Abstract
Flavin monooxygenases (FMOs) play critical roles in plant growth and development by synthesizing auxin and other signaling molecules. However, the structure and function relationship within plant FMOs is not understood. Here we defined the important residues and domains of the Arabidopsis YUC1 FMO, a key enzyme in auxin biosynthesis. We previously showed that simultaneous inactivation of YUC1 and its homologue YUC4 caused severe defects in vascular and floral development. We mutagenized the yuc4 mutant and screened for mutants with phenotypes similar to those of yuc1 yuc4 double mutants. Among the isolated mutants, five of them contained mutations in the YUC1 gene. Interestingly, the mutations identified in the new yuc1 alleles were concentrated in the two GXGXXG motifs that are highly conserved among the plant FMOs. One such motif presumably binds to flavin adenine dinucleotide (FAD) cofactor and the other binds to nicotinamide adenine dinucleotide phosphate (NADPH). We also identified the Ser139 to Phe conversion in yuc1, a mutation that is located between the two nucleotide-binding sites. By analyzing a series of yuc1 mutants, we identified key residues and motifs essential for the functions of YUC1 FMO.
Introduction
Flavin-containing monooxygenases (FMOs, EC 1.14.13.8) are a group of enzymes that catalyze the oxygenation of heteroatoms, particularly the soft nucleophilic atoms including nitrogen and sulfur in small organic molecules (Ziegler 1990, 2002). The common feature of all FMOs is that they use NADPH as the electron donor and FAD as a cofactor to transfer one atom of molecular oxygen to an organic substrate (Ziegler 1990, 2002).
Flavin-containing monooxygenases have been identified in bacteria, yeast, plants, and mammals, but much of our current understanding of the catalytic mechanisms of FMOs are derived mainly from studies on mammalian FMOs, which were the first identified FMOs (Poulsen and Ziegler 1979; Ziegler 1988, 2002; Chen and Ziegler 1994; Kim and Ziegler 2000). A key step in FMO catalysis is the formation of a 4α-hydroperoxy flavin when the cofactor FAD is reduced by NADPH and binds to molecular oxygen (Krueger and Williams 2005). Because the reactive 4α-hydroperoxy flavin is formed before the substrate binds to the enzyme, FMOs are often promiscuous in terms of substrate specificity. As long as the substrates are not sterically prevented from entering the active site, the substrates can often by hydroxylated by FMOs, providing an ideal system for detoxifying diverse xenobiotics. Much of the early biochemical studies on animal FMOs were centered on detoxification of organic compounds. It is not clear what the endogenous substrates are for mammalian FMOs and whether FMOs play a physiological role other than detoxification in animal systems.
Plants appear to have greatly broadened the use of FMOs (Schlaich 2007). In humans, only five FMOs have been identified, whereas Arabidopsis have 29 putative FMO genes on the basis of primary sequence analysis (Krueger and Williams 2005; Schlaich 2007). Unlike the animal FMOs that have undergone extensive biochemical studies without knowing their physiological functions, plant FMOs were first functionally characterized without detailed biochemical analysis. In contrast to the animal FMOs whose main functions appear to metabolize and detoxify xenobiotics, plants use FMOs to synthesize signaling molecules that play essential roles in many aspects of plant growth and development (Miranda et al. 1991; Zhao et al. 2001; Cheng et al. 2006, 2007a). We previously identified an Arabidopsis flavin-containing monooxygenase called YUCCA1 (YUC1) as a key enzyme that catalyzes a rate-limiting step in a tryptophan-dependent auxin biosynthesis pathway (Zhao et al. 2001; Cheng et al. 2006). YUC1 is a member of an Arabidopsis FMO clade that includes 11 members, a subset of which appears to have overlapping functions (Zhao et al. 2001; Cheng et al. 2006, 2007a). The rest of the FMOs in Arabidopsis form two other distinct clades (Schlaich 2007). Overexpression of each member of the YUC family genes in Arabidopsis leads to auxin overproduction and inactivation of certain combinations of YUC genes causes defects in embryogenesis, seedling growth, vascular and floral development (Zhao et al. 2001; Cheng et al. 2006). Loss-of-function yuc mutants can be rescued by tissue-specific expression of the bacterial auxin biosynthesis gene iaaM (Cheng et al. 2006). The yuc mutants also display synergistic interactions with known auxin mutants (Cheng et al. 2007a, 2007b, 2008), demonstrating the essential roles of YUC genes in auxin biosynthesis.
Arabidopsis FMO1 that belongs to an FMO clade distinct from the YUCs was discovered to play a critical role in pathogen defense (Bartsch et al. 2006; Koch et al. 2006; Mishina and Zeier 2006). Overexpression FMO1 increased resistance to bacterial and oomycete pathogens while the fmo1 mutants showed enhanced susceptibility to pathogens (Koch et al. 2006). Furthermore, the expression of FMO1 is upregulated in response to pathogen infection (Mishina and Zeier 2006).
Genetic and physiological studies have clearly demonstrated that plant FMOs are important for normal plant growth and development. However, studies on structure and function relationships of plant FMOs are still lacking. Part of the reason is that the endogenous substrates for Arabidopsis FMOs have not been unambiguously identified. The YUC flavin monooxygenases can catalyze the hydroxylation of tryptamine in vitro (Zhao et al. 2001; Expósito-Rodríguez et al. 2007; Kim et al. 2007; Le et al. 2010), but it is still an open question whether tryptamine is the in vivo substrate for the YUC proteins (Zhao 2010). For Arabidopsis FMO1, not even a candidate substrate is proposed. Because plant FMOs displayed specific physiological functions in particular processes, it is likely that plant FMOs probably are not as promiscuous as their animal counterparts. Therefore, elucidation of the structure/function relationship among plant FMOs will help us to understand the biochemical mechanisms of FMOs in both plants and animal systems.
Here we took a genetic approach to study the structure and functional relationship among the YUC flavin monooxygenases. We previously reported that yuc1 yuc4 double mutants had severe defects in vascular and floral development while the single yuc1 or yuc4 mutant behaved like wild type plants (Cheng et al. 2006). The dramatic developmental phenotypes of yuc1 yuc4 double mutants provide an effective trait to determine the effects of YUC mutations on the in vivo activities of YUC family flavin monooxygenases. We isolated a series of yuc1 mutants that when combined with a yuc4 T-DNA insertion mutant caused phenotypes similar to the previously reported yuc1 yuc4 double mutants. Molecular characterization of the yuc1 point mutations defined the residues and motifs essential for the physiological functions of YUC flavin monooxygenases in Arabidopsis.
Results
Isolation of yuc4 enhancers
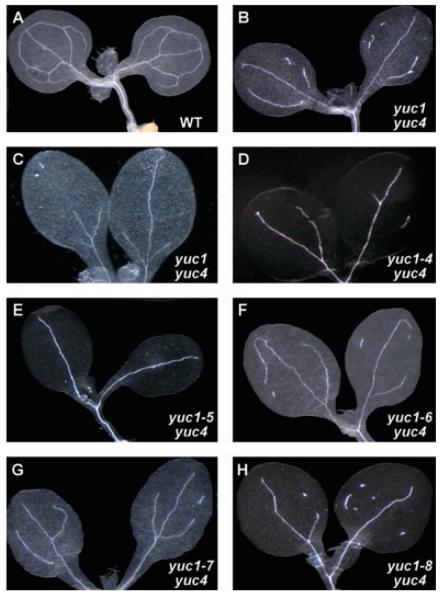
Our previous genetic work on yuc T-DNA insertion mutants established that YUC-mediated local auxin biosynthesis is essential for many developmental processes (Cheng et al. 2006, 2007a). To further dissect the mechanisms of auxin action in plant development, we conducted a genetic screen for yuc4 enhancers that display phenotypes similar to those of yuc1 yuc4 double mutants. The single yuc mutants did not display obvious developmental defects (Cheng et al. 2006). But inactivation of YUC1 and its closest homolog YUC4 greatly disrupted vascular development in cotyledons, leaves, and flowers (Cheng et al. 2006)(Figure 1). Veins in cotyledons of yuc1 yuc4 lacked complete loops and were often discontinuous, while wild type and the yuc single mutants showed more complex patterns (Figure 1). We took advantage of the dramatic vascular defects in cotyledons of yuc1 yuc4 double mutants to isolate yuc4 enhancers. We hypothesized that such a genetic screen could uncover mutations affecting auxin homeostasis, or auxin transport, or auxin signaling. Among the yuc4 enhancers, we expect to identify mutations in the YUC1 gene. The new yuc1 mutants could help us to identify the key amino acid residues and important motifs for YUC1 functions in plants.
Figure 1. Cotyledon vascular patterns of yuc4 enhancers.
(A) Wild type seedlings displayed four-loop vein pattern.
(B and C) Seedlings of yuc1 yuc4 double mutants (T-DNA insertion lines). Note that the yuc1 yuc4 double mutants made fewer veins. Some of the veins were not continuous. Vascular patterns were variable in the yuc1 yuc4 double mutants, but they never formed lobes observed in wt cotyledons.
(D to H) Seedlings of yuc4 enhancers. The enhancers were new yuc1 alleles (see text for details). Seedlings of yuc1–4 yuc4 (D), yuc1–5 yuc4 (E), yuc1–6 yuc4 (F), yuc1–7 yuc4 (G), and yuc1–8 yuc4 (H) are shown. Seedlings were grown for 7 days under long day conditions. Seedlings were cleared using acetic acid/ethanol treatments followed by chloral hydrate treatments.
We mutagenized yuc4 seeds with ethyl methane sulfonate (EMS) (see “Materials and Methods”) and grew the M1 plants to generate a mutant population. Seeds were harvested separately from each individual M1 plant. About 60 seeds from each M1 plant were grown on 0.5 × Murashige and Skoog medium (MS) media for 7 d. We cleared 20 to 30 seedlings from each M1 plant to check for vascular defects. The rest of the sister plants were used to recover the mutant. The vascular patterns in cotyledons were analyzed under a dissecting microscope under dark field background. When a putative mutant with the desired vascular defects in cotyledons was identified, we transplanted the remaining sister seedlings from the same M1 plant to soil. After screening seeds from 2 500 M1 plants, we isolated 19 putative mutants that displayed vascular phenotypes similar to those in yuc1 yuc4 double mutants. The mutants were further divided into three main groups on the basis of their adult phenotypes. The first group of mutants had dramatic floral defects similar to those observed in yuc1 yuc4 double mutants. This group of mutants is likely caused by mutations in YUC1 or other auxin genes. The second group of mutants formed pin-like inflorescences. The third group did not show obvious floral defects. In this paper, we focus on the first group of mutants.
Vascular defects in cotyledons of the yuc4 enhancers
The vascular patterns of cotyledons in wild type plants consistently formed lobes (Figure 1A). The vein patterns in cotyledons of previously characterized yuc1 yuc4 double mutants displayed slight variations; some only had a mid-vein and some formed a fork-like pattern (Figure 1B, C). Our criteria for selecting yuc4 enhancers were: (i) fewer veins; and (ii) discontinuous veins. The isolated yuc4 enhancers all displayed strong vascular defects in cotyledons (Figure 1D–H). Some of the yuc4 enhancers had vascular islands that were not connected to other veins (Figure 1D, F–H). The vascular defects of the yuc4 enhancers were only observed in the yuc4 background. The isolated enhancers had no obvious defects when the yuc4 mutation was crossed out (not shown). It is well known that disruption of auxin biosynthesis, transport, or signaling leads to defects in vascular development. For example, both monopteros (Hardtke and Berleth 1998) and axr6 (Hobbie et al. 2000) displayed dramatic vascular defects in cotyledons. Therefore, the yuc4 enhancers will be informative for understanding auxin-regulated developmental processes.
A group of yuc4 enhancers displayed strong floral defects
Among the isolated yuc4 enhancers, five of them were completely sterile and showed flower defects similar to those of yuc1 yuc4 double mutants (Figure 2). We previously reported that yuc1 yuc4 had a wide range of floral defects (Cheng et al. 2006)(Figure 2). Inactivation of YUC1 and YUC4 simultaneously affected the development of all four whorls of floral organs (Cheng et al. 2006)(Figure 2). In general, yuc1 yuc4 double mutants produced much fewer floral organs and none of the reproductive organs were functional (Figure 2). Another characteristic of yuc1 yuc4 flowers was that each flower was different in terms of the number and types of floral organs (Cheng et al. 2006). The floral defects of the yuc4 enhancers shown here (Figure 2) were very similar to those of yuc1 yuc4. The flowers in the yuc4 enhancers had fewer floral organs in each of the four whorls. It was clear that there were great variations in the flowers of yuc4 enhancers; however, none of them produced any seeds (Figure 2).
Figure 2. Floral defects of yuc4 enhancers.
(A) Adult plants of yuc4 enhancers, from left to right, wild type, yuc1 yuc4 (T-DNA insertion line), yuc1–4 yuc4, yuc1–5 yuc4, yuc1–6 yuc4, yuc1–7 yuc4, and yuc1–8 yuc4.
(B) Inflorescence apex of the yuc4 enhancers. Note that all of the new yuc1 alleles produced abnormal flowers in the yuc4 background.
Molecular lesions in the yuc4 enhancers
Because of the phenotypic similarities between the yuc4 enhancers and the yuc1 yuc4 double mutants at both seedling and adult stages, we hypothesized that the observed phenotypes of yuc4 enhancers were likely caused by mutations in the YUC1 gene or other auxin genes. We sequenced the YUC1 gene of the yuc4 enhancers to determine whether there was a mutation in YUC1. Indeed, we identified single base-pair substitutions in some of the yuc4 enhancers (Table 1). We named the new yuc1 alleles as yuc1–4, yuc1–5, yuc1–6, yuc1–7, and yuc1–8 (Table 1)(Figure 3). The mutations were either a G to A change or a C to T change (Table 1), which were consistent with the fact that the mutants were generated by EMS mutagenesis. The new yuc1 mutations all caused missense mutations except yuc1–8 in which a premature stop codon was generated by the mutation (Figure 3).
Table 1.
Molecular lesions of the yuc1 alleles
|
yuc1 alleles |
Nucleotide change |
Amino acid changes |
Reference |
|---|---|---|---|
| yuc1–1 | T-DNA insertion | – | Cheng et al. 2006 |
| yuc1–2 | T-DNA insertion | – | Not published |
| yuc1–3 | T-DNA insertion | – | Not published |
| yuc1–4 | G74 to A | Gly25 to Glu | This work |
| yuc1–5 | G89 to A | Gly30 to Asp | This work |
| yuc1–6 | G581 to A | Gly194 to Asp | This work |
| yuc1–7 | C416 to T | Ser139 to Phe | This work |
| yuc1–8 | C190 to T | Arg64 to stop | This work |
Nucleotides are numbered according to the cDNA sequence of YUC1. The nucleotide A in the start codon ATG is the first nucleotide.
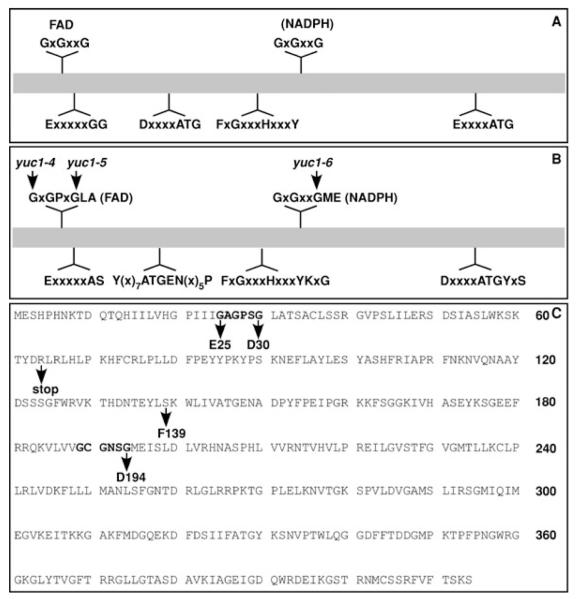
Figure 3. Conserved motifs in flavin monooxygenases (FMOs) and in YUC proteins.
(A) The conserved motifs in all of FMOs. The putative FAD and NADPH binding sites are indicated.
(B) Conserved motifs among Arabidopsis YUC proteins. In addition to the well conserved glycine residues in the FAD binding and NADPH binding sites, residues near the GXGXXG motifs appear to be conserved among YUC proteins.
(C) Mutations that disrupt YUC1 function in Arabidopsis were indicated. Note that mutations in the conserved glycine residues disrupted YUC1 functions.
The new yuc1 alleles revealed two essential motifs
There are several highly conserved motifs among the YUC flavin monooxygenases and other FMOs (Figure 3). Among the motifs in YUC proteins, the GXGPXGLA motif at the N-terminus and the GXGXXGME in the middle were postulated as nucleotide binding motifs (Figure 3). The first motif was suggested to bind FAD and the second motif was referred as NADPH binding motif. Interestingly, two of the new yuc1 alleles harbored a mutation in the putative FAD binding motif and the yuc1–6 contained a mutation in the second GXGXXG motif (Figure 3), demonstrating the importance of the conserved motifs for YUC1 functions. The yuc1–7 allele contains a mutation between the FAD domain and the NADPH binding domain (Figure 3). The yuc1–8 allele has a premature stop codon right after the FAD site (Figure 3). The yuc1–8 allele is likely a null allele because it lacked the putative NADPH binding domain.
Discussion
YUC flavin-containing monooxygenases were the first plant FMO that was assigned a biological function (Zhao et al. 2001). Here we identified several amino acid residues essential for proper functions of YUC1 flavin monooxygenases in Arabidopsis. Our data clearly indicated that the presumed FAD and NADPH binding motifs were required for YUC1 functions in Arabidopsis. This work also established an in vivo system to determine the functions of YUC flavin monooxygenase variants.
At the primary sequence level, all of the FMOs contain several highly conserved motifs and amino acid residues (Krueger and Williams 2005; Schlaich 2007)(Figure 3). A GXGXXG motif (Figure 3A) is well known to be part of the classic Rossmann fold for binding the adenosine diphosphate (ADP) moiety of dinucleotide cofactors such as FAD and NADPH (Figure 3). There are two highly conserved GXGXXG motifs in all of the FMOs (Krueger and Williams 2005; Schlaich 2007). The GXGXXG motif near the N-terminus is believed to bind the cofactor FAD and the other GXGXXG motif in the middle is the putative NADPH binding motif (Figure 3A). The less prominent GG motif near the FAD binding site is believed to further stabilize FAD binding (Figure 3A). One of the ATG containing motifs (Figure 3) probably provides linkage between the FAD site and NADPH site, while the other ATG motif at the C-terminal region may link the NADPH site to the active site (Figure 3A). The second ATG motif often has a conserved F in front of the A and a conserved Y after the G (Figure 3A). For N-hydroxylating enzymes including YUC and siderophore biosynthesis enzymes, the second ATG motif can be written as LATGY (Figure 3A and B). The FXGXXXHXXXY motif (Figure 3) that has been found in all plant FMOs including YUCs is called FMO-identifying sequence motif, which is suggested to contribute to NADPH binding (Krueger and Williams 2005).
Previous in vitro biochemical studies on recombinant rat liver FMO1 demonstrated the essential roles of the glycine residues in the FAD binding GXGXXG motif (Kubo et al. 1997). When the second or the third glycine was mutated to alanine, the resulting FMO1 mutant proteins had much lower FAD content and were catalytically inactive (Kubo et al. 1997). Interestingly, replacement of the first glycine with an alanine did not completely kill FMO1 activity, but greatly increased Km and markedly decreased Kcat values, suggesting that the first glycine was not as sensitive to mutations as the other glycine residues in the motif (Kubo et al. 1997). The yuc1 mutations presented in this paper appeared to completely abolish the YUC1 function. Mutations in the first glycine or the third glycine led to phenotypes similar to those observed in yuc1–8, a presumed null allele, indicating that the first glycine in the FAD binding motif is important for YUC1 functions. A mutation that replaced the third glycine with an arginine in the FAD binding GXGXXG motif in the maize YUC gene SPI1 also completely destroyed its function (Gallavotti et al. 2008). The discrepancy between our in vivo data and previous in vitro biochemical analysis could be due to: (i) different types of mutations being introduced. The glycine to glutamate conversion in YUC1 (Figure 3) probably leads to more structural changes than the glycine to alanine change in rat FMO1; (ii) YUC enzymes are probably quite different from animal FMOs. The NADPH binding site in FMOs has not been extensively studied by site-directed mutagenesis, but mutations in the NADPH/NADH binding motif in other enzymes have been well characterized. For example, mutating any of the glycine residues in the GXGXXG motif in S-adenosylhomocysteinase led to a complete loss of its catalytic activity (Gomi et al. 1989). Our yuc1–6 allele that contains a mutation in the third glycine residue in the putative NADPH binding GXGXXG motif also abolished YUC1 activity in Arabidopsis (Figure 3), suggesting that the NADPH binding GXGXXG motif is also essential for YUC flavin monooxygenases.
Human FMO3 is the major FMO responsible for xenobiotic metabolism in adult human liver. Mutations in FMO3 can lead the failure of trimethylamine N-oxygenation, causing the “fish-odor syndrome” (Zhou and Shephard 2006). Molecular genetic studies on patients with “fish-odor syndrome” have uncovered more than 19 causative mutations in human FMO3 (Krueger and Williams 2005; Zhou and Shephard 2006). Interestingly, none of the identified missense mutations in FMO3 lies in the GXGXXG motifs for FAD and NADPH binding. Some of the mutations are located in the highly conserved motifs other than the GXGXXG motifs. For example, Zhang et al. discovered that a mutation of E32 to K32 conversion in the EXXXXGG motif (Figure 3) abolishes FMO3 activity in vitro and is responsible for “fish-odor syndrome” in some individuals (Zhang et al. 2003; Cashman et al. 2008). Other mutations such as I199T and R205C, which are located within the Rossman fold for NADPH binding, probably affect NADPH binding (Krueger and Williams 2005). In contrast, all of the yuc1 missense alleles except yuc1–7 we identified were located in either the putative FAD binding site or the putative NADPH binding site (Figure 3). The reason that we did not hit other conserved residues and motifs in YUC1 may be due to how the genetic screen for yuc4 enhancers was conducted. We focused on the mutants that caused strong vascular defects in cotyledons in yuc4 background. Mutations in motifs other than the dinucleotide binding sites may not completely abolish yuc1 activity, thus causing weaker phenotypes that we might have missed. On the other hand, the number of mutants we have analyzed is still quite small.
The only yuc1 missense mutation located outside of the FAD and NADPH binding sites is the Ser139 to Phe139 conversion in yuc1–7. The mutation was located between the FAD site and the NADPH motif (Figure 3). Ser139 is not a highly conserved residue, but this position always has a small amino acid residue (Ser, Cys, or Ala) among the YUC family proteins. Ser139 to Phe139 mutation may disrupt the structure of YUC1 protein or the communication between the NADPH site and FAD site. Without further analysis of this mutation at the protein level, it is difficult to pin down the reason why yuc1–7 yuc4 had phenotypes as strong as the yuc1 yuc4 null.
We noticed that residues near the putative GXGXXG FAD binding site and the putative GXGXXG NADPH binding site are highly conserved in YUC flavin monooxygenases (Figure 3B). The amino acid residue after the second G in FAD binding site is always a proline and the two amino acid residues following the third G are L and A in YUCs (Figure 3B). The two amino acid residues after the third G in the NADPH binding site are always M and E in Arabidopsis YUCs (Figure 3B). It is not clear why these positions are highly conserved, but they may contribute to substrate specificity of this class of enzymes. Although flavin monooxygenases are inherently promiscuous in terms of substrate specificity due to the use of the active 4α-hydroperoxy flavin intermediate, some of the FMOs showed remarkable substrate specificity. For example, ornithine monooxygenase only reacts with ornithine and has no activity against closely related lysine, which has one more methylene group than ornithine (Mayfield et al. 2010). A recent study on A. fumigatus ornithine monooxygenase that catalyzes the conversion of ornithine to N5-hydroxyl-ornithine shed some light on how FMO substrate specificity may be achieved (Mayfield et al. 2010). Formation of the reactive 4α-hydroperoxy flavin is accelerated in the presence of the substrate ornithine (Mayfield et al. 2010). YUC flavin monooxygenases are similar to the ornithine monooxygenases at the primary sequence structure as well as the overall organization of the domain structures. Because YUC is involved in auxin biosynthesis, it is almost certain that YUC FMOs need to employ ways to ensure specific hydroxylation reactions. Detailed in vitro biochemical analysis of YUC proteins will be important for understanding the mechanisms by which YUC activities are regulated. The genetic assay used in this work also provides a powerful way to analyze variants of YUC flavin monooxygenases.
Materials and Methods
Plant materials
The yuc1 and yuc4 T-DNA insertion mutants were described previously (Cheng et al. 2006). Arabidopsis plants were grown under long-day conditions (16 h light and 8 h darkness) at 23 °C.
Mutagenesis and yuc4 enhancer screen
The yuc4 T-DNA mutant seeds were incubated in 0.3% ethyl methanesulfonate (EMS) for 11 h. The seeds were washed extensively with sterile water to remove EMS. The mutagenized seeds were then sown into soil. Plants grown from the mutagenized seeds were called M1 plants. Seeds from each M1 plant were collected as an independent pool. We generated more than 2 500 pools of M2 seeds.
Genetic screen for yuc4 enhancers were conducted using the M2 seeds. Approximately 60 seeds from each pool were surface sterilized and then put on 0.5 × MS plates. The plates were put at 4 °C for 2 d and then incubated at 23 °C for 7 d under long day conditions. About 20 to 30 seedlings from each pool were treated with a solution of acetic acid and ethanol (1:3) for 2 h. The treated seedlings were washed with water twice, and then transferred to chloral hydrate solution overnight (chloral hydrate : water : glycerol = 8:1:1) to further clear the plant tissue. Vascular patterns of cotyledons were examined and photographed under dark-field optics with a Leica dissecting microscope. The putative yuc4 enhancer lines had either fewer veins or discontinuous veins or both.
Mutant recovery and analysis
Because the cleared seedlings used in our vascular pattern screen were no longer viable after ethanol/acetic acid and chloral hydrate treatments, we used the sister plants that were from the same M1 plant to recover the putative yuc4 enhancers. We transplanted the sister plants to soil and let the M2 plants set seeds. We also crossed some of the M2 plants to Arabidopsis Columbia ecotype and Landsberg ecotype to generate backcross population and mapping populations, respectively. For plants that displayed floral phenotypes similar to those of yuc1 yuc4 double mutants, we directly sequenced the YUC1 gene to determine whether the phenotypes were caused by mutations in YUC1. If a mutation in YUC1 was identified, we designed cleaved amplified polymorphic sequences (CAPS) or derived cleaved amplified polymorphic sequences (dCAPS) markers to determine whether the phenotypes were linked to the particular yuc1 mutation (Hou et al. 2010). We also cross the new yuc1 alleles to our T-DNA reference allele to conduct complementation tests. For the yuc1 alleles that we had a mapping population, we also performed mapping analysis using molecular markers (Hou et al. 2010).
Acknowledgement
We thank Dr Genji Qin (Peking University) and Dr Youfa Cheng (Institute of Botany, the Chinese Academy of Sciences) for their contributions in generating the EMS mutagenized population when they worked in the Zhao lab. This work was supported by the National Natural Science Foundation of China (No. 30628012 to Y.Z and 30625002 to L.-J.Q) and National Institutes of Health (R01GM68631 to Y.Z.). X. H. and S. L. were partially supported by a scholarship from the China Scholarship Council.
References
- Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE. Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell. 2006;18:1038–1051. doi: 10.1105/tpc.105.039982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashman JR, Zhang J, Nelson MR, Braun A. Analysis of flavin-containing monooxygenase 3 genotype data in populations administered the anti-schizophrenia agent olanzapine. Drug Metab. Lett. 2008;2:100–114. doi: 10.2174/187231208784040942. [DOI] [PubMed] [Google Scholar]
- Chen GP, Ziegler DM. Liver microsome and flavin-containing monooxygenase catalyzed oxidation of organic selenium compounds. Arch. Biochem. Biophys. 1994;312:566–572. doi: 10.1006/abbi.1994.1346. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006;20:1790–1799. doi: 10.1101/gad.1415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell. 2007a;19:2430–2439. doi: 10.1105/tpc.107.053009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Qin G, Dai X, Zhao Y. NPY1, a BTB-NPH3-like protein, plays a critical role in auxin-regulated organogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2007b;104:18825–18829. doi: 10.1073/pnas.0708506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Qin G, Dai X, Zhao Y. NPY genes and AGC kinases define two key steps in auxin-mediated organogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2008;105:21017–21022. doi: 10.1073/pnas.0809761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Expósito-Rodríguez M, Borges AA, Borges-Pérez A, Hernández M, Pérez JA. Cloning and biochemical characterization of ToFZY, a tomato gene encoding a flavin monooxygenase involved in a tryptophan-dependent auxin biosynthesis pathway. J. Plant Growth Regul. 2007;26:329–340. [Google Scholar]
- Gallavotti A, Barazesh S, Malcomber S, Hall D, Jackson D, Schmidt RJ, McSteen P. Sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc. Natl. Acad. Sci. USA. 2008;105:15196–15201. doi: 10.1073/pnas.0805596105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomi T, Date T, Ogawa H, Fujioka M, Aksamit RR, Backlund PS, Jr, Cantoni GL. Expression of rat liver S-adenosylhomocysteinase cDNA in Escherichia coli and mutagenesis at the putative NAD binding site. J. Biol. Chem. 1989;264:16138–16142. [PubMed] [Google Scholar]
- Hardtke CS, Berleth T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998;17:1405–1411. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie L, McGovern M, Hurwitz LR, Pierro A, Liu NY, Bandyopadhyay A, Estelle M. The axr6 mutants of Arabidopsis thaliana define a gene involved in auxin response and early development. Development. 2000;127:23–32. doi: 10.1242/dev.127.1.23. [DOI] [PubMed] [Google Scholar]
- Hou X, Li L, Peng Z, Wei B, Tang S, Ding M, Liu J, Zhang F, Zhao Y, Gu H, Qu LJ. A platform of high-density INDEL/CAPS markers for map-based cloning in Arabidopsis. Plant J. 2010;63:880–888. doi: 10.1111/j.1365-313X.2010.04277.x. [DOI] [PubMed] [Google Scholar]
- Kim JI, Sharkhuu A, Jin JB, Li P, Jeong JC, Baek D, Lee SY, Blakeslee JJ, Murphy AS, Bohnert HJ, Hasegawa PM, Yun DJ, Bressan RA. yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes. Plant Physiol. 2007;145:722–735. doi: 10.1104/pp.107.104935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YM, Ziegler DM. Size limits of thiocarbamides accepted as substrates by human flavin-containing monooxygenase 1. Drug Metab. Dispos. 2000;28:1003–1006. [PubMed] [Google Scholar]
- Koch M, Vorwerk S, Masur C, Sharifi-Sirchi G, Olivieri N, Schlaich NL. A role for a flavin-containing mono-oxygenase in resistance against microbial pathogens in Arabidopsis. Plant J. 2006;47:629–639. doi: 10.1111/j.1365-313X.2006.02813.x. [DOI] [PubMed] [Google Scholar]
- Krueger SK, Williams DE. Mammalian flavin-containing monooxygenases: structure/function, genetic polymorphisms and role in drug metabolism. Pharmacol. Theor. 2005;106:357–387. doi: 10.1016/j.pharmthera.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, Itoh S, Itoh K, Kamataki T. Determination of FAD-binding domain in flavin-containing monooxygenase 1 (FMO1) Arch. Biochem. Biophys. 1997;345:271–277. doi: 10.1006/abbi.1997.0242. [DOI] [PubMed] [Google Scholar]
- Le CS, Schmelz EA, Chourey PS. Sugar levels regulate tryptophan-dependent auxin biosynthesis in developing maize kernels. Plant Physiol. 2010;153:306–318. doi: 10.1104/pp.110.155226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield JA, Frederick RE, Streit BR, Wencewicz TA, Ballou DP, Dubois JL. Comprehensive spectroscopic, steady state, and transient kinetic studies of a representative siderophore-associated flavin monooxygenase. J. Biol. Chem. 2010 doi: 10.1074/jbc.M110.157578. E pub date 2010/07/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda CL, Chung W, Reed RE, Zhao X, Henderson MC, Wang JL, Williams DE, Buhler DR. Flavin-containing monooxygenase: a major detoxifying enzyme for the pyrrolizidine alkaloid senecionine in guinea pig tissues. Biochem. Biophys. Res. Commun. 1991;178:546–552. doi: 10.1016/0006-291x(91)90142-t. [DOI] [PubMed] [Google Scholar]
- Mishina TE, Zeier J. The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol. 2006;141:1666–1675. doi: 10.1104/pp.106.081257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen LL, Ziegler DM. The liver microsomal FAD-containing monooxygenase. Spectral characterization and kinetic studies. J. Biol. Chem. 1979;254:6449–6455. [PubMed] [Google Scholar]
- Schlaich NL. Flavin-containing monooxygenases in plants: looking beyond detox. Trends Plant Sci. 2007;12:412–418. doi: 10.1016/j.tplants.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Zhang J, Tran Q, Lattard V, Cashman JR. Deleterious mutations in the flavin-containing monooxygenase 3 (FMO3) gene causing trimethylaminuria. Pharmacogenetics. 2003;13:495–500. doi: 10.1097/00008571-200308000-00007. [DOI] [PubMed] [Google Scholar]
- Zhao Y. Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 2010;61:49–64. doi: 10.1146/annurev-arplant-042809-112308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science. 2001;291:306–309. doi: 10.1126/science.291.5502.306. [DOI] [PubMed] [Google Scholar]
- Zhou J, Shephard EA. Mutation, polymorphism and perspectives for the future of human flavin-containing monooxygenase 3. Mutat. Res. 2006;612:165–171. doi: 10.1016/j.mrrev.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Ziegler DM. Flavin-containing monooxygenases: catalytic mechanism and substrate specificities. Drug Metab. Rev. 1988;19:1–32. doi: 10.3109/03602538809049617. [DOI] [PubMed] [Google Scholar]
- Ziegler DM. Flavin-containing monooxygenases: enzymes adapted for multisubstrate specificity. Trends Pharmacol. Sci. 1990;11:321–324. doi: 10.1016/0165-6147(90)90235-z. [DOI] [PubMed] [Google Scholar]
- Ziegler DM. An overview of the mechanism, substrate specificities, and structure of FMOs. Drug Metab. Rev. 2002;34:503–511. doi: 10.1081/dmr-120005650. [DOI] [PubMed] [Google Scholar]