Abstract
2-Amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) is associated with mammary carcinomas in animals and humans. PhIP is metabolized by CYP 1A1/1A2 and cytochrome b5 reductase, producing free radicals causing DNA strand breaks. Diallyl sulfide (DAS) prevents cancer in animals. We hypothesized that DAS will attenuate PhIP-induced DNA strand breaks and cell death. To test this hypothesis, we treated MCF-10A cells with PhIP, DAS and PhIP/DAS for 24, 48 and 72 h. DAS inhibited the PhIP-induced DNA strand breaks by 22% after 48 h and the strand breaks were completely inhibited at 72 h. PhIP reduced cell viability at each time point. However, DAS only attenuated this reduction in cell viability by 56% at 72 h. N-OH PhIP inhibited cell viability by 26% at 72 h. DAS completely attenuated this reduction in cell viability and may prevent PhIP-induced breast cancer via alterations in DNA damage and cell viability.
Keywords: 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine; diallyl sulfide; chemoprevention; cell viability; DNA strand breaks
Introduction
Diet is the second most preventable cause of cancer (1). Increase in well-done meat consumption is associated with an elevated risk of breast cancer (2). This correlation between increased cancer risk and meat preparation is most likely due to the production of high levels of heterocyclic amines (3). The heterocyclic amine most abundantly found in the human diet is 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP). PhIP has induced tumors in the prostate, colon and breast (3–5).
PhIP is oxidized by cytochrome P-450 1A2 (CYP1A2) to form an N-hydroxy derivative. The N-hydroxy derivative undergoes subsequent acetylation or sulphation, resulting in the formation of a nitrenium ion free radical that covalently binds to the C8 position of guanine residues. The resultant adduct is primarily responsible for GC→TA transversions and deleterious frameshift mutations of a guanine within a G-rich repetitive sequence (6). The development of these mutations are the basis for PhIP's genotoxic effects, which include chromosome aberrations, DNA damage, micronuclei and sister chromatid exchange (7,8). A failure to repair these genomic anomalies can result in cancer, especially when they occur in tumor suppressors and proto-oncogenes.
The alkaline single cell electrophoresis (comet) assay can be used to detect DNA damage, particularly single and double strand breaks that may occur as a result of exposure to genotoxic agents, such as PhIP (9,10). Under electrophoretic conditions, fragmented DNA migrates toward the anode producing a tail and giving the appearance of a comet. The amount of fragmented DNA directly correlates with the length of the tail and thus provides a quantitative assessment of DNA damage induced by a particular agent (11).
Diallyl sulfide (DAS) is a lipophilic organosulfur component of garlic that has been demonstrated to protect against chemically-induced carcinogenesis in vivo and in vitro (12). One mechanism by which DAS may exert its anti-carcinogenic properties is through its ability to inhibit DNA damage. DAS has been demonstrated to inhibit N-nitroso-dimethylamine (NDMA) and aflatoxin B1 (AFB1)-induced DNA strand breaks in the liver cells of Wistar rats (13,14). Therefore, we propose that DAS attenuates PhIP-induced cell death via the inhibition of DNA strand breaks.
Materials and methods
Chemicals and reagents
Phenol-red free DMEM/F-12 1:1 mix basal media, horse serum, trypsin, penicillin/streptomycin, Ultrapure low melting point agarose and Ultrapure normal melting point agarose were obtained from Invitrogen (Carlsbad, CA). The epidermal growth factor was obtained from BD Biosciences (San Jose, CA). Dimethly-sulfoxide (DMSO), hydrocortisone, Triton X-100, PBS, NaOH, Trizma base, NaCl, Na2EDTA, diethylpyrocarbonate (DEPC), ethanol and DAS were all purchased from Sigma (St. Louis, MO). Fully-frosted microscope slides and cover slips were purchased from Fisher Scientific (Pittsburgh, PA). PhIP was purchased from Toronto Research Chemicals. N-hydroxy PhIP was purchased from the National Cancer Institute Chemical Carcinogen Repository, Midwest Research Institute (Kansas City, MO).
Cell culture
MCF-10A human breast epithelial cells were a gift from the laboratory of Dr Thomas Kocarek at Wayne State University (Detroit, MI). The cells were cultured in a humidified incubator at 37° C under 5% CO2 atmospheric conditions in DMEM/F-12 1:1 mix basal media supplemented with 10 µg/ml insulin, 20 ng/ml epidermal growth factor, 0.5 µg/ml hydrocortisone, 5% horse serum and 1% penicillin-streptomycin (10,000 U/ml).
Cell treatment
MCF-10A human breast epithelial cells were plated and treated for 24, 48 and 72 h with PhIP (100 µM), DAS (100 µM) and N-OH PhIP (5 µM). These concentrations were chosen because PhIP (100 µM) and N-OH PhIP (1 µM) produced DNA adducts in MCF-10A cells after 24 h (15). Cells treated with combinations of PhIP/DAS and N-OH PhIP/DAS groups were pretreated with 100 µM of DAS 6 h prior to dosing with PhIP (100 µM) or N-OH PhIP (5 µM). Untreated and DMSO treated cells served as negative and vehicle controls, respectively.
Comet assay
One million MCF-10A cells were treated as indicated above in T-25 cm2 culture flasks. At 24, 48 and 72 h, the cells were trypsinized and centrifuged for 5 min at 1,000 rpm. The supernatant was discarded and the cell pellet was re-suspended in 500 µl of 1X Ca2+/Mg2+ free PBS. A 100 µl aliquot of the treated cell suspensions was added to 900 µl of low melting point agarose (0.75%). Three slides were made of each sample by adding a 100 µl aliquot of the cell/agarose mixture to the top of microscope slides precoated with a normal melting point agarose. The slides were solidified on ice and then placed in the refrigerator in ice-cold lysis buffer (pH 10) containing 1% Triton X-100 for 1 h.
After lysing, the slides were placed in alkaline electrophoresis buffer (pH >13.5) for 30 min to allow the DNA to unwind and electrophoresed at 280 Amps/25V for 30 min. Following electrophoresis, the slides were washed three times with neutralization buffer (Tris buffer, pH 7.5) and dehydrated for 5 min with 100% ethanol. The slides were stained with 100 µl of propidium iodide (20 µg/ml) and examined under a fluorescent microscope. A total of 150 cell images were examined per slide under 20 × magnification using the Kinetic Imaging comet assay software. The mean olive tail moment was used as a parameter for DNA fragmentation. Statistical analysis was performed using one-way analysis of variance and the Tukey multiple comparison test to determine significant differences amongst the treatment groups.
Cell viability
Cell viability was assessed using the CellTiter 96® AQueousOne solution cell proliferation assay. Ten thousand cells were plated per well in 96-well plates and treated as described above. At 24, 48 and 72 h, a 20 µl aliquot of AqueousOne Solution (Promega Corporation, Madison, WI) was added to each well and incubated at 37° C for 2 h to allow color development. The plates were analyzed on a Bio-Tek Ex800 microplate reader using KC Junior software, where the absorbance at 480 nm was determined for each well. In this colorimetric assay, viable cells convert the MTS tetrazolium compound to a formazan product soluble in culture media. The amount of formazan product formed is directly proportional to the number of living cells in the culture.
Statistical analysis
Results are expressed as means ± S.E.M. from a minimum of three independent experiments. Data were analyzed by one-way analysis of variance (ANOVA) and the Tukey multiple comparison test to determine significant differences (P<0.05) among the treatment groups.
Results
Alkaline single cell gel electrophoresis (comet assay)
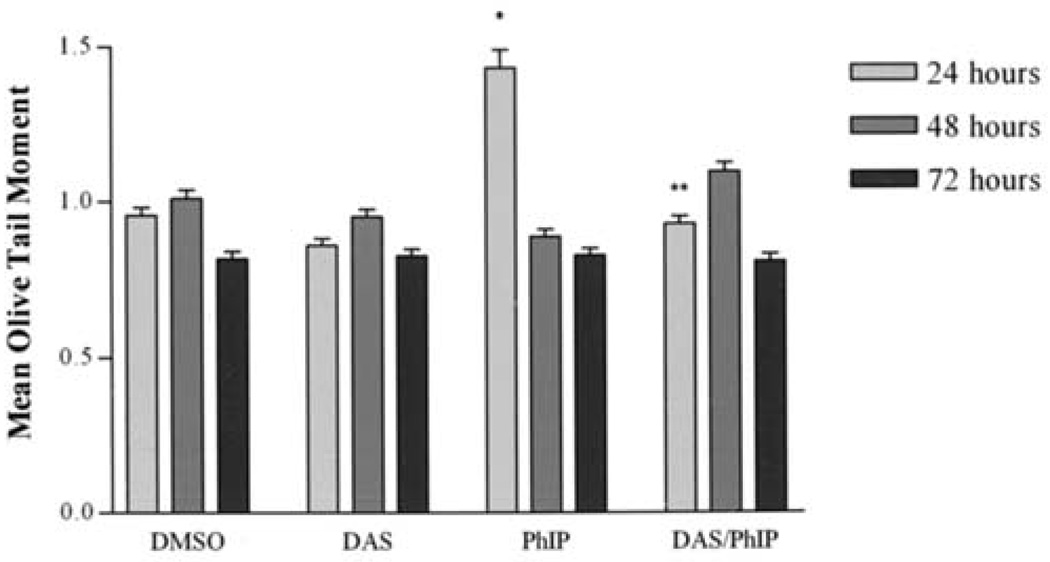
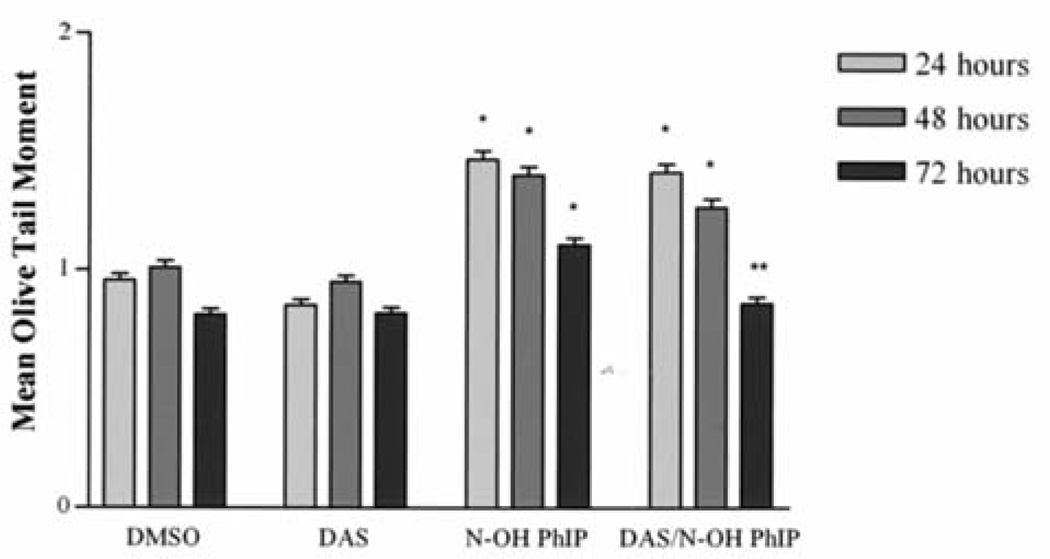
The ability of DAS to inhibit PhIP and N-OH PhIP induced DNA strand breaks in MCF-10A human breast epithelial cells was determined using the comet assay. The comet assay is frequently used to detect DNA strand breaks in individual cells. The mean olive tail moment or simply tail moment is used because it incorporates the amount of DNA that has been damaged with the size of the strands. Tail moment is defined as the product of the tail length and the fraction of total DNA in the tail. Tail moment incorporates a measure of the smallest detectable size of migrating DNA (reflected in the comet tail length) and the number of relaxed/broken pieces; represented by the intensity of DNA in the tail (16). Cells were treated with PhIP (100 µM), N-OH PhIP (5 µM) and/or DAS (100 µM) for 24, 48 and 72 h. Fig. 1 illustrates DNA strand breaks induced by PhIP and the inhibition of these strand breaks by DAS. Individual cells with no DNA strand breaks appear as spheres with no tail. However, the cells that contain DNA damage appear as spheres with tails that resemble comets. PhIP-treated cells displayed a significant increase in DNA strand breaks over that of the control at 24 h 1.4 and 0.9, respectively. The DNA strand breaks that were produced after 24 h were apparently repaired after 48 and 72 h (Fig. 2). Treatment with DAS (100 µM) showed no significant changes in the production of DNA strand breaks from that of the negative and vehicle controls at any time point. However, pretreatment with DAS significantly reduced PhIP-induced DNA strand breaks to baseline levels after 24 h. N-OH PhIP produced significant levels of DNA strand breaks at 24, 48 and 72 h having mean olive tail moments of 1.47, 1.40 and 1.11, respectively (Fig. 3). Pretreatment with DAS inhibited this DNA damage at 24, 48 and 72 h by decreasing mean olive tail moments to 1.41, 1.27 and 0.86, respectively. DAS completely inhibited the N-OH PhIP-induced DNA strand breaks by 72 h. This suggests that DAS does have an inhibitory effect on PhIP and N-OH PhIP-induced DNA strand breaks in human breast epithelial cells.
Figure 1.
Inhibition of the formation of DNA strand breaks in PhIP-treated MCF-10A cells. (A) A cell is shown that was treated with 0.1% DMSO as a control. In this cell, there are no detectable DNA strand breaks as indicated by the absence of a comet tail. (B) A cell that was treated with PhIP (100 µM) is shown. In this cell, the amount of DNA strand breaks was extensive as indicated by the large comet tail. (C) A cell is shown that was treated with PhIP and DAS, 100 µM each. In this cell, DAS inhibited the PhIP-induced DNA strand breaks as indicated by the reduction in the size of the comet tail.
Figure 2.
The inhibition of PhIP-induced DNA strand breaks by DAS at 24, 48 and 72 h. MCF-10A cells were treated, harvested and analyzed as described in Materials and methods. Each bar on the graph represents the mean olive tail moment ± S.E.M. of three independent experiments. *Data significantly different from the DMSO treatment group (P<0.05). **Data significantly different from the PhIP treatment group (P<0.05).
Figure 3.
The inhibition of N-OH PhIP-induced DNA strand breaks by DAS at 24, 48 and 72 h. MCF-10A cells were treated, harvested and analyzed as described in Materials and methods. Each bar on the graph represents the mean olive tail moment ± S.E.M. of three independent experiments. *Data significantly different from the DMSO treatment group (P<0.05). **Data significantly different from the N-OH PhIP treatment group (P<0.01).
Cell viability
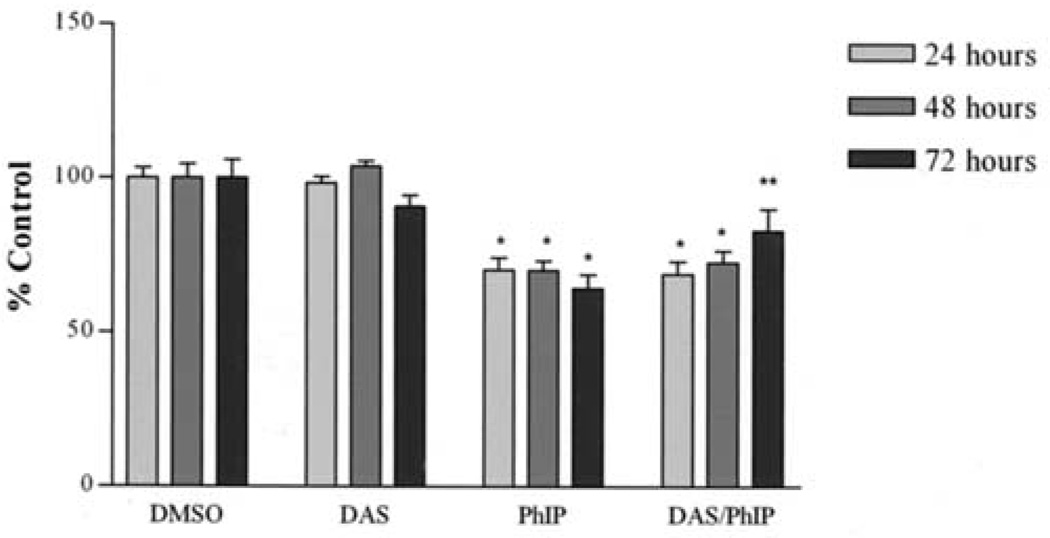
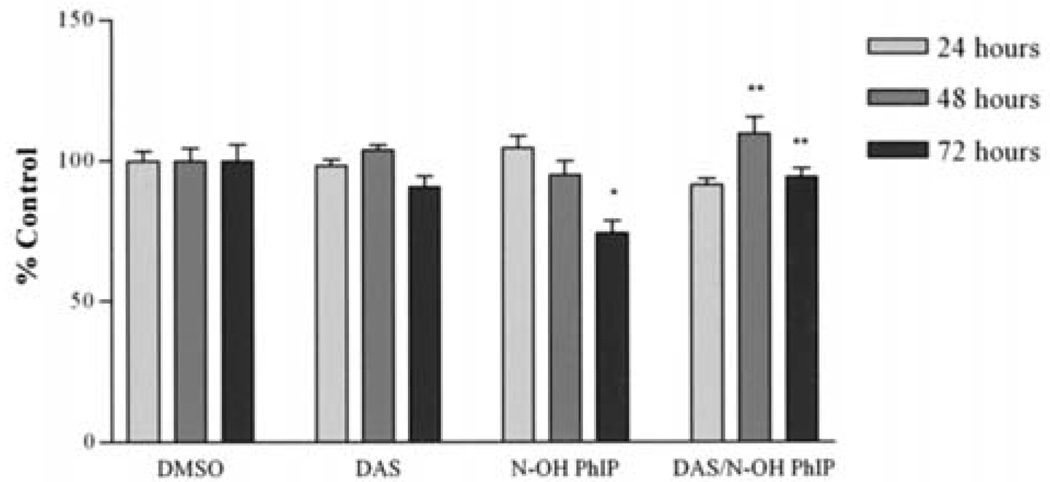
The effect of PhIP and N-OH PhIP on the viability of MCF-10A cells was evaluated using the MTS assay. After PhIP treatment for 24, 48 and 72 h, cell viability was reduced to 70, 70 and 64, respectively. Pre-treatment with DAS (100 µM) attenuated a reduction in cell viability at 48 and 72 h resulting in cell viabilities of 72 and 84%, respectively (Fig. 4). Cells treated with DAS alone showed no significant changes from the control at any time point. This suggests that DAS significantly inhibited PhIP-induced reductions in viability at 72 h. Unlike PhIP, N-OH PhIP only showed a significant reduction in cell viability at 72 h. After 72 h, N-OH PhIP reduced cell viability to 74%. DAS attenuated this reduction in cell viability resulting in cell viability of 94% (Fig. 5). These results suggest that the inhibitory effects of DAS on PhIP and N-OH PhIP-induced cytotoxicity in human breast epithelial cells may require 72 h to manifest.
Figure 4.
The attenuation of PhIP-induced growth arrest by DAS at 24, 48 and 72 h. Cells were treated, harvested and analyzed as described in Materials and methods. Each bar on the graph represents the mean percentage of viable cells ± S.E.M. of three independent experiments. *Data significantly different from the DMSO treatment group (P<0.05). **Data significantly different from the PhIP treatment group (P<0.01).
Figure 5.
The attenuation of N-OH PhIP-induced growth arrest by DAS at 24, 48 and 72 h. Cells were treated, harvested and analyzed as described in Materials and methods. Each bar on the graph represents the mean percentage of viable cells ± S.E.M. of three independent experiments. *Data significantly different from the DMSO treatment group (P<0.05). **Data significantly different from the N-OH PhIP treatment group (P<0.01).
Discussion
The breast has been identified as a target for PhIP-induced carcinogenesis, DNA adduct formation and mutagenesis in animals as well as humans (3,4,17–19). Furthermore, the positive correlation between red meat consumption and breast cancer demonstrated in epidemiological studies support the hypothesis that PhIP plays a role in the etiology of breast cancer (2). Studies have shown that the breast possesses sufficient enzymatic machinery to support PhIP bioactivation and generate DNA damage (15,20), thereby identifying a need for further investigation into the genotoxic effects of PhIP and its metabolites. While much research has been performed on the production of DNA adducts, there is minimal information available on the production of PhIP-induced DNA strand breaks in breast epithelial cells.
One proposed mechanism by which PhIP produces strand breaks is through the production of reactive oxygen species during its metabolism. An imbalance in the production/detoxification of reactive oxygen species results in oxidative stress, which is a known mediator of cancer. While the oxidative mechanisms of cancer have yet to be fully elucidated, DNA mutation as a result of oxidative damage and alterations in cell signaling pathways that control gene expression are postulated to cause cancer (21,23). We have demonstrated in previous studies a significant dose-dependent increase in lipid peroxide formation following PhIP treatment via the induction of Phase I metabolizing enzymes cytochrome P4501A1 and 1A2 (CYP1A1 and CYP1A2) (15). Therefore, this generation of lipid peroxides correlates to the production of reactive PhIP metabolites. This increase in free radicals explains the production of PhIP-induced DNA strand breaks observed at the 24 h time point in this study. Over the course of time, PhIP-induced strand breaks return to baseline levels presumably as a result of DNA repair.
N-OH PhIP is a metabolite of PhIP that can be further metabolized into DNA binding mutagens by a number of phase II enzyme systems (24–26). During its metabolism, N-OH PhIP has been shown to produce free radicals and DNA adducts (27). Unlike PhIP, the DNA strand breaks caused by N-OH PhIP persisted throughout the length of the study and had no significant effect on cell viability until 72 h. This indicates that the cells remained viable with obvious damage, which is believed to be a mechanism for cancer induction.
Cell viability in PhIP-treated cells was significantly reduced at 24, 48 and 72 h, even after DNA strand breaks returned to baseline levels. We demonstrated that PhIP (100 µM) generated DNA adducts at the same time points and under similar conditions employed in this study (15). Therefore, the production of DNA adducts may be responsible for the reduced cell viability after the level of DNA strand breaks returned to baseline levels. We propose that the cells were arrested in an attempt to repair the DNA adducts, which would result in a fewer number of viable cells if the other treatment groups continued to proliferate normally. This proposition is substantiated by the research conducted by Creton et al (28) in which PhIP-treated MCF-10A cells displayed a substantial reduction in cell numbers not associated with cytotoxicity. Under normal circumstances, cells respond to genotoxicity through the propagation of cellular defense mechanisms that attempt to mitigate genomic injury (29). Failure to repair DNA adducts should result in cell death. However, those cells that evade repair and death have the potential to cause breast cancer (19,30).
DAS is a garlic organosulfur compound that reduces chemically-induced genotoxicity and DNA damage (31,32). Pretreatment with DAS was able to inhibit PhIP-induced damage at 24 h. However, cell viability remained significantly reduced up until 72 h. We propose that DAS inhibited cell growth until the damage could be repaired, presumably by 72 h when cell viability was restored to baseline levels. DAS also attenuated N-OH PhIP DNA damage and reduction of cell viability at 72 h. The data are in agreement with research reviewed by Shukla and Kalra (12), where DAS was shown to enhance SOD activity and reduce cell proliferation of Hep-G2 cells. The induction of SOD could explain the mechanism by which DAS inhibits DNA strand breaks.
We presented herein novel evidence that DAS inhibits PhIP and N-OH PhIP-induced DNA strand breaks and attenuates cell viability in MCF-10A human breast epithelial cells. DAS may be used as a chemopreventive agent based on its ability to inhibit DNA damage induced by heterocyclic amines. Further research is needed to fully elucidate the mechanisms by which DAS exerts these anticarcinogenic properties.
Acknowledgements
This study was funded in part by NIH Programs RCMI # G12-RR-03020 and 1C06RR012512-01.
References
- 1.World Health Organization. Cancer: diet and physical activity's impact. [Accessed May 2005]; http://www.who.int/dietphysicalactivity/publications/facts/cancer/en/
- 2.Zheng W, Gustafson DR, Sinha R, Cerhan JR, Moore D, Hong CP, Anderson KE, Kushi LH, Sellers TA, Folsom AR. Well-done meat intake and the risk of breast cancer. J Natl Cancer Inst. 1998;90:4320–4324. doi: 10.1093/jnci/90.22.1724. [DOI] [PubMed] [Google Scholar]
- 3.Nagao M, Ushijima T, Wakabayashi K, Ochiai M, Kushida H, Sugimura T, Hasegawa R, Shirai T, Ito N. Dietary carcinogens and mammary carcinogenesis. Induction of rat mammary carcinomas by administration of heterocyclic amines in cooked foods. Cancer. 1994;74:1063–1069. doi: 10.1002/1097-0142(19940801)74:3+<1063::aid-cncr2820741514>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 4.Ito N, Hasegawa R, Sano M, Tamano S, Esumi H, Takayama S, Sugimura T. A new colon and mammary carcinogen in cooked food, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) Carcinogenesis. 1991;12:1503–1506. doi: 10.1093/carcin/12.8.1503. [DOI] [PubMed] [Google Scholar]
- 5.Shirai T, Sano M, Tamano S, Takahashi M, Hirose M, Futakuchi M, Hasegawa R, Imaida K, Matsumoto K, Wakabayashi K, Sugimura T, Ito N. The prostate: a target for carcinogenicity of 2-amino-1-methyl-6-phenylimidazo[4,5-b] pyridine (PhIP) derived from cooked foods. Cancer Res. 1997;57:195–198. [PubMed] [Google Scholar]
- 6.Gooderham N, Zhu H, Lauber S, Boyce A, Creton S. Molecular and genetic toxicology of 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) Mutat Res. 2002;506–507:91–99. doi: 10.1016/s0027-5107(02)00155-0. [DOI] [PubMed] [Google Scholar]
- 7.Otsuka C, Miura K, Satoh T, Hatanaka M, Wakabayashi K, Ishidate M. Cytogenic effects of a food mutagen, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) and its metabolite 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine (N-OH PhIP) on human and Chinese hamster cells in vitro. Mutat Res. 1996;359:115–121. doi: 10.1016/0165-1218(95)00082-8. [DOI] [PubMed] [Google Scholar]
- 8.Wu R, Tucker J, Sorenson K, Thompson L, Felton J. Differential effect of acetyltransferase expression on the genotoxicity of heterocyclic amines in CHO cells. Mutat Res. 1997;390:93–103. doi: 10.1016/s0165-1218(97)00005-0. [DOI] [PubMed] [Google Scholar]
- 9.Ostling O, Johanson K. Microelectrophoretic study of radiation induced DNA damages in individual mammalian cells. Biochem Biophys Res Commun. 1984;123:291–298. doi: 10.1016/0006-291x(84)90411-x. [DOI] [PubMed] [Google Scholar]
- 10.Singh N, Stephens R, Schneider E. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 11.Tice R, Agurell E, Anderson D, Burlinson B, Hartmann A, Kobayashi H, Miyamae Y, Rojas E, Ryu J, Sasaki Y. Single cell gel/comet assay: guidelines for in vitro and in vivo genetic toxicology testing. Environ Mol Mutagen. 2000;35:206–221. doi: 10.1002/(sici)1098-2280(2000)35:3<206::aid-em8>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 12.Shukla Y, Kalra N. Mini review: Cancer chemoprevention with garlic and its constitutents. Cancer Lett. 2007;247:167–181. doi: 10.1016/j.canlet.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 13.Le Bon AM, Roy C, Dupont C, Suschetet M. In vivo genotoxic effects of dietary allyl sulfides in the rat. Cancer Lett. 1997;114:131–134. doi: 10.1016/s0304-3835(97)04642-9. [DOI] [PubMed] [Google Scholar]
- 14.Guyonnet D, Belloir C, Suschetet M, Siess M, Le Bon A. Mechanisms of protection against aflatoxin B(1) genotoxicity in rats treated by organosulfur compounds from garlic. Carcinogenesis. 2002;23:1335–1341. doi: 10.1093/carcin/23.8.1335. [DOI] [PubMed] [Google Scholar]
- 15.Thomas R, Green M, Wilson C, Weckle A, Duanmu Z, Kocarek T, Runge-Morris M. Cytochrome P450 expression and metabolic activation of cooked food mutagen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in MCF10A breast epithelial cells. Chem Biol Interact. 2006;160:204–216. doi: 10.1016/j.cbi.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Duez P, Dehon G, Dubois J. Validation of raw data measurements in the comet assay. Talanta. 2004;63:879–886. doi: 10.1016/j.talanta.2003.12.035. [DOI] [PubMed] [Google Scholar]
- 17.Ghoshal A, Preisegger K, Takayama S, Thorgeirsson S, Snyderwine E. Induction of mammary tumors in female Sprague-Dawley rats by the food-derived carcionogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine and effect of dietary fat. Carcinogenesis. 1994;15:2429–2433. doi: 10.1093/carcin/15.11.2429. [DOI] [PubMed] [Google Scholar]
- 18.Fan L, Schut H, Snyderwine E. Cytotoxicity, DNA adduct formation and DNA repair induced by 2-hydroxyamino-3-methylimidazo[4,5-f]quinoline and 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine in cultured human mammary epithelial cells. Carcinogenesis. 1995;16:775–779. doi: 10.1093/carcin/16.4.775. [DOI] [PubMed] [Google Scholar]
- 19.Zhu J, Chang P, Bondy M, Sahin A, Singletary S, Takahashi S, Shirai T, Li D. Detection of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine DNA adducts in normal breast tissues and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2003;12:830–837. [PubMed] [Google Scholar]
- 20.Williams J, Phillips D. Mammary expression of xenobiotic metabolizing enzymes and their potential role in breast cancer. Cancer Res. 2000;60:4667–4677. [PubMed] [Google Scholar]
- 21.Breen A, Murphy J. Reactions of oxyl radicals with DNA. Free Radic Biol Med. 1995;18:1033–1077. doi: 10.1016/0891-5849(94)00209-3. [DOI] [PubMed] [Google Scholar]
- 22.Allen R, Tresini M. Oxidative stress and gene regulation. Free Radic Biol Med. 2000:463–499. doi: 10.1016/s0891-5849(99)00242-7. [DOI] [PubMed] [Google Scholar]
- 23.Upham B, Wagner J. Toxicant-induced oxidative stress in cancer. Toxicol Sci. 2001;64:1–3. doi: 10.1093/toxsci/64.1.1. [DOI] [PubMed] [Google Scholar]
- 24.Schut H, Snyderwine E. DNA adducts of heterocyclic amine food mutagens: implications for mutagenesis and carcinogenesis. Carcinogenesis. 1999;20:353–368. doi: 10.1093/carcin/20.3.353. [DOI] [PubMed] [Google Scholar]
- 25.Dubuisson J, Gaubatz J. Bioactivation of the proximal food mutagen 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b] pyridine (N-OH-PhIP) to DNA binding species by human mammary gland enzymes. Nutrition. 1998;14:710–712. doi: 10.1016/s0899-9007(98)00106-3. [DOI] [PubMed] [Google Scholar]
- 26.Lewis A, Walle U, King R, Kadlubar F, Falany C, Walle T. Bioactivation of the cooked food mutagen N-hydroxy-2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine by estrogen sulfotransferase in cultured human mammary epithelial cells. Carcinogenesis. 1998;19:2049–2053. doi: 10.1093/carcin/19.11.2049. [DOI] [PubMed] [Google Scholar]
- 27.Moonen H, Briede J, van Maanen J, Kleinjans J, de Kok T. Generation of free radicals and induction of DNA adducts by activation of heterocyclic aromatic amines via different metabolic pathways in vitro. Mol Carcinog. 2002;35:196–203. doi: 10.1002/mc.10089. [DOI] [PubMed] [Google Scholar]
- 28.Creton S, Zhu H, Gooderham N. A mechanistic basis for the role of cycle arrest in the genetic toxicology of the dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) Toxicol Sci. 2005;84:335–343. doi: 10.1093/toxsci/kfi075. [DOI] [PubMed] [Google Scholar]
- 29.Roos W, Kaina B. DNA damage-induced cell death by apoptosis. Trends Mol Med. 2006;12:440–450. doi: 10.1016/j.molmed.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 30.Glaab W, Kort K, Skopek T. Specificity of mutations induced by the food-associated heterocyclic amine 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine in colon cancer cell lines defective in mismatch repair. Cancer Res. 2001;60:4921–4925. [PubMed] [Google Scholar]
- 31.Huber W, McDaniel L, Kaderlik K, Teitel C, Lang N, Kadlubar F. Chemoprotection against the formation of colon DNA adducts from the food-borne carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in the rat. Mutat Res. 1997;376:115–122. doi: 10.1016/s0027-5107(97)00033-x. [DOI] [PubMed] [Google Scholar]
- 32.Belloir C, Singh V, Daurat C, Seiss M, Le Bon A. Protective effects of garlic sulfur compounds against DNA damage induced by direct and indirect acting genotoxic agents in HepG2 cells. Food Chem Toxicol. 2006;44:827–834. doi: 10.1016/j.fct.2005.11.005. [DOI] [PubMed] [Google Scholar]