by PCR a fragment located within a 3000-bp fragment next to the 5′-flanking region of the GPER gene. The primer pair used to amplify this fragment containing HRE sequences was as follows: 5′-CACAGACCTTCACAGCCATC-3′ (forward hHRE) and 5′-CCCAGCGTAGACAGTTGAGT-3′ (reverse hHRE). The amplification product obtained following 35 amplification cycles was analyzed using a 1.2% agarose gel and visualized by ethidium bromide staining. A total of 2 μl of the initial preparation of soluble chromatin was amplified to control input DNA before precipitation.
Immunofluorescence Microscopy
Fifty percent confluent cultured SkBr3 and HL-1 cells grown on coverslips were serum-deprived for 24 h and treated for 8 h with 100 μm CoCl2 or exposed for 8 h to low oxygen tension (2% O2). Then cells were fixed in 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, washed three times with PBS, and incubated overnight with a rabbit primary antibody against GPER (1:500). After incubation, the slides were extensively washed with PBS and incubated with propidium iodide (1:1000; Sigma-Aldrich) and donkey anti-rabbit IgG-FITC (1:500; purchased from Santa Cruz Biotechnology, Inc.). For SkBr3 immunofluorescence experiments, cells were previously transfected for 24 h with shHIF-1α, shGPER, and the respective negative control plasmids (as described above) and then treated for 8 h with 100 μm CoCl2 as indicated.
TUNEL Assay
70% confluent cultured SkBr3 cells grown on coverslips were serum-deprived and transfected for 24 h with shRNA plasmids for GPER or empty control vectors, pretreated for 1 h with E2 (10 nm), and then treated with CoCl2 (100 μm) for 18 h. Therefore, cells were fixed in 4% buffered paraformaldehyde (pH 7.4) for 30 min. Slides were rinsed twice in PBS, pH 7.4. For the detection of DNA fragmentation at the cellular level, cells were stained using the DeadEndTM fluorometric TUNEL system (Promega) following the manufacturer's instructions, as previously described (31). Nuclei of cells were stained with propidium iodide. The Leica AF6000 advanced fluorescence imaging system supported by quantification and image processing software (Leica Application Suite Advanced Fluorescence (Leica Microsystems CMS, GbH Mannheim, Germany)) was used for experiment evaluation.
Statistical Analysis
Statistical analysis was done using analysis of variance followed by Newman-Keuls testing to determine differences in means. p < 0.05 was considered as statistically significant.
RESULTS
Hypoxia Transactivates the Promoter of GPER and Up-regulates Its Expression
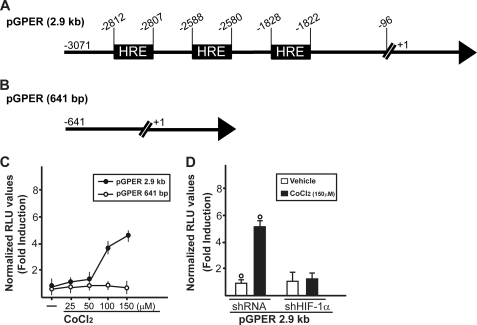
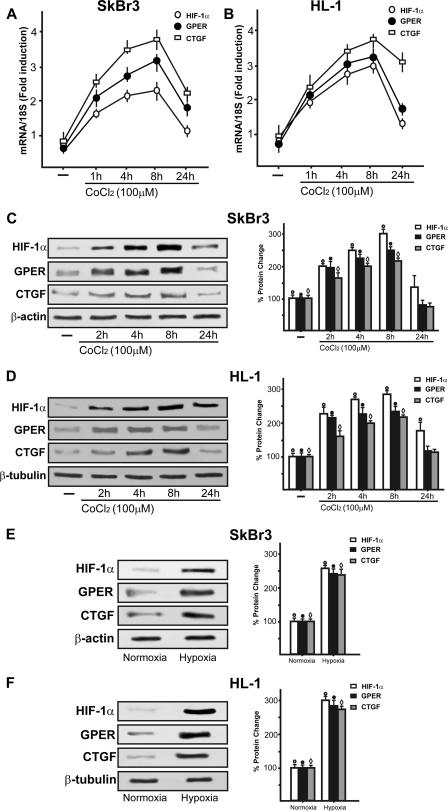
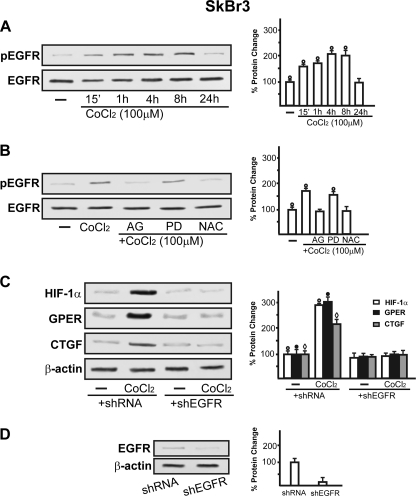
To further characterize the regulation and function of the novel estrogen receptor GPER, we evaluated its potential role in the adaptive response to hypoxia, which is a fundamental feature of both cancer and heart disease. Using a bioinformatic approach, we examined the promoter region of the human GPER gene and identified three potential core hypoxia-inducible HREs located upstream of the transcription start site (ENSG00000164850). On the basis of this observation, we cloned an expression vector encoding a long GPER 5′-flanking region (Fig. 1A), together with a short construct lacking the HREs (Fig. 1B). In order to evaluate the transcriptional responses to the well known hypoxia-mimetic agent CoCl2, we transiently transfected the GPER promoter plasmids in SkBr3 breast cancer cells, which lack ERα and ERβ but express GPER as we and others assessed in previous studies (7–9, 11, 13, 14). Starting from a concentration of 100 μm, CoCl2 transactivated the 2.9-kb GPER promoter sequence, whereas the shorter 641-bp GPER construct lacking the HRE motifs did not evidence any luciferase activity (Fig. 1C). Furthermore, we ascertained that HIF-1α is involved in the transactivation of the GPER promoter by CoCl2 because the knockdown of HIF-1α (see below) inhibited the transcriptional response (Fig. 1D). In accordance with the aforementioned results, CoCl2 was able to stimulate in SkBr3 cells the mRNA and protein expression of HIF-1α, GPER, and its downstream target CTGF (11) in a time-dependent manner (Fig. 2, A and C). In order to corroborate these findings, we turned to a completely different model system. In HL-1 cardiomyocytes (29), CoCl2 induced the up-regulation of these genes as observed in SkBr3 cells (Fig. 2, B and D). Moreover, incubating both SkBr3 and HL-1 cells in the presence of low oxygen tension (2% O2), we observed gene stimulations (Fig. 2, E and F) similar to those obtained using CoCl2.
FIGURE 1.
Hypoxia transactivates the GPER promoter in SkBr3 breast cancer cells. Shown are gene reporter constructs encoding (A) or lacking (B) putative HRE elements within the GPER-5′-flanking region. C, SkBr3 cells were transfected with the 2.9-kb GPER promoter plasmid described in A or with the 641-bp GPER promoter plasmid described in B. Then cells were treated with increasing concentrations of the hypoxia-mimetic agent CoCl2. D, the response of the 2.9-kb GPER promoter plasmid to CoCl2 was abrogated by silencing the expression of HIF-1α using a specific shHIF-1α. The luciferase activities were normalized to the internal transfection control, and values of cells receiving vehicle (−) were set as 1-fold induction, upon which the activity induced by treatments was calculated. Each data point represents the mean ± S.D. (error bars) of three independent experiments performed in triplicate. ○, p < 0.05 for cells receiving vehicle (−) versus treatment.
FIGURE 2.
Hypoxia induces HIF-1α, GPER, and CTGF expression in a time-dependent manner in SkBr3 and HL-1 cells. CoCl2 up-regulates the expression of HIF-1α, GPER, and CTGF at both the mRNA and protein level, as evaluated by real time PCR (A and B) and immunoblotting (C and D). HIF-1α, GPER, and CTGF protein expressions were also induced by exposing SkBr3 and HL-1 cells to low oxygen tension (2% O2) for 8 h (E and F). The side panels show densitometric analysis of the blots normalized to β-actin or β-tubulin. Each data point represents the mean ± S.D. (error bars) of three independent experiments. ○, ●, and ♢, p < 0.05 for cells receiving vehicle (−) versus treatments or cells exposed to 2% O2 with respect to cells incubated under normoxia. In RNA experiments, gene expression was normalized to 18 S expression, and results are shown as -fold changes of mRNA expression compared with cells treated with vehicle.
HIF-1α Is Involved in the Transcriptional Regulation of Both GPER and CTGF
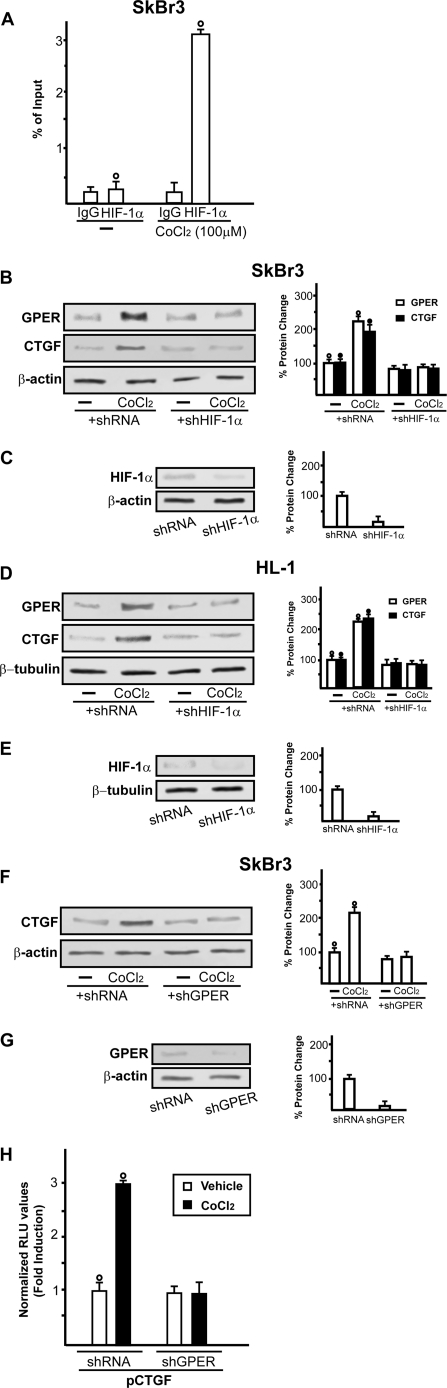
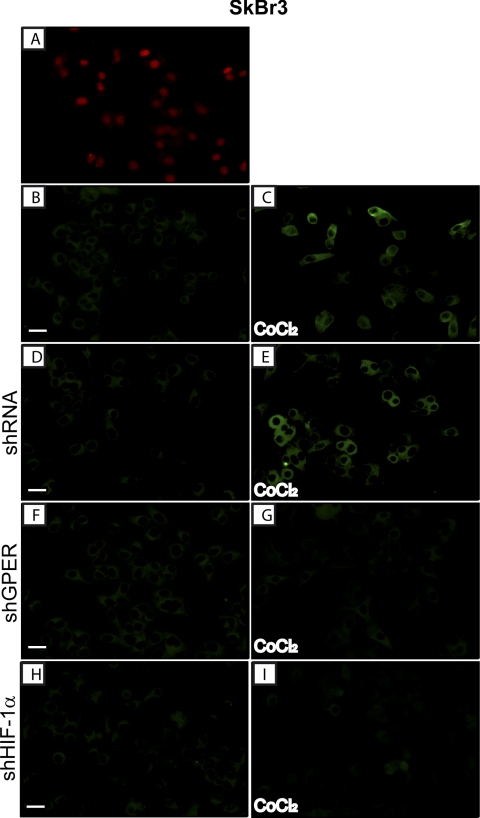
Considering that the knockdown of HIF-1α abolished the ability of CoCl2 to transactivate the GPER promoter sequence, we evaluated by ChIP assay whether HIF-1α is recruited to the GPER promoter region in SkBr3 cells. Notably, a 2-h CoCl2 treatment induced the recruitment of HIF-1α to the distal HRE region (−2812 to −2807) (Fig. 3A), indicating that HIF-1α is involved in the activation of the GPER promoter by CoCl2. In agreement with the above mentioned results, HIF-1α silencing abolished the protein expression of GPER as well as that of the GPER downstream target CTGF in both SkBr3 (Fig. 3B) and HL-1 cells (Fig. 3D). Further supporting these findings, CoCl2 was no longer able to induce CTGF protein levels and transactivate the promoter of CTGF knocking down GPER expression (Fig. 3, F and H). Next, the accumulation of GPER in the cytoplasm of both SkBr3 and HL-1 cells upon exposure to CoCl2 or hypoxia (2% O2) was evidenced by immunofluorescence studies (Fig. 4, B and C, and supplemental Figs. 1–3). The specificity of the GPER signal was ascertained using a selective shGPER, which abrogated the up-regulation of GPER by CoCl2 (Fig. 4, D–G), whereas the involvement of HIF-1α in this response was confirmed by silencing HIF-1α expression (Fig. 4, H and I).
FIGURE 3.
A, CoCl2 induces the recruitment of HIF-1α to the HRE site located in the GPER promoter sequence in SkBr3 cells. In control samples, nonspecific IgG was used instead of the primary antibody. Each data point represents the mean ± S.D. (error bars) of three independent experiments. ○, p < 0.05 for cells receiving vehicle (−) versus treatment. B and D, representative immunoblots of GPER and CTGF expression from SkBr3 and HL-1 cells transfected with control shRNA or shHIF-1α and treated for 8 h with vehicle (−) or 100 μm CoCl2. C and E, show the efficacy of HIF-1α silencing obtained using shHIF-1α. F, representative immunoblots of CTGF from SkBr3 cells transfected with control shRNA and shGPER and treated for 8 h with vehicle (−) or 100 μm CoCl2. G, shows the efficacy of GPER silencing obtained using shGPER. Side panels show densitometric analysis of the blots normalized to β-actin or β-tubulin. Each data point represents the mean ± S.D. of three independent experiments. H, SkBr3 cells were transfected with CTGF promoter construct and control shRNA or shGPER, and then cells were treated with 100 μm CoCl2. The luciferase activities were normalized to the internal transfection control, and values of cells receiving vehicle (−) were set as 1-fold induction, upon which the activity induced by treatments was calculated. Each data point represents the mean ± S.D. of three independent experiments performed in triplicate. ○ and ●, p < 0.05 for cells receiving vehicle (−) versus treatments.
FIGURE 4.
Evaluation of GPER expression by immuofluorescent microscopy in SkBr3 cells fixed, permeabilized, and stained with anti-GPER antibody. A, nuclei were stained by propidium iodide (PI; red). B and C, cells were treated for 8 h with vehicle (−) or 100 μm CoCl2, as indicated; GPER accumulation was evidenced by the green signal. SkBr3 cells were transfected with control shRNA (D and E), with shGPER (F and G) or shHIF-1α (H and I) and treated as described above and then stained with GPER antibody. Each experiment shown is representative of 20 random fields. Data are representative of three independent experiments.
The EGFR/ERK Pathway Is Involved in Hypoxia-stimulated GPER Expression
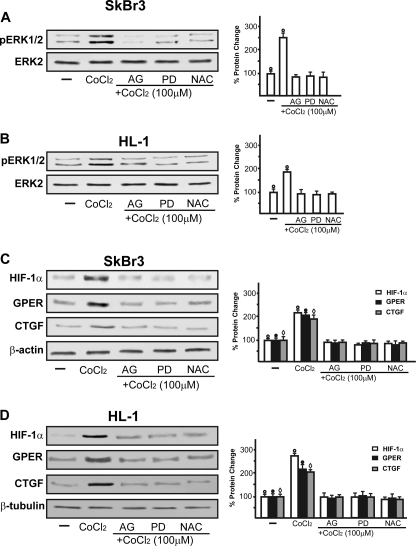
On the basis of previous studies showing that hypoxia induces mitochondrial ROS levels that may serve as second messengers in triggering signaling cascades (32–36), using SkBr3 cells, we demonstrated that CoCl2 promotes EGFR phosphorylation in a time-dependent manner (Fig. 5A). This response was abrogated in the presence of the EGFR inhibitor AG1478 and the ROS scavenger NAC but not using the ERK inhibitor PD98059 (Fig. 5B). Accordingly, the silencing of EGFR expression prevented HIF-1α, GPER, and CTGF up-regulation upon exposure to CoCl2 (Fig. 5C). Taken together, these findings suggest that EGFR transactivation acts as an upstream mediator of hypoxia-induced signaling. Next, the rapid ERK phosphorylation induced in both SkBr3 and HL-1 cells by CoCl2 was blocked in the presence of the EGFR and ERK inhibitors AG1478 and PD98059, respectively, as well as using the ROS scavenger NAC (Fig. 6, A and B). These pharmacological inhibitors also prevented the induction of HIF-1α, GPER, and CTGF by CoCl2 in SkBr3 (Fig. 6C) and HL-1 cells (Fig. 6D). Collectively, our results indicate that ROS-mediated EGFR/ERK activation by CoCl2 triggers intracellular signals that lead to the up-regulation of HIF-1α, GPER, and CTGF in both SkBr3 and HL-1 cells.
FIGURE 5.
A, exposure to CoCl2 induces EGFR phosphorylation in a time-dependent manner. B, EGFR activation by 100 μm CoCl2 was prevented by treating cells with 10 μm EGFR inhibitor AG1478 (AG) or 300 μm free radical scavenger NAC. C, cells were transfected with a control shRNA or shEGFR and then treated for 8 h with vehicle (−) or 100 μm CoCl2. D, shows the efficacy of EGFR silencing using shEGFR. All immunoblots shown are representative of three independent experiments. The side panels show densitometric analysis of the blots normalized to β-actin. ○, ●, and ♢, p < 0.05 for cells receiving vehicle (−) versus treatments. Error bars, S.D.
FIGURE 6.
A and B, immunoblots of pERK1/2 from SkBr3 (A) or HL-1 cells (B) treated for 15 min with vehicle (−) or 100 μm CoCl2 alone and in combination with 10 μm EGFR inhibitor AG1478 (AG), 10 μm ERK inhibitor PD98059 (PD), and 300 μm free radical scavenger NAC. Shown are immunoblots of HIF-1α, GPER, and CTGF from SkBr3 (C) and HL-1 (D) cells treated for 8 h with vehicle (−) or 100 μm CoCl2 alone and in combination with 10 μm EGFR inhibitor AG1478, 10 μm ERK inhibitor PD98059, and 300 μm free radical scavenger NAC. All immunoblots shown are representative of experiments performed in triplicate. The side panels show densitometric analysis of the blots normalized to total ERK/2, β-actin, or β-tubulin. ○, ●, and 224, p < 0.05 for cells receiving vehicle (−) versus treatments. Error bars, S.D.
Hypoxia-induced Apoptosis Is Prevented by E2 through GPER
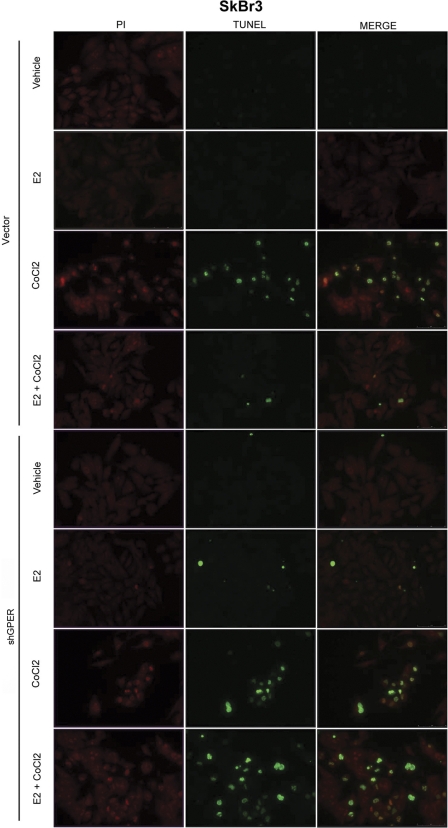
It has been shown that in different cell contexts, E2 may protect cells from the apoptosis induced by hypoxia (37–39), which, interestingly, herein we have for the first time linked to GPER induction. In order to evaluate the involvement of GPER in mediating the protective effects elicited by E2 in hypoxic conditions, we performed TUNEL assays in SkBr3 cells following treatment with CoCl2, a known inducer of apoptosis (40) (Fig. 7). Approximately 70% of SkBr3 cells were positive for TUNEL staining upon exposure to CoCl2 (Fig. 7 and supplemental Fig. 4); however, E2 significantly reduced the percentage of apoptotic cells (Fig. 7). It is worth noting that these protective effects elicited by E2 were no longer evident knocking down GPER expression (Fig. 7 and supplemental Fig. 4). Altogether, our results indicate that the adaptive cell response to hypoxia may include the regulation of GPER, which in turn mediates the protective action exerted by E2 against apoptosis.
FIGURE 7.
E2 prevented CoCl2-induced apoptosis through GPER in SkBr3 cells. Apoptotic changes were detected using TUNEL (green) and propidium iodide (PI; red) staining after an 18-h treatment with vehicle, 10 nm E2, or 100 μm CoCl2 alone and in combination as well as in the presence of control shRNA or shGPER. Each experiment shown is representative of 20 random fields. Data are representative of three independent experiments.
DISCUSSION
The adaptation to low oxygen availability in the tumor mass mainly occurs through the induction of HIF-1α, which binds to and activates the transcription of target genes containing HRE sequences within their promoter regions (17, 41–43). Moreover, ROS generation in hypoxic conditions may contribute to cancer progression through the EGFR/ERK transduction pathway (44, 45). As it concerns breast cancer, several studies have shown that estrogens induce growth effects by activating EGFR/ERK-mediated signaling (46–49), which is also involved by hypoxia in stimulating tumor progression (50–52). An increasing body of data, including our own, has recently suggested that estrogen action is also mediated by GPER in both normal and cancer cells (7, 8, 53, 54).
In order to evaluate whether the biological responses to hypoxic conditions may involve GPER, we performed a bioinformatic analysis of the GPER 5′-promoter region, identifying three putative HRE binding sites. Then, exposing ER-negative SkBr3 breast cancer cells to the hypoxia-mimetic agent CoCl2, we observed that HIF-1α mediates the transactivation of the GPER promoter sequence. It is noteworthy that the up-regulation of HIF-1α, GPER, and CTGF expression was also determined in both SkBr3 and HL-1 cells after exposure to low oxygen tension (2% O2). Interestingly, the up-regulation of GPER by CoCl2 occurred through a ROS- and EGFR/ERK-dependent increase of HIF-1α, which was recruited to an HRE site located within the GPER promoter region.
Although the role of HIF-1α in apoptosis remains to be completely understood (55–57), its involvement in programmed cell death in hypoxic conditions has been largely reported (58, 59). On the basis of the results obtained in the present study showing on one hand the up-regulation of GPER by hypoxia and the requirement of GPER by estradiol to protect cells from apoptosis on the other, GPER may be considered a novel target for hypoxia-induced and HIF-1α-mediated effects.
Following tumor development, cancer cells are exposed to increasingly hypoxic conditions, which stimulate the transcription of diverse HIF-1α target genes, including CTGF (21, 60–62). Accordingly, we found that hypoxia up-regulates CTGF expression in an HIF-1α-dependent manner; however, we also provided evidence that GPER expression is required for the HIF-1α-mediated induction of CTGF by hypoxia.
Recently, we have shown that in ER-negative breast cancer cells, the activation of GPER signaling by estrogens or even by the antiestrogen hydroxytamoxifen triggers a transcription factor network resembling the stimulation of serum in fibroblasts (11). In this regard, we showed that CTGF not only contributes to cell proliferation but also mediates the stimulation of cell migration induced through GPER (11). Given that CTGF primarily modulates and coordinates signaling responses involving components of the extracellular matrix, the hypoxia-induced GPER/CTGF pathway might contribute to cell motility and invasion following cancer progression. In this regard, it is worth noting that an aggressive tumor phenotype and poor survival rates were recently associated with GPER expression (63–65).
Overall, our results indicate that the adaptive cell response to hypoxia may include the regulation of GPER, which in turn mediates the protective action exerted by E2 against apoptosis. In this regard, previous studies have shown that estrogen favors pancreatic islet survival by preventing apoptosis via GPER-dependent mechanisms (66), and E2/GPER signaling can also counteract cytokine-induced apoptosis in female pancreatic islets (67). As it concerns the role of CTGF as target of the estrogenic GPER signaling in relation to apoptosis, it remains to be further evaluated. However, it should be noted that CTGF protected pancreatic tumor cells from hypoxia-induced apoptosis (62), whereas CTGF inhibition increased apoptosis of rhabdomyosarcoma cells (68).
In previous reports, hypoxia-induced ROS generation has been shown to be both necessary and sufficient to stabilize and activate HIF-1α (32–34). In this context, it is worth mentioning that increased ROS levels activate upstream HIF-1α pathways, such as EGFR/ERK signaling, which can in turn induce HIF-1α-dependent transcriptional activity even in cancer cells (17, 35, 69). Interestingly, the hypoxic microenvironment in breast tumors triggers HIF-1α and its downstream effectors to promote estrogen-independent growth and a more aggressive phenotype (reviewed in Refs. 17, 41, and 70). These findings are in agreement with the results from the present study indicating that ROS/EGFR/ERK signaling is active in hormone-independent SkBr3 breast cancer cells under hypoxic conditions. In particular, our data include the regulation of GPER and CTGF expression by the hypoxia-mediated transduction pathway, providing new evidence regarding a further mechanism whereby estrogen may protect breast cancer cells from hypoxia-induced apoptosis. In this vein, it remains to be determined whether these molecular events could lead to an increased risk of recurrence and/or adverse clinical outcome. Nevertheless, previous reports have shown that higher GPER levels were associated with worse clinical pathological features and lower survival rates in endometrial (64), breast (71), and ovarian (65) cancer patients. Here, we have shown that GPER is an HIF-1α target gene, providing evidence for a new mechanism by which estrogens may exert biological effects under hypoxic conditions.
This work was supported by Associazione Italiana per la Ricerca sul Cancro Project 8925/2009 and Ministero dell'Università e Ricerca Scientifica e Tecnologica Cofin Project Prot. 2008PK2WCW/2008.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- ROS
- reactive oxygen species
- GPER
- G protein-coupled estrogen receptor
- CTGF
- connective tissue growth factor
- EGFR
- epidermal growth factor receptor
- NAC
- N-acetylcysteine
- HRE
- hypoxia-responsive element
- E2
- 17β-estradiol.
REFERENCES
- 1. Risbridger G. P., Davis I. D., Birrell S. N., Tilley W. D. (2010) Nat. Rev. Cancer 10, 205–212 [DOI] [PubMed] [Google Scholar]
- 2. Bai Z., Gust R. (2009) Arch. Pharm. 342, 133–149 [DOI] [PubMed] [Google Scholar]
- 3. Stice J. P., Lee J. S., Pechenino A. S., Knowlton A. A. (2009) Future Cardiol. 5, 93–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Koledova V. V., Khalil R. A. (2007) Expert Rev. Cardiovasc. Ther. 5, 777–789 [DOI] [PubMed] [Google Scholar]
- 5. Xing D., Nozell S., Chen Y. F., Hage F., Oparil S. (2009) Arterioscler. Thromb. Vasc. Biol. 29, 289–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhao Z. Q. (2004) Curr. Opin. Pharmacol. 4, 159–165 [DOI] [PubMed] [Google Scholar]
- 7. Maggiolini M., Picard D. (2010) J. Endocrinol. 204, 105–114 [DOI] [PubMed] [Google Scholar]
- 8. Maggiolini M., Vivacqua A., Fasanella G., Recchia A. G., Sisci D., Pezzi V., Montanaro D., Musti A. M., Picard D., Andò S. (2004) J. Biol. Chem. 279, 27008–27016 [DOI] [PubMed] [Google Scholar]
- 9. Prossnitz E. R., Maggiolini M. (2009) Mol. Cell. Endocrinol. 308, 1–2 [DOI] [PubMed] [Google Scholar]
- 10. Olde B., Leeb-Lundberg L. M. (2009) Trends Endocrinol. Metab. 20, 409–416 [DOI] [PubMed] [Google Scholar]
- 11. Pandey D. P., Lappano R., Albanito L., Madeo A., Maggiolini M., Picard D. (2009) EMBO J. 28, 523–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chu C. Y., Chang C. C., Prakash E., Kuo M. L. (2008) J. Biomed. Sci. 15, 675–685 [DOI] [PubMed] [Google Scholar]
- 13. Albanito L., Sisci D., Aquila S., Brunelli E., Vivacqua A., Madeo A., Lappano R., Pandey D. P., Picard D., Mauro L., Andò S., Maggiolini M. (2008) Endocrinology 149, 3799–3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vivacqua A., Lappano R., De Marco P., Sisci D., Aquila S., De Amicis F., Fuqua S. A., Andò S., Maggiolini M. (2009) Mol. Endocrinol. 23, 1815–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kizaka-Kondoh S., Tanaka S., Harada H., Hiraoka M. (2009) Adv. Drug. Deliv. Rev. 61, 623–632 [DOI] [PubMed] [Google Scholar]
- 16. Calzada M. J., del Peso L. (2007) Clin. Transl. Oncol. 9, 278–289 [DOI] [PubMed] [Google Scholar]
- 17. Semenza G. L. (2010) Oncogene 29, 625–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goldberg M. A., Dunning S. P., Bunn H. F. (1988) Science 242, 1412–1415 [DOI] [PubMed] [Google Scholar]
- 19. Wang G. L., Semenza G. L. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 4304–4308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Semenza G. L. (2004) Physiology 19, 176–182 [DOI] [PubMed] [Google Scholar]
- 21. Higgins D. F., Biju M. P., Akai Y., Wutz A., Johnson R. S., Haase V. H. (2004) Am. J. Physiol. Renal Physiol. 287, F1223–F1232 [DOI] [PubMed] [Google Scholar]
- 22. Ke Q., Costa M. (2006) Mol. Pharmacol. 70, 1469–1480 [DOI] [PubMed] [Google Scholar]
- 23. Lendahl U., Lee K. L., Yang H., Poellinger L. (2009) Nat. Rev. Genet. 10, 821–832 [DOI] [PubMed] [Google Scholar]
- 24. Poon E., Harris A. L., Ashcroft M. (2009) Expert Rev. Mol. Med. 11, e26. [DOI] [PubMed] [Google Scholar]
- 25. Ruan K., Song G., Ouyang G. (2009) J. Cell. Biochem. 107, 1053–1062 [DOI] [PubMed] [Google Scholar]
- 26. Zhong H., De Marzo A. M., Laughner E., Lim M., Hilton D. A., Zagzag D., Buechler P., Isaacs W. B., Semenza G. L., Simons J. W. (1999) Cancer Res. 59, 5830–5835 [PubMed] [Google Scholar]
- 27. Talks K. L., Turley H., Gatter K. C., Maxwell P. H., Pugh C. W., Ratcliffe P. J., Harris A. L. (2000) Am. J. Pathol. 157, 411–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Goonewardene T. I., Sowter H. M., Harris A. L. (2002) Microsc. Res. Tech. 59, 41–48 [DOI] [PubMed] [Google Scholar]
- 29. Claycomb W. C., Lanson N. A., Jr., Stallworth B. S., Egeland D. B., Delcarpio J. B., Bahinski A., Izzo N. J., Jr. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2979–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chaqour B., Yang R., Sha Q. (2006) J. Biol. Chem. 281, 20608–20622 [DOI] [PubMed] [Google Scholar]
- 31. Madeo A., Vinciguerra M., Lappano R., Galgani M., Gasperi-Campani A., Maggiolini M., Musti A. M. (2010) Oncogene 29, 978–991 [DOI] [PubMed] [Google Scholar]
- 32. Bell E. L., Klimova T. A., Eisenbart J., Moraes C. T., Murphy M. P., Budinger G. R., Chandel N. S. (2007) J. Cell Biol. 177, 1029–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chandel N. S., Maltepe E., Goldwasser E., Mathieu C. E., Simon M. C., Schumacker P. T. (1998) Proc. Natl. Acad. Sci. 95, 11715–11720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chandel N. S., McClintock D. S., Feliciano C. E., Wood T. M., Melendez J. A., Rodriguez A. M., Schumacker P. T. (2000) J. Biol. Chem. 275, 25130–25138 [DOI] [PubMed] [Google Scholar]
- 35. Galanis A., Pappa A., Giannakakis A., Lanitis E., Dangaj D., Sandaltzopoulos R. (2008) Cancer Lett. 266, 12–20 [DOI] [PubMed] [Google Scholar]
- 36. Lander H. M. (1997) FASEB J. 11, 118–124 [PubMed] [Google Scholar]
- 37. Patten R. D., Karas R. H. (2006) Trends Cardiovasc. Med. 16, 69–75 [DOI] [PubMed] [Google Scholar]
- 38. Zhao L., Brinton R. D. (2006) BMC Neurosci. 7, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gerstner B., Lee J., DeSilva T. M., Jensen F. E., Volpe J. J., Rosenberg P. A. (2009) J. Neurosci. Res. 87, 2078–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Guo M., Song L. P., Jiang Y., Liu W., Yu Y., Chen G. Q. (2006) Apoptosis 11, 67–77 [DOI] [PubMed] [Google Scholar]
- 41. Harris A. L. (2002) Nat. Rev. Cancer 2, 38–47 [DOI] [PubMed] [Google Scholar]
- 42. Brahimi-Horn M. C., Chiche J., Pouysségur J. (2007) J. Mol. Med. 85, 1301–1307 [DOI] [PubMed] [Google Scholar]
- 43. Semenza G. L. (2009) Syst. Biol. Med. 2, 336–361 [DOI] [PubMed] [Google Scholar]
- 44. Gao P., Zhang H., Dinavahi R., Li F., Xiang Y., Raman V., Bhujwalla Z. M., Felsher D. W., Cheng L., Pevsner J., Lee L. A., Semenza G. L., Dang C. V. (2007) Cancer Cell. 12, 230–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dewhirst M. W., Cao Y., Moeller B. (2008) Nat. Rev. Cancer 8, 425–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Knowlden J. M., Hutcheson I. R., Jones H. E., Madden T., Gee J. M., Harper M. E., Barrow D., Wakeling A. E., Nicholson R. I. (2003) Endocrinology 144, 1032–1044 [DOI] [PubMed] [Google Scholar]
- 47. Kurokawa H., Arteaga C. L. (2003) Clin. Cancer Res. 9, 511S–515S [PubMed] [Google Scholar]
- 48. Nicholson R. I., Hutcheson I. R., Harper M. E., Knowlden J. M., Barrow D., McClelland R. A., Jones H. E., Wakeling A. E., Gee J. M. (2002) Ann. N.Y. Acad. Sci. 963, 104–115 [DOI] [PubMed] [Google Scholar]
- 49. Davoli A., Hocevar B. A., Brown T. L. (2010) Cancer Chemother. Pharmacol. 65, 611–623 [DOI] [PubMed] [Google Scholar]
- 50. Lee M. (2009) Adv. Drug. Deliv. Rev. 61, 842–849 [DOI] [PubMed] [Google Scholar]
- 51. Yi J. M., Kwon H. Y., Cho J. Y., Lee Y. J. (2009) Biochem. Biophys. Res. Commun. 378, 842–846 [DOI] [PubMed] [Google Scholar]
- 52. Generali D., Buffa F. M., Berruti A., Brizzi M. P., Campo L., Bonardi S., Bersiga A., Allevi G., Milani M., Aguggini S., Papotti M., Dogliotti L., Bottini A., Harris A. L., Fox S. B. (2009) J. Clin. Oncol. 27, 227–234 [DOI] [PubMed] [Google Scholar]
- 53. Revankar C. M., Cimino D. F., Sklar L. A., Arterburn J. B., Prossnitz E. R. (2005) Science 307, 1625–1630 [DOI] [PubMed] [Google Scholar]
- 54. Madeo A., Maggiolini M. (2010) Cancer Res. 70, 6036–6046 [DOI] [PubMed] [Google Scholar]
- 55. Malhotra R., Tyson D. W., Rosevear H. M., Brosius F. C., 3rd (2008) BMC Cardiovasc. Disord. 8, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chen J., Zhao S., Nakada K., Kuge Y., Tamaki N., Okada F., Wang J., Shindo M., Higashino F., Takeda K., Asaka M., Katoh H., Sugiyama T., Hosokawa M., Kobayashi M. (2003) Am. J. Pathol. 162, 1283–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Piret J. P., Mottet D., Raes M., Michiels C. (2002) Biochem. Pharmacol. 64, 889–892 [DOI] [PubMed] [Google Scholar]
- 58. Krick S., Eul B. G., Hänze J., Savai R., Grimminger F., Seeger W., Rose F. (2005) Am. J. Respir. Cell Mol. Biol. 32, 395–403 [DOI] [PubMed] [Google Scholar]
- 59. Greijer A. E., van der Wall E. (2004) J. Clin. Pathol. 57, 1009–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rimon E., Chen B., Shanks A. L., Nelson D. M., Sadovsky Y. (2008) Endocrinology 149, 2952–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hong K. H., Yoo S. A., Kang S. S., Choi J. J., Kim W. U., Cho C. S. (2006) Clin. Exp. Immunol. 146, 362–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bennewith K. L., Huang X., Ham C. M., Graves E. E., Erler J. T., Kambham N., Feazell J., Yang G. P., Koong A., Giaccia A. J. (2009) Cancer Res. 69, 775–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Arias-Pulido H., Royce M., Gong Y., Joste N., Lomo L., Lee S. J., Chaher N., Verschraegen C., Lara J., Prossnitz E. R., Cristofanilli M. (2010) Breast Cancer Res. Treat. 123, 51–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Smith H. O., Leslie K. K., Singh M., Qualls C. R., Revankar C. M., Joste N. E., Prossnitz E. R. (2007) Am. J. Obstet. Gynecol. 196, 386.e1–9; discussion 386.e9–11 [DOI] [PubMed] [Google Scholar]
- 65. Smith H. O., Arias-Pulido H., Kuo D. Y., Howard T., Qualls C. R., Lee S. J., Verschraegen C. F., Hathaway H. J., Joste N. E., Prossnitz E. R. (2009) Gynecol. Oncol. 114, 465–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Liu S., Mauvais-Jarvis F. (2009) Islets 1, 273–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Balhuizen A., Kumar R., Amisten S., Lundquist I., Salehi A. (2010) Mol. Cell. Endocrinol. 320, 16–24 [DOI] [PubMed] [Google Scholar]
- 68. Croci S., Landuzzi L., Astolfi A., Nicoletti G., Rosolen A., Sartori F., Follo M. Y., Oliver N., De Giovanni C., Nanni P., Lollini P. L. (2004) Cancer Res. 64, 1730–1736 [DOI] [PubMed] [Google Scholar]
- 69. Richard D. E., Berra E., Gothié E., Roux D., Pouysségur J. (1999) J. Biol. Chem. 274, 32631–32637 [DOI] [PubMed] [Google Scholar]
- 70. Kurebayashi J., Otsuki T., Moriya T., Sonoo H. (2001) Jpn. J. Cancer Res. 92, 1093–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Filardo E. J., Graeber C. T., Quinn J. A., Resnick M. B., Giri D., DeLellis R. A., Steinhoff M. M., Sabo E. (2006) Clin. Cancer Res. 12, 6359–6366 [DOI] [PubMed] [Google Scholar]