Abstract
During the last 15 years, the perception of the cardiac z-disc has undergone substantial changes. Initially viewed as a structural component at the lateral boundaries of the sarcomere, the cardiac z-disc has increasingly become recognized as a nodal point in cardiomyocyte signal transduction and disease. This minireview thus focuses on novel components and recent developments in z-disc biology and their role in cardiac signaling and disease.
Keywords: Calcineurin, Cardiac Muscle, Cytoskeleton, Nuclear Translocation, Signal Transduction, Sarcomere, Z-disc
Overview
Anatomy
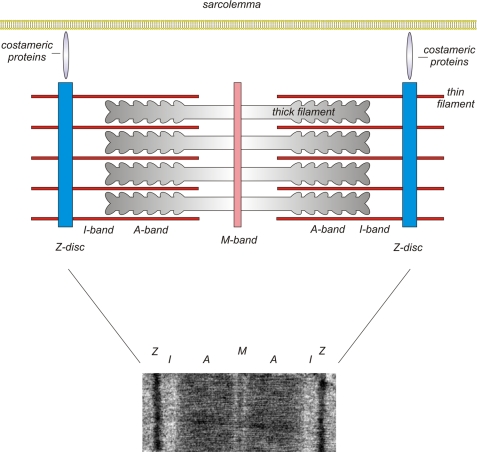
The z-discs (z-lines, z-bands) are the lateral boundaries of the basal contractile unit of the myocyte, the sarcomere. Three of the four filament systems of the sarcomere, filamentous F-actin, titin, and nebulin/nebulette, interact with z-disc structures (reviewed in Ref. 1). Only the myosin-based thick filaments do not directly interact with the z-disc (a schematic drawing of a sarcomere is shown in Fig. 1). The z-discs of individual sarcomeres are aligned in parallel and connected by the intermediate filament desmin, thereby providing a link to the intermediate filaments. In addition, costameres (Latin, costa, rib; Greek, meros, part), which consist of peripheral z-disc and subsarcolemmal proteins, ensure force transmission from the sarcomere to the sarcolemma (reviewed in Ref. 2).
FIGURE 1.
EM photography and schematic overview of the cardiac sarcomere. The z-discs represent the lateral boundary of the sarcomere. The F-actin-containing thin filaments anchor at the z-disc and interdigitate with the myosin-containing thick filaments at the level of the A-band. Costameric proteins ensure lateral force transduction and linkage to the sarcolemma and its associated protein complexes.
The backbone of the z-disc consists of layers of α-actinin aligned in an antiparallel fashion (Fig. 2). The amount of α-actinin layers (each measuring ∼19 nm) thereby determines the width of the z-disc, which in cardiac and slow skeletal muscle is typically 100–140 nm, whereas fast white muscle fibers have narrower z-discs of 30–50 nm (3). α-Actinin cross-links the interdigitating barbed ends of the thin filament F-actin of adjacent sarcomeres (3). Although several excellent reviews have been written on z-disc structure (1, 3), we focus here on the emerging role of the sarcomeric z-disc as a nodal point and hub of cardiomyocyte signaling.
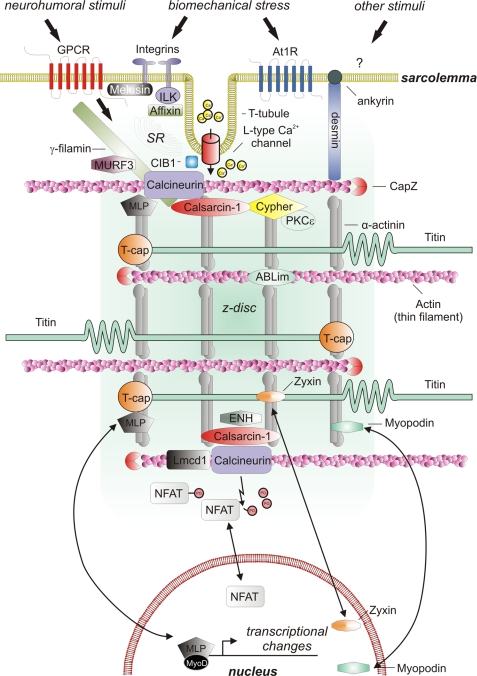
FIGURE 2.
Signaling molecules localizing to the cardiac z-disc. Shown is a schematic depiction of a cardiac z-disc and its adjacent structures involved in signaling, including sarcolemmal and nuclear structures. GPCR, G-protein-coupled receptor; At1R, angiotensin II type 1 receptor; SR, sarcoplasmic reticulum.
Signaling
This dual role of the z-disc places it in an ideal position to sense, integrate, and transduce biomechanical stress signals. Specifically, multiple upstream signals from the sarcomere as well as the membrane converge on the z-disc. Likewise, several components of downstream signaling, including bona fide signaling molecules such as kinases and phosphatases (including the phosphatase calcineurin and protein kinases like PKC) and their positive and negative modulators, are localized at or in immediate proximity of the z-disc. Moreover, several z-disc molecules share the ability to shuttle to the nucleus, where they can act as transcriptional co-modulators.
Mechanotransduction and Hypertrophy
An important component of cardiac signaling is a process termed mechanotransduction, which describes how mechanical stress is sensed by the cardiac myocyte and translated into a transcriptional response (4). Biomechanical stress is one of the most important stimuli leading to cardiac hypertrophy. Moreover, sustained pressure and/or volume overload ultimately leads to congestive heart failure, arrhythmias, and sudden cardiac death. Z-disc-associated proteins appear to play a critical role in this process (reviewed in Refs. 5–8). Several mouse knock-out studies of z-disc proteins, including muscle LIM protein (MLP)2 (9), calsarcin-1 (10), and the costameric protein melusin (11), revealed an indispensable role for these molecules in the adaptation to increased cardiac stress.
Molecular Architecture of Z-disc Proteins
Despite the fact that z-disc proteins are hallmarked by a remarkable heterogeneity, some molecular domains appear to be overrepresented. In particular, much attention has been paid to LIM proteins (originally described in the LIM proteins LIN-11, Isl1m, and MEC-3). LIM domains are tandem zinc finger structures that consist of the cysteine-rich consensus sequence CX2CX16–23HX2CX2CX2CX16–21CX2(C/H/D) (12) and that are known to mediate diverse protein-protein interactions (13). At least 10 LIM domain-containing proteins have been reported to localize to the z-disc (zyxin, Lmcd1 (LIM and cysteine-rich domains-1)/dyxin, MLP (CSRP3), ALP, Cypher/ZASP/Oracle, ENH (Enigma-like homolog), Enigma, CLP36/Elfin/CLIM1, ABLim, and the FHL family (which is also known to localize to the I-band (5)). As several LIM domain proteins have been shown to shuttle to the nucleus, they are attractive candidates to provide a connection between stretch sensing at the level of the sarcomere and signaling modules leading to transcriptional changes and cardiomyocyte growth.
Another important domain present at the z-disc is the PDZ domain (postsynaptic density 95, discs large, and zonula occludens-1). PDZ domains are protein-protein interaction modules that can mediate multiple biological processes such as vesicle transport, ion channel signaling, and signal transduction in several tissues (14). Interestingly, several proteins at the z-disc share both PDZ and LIM domains (recently reviewed by Zheng et al. (15)). Again, at least five of the 10 known PDZ-LIM proteins localize to the z-disc (including Enigma, ENH, Cypher/ZASP/Oracle, CLP36, and ALP). Recently, van Nandelstadh et al. (16) proposed a new PDZ type III domain in proteins of the myotilin family (including myotilin, palladin, and myopalladin) and the calsarcin/FATZ/myozenin family that mediates the interaction of both families with proteins of the Enigma family.
In conclusion, many z-disc proteins share “sticky” domains. These molecules mediate multiple protein-protein interactions, thereby playing a major role in integrating structure and signaling in this complex three-dimensional network.
Role of the Z-disc in Pathogenesis of Primary Cardiomyopathies
Given the importance of z-disc components in mechanotransduction and modulation of hypertrophic cardiac growth, it is not surprising that z-disc proteins have increasingly been shown to be causally involved in the pathogenesis of primary cardiomyopathies (for review, see Ref. 17). Our group (18) and others have shown that up to 35% of patients suffering from dilated cardiomyopathy (DCM) have relatives with reduced cardiac contractility and/or left ventricular dilation, suggesting that a substantial amount of dilated cardiomyopathies might be caused by inherited gene mutations. In contrast to hypertrophic cardiomyopathy, which is often termed a “disease of the sarcomere” because nearly all known mutations are located in sarcomeric proteins (reviewed in Ref. 19), DCM-causing mutations are more heterogeneous. DCM-associated proteins include sarcolemmal proteins, proteins of the nuclear membrane, or proteins implicated in calcium metabolism but also a steadily growing number of z-disc proteins. These DCM-associated z-disc proteins include Cypher/ZASP/Oracle (20), MLP (9), calsarcin,3 T-cap (telethonin cap)/telethonin (21), α-actinin-2 (22), myopalladin (23), myotilin (24), nexilin (25), and titin (26). Interestingly, mutations in several z-disc proteins can also cause hypertrophic cardiomyopathy, including MLP (27), calsarcin (28), T-cap/telethonin (21), and titin (29).
The z-disc integrates structural and signaling molecules, and mutations in either component can lead to severe functional failure, resulting in cardiac disease. We will thus highlight below a selected subset of important molecules involved in z-disc signaling.
Upstream Signaling Targeting the Cardiac Z-disc
The z-disc is structurally linked to the sarcolemma via the costameres, which circumferentially surround the z-disc (2). The costameres are in many regards analogous to focal adhesions in other cell types (30). Several costameric proteins directly interact with z-disc molecules and thus may mediate upstream signals.
Desmin
Desmin is the major intermediate filament in cardiac muscle, accounting for ∼2% of cardiac protein content. It mechanically links the z-discs to the costameres (31). Desmin is characterized by numerous protein-protein interactions ensuring cellular integrity, force transmission, and biomechanical signaling (reviewed in Ref. 32). Given this crucial localization, it is not surprising that desmin knock-out mice develop a multisystem disorder involving cardiac, skeletal, and smooth muscle, with the most prominent pathological processes appearing in the heart, displaying severe cardiomyopathy accompanied by extensive fibrosis and calcification (33). Consistently, up to now, at least 45 mutations in the desmin gene have been identified that lead to a skeletal and cardiac myopathy termed desminopathy (recently reviewed in Ref. 32).
Integrins
Integrins are important transmembrane proteins located at the periphery of the costamere, linking the contractile apparatus to the extracellular matrix, thereby playing an essential role in mechanotransduction (for review, see Ref. 34). Mechanical deformation of integrins is thought to be one of the most important upstream steps in sensing of mechanical stress (35). Integrins are heterodimers consisting of an α- and a β-subunit without any enzymatic activity. Hence, their cytoplasmic domains mediate numerous protein-protein interactions. The interacting signaling molecules include integrin-linked kinase (ILK) and melusin (see below).
ILK
ILK is a widely expressed serine/threonine kinase that binds to the C terminus of β1-integrin (36). ILK links extracellular matrix interactions to cellular processes such as remodeling of cytoskeletal proteins, growth, proliferation, survival, and differentiation (reviewed in Ref. 30). It binds to α-actinin via β-parvin/affixin and forms a complex with PINCH and thymosin β4 (for review, see Ref. 30). It has been shown to phosphorylate myosin light chain, GSK-3β (glycogen synthase kinase-3β), and AKT/PKB (37). Several genetic loss-of-function studies in flies, worms, and mice have revealed embryonic death due to cell adhesion and cytoskeletal defects (reviewed in Ref. 30). The conditional cardiac knock-out in mice leads to DCM and sudden cardiac death (38). Bendig et al. (39) applied a forward genetic screen in zebrafish and identified an L308P mutation in the zILK gene causing progressive loss of contractility in zebrafish hearts. This mutation disrupted the interaction with β-parvin/affixin, suggesting that its presence is essential for normal cardiac function and potentially cardiac stress sensing (39). Likewise, in another zebrafish study, a nonsense mutation (Y319X) led to a dysmorphic ventricle with reduced cardiac function combined with severe endothelial defects, similar to alterations observed in mice lacking the integrin-binding extracellular matrix protein laminin α4 (40). Cardiac-restricted overexpression of ILK induces cardiac hypertrophy via activation of ERK and p38 MAPK, hence suggesting ILK to be a proximal prohypertrophic signaling activator (41).
Melusin
Melusin has been identified as an integrin-interacting protein in a yeast two-hybrid experiment using the β1A- and β1D-integrin subunits as bait (42). Mice with a homozygous genetic deletion of melusin showed no pathological phenotype at base-line conditions, but when subjected to biomechanical stress due to aortic banding, the animals developed DCM and contractile dysfunction. At the molecular level, phosphorylation of GSK-3β was blunted, although the precise mechanism remains elusive (11). The functional deterioration did not occur when prohypertrophic stimuli (low dose angiotensin-2 or phenylephrine infusion without an increase in blood pressure) without biomechanical stress were applied, suggesting a selective role for melusin in mechanotransduction. In turn, cardiac-restricted overexpression of melusin protected banded mice from the transition from “compensatory” to pathological hypertrophy with apoptosis, fibrosis, and progressive ventricular dilation (43).
L-type Calcium Channel
The L-type calcium channel (LTCC) is the major mediator of calcium influx in cardiomyocytes, regulating both excitation-contraction coupling and the activation of several signaling cascades. The calcium channel current itself is regulated in turn by several pathways, including β-adrenergic, Ca2+/calmodulin-dependent protein kinase, and calcineurin signaling (44). It localizes to the T-tubules, which are membrane invaginations closely interdigitating with the z-disc (45–47). In fact, immunofluorescence imaging revealed a striated pattern for the LTCC, which is indistinguishable from z-disc staining (48). Interestingly, there are even examples of z-disc proteins that simultaneously bind to membrane proteins of T-tubules, e.g. T-cap/telethonin, which associates with the potassium channel MinK (49).
Calcineurin is linked to the z-disc via calsarcin and MLP (see below) but also directly binds to and dephosphorylates the LTCC. Dephosphorylation of the LTCC leads to an increase in channel function, making calcineurin an important modulator of pathological electrical remodeling in hypertrophy. This appears to be a feed-forward mechanism because calcineurin is itself activated via calcium/calmodulin (50). Molkentin et al. (51) very recently identified the widely expressed EF-hand domain-containing protein CIB1 (Ca2+- and integrin-binding protein-1) as a novel calcineurin B-interacting protein that localizes to the sarcolemma in proximity to the LTCC. Homozygous genetic deletion of CIB1 rendered mice resistant to pathological hypertrophy accompanied by marked reduction of calcineurin activation. Conversely, inducible cardiac transgenic overexpression enhanced hypertrophy and calcineurin activity upon exposure to pathological stimuli. Thus, CIB1 is a novel upstream regulator of calcineurin activity in pathological hypertrophy and might represent an important link between the upstream integrin and calcineurin “stretch” sensors (51).
Downstream Signaling of the Cardiac Z-disc
Calcineurin Signaling
Several phosphatases and kinases reside at the z-disc (Fig. 2). During the last decade, it has been shown that the calcium/calmodulin-dependent phosphatase calcineurin plays a central role in cardiomyocyte signal integration. Calcineurin dephosphorylates and thereby activates NFAT (nuclear factor of activated T-cells) family transcription factors, which are major positive regulators of pathological cardiac hypertrophy and remodeling (reviewed in Ref. 5). We (47) and others (45, 46, 52, 53) have shown that at least part of the cardiomyocyte calcineurin pool localizes to the z-disc, where it resides in close proximity to several important modulators of its activity.
Calsarcins
Initially found in yeast two-hybrid screening using calcineurin as bait (47), the calsarcin family (also termed myozenin or FATZ) comprises three members, all localizing to the z-disc: calsarcin-1 is the only isoform expressed in the adult heart and slow skeletal muscle, whereas calsarcin-2 (54) and calsarcin-3 (55) are expressed in fast skeletal muscle. In cardiac tissue, calsarcin-1 is a negative regulator of calcineurin. Despite the lack of an obvious base-line phenotype, calsarcin-1 knock-out mice were sensitized to pathological stimuli such as pressure overload, resulting in excessive calcineurin activation and exacerbated hypertrophy as well as accelerated cardiomyopathy (10). Conversely, transgenic mice overexpressing calsarcin-1 in a cardiac-specific fashion were protected against angiotensin II-induced cardiac hypertrophy (56). Interestingly, calsarcin-1 was recently identified as a novel disease gene for human hypertrophic cardiomyopathy (28). Like many z-disc proteins, calsarcins are characterized by a multitude of binding partners, including Cypher/ZASP/Oracle, T-cap/telethonin, α-actinin, and γ-filamin (47, 55); myotilin (57); MuRF1 and MuRF2 (58); and ENH and other members of the Enigma family (16, 59). However, the precise mechanism by which calsarcins modulate calcineurin activity still remains unclear. Interesting, new results focus on post-transcriptional modification of calsarcins by phosphorylation (16, 60) and potential changes in their subcellular localization (60); however, the biological relevance of these processes remains to be determined.
Lmcd1/Dyxin
Lmcd1 or dyxin is a member of the large LIM domain-containing protein family localizing to the z-disc. Besides two C-terminal LIM domains, Lmcd1 contains a PET domain, which is also involved in protein-protein interactions (61). In cardiomyocytes, Lmcd1 is strongly up-regulated by prohypertrophic stimuli such as biomechanical stress both in vitro and in vivo (62, 63). When overexpressed in neonatal cardiomyocytes, Lmcd1 induced hypertrophy accompanied by strong activation of calcineurin signaling. Similar effects could be observed in a transgenic mouse model with cardiac-restricted overexpression of Lmcd1 (62). Conversely, knockdown of Lmcd1 attenuated the hypertrophic response to phenylephrine or stretch. Moreover, phenylephrine-induced calcineurin activation was completely abolished when Lmcd1 was knocked down (62). Thus, Lmcd1 appears be required as a coactivator for calcineurin activation. In non-cardiac cells, Lmcd1 has also been implicated in GATA6-dependent signaling as well as cytoplasmic-nuclear shuttling, similar to its relative zyxin (see below) (64).
PICOT
Another interesting negative modulator of calcineurin activity at the z-disc is PICOT (protein kinase C-interacting cousin of thioredoxin). PICOT binds to and colocalizes with MLP at the z-disc. It thereby interferes with the MLP-calcineurin interaction, causing a dose-dependent displacement of calcineurin from the z-disc. As a consequence, pharmacological agonist- or pressure overload-mediated calcineurin activation is abrogated, which in turn prevents cardiomyocyte hypertrophy (45).
PAK1
PAK1 (p21-activated kinase-1) is a serine/threonine protein kinase activated by the small GTPases Cdc42 and Rac1 (65). Solaro and co-workers (66) showed that PAK1 is highly expressed in different regions of the cardiomyocyte, including the z-disc as well as the cell membrane, intercalated disc, and nuclear membrane. PAK1 is upstream of PP2A (protein phosphatase 2A), inducing its post-translational modification and subsequent dephosphorylation of troponin I, suggesting that both form a functional complex (66). PPA2 is one of the major phosphatases of PKA-dependent phosphorylation sites, suggesting a role for the PAK1-PP2A complex in modulating the β-adrenergic effects in cardiac contractility (reviewed in Ref. 67).
PKC
Considerable attention has been paid to PKC signaling at the level of the z-disc. PKCϵ, a known modulator of cardiac hypertrophy that belongs to the group of “novel” or “unconventional” PKCs (characterized by the fact that they do not require Ca2+ for their activation) (reviewed in Ref. 68), has been shown to translocate to the z-disc in cardiomyocytes upon stimulation (69), in particular in response to mechanical stress (70). In vivo, PKCϵ (as well as PKCδ) activation induces “physiological” hypertrophy rather than maladaptive hypertrophy (71). PKCϵ can also mediate prohypertrophic stimuli by forming a complex with PKD1 at the z-disc. PKCϵ-dependent activation of PKD1 appears to be necessary for cardiac hypertrophy induced by G-protein-coupled receptor agonists (72). An important anchoring protein for several PKCs (including PKCϵ) is the PDZ-LIM domain protein Cypher/ZASP/Oracle, which tethers them to the α-actinin skeleton of the z-disc (73). The potential importance of this interaction is further emphasized by the finding that a human cardiomyopathy-causing mutation in Cypher strengthens its binding affinity for PKCϵ (74). It was speculated that Cypher competes with RACK2 (receptor for activated C kinase 2) for binding to PKCϵ, thereby promoting progression to heart failure (74). Another important PKC-regulating molecule at the z-disc is the capping molecule for the barbed ends of actin, CapZ. CapZ is a heterodimer consisting of an α- and a β-subunit. The β2-isoform is ubiquitously expressed, whereas the β1-isoform is expressed in striated muscle and localizes to the z-disc (75). CapZ interacts with α-actinin (76) and regulates actin dynamics and tethers the thin filaments to the z-disc (77). Pyle et al. (78) demonstrated that down-regulation of CapZ leads to a decrease in PKCβ and alteration in PKC-dependent signaling. Cardiac CapZ regulates the binding of PKCβII to the myofilaments, with subsequent effects on cardiac contractility (79).
PDE5
Another interesting signaling molecule that localizes to the z-disc (80) is PDE5 (phosphodiesterase 5), which mediates cGMP degradation and is up-regulated to high levels in failing hearts (81). Interestingly, in mice subjected to chronic pressure overload, selective PDE5 inhibition inhibited hypertrophy and prevented functional deterioration (82).
Acetylases and Deacetylases
In addition to phosphorylation, reversible acetylation of lysine residues is an important functional modification of protein function. Interestingly, both a histone acetyltransferase, PCAF (p300/CBP-associated factor), and histone deacetylase, HDAC4, localize to the cardiac sarcomere at the z-disc (A- and I-bands), where the z-disc protein MLP is a target for both molecules (83). Inhibition of histone deacetylases increased myofilament sensitivity in wild-type but not MLP knock-out mice, providing evidence for a role of z-disc protein acetylation in the regulation of cardiac contractility.
Shuttling Z-disc Molecules
Zyxin
Zyxin is a ubiquitously expressed LIM protein that has been implicated in cytoskeletal organization and cell motility (84). In addition to its localization to the focal adhesion complex, zyxin interacts with several z-disc molecules, including α-actinin, MLP, and myopodin (5, 85). In a non-cardiomyocyte cell system (vascular smooth muscle), Cattaruzza et al. (86) showed that zyxin shuttles to the nucleus selectively upon mechanical stress, thereby modulating gene expression. In cardiomyocytes, zyxin translocates to the nucleus in response to atrial natriuretic peptide/cGMP-dependent upstream signals, where it associates with AKT and enhances cell survival (87).
MLP
MLP (CSRP3/CRP3) is a small 198-amino acid protein that is highly expressed in striated muscle cells. A mouse model with deletion of MLP was the first genetically engineered animal model of DCM (88). Although at least part of the MLP appears to localize to the cardiac z-disc, its exact subcellular localization remains controversial (89). MLP interacts with several z-disc proteins, including T-cap/telethonin, α-actinin, the phosphatase calcineurin, and zyxin, but also with N-RAP at the intercalated disc and β-spectrin as well as ILK at the costamere (reviewed in Refs. 5, 17, and 90). Moreover, MLP has been shown to shuttle to the nucleus, where it interacts with the muscle basic helix-loop-helix transcription factors MyoD, MRF4, and myogenin, thereby modulating gene expression (reviewed in Ref. 5). Knöll et al. (9) proposed that MLP might be part of the elusive cardiac stretch receptor, an idea that is supported by data that cyclic stretch could drive nuclear translocation of MLP (91) and that MLP is up-regulated in several models of cardiac hypertrophy and remodeling (46, 92). MLP also regulates calcineurin activity (see above) by direct binding. Heterozygous loss of MLP thereby leads to dissociation of MLP from the z-disc and subsequently to reduced calcineurin/NFAT activation during post-infarction remodeling (46).
Myopodin
Myopodin, an actin- and α-actinin-binding member of the synaptopodin family, also shows a two-compartment redistribution behavior. It localizes to the z-disc under base-line conditions but shuttles to the nucleus in stressed cardiomyocytes and during myoblast development (93). The importin-α-mediated nuclear import of myopodin relies on serine/threonine phosphorylation-dependent binding of myopodin to 14-3-3 (94). Phosphorylation via PKA and Ca2+/calmodulin-dependent protein kinase II promotes nuclear import, whereas dephosphorylation by calcineurin abrogates 14-3-3 binding and supports myopodin binding to α-actinin (95). Although its biological function is still undefined, recent findings support a role for myopodin as a z-disc multiadaptor protein (96).
Ubiquitin Ligases
The ubiquitin-proteasome system controls a wide spectrum of physiological and pathophysiological processes in cardiac muscle, including degradation of misfolded proteins, but also several signaling pathways involved in stress response, hypertrophy, the cell cycle, and cell death (recently reviewed in Ref. 97). At least two important participants in the ubiquitin-proteasome system localize to the cardiac z-disc: the MAFbx/atrogin-1 and MuRF (muscle-specific ring finger) protein family E3 ligases. MAFbx/atrogin-1 has been shown to directly bind to α-actinin and calcineurin at the z-disc. It reduces the amount (and activity) of calcineurin via ubiquitination and subsequent degradation (52). Consistently, mice overexpressing MAFbx/atrogin-1 showed a blunted hypertrophic response and reduced calcineurin signaling upon pressure overload. The MuRF family comprises three members, MuRF1–3, but only MuRF3 reveals a z-disc localization (98). MuRF3 knock-out mice showed normal cardiac function under base-line conditions but were prone to ventricular rupture after myocardial infarction. Interestingly, the MuRF3 binding partners (and targets) γ-filamin and FHL-2 (both also at least partially localizing to the z-disc) accumulated, suggesting a cardioprotective role for MuRF3 (99). In line with these findings, MuRF1/3 double knock-out mice showed a severe myopathy and hypertrophic cardiomyopathy accompanied by subsarcolemmal myosin heavy chain accumulation, further emphasizing the crucial role for MuRF proteins in proper sarcomeric protein turnover (100).
Perspectives
Z-disc biology is a rapidly evolving field. The view of the z-disc has been shifted from a simple structural lateral margin of sarcomeres to a hot spot in signaling and disease, where a plethora of different proteins reside, the majority of which have been implicated in signal transduction. Much progress has been made to unravel this intricate network of proteins.
A prominent feature shared by most z-disc proteins is their ability to mediate multiple protein-protein interactions. Thus, it is likely that the number of signaling proteins localizing to the z-disc will continue to grow. Given the relevance of many z-disc proteins in inherited and acquired cardiac disease, it will be crucial to further investigate and understand the underlying pathomechanisms to develop effective therapeutic strategies.
This work was supported by a grant from the Bundesministerium für Bildung und Forschung (Nationales Genomforschungsnetz NGFNplus; to N. F.) and by a grant from the Hengstberger Stiftung/German Society of Cardiology (DGK; to D. F.). This is the first article in the Thematic Minireview Series on Signaling in Cardiac Sarcomeres in Health and Disease. This minireview will be reprinted in the 2011 Minireview Compendium, which will be available in January, 2012.
D. Frank and N. Frey, unpublished data.
- MLP
- muscle LIM protein
- DCM
- dilated cardiomyopathy
- ILK
- integrin-linked kinase
- LTCC
- L-type calcium channel.
REFERENCES
- 1. Clark K. A., McElhinny A. S., Beckerle M. C., Gregorio C. C. (2002) Annu. Rev. Cell Dev. Biol. 18, 637–706 [DOI] [PubMed] [Google Scholar]
- 2. Ervasti J. M. (2003) J. Biol. Chem. 278, 13591–13594 [DOI] [PubMed] [Google Scholar]
- 3. Luther P. K. (2009) J. Muscle Res. Cell Motil. 30, 171–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Knöll R., Hoshijima M., Chien K. (2003) J. Mol. Med. 81, 750–756 [DOI] [PubMed] [Google Scholar]
- 5. Frank D., Kuhn C., Katus H. A., Frey N. (2006) J. Mol. Med. 84, 446–468 [DOI] [PubMed] [Google Scholar]
- 6. Hoshijima M. (2006) Am. J. Physiol. Heart Circ. Physiol. 290, H1313–H1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Epstein N. D., Davis J. S. (2003) Cell 112, 147–150 [DOI] [PubMed] [Google Scholar]
- 8. Pyle W. G., Solaro R. J. (2004) Circ. Res. 94, 296–305 [DOI] [PubMed] [Google Scholar]
- 9. Knöll R., Hoshijima M., Hoffman H. M., Person V., Lorenzen-Schmidt I., Bang M. L., Hayashi T., Shiga N., Yasukawa H., Schaper W., McKenna W., Yokoyama M., Schork N. J., Omens J. H., McCulloch A. D., Kimura A., Gregorio C. C., Poller W., Schaper J., Schultheiss H. P., Chien K. R. (2002) Cell 111, 943–955 [DOI] [PubMed] [Google Scholar]
- 10. Frey N., Barrientos T., Shelton J. M., Frank D., Rütten H., Gehring D., Kuhn C., Lutz M., Rothermel B., Bassel-Duby R., Richardson J. A., Katus H. A., Hill J. A., Olson E. N. (2004) Nat. Med. 10, 1336–1343 [DOI] [PubMed] [Google Scholar]
- 11. Brancaccio M., Fratta L., Notte A., Hirsch E., Poulet R., Guazzone S., De Acetis M., Vecchione C., Marino G., Altruda F., Silengo L., Tarone G., Lembo G. (2003) Nat. Med. 9, 68–75 [DOI] [PubMed] [Google Scholar]
- 12. Gill G. N. (1995) Structure 3, 1285–1289 [DOI] [PubMed] [Google Scholar]
- 13. Kadrmas J. L., Beckerle M. C. (2004) Nat. Rev. Mol. Cell Biol. 5, 920–931 [DOI] [PubMed] [Google Scholar]
- 14. Lee H. J., Zheng J. J. (2010) Cell Commun. Signal. 8, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zheng M., Cheng H., Banerjee I., Chen J. (2010) J. Mol. Cell. Biol. 2, 96–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. von Nandelstadh P., Ismail M., Gardin C., Suila H., Zara I., Belgrano A., Valle G., Carpen O., Faulkner G. (2009) Mol. Cell. Biol. 29, 822–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Frank D., Kuhn C., Katus H. A., Frey N. (2007) Future Cardiol. 3, 611–622 [DOI] [PubMed] [Google Scholar]
- 18. Grünig E., Tasman J. A., Kücherer H., Franz W., Kübler W., Katus H. A. (1998) J. Am. Coll. Cardiol. 31, 186–194 [DOI] [PubMed] [Google Scholar]
- 19. Ahmad F., Seidman J. G., Seidman C. E. (2005) Annu. Rev. Genomics Hum. Genet. 6, 185–216 [DOI] [PubMed] [Google Scholar]
- 20. Vatta M., Mohapatra B., Jimenez S., Sanchez X., Faulkner G., Perles Z., Sinagra G., Lin J. H., Vu T. M., Zhou Q., Bowles K. R., Di Lenarda A., Schimmenti L., Fox M., Chrisco M. A., Murphy R. T., McKenna W., Elliott P., Bowles N. E., Chen J., Valle G., Towbin J. A. (2003) J. Am. Coll. Cardiol. 42, 2014–2027 [DOI] [PubMed] [Google Scholar]
- 21. Hayashi T., Arimura T., Itoh-Satoh M., Ueda K., Hohda S., Inagaki N., Takahashi M., Hori H., Yasunami M., Nishi H., Koga Y., Nakamura H., Matsuzaki M., Choi B. Y., Bae S. W., You C. W., Han K. H., Park J. E., Knöll R., Hoshijima M., Chien K. R., Kimura A. (2004) J. Am. Coll. Cardiol. 44, 2192–2201 [DOI] [PubMed] [Google Scholar]
- 22. Mohapatra B., Jimenez S., Lin J. H., Bowles K. R., Coveler K. J., Marx J. G., Chrisco M. A., Murphy R. T., Lurie P. R., Schwartz R. J., Elliott P. M., Vatta M., McKenna W., Towbin J. A., Bowles N. E. (2003) Mol. Genet. Metab. 80, 207–215 [DOI] [PubMed] [Google Scholar]
- 23. Duboscq-Bidot L., Xu P., Charron P., Neyroud N., Dilanian G., Millaire A., Bors V., Komajda M., Villard E. (2008) Cardiovasc. Res. 77, 118–125 [DOI] [PubMed] [Google Scholar]
- 24. Selcen D., Engel A. G. (2004) Neurology 62, 1363–1371 [DOI] [PubMed] [Google Scholar]
- 25. Hassel D., Dahme T., Erdmann J., Meder B., Huge A., Stoll M., Just S., Hess A., Ehlermann P., Weichenhan D., Grimmler M., Liptau H., Hetzer R., Regitz-Zagrosek V., Fischer C., Nürnberg P., Schunkert H., Katus H. A., Rottbauer W. (2009) Nat. Med. 15, 1281–1288 [DOI] [PubMed] [Google Scholar]
- 26. Gerull B., Gramlich M., Atherton J., McNabb M., Trombitás K., Sasse-Klaassen S., Seidman J. G., Seidman C., Granzier H., Labeit S., Frenneaux M., Thierfelder L. (2002) Nat. Genet. 30, 201–204 [DOI] [PubMed] [Google Scholar]
- 27. Geier C., Perrot A., Ozcelik C., Binner P., Counsell D., Hoffmann K., Pilz B., Martiniak Y., Gehmlich K., van der Ven P. F., Fürst D. O., Vornwald A., von Hodenberg E., Nürnberg P., Scheffold T., Dietz R., Osterziel K. J. (2003) Circulation 107, 1390–1395 [DOI] [PubMed] [Google Scholar]
- 28. Osio A., Tan L., Chen S. N., Lombardi R., Nagueh S. F., Shete S., Roberts R., Willerson J. T., Marian A. J. (2007) Circ. Res. 100, 766–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Satoh M., Takahashi M., Sakamoto T., Hiroe M., Marumo F., Kimura A. (1999) Biochem. Biophys. Res. Commun. 262, 411–417 [DOI] [PubMed] [Google Scholar]
- 30. Hannigan G. E., Coles J. G., Dedhar S. (2007) Circ. Res. 100, 1408–1414 [DOI] [PubMed] [Google Scholar]
- 31. Lazarides E., Hubbard B. D. (1976) Proc. Natl. Acad. Sci. U.S.A. 73, 4344–4348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Goldfarb L. G., Dalakas M. C. (2009) J. Clin. Invest. 119, 1806–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Milner D. J., Taffet G. E., Wang X., Pham T., Tamura T., Hartley C., Gerdes A. M., Capetanaki Y. (1999) J. Mol. Cell. Cardiol. 31, 2063–2076 [DOI] [PubMed] [Google Scholar]
- 34. Russell B., Curtis M. W., Koshman Y. E., Samarel A. M. (2010) J. Mol. Cell. Cardiol. 48, 817–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang N., Butler J. P., Ingber D. E. (1993) Science 260, 1124–1127 [DOI] [PubMed] [Google Scholar]
- 36. Hannigan G. E., Leung-Hagesteijn C., Fitz-Gibbon L., Coppolino M. G., Radeva G., Filmus J., Bell J. C., Dedhar S. (1996) Nature 379, 91–96 [DOI] [PubMed] [Google Scholar]
- 37. Delcommenne M., Tan C., Gray V., Rue L., Woodgett J., Dedhar S. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 11211–11216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. White D. E., Coutu P., Shi Y. F., Tardif J. C., Nattel S., St Arnaud R., Dedhar S., Muller W. J. (2006) Genes Dev. 20, 2355–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bendig G., Grimmler M., Huttner I. G., Wessels G., Dahme T., Just S., Trano N., Katus H. A., Fishman M. C., Rottbauer W. (2006) Genes Dev. 20, 2361–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Knöll R., Postel R., Wang J., Krätzner R., Hennecke G., Vacaru A. M., Vakeel P., Schubert C., Murthy K., Rana B. K., Kube D., Knöll G., Schäfer K., Hayashi T., Holm T., Kimura A., Schork N., Toliat M. R., Nürnberg P., Schultheiss H. P., Schaper W., Schaper J., Bos E., Den Hertog J., van Eeden F. J., Peters P. J., Hasenfuss G., Chien K. R., Bakkers J. (2007) Circulation 116, 515–525 [DOI] [PubMed] [Google Scholar]
- 41. Lu H., Fedak P. W., Dai X., Du C., Zhou Y. Q., Henkelman M., Mongroo P. S., Lau A., Yamabi H., Hinek A., Husain M., Hannigan G., Coles J. G. (2006) Circulation 114, 2271–2279 [DOI] [PubMed] [Google Scholar]
- 42. Brancaccio M., Guazzone S., Menini N., Sibona E., Hirsch E., De Andrea M., Rocchi M., Altruda F., Tarone G., Silengo L. (1999) J. Biol. Chem. 274, 29282–29288 [DOI] [PubMed] [Google Scholar]
- 43. De Acetis M., Notte A., Accornero F., Selvetella G., Brancaccio M., Vecchione C., Sbroggiò M., Collino F., Pacchioni B., Lanfranchi G., Aretini A., Ferretti R., Maffei A., Altruda F., Silengo L., Tarone G., Lembo G. (2005) Circ. Res. 96, 1087–1094 [DOI] [PubMed] [Google Scholar]
- 44. Benitah J. P., Alvarez J. L., Gómez A. M. (2010) J. Mol. Cell. Cardiol. 48, 26–36 [DOI] [PubMed] [Google Scholar]
- 45. Jeong D., Kim J. M., Cha H., Oh J. G., Park J., Yun S. H., Ju E. S., Jeon E. S., Hajjar R. J., Park W. J. (2008) Circ. Res. 102, 711–719 [DOI] [PubMed] [Google Scholar]
- 46. Heineke J., Ruetten H., Willenbockel C., Gross S. C., Naguib M., Schaefer A., Kempf T., Hilfiker-Kleiner D., Caroni P., Kraft T., Kaiser R. A., Molkentin J. D., Drexler H., Wollert K. C. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1655–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Frey N., Richardson J. A., Olson E. N. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 14632–14637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Song L. S., Sobie E. A., McCulle S., Lederer W. J., Balke C. W., Cheng H. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4305–4310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Furukawa T., Ono Y., Tsuchiya H., Katayama Y., Bang M. L., Labeit D., Labeit S., Inagaki N., Gregorio C. C. (2001) J. Mol. Biol. 313, 775–784 [DOI] [PubMed] [Google Scholar]
- 50. Tandan S., Wang Y., Wang T. T., Jiang N., Hall D. D., Hell J. W., Luo X., Rothermel B. A., Hill J. A. (2009) Circ. Res. 105, 51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Heineke J., Auger-Messier M., Correll R. N., Xu J., Benard M. J., Yuan W., Drexler H., Parise L. V., Molkentin J. D. (2010) Nat. Med. 16, 872–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li H. H., Kedar V., Zhang C., McDonough H., Arya R., Wang D. Z., Patterson C. (2004) J. Clin. Invest. 114, 1058–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zou Y., Yao A., Zhu W., Kudoh S., Hiroi Y., Shimoyama M., Uozumi H., Kohmoto O., Takahashi T., Shibasaki F., Nagai R., Yazaki Y., Komuro I. (2001) Circulation 104, 102–108 [DOI] [PubMed] [Google Scholar]
- 54. Frey N., Frank D., Lippl S., Kuhn C., Kögler H., Barrientos T., Rohr C., Will R., Müller O. J., Weiler H., Bassel-Duby R., Katus H. A., Olson E. N. (2008) J. Clin. Invest. 118, 3598–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Frey N., Olson E. N. (2002) J. Biol. Chem. 277, 13998–14004 [DOI] [PubMed] [Google Scholar]
- 56. Frank D., Kuhn C., van Eickels M., Gehring D., Hanselmann C., Lippl S., Will R., Katus H. A., Frey N. (2007) Circulation 116, 2587–2596 [DOI] [PubMed] [Google Scholar]
- 57. Gontier Y., Taivainen A., Fontao L., Sonnenberg A., van der Flier A., Carpen O., Faulkner G., Borradori L. (2005) J. Cell Sci. 118, 3739–3749 [DOI] [PubMed] [Google Scholar]
- 58. Moriscot A. S., Baptista I. L., Bogomolovas J., Witt C., Hirner S., Granzier H., Labeit S. (2010) J. Struct. Biol. 170, 344–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cheng H., Kimura K., Peter A. K., Cui L., Ouyang K., Shen T., Liu Y., Gu Y., Dalton N. D., Evans S. M., Knowlton K. U., Peterson K. L., Chen J. (2010) Circ. Res. 6, 348–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Paulsson A. K., Franklin S., Mitchell-Jordan S. A., Ren S., Wang Y., Vondriska T. M. (2010) J. Mol. Cell. Cardiol. 48, 1206–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bespalova I. N., Burmeister M. (2000) Genomics 63, 69–74 [DOI] [PubMed] [Google Scholar]
- 62. Frank D., Frauen R., Hanselmann C., Kuhn C., Will R., Gantenberg J., Fuzesi L., Katus H. A., Frey N. (2010) J. Mol. Cell. Cardiol. 49, 673–682 [DOI] [PubMed] [Google Scholar]
- 63. Frank D., Kuhn C., Brors B., Hanselmann C., Lüdde M., Katus H. A., Frey N. (2008) Hypertension 51, 309–318 [DOI] [PubMed] [Google Scholar]
- 64. Rath N., Wang Z., Lu M. M., Morrisey E. E. (2005) Mol. Cell. Biol. 25, 8864–8873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Brown J. L., Stowers L., Baer M., Trejo J., Coughlin S., Chant J. (1996) Curr. Biol. 6, 598–605 [DOI] [PubMed] [Google Scholar]
- 66. Ke Y., Wang L., Pyle W. G., de Tombe P. P., Solaro R. J. (2004) Circ. Res. 94, 194–200 [DOI] [PubMed] [Google Scholar]
- 67. Ke Y., Lei M., Solaro R. J. (2008) Prog. Biophys. Mol. Biol. 98, 238–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Dorn G. W., 2nd, Force T. (2005) J. Clin. Invest. 115, 527–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Robia S. L., Ghanta J., Robu V. G., Walker J. W. (2001) Biophys. J. 80, 2140–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gu X., Bishop S. P. (1994) Circ. Res. 75, 926–931 [DOI] [PubMed] [Google Scholar]
- 71. Wu G., Toyokawa T., Hahn H., Dorn G. W., 2nd (2000) J. Biol. Chem. 275, 29927–29930 [DOI] [PubMed] [Google Scholar]
- 72. Iwata M., Maturana A., Hoshijima M., Tatematsu K., Okajima T., Vandenheede J. R., Van Lint J., Tanizawa K., Kuroda S. (2005) Biochem. Biophys. Res. Commun. 327, 1105–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhou Q., Ruiz-Lozano P., Martone M. E., Chen J. (1999) J. Biol. Chem. 274, 19807–19813 [DOI] [PubMed] [Google Scholar]
- 74. Arimura T., Hayashi T., Terada H., Lee S. Y., Zhou Q., Takahashi M., Ueda K., Nouchi T., Hohda S., Shibutani M., Hirose M., Chen J., Park J. E., Yasunami M., Hayashi H., Kimura A. (2004) J. Biol. Chem. 279, 6746–6752 [DOI] [PubMed] [Google Scholar]
- 75. Schafer D. A., Korshunova Y. O., Schroer T. A., Cooper J. A. (1994) J. Cell Biol. 127, 453–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Papa I., Astier C., Kwiatek O., Raynaud F., Bonnal C., Lebart M. C., Roustan C., Benyamin Y. (1999) J. Muscle Res. Cell Motil. 20, 187–197 [DOI] [PubMed] [Google Scholar]
- 77. Schafer D. A., Hug C., Cooper J. A. (1995) J. Cell Biol. 128, 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Pyle W. G., Hart M. C., Cooper J. A., Sumandea M. P., de Tombe P. P., Solaro R. J. (2002) Circ. Res. 90, 1299–1306 [DOI] [PubMed] [Google Scholar]
- 79. Pyle W. G., La Rotta G., de Tombe P. P., Sumandea M. P., Solaro R. J. (2006) J. Mol. Cell. Cardiol. 41, 537–543 [DOI] [PubMed] [Google Scholar]
- 80. Senzaki H., Smith C. J., Juang G. J., Isoda T., Mayer S. P., Ohler A., Paolocci N., Tomaselli G. F., Hare J. M., Kass D. A. (2001) FASEB J. 15, 1718–1726 [DOI] [PubMed] [Google Scholar]
- 81. Pokreisz P., Vandenwijngaert S., Bito V., Van den Bergh A., Lenaerts I., Busch C., Marsboom G., Gheysens O., Vermeersch P., Biesmans L., Liu X., Gillijns H., Pellens M., Van Lommel A., Buys E., Schoonjans L., Vanhaecke J., Verbeken E., Sipido K., Herijgers P., Bloch K. D., Janssens S. P. (2009) Circulation 119, 408–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Takimoto E., Champion H. C., Li M., Belardi D., Ren S., Rodriguez E. R., Bedja D., Gabrielson K. L., Wang Y., Kass D. A. (2005) Nat. Med. 11, 214–222 [DOI] [PubMed] [Google Scholar]
- 83. Gupta M. P., Samant S. A., Smith S. H., Shroff S. G. (2008) J. Biol. Chem. 283, 10135–10146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Fraley S. I., Feng Y., Krishnamurthy R., Kim D. H., Celedon A., Longmore G. D., Wirtz D. (2010) Nat. Cell Biol. 12, 598–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yu Y. P., Luo J. H. (2006) Cancer Res. 66, 7414–7419 [DOI] [PubMed] [Google Scholar]
- 86. Cattaruzza M., Lattrich C., Hecker M. (2004) Hypertension 43, 726–730 [DOI] [PubMed] [Google Scholar]
- 87. Kato T., Muraski J., Chen Y., Tsujita Y., Wall J., Glembotski C. C., Schaefer E., Beckerle M., Sussman M. A. (2005) J. Clin. Invest. 115, 2716–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Arber S., Hunter J. J., Ross J., Jr., Hongo M., Sansig G., Borg J., Perriard J. C., Chien K. R., Caroni P. (1997) Cell 88, 393–403 [DOI] [PubMed] [Google Scholar]
- 89. Gehmlich K., Ehler E., Perrot A., Fürst D. O., Geier C. (2010) J. Mol. Cell. Cardiol. 48, 424–425; author reply 426–427 [DOI] [PubMed] [Google Scholar]
- 90. Gehmlich K., Geier C., Milting H., Fürst D., Ehler E. (2008) J. Muscle Res. Cell Motil. 29, 155–158 [DOI] [PubMed] [Google Scholar]
- 91. Boateng S. Y., Senyo S. E., Qi L., Goldspink P. H., Russell B. (2009) J. Mol. Cell. Cardiol. 47, 426–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Boateng S. Y., Belin R. J., Geenen D. L., Margulies K. B., Martin J. L., Hoshijima M., de Tombe P. P., Russell B. (2007) Am. J. Physiol. Heart Circ. Physiol. 292, H259–H269 [DOI] [PubMed] [Google Scholar]
- 93. Weins A., Schwarz K., Faul C., Barisoni L., Linke W. A., Mundel P. (2001) J. Cell Biol. 155, 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Faul C., Hüttelmaier S., Oh J., Hachet V., Singer R. H., Mundel P. (2005) J. Cell Biol. 169, 415–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Faul C., Dhume A., Schecter A. D., Mundel P. (2007) Mol. Cell. Biol. 27, 8215–8227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Linnemann A., van der Ven P. F., Vakeel P., Albinus B., Simonis D., Bendas G., Schenk J. A., Micheel B., Kley R. A., Fürst D. O. (2010) Eur. J. Cell Biol. 89, 681–692 [DOI] [PubMed] [Google Scholar]
- 97. Willis M. S., Townley-Tilson W. H., Kang E. Y., Homeister J. W., Patterson C. (2010) Circ. Res. 106, 463–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Spencer J. A., Eliazer S., Ilaria R. L., Jr., Richardson J. A., Olson E. N. (2000) J. Cell Biol. 150, 771–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Fielitz J., van Rooij E., Spencer J. A., Shelton J. M., Latif S., van der Nagel R., Bezprozvannaya S., de Windt L., Richardson J. A., Bassel-Duby R., Olson E. N. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 4377–4382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Fielitz J., Kim M. S., Shelton J. M., Latif S., Spencer J. A., Glass D. J., Richardson J. A., Bassel-Duby R., Olson E. N. (2007) J. Clin. Invest. 117, 2486–2495 [DOI] [PMC free article] [PubMed] [Google Scholar]