Abstract
Outer membrane protein A (OmpA) is a class of proteins highly conserved among the Enterobacteriaceae family and throughout evolution. Klebsiella pneumoniae is a capsulated Gram-negative pathogen. It is an important cause of community-acquired and nosocomial pneumonia. Evidence indicates that K. pneumoniae infections are characterized by a lack of an early inflammatory response. Data from our laboratory indicate that K. pneumoniae CPS helps to suppress the host inflammatory response. However, it is unknown whether K. pneumoniae employs additional factors to modulate host inflammatory responses. Here, we report that K. pneumoniae OmpA is important for immune evasion in vitro and in vivo. Infection of A549 and normal human bronchial cells with 52OmpA2, an ompA mutant, increased the levels of IL-8. 52145-ΔwcaK2ompA, which does not express CPS and ompA, induced the highest levels of IL-8. Both mutants could be complemented. In vivo, 52OmpA2 induced higher levels of tnfα, kc, and il6 than the wild type. ompA mutants activated NF-κB, and the phosphorylation of p38, p44/42, and JNK MAPKs and IL-8 induction was via NF-κB-dependent and p38- and p44/42-dependent pathways. 52OmpA2 engaged TLR2 and -4 to activate NF-κB, whereas 52145-ΔwcaK2ompA activated not only TLR2 and TLR4 but also NOD1. Finally, we demonstrate that the ompA mutant is attenuated in the pneumonia mouse model. The results of this study indicate that K. pneumoniae OmpA contributes to attenuate airway cell responses. This may facilitate pathogen survival in the hostile environment of the lung.
Keywords: Cellular Immune Response, Epithelial Cell, Innate Immunity, MAP Kinases (MAPKs), NF-κB Transcription Factor, Toll-like Receptors (TLR), Klebsiella, OmpA
Introduction
The OM3 of Gram-negative bacteria is composed of phospholipids, LPS, and outer membrane proteins. These proteins are important for membrane integrity and transport of molecules across the membrane (for a review, see Ref. 1). OmpA (outer membrane protein A) is one of the best characterized OM proteins. Its crystal structure has been determined (at a resolution of 1. 65 Å), and it consists of an eight-stranded all-next-neighbor antiparallel β-barrel with short turns at the periplasmic barrel (2). A number of studies have highlighted the role of OmpA in pathogenesis. Escherichia coli OmpA mediates adhesion and/or invasion to epithelial cells and macrophages (3, 4). It also mediates serum resistance and may protect bacteria against the bactericidal action of lung collectins SP-D and SP-A (5, 6). However, OmpA is also targeted by the innate immune system. Neutrophil elastase, a protein included in the array of oxygen-independent weapons of the innate immune system, degrades OmpA, resulting in cell death (7). The acute-phase protein serum amyloid protein A binds to OmpA, and this is associated with an increased uptake of bacteria by neutrophils and macrophages (8, 9).
Airway epithelial cells play a pivotal role in lung defense against infections by detecting pathogens, thereby leading to the activation of signaling pathways resulting in the production of antimicrobial molecules, the expression of co-stimulatory molecules, and the release of cytokines and chemokines (10, 11). In particular, IL-8 is a major secreted product of infected cells independent of the infecting microorganism (12, 13). This chemokine is a potent chemoattractant for polymorphonuclear cells into the infected tissue (12, 13). To launch these responses, airway epithelial cells recognize conserved molecules expressed by pathogens, the so-called PAMPs, through a set of germ line-encoded receptors referred to as PRRs (14, 15). The best characterized PRRs belong to the families of Toll-like receptors (TLRs) and nucleotide binding and oligomerization domain-like receptors (NLRs) (16–18). Among TLRs, most of the studies focus on TLR4, mainly involved in the detection of LPS, and on TLR2, which responds to a variety of Gram-positive PAMPs (10, 14, 15). Among NLRs, NOD1 has received increasing attention. NOD1 is located intracellularly, and evidence indicates that it recognizes a peptidoglycan motif from Gram-negative bacteria (16, 17).
Klebsiella pneumoniae is a capsulated Gram-negative pathogen that causes a wide range of infections, from urinary tract infections to pneumonia, the latter being particularly devastating among immunocompromised patients with mortality rates between 25 and 60% (19). The best characterized virulence factor of this species is CPS, which is responsible for protection against complement and antimicrobial peptide-mediated killing (20–22). In addition, isogenic CPS mutant strains are avirulent, being unable to cause pneumonia and urinary tract infections (23–25).
A wealth of evidence indicates that activation of inflammatory responses is essential to clear Klebsiella infections (26–28) and that TLRs seem to play a major role in detecting K. pneumoniae (29, 30). Conversely, this suggests that K. pneumoniae may somehow try to counteract the induction of these host defense responses. Indeed, we and others (31, 32) have shown that in sharp contrast to wild-type strains, avirulent CPS mutants activate an inflammatory program through TLR-dependent pathways. In fact, it has been postulated that CPS helps to suppress the host inflammatory response, thereby allowing the bacteria to replicate in a more permissive niche. Whether K. pneumoniae employs additional factors to modulate host inflammatory responses is still unknown. In this context, several studies have shown that recombinant purified OmpA from K. pneumoniae induces the expression of inflammatory molecules in a TLR2-dependent manner in various cell types (33–35). Hence, it has been postulated that detection of K. pneumoniae OmpA may contribute to the activation of host responses leading to the clearance of K. pneumoniae (36). However, there might be differences between the cellular recognition of recombinant purified OmpA and OmpA expressed in the complex lipid environment of the bacterial OM, and hence the cellular responses could be different.
The purpose of this study was to analyze whether OmpA may contribute to the activation of inflammatory responses when expressed in the bacterial OM. To this end, we compared the host responses to wild-type K. pneumoniae and an isogenic ompA mutant. We report that K. pneumoniae OmpA is important for immune evasion of K. pneumoniae in vitro and in vivo. Mechanistically, we demonstrate that the ompA mutant induces the secretion of inflammatory mediators via activation of NF-κB and MAPKs p38 and p44/42. In addition, we show that ompA mutant induction of inflammatory responses is dependent on TLR activation. Finally, we demonstrate that the K. pneumoniae ompA mutant is attenuated in a mouse model of pneumonia.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Growth Conditions, and Reagents
K. pneumoniae 52145 is a clinical isolate (serotype O1:K2) described previously (24, 37). The isogenic mutants 52OmpA2, which does not express ompA; 52145-ΔwcaK2, which does not express CPS; and 52145-ΔwcaK2ompA, which does not express CPS and ompA, have been described previously (38, 39). Analysis of OM proteins from these strains as well as the description of the complementing strains have been reported previously (39). Bacteria were grown in Luria-Bertani medium at 37 °C. When appropriate, antibiotics were added to the growth medium at the following concentrations: rifampicin (50 μg/ml), kanamycin (50 μg/ml), and chloramphenicol (25 μg/ml). Caffeic acid phenethyl ester (CAPE), an NF-κB inhibitor, and SB203580, a p38 MAPK inhibitor, were purchased from Sigma, whereas U0126, a p44/42 MAPK inhibitor, and SP600125, a JNK inhibitor, were purchased from Calbiochem.
CPS Purification and Quantification
Cell-associated CPSs from K. pneumoniae strains, grown in 5 ml of LB, were obtained using the hot phenol-water method exactly as described previously (20). CPS was quantified by determining the concentration of uronic acid in the samples, using a modified carbazole assay (40), exactly as described by Rahn and Whitfield (41).
Cell Culture and Infection
Monolayers of A549 (ATCC CCL185) and NHBE (Lonza) cells were grown as described previously (32). For infections, A549 cells were seeded to 90% confluence (3 × 105 cells/well) in 24-well tissue culture plates. Cells were serum-starved for 16 h before infection. For NHBE cells, cells were seeded to 80% confluence (2 × 105 cells/well) in collagen-coated 24-well tissue culture plates using 1 ml of bronchial epithelial cell growth medium (Lonza) per well. Bacteria were prepared as described (32, 42), and infections were performed using a multiplicity of infection of 100 bacteria/cell, unless indicated otherwise. To synchronize infection, plates were centrifuged at 200 × g for 5 min. For incubation times longer than 120 min, bacteria were killed by the addition of gentamicin (100 μg/ml), which was not removed until the end of the experiment. For experiments reported in Fig. 2, infections were performed for 3 h. Cell viability was assessed by trypan blue dye exclusion, and it was >95% even after 9 h of infection.
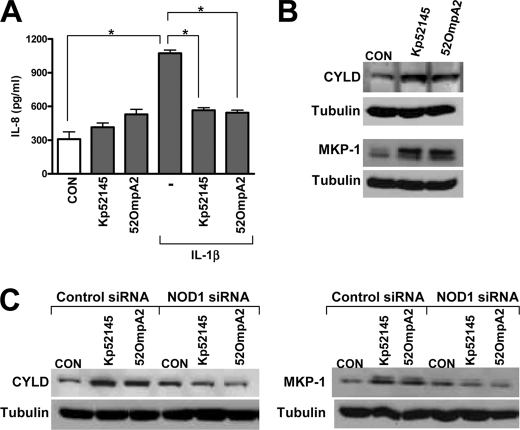
FIGURE 2.
K. pneumoniae ompA attenuates IL-1β-induced IL-8 expression. A, ELISA of IL-8 released by A549 left untreated (CON) or infected for 1 h with Kp52145 or 52OmpA2 and then stimulated with IL-1β (50 ng/ml) for 2 h (n = 3). *, p < 0.05 (for the indicated comparisons; one-way ANOVA). B, immunoblot analysis of CYLD, MKP-1, and tubulin levels in A549 cells left uninfected (CON) or infected with Kp52145 or 52OmpA2 for 3 h. Data are representative of three independent experiments. C, immunoblot analysis of CYLD, MKP-1, and tubulin levels in A549 cells transfected with either control or NOD1 siRNA, which were left uninfected (CON) or infected with Kp52145 or 52OmpA2 for 3 h. Data are representative of three independent experiments.
Adhesion and Internalization Assays
To determine the adhesion of Klebsiella strains to A549 cells, after 2 h of infection, monolayers were washed five times with PBS, and cells were lysed with 300 μl of 0.5% saponin-PBS for 5 min at room temperature, and serial dilutions were plated on LB agar plates for viable counts of bacteria. The results are expressed as log cfu/well. For invasion assays, cells were infected for 2 h and washed three times with PBS and then incubated for an additional 2 h with fresh medium plus gentamicin (100 μg/ml) to kill extracellular bacteria. This treatment was long enough to kill all extracellular bacteria. Epithelial monolayers were washed three times with PBS and lysed as described before. The results are expressed as log cfu/well. Experiments were carried out in triplicate on at least two independent occasions.
IL-8 Stimulation Assay
Epithelial monolayers were infected for different time periods. Supernatants were removed from the wells, cell debris was removed by centrifugation, and samples were frozen at −80 °C. IL-8 in the supernatants was determined by a commercial ELISA (Bender MedSystems) with a sensitivity of <2 pg/ml. Experiments were run in duplicate and repeated at least three times.
Transfections and Luciferase Assays
A549 cells were seeded into 24-well tissue culture plates to obtain a 40–60% confluence 24 h later. Cells were washed twice with PBS before transfection. Transfection experiments were carried out in 500 μl of Opti-MEM reduced serum medium (Invitrogen) using LipofectamineTM 2000 transfection reagent following the manufacturer's recommendations (Invitrogen).
For luciferase experiments, the PathDetect® NF-κB cis-reporting plasmid (250 ng; Stratagene) was co-transfected with the pRL-TK Renilla luciferase control reporter vector (20 ng; Promega). For luciferase assays, cells were lysed with passive lysis buffer (Promega). Luciferase activity was assayed using the dual luciferase assay kit according to the manufacturer's instructions (Promega). Firefly luciferase values were normalized to Renilla control values. Results were plotted as relative luciferase activity compared with activity measured in non-stimulated control cells. The luciferase assay was carried out in triplicate on at least three independent occasions.
Small Interfering RNA (siRNA)
Interference RNA for TLR4 (catalogue number HSS110818), TLR2 (catalogue number HSS110813), and CARD4/NOD1 (catalogue number HSS115906) were purchased from Invitrogen. StealthTM RNAi negative control low GC was used as control interference RNA for TLR2 and TLR4, whereas StealthTM RNAi negative control medium GC was used as control interference RNA for NOD1. RNA-mediated interference for down-regulating MyD88 was done by the transfection of MyD88 siRNA (5′-AACTGGAACAGACAAACTATC-3′) purchased from Qiagen. The AllStars negative control siRNA (Qiagen) was used as non-silencing control interference RNA. In all cases, 20 nm siRNA/well was used for transfection using LipofectamineTM 2000 transfection reagent and following the manufacturer's recommendations (Invitrogen). Cells were infected 48 h post-transfection as described above.
Real-time Quantitative PCR (RT-qPCR)
Quantification of mRNA levels was conducted by RT-qPCR. RNA was purified using a Nucleospin RNAII kit (Macherey-Nagel) exactly as recommended by the manufacturer. cDNA was obtained by retrotranscription of 1.5–2 μg of total RNA using a commercial RT2 first strand kit as recommended by the manufacturer (SABioscience). The reaction included one step to eliminate traces of genomic DNA. Real-time PCR (RT-PCR) analyses were performed with a Smart Cycler real-time PCR instrument (Cepheid, Sunnyvale, CA). To amplify human TLR2, TLR4, MyD88, and NOD1, 500 ng of cDNA were used as a template in a 25-μl reaction containing 1× QuantiTect SYBR Green PCR mix (Qiagen) and QuantiTect primer assays (Qiagen; catalogue numbers QT00236131 (TLR2) and QT00035238 (TLR4)) or the intron-spanning primers: 5′-GGCATCACCACACTTGATGAC-3′ (sense) and 5′-ATAGACCAGACACAGGTGCCAG-3′ (antisense) for MyD88 and 5′-TCAAGTTGGGGATGAAGGAG-3′ (sense) and 5′-GCCAAACTCTCTGCCACTTC-3′ (antisense) for NOD1. As an internal control, we amplified the human housekeeping gene GAPDH using 50 ng of cDNA and the following intron spanning primers: 5′-GAAGGTGAAGGTCGGAGTC-3′ (sense); 5′-GAAGATGGTGATGGGATTTC-3′ (antisense). The thermocycling protocol was as follows: 95 °C for 15 min for hot start polymerase activation, followed by 45 cycles of denaturation at 95 °C for 30 s, annealing at 60 °C for TLRs, MyD88, and NOD1 or 54 °C for GADPH for 30 s; and extension at 72 °C for 30 s. The SYBR Green dye was measured at 521 nm during the annealing phase. The threshold cycle (Ct) value reflects the cycle number at which the fluorescence generated within a reaction crosses a given threshold. The Ct value assigned to each well thus reflects the point during the reaction at which a sufficient number of amplicons have been accumulated. The relative mRNA amount in each sample was calculated based on its Ct in comparison with the Ct of GAPDH. Specificity of the PCR products was determined by melting curve analysis, and amplification products were resolved on a 1.5% agarose gel to confirm the correct size of the amplicons (92, 102, 185, 201, and 226 bp for TLR2, TLR4, MyD88, NOD1, and GAPDH, respectively).
Immunoblotting
Proteins from lysed cells were separated by 10% SDS-PAGE, electrotransferred to nitrocellulose membrane, and blocked with 4% skimmed milk in PBS. Immunostainings for MKP-1, CYLD, and IκBα were performed using polyclonal rabbit antibodies anti-MKP-1 (1:1,000; Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)), anti-IκBα (1:1,000; Cell Signaling), and anti-CYLD (1:1,000) (Imgenex Corp.) Immunostainings to assess phosphorylation of NF-κB p65, p38, p44/42, and JNK MAPKs were performed with polyclonal rabbit anti-phospho-p65, anti-phospho-p38, anti-phospho-p44/42, and anti-phospho-JNK antibodies, respectively (all used at 1:1,000; Cell Signaling). Immunoreactive bands were visualized by incubation with swine anti-rabbit immunoglobulins conjugated to horseradish peroxidase (Dako P0217) using the SuperSignal West-dura system (Pierce). Blots were reprobed with polyclonal anti-human tubulin antibody (1:3,000; Santa Cruz Biotechnology, Inc.) to control that equal amounts of proteins were loaded in each lane.
NOD1 NF-κB Activation Assay
HEK293T, seeded in 24-well plates, were transfected overnight with pCI-Nod1 (0.3 ng; kindly donated by T. Kufer), PathDetect® NF-κB cis-reporting plasmid (75 ng), pRL-TK Renilla luciferase (7.5 ng), and 120 ng of pcDNA3 (120 ng) to balance the DNA concentration. Cells were transfected using calcium chloride. 24 h post-transfection, cells were washed once with PBS and infected for 2 h, washed once with PBS, and then incubated for an additional 6 h with fresh medium plus gentamicin (100 μg/ml) before performing luciferase measurements.
Intranasal Infection Model
5–7-week-old female C57BL/6JOlaHsd mice (Harlan) were anesthetized by intraperitoneal injection with a mixture containing ketamine (50 mg/kg) and xylazine (5 mg/kg). Overnight bacterial cultures were centrifuged (2,500 × g, 20 min, 22 °C), resuspended in PBS, and adjusted to 5 × 106 cfu/ml for analysis of cytokine expression and to 5 × 104 cfu/ml for determination of bacterial loads. 20 μl of the bacterial suspension were inoculated intranasally in four 5-μl aliquots. Non-infected mice were inoculated intranasally with 20 μl of PBS in four 5-μl aliquots. To facilitate consistent inoculations, mice were held vertically during inoculation and placed on a 45° incline while recovering from anesthesia. At the indicated times after infection, mice were euthanized by cervical dislocation, and lungs were either rapidly dissected for bacterial load determination or immediate frozen in liquid nitrogen and stored at −80 °C until purification of RNA was carried out. To this end, lungs were quickly weighted and homogenized with 1 ml of TRI reagent (Ambion) using an Ultra-Turrax TIO basic (IKA) on ice. Total RNA was purified first using a standard chloroform/isopropyl alcohol protocol, and the obtained RNA was further purified using a Nucleospin RNAII kit (Macherey-Nagel) exactly as recommended by the manufacturer. RNA integrity was verified using a formaldehyde-agarose gel, quantified spectrophotometrically with a NanoDrop spectrophotometer, and stored at −80 °C. mRNA expression was measured by RT-qPCR analysis as described above. The following intron-spanning primers were used: 5′-CCACATCTCCCTCCAGAAAA-3′ (sense) and 5′-AGGGTCTGGGCCATAGAACT-3′ (antisense) for tnf-α; 5′-CCGGAGAGGAGACTTCACAG-3′ (sense) and 5′-CAGAATTGCCATTGCACAAC-3′ (antisense) for il-6; and 5′-GACAGACTGCTCTGATGGCA-3′ (sense) and 5′-TGCACTTCTTTTCGCACAAC-3′ (antisense) for kc. As internal controls, we amplified the mouse housekeeping actin and gapdh (5′-TGTTACCAACTGGGACGACA-3′ (sense) and 5′-CTGGGTCATCTTTTCACGGT-3′ (antisense) for actin and 5′-CCCACTAACATCAAATGGGG-3′ (sense) and 5′-CCTTCCACAATGCCAAAGTT-3′ (antisense) for gapdh. The sizes of the amplicons are 259, 134, 292, 139, and 274 bp for tnf-α, il-6, kc, actin, and gapdh, respectively.
Dissected lungs from those animals infected to determine bacterial loads were homogenized in 500 μl of PBS using an Ultra-Turrax TIO basic disperser (IKA) on ice. Bacteria from the homogenates and serial dilutions thereof were recovered in LB agar plates containing rifampicin for the wild-type strain or rifampicin plus kanamicin for the ompA mutant. Results were reported as log cfu/g of tissue. Mice were treated in accordance with the European Convention for the Protection of Vertebrate Animals used for Experimental and other Scientific Purposes (Directive 86/609/EEC) and in agreement with the Bioethical Committee of the University of the Balearic Islands (Spain).
Statistical Methods
Statistical analyses were performed using one-way analysis of variance (ANOVA) with Bonferroni contrasts or the one-tailed t test or, when the requirements were not met, by the Mann-Whitney U test. p < 0.05 was considered statistically significant. The analyses were performed using Prism4 for PC (GraphPad Software).
RESULTS
Infection of Airway Epithelial Cells by K. pneumoniae ompA Mutant Induces the Secretion of IL-8
Previous data (32), further confirmed here, had shown that wild-type K. pneumoniae strain 52145 (hereafter referred to as Kp52145) does not induce the secretion of IL-8 by A549 cells (Fig. 1A). We asked whether infection with strain 52OmpA2, an isogenic ompA mutant, would induce the secretion of IL-8. Time course experiments showed a correlation between the duration of the infection with 52OmpA2 and the levels of IL-8 secreted by A549 cells, the levels of IL-8 being higher at 8 than at 4 h postinfection (Fig. 1A). Moreover, at 6 and 8 h postinfection, 52OmpA2-infected cells secreted levels of IL-8 significantly higher than those of Kp52145-infected cells, which were similar to those of non-infected cells (Fig. 1A) (p > 0.05; one-tailed Student's t test).
FIGURE 1.
K. pneumoniae ompA mutants induce higher levels of IL-8 than wild-type strains by airway epithelial cells. A, ELISA of IL-8 released by A549 cells left untreated (CON) or infected for different time points with Kp52145 (gray bars) and 52OmpA2 (black bars) (n = 3). B, ELISA of IL-8 released by A549 cells left untreated (CON) or infected for 8 h with different K. pneumoniae strains (n = 3). C, ELISA of IL-8 released by A549 cells left untreated (CON) or infected for different time points with 52OmpA2 (gray bars) and 52145-ΔwcaK2ompA (black bars) (n = 3). D, ELISA of IL-8 released by NHBE cells left untreated (CON) or infected for 8 h with different K. pneumoniae strains (n = 3). E, adhesion (left) and internalization (right) of Klebsiella strains to A549 cells (n = 3). Gray bars, CPS-expressing strains; black bars, CPS negative strains. The data (A–E) are means and S.E. (error bars). A–D, *, p < 0.05 (for the indicated comparisons; one-way ANOVA). E, *, p < 0.05 (results are significantly different from the results for Kp52145; one-tailed t test).
Evidence indicates that K. pneumoniae CPS mutants induce the secretion of IL-8 by airway epithelial cells (32). Therefore, the induction of IL-8 by 52OmpA2 could be due to a reduction in the amount of CPS expressed by this mutant. However, Kp52145 and 52OmpA2 expressed similar amounts of cell-bound CPS (360 ± 15 μg/106 cells and 339 ± 23 μg/106 cells, respectively; p > 0.05; one-tailed Student's t test). We tested whether the absence of OmpA further increases the levels of IL-8 induced by 52145-ΔwcaK2, an isogenic CPS mutant from Kp52145. Indeed, at 8 h postinfection, 52145-ΔwcaK2ompA-infected cells secreted higher levels of IL-8 than 52145-ΔwcaK2- or 52OmpA2-infected cells (Fig. 1B). IL-8 levels induced by 52OmpA2Com- and 52145-ΔwcaK2ompACom-complemented strains of 52OmpA2 and 52145-ΔwcaK2, respectively, were not significantly different from those triggered by Kp52145 and 52145-ΔwcaK2, hence indicating that the absence of OmpA is responsible for the induction of IL-8 secretion by OmpA mutants. For the sake of comparison, IL-8 levels induced by 52OmpA2 and 52145-ΔwcaK2ompA were compared in a time course experiment (Fig. 1C). At all time points, 52145-ΔwcaK2ompA-infected cells secreted higher levels of IL-8 than 52OmpA2-infected ones. ompA mutants also induced higher levels of IL-8 than parental strains in normal human bronchial cells (Fig. 1D). At 8 h postinfection, ompA mutants induced higher levels of IL-8 than the parental strains, 52145-ΔwcaK2ompA being the strain inducing the highest levels. Both mutants were complemented (Fig. 1D). Collectively, these data show that CPS and OmpA are bacterial factors required to reduce the secretion of IL-8 by airway epithelial cells upon K. pneumoniae infection.
Control experiments revealed that the adhesion and internalization to cells were not significantly different between each ompA mutant and its corresponding parental strain (Fig. 1E). As reported previously (43, 44), strains lacking CPS adhered and were internalized in higher numbers than CPS-expressing strains (for each comparison between CPS-expressing and CPS-negative strains, p < 0.05 (one-tailed Student's t test) (Fig. 1E).
ompA Mutant Attenuates IL-1β-induced IL-8 Secretion
Recently, we have shown that Kp52145 attenuates proinflammatory mediator-induced IL-8 secretion (45). This process requires bacteria-cell contact, and removal of bacteria by washing followed by 1-h gentamicin treatment rendered cells responsive to agonist-induced IL-8 secretion (45). To exert this anti-inflammatory effect, Kp52145 up-regulates the expression of the deubiquitinase CYLD and the phosphatase MKP-1 (DUSP1) in a NOD1-dependent manner (45). To explore whether OmpA could account for the Kp52145 anti-inflammatory effect, we determined the effect of 52OmpA2 on IL-1β-induced IL-8 secretion by A549 cells using the assay that we have described (45). Similarly to Kp52145, 52OmpA2 did attenuate IL-1β-induced IL-8 secretion (Fig. 2A). Furthermore, Western blot analysis revealed that 52OmpA2 induced the expression of CYLD and MKP-1 (Fig. 2B). This was dependent on NOD1 because 52OmpA2 did not increase the expression of CYLD and MKP-1 in NOD1 knockdown cells (Fig. 2C).
K. pneumoniae ompA Mutant Induces Higher Levels of Cytokines than Wild-type Strain in Vivo
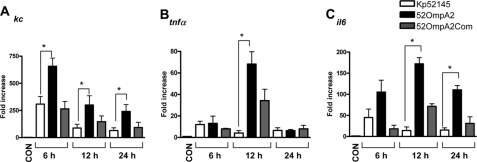
We investigated whether 52OmpA2 induces higher levels of cytokines than Kp52145 in the lungs of infected mice. Mice were infected intranasally and euthanized at different time points. Whole lungs were dissected, and the expressions of kc, tnfα, and il6 were measured by RT-qPCR at 6, 12, and 24 h postinfection (Fig. 3). Levels of kc, tnfα, and il6 were higher in lungs of infected mice than in lungs of non-infected animals (p < 0.05 for all comparisons versus non-infected mice (one-tailed Student's t test)) (Fig. 3). However, levels of kc were higher in lungs of mice infected with 52OmpA2 than in those infected with Kp52145 at all time points analyzed. 52OmpA2Com induced similar levels of kc than Kp52145. Only at 12 h postinfection, levels of tnfα were significantly different between Kp52145- and 52OmpA2-infected mice. At 12 and 24 h postinfection, levels of il6 were higher in lungs of mice infected with 52OmpA2 than in those infected with Kp52145. Levels of il6 were not significantly different between mice infected with Kp52145 and animals infected with 52OmpA2Com at all time points. In summary, these data show that OmpA plays a role in vivo contributing to reduce host inflammatory responses.
FIGURE 3.
Proinflammatory cytokine expressions in mice lungs after K. pneumoniae infections. Mice were non-infected (white bars, n = 5) or infected with wild-type K. pneumoniae 52145 (black bars, n = 15), 52OmpA2 (black bars; n = 15), or 52OmpA2Com (gray bars; n = 15). kc (A), tnfα (B), and il6 (C) mRNA expressions in whole lungs at the indicated time points postinfection were assessed by RT-qPCR (5 mice/time point). Bars, mean ± S.E. (error bars). *, p < 0.05 (for the indicated comparisons; one-way ANOVA).
Activation of NF-κB Is Required for K. pneumoniae ompA Mutant-induced IL-8 Expression
We next aimed to identify the signaling pathways activated by the ompA mutant to induce an inflammatory response. Several studies show that NF-κB participates in the inducible expression of genes involved in the inflammatory response, including IL-8 (46). Therefore, we analyzed the effect of 52OmpA2 on the NF-κB activation pathway by studying the activation of a reporter construct controlled by synthetic NF-κB response elements. A549 cells were transiently transfected with a NF-κB-dependent luciferase reporter followed by infection, and infection-induced NF-κB activation was measured as relative luciferase activity (Fig. 4A). In contrast to Kp52145, 52OmpA2 induced the activation of the reporter construct. Complemented strain 52OmpA2Com induced a NF-κB-dependent luciferase activity similar to Kp52145-infected cells. Supporting previous findings (22, 32), 52145-ΔwcaK2 induced the reporter construct, which was further induced by 52145-ΔwcaK2ompA (Fig. 4A). Complementation of the mutant, strain 52145-ΔwcaK2ompACom, restored NF-κB-dependent luciferase activity to 52145-ΔwcaK2-induced levels (Fig. 4A).
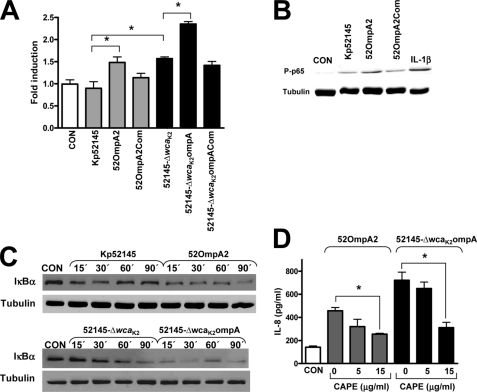
FIGURE 4.
Activation of NF-κB is required for K. pneumoniae ompA mutant-induced IL-8 expression. A, activation of a NF-κB luciferase reporter plasmid in A549 cells left untreated (CON) or infected for 8 h with K. pneumoniae strains. Activity is normalized by correction of Renilla expression and is presented relative to the cells untreated (n = 3). *, p < 0.05 (for the indicated comparisons; one-way ANOVA). B, immunoblot analysis showing phospho-p65 and tubulin levels in lysates of A549 infected with K. pneumoniae strains for 90 min IL-1β (20 ng/ml for 30 min) was used as a positive control for NF-κB activation. The results are representative of three independent experiments. C, immunoblots of IκBα and tubulin levels in lysates of A549 cells left untreated (CON; time 0) or infected for different time periods with K. pneumoniae strains. Data are representative of three independent experiments. D, ELISA of IL-8 released by A549 cells left untreated (CON; white bar) or infected for 6 h with 52OmpA2 (gray bars) or 52145-ΔwcaK2ompA (black bars) in the absence or presence of different concentrations of CAPE, an inhibitor of NF-κB, which was added 1 h before infecting the cells and kept until the end of the experiment (n = 3). The data (A and C) are means and S.E. (error bars).
Upon cellular stimulation, in the canonical pathway of NF-κB activation, IκBα, which is normally present in the cytosol complexed to NF-κB dimers preventing nuclear translocation, becomes phosphorylated, leading to its ubiquitination and subsequent degradation by the proteosome, thereby allowing nuclear translocation of NF-κB, a process linked to phosphorylation of the p65 subunit (47). We analyzed the levels of phosphorylated p65 by Western blot. At 60 min postinfection, phosphorylated p65 was detected in extracts from infected cells (Fig. 4B). However, these levels were higher in 52OmpA2-infected cells than in Kp52145-infected ones. Complementation of the mutant restored phoshorylated p65 levels to those of Kp52145-infected cells (Fig. 4B). IκBα levels in cytoplasmic extracts were analyzed by immunoblot. IκBα degradation was apparent in extracts from cells infected with 52OmpA2 already 15 min postinfection, and it was still apparent at 90 min postinfection. In contrast, IκBα degradation was only detected at 15 and 30 min postinfection in extracts from Kp52145-infected cells, and at later time points, IκBα levels were similar to those of non-infected cells. For the sake of comparison, the levels of IκBα in cell extracts infected with the CPS mutants were also analyzed. In 52145-ΔwcaK2ompA-infected cells, IκBα degradation was clearly detected at 15 min postinfection, and it was almost total at 90 min postinfection. IκBα degradation in 52145-ΔwcaK2-infected cells was apparent at 30 min postinfection and was still detected at 90 min postinfection.
To determine the contribution of NF-κB activation to ompA mutant-induced IL-8 expression, we asked whether CAPE, a chemical inhibitor used to block the NF-κB signaling pathway (48), alters infection-induced IL-8 expression. As shown in Fig. 4D, CAPE (15 μg/ml) reduced 52OmpA2- and 52145-ΔwcaK2ompA-triggered IL-8 levels. Control experiments showed that addition of DMSO (the solvent used for CAPE) to either 52OmpA2- or 52145-ΔwcaK2ompA-infected cells did not affect IL-8 levels (in the absence of DMSO, 457 ± 28 and 721 ± 70 pg/ml, respectively; in the presence of DMSO, 490 ± 35 and 747 ± 64 pg/ml, respectively). CAPE-treated cells (15 μg/ml) expressed levels of IL-8 (110 ± 85 pg/ml) similar to those of non-treated cells (97 ± 50 pg/ml).
Collectively, these data demonstrated that IκBα-dependent activation of NF-κB is required for ompA induction of IL-8 expression in A549 cells. Moreover, CPS and OmpA contribute to limit the activation of the canonical NF-κB signaling pathway.
Activation of MAPKs Is Required for K. pneumoniae ompA Mutant-induced IL-8 Expression
In addition to the NF-κB signaling cascade, many cellular stimuli also activate MAPK pathways (49). The activation of the three MAPKs p38, JNK, and p44/42 occurs through phosphorylation of serine and threonine residues. We sought to determine the phosphorylation levels of the three MAPKs in 52OmpA2-A549-infected cells. Western blot analysis shown in Fig. 5A revealed that infection with 52OmpA2 triggered the phosphorylation of the three MAPKs. Phosphorylation of the three MAPKs occurred at earlier time points in 52OmpA2-infected cells than in Kp52145-infected ones. MAPKs phosphorylation levels were also studied in 52145-ΔwcaK2ompA- and 52145-ΔwcaK2-infected cells. In 52145-ΔwcaK2ompA-infected cells, phosphorylation of p38 was apparent at 30 min postinfection and maximum levels were observed at 120 min postinfection, whereas in 52145-ΔwcaK2-infected cells, p38 phosphorylation was apparent at 60 min postinfection, and maximum levels were observed at 120 min postinfection. No clear differences in the phosphorylation of p44/42 were observed between 52145-ΔwcaK2ompA- and 52145-ΔwcaK2-infected cells. In 52145-ΔwcaK2ompA-infected cells, maximum levels of phosphorylated JNK were observed at 30 min postinfection, whereas in 52145-ΔwcaK2-infected cells, phosphorylation of JNK was apparent at 30 min postinfection, and maximum levels were observed at 120 postinfection.
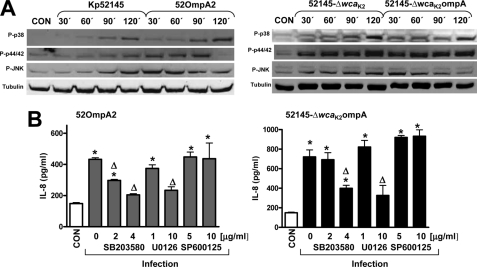
FIGURE 5.
Activation of MAPKs is required for K. pneumoniae ompA mutant-induced IL-8 expression. A, immunoblots showing phospho-p38 (P-p38), phospho-p44/42 (P-p44/42), phospho-JNK (P-JNK), and tubulin levels in cell extracts of A549 cells left uninfected (CON; time 0) or infected with K. pneumoniae strains for different times. The results are representative of three independent experiments. B, ELISA of IL-8 released by A549 cells left untreated (CON; white bar) or infected for 6 h with either 52OmpA2 (gray bars) or 52145-ΔwcaK2ompA (black bars) in the absence or presence of different concentrations of SB203580 (2 and 4 μg/ml; p38 MAPK inhibitor), U0126 (1 and 10 μg/ml; p44/42 MAPK inhibitor), or SP600125 (5 and 10 μg/ml; JNK MAPK inhibitor), which were added 2 h before infecting the cells and kept until the end of the experiment (n = 3). The data (B) are means and S.E. (error bars). *, p < 0.05 (results are significantly different from the results for untreated cells; one-way ANOVA). Δ, p < 0.05 (results are significantly different from the results for infected cells in the absence of inhibitor).
To explore whether the activation of MAPKs is involved in 52OmpA2-induced IL-8 expression, infections were carried out in the presence of chemical inhibitors for each MAPK. Fig. 5B shows that SB203580, a specific inhibitor of p38 MAPK, reduced 52OmpA2-induced IL-8 expression. The p44/42 inhibitor, U0126, also reduced 52OmpA2-dependent IL-8 expression but only at the highest dose tested (Fig. 5C). Finally, SP600125, a JNK inhibitor, did not alter 52OmpA2-induced IL-8 expression (Fig. 5B). We sought to determine whether activated MAPKs are involved in 52145-ΔwcaK2ompA-triggered IL-8 levels. SB203580 and U0126 at the highest dose tested also reduced 52145-ΔwcaK2ompA-induced IL-8 expression, whereas SP600125 did not reduce IL-8 levels (Fig. 5B). SB203580-, U0126-, and SP600125-treated cells secreted similar amounts of IL-8 (120 ± 95, 95 ± 67, and 135 ± 87 pg/ml, respectively) as non-treated cells (130 ± 75 pg/ml). Taken together, these findings indicate that the activation of MAPKs p38 and p44/42 is involved in K. pneumoniae ompA mutant-induced expression of IL-8 in A549 cells.
Dissection of Host Cell Receptors Required for Activation of Inflammatory Responses by K. pneumoniae ompA Mutant Strains
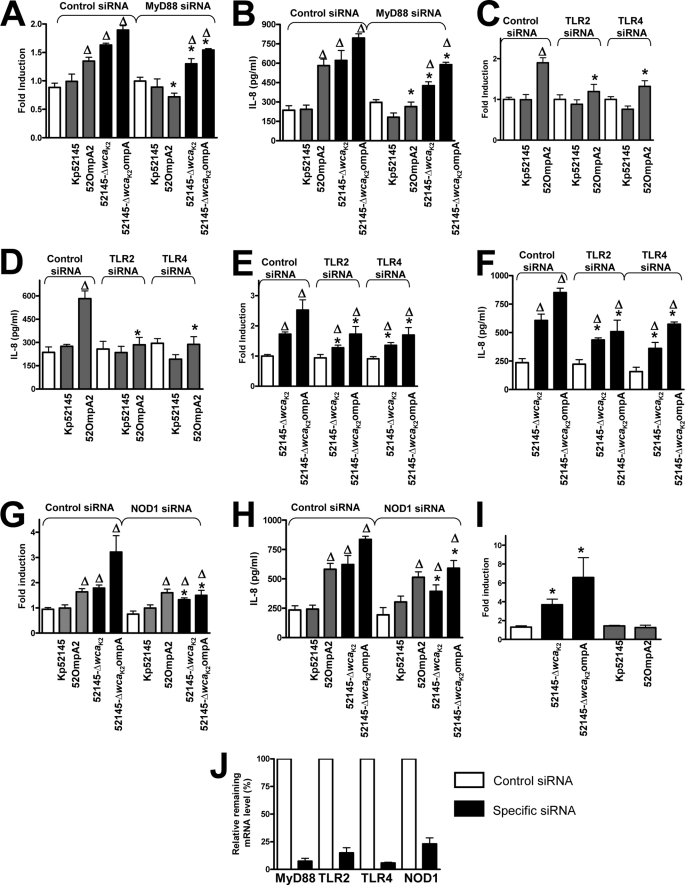
PRRs have a central role in pathogen recognition by airway epithelial cells leading to the activation of NF-κB- and MAPK-dependent signaling pathways; we therefore investigated the involvement of PRRs in recognition of K. pneumoniae ompA mutants by using as cellular read-outs the activation of NF-κB and the secretion of IL-8. Almost all TLRs activate cellular signaling pathways through TIR domain-mediated interactions with the adaptor molecule MyD88 (50). To explore the involvement of TLRs in ompA mutant-induced cell activation, the function of the MyD88 adaptor molecule was interrupted by siRNA. In MyD88 knocked down cells, 52OmpA2 induced neither the activation of the NF-κB reported construct nor the secretion of IL-8 (Fig. 6, A and B, respectively). 52145-ΔwcaK2ompA still activated the NF-κB reported construct and induced the secretion of IL-8, although the levels were significantly lower than those of infected cells transfected with control siRNA (Fig. 6, A and B). Similar results were obtained when cells were infected with 52145-ΔwcaK2 (Fig. 6, A and B). To further dissect the contribution of TLR-dependent signaling to ompA mutant-induced cell activation, TLR2 and TLR4 were knocked down by siRNA. Results shown in Fig. 6, C–F, indicate that both receptors contributed to 52OmpA2- and 52145-ΔwcaK2ompA-induced cell activation. However, 52145-ΔwcaK2ompA still triggered the activation of NF-κB and induced the secretion of IL-8 in TLR knocked down cells (Fig. 6, E and F, respectively). On the whole, these data suggest that 52OmpA2-induced NF-κB activation and IL-8 secretion is mediated by the TLR4-TLR2-MyD88 pathway and that 52145-ΔwcaK2ompA may also activate additional MyD88-independent signaling pathway(s).
FIGURE 6.
Role of MyD88, TLR2, TLR4, and NOD1 in K. pneumoniae ompA mutant-induced cell activation. A, C, E, and G, activation of an NF-κB luciferase reporter plasmid in A549 cells transfected with either control or the indicated siRNA for different pattern recognition receptors, which were left untreated (white bars) or infected for 8 h with different strains. Activity is normalized by correction of Renilla expression and is presented relative to the cells untreated (n = 3). B, D, F, and H, ELISA of IL-8 released by A549 cells transfected with either control or the indicated siRNA for different pattern recognition receptors, which were left untreated (white bars) or infected for 8 h with different strains (n = 3). I, activation of an NF-κB luciferase reporter plasmid in HEK293T cells transfected with HA-NOD1 plasmid, which were left untreated (white bar) or infected with the indicated strains for 8 h. Activity is normalized by correction of Renilla expression and is presented relative to untreated cells (data are means and S.E. (error bars); n = 3). J, siRNA efficiency was quantified by RT-qPCR in samples from the same experiment shown in A–H. mRNA level was normalized to GADPH, and then relative mRNA levels in cells transfected with control siRNA or specific siRNA were compared. mRNA levels in cells transfected with control siRNA were set to 100% (data are means and S.E.; n = 3). Gray bars, CPS-expressing strains; black bars, CPS-negative strains. A–H, Δ, p < 0.05 (results are significantly different from the results for untreated cells; one-way ANOVA); *, p < 0.05 (results are significantly different from those for transfected cells with control siRNA infected with the same strain). I, *, p < 0.05 (results are significantly different from the results for untreated cells; one-way ANOVA).
Another PRR constitutively expressed by airway epithelial cells is NOD1 (51–54), and its engagement also results in the activation of NF-κB and MAPK signaling cascades (53). This led us to explore whether NOD1 contributes to 52145-ΔwcaK2ompA-induced cell activation by using siRNA to knock down its expression. As shown in Fig. 6, G and H, there was a reduction in 52145-ΔwcaK2ompA-triggered NF-κB activation and IL-8 secretion in NOD1 knockdown cells compared with those found in control siRNA-treated cells. Similar results were obtained when cells were infected with 52145-ΔwcaK2 (Fig. 6, G and H). 52OmpA2-induced NF-κB activation and IL-8 secretion in NOD1 knockdown cells were similar to those found in control siRNA-treated cells (Fig. 6, G and H). In contrast to Kp52145 and 52OmpA2, 52145-ΔwcaK2ompA and 52145-ΔwcaK2 did trigger NF-κB-dependent reporter activity in NOD1-expressing HEK293T cells (Fig. 6I). Altogether, these data indicate that the other signaling pathway engaged by 52145-ΔwcaK2ompA to activate NF-κB and induce IL-8 secretion is NOD1-dependent. The efficiency of siRNA-mediated down-regulation of target gene mRNA levels was confirmed by RT-qPCR (Fig. 6J).
Virulence of K. pneumoniae ompA Mutant
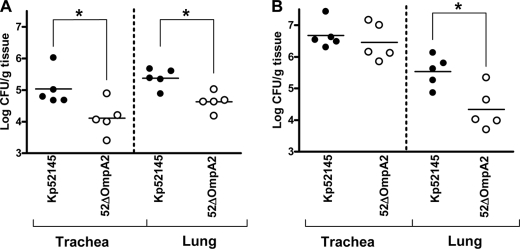
To determine the ability of 52OmpA2 to cause pneumonia, C57BL/6JOlaHsd mice were infected intranasally, and 24 and 96 h postinfection, bacterial loads in trachea and lung homogenates were determined. At 24 h postinfection, Kp52145 and 52OmpA2 colonized trachea and lungs, although bacterial loads of 52OmpA2 were lower than those of the wild type (Fig. 7A). At 96 h postinfection, an increase of bacterial loads was found in tracheas (Fig. 7, compare A and B) that were not significantly different between both strains (Fig. 7B). In contrast, bacterial loads of 52OmpA2 in lungs were significantly lower than those of the wild-type strain (Fig. 7B).
FIGURE 7.
Bacterial counts in mouse organs at 24 h postinfection (A) or 96 h postinfection (B). Mice were infected intranasally with a bacterial mixture containing 5 × 104 bacteria of wild type (Kp52145) (●) or ompA mutant (52OmpA2) (○). Results were reported as log cfu/g of tissue. *, results are significantly different (p < 0.05; one-tailed t test) from the results for Kp52145.
DISCUSSION
In this study, we provide new insights into the role of OmpA as a virulence factor. Our findings revealed that K. pneumoniae OmpA plays a role in the progression of the infection and that it is important for immune evasion. Thus, we show that the K. pneumoniae ompA mutant, strain 52OmpA2, induces higher levels of inflammatory mediators than the wild-type strain in vitro and in vivo. Mechanistically, we present evidence showing that 52OmpA2 activates NF-κB-dependent and MAPK p38- and p44/42-dependent signaling pathways. In addition, ompA mutant induction of inflammatory responses is dependent on TLR2-TLR4-MyD88 activation. Finally, we demonstrate that the ompA mutant is attenuated in the pneumonia mouse model.
Innate immune responses, particularly inflammation, are essential for effective lung defense against pathogens. To activate these responses, the host recognizes conserved molecules uniquely expressed by pathogens, the so-called PAMPs, which are characterized by being expressed among entire classes of pathogens, are important for the survival/virulence of the pathogen, and are distinguishable from “self.” These PAMPs are also targeted by weapons of the innate system with antibacterial properties, such as antimicrobial peptides and the complement system. In turn, many PAMPs play an important role in immune evasion by the pathogen. Our findings, together with others, give support to the notion that K. pneumoniae OmpA should be considered a bona fide PAMP. OmpA is targeted by neutrophil elastase and serum amyloid protein A, hence leading to cell death and increased bacterial phagocytosis, respectively (7–9). However, OmpA protects bacteria against the bactericidal action of lung collectins and antimicrobial peptides (6, 39), and here we have shown that it acts as immune evasin. On the whole, this evidence points out that an essential attribute of K. pneumoniae OmpA is to thwart the innate immune system.
Other studies have shown that recombinant purified K. pneumoniae OmpA induces the expression of proinflammatory molecules in various cell types, including airway epithelial cells (for a review, see Ref. 36). Hence, it has been postulated that recognition of OmpA contributes to the activation of host responses leading to the clearance of K. pneumoniae (36). Given that in this work we used a bacterial mutant instead of a recombinant protein, our data, far from challenging these findings, shed new light on OmpA functions in its physiological context, the OM. In fact, we emphasize that care should be taken to extrapolate findings obtained using purified products to the outcome of an infection process and that using whole live bacteria is a more likely in vivo scenario. It should be noted that the ompA mutant strain was shown to induce more inflammation in vivo and in vitro than the wild-type strain. Interestingly, and in agreement with our findings, E. coli ompA mutant strain induces higher expression of proinflammatory mediators than the wild type in vitro (55).
Recently, we have demonstrated that K. pneumoniae counteracts the activation of inflammatory responses by inducing the deubiquitinase CYLD and the phosphatase MKP-1 (DUSP1) in a NOD1-dependent manner (45). This action was strictly dependent on bacteria-cell contact (45). Furthermore, bacterial removal by gentamicin treatment rendered cells again responsive to proinflammatory mediators (45). Considering that CPS was necessary but not sufficient to attenuate inflammation (45), in this work we asked whether OmpA could mediate this attenuation. However, findings shown in Fig. 2 do not support this. These results do not contradict the other findings reported in this work showing that 52OmpA2 does activate inflammatory responses. It should be noted that in most experiments of this study, gentamicin is used to remove bacteria after 2 h of infection, and, for example, IL-8 in the supernatants is measured at 4 or 6 h postinfection. In turn, the fact that 52OmpA2-induced responses were dependent on TLR2-TLR4-MyD88 activation suggests that OmpA, together with CPS, perturbs TLR-dependent recognition of K. pneumoniae. On the whole, this evidence is consistent with a model in which the reduced inflammatory response characteristic of K. pneumoniae infections is the sum of prevention of TLR-dependent recognition of Klebsiella (mediated at least by CPS and OmpA) and the anti-inflammatory effect (K. pneumoniae effectors yet to be identified) recently described (45).
Our findings further sustain the notion that airway epithelial cells orchestrate a defense response upon pathogen challenge by activating NF-κB- and MAPK-dependent signaling pathways (56). In fact, activation of these pathways seems to be a central event of pathogen-exposed epithelial cells lining mucosal surfaces. Interestingly, this is true regardless of the tissue local environment, either constantly exposed to microbes, like the intestinal tract, or essentially sterile, like the urinary and respiratory tracts. Although p38 and p44/42 MAPKs were required for ompA mutant strain-induced IL-8, the mutants also activated JNK MAPK, hence suggesting that there may be additional host cell responses activated in a JNK MAPK-dependent manner. Future studies will aim to identify these responses.
The ompA mutant strains tested in this study engaged different PRRs, depending on the expression of CPS on their surfaces. Strain 52OmpA2, expressing CPS, engaged TLR2 and TLR4, whereas strain 52145-ΔwcaK2ompA, lacking CPS, activated not only TLR2 and TLR4 but also NOD1. These and our previous findings implicating TLR2 and TLR4 in the recognition of a K. pneumoniae CPS mutant (22, 32) suggest that TLR-mediated recognition of K. pneumoniae is a key event for the induction of host defense responses against this pathogen. Supporting this, MyD88- and TLR4-dependent responses are essential for successful mouse defense against K. pneumoniae pneumonia (29, 30). Our results implicate NOD1 in the recognition of K. pneumoniae CPS mutants, strains 52145-ΔwcaK2ompA (this work) and 52145-ΔwcaK2 (this work and Ref. 22). Taking into account that K. pneumoniae CPS mutants are internalized by epithelial cells (43, 44), a NOD1-dependent recognition is consistent with the idea that this PRR participates in the recognition of internalized pathogens (53, 57, 58). Intracellular 52145-ΔwcaK2ompA and 52145-ΔwcaK2 reside in vacuoles displaying features of acidic late endosomes,4 thereby raising the question of how bacteria that do not reach the cytosol could activate NOD1. However, recent studies indicate that there is a link between endosomal acidification/maturation and NOD-dependent responses (59–61). Furthermore, data indicate that bacterial products are actively transported from the phagosome to the cytosol to engage NOD receptors (59–61).
Here, we demonstrate for the first time, using a pneumonia mouse model, that OmpA is important for bacterial survival in the lung. Research over the last 20 years has demonstrated a correlation between activation of inflammatory responses and Klebsiella clearance from the airways (26–28). Hence, the higher inflammatory response induced by 52OmpA2 may contribute to the mutant's clearance. The in vivo scenario is complex, and the final outcome of pneumonia is the combination of the action of antimicrobial factors (among others complement and antimicrobial peptides) and several types of cells, including alveolar macrophages, epithelial cells, and neutrophils. It should be noted that cytokines and chemokines released by epithelial cells do increase the bactericidal activity of professional phagocytes. Studies are ongoing to determine whether OmpA plays any role in the interaction of K. pneumoniae with alveolar macrophages and neutrophils.
Finally, it is worthwhile commenting on the clinical implications of our findings. We put forward the idea that compounds directed to block OmpA may be new promising therapeutic molecules to treat infections caused by Gram-negative bacteria, at least those caused by Klebsiella. Data from our laboratory suggest that these compounds will render bacteria susceptible to the bactericidal elements of the innate immune system (39) and will also increase TLR-mediated defense responses (this work). Both actions should facilitate the clearance of the pathogen from the airways, which might be further enhanced with the aid of antibiotics.
Acknowledgments
We are grateful to members of the Bengoechea laboratory for helpful discussions and Christian Frank for critical reading of the manuscript.
This work was supported by Fondo de Investigación Sanitaria Grant PI06/1629, ERA-NET Pathogenomics Grants GEN2006-27776-C2–2-E/PAT and SAF2008-04353-E, and Biomedicine Program Grant SAF2009-07885 from the Ministerio de Ciencia e Innovación (to J. A. B.). CibeRes is an initiative from Instituto de Salud Carlos III.
C. March and J. A. Bengoechea, unpublished data.
- OM
- outer membrane
- CPS
- capsule polysaccharide
- PAMP
- pathogen-associated molecular pattern
- PRR
- pattern recognition receptor
- TLR
- Toll-like receptor
- NLR
- nucleotide binding and oligomerization domain-like receptor
- CAPE
- caffeic acid phenethyl ester
- cfu
- colony-forming units
- RT-qPCR
- real-time quantitative PCR
- ANOVA
- analysis of variance.
REFERENCES
- 1. Lin J., Huang S., Zhang Q. (2002) Microbes Infect. 4, 325–331 [DOI] [PubMed] [Google Scholar]
- 2. Pautsch A., Schulz G. E. (2000) J. Mol. Biol. 298, 273–282 [DOI] [PubMed] [Google Scholar]
- 3. Prasadarao N. V., Wass C. A., Weiser J. N., Stins M. F., Huang S. H., Kim K. S. (1996) Infect. Immun. 64, 146–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sukumaran S. K., Shimada H., Prasadarao N. V. (2003) Infect. Immun. 71, 5951–5961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Prasadarao N. V., Blom A. M., Villoutreix B. O., Linsangan L. C. (2002) J. Immunol. 169, 6352–6360 [DOI] [PubMed] [Google Scholar]
- 6. Wu H., Kuzmenko A., Wan S., Schaffer L., Weiss A., Fisher J. H., Kim K. S., McCormack F. X. (2003) J. Clin. Invest. 111, 1589–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Belaaouaj A., Kim K. S., Shapiro S. D. (2000) Science 289, 1185–1188 [DOI] [PubMed] [Google Scholar]
- 8. Hari-Dass R., Shah C., Meyer D. J., Raynes J. G. (2005) J. Biol. Chem. 280, 18562–18567 [DOI] [PubMed] [Google Scholar]
- 9. Shah C., Hari-Dass R., Raynes J. G. (2006) Blood 108, 1751–1757 [DOI] [PubMed] [Google Scholar]
- 10. Mogensen T. H. (2009) Clin. Microbiol. Rev. 22, 240–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hippenstiel S., Opitz B., Schmeck B., Suttorp N. (2006) Respir. Res. 7, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Craig A., Mai J., Cai S., Jeyaseelan S. (2009) Infect. Immun. 77, 568–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoffmann E., Dittrich-Breiholz O., Holtmann H., Kracht M. (2002) J. Leukoc. Biol. 72, 847–855 [PubMed] [Google Scholar]
- 14. Kumar H., Kawai T., Akira S. (2009) Biochem. J. 420, 1–16 [DOI] [PubMed] [Google Scholar]
- 15. Medzhitov R. (2007) Nature 449, 819–826 [DOI] [PubMed] [Google Scholar]
- 16. Chamaillard M., Girardin S. E., Viala J., Philpott D. J. (2003) Cell Microbiol. 5, 581–592 [DOI] [PubMed] [Google Scholar]
- 17. Inohara N., Chamaillard M., McDonald C., Nuñez G. (2005) Annu. Rev. Biochem. 74, 355–383 [DOI] [PubMed] [Google Scholar]
- 18. Kumar H., Kawai T., Akira S. (2009) Biochem. Biophys. Res. Commun. 388, 621–625 [DOI] [PubMed] [Google Scholar]
- 19. Sahly H., Podschun R. (1997) Clin. Diagn. Lab. Immunol. 4, 393–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Campos M. A., Vargas M. A., Regueiro V., Llompart C. M., Albertí S., Bengoechea J. A. (2004) Infect. Immun. 72, 7107–7114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Merino S., Camprubí S., Albertí S., Benedí V. J., Tomás J. M. (1992) Infect. Immun. 60, 2529–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moranta D., Regueiro V., March C., Llobet E., Margareto J., Larrate E., Garmendia J., Bengoechea J. A. (2010) Infect. Immun. 78, 1135–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Camprubí S., Merino S., Benedí V. J., Tomás J. M. (1993) FEMS Microbiol. Lett. 111, 9–13 [DOI] [PubMed] [Google Scholar]
- 24. Cortés G., Borrell N., de Astorza B., Gómez C., Sauleda J., Albertí S. (2002) Infect. Immun. 70, 2583–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lawlor M. S., Hsu J., Rick P. D., Miller V. L. (2005) Mol. Microbiol. 58, 1054–1073 [DOI] [PubMed] [Google Scholar]
- 26. Greenberger M. J., Kunkel S. L., Strieter R. M., Lukacs N. W., Bramson J., Gauldie J., Graham F. L., Hitt M., Danforth J. M., Standiford T. J. (1996) J. Immunol. 157, 3006–3012 [PubMed] [Google Scholar]
- 27. Standiford T. J., Wilkowski J. M., Sisson T. H., Hattori N., Mehrad B., Bucknell K. A., Moore T. A. (1999) Hum. Gene. Ther. 10, 899–909 [DOI] [PubMed] [Google Scholar]
- 28. Ye P., Garvey P. B., Zhang P., Nelson S., Bagby G., Summer W. R., Schwarzenberger P., Shellito J. E., Kolls J. K. (2001) Am. J. Respir. Cell Mol. Biol. 25, 335–340 [DOI] [PubMed] [Google Scholar]
- 29. Cai S., Batra S., Shen L., Wakamatsu N., Jeyaseelan S. (2009) J. Immunol. 183, 6629–6638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schurr J. R., Young E., Byrne P., Steele C., Shellito J. E., Kolls J. K. (2005) Infect. Immun. 73, 532–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lawlor M. S., Handley S. A., Miller V. L. (2006) Infect. Immun. 74, 5402–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Regueiro V., Campos M. A., Pons J., Albertí S., Bengoechea J. A. (2006) Microbiology 152, 555–566 [DOI] [PubMed] [Google Scholar]
- 33. Jeannin P., Renno T., Goetsch L., Miconnet I., Aubry J. P., Delneste Y., Herbault N., Baussant T., Magistrelli G., Soulas C., Romero P., Cerottini J. C., Bonnefoy J. Y. (2000) Nat. Immunol. 1, 502–509 [DOI] [PubMed] [Google Scholar]
- 34. Jeannin P., Magistrelli G., Herbault N., Goetsch L., Godefroy S., Charbonnier P., Gonzalez A., Delneste Y. (2003) Eur. J. Immunol. 33, 326–333 [DOI] [PubMed] [Google Scholar]
- 35. Jeannin P., Bottazzi B., Sironi M., Doni A., Rusnati M., Presta M., Maina V., Magistrelli G., Haeuw J. F., Hoeffel G., Thieblemont N., Corvaia N., Garlanda C., Delneste Y., Mantovani A. (2005) Immunity 22, 551–560 [DOI] [PubMed] [Google Scholar]
- 36. Jeannin P., Magistrelli G., Goetsch L., Haeuw J. F., Thieblemont N., Bonnefoy J. Y., Delneste Y. (2002) Vaccine 20, Suppl. 4, A23–A27 [DOI] [PubMed] [Google Scholar]
- 37. Nassif X., Fournier J. M., Arondel J., Sansonetti P. J. (1989) Infect. Immun. 57, 546–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Llobet E., Tomás J. M., Bengoechea J. A. (2008) Microbiology 154, 3877–3886 [DOI] [PubMed] [Google Scholar]
- 39. Llobet E., March C., Giménez P., Bengoechea J. A. (2009) Antimicrob. Agents Chemother. 53, 298–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bitter T., Muir H. M. (1962) Anal. Biochem. 4, 330–334 [DOI] [PubMed] [Google Scholar]
- 41. Rahn A., Whitfield C. (2003) Mol. Microbiol. 47, 1045–1060 [DOI] [PubMed] [Google Scholar]
- 42. Regueiro V., Moranta D., Campos M. A., Margareto J., Garmendia J., Bengoechea J. A. (2009) Infect. Immun. 77, 714–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cortés G., Alvarez D., Saus C., Albertí S. (2002) Infect. Immun. 70, 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sahly H., Podschun R., Oelschlaeger T. A., Greiwe M., Parolis H., Hasty D., Kekow J., Ullmann U., Ofek I., Sela S. (2000) Infect. Immun. 68, 6744–6749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Regueiro V., Moranta D., Frank C. G., Larrarte E., Margareto J., March C., Garmendia J., Bengoechea J. A. (2011) Cell Microbiol. 13, 135–153 [DOI] [PubMed] [Google Scholar]
- 46. Ghosh S., May M. J., Kopp E. B. (1998) Annu. Rev. Immunol. 16, 225–260 [DOI] [PubMed] [Google Scholar]
- 47. Hayden M. S., Ghosh S. (2008) Cell 132, 344–362 [DOI] [PubMed] [Google Scholar]
- 48. Natarajan K., Singh S., Burke T. R., Jr., Grunberger D., Aggarwal B. B. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 9090–9095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dong C., Davis R. J., Flavell R. A. (2002) Annu. Rev. Immunol. 20, 55–72 [DOI] [PubMed] [Google Scholar]
- 50. Kenny E. F., O'Neill L. A. (2008) Cytokine 43, 342–349 [DOI] [PubMed] [Google Scholar]
- 51. Girardin S. E., Tournebize R., Mavris M., Page A. L., Li X., Stark G. R., Bertin J., DiStefano P. S., Yaniv M., Sansonetti P. J., Philpott D. J. (2001) EMBO Rep. 2, 736–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hisamatsu T., Suzuki M., Podolsky D. K. (2003) J. Biol. Chem. 278, 32962–32968 [DOI] [PubMed] [Google Scholar]
- 53. Inohara N., Nuñez G. (2003) Nat. Rev. Immunol. 3, 371–382 [DOI] [PubMed] [Google Scholar]
- 54. Strober W., Murray P. J., Kitani A., Watanabe T. (2006) Nat. Rev. Immunol. 6, 9–20 [DOI] [PubMed] [Google Scholar]
- 55. Selvaraj S. K., Prasadarao N. V. (2005) J. Leukoc. Biol. 78, 544–554 [DOI] [PubMed] [Google Scholar]
- 56. Gribar S. C., Richardson W. M., Sodhi C. P., Hackam D. J. (2008) Mol. Med. 14, 645–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Girardin S. E., Sansonetti P. J., Philpott D. J. (2002) Trends Microbiol. 10, 193–199 [DOI] [PubMed] [Google Scholar]
- 58. Kim J. G., Lee S. J., Kagnoff M. F. (2004) Infect. Immun. 72, 1487–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Herskovits A. A., Auerbuch V., Portnoy D. A. (2007) PLoS Pathog. 3, e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lee J., Tattoli I., Wojtal K. A., Vavricka S. R., Philpott D. J., Girardin S. E. (2009) J. Biol. Chem. 284, 23818–23829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Marina-García N., Franchi L., Kim Y. G., Hu Y., Smith D. E., Boons G. J., Núñez G. (2009) J. Immunol. 182, 4321–4327 [DOI] [PMC free article] [PubMed] [Google Scholar]