Abstract
Myosin phosphatase is a heterotrimeric holoenzyme consisting of myosin phosphatase-targeting subunit 1 (MYPT1), a catalytic subunit of PP1Cβ, and a 20-kDa subunit of an unknown function. We have previously reported that myosin phosphatase also controls mitosis, apparently by antagonizing polo-like kinase 1 (PLK1). Here we found that depletion of MYPT1 by siRNA led to precocious chromatid segregation when HeLa cells were arrested at metaphase by a proteasome inhibitor, MG132, or by Cdc20 depletion. Consistently, cyclin B1 and securin were not degraded, indicating that the chromatid segregation is independent of the anaphase-promoting complex/cyclosome. Precocious segregation induced by MYPT1 depletion requires PLK1 activity because a PLK1 inhibitor, BI-2536, blocked precocious segregation. Furthermore, the expression of an unphosphorylatable mutant of SA2 (SCC3 homologue 2), a subunit of the cohesin complex, prevented precocious chromatid segregation induced by MYPT1 depletion. It has been shown that SA2 at centromeres is protected from phosphorylation by PP2A phosphatase recruited by Shugoshin (Sgo1), whereas SA2 along chromosome arms is phosphorylated by PLK1, leading to SA2 dissociation at chromosome arms. Taken together, our results suggest that hyperactivation of PLK1 caused by MYPT1 reduction could override the counteracting PP2A phosphatase, resulting in precocious chromatid segregation. We propose that SA2 at the centromeres is protected by two phosphatases. One is PP2A directly dephosphorylating SA2, and the other is myosin phosphatase counteracting PLK1.
Keywords: Cell Cycle, Chromosomes, Myosin, Phosphatase, Protein Kinases, Chromatid Segregation, Myosin Phosphatase, Polo-like Kinase
Introduction
It has become increasingly clear that Ser/Thr protein phosphatases play active roles in regulating protein phosphorylation during mitosis (1). Protein phosphatase 1 (PP1) belongs to the phosphoprotein phosphatase (PPP) family of Ser/Thr protein phosphatases, and its catalytic subunits (PP1C) are known to bind to a variety of targeting subunits (2, 3). The association of PP1C with its targeting subunits is physiologically important for temporal and spatial regulation of protein phosphorylation because such binding provides the limited number of PP1C a means to counteract a wide array of Ser/Thr kinases by targeting PP1C either to a specific subcellular site or to a particular substrate at a specific stage of cell cycle and/or by allowing them to directly counteract a Ser/Thr kinase (2, 3). For example, it has been demonstrated that Repo-Man, a novel PP1C-targeting protein, is essential for recruiting PP1Cγ, one of three mammalian PP1C isoforms, to chromatin during anaphase. This targeting plays a critical role in the maintenance of chromosome architecture (4, 5). However, judging from the complexity of cell cycle control, multiple targeting subunits must be involved in various mitotic events. Thus, major questions remaining to be addressed are how and which mitotic event each targeting subunit controls and which kinase each targeting subunit counteracts.
Myosin phosphatase-targeting subunit 1 (MYPT1,2 also called myosin binging subunit (MBS) or M130) is a known targeting subunit of PP1C (2, 6). We have recently demonstrated that MYPT1 also controls mitosis, apparently by antagonizing polo-like kinase 1 (PLK1) (7). MYPT1 binds to PP1Cβ at its N terminus and to a 20-kDa subunit of unknown function at its C terminus to form a heterotrimeric holoenzyme. As the holoenzyme is known to dephosphorylate the regulatory light chains of myosin II and control actomyosin contractility in smooth muscle and nonmuscle cells, it is often called myosin phosphatase (MP) (2, 6). The phenotype of mice lacking MYPT1, however, indicates that MYPT1 plays a fundamental role in cell proliferation beyond the regulation of actomyosin contractility. Deletion of MYPT1 is embryonic lethal; the mice die at a very early stage of development (7.5 days postcoitus), and no MYPT1 null cells have been isolated so far (8). In contrast, mice lacking myosin light chain kinase (MLCK), a major kinase that phosphorylates the regulatory myosin light chains, are able to survive until birth, although they show postnatal lethality (9). Our previous finding that MYPT1 controls mitosis by counteracting PLK1 (7) explains, at least in part, the lethal phenotype of MYPT1 KO mice.
PLK1 is an essential mitotic kinase that controls a variety of critical mitotic events including the G2/M transition, centrosome maturation, bipolar mitotic spindle assembly, chromatid segregation, mitotic exit, and cytokinesis (10–15). We previously showed that depletion of MYPT1 resulted in increased phosphorylation of PLK1 at its activation site of Thr-210, and consistently, simultaneous reduction of both MYPT1 and PLK1 levels by siRNA rescued the mitotic arrest phenotype caused by single reduction of PLK1 levels. Due to this antagonism between MP and PLK1, loss of MYPT1 would be predicted to deleteriously activate PLK1 at incorrect times and/or places, disrupting one or more PLK1-mediated mitotic events. In this work, we examined the effects of MYPT1 depletion on mitotic exit, one of the PLK1-mediated mitotic events. We found that MYPT1 depletion resulted in premature chromatid segregation.
EXPERIMENTAL PROCEDURES
Cells and Antibodies
SW962 human vulva carcinoma (HTB-118, ATCC, Manassas, VA) and HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum. HeLa Tet-On cells that were able to induce Myc-tagged, wild type SA2 (called Myc-SA2-wild), as well as an unphosphorylatable mutant of SA2 (called Myc-SA2-12xA, mitosis-specific, 12 Ser and Thr sites replaced with Ala) were kindly provided by Dr. J.-M. Peters (Research Institute of Molecular Pathology, Vienna, Austria) (16). Myc-tagged SA2 was induced by 2 μg/ml doxycycline for 2 days. In some experiments, HeLa cells were synchronized by a double thymidine block. Treatment of MG132 (5 μm) was performed for 1–5.5 h. We confirmed that HeLa cells were kept arrested at mitosis during this time window of MG132 treatment. As live cell imaging (supplemental Fig. 1A) shows, HeLa cells stably expressing histone H2B-GFP were arrested at the metaphase during a 6-h treatment of MG132, and the metaphase chromatid morphology appeared intact without any sign of mitotic exit or apoptosis (see supplemental Movie 1). Furthermore, Western blotting revealed that the levels of cyclin B1, as well as those of securin and phospho-(Ser-10) histone H3, remained unaltered during 6 h of MG132 treatment (supplemental Fig. 1B).
The following antibodies were used: rabbit polyclonal antibodies against MYPT1 (17); a mouse polyclonal antibody against Ser-473-phopshorylated MYPT1 (7); a monoclonal antibody against Myc (9E10, Covance, Richmond, CA); human autoimmune serum against CREST (B. Brinkley, Baylor College of Medicine); a rabbit polyclonal antibody against Shugoshin (Sgo1) (Y. Watanabe, University of Tokyo); a rabbit polyclonal antibody against cyclin B1 (BioLegend, San Diego, CA); a mouse monoclonal antibody against cyclin B1 (GNS1, Santa Cruz Biotechnology); a rabbit polyclonal antibody against phospho-(Ser-10) histone H3 (Cell Signaling Technology, Boston MA); a rabbit polyclonal antibody against Cdc20 (Bethyl Laboratories, Montgomery TX); and a rabbit polyclonal antibody against securin (Invitrogen).
Immunofluorescence
Immunofluorescence was performed as described (17). Briefly, cells were fixed with 3.7% formaldehyde in the PHEM buffer (54 mm Pipes, 22.5 mm Hepes, 10 mm EGTA, and 8 mm MgSO4, pH 7.0) for 10 min and permeabilized with 0.2% Triton X-100 in PHEM for 5 min. After blocking with normal goat serum (The Jackson Laboratory, Bar Harbor, ME), cells were labeled with primary antibodies. Chromosomes were detected by DAPI staining (Molecular Probes). Mitotic cells were obtained by the shake-off method, cytospun onto poly-d-lysine-coated coverslips, and analyzed by immunofluorescence as described above. Immunofluorescent images were taken as Z-stacks with a DeltaVision image restoration microscope system (Applied Precision LLC, Issaquah, WA) and deconvoluted with the SoftWoRx software (Applied Precision LLC) or the Huygen software (Scientific Volume Imaging, Hilversum, The Netherlands), and projected images were generated. Exposure times and settings for deconvolution were constant for all samples to be compared within any given experiment. Image contrast and brightness were adjusted by Photoshop (Adobe, San Jose, CA). Immunofluorescent images were also taken by a Nikon TE300 microscope with a ×60 plan Apo lens (NA 1.40).
Live Cell Imaging
Live cell imaging was performed with HeLa cells or SW962 cells stably expressing histone H2B GFP as described previously (7). HeLa cells stably expressing histone H2B-GFP were kindly provided by Dr. G. M. Wahl (Salk Institute for Biological Studies, La Jolla, CA) (18), and SW962 cells stably expressing histone H2B-GFP were isolated by transfection with a pBOS-H2BGFP vector (BD Biosciences). After transfection with siRNAs for 72 h, cells were imaged at 37 °C using a DeltaVision Microscope system with a Narishige temperature control system (MS-200D, Narishige International USA, East Meadow, NY). Five Z-section images were taken in 1-min intervals and deconvoluted, and projected images were generated with the SoftWoRx software (Applied Precision LLC).
siRNA Treatment
MYPT1 was depleted by siRNA treatment as described (7), and the effects of depletion were analyzed at either 48 or 72 h after transfection. Sgo1 depletion was performed according to Ref. 19. Cdc20 depletion was performed using an siRNA sequence described in Ref. 20, which has been demonstrated to deplete Cdc20 levels less than 5% and to arrest HeLa cells at mitosis for an average of 18 h (20). Single-strand, sense RNA was used for a mock control.
Generation of MYPT1-depleted, Stable HeLa Cells
Lentiviral-based shRNA constructs (Mission shRNA, Sigma-Aldrich) were used to generate stable, MYPT1-depleted HeLa cells as described (21). Of three shRNA constructs (TRCN0000231493; 0000002445; 0000002444) we tested, an shRNA construct (TRCN0000231493) was found to show strongest reduction of MYPT1. The MYPT1-depleted, stable HeLa cells exhibited oblique chromatid segregation (data not shown), the phenotype characteristic to MYPT1-depleted HeLa cells by siRNA transfection (7), indicating that the shRNA-based depletion of MYPT1 replicated the phenotype observed with siRNA-based MYPT1 depletion. Control cells were isolated using a nontargeted shRNA construct.
Giemsa Staining and Immunofluorescence of Chromosome Spreads
Chromosome spreading was performed as described (22) with minor modifications. Briefly, mitotic cells were diluted four times with water for 7 min at room temperature. After centrifugation, swollen cells were fixed in Camoy's solution (75% methanol, 25% acetic acid). After washing three times with the same solution, cells were dropped onto coverslips, dried, and stained either with DAPI or with 5% Giemsa solution. Seventy to hundred chromatids were analyzed to determine chromatid segregation, and analyses were repeated three times.
Immunofluorescence of chromosome spreads was performed as described (19) with slight modification. Briefly, cells swollen as described above were cytospun on a coverslip, fixed with 4% formaldehyde containing 0.2% Triton X-100, and stained with the anti-Myc, anti-CREST antibodies together with DAPI.
Statistical Analysis
Statistical analyses were performed with two-tailed Student's t test. To determine the statistical significance of precocious segregation, fractions of mostly detached and completely scattered chromatids (see Fig. 1 for definition) were combined and analyzed by Student's t test. Means and standard deviations for the analyses of precocious chromatid segregation are listed in supplemental Table 1.
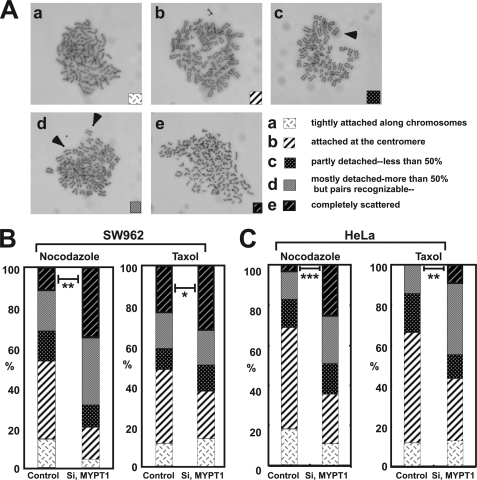
FIGURE 1.
Precocious chromatid segregation by MYPT1 depletion in the presence of nocodazole or taxol. A, Giemsa staining images representing five categories of the chromatid morphology. Arrowheads indicate segregated chromatids. B and C, MYPT1 depletion (Si) increases the fraction of segregated chromatids in the presence of either nocodazole or taxol. SW962 (B) or HeLa (C) cells were treated either with nocodazole or with taxol overnight, and then chromatid morphology was examined by Giemsa staining and categorized according to the groups shown in A. One asterisk, p < 0.05; two asterisks; p < 0.01; three asterisks, p < 0.001.
RESULTS
MYPT1 Is Required to Prevent Premature Chromatid Segregation in the Presence of Nocodazole or Taxol
Using Giemsa staining, we examined whether MYPT1 depletion affects chromatid segregation when cells are arrested overnight by nocodazole (0.25 μg/ml) or taxol (5 μm). We found that MYPT1 depletion resulted in premature chromatid segregation. To quantitatively determine the effects of MYPT1 depletion on chromatid segregation, we categorized chromatid morphology into the following five groups (Fig. 1A, panels a–e): (a) prophase chromatids; (b) metaphase chromatids; (c) partially detached chromatids (arrowheads, less than 50% detached at the centromere); (d) mostly detached chromatids (more than 50% sister detachment); and (e) completely scattered chromatids with no identifiable pairs of sister chromatids representing completely and prematurely segregated chromatids. Although categories “a” and “b” represent normal chromatids, categories “c” and “d” represent the process of premature chromatid segregation.
Figs. 1, B and C, show quantitative analyses of chromatid segregation of SW962 and HeLa cells, respectively. In nocodazole-treated SW962 cells, MYPT1 depletion greatly increased the fraction of category “e” (completely scattered chromatids) from 12 (control) to 35% (MYPT1-depleted). The fraction of mostly detached chromatids (category d) also increased from 17 to 34%. On the other hand, the percentage of sister chromatids attached at the centromeres (category b) was reduced from 43 (control) to 18% (MYPT1-depleted). Similar results were obtained when SW962 cells were treated with taxol.
HeLa cells also showed premature segregation of sister chromatids when MYPT1 was depleted. After overnight treatment of nocodazole or taxol, MYPT1 depletion increased the category of completely scattered chromatids (from 3 to 25% by nocodazole treatment and from 0 to 9% by taxol treatment). The category of mostly detached chromatids was also increased from 13 to 25% by nocodazole treatment and from 15 to 35% by taxol treatment. These results indicate that MYPT1 depletion results in precocious chromatid segregation when cells are arrested at mitosis.
Premature Chromatid Segregation Occurs Independently of the Anaphase-promoting Complex/Cyclosome (APC/C)
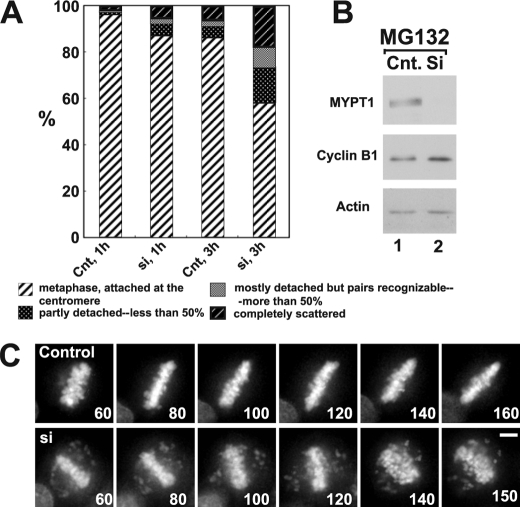
One possibility for precocious chromatid segregation is premature activation of APC/C. To test this possibility, we examined whether the inhibition of APC/C by a proteasome inhibitor, MG132, can block the precocious chromatid segregation of MYPT1-depleted HeLa cells. MYPT1-depleted cells were treated with MG132 for 1 and 3 h, and chromatid segregation was examined by Giemsa staining. Because MG132-arrested mitotic cells had few prophase cells, chromatid morphology was categorized into the following four groups including: (a) metaphase chromatids; (b) partially detached chromatids (less than 50% detached at the centromere); (c) mostly detached chromatids (more than 50% sister detachment); and (d) completely scattered chromatids.
As Fig. 2A shows, control mock-transfected cells showed no obvious chromatid segregation at 1 h, whereas MYPT1-depleted cells exhibited slightly increased chromatid segregation. At 3 h of MG132 treatment, however, the effects of MYPT1 depletion became obvious. About 19% of MYPT1-depleted cells exhibited completely scattered chromatids, whereas only 6% of control cells showed such chromatid morphology. Western blots with MG132-treated cells confirmed that cyclin B1 was not degraded in both control and MYPT1-depleted cells (B), indicating that APC/C was effectively blocked by MG132. Precocious segregation in the presence of MG132 was confirmed by live cell imaging using HeLa cells stably expressing GFP-histone H2B (C). We found that about one-fifth of MYPT1-depleted cells (4 out of 20 cells) exhibited precocious chromatid segregation (Fig. 2C, lower panel; see supplemental Movie 1), whereas virtually all control cells were arrested at metaphase (upper panel; see supplemental Movie 2).
FIGURE 2.
Precocious chromatid segregation in the presence of MG132. A, control (Cnt) and MYPT1-depleted (si) HeLa cells were treated with 5 μm MG132 for 1 or 3 h. Mitotic cells were isolated, and chromatid morphology was categorized into the four groups shown below the figure. One asterisk, p < 0.05; two asterisks, p < 0.01. B, Western blot analyses of control (lane 1) and MYPT1-depleted (lane 2) mitotic cells to indicate that cyclin B1 was not degraded in the presence of MG132. C, live cell imaging of control (upper panel) and MYPT1-depleted (lower panel) mitotic cells in the presence of MG132. Time-lapse imaging was started 1 h after the treatment. Examples of control and precocious segregation are shown. Bar, 10 μm. Time is in minutes.
Shugoshin (Sgo1) depletion has been demonstrated to cause precocious chromatid segregation (19, 23). It was thus possible that MYPT1 depletion might cause dissociation of Sgo1 from the centromeres, leading to precocious segregation. To test this possibility, Sgo1 localization was examined in MYPT1-depleted cells. We found that Sgo1 localization was not affected by MYPT1 depletion (see supplemental Fig. 2), indicating that precocious chromatid segregation by MYPT1 depletion is independent of Sgo1. In reciprocal experiments, we found that Sgo1 depletion did not affect MYPT1 localization. These results indicate that the localization of Sgo1 and MYPT1 is not interdependent.
Double Depletion of MYPT1 and Cdc20 Resulted in Increased Premature Chromatid Segregation
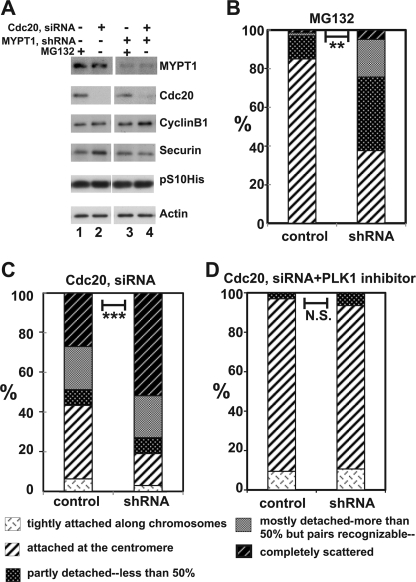
Because MG132 is a general inhibitor of proteasomes, we asked whether MYPT1-depleted cells show precocious segregation when APC/C is specifically inhibited by depletion of Cdc20, an activator of APC/C. To this end, MYPT1-depleted, stable HeLa cells generated with a lentiviral-based shRNA construct were transfected with siRNA to deplete Cdc20. Eight hours after transfection, control cells, as well as MYPT1-depleted, stable HeLa cells, were treated with thymidine (2 mm) for 24 h and released for synchronization. Cells entered mitosis at about 11 h after the release and stayed arrested at mitosis as reported (20, 24, 25). We collected mitotic cells at 14 h after thymidine block (at which time cells were arrested at mitosis for 4 h) and analyzed by Giemsa staining, as well as by Western blotting, to determine chromatid morphology and states of mitosis, respectively.
Western blots (Fig. 3A) revealed that Cdc20 depletion effectively blocked APC/C in both control cells (lanes 1 and 2) and MYPT1-depleted stable HeLa cells (lanes 3 and 4). The levels of cyclin B1 and securin (another substrate of APC/C) of Cdc20-depleted cells (lanes 2 and 4) were equivalent to those of MG132-arrested (for 2 h) mitotic cells (lanes 1 and 3). The levels of phospho-(Ser10)-histone H3 are also similar, indicating that Cdc20-depleted cells were arrested at mitosis in this time window.
FIGURE 3.
Double depletion of MYPT1 and Cdc20 results in severe precocious chromatid segregation. A, Western blots showing that Cdc20 depletion arrested both control and MYPT1-depleted HeLa cells at mitosis. Control HeLa cells (lanes 1 and 2) or HeLa cells stably expressing shRNA for MYPT1 depletion (lanes 3 and 4) were transfected with siRNA to deplete Cdc20 (lanes 2 and 4). Both cyclin B1 and securin are not degraded, indicating that APC/C is kept inactive by depletion of Cdc20. Lanes 1 and 2, no Cdc20 depletion. pS10His, phospho-(Ser10)-histone. B, precocious segregation shown by MYPT1-depleted, stable HeLa cells in the presence of MG132 for 2 h. C, severe precocious chromatid segregation by double depletion of MYPT1 and Cdc20. Although Cdc20 depletion alone resulted in precocious chromatid segregation of control HeLa cells to a certain extent, simultaneous depletion of MYPT1 and Cdc20 greatly increased the fraction of completely scattered chromatids. D, PLK1 inhibition blocked precocious chromatid segregation of Cdc20-depleted control or Cdc20-depleted, MYPT1-depleted HeLa cells. Two asterisks, p < 0.01; three asterisks, p < 0.001; N.S., statistically not significant.
We first checked whether shRNA-based MYPT1 depletion replicated the precocious chromatid segregation phenotype observed with siRNA-based MYPT1 depletion. Giemsa staining (B) revealed that MYPT1-depleted, stable HeLa cells, when arrested at mitosis for 2 h by MG132, showed precocious chromatid segregation. The fraction of completely scattered chromatids in MYPT1-depleted, stable HeLa cells was increased from 1.4 to 5%. The fraction of mostly detached chromatids also increased from 1.4 to 19%. The extent of precocious segregation is equivalent to that observed with siRNA-based MYPT1 depletion (compare Fig. 3B with Fig. 2A).
MYPT1 depletion also resulted in increased precocious segregation when cells were arrested at mitosis by Cdc20 depletion (C). Although Cdc20-depleted cells exhibited precocious chromatid segregation to a certain extent, double depletion of both Cdc20 and MYPT1 greatly increased the fraction of completely scattered chromatids from 27 to 52%. These findings, together with the precocious segregation of MYPT1-depleted cells in the presence of MG132, support the notion that precocious chromatid segregation by MYPT1 depletion is APC/C-independent. We do not know why Cdc20 depletion alone resulted in precocious segregation. Cdc20 has been reported to localize at kinetochores (25, 26) and affect kinetochore-microtubule attachments (25). It is thus possible that Cdc20 is involved in sister chromatid cohesion.
Premature Chromatid Segregation Requires PLK1 Activity
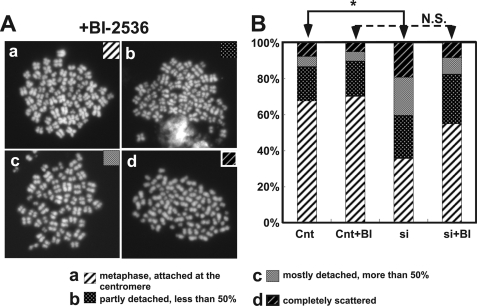
Our previous results indicate that MYPT1 depletion increased PLK1 activity (7). If the precocious chromatid segregation is caused by hyperactivated PLK1, then inhibition of PLK1 should block the precocious segregation. To test this idea, we examined whether MYPT1-depleted cells showed precocious segregation in the presence of a PLK1 inhibitor of BI-2536. After short time (1.5-h) treatment with MG132, metaphase cells were isolated by the shake-off method from control, as well as MYPT1-depleted cells (in this short time treatment, precocious chromatid segregation was minimum in both control and MYPT1-depleted cells, Fig. 2). The metaphase cells were then transferred into MG132-containing medium with or without BI-2536 for another 4 h, and their chromosome morphology was categorized into the same four groups as described in Fig. 2A, i.e. (a) metaphase chromatids; (b) partially detached chromatids (less than 50% detached at the centromere); (c) mostly detached chromatids (more than 50% sister detachment); and (d) completely scattered chromatids. It was noted that PLK1 inhibition altered chromosome appearance. As Fig. 4A shows, chromosomes in the presence of BI-2536 are more condensed and shorter than those in the absence of the inhibitor. Nonetheless, we were able to categorize the chromatid morphology of PLK1-inhibited cells into the above four groups (Fig. 4A, panels a–d).
FIGURE 4.
Precocious chromatid segregation requires PLK1 activity. A, representative images of chromatids in the presence of both MG132 and BI-2536. They are categorized into four groups including chromatids at metaphase (panel a), as well as partially separated chromatids (panel b), mostly separated chromatids (panel c), and completely scattered chromatids (panel d). Note that although chromatids are shorter and more condensed, chromosome arms in panel a are clearly separated as expected from metaphase arrested cells. B, quantitative measurements of chromatid morphologies of control (Cnt) and MYPT1-depleted (si) mitotic cells in the absence or presence of BI-2536. One asterisk, p < 0.05; N.S., statistically not significant.
Quantitative analyses (Fig. 4B) revealed that PLK1 inhibition blocked precocious segregation caused by MYPT1 depletion. In the absence of BI-2536, MYPT1 depletion causes precocious chromatid segregation. Forty-one percent of MYPT1-depleted cells exhibited either completely scattered chromatids or mostly detached chromatids, whereas only 13% of control cells showed such detached chromatids (the extent of precocious segregation in this experimental setting was more severe than that observed in Fig. 2, which is probably due to the longer treatment with MG132 (5.5 versus 3 h)). PLK1 inhibition, however, greatly reduced the fractions of the precociously segregated chromatids of MYPT1-depleted cells. Although 20% of MYPT1-depleted cells showed completely scattered chromatids in the absence of BI-2536, PLK1 inhibition decreased this fraction to 8%. The fraction of mostly detached chromatids was also lessened from 21 to 9%. PLK1 inhibition in control cells also reduced the fraction of completely scattered chromatids from 8 to 5%. These findings indicate that PLK1 activity is required for the precocious dissociation of cohesin at the centromeres in MYPT1-depleted cells and are consistent with our previous result that MP antagonizes PLK1 (7).
PLK1 activity was also required for the precocious segregation in MYPT1-depleted cells that were arrested at mitosis by Cdc20 depletion. As Fig. 3D shows, mostly detached and completely scattered sister chromatids were virtually eliminated in the presence of BI-2536, further supporting the requirement of PLK1 for precocious segregation caused by MYPT1 depletion.
An Unphosphorylatable Mutant of SA2 Blocks Premature Chromatid Segregation
One possible mechanism for the precocious segregation by MYPT1 depletion is via phosphorylation of a cohesin subunit of SA2. It has been reported that PLK1 phosphorylates SA2, leading to the dissociation of most cohesin complexes along chromosome arms during prometaphase. The cohesin complex at the centromeres, however, was protected from phosphorylation by PP2A protein phosphatase recruited by Sgo1, preventing untimely chromatid segregation (1, 16, 27). We therefore hypothesized that excessive activation of PLK1 caused by MYPT1 depletion would override PP2A phosphatase, leading to SA2 phosphorylation at the centromeres, thereby dissociating the cohesin complex and causing precocious chromatid segregation.
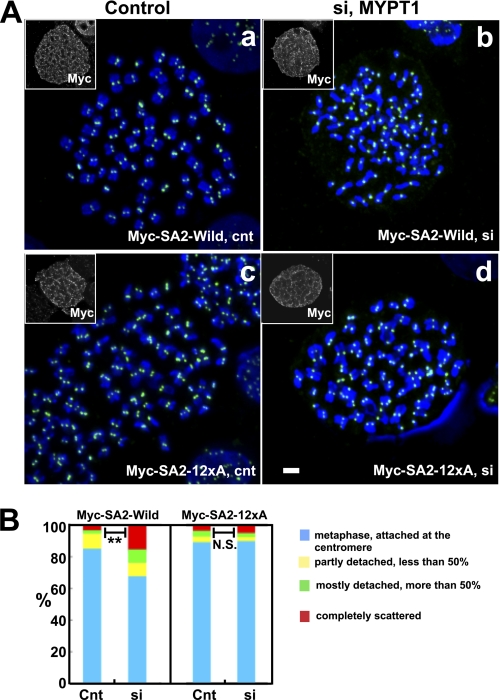
To test this hypothesis, we examined whether the unphosphorylatable mutant of SA2 could prevent precocious chromatid segregation of MYPT1-depleted cells. We used Tet-On HeLa cell lines developed by Hauf et al. (16), who demonstrated that the SA2 unphosphorylatable mutant stayed associated with centromeres, as well as along chromosome arms. The Tet-On HeLa cells can induce Myc-tagged, unphosphorylatable mutant (Myc-SA2-12xA) or wild type SA2 (see Ref. 16) upon the addition of doxycycline. Because about 30–40% of cells of these cell lines lost expression of Myc-tagged SA2, chromosome spreads were stained with an anti-Myc antibody to identify cells expressing the Myc-tagged SA2. They were counterstained with DAPI and an anti-CREST antibody to determine chromatid morphology and kinetochore segregation, respectively. Fig. 5A shows typical immunofluorescence of chromosome spreads from mock-transfected, control cells expressing wild type SA2 (a) or mutant SA2 (c), as well as from MYPT1-depleted cells expressing wild type (b) or mutant SA2 (d). CREST staining of MYPT1-depleted cells expressing the mutant SA2 revealed that all kinetochores were present as closely spaced pairs (d), indicating that the unphosphorylatable mutant of SA2 blocked precocious segregation of chromatids. In contrast, MYPT1-depleted cells expressing wild type SA2 showed scattered CREST staining (b), indicating chromatid segregation. Quantitative measurement (B) confirmed the above conclusion. The unphosphorylatable mutant of SA2, but not wild type SA2, effectively protected MYPT1-depleted cells from precocious chromatid segregation. Without induction with doxycycline, both types of cells show precocious chromatid segregation to a similar extent when MYPT1 was depleted (supplemental Fig. 3). Taken together, these results support our hypothesis that MP, by antagonizing PLK1, prevents SA2 phosphorylation at the centromeres.
FIGURE 5.
The unphosphorylatable SA2 mutant rescues precocious chromatid segregation. A, representative immunofluorescence images of chromatid spreads isolated from control (Cnt) (panel a and c) and MYPT1-depleted (si) (panels b and d) Tet-On HeLa cells expressing Myc-tagged, wild type (panels a and b) or the unphosphorylatable mutant (panels c and d) of SA2. Merged images show CREST (green) and DAPI (blue) localization. Insets, Myc staining. Bar, 10 μm. B, quantitative measurements of chromatid segregation of control or MYPT1-depleted Tet-On HeLa cells expressing wild type or the mutant SA2. Two asterisks, p < 0.01; N.S., statistically not significant.
MYPT1-depleted SW962 Cells Exit Mitosis Prematurely under Normal Conditions
Our result that MYPT1 depletion causes premature chromatid segregation in mitotically arrested cells suggests that MYPT1 depletion could also affect mitotic exit under normal conditions. We found that this is the case with SW962 cells.
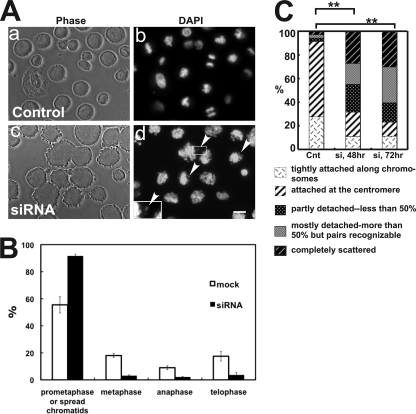
Fig. 6A shows phase-contrast (panels a and c) and DAPI staining (panels b and d) images of control (panels a and b) and MYPT1-depleted (panels c and d) mitotic cells isolated from asynchronous culture of SW962 cells. As expected from asynchronous culture, DAPI staining of control mitotic cells (A, panel b) exhibited varying stages of mitosis including prophase, prometaphase, metaphase, anaphase, and telophase. In contrast, MYPT1-depleted chromatids were mostly abnormal, showing few chromosomes at a metaphase, anaphase, or telophase stage (A, panel d). They were clearly more spread than normal prometaphase chromatids and did not seem to represent normal prophase chromosomes. Quantitative analyses (B) revealed that about 90% of MYPT1-depleted chromatids (n = 287) had such spread morphology. Box plot analyses of area measurements (supplemental Fig. 4) revealed that the median value of the area of MYPT1-depleted chromatids was twice as large as that of control prophase chromatids (p < 0.0001). This large increase of the area could not be accounted by cytokinesis failure alone because MYPT1 depletion resulted in only 16% increase in multinuclear cells (7). Importantly, these spread chromatids were often associated with orphan chromatids (A, panel d, arrowheads), suggesting that chromatids are precociously segregated and/or incorrectly connected to kinetochore microtubules.
Giemsa staining (Fig. 6C) confirmed that the chromatids of MYPT1-depleted SW962 cells were indeed precociously segregated without mitotic arrest. The morphology of mitotic chromatids was categorized into five groups according to the same criteria described for Fig. 1 (anaphase and telophase chromatids were excluded from the measurements). Although more than 90% of control cells exhibited normal chromatids with either the prophase (category a) or the prometaphase/metaphase (category b) morphology, only 33% of MYPT1-depleted chromatids showed such normal morphology. The fraction of category e (completely scattered chromatids) in MYPT1-depleted mitotic cells was increased from 3 to 30% after 72 h of MYPT1 depletion. In addition, partially (category c) and mostly (category d) detached chromatids were also increased from 3 to 16% and from 3 to 30%, respectively.
FIGURE 6.
MYPT1 depletion results in precocious chromatid segregation under normal conditions. A, MYPT1-depleted cells accumulate mitotic cells with spread chromatid morphology. Control (panels a and b) or MYPT1-depleted (panels c and d) mitotic cells were collected by the shake-off method and observed with phase-contrast (panels a and c) or fluorescence microscopy with DAPI staining (panels b and d). In panel d, arrowheads indicate orphan chromosomes in MYPT1-depleted cells (inset, the enlarged image of the white rectangle showing an orphan chromosome). Bar, 20 μm. Representative images from three independent experiments are shown. B, distribution of prometaphase, metaphase, anaphase, and telophase cells in mock-transfected (white bar) and MYPT1-depleted (black bar) mitotic cells. Chromatids with spread morphology of MYPT1-depleted cells are not clearly distinguished from prophase cells and thus are categorized into spread chromatids. C, precocious chromatid segregation by MYPT1 depletion. Chromatids from control (Cnt) and MYPT1-depleted (si) mitotic cells (48- or 72-h depletion) were Giemsa-stained and categorized into the five groups according to the criteria shown in Fig. 1A. Two asterisks, p < 0.01.
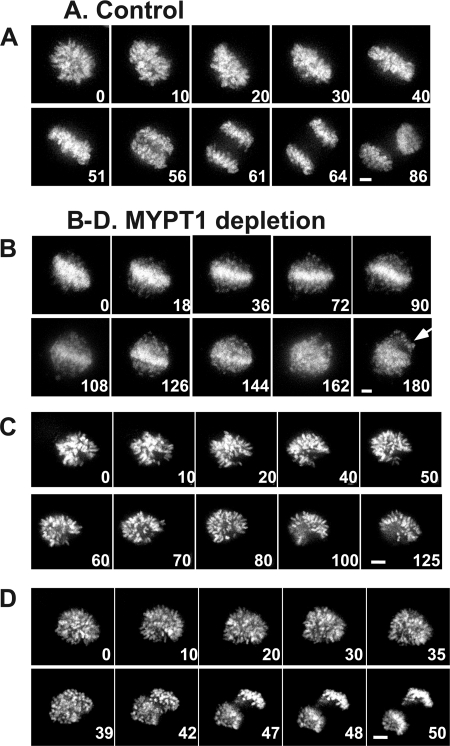
Precocious chromatid segregation was confirmed by live cell imaging with SW962 cells stably expressing GFP-tagged histone H2B. Fig. 7 exhibits representative still images of mitotic progression for control (A) and three typical examples of MYPT1-depleted (B–D) SW962 cells. In control cells (15/16), sister chromatids were able to align at the metaphase plate around 40–50 min and segregated at about 50–60 min after chromatid condensation (A; see supplemental Movie 3). In contrast, most (30/32) MYPT1-depleted cells showed abnormal mitosis of three phenotypes (B–D). The largest fraction (53%, 16 out of 30) was unable to completely align the chromatids at the metaphase plate and showed apparent premature segregation of chromatids (B; see supplemental Movie 4). The precocious segregation resulted in spread chromatids with orphan chromosomes (B, arrows at 180 min), explaining the image obtained by DAPI staining of MYPT1-depleted chromatids (Fig. 6A, panel d). In the second phenotype (13%, 4/30), chromatids were unable to align, did not show apparent segregation (C; see supplemental Movie 5), and stayed condensed for a prolonged time (over 2 h). The third phenotype (6.7%, 2/30) observed in MYPT1-depleted cells was oblique segregation of chromatids (D; see supplemental Movie 6). These results strongly suggest that the majority of MYPT1-depleted chromatids were precociously segregated.
FIGURE 7.
Live cell imaging of precocious chromatid segregation shown by of MYPT1-depleted SW962 cells. A, control. B–D, MYPT1-depleted SW962 cells stably expressing histone H2B-GFP. Examples of precocious segregation (B), prolonged mitosis (C), and oblique segregation (D) are shown. B, major phenotype of MYPT1-depleted cells; C and D, minor phenotype. Bar, 5 μm. Time is in minutes.
DISCUSSION
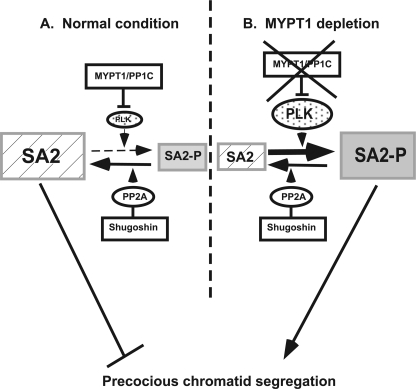
We have shown that MYPT1 depletion induced precocious chromatid segregation in the absence of APC/C activity. This precocious segregation requires PLK1 activity (Fig. 4) and is blocked by expression of the unphosphorylatable mutant of SA2 (Fig. 5), suggesting that SA2 phosphorylation by PLK1 at the centromeres is likely to cause the precocious segregation. SA2 phosphorylation is controlled by a balance between kinases (including PLK1) and PP2A phosphatase. This enzymatic balance is inherently delicate and could be error-prone. If PLK1 is hyperactivated, it could override PP2A phosphatase. Such hyperactivation of PLK1 could happen, in particular, at the kinetochores and their vicinities because PLK1 activity needs to be kept active for the attachment between kinetochores and kinetochore microtubules, as well as stabilization of kinetochore microtubules (28, 29). We propose that MP keeps PLK1 in check, thereby acting as a safeguard to protect the cohesin complexes at the centromeres from deleterious dissociation before the onset of the anaphase. Fig. 8 shows our model. Under normal conditions (A), the cohesin complexes at the centromeres are protected by two phosphatases; one is Sgo1-associated PP2A, directly dephosphorylating SA2, and the other is MP (MYPT1-PP1C), keeping PLK1 in check. In MYPT1-depleted cells (B), PLK1 is hyperactivated, overriding dephosphorylation by PP2A, leading to precocious chromatid segregation.
FIGURE 8.
A working model for the role of MP in antagonizing PLK1 in the regulation of SA2 phosphorylation at the centromeres.
It has been reported that depletion of PLK1 or inhibition of AuroraB did not rescue precocious segregation of chromatids observed in Sgo1-depleted cells, suggesting that there is yet another kinase that phosphorylates SA2 (19). This result does not seem to be consistent with our result that inhibition of PLK1 blocked precocious segregation caused by MYPT1 depletion (Fig. 4). Although we do not know the reason for the discrepancy, we speculate that double depletion of Sgo1 and PLK1 might activate some other kinases that could phosphorylate SA2. This speculation is based on the following reports. PLK1 depletion alone is known to block the dissociation of the cohesin complex along chromosome arms (16, 30), indicating that PLK1 is mainly responsible for the dissociation of chromosome arms. However, cells depleted for both Sgo1 and PLK1 have been shown to exhibit chromatid morphology with open arms (19). Thus, it is possible that other kinases that are not usually activated in normal, as well as PLK1 single-depleted cells, might be activated in these double-depleted cells.
In contrast to MYPT1-depleted SW962 cells, MYPT1-depleted HeLa cells showed no obvious precocious chromatid segregation under normal conditions. Instead, MYPT1-depleted HeLa cells frequently underwent oblique chromatid segregation (7). The reason for this difference in the phenotypes is not clear at this time. One reason could be due to lower levels of MYPT1 in SW962 cells. The MYPT1 level in SW962 cells is 3-fold less than that in HeLa cells (data not shown), and thus, MYPT1 depletion in SW962 cells is more complete than that in HeLa cells. The correlation between more severe effects and more complete depletion of MYPT1 appears to be consistent with the embryonic lethal phenotype of MYPT1 KO mice (8).
We and others have demonstrated that MYPT1 and PLK1 are associated in a mitosis-specific way (7, 31). Such physical association would allow rapid regulation of PLK1. In addition, because MP is controlled by phosphorylation of MYPT1 with upstream kinases including ROCK (for review, see Ref. 6), such upstream kinases could form a new regulatory pathway for PLK1. For example, because ROCK is present in centrosomes (32), it might be possible that ROCK negatively regulates MYPT1, allowing PLK1 to be more active at the centrosomes. Furthermore, PLK1 is known to phosphorylate and activate ROCK (31), possibly generating a positive feedback system for further activation of PLK1. Such activation of PLK1 may be necessary for the centrosomal functions including centrosome maturation. On the other hand, the apparent absence of ROCK at the kinetochores might keep MP active, controlling PLK1 for the protection of the centromeric cohesin.
Acknowledgments
We thank Dr. G. M. Wahl for HeLa cells stably expressing histone H2B-GFP, Dr. J.-M. Peters for Tet-On HeLa cells expressing Myc-tagged SA2 and SA2 mutant, Dr. B. Brinkley for the anti-CREST antibody, Dr. Y. Watanabe for the anti-Shugoshin antibody, and Drs Barth Grant and Frank Deis for critical reading.
This work was supported, in whole or in part, by National Institutes of Health Grant CA42742 throughout the NCI (to F. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1, Figs. 1–4, and movies 1–6.
- MYPT1
- myosin phosphatase-targeting subunit 1
- PLK1
- polo-like kinase 1
- MP
- myosin phosphatase
- APC/C
- anaphase-promoting complex/cyclosome
- ROCK
- Rho-associated protein kinase.
REFERENCES
- 1. Trinkle-Mulcahy L., Lamond A. I. (2006) Curr. Opin. Cell Biol. 18, 623–631 [DOI] [PubMed] [Google Scholar]
- 2. Cohen P. T. (2002) J. Cell Sci. 115, 241–256 [DOI] [PubMed] [Google Scholar]
- 3. Ceulemans H., Bollen M. (2004) Physiol. Rev. 84, 1–39 [DOI] [PubMed] [Google Scholar]
- 4. Vagnarelli P., Hudson D. F., Ribeiro S. A., Trinkle-Mulcahy L., Spence J. M., Lai F., Farr C. J., Lamond A. I., Earnshaw W. C. (2006) Nat. Cell Biol. 8, 1133–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Trinkle-Mulcahy L., Andersen J., Lam Y. W., Moorhead G., Mann M., Lamond A. I. (2006) J. Cell Biol. 172, 679–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hartshorne D. J., Ito M., Erdödi F. (2004) J. Biol. Chem. 279, 37211–37214 [DOI] [PubMed] [Google Scholar]
- 7. Yamashiro S., Yamakita Y., Totsukawa G., Goto H., Kaibuchi K., Ito M., Hartshorne D. J., Matsumura F. (2008) Dev. Cell 14, 787–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Okamoto R., Ito M., Suzuki N., Kongo M., Moriki N., Saito H., Tsumura H., Imanaka-Yoshida K., Kimura K., Mizoguchi A., Hartshorne D. J., Nakano T. (2005) Transgenic Res. 14, 337–340 [DOI] [PubMed] [Google Scholar]
- 9. Somlyo A. V., Wang H., Choudhury N., Khromov A. S., Majesky M., Owens G. K., Somlyo A. P. (2004) J. Muscle Res. Cell Motil. 25, 241–242 [DOI] [PubMed] [Google Scholar]
- 10. Blagden S. P., Glover D. M. (2003) Nat. Cell Biol. 5, 505–511 [DOI] [PubMed] [Google Scholar]
- 11. Barr F. A., Silljé H. H., Nigg E. A. (2004) Nat. Rev. Mol. Cell Biol. 5, 429–440 [DOI] [PubMed] [Google Scholar]
- 12. Dai W., Cogswell J. P. (2003) Prog. Cell Cycle Res. 5, 327–334 [PubMed] [Google Scholar]
- 13. Archambault V., Glover D. M. (2009) Nat. Rev. Mol. Cell Biol. 10, 265–275 [DOI] [PubMed] [Google Scholar]
- 14. Petronczki M., Lénárt P., Peters J. M. (2008) Dev. Cell 14, 646–659 [DOI] [PubMed] [Google Scholar]
- 15. Lowery D. M., Lim D., Yaffe M. B. (2005) Oncogene 24, 248–259 [DOI] [PubMed] [Google Scholar]
- 16. Hauf S., Roitinger E., Koch B., Dittrich C. M., Mechtler K., Peters J. M. (2005) PLoS Biol. 3, e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Totsukawa G., Yamakita Y., Yamashiro S., Hartshorne D. J., Sasaki Y., Matsumura F. (2000) J. Cell Biol. 150, 797–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kanda T., Sullivan K. F., Wahl G. M. (1998) Curr. Biol. 8, 377–385 [DOI] [PubMed] [Google Scholar]
- 19. McGuinness B. E., Hirota T., Kudo N. R., Peters J. M., Nasmyth K. (2005) PLoS Biol. 3, e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang H. C., Shi J., Orth J. D., Mitchison T. J. (2009) Cancer Cell 16, 347–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zagórska A., Deak M., Campbell D. G., Banerjee S., Hirano M., Aizawa S., Prescott A. R., Alessi D. R. (2010) Sci. Signal. 3, ra25. [DOI] [PubMed] [Google Scholar]
- 22. Giménez-Abián J. F., Clarke D. J., Mullinger A. M., Downes C. S., Johnson R. T. (1995) J. Cell Biol. 131, 7–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Salic A., Waters J. C., Mitchison T. J. (2004) Cell 118, 567–578 [DOI] [PubMed] [Google Scholar]
- 24. Wolthuis R., Clay-Farrace L., van Zon W., Yekezare M., Koop L., Ogink J., Medema R., Pines J. (2008) Mol. Cell 30, 290–302 [DOI] [PubMed] [Google Scholar]
- 25. Malureanu L., Jeganathan K. B., Jin F., Baker D. J., van Ree J. H., Gullon O., Chen Z., Henley J. R., van Deursen J. M. (2010) J. Cell Biol. 191, 313–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kallio M. J., Beardmore V. A., Weinstein J., Gorbsky G. J. (2002) J. Cell Biol. 158, 841–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kitajima T. S., Sakuno T., Ishiguro K., Iemura S., Natsume T., Kawashima S. A., Watanabe Y. (2006) Nature 441, 46–52 [DOI] [PubMed] [Google Scholar]
- 28. Lénárt P., Petronczki M., Steegmaier M., Di Fiore B., Lipp J. J., Hoffmann M., Rettig W. J., Kraut N., Peters J. M. (2007) Curr. Biol. 17, 304–315 [DOI] [PubMed] [Google Scholar]
- 29. Peters U., Cherian J., Kim J. H., Kwok B. H., Kapoor T. M. (2006) Nat. Chem. Biol. 2, 618–626 [DOI] [PubMed] [Google Scholar]
- 30. Giménez-Abián J. F., Sumara I., Hirota T., Hauf S., Gerlich D., de la Torre C., Ellenberg J., Peters J. M. (2004) Curr. Biol. 14, 1187–1193 [DOI] [PubMed] [Google Scholar]
- 31. Lowery D. M., Clauser K. R., Hjerrild M., Lim D., Alexander J., Kishi K., Ong S. E., Gammeltoft S., Carr S. A., Yaffe M. B. (2007) EMBO J. 26, 2262–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chevrier V., Piel M., Collomb N., Saoudi Y., Frank R., Paintrand M., Narumiya S., Bornens M., Job D. (2002) J. Cell Biol. 157, 807–817 [DOI] [PMC free article] [PubMed] [Google Scholar]