Abstract
On first impression the ‘whole-istic approach to understanding biology’ that has been used to describe Systems Biology bears a striking resemblance to what many of us know as Integrative Physiology. However, closer scrutiny reveals that at the present time Systems Biology is rooted in processes operating at a cellular level (‘the study of an organism, viewed as an integrated and interacting network of genes, proteins and biochemical reactions which give rise to life … ultimately responsible for an organism's form and functions’; http://www.systemsbiology.org), and appears to have evolved as a direct result of advances in high throughput molecular biology platforms (and associated bioinformatics) over the past decade. The Systems Biology approach is in many ways laudable, but it will be immediately apparent to most exercise or integrative physiologists that the challenge of understanding the whole-animal response to exercise as a network of integrated and interacting genes, proteins and biochemical reactions is unlikely to be realized in the near future. This short review will attempt to clarify conceptual inconsistencies between the fields of Systems Biology and Integrative Physiology in the context of exercise science, and will attempt to identify the challenges to whole-body physiologists wishing to harness the tools of Systems Biology.

Paul Greenhaff (left) is Professor of Muscle Metabolism in the School of Biomedical Sciences at the University of Nottingham. His main research interests are centred on the regulation of skeletal muscle fuel and protein metabolism, principally in humans. His group's research work is of basic and clinical physiological relevance, but also has practical application in relation to athletic performance. Mark Hargreaves (right) is Professor and Head, Department of Physiology at The University of Melbourne in Australia. His research interests focus on the physiological and metabolic responses to acute and chronic exercise, with an emphasis on skeletal muscle carbohydrate metabolism and GLUT4 expression.
On first appearance systems biology is integrative physiology
The discipline of Systems Biology has emerged over the past decade and has been defined as ‘a whole-istic approach to understanding biology’ (Chong & Ray, 2002), or put another way ‘a holistic approach to the study of biology with the objective of simultaneously monitoring all biological processes operating as an integrated system’ (http://www.epa.gov/opp00001/science/comptox-glossary.html). To scientists working in the field of Systems Biology, this encompasses ‘examining the structure and dynamics of cellular and organism function, rather than the characteristics of isolated parts of a cell or organism’ (Kitano, 2002). To the authors the above account describes very succinctly the field of Integrative Physiology, particularly during exercise where striking organ-systems integration is required to acutely match demands, or during chronic adaptation (training) as systems respond to repeated exercise stresses. Importantly, this field is not an emerging area of research, e.g. in the 1920s Krogh and Lindhard used pulmonary gas exchange measures to assess energy metabolism and the relative contribution of carbohydrate and fat to whole body substrate oxidation during exercise (Krogh & Lindhard, 1920). Today, contemporary approaches like transcranial Doppler ultrasound and functional magnetic resonance imaging, coupled with more established approaches such as the muscle biopsy technique and limb/organ arterial-venous (a-v) balance, are allowing exercise scientists far-reaching insight into the regulation of physiological integration during exercise and what limits metabolic flux and performance during exercise. For example, our colleagues have presented evidence that changes in brain redox state and oxygenation, occurring secondary to brain activation-induced increases in cerebral blood flow and metabolic rate, modulate the increase in cerebral blood flow at the onset of exercise in humans (Rasmussen et al. 2009). Moreover, they have gone on to elegantly demonstrate that reduced cerebral oxygenation may play an important role in the aetiology of fatigue during maximal exercise. Indeed, concluding that because no evidence of muscle fatigue during maximal elbow flexor exercise could be found, central fatigue may play a bigger role than peripheral muscle fatigue in limiting maximal exercise performance (Rasmussen et al. 2010). It appears therefore that physical activity may create competition between the brain and skeletal muscle during exercise, and perhaps even more so in patients with compromised cardiac function. Similarly, it has also been demonstrated that feedback from contracting skeletal muscles can modify central motor drive and limit muscle fatigue development (Amann et al. 2009), as well as regulating the ventilatory and cardiovascular responses to exercise (Amann et al. 2010). Perhaps more importantly in the context of this special edition of The Journal of Physiology, however, is the realisation that it is possible for exercise physiologists to study integration in vivo in humans in an increasingly insightful manner. As such we are increasingly ‘examining the structure and dynamics of cellular and organism function’ in humans, said to be at the core of the emerging field of Systems Biology.
Is there a fundamental difference of opinion?
One may rightly question therefore how the field of Systems Biology has evolved to the extent that it has today. Closer scrutiny demonstrates that, despite claims of ‘a whole-istic approach to understanding biology’, at this moment in time Systems Biology invariably focuses on systems operating at a cellular level, and this cell-based focus appears to have evolved as a direct result of advances in high throughput molecular biology platforms (and associated bioinformatics) over the past decade (viewed by many as the ‘omics era’). In keeping with this, Systems Biology has been described as:
‘the study of an organism, viewed as an integrated and interacting network of genes, proteins and biochemical reactions which give rise to life. Instead of analysing individual components or aspects of the organism, such as sugar metabolism or a cell nucleus, systems biologists focus on all the components and the interactions among them, all as part of one system. These interactions are ultimately responsible for an organism's form and functions’ (from http://www.systemsbiology.org).
Whilst such an approach is laudable, it will be immediately apparent to an exercise or integrative physiologist that the challenge of understanding the whole-animal response to exercise and/or fatigue as a network of integrated and interacting genes, proteins and biochemical reactions is unlikely to be realised soon. Indeed, the idea that any complex organism is an ‘integrated and interacting network of genes, proteins and biochemical reactions … that are ultimately responsible for form and functions’ seems largely misplaced in the context of physical activity, as it is the physical exercise that will largely dictate the magnitude and pattern of how networks of genes, proteins and biochemical reactions will integrate and interact. This is an important point, because to the exercise physiologist most, if not all, cellular network based change will be secondary to the physiological stimulus causing that change, e.g. muscle contraction, rather than originating at the level of the network per se. This in essence is one important difference between the molecular biology focus at the core of Systems Biology and functional feedback approach of Integrative Physiology, a difference eloquently described by Denis Noble (Noble, 2006). Indeed, accepting this viewpoint helps to at least partly explain why ‘omics’ and animal knock-in/out based approaches have to date failed to deliver the level of insight originally hoped for in elucidating the aetiology of ‘lifestyle related’ chronic diseases, i.e. there is no defect in the network per se and it will merely respond to physiological stimuli experienced. Therefore focusing on changes in networks of genes, proteins and/or metabolic pathways at a cell-based level in the absence of quantified physiological inputs and outcomes will fail to provide comprehensive insight of whole tissue or whole-body function and dysfunction.
The future requires a coalescence of systems biology and integrative physiology approaches
As already stated, the goal of understanding biology as an ‘integrated and interacting network of genes, proteins and biochemical reactions which give rise to life … which is ultimately responsible for an organism's form and functions’ is a laudable objective, but in the context of human exercise physiology, it is a demanding challenge. We have arrived at a position where ‘omics’ based approaches have been at the forefront of science, but have not delivered in the way many thought they would, not least in our mind because Integrative Physiology has largely been missing from the scientific strategy. However, it is now clear to many that to yield effective research a melding of the two approaches is critical if scientific goals are to be realized. It will not be easy, but it is a goal worth pursuing!
At present we are at a state of play where descriptive data from ‘omics’ based approaches in humans during exercise have been generated. For example, gene transcript profiling has been used to show that a single bout of eccentric exercise induces greater inflammatory gene network responses in human quadriceps muscle than concentric exercise (Chen et al. 2003), and that these inflammatory network responses are maintained when a second bout of eccentric exercise is performed (Hubal et al. 2008), that a single endurance exercise bout produces time-dependent changes in global mRNA expression (Mahoney et al. 2005), that resistance training can reverse ageing-related defects in gene expression, notably those associated with mitochondrial function (Melov et al. 2007), and long-term strength or endurance training produces differences in skeletal muscle gene expression relative to each other and untrained control subjects (Stepto et al. 2009). However, it is clear that functional, physiological relevance, e.g. measures of protein synthesis rates, glucose oxidation rates, etc., is not always concurrent with ‘omics’ based approaches. These are obviously major challenges, but some headway is being made. For example, the HERITAGE family study (Bouchard et al. 2000, Bouchard & Rankinen, 2001) and the recent work of Timmons et al. (2010) have used high throughput genomic based approaches in an attempt to produce a molecular ‘fingerprint’ that can predict the heritability of training and magnitude of response of maximal oxygen uptake to training in humans. Notably, this research has revealed that about 50% of the gain in maximal oxygen consumption with endurance training is attributable to heritability. Furthermore, the authors identified a RNA signature that predicted the magnitude of aerobic training response, which perhaps surprisingly did not change in abundance with training. At the very heart of this work is the detailed physiological phenotyping of volunteers providing sensitive and relevant landmarks to guide and inform the systems biology based analytical approaches used by the authors. Another example is the use of high throughput mass spectrometry platforms to obtain ‘metabolic signatures’ in biological samples, as demonstrated in a recent study (Lewis et al. 2010). Although the conclusion that exercise resulted in ‘rapid activation of a catabolic program consisting of heightened lipolysis, glycolysis and glycogenolysis, as well as amino acid and purine catabolism that largely persists for at least 60 min after completion of exercise’ was not particularly novel, the identification of a large number of metabolites in human plasma after exercise informed further research directions. Indeed, in additional experiments the authors demonstrated that a combination of selected metabolites, but interestingly not individual metabolites, induced expression of Nur77, a transcription factor implicated in skeletal muscle glucose and lipid metabolism, in C2C12 myocytes in culture. Given that exercise induces large changes in plasma and muscle metabolites, as well as in the plasma levels of an ever expanding list of adipokines, myokines and other bioactive molecules, these metabolite and proteomic profiling technologies provide new tools to examine metabolic and physiological responses to exercise in humans, perhaps eventually offering insight into inter-organ integration during exercise.
Whilst we can build upon these leads, to make significant scientific progress more efficiently, the following points are worthy of consideration when developing research strategy.
Our limited understanding of gene and protein function in vivo
Scientific goals need to be moderated with the knowledge that as yet we are still some way from knowing the physiological function of the majority of genes and proteins that comprise the human genome and proteome. Importantly, advances will be difficult because it is unlikely that genes and proteins have single discrete functions – hence the need for network based strategies towards understanding physiological function (Noble, 2006). It is also likely that redundancy exists in any network, such that the same functional outcome can be achieved in a number of ways (Noble, 2006). Furthermore, particularly in the context of human and exercise physiology, progress will be further slowed because insight gleaned from cell and animal based investigation does not necessarily translate directly to the in vivo condition in humans. By way of an example of this latter point, protein translation initiation has been identified as being central to the regulation of muscle protein synthesis. More specifically, the AKT/mTOR signalling pathway has been identified in animal and cell based research as being an important regulator of this process (Vary et al. 1994, Vary & Kimball, 2000), such that it is considered to occupy a pivotal role in the regulation of muscle protein synthesis. Accordingly, muscle specific over-expression of AKT in a transgenic mouse model has been shown to result in profound muscle hypertrophy (Bodine et al. 2001). Likewise, increased muscle protein synthesis in response to contraction in rodent muscle occurs in parallel with increased phosphorylation of AKT, mTOR and several downstream signalling proteins in the pathway (Atherton et al. 2005). However, recent studies involving healthy human volunteers question the established notion that the AKT/mTOR signalling pathway drives muscle protein synthesis in the same manner as gleaned from cell and rodent studies. For example, a clear disassociation between AKT and mTOR phosphorylation (and several downstream targets) and protein synthesis has been reported in human skeletal muscle under controlled experimental conditions by at least three laboratories (Greenhaff et al. 2008, Wilkinson et al. 2008, Mayhew et al. 2009). These human volunteer studies all involved protocols expected to result in an anabolic response, and therefore strongly suggest that the mere phosphorylation of AKT and its downstream targets is insufficient to increase muscle protein synthesis. This does not preclude the AKT/mTOR signalling pathway from having an important function in the processes that govern human muscle protein synthesis, but does suggest key signalling events and/or temporal changes in known events have yet to be fully elucidated in humans. Indeed, in the case of temporal resolution it has recently been demonstrated that whilst changes in the phosphorylation status of several proteins in the AKT/mTOR pathway can be connected with the rise in muscle protein synthesis seen following dietary protein ingestion in humans, the same cannot be said of the subsequent decline in muscle protein synthesis (Atherton et al. 2010). Clearly therefore the processes governing muscle protein synthesis in the human are more complex than currently perceived. A major goal therefore should be to perform studies in humans under conditions that enable the function of the genome and proteome to be realised in vivo, and use animal and cell based research retrospectively to underpin this approach (back translation). Certainly, genetic models used in isolation have failed to deliver the level of scientific progress originally anticipated perhaps because of the lack of realization that it is the physiological stress (not the genetic environment per se) that will dictate the phenotypic response. Exercise is an excellent physiological stressor to facilitate this process. However, undertaking detailed in vivo physiological measurements in human volunteers is not straightforward as this realization comes at a time when institutions have lost the ability to perform detailed in vivo human research, and funding bodies in the main still view cell based molecular biology to be at the forefront of science and better value for money, so the problem is not easily remedied. For sure, however, human researchers can, and are, using systems based approaches, which funding bodies are hopefully increasingly realizing.
Ascertaining temporal resolution
Establishing temporal resolution both from a physiology perspective (i.e. quantifying physiological adaptation with training, immobilisation, ageing, etc.) and frequency of tissue sampling is critical as any network regulating physiological function by necessity should be highly dynamic. Human volunteer studies to date have usually quantified the magnitude of physiological adaptation to a given stressor (e.g. 10 weeks of exercise training) and collected tissue samples (e.g. quadriceps muscle biopsy samples) at pre- and postintervention time points (i.e. 2 point investigation). However, it is unquestionable that the responses of gene and protein networks will vary considerably over time. By way of example, Fig. 1 (Menetski et al. 2009) shows the mean fold change in mRNA abundance from basal (determined using gene expression profiling) in muscle biopsy samples obtained from a group of healthy, male volunteers (n= 32) who performed supervised resistance training on three occasions each week for 10 weeks. As the figure shows, muscle biopsy samples were obtained at six time points (baseline and on day 2, 10, 35, 68 and 70, all in the fasted resting state, 24 h post-training) and genes were assigned to clusters based upon their function gleaned from databases of published literature (clusters 1–7). It is clear therefore that mRNA abundance, and presumably gene function, changes substantially over time within any given cluster and also between clusters. Furthermore, the physiological stimulus created by the same bout of maximal resistance training at the outset of the training regimen is perceived very differently by the network at the end of the training when adaptation has occurred.
Figure 1. Fold change in mRNA abundance from basal (determined using gene expression profiling) in muscle biopsy samples obtained from healthy male volunteers who performed supervised resistance training on 3 occasions per week for 10 weeks.
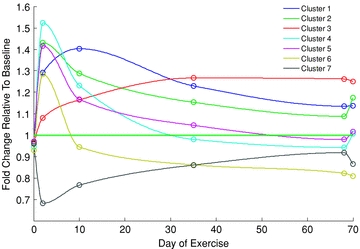
Muscle biopsy samples were obtained at baseline and after 2, 10, 35, 68 and 70 days of training (in the fasted and resting state, 24 h following a bout of training). Clusters 1–7 depict different cellular functions (Menetski et al. 2009).
Having an open mind
Along with advances in technology and knowledge must also come the adoption of a more open-minded view by physiologists that physiological processes may not necessarily be controlled in the manner currently perceived, e.g. the molecular mechanisms regulating muscle mass loss in vivo may not simply be the same as those driving muscle mass accrual, but operating in reverse.
Furthermore, as ‘omics’ technologies continue to develop and offer greater molecular insight, exercise physiologists need to be able to recognise the limitations of these approaches, e.g. identification of genomic markers of complex human traits using single nucleotide polymorphism (SNP) analysis is likely to require enormous sample sizes (Frazer et al. 2009, Timmons et al 2010), but at the same time be prepared to adopt and dovetail these new technologies to underpin physiological based investigation, thereby enabling the understanding of the molecular regulation of in vivo physiological function to be realised. In this respect, it has very recently been shown that RUNX1, SOX9 and PAX3 transcription factor binding sites are increased in abundance in muscle in response to endurance training in humans, but moreover microRNA profiling revealed microRNAs targeting RUNX1, SOX9 and PAX3 were down-regulated in the same muscle biopsy samples (Keller et al. 2011). Similarly, the introduction of phosphoprotein array based approaches in human exercise physiology will provide greater advances than has been realised to date from proteomics.
To sum up, at the present time it would seem Systems Biology and Integrative Physiology are very different beasts, but they strive for the same goal, i.e. a ‘whole-istic approach to understanding biology’. From the standpoint of the exercise physiologist, it would appear that ‘systems biology is an approach rather than a field of research’ (Kohl et al. 2010). However, perhaps the tools of systems biology should be viewed increasingly as a valuable addition to the arsenal that exercise scientists can use to interrogate physiological function and adaptation.
References
- Amann M, Blain GM, Proctor LT, Sebranek JJ, Peglow DF, Dempsey JA. Group III and IV muscle afferents contribute to ventilatory and cardiovascular response to rhythmic exercise in humans. J Appl Physiol. 2010;109:966–976. doi: 10.1152/japplphysiol.00462.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann M, Proctor LT, Sebranek JJ, Peglow DF, Dempsey JA. Opioid-mediated muscle afferents inhibit central motor drive and limit peripheral muscle fatigue development in humans. J Physiol. 2009;587:271–283. doi: 10.1113/jphysiol.2008.163303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atherton PJ, Babraj J, Smith K, Singh J, Rennie MJ, Wackerhage H. Selective activation of AMPK-PGC-1α or PKB-TSC2-mTOR signaling can explain specific adaptive responses to endurance or resistance training-like electrical muscle stimulation. FASEB J. 2005;19:786–788. doi: 10.1096/fj.04-2179fje. [DOI] [PubMed] [Google Scholar]
- Atherton PJ, Etheridge T, Watt PW, Wilkinson D, Selby A, Rankin D, Smith K, Rennie MJ. Muscle full effect after oral protein: time-dependent concordance and discordance between human muscle protein synthesis and mTORC1 signaling. Am J Clin Nutr. 2010;92:1080–1088. doi: 10.3945/ajcn.2010.29819. [DOI] [PubMed] [Google Scholar]
- Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, Yancopoulos GD. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol. 2001;3:1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- Bouchard C, Rankinen T, Chagnon YC, Rice T, Pérusse L, Gagnon J, Borecki I, An P, Leon AS, Skinner JS, Wilmore JH, Province M, Rao DC. Genomic scan for maximal oxygen uptake and its response to training in the HERITAGE family study. J Appl Physiol. 2000;88:551–559. doi: 10.1152/jappl.2000.88.2.551. [DOI] [PubMed] [Google Scholar]
- Bouchard C, Rankinen T. Individual differences in response to regular physical activity. Med Sci Sports Exerc. 2001;33:S446–451. doi: 10.1097/00005768-200106001-00013. [DOI] [PubMed] [Google Scholar]
- Chen YW, Hubal MJ, Hoffman EP, Thompson PD, Clarkson PM. Molecular responses of human muscle to eccentric exercise. J Appl Physiol. 2003;95:2485–2494. doi: 10.1152/japplphysiol.01161.2002. [DOI] [PubMed] [Google Scholar]
- Chong L, Ray LB. Whole-istic biology. Science. 2002;295:1661. [Google Scholar]
- Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10:241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- Greenhaff PL, Karagounis L, Peirce N, Simpson EJ, Hazell M, Layfield R, Wackerhage H, Smith K, Atherton P, Selby A, Rennie MJ. Disassociation between the effects of amino acids and insulin on signalling, ubiquitin-ligases and protein turnover in human muscle. Am J Physiol Endocrinol Metab. 2008;295:E595–604. doi: 10.1152/ajpendo.90411.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubal MJ, Chen TC, Thompson PD, Clarkson PM. Inflammatory gene changes associated with the repeated-bout effect. Am J Physiol Regul Integr Comp Physiol. 2008;294:R1628–1637. doi: 10.1152/ajpregu.00853.2007. [DOI] [PubMed] [Google Scholar]
- Keller P, Vollaard NB, Gustafsson T, Gallagher IJ, Sundberg CJ, Rankinen T, Britton SL, Bouchard C, Koch LG, Timmons JA. A transcriptional map of the impact of endurance exercise training on skeletal muscle phenotype. J Appl Physiol. 2011;110:46–59. doi: 10.1152/japplphysiol.00634.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- Kohl P, Crampin EJ, Quinn TA, Noble D. Systems biology: an approach. Clin Pharmacol Ther. 2010;88:25–33. doi: 10.1038/clpt.2010.92. [DOI] [PubMed] [Google Scholar]
- Krogh A, Lindhard J. The relative value of fat and carbohydrate as sources of muscular energy: with appendices on the correlation between standard metabolism and the respiratory quotient during rest and work. Biochem J. 1920;14:290–363. doi: 10.1042/bj0140290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis GD, Farrell L, Wood MJ, Martinovic M, Arany Z, Rowe GC, Souza A, Cheng S, McCabe EL, Yang E, Shi X, Deo R, Roth FP, Asnani A, Rhee EP, Systrom DM, Semigran MJ, Vasan RS, Carr SA, Wang TJ, Sabatine MS, Clish CB, Gerszten RE. Metabolic signatures of exercise in human plasma. Science Trans Med. 2010;2:33–37. doi: 10.1126/scitranslmed.3001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney DJ, Parise G, Melov S, Safdir A, Tarnopolsky MA. Analysis of global mRNA expression in human skeletal muscle during recovery from endurance exercise. FASEB J. 2005;19:1498–1500. doi: 10.1096/fj.04-3149fje. [DOI] [PubMed] [Google Scholar]
- Mayhew DL, Kim JS, Cross JM, Ferrando AA, Bamman MM. Translational signaling responses preceding resistance training-mediated myofiber hypertrophy in young and old humans. J Appl Physiol. 2009;107:1655–1662. doi: 10.1152/japplphysiol.91234.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menetski JP, Hu R, Opiteck GJ, Constantin D, Constantin-Teodosiu D, Pittas G, Karagounis L, Greenhaff PL. Exercise Induced Time-dependent Transcription in Human Skeletal Muscle. Barcelona: 2009. 5th Cachexia Conference 185, 156. [Google Scholar]
- Melov S, Tarnopolsky MA, Beckman K, Felkey K, Hubbard A. Resistance exercise reverses aging in human skeletal muscle. PLoS One. 2007;5:e465. doi: 10.1371/journal.pone.0000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble D. The Music of Life (Biology Beyond the Genome) Oxford: Oxford University Press; 2006. [Google Scholar]
- Rasmussen P, Madsen CA, Nielsen HB, Zaar M, Gjedde A, Secher NH, Quistorff B. Coupling between the blood lactate-to-pyruvate ratio and MCA Vmean at the onset of exercise in humans. J Appl Physiol. 2009;107:1799–1805. doi: 10.1152/japplphysiol.00468.2009. [DOI] [PubMed] [Google Scholar]
- Rasmussen P, Nielsen J, Overgaard M, Krogh-Madsen R, Gjedde A, Secher NH, Petersen NC. Reduced muscle activation during exercise related to brain oxygenation and metabolism in humans. J Physiol. 2010;588:1985–1995. doi: 10.1113/jphysiol.2009.186767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepto NK, Coffey VG, Carey AL, Ponnampalam AP, Canny BJ, Powell D, Hawley JA. Global gene expression in skeletal muscle from well-trained strength and endurance athletes. Med Sci Sports Exerc. 2009;41:546–565. doi: 10.1249/MSS.0b013e31818c6be9. [DOI] [PubMed] [Google Scholar]
- Timmons JA, Knudson S, Rankinen T, Koch LG, Sarzynski M, Jensen T, Keller P, Scheele C, Vollaard NBJ, Nielsen S, Åkerstrom T, MacDougald OA, Jansson E, Greenhaff PL, Tarnopolsky MA, van Loon LJC, Pedersen BK, Sundberg CJ, Wahlestedt C, Britton SL, Bouchard C. Using molecular classification to predict gains in maximal aerobic capacity following endurance exercise training exercise training in humans. J Appl Physiol. 2010;108:1487–1496. doi: 10.1152/japplphysiol.01295.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vary TC, Jurasinski CV, Karinch AM, Kimball SR. Regulation of eukaryotic initiation factor-II expression during sepsis. Am J Physiol Endocrinol Metab. 1994;266:E193–E201. doi: 10.1152/ajpendo.1994.266.2.E193. [DOI] [PubMed] [Google Scholar]
- Vary TC, Kimball SR. Effect of sepsis on eIE4E availability in skeletal muscle. Am J Physiol Endocrinol Metab. 2000;279:E1178–1184. doi: 10.1152/ajpendo.2000.279.5.E1178. [DOI] [PubMed] [Google Scholar]
- Wilkinson SB, Phillips SM, Atherton PJ, Patel R, Yarasheski KE, Tarnopolsky MA, Rennie MJ. Differential effects of resistance and endurance exercise in the fed state on signalling molecule phosphorylation and protein synthesis in human muscle. J Physiol. 2008;586:3701–3717. doi: 10.1113/jphysiol.2008.153916. [DOI] [PMC free article] [PubMed] [Google Scholar]


