Abstract
Parkinson disease is characterized by the accumulation of aggregated α-synuclein as the major component of the Lewy bodies. α-Synuclein accumulation in turn leads to compensatory effects that may include the up-regulation of autophagy. Another common feature of Parkinson disease (PD) is mitochondrial dysfunction. Here, we provide evidence that the overactivation of autophagy may be a link that connects the intracellular accumulation of α-synuclein with mitochondrial dysfunction. We found that the activation of macroautophagy in primary cortical neurons that overexpress mutant A53T α-synuclein leads to massive mitochondrial destruction and loss, which is associated with a bioenergetic deficit and neuronal degeneration. No mitochondrial removal or net loss was observed when we suppressed the targeting of mitochondria to autophagosomes by silencing Parkin, overexpressing wild-type Mitofusin 2 and dominant negative Dynamin-related protein 1 or blocking autophagy by silencing autophagy-related genes. The inhibition of targeting mitochondria to autophagosomes or autophagy was also partially protective against mutant A53T α-synuclein-induced neuronal cell death. These data suggest that overactivated mitochondrial removal could be one of the contributing factors that leads to the mitochondrial loss observed in PD models.
Keywords: Autophagy, Bioenergetics, Energy Metabolism, Mitochondria, Synuclein
Introduction
Mitochondrial dysfunction is one of the hallmarks of Parkinson disease (PD).2 The link between mitochondrial dysfunction and PD was made after the discovery of mitochondrial complex I deficiency in the substantia nigra (1). This connection has been supported by the finding that the products of several PD-related genes show mitochondrial localization under certain conditions, including SNCA, Parkin, PINK1, DJ-1, LRRK2, and HTR2A (2), and that the mitochondrial toxins MPTP, rotenone, and acetogenins can cause PD (3). A variety of mechanisms have been proposed to explain mitochondrial dysfunction. Oxidative stress, mitochondrial DNA deletions, pathological mutations in genes encoding mitochondrial proteins, altered mitochondrial morphology, and the interaction of pathogenic proteins with mitochondria can all lead to mitochondrial dysfunction and neuronal demise (4).
In this study, we propose an intriguing possibility whereby mitochondrial dysfunction may arise from the loss of mitochondria because of the overactivation of autophagy. Signs of autophagy have been detected in the brains of PD patients, whereas autophagosomes are rarely detected in normal brain (5–6). Moreover, several studies have also demonstrated that the overexpression of mutant A53T α-synuclein in PC12 cells, cultured neurons, and nigrostriatal systems activates autophagy (7–10). Here, we provide evidence that shows that the up-regulation of macroautophagy by mutant A53T α-synuclein can augment mitochondrial removal, which results in a net mitochondrial loss, energetic failure, and neuronal cell death.
EXPERIMENTAL PROCEDURES
Neuronal Cultures
Primary cultures of rat cortical cells were prepared from neonatal Wistar rats. Briefly, cortices were dissected in ice-cold Krebs-Ringer solution (135 mm NaCl, 5 mm KCl, 1 mm MgSO4, 0.4 mm K2HPO2, 15 mm glucose, 20 mm HEPES, pH 7.4) containing 0.3% BSA and trypsinized in 0.8% trypsin for 10 min at 37 °C. This was followed by trituration in a 0.008% DNase solution containing 0.05% soybean trypsin inhibitor. Cells were resuspended in Basal Medium Eagle with Earle's Salts (BME) containing 10% heat-inactivated FBS, 25 mm KCl, 2 mm glutamine, and 100 μg/ml gentamicin and plated onto 35 mm glass-bottom (MatTek) or plastic dishes (Nunc, Thermo Fisher Scientific) precoated with poly-l-lysine at a density of 1 × 106 cells/ml (2 ml of cell suspension per dish) or 170 μl per well into white microwell plates (96 F Nuclon Delta, Nunc). After a 3-h incubation, the medium was changed to NeurobasalTM-A medium containing B-27 supplement, 2 mm GlutaMAXTM-I, and 100 μg/ml gentamicin. The majority of the cells (over 60%) had a neuronal phenotype.
Primary cultures of mesencephalic neurons were prepared from the mesencephali of mouse embryos at embryonic day 15. Midbrain anlages were dissected, cleaned from the meninges, mechanically triturated through a 1-ml pipette tip in BME containing 10% FBS, and plated as droplets containing 0.2 × 105 cells directly onto poly-l-lysine precoated glass bottom dishes. One hour after plating, 2 ml of NeurobasalTM-B medium containing B-27 supplement, 2 mm GlutaMAXTM-I, and 100 μg/ml gentamicin was added. All of the cultures were grown in a humidified 5% CO2/95% air incubator at 37 °C. The mouse fetal mesencephalic cells contained 95% neurons, 20% of which were tyrosine hydroxylase positive.
The PC12 cell line was maintained in RPMI medium supplemented with 10% horse serum and 5% FBS on collagen IV-coated plastic dishes. All of the culture media and supplements, were obtained from Invitrogen (Carlsbad).
Plasmids
The plasmid expressing EGFP-LC3 (plasmid no. 11546) was obtained from ADDGENE (Cambridge), mito-CFP and mito-KillerRed from Evrogen (Moscow, Russia), and GFP and mitochondrial-pDsRed2 from Clontech. Plasmids expressing shRNA against Parkin, ATG12, and Beclin1 were from SABiosciences (Frederick, MD). The plasmids expressing WT α-synuclein and A53T α-synuclein were kind gifts from Dr. S. Kõks. mRFP-GFP-LC3 was a generous gift from Dr. E. Bampton, WT Mfn2 from Dr. S. Hirose and Dr. N. Nakamura (11), dominant negative Drp1 K38A from Dr. A. van der Bliek (12), EGFP-p62 from Dr. T. Johansen (13), APPArc-Swe from Dr. C. Sahlin (14), SOD1G93A from Dr. H. D. Durham (15), and YFP-Parkin from Dr. R. Youle (16). The expression of the α-synuclein proteins, WT Mfn2, and dominant negative Drp1 were confirmed by immunohistochemistry.
Transfections
Cultures were transiently transfected on the 2nd day after plating with LipofectamineTM 2000 (Invitrogen). Briefly, conditioned medium was collected and 100 μl of Opti-MEM® I medium containing 2% LipofectamineTM 2000 and 1–2 μg of total DNA was added directly to cells grown on glass-bottom dishes with a 14-mm diameter and incubated for 3–4 h in a humidified 5% CO2/95% air incubator. In multi-well plates, 50 μl of Opti-MEM® I medium with 2% LipofectamineTM 2000 and 0.6–0.7 μg of DNA were added to each well. Conditioned medium mixed 1:1 with fresh NeurobasalTM medium was added at the end of the incubation.
The experiments were performed 6 days after transfection unless otherwise specified. Approximately 1 × 105 PC12 cells were seeded on glass-bottom dishes 12–24 h before the transfection. The transfections were conducted as described above.
Mitochondrial-pDsRed2 was used as a compensation control in the LC3 experiments and EGFP in the mitochondrial density experiments when necessary. In the shRNA experiments, a plasmid encoding scrambled shRNA was used as a control. In addition, EGFP was cotransfected with the desired shRNA to calculate the transfection efficiency.
Image Acquisition and Analysis
For autophagy measurements, the neuronal cultures were transfected with EGFP-LC3 (or EGFP-LC3-RFP), mito-pDsRed2, and the plasmids of interest. Fluorescence images were randomly captured 6 days later from each dish using a LSM 510 META Zeiss confocal microscope equipped with LCI Plan-Neofluar 63×/1.33 water immersion objective. The temperature was maintained at 37 °C using a climate chamber. The distribution and colocalization of GFP-LC3 punctates (for autophagosome visualization) and pDsRed2 (for mitochondrial visualization) were analyzed using LSM5 Duo version 4.2 software (Carl Zeiss MicroImaging Gmbh and EMBL Heidelberg, Germany) and software (Media Cybernetics Inc, Bethesda, MD). At least 15 cell bodies or 18 axonal fragments were analyzed per group. The duration of the colocalization was measured using time-lapse microscopy from the time point when the autophagosome and mitochondria started to colocalize until the moment where the green LC3 signal faded (indicating fusion with the lysosome).
For the mitochondrial density measurements in axons, the neuronal cultures were transfected with GFP, mito-pDsRed2, and the plasmids of interest. Ten fluorescence images were randomly captured 6 days later from each dish using an Olympus IX70 inverted microscope equipped with WLSM PlanApo 40×/0.90 water immersion objective and Olympus DP70 CCD camera (Olympus Corp.). Morphometric analysis was performed using MicroImage software. For the mitochondrial density calculation (mitochondrial length per axonal length) at least 40 axons per group were analyzed (1 axon per image).
For mitochondrial density measurements in the cell body, 512 × 512 pixel sections were acquired by an LSM 510 META Zeiss confocal microscope equipped with Plan-Apo 100x/1.4 oil immersion objective. Voxels were collected at 43–74-nm lateral and 100-nm axial intervals. Raw images were three-dimensional deconvolved and reconstructed with the AutoDeblur and Autovisualize X software package (Media Cybernetics). Reconstructed images were then subjected to stereologic analysis using the Computer Assisted Stereology Toolbox software (Olympus, Denmark). The mitochondrial volume and cell body volume were estimated using the Cavalieri principle. A grid of points was superimposed on the images and the points that overlaid the fluorescent signal were counted. Mitochondrial density in the cell body was calculated by dividing the mitochondrial volume by the volume of the cell body. Seven to eight cell bodies were analyzed in each group.
For the Parkin visualization experiments, PC12 cells or cortical neurons were transfected with YFP-Parkin, mito-CFP, and mito-KillerRed. Mito-KillerRed was activated with a 561 nm laser and the images were collected before bleaching, immediately after bleaching, and two hours after bleaching (excitations 458, 514, and 561 nm with emissions separated by filters BP465–510, BP520–525, and LP575, respectively). Four images were collected from each dish and a total of 6 dishes were analyzed. For mitochondrial membrane potential visualization, the cultures were stained with mitochondrial membrane potential sensitive dye Tetramethylrhodamine, ethyl ester (TMRE) at a concentration of 50 nm in complete NeurobasalTM-B medium at 37 °C for 30 min.
Western Blot
For Western blot analysis, the cells were lysed in a buffer containing 50 mm Tris-Cl, 1 mm EDTA, 150 mm NaCl, 1% Nonidet P-40, 1 mm Na3VO4, 1 mm NaF, 0.25% sodium deoxycholate, and 4% protease inhibitor mixture for 30 min on ice. Equivalent amounts of total protein were separated by SDS-PAGE on 10% or 12% polyacrylamide gels and then transferred to Hybond-P PVDF transfer membranes (Amersham Biosciences) in 0.1 m Tris-base, 0.192 m glycine, and 10% (v/v) methanol using an electrophoretic transfer system. The membranes were blocked with 5% (w/v) nonfat dried milk in TBS containing 0.1% (v/v) Tween-20 at room temperature for 1 h. After blocking, the membranes were incubated overnight with primary antibodies (mouse anti-β-actin 1:4000, with rabbit (Rb) anti-Parkin 1:2000, Rb anti-Atg12 1:2000, Rb anti-Beclin1 1:2000, Rb anti-GFP 1:10000, or Rb anti-His 1:3000) followed by washing and subsequent incubation with appropriate HRP-conjugated secondary antibodies (1:4000, Pierce) for 1 h at room temperature. Immunoreactive bands were detected by enhanced chemiluminescence (ECL, Amersham Biosciences) using medical x-ray film blue (Agfa, Belgium). The probed blots were then densitometrically analyzed using a QuantityOne 710 System (Bio-Rad).
PGC-1α Activity Assay
Neurons were co-transfected with a fusion protein that connected the yeast Gal4 DNA binding domain with full-length PGC-1α on the second day after plating, together with an appropriate Gal4-UAS-luciferase reporter and A53T α-synuclein. All transfections also included 0.1 μg of Renilla luciferase (pRL-CMV) plasmid. The luciferase assays were performed 24 h post-transfection using Dual-Glo Luciferase Assay reagent (Promega) according to the manufacturer's instructions. PGC-1α activity was determined as relative firefly luciferase luminescence normalized to the Renilla luciferase signal measured by a MicroBeta® TriLux luminescence counter (Perkin Elmer). At least eight independent samples were analyzed per group.
ATP Level
Cortical neurons were transfected with plasmids expressing fire-fly luciferase, Renilla luciferase, and the plasmids of interest. To measure fire-fly luciferase activity, cells were incubated for 10 min with 25 μm 1-(4,5-dimethoxy-2-nitrophenyl)ethyl (DMNPE) caged luciferin at 37 °C and the luminescence was measured by MicroBeta® TriLux. For the measurement of Renilla luciferase activity, the neurons were lysed and the luminescence was measured using Dual-Glo Luciferase Assay reagent. Firefly luciferase activity from living cells normalized to Renilla luciferase activity from lysed cells was used to estimate the ATP level in transfected cells. Sixteen independent samples were analyzed in all of the experiments.
Respiration
Oxygen consumption was monitored with the phosphorescent oxygen sensitive probe MitoXpressTM (Luxcel Biosciences, Cork, Ireland). Neurons were harvested in Krebs-Ringer solution at 37 °C containing 1 mm CaCl2, centrifuged for 5 min at 1000 rpm, resuspended in fresh Krebs buffer, and dispensed in 150-μl aliquots into the wells of a 96-well plate. The MitoXpressTM probe was then added at a final concentration of 100 nm. Each well was covered with 100 μl of heavy mineral oil and the time resolved fluorescence was monitored over 90 min at intervals of 2 min (excitation 380 nm, cut-off 530 nm, emission 655 nm, filters, delay time 50 μs, and measurement window 200 μs) at 37 °C using FlexStationTM (Molecular Devices, Sunnyvale, CA). Oxygen consumption rates were assessed by determining the rate of increase of probe fluorescent signal.
Citrate Synthase Activity
Neurons were washed once with ice-cold PBS and lysed in 0.25% Triton X-100. Citrate synthase activity was measured in the reaction mixture containing 86 μm 5,5′-dithiobis (2-nitrobenzoic acid), 0.42 mm oxaloactetic acid, 0.17 mm acetyl CoA, 86 mm Tris-HCl (pH 8.0), and 100 μl of cell lysate. The reaction was initiated with the addition of acetyl CoA, and the change in absorbance was followed at 412 nm for 5 min at 30 °C.
Neuronal Viability
Cortical neurons were cotransfected with EGFP, A53T α-synuclein, and/or the plasmids of interest. Fluorescent living cells with visible neurites were manually count in 70 fields per dish under an Olympus IX70 inverted microscope equipped with a WLSM PlanApo 40x/0.90 water immersion objective. At least 3 dishes per group were used for comparisons.
Statistics
The D'Agostino-Pearson omnibus test was used to test the normality of distribution. The Mann-Whitney or Student's t test and one-way ANOVA, followed by a Newman-Keuls posthoc test or Kruskal-Wallis test, followed by a Dunn's test were then used to compare the differences between the samples and the control group. A two-way ANOVA was used to analyze the interaction between treatments. All of the counting analyses (colocalization, mitochondrial density, and toxicity analyses) were performed in a blinded manner.
RESULTS
Overexpression of Mutant A53T α-Synuclein Increases Autophagic Activity and Mitochondrial Autophagy
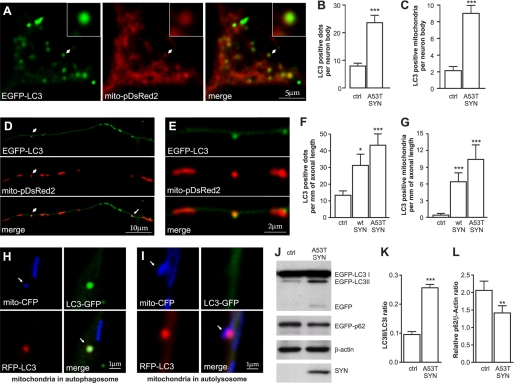
Cortical neurons were cotransfected with mutant A53T α-synuclein, EGFP-LC3 as the autophagosome marker, and mitochondrial-pDsRed2. Fig. 1 shows that the overexpression of A53T α-synuclein led to an increase in the number of EGFP-LC3 positive autophagosomes in both the cell body (A and B) and the neurites (D and F). A53T α-synuclein overexpression also enhanced LC3-I cleavage into LC3-II (Fig. 1, J and K) and increased the turnover of p62/SQSTM1 (Fig. 1, J and L). Moreover, the increased autophagic activity in response to mutant A53T α-synuclein overexpression was associated with an increased number of mitochondria that colocalized with autophagosomes both in the neuron body (Fig. 1, A and C) and in neurites (Fig. 1, D, E, and G). In addition, the overexpression of WT α-synuclein increased the number of EGFP-LC3-positive autophagosomes as well as the number of mitochondria that colocalized with autophagosomes, although to a lesser extent than mutant A53T α-synuclein (Fig. 1, F and G). In contrast, the overexpression of several other disease-related proteins (APPArc-Swe and SOD1G93A) did not increase the number of EGFP-LC3 puncta in primary cortical neurons. For all experiments, the mitochondrial-pDsRed2 encoding plasmid was used to compensate for differences in the DNA concentration of the effector plasmid.
FIGURE 1.
Overexpression of mutant A53T α-synuclein induces mitochondrial removal. Cortical neurons were transfected with plasmids expressing WT or mutant A53T α-synuclein, EGFP-LC3, and mito-pDsRed2 at day 2 after plating and visualized 6 days later. A, colocalization of EGFP-LC3 with mito-pDsRed2 in the cell body of neurons overexpressing mutant A53T α-synuclein. The left panel shows the EGFP-LC3 signal, the middle panel shows the mito-pDsRed2 signal, and the right panel shows the merged signals. The magnified inset shows the colocalization between the mitochondrial and autophagosome signal. Further analysis demonstrated an increased number of autophagosomes (B) as well as an increased number of mitochondria that colocalized with autophagosomes (C) per neuron body in neurons overexpressing mutant A53T α-synuclein overexpressing neurons (Mann-Whitney test). D, upper part of the panel shows the EGFP-LC3 signal, the middle panel shows the mito-pDsRed2 signal, and the lower panel shows the merged signals in neurites of neurons overexpressing mutant A53T α-synuclein. A magnified portion of the picture (E) demonstrates the exact colocalization between the mitochondrial and autophagosome signals. F and G, an increased number of autophagosomes (F) as well as increased number of mitochondria that colocalize with autophagosomes (G) per mm of axonal length in both the WT and mutant A53T α-synuclein overexpressing neurons (Kruskal-Wallis test followed by Dunn's test). H and I, neurons were transfected with A53T α-synuclein, mito-CFP, and RFP-LC3-GFP. H, Mito-CFP and RFP-LC3-GFP colocalize in the autophagosome while in the autolysosome (I) the GFP signal had faded in the acidic environment. J–L, overexpression of A53T α-synuclein enhances LC3-I conversion into LC3-II and decreases the levels of p62 (paired t test). *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus control.
These experiments were repeated in midbrain neurons that are known to be the most vulnerable in Parkinson disease. Midbrain neurons responded to mutant A53T α-synuclein overexpression in a similar manner to cortical neurons. The number of autophagosomes per mm of axonal length increased from 10 ± 3 in the control to 26 ± 6 in the mutant A53T α-synuclein overexpression group (p < 0.05). Moreover, the number of mitochondria that colocalized with autophagosomes per mm of axonal length increased from 0.8 ± 0.6 in the control to 6.1 ± 1.7 in the mutant A53T α-synuclein overexpression group (p < 0.05).
We next determined whether the observed increase in autophagosome number was due to an increase in autophagosome formation or whether it was a consequence of an inhibition in autolysosome formation. We employed double-labeled LC3 (mRFP-GFP-LC3) to differentiate between autophagosomes and autolysosomes. RFP has been shown to be less sensitive to an acidic environment when compared with GFP and retains longer fluorescence in acidic lysosomal environments (17). Fig. 1, H–I depicts the colocalization of mitochondria with autophagosomes (colocalization of all markers) and with autolysosomes (no GFP signal). Our results demonstrate that mutant A53T α-synuclein increased the number of autophagosomes as well as the number of autolysosomes. The ratio of autophagosomes and autolysosomes remained the same between both groups (1.3 versus 1.3 in the control and mutant A53T α-synuclein groups, respectively), which suggested that autophagosome fusion with lysosomes was not altered. We also did not observe any increase in the duration of colocalization between neurons overexpressing mutant A53T α-synuclein and control neurons (72 ± 3 min versus 77 min, respectively), which was measured from the moment when the autophagosome and mitochondria started to colocalize until the moment when the green LC3 signal faded. These results suggested that the increase in autophagosome number was due to increased formation rather than an inhibition of fusion with lysosomes.
Increased Removal of Polarized Mitochondria Leads to Mitochondrial Loss and a Bioenergetic Deficit
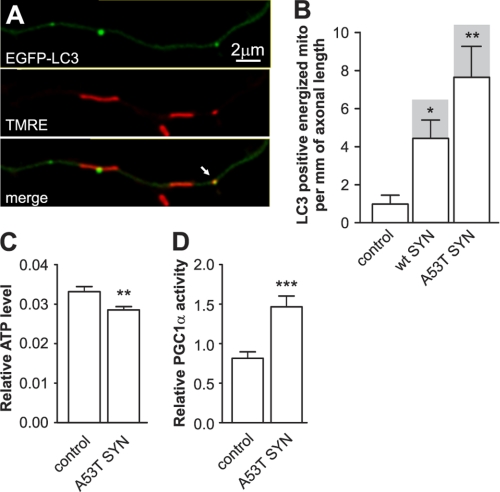
We next explored whether the autophagosomes could only colocalize with unhealthy, depolarized mitochondria or whether they were also able to colocalize with healthy mitochondria that remained polarized. Cortical neurons were co-transfected with WT α-synuclein or mutant A53T α-synuclein and EGFP-LC3 and stained with TMRE to monitor the membrane potential of the mitochondria (Fig. 2A). Interestingly, the majority of the mitochondria that colocalized were TMRE positive (Fig. 2B). Moreover, the number of mitochondria that colocalized per mm of neurite length was significantly increased for both WT and mutant α-synuclein, however the extent of colocalization was significantly higher in neurons overexpressing mutant A53T α-synuclein. The observed increase in the utilization of the polarized mitochondria through the autophagic pathway should lead to a bioenergetic loss in neurons overexpressing mutant A53T α-synuclein. Therefore, we compared the relative ATP levels in the control and neurons overexpressing mutant A53T α-synuclein. Neurons were cotransfected with firefly and Renilla luciferase expressing plasmids and the firefly luciferase activity was measured 24 h later in living cells using DMNPE-caged luciferin as a substrate. The results were normalized to Renilla luciferase activity measured after the lysis of cells and demonstrated a 15% decrease in relative ATP levels in the neurons overexpressing mutant A53T α-synuclein (Fig. 2C). We also measured the transcriptional activity of PGC-1α, which is known to be activated during energy deficit and can therefore be used as a sensor for the energetic state (18). Neurons were cotransfected with a fusion protein that consisted of the yeast Gal4 DNA binding domain fused with PGC-1α, the Gal4-UAS-luciferase reporter, and A53T α-synuclein. Luciferase assays conducted 24 h post-transfection revealed that A53T α-synuclein increased Gal4-PGC1α transcriptional activity by ∼80% (Fig. 2D).
FIGURE 2.
Overexpression of mutant A53T α-synuclein induces a bioenergetic deficit. Cortical neurons were transfected with plasmids expressing WT or mutant A53T α-synuclein and EGFP-LC3 and the mitochondria were stained with the mitochondrial membrane potential sensitive dye TMRE. A, colocalization of EGFP-LC3 (upper), TMRE (middle), and the superimposed channel (lower). B, colocalization analysis demonstrated a considerable increase of LC3-positive energized mitochondria in neurons overexpressing α-synucleins. Also note that the majority of LC3 positive mitochondria were energized (shaded columns show the total number of colocalizations; Kruskal-Wallis test followed by Dunn's test). C, neurons were cotransfected with plasmids expressing firefly luciferase and Renilla luciferase with or without mutant A53T α-synuclein at day 2 after plating. Firefly luciferase activity was measured in living neurons 24 h post-transfection using DMNPE-caged luciferin as a substrate followed by Renilla luciferase measurement (for normalization) in lysed neurons. The expression of mutant A53T α-synuclein decreased the ratio of firefly to Renilla luciferase activities, which indicated a decline in ATP levels. D, neurons were cotransfected with plasmids coding the Gal4-PGC-1α fusion protein, Gal4-UAS-luciferase reporter, and mutant A53T α-synuclein. Increased luciferase activity demonstrates that mutant A53T α-synuclein increases PGC-1α transcriptional activity (t test). *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus control.
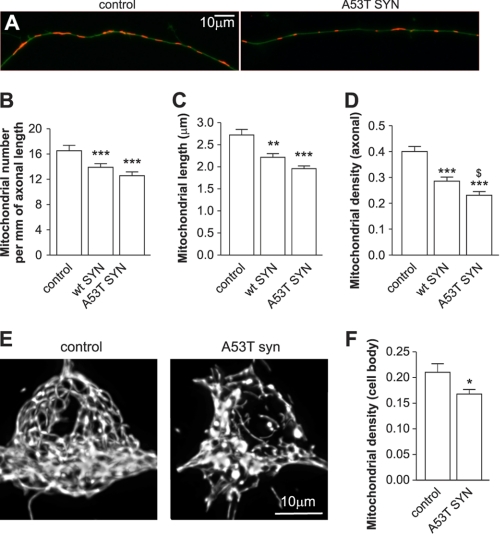
We further determined whether an increase in mitochondrial removal is associated with a net mitochondrial loss. As shown in Fig. 3, A–D, overexpression of WT and mutant A53T α-synuclein decreased the mitochondrial number and length, which led to a considerable decrease in mitochondrial density in the axons. Similar results were obtained when mutant A53T α-synuclein was overexpressed in midbrain neurons. Mitochondrial density decreased from 0.366 ± 0.022 (mitochondrial length per axonal length) in the control to 0.275 ± 0.011 in midbrain neurons overexpressing mutant A53T α-synuclein (p < 0.001). To test whether mitochondrial loss in the axons was accompanied with mitochondrial loss in the cell body, we created three-dimensional reconstructions of mitochondria that resided in the cell body (Fig. 3E) and analyzed mitochondrial volume using a stereological approach. As shown in Fig. 3F, a 20% loss of mitochondria was also observed in the cell body of neurons overexpressing mutant A53T α-synuclein compared with the control.
FIGURE 3.
Overexpression of α-synuclein leads to mitochondrial loss. Cortical neurons were cotransfected with plasmids expressing WT or A53T mutated α-synuclein, GFP, and mitochondrially targeted pDsRed2 at day 2 after plating and visualized 6 days later. A, morphometric analysis of axonal mitochondria demonstrated that the mitochondrial number per mm axonal length (B), average mitochondrial length (C), and mitochondrial density (mitochondrial length per axonal length) (D) decreased in WT or mutant A53T α-synuclein overexpressing neurons. Note that the effect of mutant A53T α-synuclein is more pronounced than the WT protein (one way ANOVA followed by Newman Keuls test). To compare the mitochondrial density in the neuronal body between the control and mutant A53T α-synuclein expressing neurons, we performed a three-dimensional scan using a confocal microscope followed by three-dimensional reconstruction of the mitochondrial network in the neuronal body. E, three-dimensional reconstruction of the mitochondrial network from control neurons or mutant A53T α-synuclein expressing neurons. F, further stereological analysis of three-dimensional reconstructed mitochondrial networks in control neurons or in neurons expressing mutant A53T α-synuclein revealed a clear decrease in mitochondrial volume per cell volume (t test). *, p < 0.05; **, p < 0.01; and ***, p < 0.001 versus control; and $, p < 0.05 versus WT α-synuclein.
Induction of Autophagy by Rapamycin Leads to the Removal of Functional Mitochondria
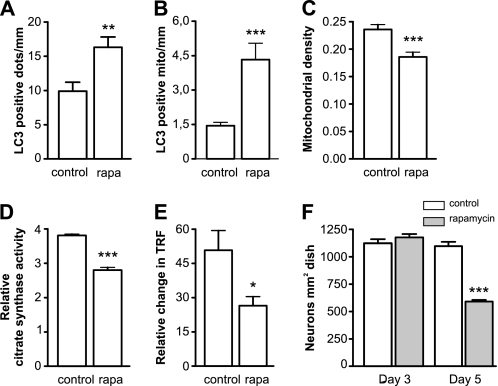
Autophagy was induced by the treatment of neurons with 100 nm rapamycin for 3 days. This led to 1.6-fold increase in the number of LC3 positive autophagosomes (Fig. 4A) and 3-fold increase in the number of autophagosomes that colocalized with mitochondria or their fragments (Fig. 4B). Similar to neurons overexpressing mutant A53T α-synuclein, the majority of the mitochondria that colocalized with autophagosomes were TMRE positive. The increase in mitochondrial removal after treatment with rapamycin for 3 days was also associated with a net mitochondrial loss of 20% in neurites (Fig. 4C). The activity of the mitochondrial matrix enzyme citrate synthase decreased by ∼25% in the rapamycin-treated group compared with the control (Fig. 4D). We also compared oxygen consumption of rapamycin-treated and non-treated neurons using the phosphorescent oxygen-sensitive probe MitoXpressTM. As shown in Fig. 4E, a considerable reduction in oxygen consumption was observed in the neurons treated with rapamycin. These changes were not related to rapamycin-mediated toxicity, since 3 days of rapamycin treatment failed to induce a measurable reduction in neuronal viability (Fig. 4F). However, when the rapamycin exposure time was increased up to 5 days, a reduction in viability was observed. The rapamycin-mediated toxicity was partially inhibited by transfection with shRNA against Parkin and Beclin1 (38 ± 1% and 32 ± 1% protection, respectively; p < 0.0001). Together, these data demonstrate that the specific induction of autophagy by rapamycin leads to the removal of functional mitochondria.
FIGURE 4.
Rapamycin-induced mitochondrial removal and bioenergetic deficit. A and B, neurons were transfected with EGFP-LC3 and mitochondria-targeted pDsRed2 and treated with 100 nm rapamycin at day 2 after plating. Three days post-transfection, the LC3-positive dots were visualized. Rapamycin treatment led to a significant increase in LC3 positive dots (A) as well as LC3 positive mitochondria (B) per mm of axonal length (Mann Whitney test). C, neurons were transfected with GFP and mitochondria-targeted pDsRed2 and treated with rapamycin for 3 days, which led to a decrease in mitochondrial density. Rapamycin-treated neurons had decreased citrate synthase activity (D) and diminished oxygen consumption (E), both of which were normalized per number of intact neurons. Oxygen consumption was estimated by measuring the relative change in time resolved fluorescence (TRF) of the MitoXpressTM probe. F, rapamycin had no effect on neuronal survival after 3 days of treatment, but was clearly toxic after 5 days of treatment. Student's t test, *, p < 0.05; **, p < 0.01; and ***, p < 0.001 versus control.
Based on these findings, we wished to determine whether the blockage of mitochondrial removal could protect against mitochondrial loss and promote neuronal survival. We decided to interfere with mitochondria removal at three different stages: 1) by suppressing Parkin expression to inhibit mitochondrial removal by autophagy; 2) by manipulating mitochondrial size to make them less attractive for autophagosomes; and 3) by blocking the autophagic machinery directly.
Parkin Is Required for Mutant A53T α-Synuclein-induced Mitochondrial Removal
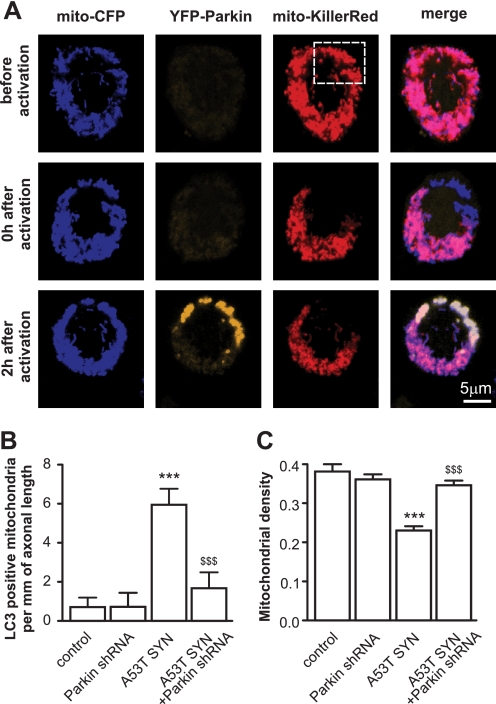
A recent study by Narendra et al. (16) elegantly showed that the treatment of cells with mitochondrial depolarizing agents led to the relocation of Parkin from the cytoplasm to mitochondria, and suggested that mitochondrial-localized Parkin could mark mitochondria for autophagic removal. Therefore, we tested whether Parkin could also translocate to mitochondria in neuronal cells and whether this localization was selective (e.g. whether the translocation of Parkin was only limited to damaged mitochondria). PC12 neuronal cells were transfected with mitochondrial-targeted CFP, YFP-Parkin, and mitochondrial-targeted Killer Red, which is a genetically-encoded photosensitizer that was developed on the basis of the Anthomedusae chromoprotein anm2CP that can generate reactive oxygen species (ROS) upon light irradiation (19). A subpopulation of mitochondria was then irradiated with yellow-green laser at 561 nm. As shown in Fig. 5A, Parkin only translocated from the cytoplasm to irradiated mitochondria. A similar relocation, although to a lesser extent, was also observed in primary cortical neurons. Parkin-positive mitochondria were also observed in neurons overexpressing mutant A53T α-synuclein and frequently colocalized with the autophagosome marker LC3. We hypothesized that Parkin may be also involved in α-synuclein-induced mitochondrial removal and that the silencing of Parkin may provide some protection against mitochondrial loss. To test this hypothesis, we assessed mitochondrial autophagy in Parkin-silenced cortical neurons. The knockdown efficiency of shRNA-encoding plasmids was validated by Western blot (supplemental Fig. S1). Fig. 5B shows that the knockdown of Parkin suppressed mitochondrial removal by 72% in neurons overexpressing mutant A53T α-synuclein, but had no effect at control neurons (p = 0.0042). Moreover, silencing of Parkin restored the axonal mitochondrial density in neurons overexpressing mutant A53T α-synuclein (Fig. 5C, p < 0.0001). Importantly, Parkin shRNA as well as other treatments did not alter the protein levels of A53T α-synuclein (supplemental Fig. S2).
FIGURE 5.
The role of Parkin in mutant A53T α-synuclein-induced mitochondrial removal and mitochondrial loss. A, PC12 cells were transfected with plasmids expressing mito-CFP, YFP-Parkin, and mito-KillerRed. A subpopulation of mitochondria was then irradiated with 561 laser line, which caused a bleaching of the KillerRed signal. Two hours later, YFP-Parkin was localized to these mitochondria. For panels B and C, cortical neurons were transfected with plasmids expressing mutant A53T α-synuclein, EGFP or EGFP-LC3, mito-pDsRed2, and plasmids expressing shRNA against Parkin per the indicated combinations. B, suppression of Parkin inhibits mutant A53T α-synuclein-induced mitochondrial removal (Kruskal-Wallis test followed by Dunn's test). A plasmid encoding scrambled shRNA was used as a control for all of the shRNA experiments. C, suppression of Parkin also protects neurons from mutant A53T α-synuclein-induced mitochondrial loss (one way ANOVA followed by Newman Keuls test; interaction in all cases p < 0.0001, two way ANOVA). ***, p < 0.001 versus control, and $$, p < 0.01; $$$, p < 0.001 versus mutant A53T α-synuclein.
Mitochondrial Fragmentation Is Required for Mutant A53T α-Synuclein-induced Mitochondrial Removal
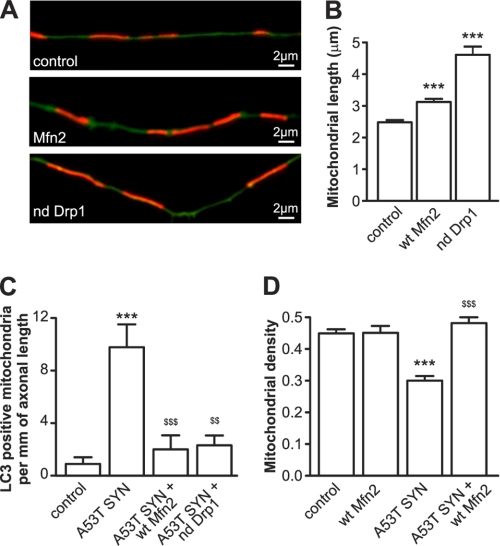
Mitochondrial fragmentation precedes mitochondrial removal, and the inhibition of fragmentation has been shown to impede mitochondrial removal (20). Therefore, we decided to increase the average mitochondrial length in order to reduce the accessibility to autophagosomes. Either WT Mitofusin 2 (Mfn2), which activates mitochondrial fusion, or a dominant negative form of Dynamin-related protein (Drp1 K38A), which inhibits mitochondrial fission, were overexpressed in order to increase mitochondrial length (Fig. 6, A and B). The overepxression of each protein led to marked inhibition in mutant A53T α-synuclein-induced mitochondrial removal (Fig. 6C). The overexpression of either protein also decreased the total number of autophagosomes to a level similar to that of the control. Moreover, WT Mfn2 protected neurons from mutant A53T α-synuclein-induced net mitochondrial loss (Fig. 6D). The mitochondria protected by WT Mfn2 and dominant negative Drp1 overexpression were also energized (data not shown).
FIGURE 6.
Inhibition of mitochondrial fragmentation protects against mutant A53T α-synuclein-induced mitochondrial loss. A–D, cortical neurons were transfected with plasmids expressing mutated A53T α-synuclein, mito-pDsRed2, EGFP (A, B, D) or EGFP-LC3 (C) and WT Mfn2 or a dominant negative Drp1. A and B, overexpression of WT Mfn2 and dominant negative Drp1 increase mitochondrial length. C, overexpression of WT Mfn2 or expression of dominant negative Drp1 inhibits mutant A53T α-synuclein-induced mitochondrial removal (Kruskal-Wallis test followed by Dunn's test; interaction in all cases p < 0.0001, two way ANOVA. D, overexpression of WT Mfn2 also protects neurons from mitochondrial loss induced by mutant A53T α-synuclein. ***, p < 0.001 versus control, and $$, p < 0.01; $$$, p < 0.001 versus A53T α-synuclein.
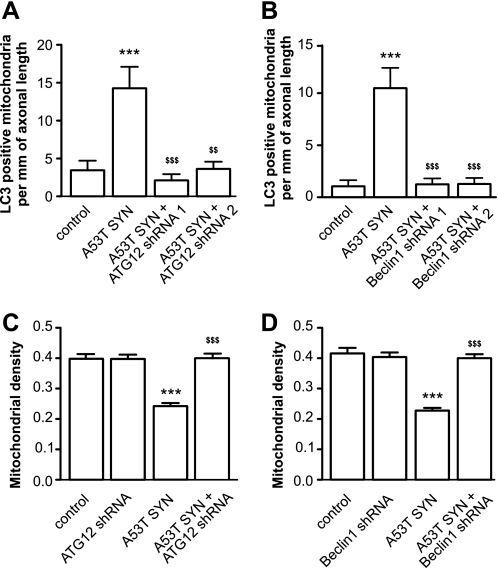
Silencing of Beclin1 or ATG12 Inhibits Mitochondrial Removal by Autophagosomes and Restores Mitochondrial Mass
To address the question of whether mutant A53T α-synuclein-induced autophagy is the primary reason for mutant A53T α-synuclein-induced mitochondrial loss, we suppressed autophagy by reducing the levels of the essential components of autophagy ATG12 or Beclin1 using specific shRNA plasmids. As shown in Fig. 7, A and B, mutant A53T α-synuclein-induced mitochondrial removal was significantly inhibited in neurons where ATG12 or Beclin1 levels were reduced by different shRNA plasmids. Moreover, in the presence of ATG12- or Beclin1-directed shRNA, mutant A53T α-synuclein failed to induce any mitochondrial loss (Fig. 7, C–D). ATG12- and Beclin1-directed shRNA alone had no effect on the mitochondrial number or density at control conditions.
FIGURE 7.
Inhibition of autophagy protects against mutant A53T α-synuclein-induced mitochondrial removal and mitochondrial loss. Cortical neurons were transfected with plasmids expressing mutant A53T α-synuclein, EGFP or EGFP-LC3, mito-pDsRed2, and plasmids expressing shRNA against Beclin1 or ATG12 as indicated. A and B, suppression of ATG12 and Beclin1 by specific shRNA decreases mutant A53T α-synuclein-induced mitochondrial removal. C and D, suppression of Beclin1 or ATG12 protects neurons from mutant A53T α-synuclein-induced mitochondrial loss (one way ANOVA followed by Newman Keuls test; interaction in all cases p < 0.0001, two way ANOVA). ***, p < 0.001 versus control and $$, p < 0.01; $$$, p < 0.001 versus mutant A53T α-synuclein.
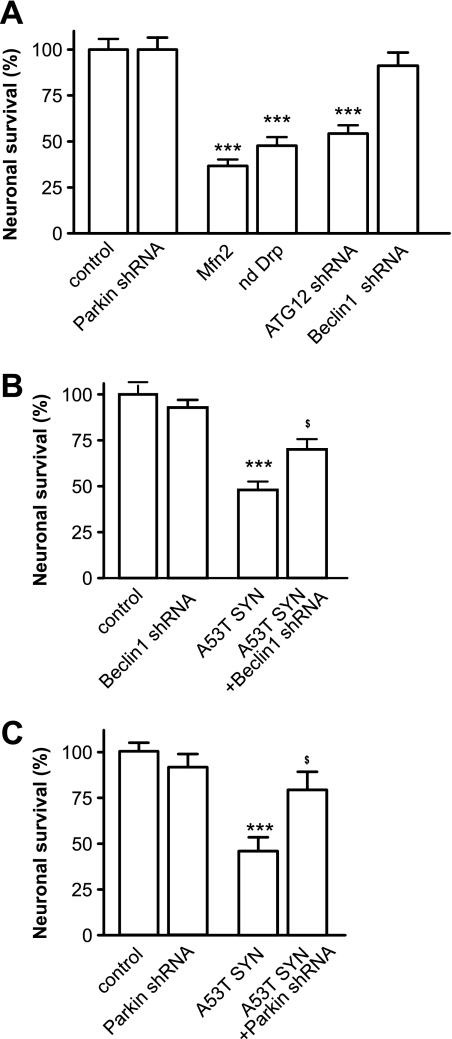
Suppression of Autophagy Provides Partial Protection against Neuronal Death
Finally, we tested whether the inhibition of mitochondrial autophagy could protect neurons from mutant A53T α-synuclein-induced cell death. Overexpression of mutant A53T α-synuclein led to visible neuronal loss. We first tested whether potential protective treatments were nontoxic in control conditions. As shown in Fig. 8A, the suppression of Parkin or Beclin1 was relatively nontoxic, while the suppression of ATG12 or the overexpression of Mfn2 and dominant negative Drp1 were markedly toxic. Therefore, we performed rescue experiments with only Beclin1- and Parkin-directed shRNA plasmids. Suppression of either Beclin1 or Parkin provided partial protection against mutant A53T α-synuclein-induced toxicity and increased the number of viable neurons (Fig. 8, B and C). Overexpression of mutant A53T α-synuclein did not alter EGFP expression (supplemental Fig. S3). Moreover, suppression of Beclin1 also partially restored the ATP pool (data not shown).
FIGURE 8.
Suppression of Parkin and Beclin1 protects neurons from mutant α-synuclein-induced neuronal death. Cortical neurons were transfected with plasmids expressing mutant A53T α-synuclein, EGFP, mito-pDsRed2, and the plasmids of interest at day 2 after plating. A, overexpression of WT Mfn2, dominant negative Drp1, or the suppression of ATG12 decreases the number of EGFP-expressing, viable neurons 6 days post-transfection, while the suppression of Parkin or Beclin1 was relatively nontoxic. B and C, suppression of Beclin1 (B) and Parkin (C) partially protects against mutant A53T α-synuclein-induced neuronal loss. EGFP-positive viable neurons from the control at 6 days post-transfection were set as 100%. One way ANOVA followed by Newman Keuls test. **, p < 0.01; ***, p < 0.001 versus control; and $, p < 0.05; $$$, p < 0.001 versus mutant A53T α-synuclein.
DISCUSSION
Mitochondrial dysfunction has long been recognized as a possible causative factor for Parkinson disease (4). In contrast, increased autophagy has been considered to be a beneficial compensatory response through which neurons attempt to remove accumulating aggregates (20–21). Based on this, we hypothesized that mitochondrial dysfunction may be a by-product of the overactivation of intracellular degradation machinery. We took advantage of modern visualization techniques to investigate such a possibility. The results presented in this study showed that overexpression of A53T α-synuclein leads to excessive autophagy, mitochondrial removal, and net mitochondrial loss. The mitochondrial loss was accompanied by excessive neuronal death, which was significantly reversed by inhibiting mitochondrial autophagy.
Increased Mitochondrial Autophagy, a Reason for Mitochondrial Loss or the Result of Mitochondrial Damage?
Accumulation of α-synuclein may directly or indirectly damage mitochondria, thereby inducing the autophagic removal of defective mitochondria. Indeed, α-synuclein has been shown to directly interact with mitochondria, induce cytochrome c release or inhibit complex I, and increase the production of ROS (22). However, mitochondrial removal may also be the unwanted “side-effect” of an otherwise beneficial cellular attempt to remove accumulating α-synuclein. Several lines of evidence shown here argue for autophagy being a cause rather than a consequence.
In our experiments, the majority of mitochondria that colocalized with autophagosomes in neurons overexpressing mutant A53T α-synuclein were originally energized (polarized), suggesting that at least under our experimental settings, the macroautophagy eliminated polarized mitochondria. Although depolarized and damaged mitochondria have been suggested to be the preferred targets of destruction by autophagy (23–26), several reports have shown the sequestration of polarized mitochondria as well (27–29).
Suppression of autophagy by silencing Beclin1 or ATG12 protected mitochondria from removal and reversed mitochondrial loss. Suppression of Beclin1 was also found to be partially neuroprotective and protected a significant number of neurons from mutant A53T α-synuclein-induced cell death. Similar to our findings, suppression of autophagy by the silencing of ATG5 was also shown to be protective against A53T α-synuclein-induced toxicity in SH-SH5Y cells as well as in cortical neurons (9). It is important to note that the supression of Beclin 1 or Atg12 did not increase the mitochondrial density in control cells. These data suggest that the levels of Beclin 1 or Atg12-dependent mitophagy are markedly low under control conditions.
A causal link between the induction of autophagy and the removal of functional mitochondria is further supported by experiments with the autophagy inducer rapamycin. A 3 day treatment with rapamycin increased mitochondrial clearance and net mitochondrial loss, similar to mutant A53T α-synuclein overexpression, and decreased the oxidative capacity of the neurons.
The finding that mitochondrial elongation, which was induced by the overexpression of WT Mfn2 or dominant negative Drp1, inhibits A53T α-synuclein-induced mitochondrial autophagy is consistent with the hypotheses proposed earlier. Mitochondrial fission, and specifically fragmentation, is required for the progression of mitochondrial autophagy (30–32), which suggests that promoting mitochondrial elongation may suppress excessive mitochondrial autophagy. Indeed, Tanaka et al. (33) demonstrated that inhibition of Drp1-mediated mitochondrial fission could prevent Parkin-induced mitochondrial autophagy in mouse embryonic fibroblasts and in HeLa cells. Kamp et al. (34) recently reported that α-synuclein could promote mitochondrial fission, which could also be one of the factors that further promotes A53T α-synuclein-induced mitochondrial autophagy.
Activated Autophagy, Protective or Toxic?
It is believed that boosting the intracellular degradation machinery is beneficial for protein aggregation-related neurodegenerative diseases. For example, the deletion of Cystatin B, which is an endogenous inhibitor of intracellular cathepsins, significantly reduced the brain amyloid plaque load in a mouse model of Alzheimer disease (35). Activation of autophagy by Beclin 1 injections ameliorated the synaptic and dendritic pathology in α-synuclein transgenic mice and reduced the accumulation of α-synuclein in the limbic system (36). It has been also demonstrated that activators of autophagy enhance the clearance of mutant huntingtin and attenuate toxicity in cellular and Drosophila models of Huntington disease (37). Autophagy induction with rapamycin also reduces mutant Ataxin-3 levels and toxicity in a mouse model of spinocerebellar ataxia type 3 (38).
However, evidence also suggests that overactivation of the degradative system may have deleterious effects. For example, even the partial loss of Cystatin B leads to neuronal loss, ataxia, and seizures in mice (39). We and others have shown that both cathepsin and autophagy inhibitors are protective in several cellular neurotoxicity models (40–42). A recent study by Xilouri et al. (9) demonstrated that the inhibition of autophagy by down regulating ATG5 mitigates mutant A53T α-synuclein-induced toxicity. In addition, it has also been shown that the inhibition of autophagy induction delays neuronal loss in a cellular model of frontotemporal dementia (43). Consistent with our results, Xilouri et al. (9) also demonstrated that mutant A53T α-synuclein overactivates the macroautophagic degradation of long-lived proteins. The authors suggested that overexpression of mutant A53T α-synuclein inhibits chaperone-mediated autophagy, which is followed by a compensatory increase in macroautophagy. These results are inconsistent with a recent report by Winslow et al. (44), which showed that α-synuclein could impair macroautophagy. However, this effect was specific to the WT protein, and mutant A53T α-synuclein had no inhibitory effect on the autophagy level. Still, the authors suggested that mutant synucleins could lead to a compensatory increase in autophagosome formation. Moreover, the human neuroblastoma cells (SKNSH cell line) used by Winslow et al. could also respond differently to α-synuclein when compared with the primary neurons used in our current study, which are known to be more sensitive to different cellular stresses.
Although these two sets of findings could be considered contradictory, they also suggest that the activation of autophagy has both yin and yang sides. The positive side is related to an increased clearance of aggregated proteins, as described in several studies (32, 45–49), while the negative side could be associated with too much clearance of “innocent” intracellular components. However, the question of how this activation leads to mitochondrial removal remains unclear. Compensatory macroautophagy could be nonspecific enough to engulf both polarized and healthy mitochondria. In contrast, there could also be some specificity toward mitochondrial autophagy or additional factors that make mitochondria more prone to removal.
Parkin: Toxic, Protective, or Both?
One of the currently known factors that targets defective mitochondria for autophagosomal removal is Parkin. Recent reports (16, 50–51) have demonstrated that Parkin is recruited to impaired mitochondria and promotes their autophagy. In addition to these experiments, we have demonstrated here that Parkin translocation is specific to damaged mitochondria. The irradiation of individual mitochondria expressing KillerRed to boost the ROS production led to selective Parkin translocation to these mitochondria. We also observed the frequent colocalization of Parkin positive mitochondria and LC3 in cells overexpressing mutant A53T α-synuclein. Furthermore, our results demonstrate that the suppression of Parkin inhibited mitochondrial removal, reversed the net mitochondrial loss, and partially protected cells from mutant A53T α-synuclein-induced neuronal death. These results were surprising, since it has been shown that Parkin overexpression is protective in a variety of models, including α-synuclein overexpressing neurons (52–53), and it has been suggested that the loss of Parkin function damages neurons through the accumulation of toxic proteins. Nevertheless, there are also several reports that are in apparent contradiction with this theory. A recent publication by Fournier et al. (54) demonstrated that a Parkin deficiency delays the progression of neurodegeneration in mice overexpressing mutant A30P α-synuclein. In another study, the Parkin deficiency had no impact on the onset and progression of the lethal phenotype induced by the overexpression of human A53T α-synuclein (55). These findings could again be explained by the dual role of Parkin. On one hand, its ubiquitin ligase activity may be essential for the removal of misfolded proteins, and the loss of Parkin could lead to the accumulation of these proteins. On the other hand, Parkin participates in the targeting of mitochondria for autophagic removal, and therefore the inhibition of the Parkin could suppress mitochondrial loss. This hypothesis suggests that a certain balance between Parkin-mediated degradation of misfolded proteins and mitochondrial removal is required for normal cell metabolism.
Taken together, our data suggest that overactivated mitochondrial removal could be one of the main contributing factors for mitochondrial loss. These results also demonstrate that mitochondrial loss has a prominent role in the observed mitochondrial defect. We propose that increased mitochondrial removal could be one of the links that connects intracellular accumulation of α-synuclein with the mitochondrial loss observed in PD. These results also show that the balanced activity of Parkin and macroautophagy is required for the proper functioning of cells, and therefore their overactivation may have both positive and negative consequences.
Acknowledgments
We thank Dr. E. Bampton, Dr. S. Kõks, Dr. S. Hirose, Dr. N. Nakamura, Dr. R. Youle, Dr. T. Johansen, Dr. C. Sahlin, Dr. H. D. Durham, and Dr. A. van der Bliek for providing the plasmids used in this study.
This work was supported by Grants 7991, 7175, and 8125 from the Estonian Science Foundation, the European Community (Projects BRAIN BIOENERGETICS and ESTBIOREG), and by the European Regional Development Fund.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- PD
- Parkinson disease
- BME
- Basal Medium Eagle with Earle's Salts
- DMNPE
- 1-(4,5-dimethoxy-2-nitrophenyl)ethyl ester
- Drp1
- dynamin-related protein
- Mfn2
- Mitofusin 2
- ROS
- reactive oxygen species
- TMRE
- tetramethylrhodamine ethyl ester
- TRF
- time resolved fluorescence.
REFERENCES
- 1. Schapira A. H., Cooper J. M., Dexter D., Jenner P., Clark J. B., Marsden C. D. (1989) Lancet 1, 1269. [DOI] [PubMed] [Google Scholar]
- 2. Henchcliffe C., Beal M. F. (2008) Nat. Clin. Pract. Neurol. 4, 600–609 [DOI] [PubMed] [Google Scholar]
- 3. Ayala A., Venero J. L., Cano J., Machado A. (2007) Front Biosci. 12, 986–1007 [DOI] [PubMed] [Google Scholar]
- 4. Schapira A. H. (2008) Lancet Neurol. 7, 97–109 [DOI] [PubMed] [Google Scholar]
- 5. Anglade P., Vyas S., Javoy-Agid F., Herrero M. T., Michel P. P., Marquez J., Mouatt-Prigent A., Ruberg M., Hirsch E. C., Agid Y. (1997) Histol. Histopathol. 12, 25–31 [PubMed] [Google Scholar]
- 6. Zhu J. H., Guo F., Shelburne J., Watkins S., Chu C. T. (2003) Brain Pathol. 13, 473–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kirik D., Rosenblad C., Burger C., Lundberg C., Johansen T. E., Muzyczka N., Mandel R. J., Björklund A. (2002) J. Neurosci. 22, 2780–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stefanis L., Larsen K. E., Rideout H. J., Sulzer D., Greene L. A. (2001) J. Neurosci. 21, 9549–9560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xilouri M., Vogiatzi T., Vekrellis K., Park D., Stefanis L. (2009) PLoS One 4, e5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang Q., She H., Gearing M., Colla E., Lee M., Shacka J. J., Mao Z. (2009) Science 323, 124–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Honda S., Hirose S. (2003) Biochem. Biophys. Res. Commun. 311, 424–432 [DOI] [PubMed] [Google Scholar]
- 12. Smirnova E., Shurland D. L., Ryazantsev S. N., van der Bliek A. M. (1998) J. Cell Biol. 143, 351–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lamark T., Perander M., Outzen H., Kristiansen K., Ǿvervatn A., Michaelsen E., Bjorkoy G., Johansen T. (2003) J. Biol. Chem. 278, 34568–34581 [DOI] [PubMed] [Google Scholar]
- 14. Sahlin C., Lord A., Magnusson K., Englund H., Almeida C. G., Greengard P., Nyberg F., Gouras G. K., Lannfelt L., Nilsson L. N. (2007) J. Neurochem. 101, 854–862 [DOI] [PubMed] [Google Scholar]
- 15. Batulan Z., Taylor D. M., Aarons R. J., Minotti S., Doroudchi M. M., Nalbantoglu J., Durham H. D. (2006) Neurobiol. Dis. 24, 213–225 [DOI] [PubMed] [Google Scholar]
- 16. Narendra D., Tanaka A., Suen D. F., Youle R. J. (2008) J. Cell Biol. 183, 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kimura S., Noda T., Yoshimori T. (2007) Autophagy 3, 452–460 [DOI] [PubMed] [Google Scholar]
- 18. Cantó C., Auwerx J. (2009) Curr. Opin. Lipidol 20, 98–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bulina M. E., Chudakov D. M., Britanova O. V., Yanushevich Y. G., Staroverov D. B., Chepurnykh T. V., Merzlyak E. M., Shkrob M. A., Lukyanov S., Lukyanov K. A. (2006) Nat. Biotechnol. 24, 95–99 [DOI] [PubMed] [Google Scholar]
- 20. Cheung Z. H., Ip N. Y. (2009) Mol. Brain 2, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moreau K., Luo S., Rubinsztein D. C. (2010) Curr. Opin. Cell Biol. 22, 206–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vila M., Ramonet D., Perier C. (2008) J. Neurochem. 107, 317–328 [DOI] [PubMed] [Google Scholar]
- 23. Chu C. T., Zhu J., Dagda R. (2007) Autophagy 3, 663–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Elmore S. P., Qian T., Grissom S. F., Lemasters J. J. (2001) Faseb. J. 15, 2286–2287 [DOI] [PubMed] [Google Scholar]
- 25. Lemasters J. J. (2005) Rejuvenation Res. 8, 3–5 [DOI] [PubMed] [Google Scholar]
- 26. Priault M., Salin B., Schaeffer J., Vallette F. M., di Rago J. P., Martinou J. C. (2005) Cell Death Differ. 12, 1613–1621 [DOI] [PubMed] [Google Scholar]
- 27. Kim I., Rodriguez-Enriquez S., Lemasters J. J. (2007) Arch. Biochem. Biophys. 462, 245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim I., Rodriguez-Enriquez S., Mizushima N., Ohsumi Y., Lemasters J. J. (2004) Hepatology 40, 291A [Google Scholar]
- 29. Kissová I., Deffieu M., Manon S., Camougrand N. (2004) J. Biol. Chem. 279, 39068–39074 [DOI] [PubMed] [Google Scholar]
- 30. Nowikovsky K., Reipert S., Devenish R. J., Schweyen R. J. (2007) Cell Death Differ. 14, 1647–1656 [DOI] [PubMed] [Google Scholar]
- 31. Twig G., Elorza A., Molina A. J., Mohamed H., Wikstrom J. D., Walzer G., Stiles L., Haigh S. E., Katz S., Las G., Alroy J., Wu M., Py B. F., Yuan J., Deeney J. T., Corkey B. E., Shirihai O. S. (2008) EMBO J. 27, 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kanki T., Wang K., Baba M., Bartholomew C. R., Lynch-Day M. A., Du Z., Geng J., Mao K., Yang Z., Yen W. L., Klionsky D. J. (2009) Mol. Biol. Cell 20, 4730–4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tanaka A., Cleland M. M., Xu S., Narendra D. P., Suen D. F., Karbowski M., Youle R. J. (2010) J. Cell Biol. 191, 1367–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kamp F., Exner N., Lutz A. K., Wender N., Hegermann J., Brunner B., Nuscher B., Bartels T., Giese A., Beyer K., Eimer S., Winklhofer K. F., Haass C. (2010) EMBO J. 29, 3571–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang D.-S., Stavrides P., Mohan P., Kumar A., Schmidt S. D., Pawlik M., Bandyopadhyay U., Mathews P. M., Levy E., Cuervo A. M., Nixon R. A. (2008) Alzheimer's Dementia 4, T223–T223 [Google Scholar]
- 36. Spencer B., Potkar R., Trejo M., Rockenstein E., Patrick C., Gindi R., Adame A., Wyss-Coray T., Masliah E. (2009) J. Neurosci. 29, 13578–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sarkar S., Davies J. E., Huang Z., Tunnacliffe A., Rubinsztein D. C. (2007) J. Biol. Chem. 282, 5641–5652 [DOI] [PubMed] [Google Scholar]
- 38. Menzies F. M., Huebener J., Renna M., Bonin M., Riess O., Rubinsztein D. C. (2010) Brain 133, 93–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kaasik A., Kuum M., Aonurm A., Kalda A., Vaarmann A., Zharkovsky A. (2007) Epilepsia 48, 752–757 [DOI] [PubMed] [Google Scholar]
- 40. Canu N., Tufi R., Serafino A. L., Amadoro G., Ciotti M. T., Calissano P. (2005) J. Neurochem. 92, 1228–1242 [DOI] [PubMed] [Google Scholar]
- 41. Kaasik A., Rikk T., Piirsoo A., Zharkovsky T., Zharkovsky A. (2005) Eur. J. Neurosci. 22, 1023–1031 [DOI] [PubMed] [Google Scholar]
- 42. Wang Y., Dong X. X., Cao Y., Liang Z. Q., Han R., Wu J. C., Gu Z. L., Qin Z. H. (2009) Eur. J. Neurosci. 30, 2258–2270 [DOI] [PubMed] [Google Scholar]
- 43. Lee J. A., Gao F. B. (2009) J. Neurosci. 29, 8506–8511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Winslow A. R., Chen C. W., Corrochano S., Acevedo-Arozena A., Gordon D. E., Peden A. A., Lichtenberg M., Menzies F. M., Ravikumar B., Imarisio S., Brown S., O'Kane C. J., Rubinsztein D. C. (2010) J. Cell Biol. 190, 1023–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Atwal R. S., Xia J., Pinchev D., Taylor J., Epand R. M., Truant R. (2007) Hum. Mol. Genet. 16, 2600–2615 [DOI] [PubMed] [Google Scholar]
- 46. Cuervo A. M. (2004) Trends Cell Biol. 14, 70–77 [DOI] [PubMed] [Google Scholar]
- 47. Cuervo A. M., Stefanis L., Fredenburg R., Lansbury P. T., Sulzer D. (2004) Science 305, 1292–1295 [DOI] [PubMed] [Google Scholar]
- 48. Iwata A., Christianson J. C., Bucci M., Ellerby L. M., Nukina N., Forno L. S., Kopito R. R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13135–13140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Webb J. L., Ravikumar B., Atkins J., Skepper J. N., Rubinsztein D. C. (2003) J. Biol. Chem. 278, 25009–25013 [DOI] [PubMed] [Google Scholar]
- 50. Geisler S., Holmstrom K. M., Skujat D., Fiesel F. C., Rothfuss O. C., Kahle P. J., Springer W. (2010) Nat. Cell Biol. 12, 119–131 [DOI] [PubMed] [Google Scholar]
- 51. Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R. L., Kim J., May J., Tocilescu M. A., Liu W., Ko H. S., Magrané J., Moore D. J., Dawson V. L., Grailhe R., Dawson T. M., Li C., Tieu K., Przedborski S. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 378–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lo Bianco C., Schneider B. L., Bauer M., Sajadi A., Brice A., Iwatsubo T., Aebischer P. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17510–17515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Petrucelli L., O'Farrell C., Lockhart P. J., Baptista M., Kehoe K., Vink L., Choi P., Wolozin B., Farrer M., Hardy J., Cookson M. R. (2002) Neuron. 36, 1007–1019 [DOI] [PubMed] [Google Scholar]
- 54. Fournier M., Vitte J., Garrigue J., Langui D., Dullin J. P., Saurini F., Hanoun N., Perez-Diaz F., Cornilleau F., Joubert C., Ardila-Osorio H., Traver S., Duchateau R., Goujet-Zalc C., Paleologou K., Lashuel H. A., Haass C., Duyckaerts C., Cohen-Salmon C., Kahle P. J., Hamon M., Brice A., Corti O. (2009) PLoS One 4, e6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. von Coelln R., Thomas B., Andrabi S. A., Lim K. L., Savitt J. M., Saffary R., Stirling W., Bruno K., Hess E. J., Lee M. K., Dawson V. L., Dawson T. M. (2006) J. Neurosci. 26, 3685–3696 [DOI] [PMC free article] [PubMed] [Google Scholar]