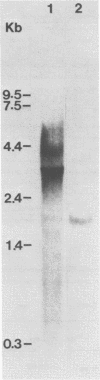
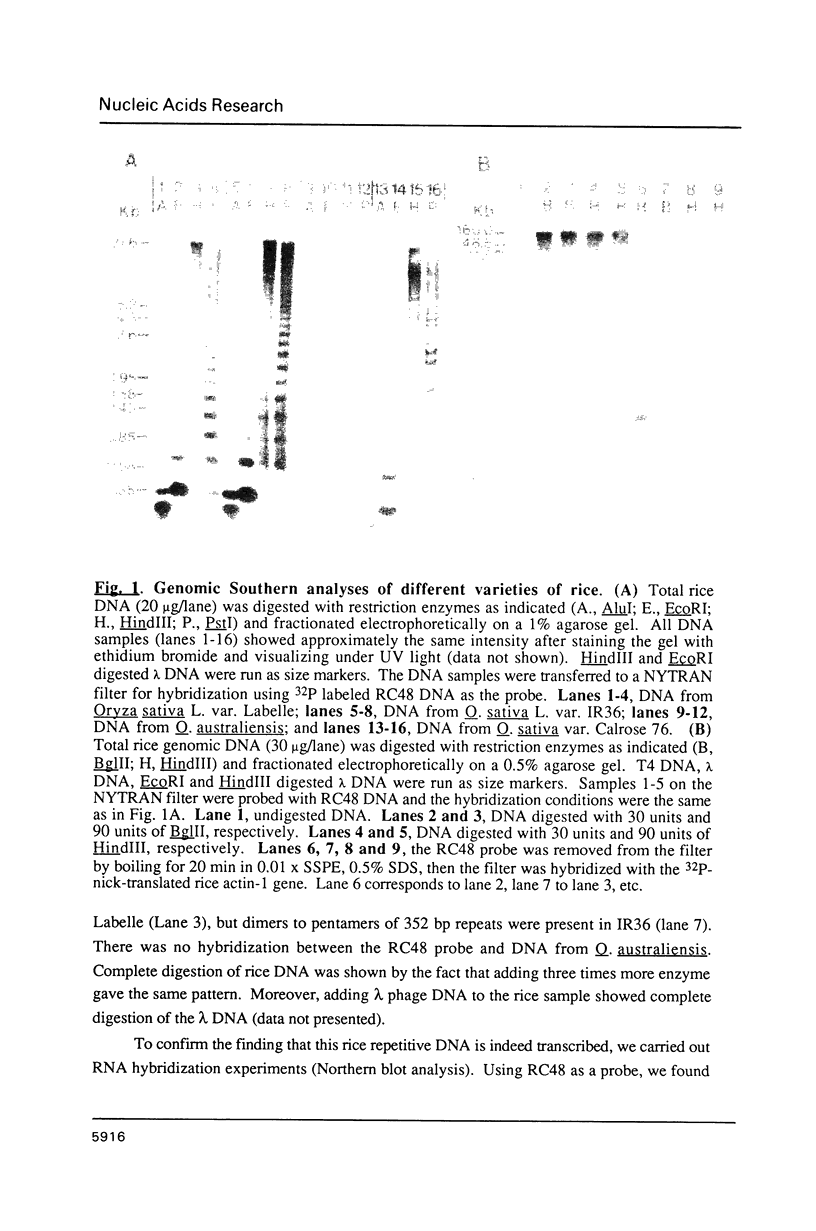
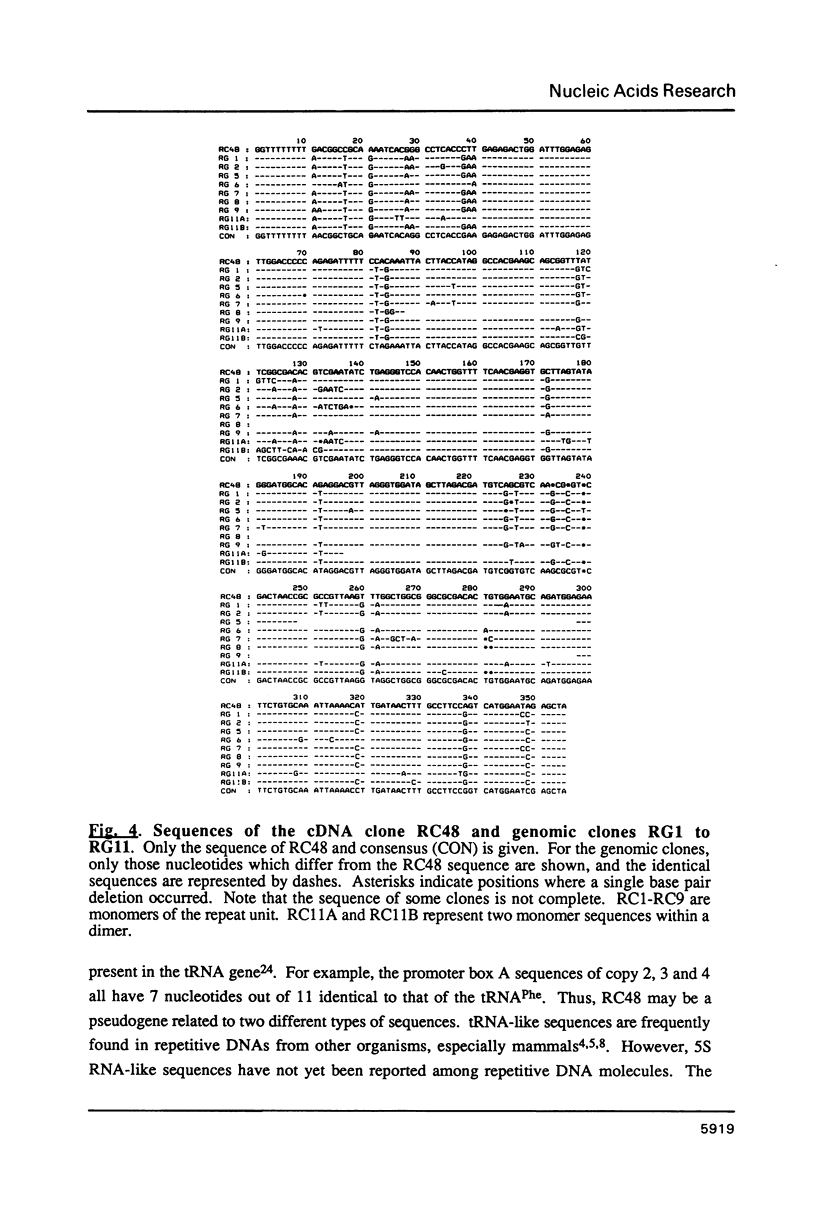
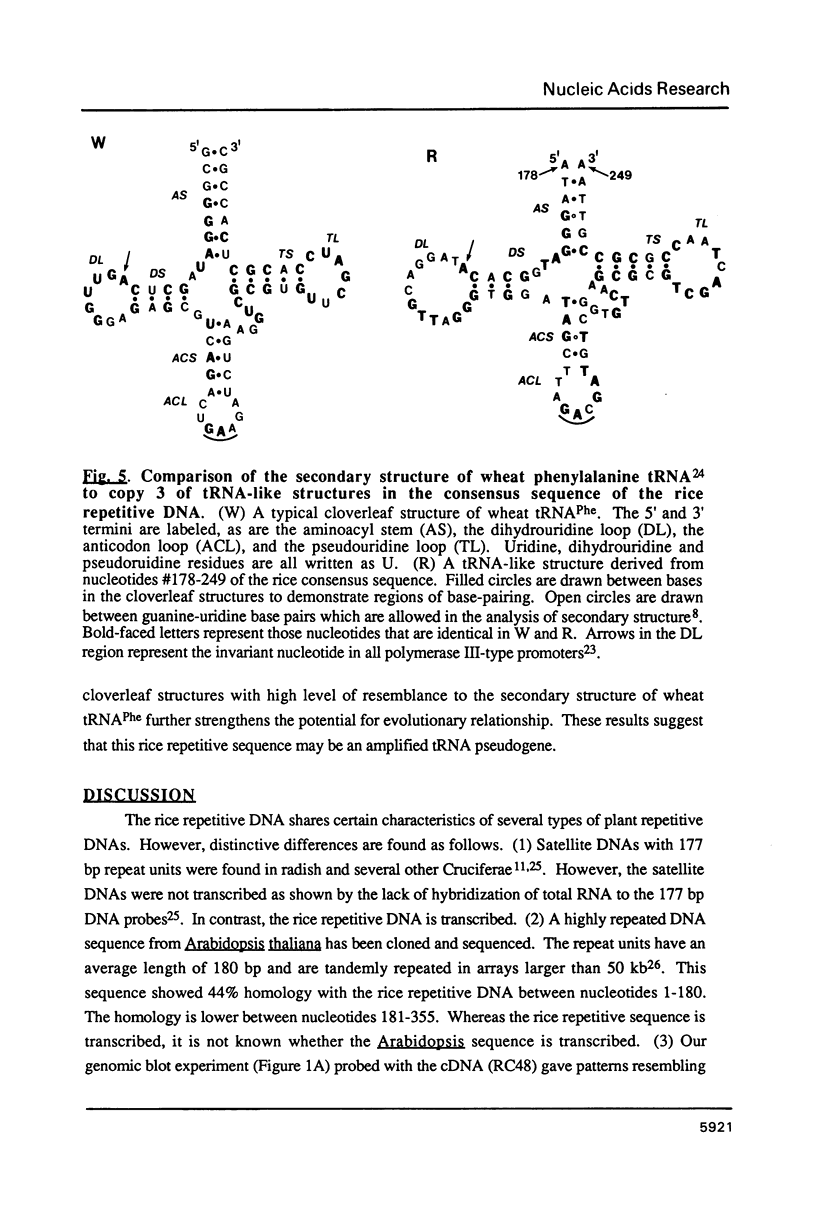
Abstract
Moderately repetitive DNA sequences are found in the genomes of all eucaryotes that have been examined. We now report the discovery of a novel, transcribed, moderately repetitive DNA sequence in a higher plant which is different from any of the known repetitive DNA sequences from any organism. We isolated a rice cDNA clone which hybridizes to multiple bands on genomic blot analysis. The sequence of this 352 bp cDNA contains four regions of homology to the wheat phenylalanine tRNA, including the polymerase III-type promoter. Unexpectedly, two regions of the same 352 bp sequence also show homology to the wheat 5S RNA sequence. Using the cDNA as a probe, we have isolated six genomic clones which contain long tandem repeats of 355 bp sequence, and have sequenced nine repeat units. Our findings suggest that the rice repetitive sequence may be an amplified pseudogene with sequence homology to both 5S RNA and tRNA, but organized as long tandem repeats resembling 5S RNA genes. This is the first example showing homology between the sequences of a moderately repetitive DNA with unknown function and 5S RNA.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allan M., Paul J. Transcription in vivo of an Alu family member upstream from the human epsilon-globin gene. Nucleic Acids Res. 1984 Jan 25;12(2):1193–1200. doi: 10.1093/nar/12.2.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benslimane A. A., Dron M., Hartmann C., Rode A. Small tandemly repeated DNA sequences of higher plants likely originate from a tRNA gene ancestor. Nucleic Acids Res. 1986 Oct 24;14(20):8111–8119. doi: 10.1093/nar/14.20.8111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels G. R., Deininger P. L. A second major class of Alu family repeated DNA sequences in a primate genome. Nucleic Acids Res. 1983 Nov 11;11(21):7595–7610. doi: 10.1093/nar/11.21.7595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels G. R., Deininger P. L. Repeat sequence families derived from mammalian tRNA genes. 1985 Oct 31-Nov 6Nature. 317(6040):819–822. doi: 10.1038/317819a0. [DOI] [PubMed] [Google Scholar]
- Daniels G. R., Fox G. M., Loewensteiner D., Schmid C. W., Deininger P. L. Species-specific homogeneity of the primate Alu family of repeated DNA sequences. Nucleic Acids Res. 1983 Nov 11;11(21):7579–7593. doi: 10.1093/nar/11.21.7579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli G., Hofstetter H., Birnstiel M. L. Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature. 1981 Dec 17;294(5842):626–631. doi: 10.1038/294626a0. [DOI] [PubMed] [Google Scholar]
- Gauss D. H., Sprinzl M. Compilation of tRNA sequences. Nucleic Acids Res. 1983 Jan 11;11(1):r1–53. [PMC free article] [PubMed] [Google Scholar]
- Grellet F., Delcasso D., Panabieres F., Delseny M. Organization and evolution of a higher plant alphoid-like satellite DNA sequence. J Mol Biol. 1986 Feb 20;187(4):495–507. doi: 10.1016/0022-2836(86)90329-3. [DOI] [PubMed] [Google Scholar]
- Jagadeeswaran P., Forget B. G., Weissman S. M. Short interspersed repetitive DNA elements in eucaryotes: transposable DNA elements generated by reverse transcription of RNA pol III transcripts? Cell. 1981 Oct;26(2 Pt 2):141–142. doi: 10.1016/0092-8674(81)90296-8. [DOI] [PubMed] [Google Scholar]
- Jelinek W. R., Schmid C. W. Repetitive sequences in eukaryotic DNA and their expression. Annu Rev Biochem. 1982;51:813–844. doi: 10.1146/annurev.bi.51.070182.004121. [DOI] [PubMed] [Google Scholar]
- Moon E., Kao T. H., Wu R. Rice chloroplast DNA molecules are heterogeneous as revealed by DNA sequences of a cluster of genes. Nucleic Acids Res. 1987 Jan 26;15(2):611–630. doi: 10.1093/nar/15.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J. H. The origin and evolution of retroposons. Int Rev Cytol. 1985;93:187–279. doi: 10.1016/s0074-7696(08)61375-3. [DOI] [PubMed] [Google Scholar]
- Schmid C. W., Jelinek W. R. The Alu family of dispersed repetitive sequences. Science. 1982 Jun 4;216(4550):1065–1070. doi: 10.1126/science.6281889. [DOI] [PubMed] [Google Scholar]
- Singer M. F. Highly repeated sequences in mammalian genomes. Int Rev Cytol. 1982;76:67–112. doi: 10.1016/s0074-7696(08)61789-1. [DOI] [PubMed] [Google Scholar]
- Ullu E., Tschudi C. Alu sequences are processed 7SL RNA genes. Nature. 1984 Nov 8;312(5990):171–172. doi: 10.1038/312171a0. [DOI] [PubMed] [Google Scholar]
- Van Arsdell S. W., Denison R. A., Bernstein L. B., Weiner A. M., Manser T., Gesteland R. F. Direct repeats flank three small nuclear RNA pseudogenes in the human genome. Cell. 1981 Oct;26(1 Pt 1):11–17. doi: 10.1016/0092-8674(81)90028-3. [DOI] [PubMed] [Google Scholar]
- Weiner A. M., Deininger P. L., Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- Wu R., Wu T., Ray A. Adaptors, linkers, and methylation. Methods Enzymol. 1987;152:343–349. doi: 10.1016/0076-6879(87)52041-9. [DOI] [PubMed] [Google Scholar]