Abstract
Polyadenylation of mRNA precursors is frequently coupled to transcription by RNA polymerase II. Although this coupling is known to involve interactions with the C-terminal domain of the RNA polymerase II largest subunit, the possible role of other factors is not known. Here we show that a prototypical transcriptional activator, GAL4-VP16, stimulates transcription-coupled polyadenylation in vitro. In the absence of GAL4-VP16, specifically initiated transcripts accumulated but little polyadenylation was observed, while in its presence polyadenylation was strongly enhanced. We further show that this stimulation requires the transcription elongation-associated PAF complex (PAF1c), as PAF1c depletion blocked GAL4-VP16-stimulated polyadenylation. Furthermore, knockdown of PAF subunits by siRNA resulted in decreased 3′ cleavage, and nuclear export, of mRNA in vivo. Finally, we show that GAL4-VP16 interacts directly with PAF1c and recruits it to DNA templates. Our results indicate that a transcription activator can stimulate transcription-coupled 3′ processing and does so via interaction with PAF1c.
INTRODUCTION
mRNA synthesis in eukaryotic cells is a highly complex process, involving transcription of the mRNA precursor followed by its capping, splicing and polyadenylation. Considerable evidence now indicates that transcription and the subsequent pre-mRNA processing reactions occur cotranscriptionally (Hirose and Manley 2000; Bentley 2002; Maniatis and Reed 2002; Proudfoot et al. 2002). An important factor in linking transcription to pre-mRNA processing is the carboxy-terminal domain of the RNA polymerase II (RNAP II) largest subunit (CTD), which has a significant role in enhancing the efficiency of all the processing reaction (e.g., McCracken et al. 1997a, b; Hirose and Manley 1998; Hirose et al. 1999). How the CTD functions is not entirely understood, but a number of interactions with specific processing factors have been reported (Phatnani and Greenleaf 2006), and these likely serve to help recruit the processing machinery to the pre-mRNA and then to stabilize or enhance the activity of these factors. The coupling of transcription to mRNA processing is believed to ensure accurate, efficient, and rapid processing of nascent pre-mRNAs.
In addition to the CTD, the general transcription factor (GTF) TFIID helps to couple transcription and polyadenylation. Cleavage-polyadenylation specificity factor (CPSF), an essential polyadenylation factor, associates with TFIID and, based on in vitro transcription experiments, is recruited to the preinitiation complex by TFIID. After transcription initiates, CPSF dissociates from TFIID and becomes associated with the elongating RNAP II, likely via the CTD (Dantonel et al. 1997). Consistent with the idea that polyadenylation factors are present at promoters, chromatin immunoprecipitation experiments have localized 3′ processing factors to promoter regions in yeast and mammals (e.g., Licatalosi et al. 2002; Calvo and Manley 2005; Venkataraman et al. 2005; Rozenblatt-Rosen et al. 2009).
Sequence-specific transcription factors function to facilitate recruitment of GTFs, including TFIID, to promoter regions. Evidence suggests that they may also play a role in recruiting polyadenylation factors. For example, transient cotransfection assays have suggested that such transcription factors can increase not only transcription but also splicing and 3′-end processing (Rosonina et al. 2003). This activator-dependent pre-mRNA processing was found to be independent of the overall levels of the transcript generated and to require the CTD of RNAP II. The multifunctional protein PSF was found to facilitate activator-dependent pre-mRNA processing (Rosonina et al. 2005). PSF is localized across the length of transcribed genes, and is thought to function in essentially all steps of transcription and processing (Kaneko et al. 2007 and references therein). Further evidence suggesting an association between transcriptional activators and polyadenylation comes from the observation that the strong viral activator VP16 recruits 3′ processing factors CPSF and CstF to promoter regions in vivo (Uhlmann et al. 2007). But whether or not this involves direct interactions and/or additional intermediary factors, or whether it affects the efficiency of 3′ processing, is not known.
One potentially important factor in coupling transcription and polyadenylation is the PAF1 complex (PAF1c). PAF1c was first identified in yeast as an RNAP II-associated factor (Shi et al. 1996). Genetic studies of PAF subunits revealed transcript elongation phenotypes (Costa and Arndt 2000; Mueller and Jaehning 2002; Squazzo et al. 2002), and PAF1c has also been demonstrated to cross-link along the entire length of several genes, consistent with its functioning in some way as an elongation factor (Krogan et al. 2002; Pokholok et al. 2002). PAF1c is also known to facilitate certain histone modifications on active genes, such as methylation of histone H3K4 and K36 (Krogan et al. 2003a, b), as well as CTD phosphorylation at the Ser2 position (Mueller et al. 2004). Importantly, poly(A) tail length (Mueller et al. 2004) and poly(A) site selection (Penheiter et al. 2005) were found to be altered in PAF mutant cells, suggesting a role in formation of mRNA 3′ ends. PAF1c is conserved in mammalian cells (Rozenblatt-Rosen et al. 2005; Yart et al. 2005; Zhu et al. 2005a), and also plays a role in transcription elongation (e.g., Kim et al. 2010). Additionally, components of the polyadenylation machinery have recently been found to associate with the yeast and human PAF complex (Nordick et al. 2008; Rozenblatt-Rosen et al. 2009). Depletion of PAF1c from cell extracts in vitro inhibited polyadenylation but not transcription or splicing, and from cells in vivo reduced expression and extended transcription of a target gene (Rozenblatt-Rosen et al. 2009). These findings all support a role for PAF1c in helping to couple transcription to 3′ processing.
In this study, we provide evidence that transcriptional activators can directly stimulate mRNA 3′ processing, as long as it is coupled to transcription, and that this stimulation is mediated by PAF1c. We first show that the model activators such as GAL4-VP16 strongly induce transcription-coupled polyadenylation in HeLa nuclear extract (NE). In the absence of GAL4-VP16, specifically initiated transcripts accumulate but little polyadenylation occurs. We next show that depletion of PAF1c markedly reduces VP16-mediated polyadenylation, but not transcription, in vitro. Furthermore, depletion of PAF subunits in vivo by siRNA results in decreased 3′-end cleavage, and nuclear export, of mRNA produced from a GAL4-VP16 driven reporter plasmid. Finally, we found GAL4-VP16 interacts directly with PAF1c and stimulates recruitment of PAF1c to DNA templates in NE. Thus, our results suggest that a transcriptional activator can stimulate transcription-coupled polyadenylation via recruitment of PAF1c to DNA.
RESULTS
Transcriptional activators stimulate transcription-coupled polyadenylation in vitro
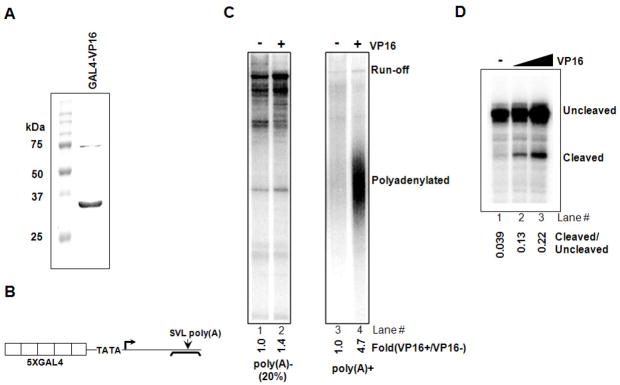
Our previous studies utilized a coupled transcription-3′ processing system in HeLa NE to provide evidence that PAF1c plays a role in 3′ processing (Rozenblatt-Rosen et al. 2009). To extend these studies, and also to investigate the possible role of transcriptional activators in transcription-coupled polyadenylation, we established an in vitro assay utilizing the chimeric transcriptional activator GAL4-VP16 (Fig. 1A). The DNA template (pG5E4SVL) contained five GAL4 binding sites upstream of a minimal adenovirus E4 core promoter and the SVL poly(A) site downstream (Fig. 1B). Transcription reactions were performed in NE with or without purified GAL4-VP16 and linearized pG5E4SVL. Following transcription, RNA was separated into poly (A+) and poly (A−) fractions and analyzed by denaturing PAGE. As shown in Figure 1C, run-off transcript accumulated in the absence of GAL4-VP16, but little polyadenylation was observed (lanes 1 and 3). In the presence of GAL4-VP16, total transcription was increased, although relatively modestly (compare lanes 1 and 2), consistent with the use of a naked DNA template (e.g., Pazin et al. 1998). In contrast, polyadenylation was strongly induced when transcription was activated by GAL4-VP16 (compare lanes 3 and 4). To examine 3′ cleavage, and also allow more precise quantitation of the stimulation, we analyzed total synthesized RNA by RNase protection, using RNA synthesized in the absence of labeled UTP and a 32P RNA probe spanning the poly(A) addition site, and determined the ratio of cleaved to uncleaved (read-through) RNA. The results (Fig. 1D) confirm that GAL4-VP16 enhances 3′ processing, increasing the cleaved to uncleaved ratio more than five fold (lanes 1–3).
Figure 1. GAL4-VP16 activates transcription-coupled polyadenylation.
(A) Purification of bacterially expressed GAL4-VP16. His-tagged GAL4-VP16 was purified by using talon resin (clontech), and 5 μg resolved by SDS-PAGE.
(B) Schematic of the DNA template used for transcription-coupled polyadenylation assays. The template contained tandem repeats of GAL4 binding sites upstream of the adenovirus E4 core promoter region and SVL poly(A) site downstream. The position of the RNA probe used to analyze cleavage levels is also indicated.
(C) Transcription-polyadenylation assay with or without GAL4-VP16. After reactions in HeLa NE, RNAs were purified, separated by oligo(dT) selection into poly(A)- and poly(A)+ fraction, and analyzed on 5% denaturing gel. Run-off and polyadenylated products were quantitated with ImageJ and results are shown at the bottom of each lane.
(D) RNase protection assay to examine cleavage level. After transcription-polyadenylation was carried out as in (C) without radioactive α-32P UTP, RNAs were isolated, treated with turbo-DNase (ambion) and subject to RNase protection analysis. Quantitation of cleaved and uncleaved products were done with ImageJ and the results are shown at the bottom of each lane.
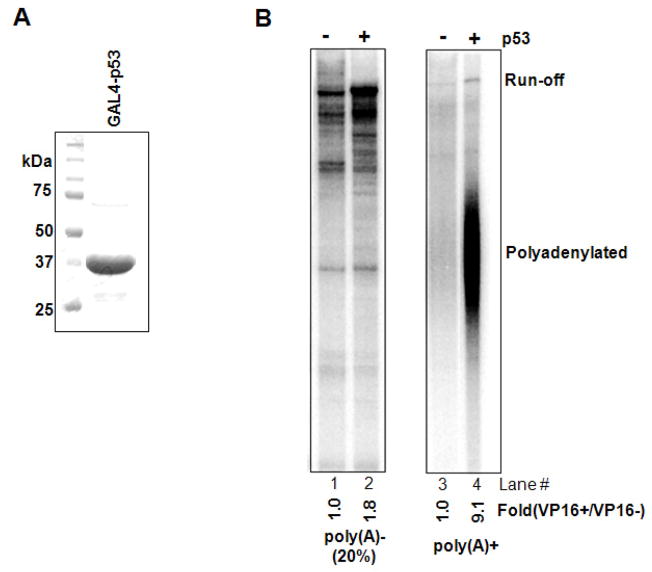
We next characterized several parameters of GAL4-VP16-mediated stimulation of 3′ processing. We first verified that the transcription observed was specific, and showed that a DNA template containing a mutated TATA box did not produce run-off or polyadenylated RNA (Fig. S1A; lanes 3, 4, 7, 8). To show that the effect was not specific to the SV40L poly(A) site, we performed transcription-coupled polyadenylation with another DNA template (pG3G5E4L3) containing the adenovirus L3 poly(A) site (Fig. S1B). A similar stimulation of 3′ processing was observed with this DNA template: GAL4-VP16 activated polyadenylation more strongly than transcription (Fig. S1C; lanes 3, 4, 11, 12). The polyadenylated RNA detected was AAUAAA-dependent, as no polyadenylation was detected with an L3 poly(A) site mutant (AAUAAA→AAAAAA, lanes 15 and 16). Furthermore, polyadenylation was not induced using DNA templates lacking GAL4 binding sites (lanes 9 and 10), indicating GAL4-VP16 needs to be recruited to the DNA template to stimulate transcription-coupled polyadenylation. Likewise, when a presynthesized SVL RNA was incubated in NE under conditions similar to those used in the coupled reaction, GAL4-VP16 was without effect (Fig. S2). Finally, the ability to enhance transcription-coupled polyadenylation was not unique to GAL4-VP16, as a GAL4 fusion protein containing the p53 activation domain (Fig. 2A) also strongly activated polyadenylation in an assay similar to that shown in Figure 1 (Fig. 2B).
Figure 2. GAL4-p53 stimulates transcription-coupled polyadenylation.
(A) Purification of bacterially expressed GAL4-p53. His-tagged GAL4-p53 was purified by using talon resin and 5 μg resolved by SDS-PAGE.
(B) Transcription-polyadenylation assay with or without GAL4-p53. Transcripts produced in NE were analyzed on 5% denaturing gel and quantitated as in Fig. 1C.
Depletion of PAF complex blocks VP16-induced polyadenylation in vitro and vivo
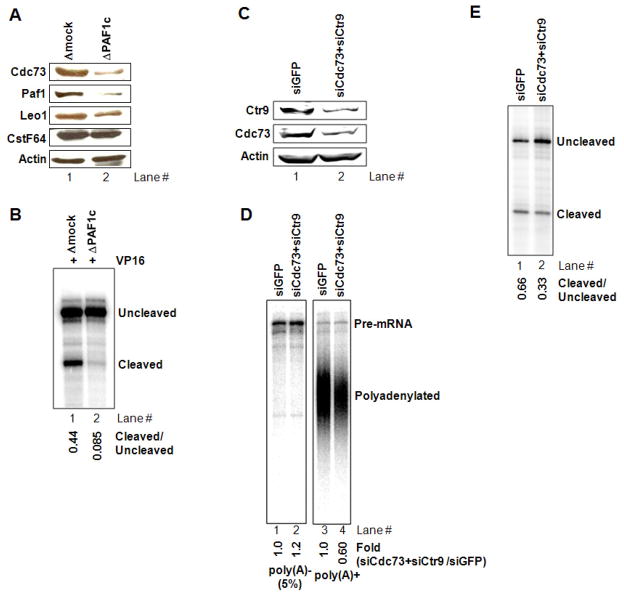
We showed previously that the multisubunit PAF complex (PAF1c) associates with 3′ processing factors and functions in transcription-coupled polyadenylation (Rozenblatt-Rosen et al. 2009). To investigate the possible role of PAF1c in GAL4-VP16-mediated enhancement of transcription-coupled 3′ processing, PAF1c was immunodepleted from NE using anti-Cdc73 antibody as described previously (Rozenblatt-Rosen et al. 2009), and PAF depletion was confirmed by western blot. PAF1c was specifically depleted while levels of other proteins tested (e.g., CstF64 and actin) were unaffected (Fig. 3A; although PAF1c associates with CstF, it was not codepleted under the conditions used). The PAF1c-depleted NE and a mock-depleted control were used in coupled transcription-polyadenylation reactions with GAL4-VP16 and transcripts were analyzed by RNase protection as above. Significantly, 3′ processing was reduced in the PAF1c-depleted NE relative to the control, whereas accumulation of run-off, unprocessed transcript was not affected (Fig. 3B).
Figure 3. PAF1c depletion diminished VP16-dependent polyadenylation but not transcription.
(A) Immunodepletion of PAF complex from NE. Depletion was performed with anti-Cdc73 antibody and was confirmed by Western blot.
(B) Transcription-polyadenylation assay with PAF depleted NE in the presence of GAL4-VP16. RNAs were analyzed by RNase protection assays and quantitated with ImageJ as in Fig. 1D.
(C) PAF depletion by siRNA knockdown. NE was prepared from 293T cells treated with siRNA targeting Cdc73 and Ctr9. Depletion was confirmed by Western blot. siRNA targeting GFP served as a control.
(D) Transcription-polyadenylation assay with PAF depleted NE prepared in (C), and RNAs were analyzed and quantitated as in Fig. 1C.
(E) Transcription-polyadenylation was done as in (D) except that α-32P UTP was omitted from reaction mixtures. RNAs were analyzed by RNase protection assays as in (B). Quantitation was done with ImageJ.
To extend these results, we also performed siRNA-mediated knockdown of PAF subunits in 293T cells and prepared NEs from cells treated with Cdc73-siRNA and Ctr9-siRNA or from control cells treated with GFP-siRNA. Knockdown efficiency was confirmed by Western (Fig. 3C). When these NEs were tested in transcription-coupled polyadenylation reactions with GAL4-VP16, polyadenylation was decreased in PAF-depleted NE, while pre-mRNA accumulation was increased (Fig. 3D). RNase protection assays confirmed that cleavage efficiency was reduced (by ~50%) in the PAF-depleted NE (Fig. 3E). RNAi-mediated depletion of PAF1c gave a modest effect on 3′ processing compared to immunodepletion, perhaps because only two subunits (Cdc73 and Ctr9) of the complex were depleted by siRNA knockdown and the knockdown was partial, while the entire PAF1c was effectively removed by immunodepletion (Fig. 3A). Since it was possible that PAF1c functions in cleavage-polyadenylation in a transcription-independent manner, we performed an uncoupled reaction using SVL pre-mRNA as a substrate. Importantly, no significant effect on cleavage or polyadenylation was observed (Fig. S3A and B), suggesting that PAF1c acts in transcription-polyadenylation coupling but not in uncoupled 3′ end processing.
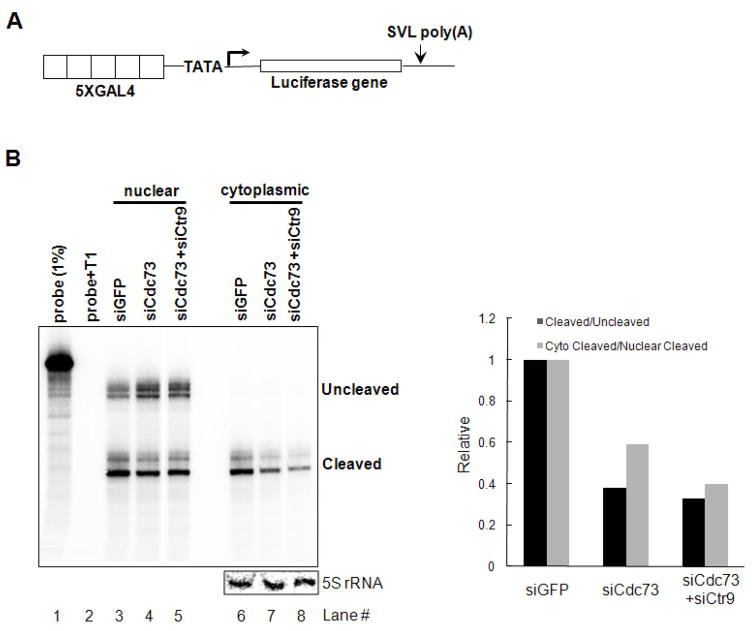
We next asked whether PAF1c functions in VP16-dependent transcription-3′ processing in vivo. To this end, transfection assays were carried out with a GAL4-VP16 expression plasmid and the reporter plasmid pGL3G5E4 containing GAL4 binding sites upstream of the E4 core promoter and the SVL poly(A) site downstream (Fig. 4A). The plasmids were transfected into 293T cells treated with siRNA targeting GFP or PAF1c subunits (Cdc73 and/or Ctr9). Cells were harvested, separated into cytoplasmic and nuclear fractions (Wang et al. 2006) and RNA isolated and analyzed by RNase protection to analyze 3′ end cleavage levels. As shown in Fig. 4B, cleavage was decreased upon siRNA knockdown of the PAF subunits (see lanes 3–8) whereas pre-mRNA levels were increased. Notably, the amount of the mRNA in the cytoplasm was decreased significantly when PAF1c was depleted (compare lanes 6–8 and lanes 3–5). This finding suggests PAF1c functions in mRNA nuclear export (see Discussion). The decrease in 3′-end cleavage and accumulation of cytoplasmic mRNA was also obsesrved when Paf1 or Ctr9 was depleted with siRNAs (Fig. S4). Taken together, these results indicate that PAF1c functions in VP16-induced polyadenylation both in vitro and vivo.
Figure 4. siRNA knockdown of PAF1c inhibits 3′end processing of reporter mRNA.
(A) Schematic of reporter plasmid pGL3G5E4 containing GAL4 binding sites upstream of E4 core promoter, Luciferase coding sequences and SVL poly(A) site downstream.
(B) RNase protection assay of transcripts isolated from 293T cells treated with siRNA targeting PAF1c subunits. The reporter plasmid was transfected into 293T cells pre-treated with siRNAs targeting PAF1c subunit(s) as indicated. The next day, cells were harvested and total RNAs were obtained using Trizol (Invitrogen), fractionated into nuclear and cytoplasmic fractions and 3′ cleavage efficiency was examined by RNase protection assays. Quantitation was performed with ImageJ. Ethidium bromide staining of 5S rRNA (lower panel) indicates uniform recovery between samples.
The VP16 activation domain directly interacts with the PAF1c subunit Paf1
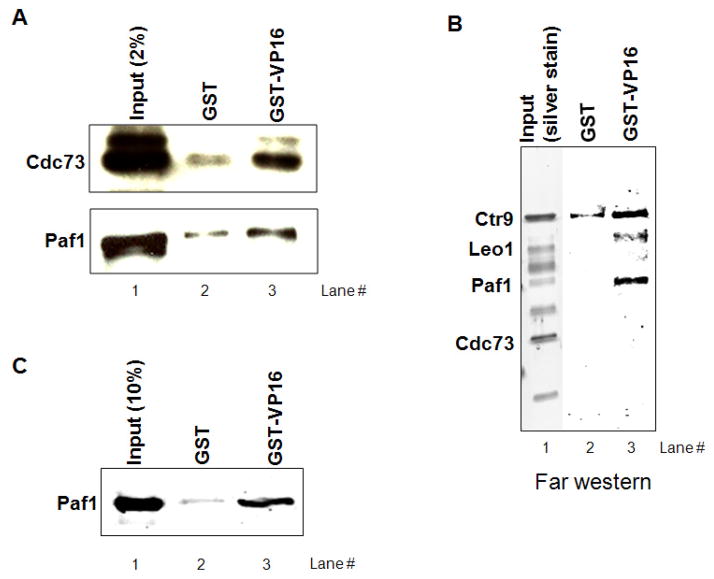
We next asked whether the GAL4-VP16 functions in enhancing transcription-coupled 3′processing might involve an interaction with PAF1c. First, we carried out GST-pulldown assays to determine whether the VP16 activation domain can associate with PAF1c in NE. A GST-VP16 fusion protein indeed interacted with PAF1c, as measured by Western blot with anti-Cdc73 and -Paf1 antibodies (Fig. 5A). Next, to elucidate which PAF subunit(s) is (are) involved in the interaction, PAF1c was purified from 293T cells stably expressing Flag-tagged Paf1. The purified PAF1c was then used in Far-western analysis using GST-VP16 as probe. As shown in Figure 5B, VP16 appeared to interact strongly and directly with the Paf1 subunit, although weak association was also detected with Ctr9 and Leo1. To confirm the Paf1 interaction, a GST-pull down assay was carried out with purified proteins, His-tagged Paf1 and GST-VP16. As shown in Figure 5C, GST-VP16 specifically associated with His-Paf1. Together, these results indicate that the VP16 activation domain interacts with the PAF1c, and does so via a direct interaction with the Paf1 subunit.
Figure 5. VP16 interacts with PAF1c.
(A) GST-pulldown assay of PAF1c subunits (Cdc73 and Paf1) from NE. The assay was performed using 5 μg immobilized GST-VP16 together with NE. After extensive washing, bound proteins were eluted by boiling and analyzed by western blotting. For comparison, 2% of input NE is shown in lane 1.
(B) Far-western analysis of purified PAF1c. PAF1c purified from 293T cells stably expressing Flag-tagged paf1 was resolved on SDS-PAGE and transferred to a nitrocellulose membrane. After denaturating-renaturation using guanidine hydrochloride, the membrane was probed with either GST or GST-VP16 and subject to western blotting with anti-GST antibodies.
(C) GST-pulldown of bacterially expressed His-Paf1. The assay was carried out using GST-VP16 and purified His-tagged Paf1. After extensive washing, bound His-Paf1 were eluted and analyzed by western blotting. 10% of input His-Paf1 is also shown in lane 1.
GAL4-VP16 enhances binding of PAF1c to DNA templates
The above results suggest that GAL4-VP16 interacts with the PAF1 complex, perhaps to recruit it to promoter regions and facilitate transcription-coupled polyadenylation. To examine this possibility, recruitment assays using immobilized DNA templates were performed (Johnson et al. 2004). Conditions were the same as for transcription-coupled polyadenylation, except that PEG was omitted to reduce nonspecific protein binding (results not shown). As shown in Figure 6A, in the presence of GAL4-VP16, but not nucleotide triphosphates (NTPs), PAF1c association with the DNA template was significantly stimulated (Fig. 6A, compare lane 1 and 2, upper panels indicated by PAF). Template association of all PAF subunits tested (Cdc73, Paf1, Leo1 and Ctr9) was enhanced by GAL4-VP16 (Fig. 6A; quantitation of multiple experiments shown in right upper panel). In the presence of NTPs, RNAPII was detected in reduced amounts, reflecting its elongation on the template, while GAL4-VP16 remained associated to the DNA (Fig. 6A), consistent with previous results (Yudkovsky et al. 2000; Uhlmann et al. 2007; Ahn et al. 2009). PAF1c subunits were also detected in reduced amounts in the presence of NTPs (Fig. 6A). Considering that PAF1c is a RNAPII associated complex, this suggests that PAF1c dissociates from preinitiaton complex, is integrated into the elongation complex and travels along the DNA template with RNAPII. The enhancement of PAF1c recruitment by GAL4-VP16 depends on the presence of GAL4 binding sites (Fig. 6B; lanes 1, 2, 5, 6), demonstrating that GAL4-VP16 interacts with and recruits PAF1c to the promoter region of the DNA template. Significantly, an intact TATA box was not required for GAL4-VP16 to stimulate binding of PAF1c to the template (Figure S5). This is consistent with the direct interaction between the VP16 activation domain and PAF1c (Fig. 5), and provides evidence that this interaction is sufficient for PAF1c promoter recruitment, and that interactions with the general transcription machinery are not required.
Figure 6. GAL4-VP16 recruits PAF1c to DNA templates.
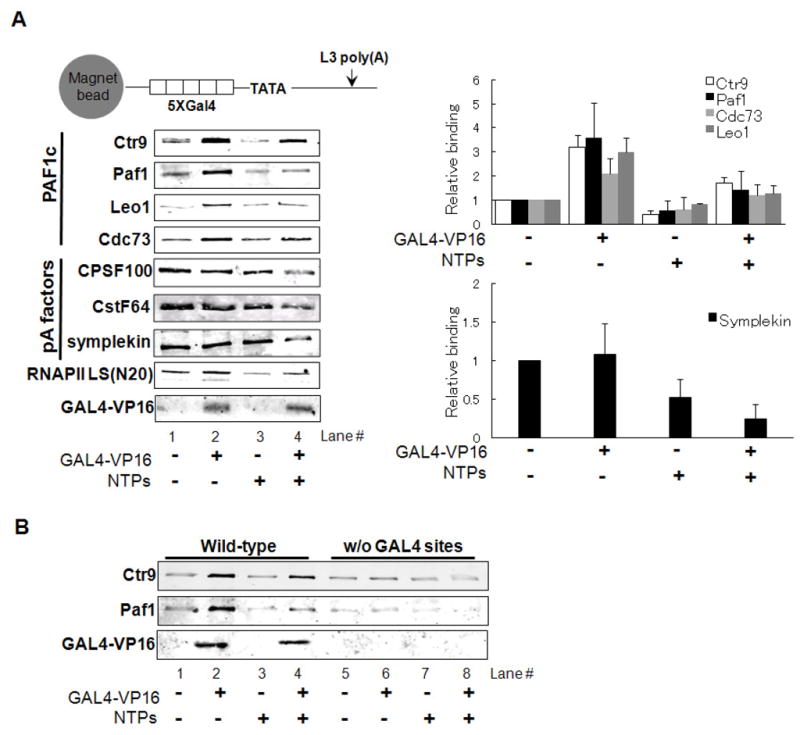
(A) Immobilized template assay to analyze protein recruitment. Biotinylated DNA templates were immobilized using streptoavidin magnetic beads (Invitrogen) and incubated with NE alone or supplemented with GAL4-VP16 in the presence or absence of NTPs as indicated under transcription-coupled polyadenylation conditions. After extensive washing, proteins bound to the templates were eluted and analyzed by western blotting with the indicated antibodies. Protein bands were detected and quantitated with LI-COR Odyssey. Error bars represent standard deviations from three independent experiments.
(B) Immobilized template assay with mutant templates lacking GAL4 binding sites. Assay were done and analyzed as in (A).
We also examined association of several polyadenylation factors with the template. Perhaps unexpectedly, GAL4-VP16 did not enhance binding of the polyadenylation factors tested (Fig. 6A). However, one of the factors tested, symplekin, which functions as a scaffolding protein for 3′ processing (Takagaki and Manley, 2000), was consistently detected in reduced amounts in the presence of GAL4-VP16 and NTPs (Fig. 6A; quantitation in right bottom panel), analogous to the behavior of RNAP II and the PAF1c. Together, these results suggest a possible mechanism by which GAL4-VP16, together with PAF1c, functions to facilitate transcription-coupled 3′ processing (see Discussion).
DISCUSSION
VP16, due to its strong activation domain, has long been used as a model transcription factor (often as a GAL4 fusion protein) to analyze mechanisms of transcription activation in vitro and in vivo (e.g., Sadowski et al. 1988; Triezenberg et al. 1988; Cousens et al. 1989). In this paper, we showed that addition of GAL4-VP16 to nuclear extract stimulates pre-mRNA polyadenylation, but only when the processing reaction is coupled to transcription. Furthermore, immunodepletion and siRNA knockdown experiments revealed that PAF1c is required for GAL4-VP16-induced polyadenylation in vitro and in vivo. The VP16 activation domain interacts directly with a PAF1c subunit and stimulates association of PAF1c with DNA templates, suggesting that GAL4-VP16 enhances polyadenylation by recruiting PAF1c.
How do GAL4-VP16 and PAF1c function in transcription-coupled polyadenylation? We initially postulated that GAL4-VP16 recruits polyadenylation factors directly to the promoter region, but we were unable to obtain any evidence for this. We therefore considered instead the possibility that recruitment might be via PAF1c, based on our earlier findings that PAF1c associates with CPSF, CstF and symplekin (Rozenblatt-Rosen et al. 2009). However, as judged by our immobilized template experiments, recruitment of these factors was not stimulated by GAL4-VP16. These findings were somewhat unexpected, both because this would seem the most straightforward way that GAL4-VP16 could function to influence 3′ processing, and also because previous in vivo experiments had suggested that GAL4-VP16 can recruit CPSF/CstF to promoter regions (Uhlmann et al. 2007). It is possible that this difference reflects differences between in vitro and in vivo conditions, and/or the assay methods employed (immobilized template vs. ChIP, respectively). For example, we showed previously that CPSF can be recruited to promoters in vitro by association with TFIID (Dantonel et al. 1997), and it may be that this interaction predominates in vitro. It is also possible that the association of the polyadenylation factors at the promoter is relatively unstable, and any increase brought about by GAL4-VP16 might be difficult to detect with the immobilized template assay.
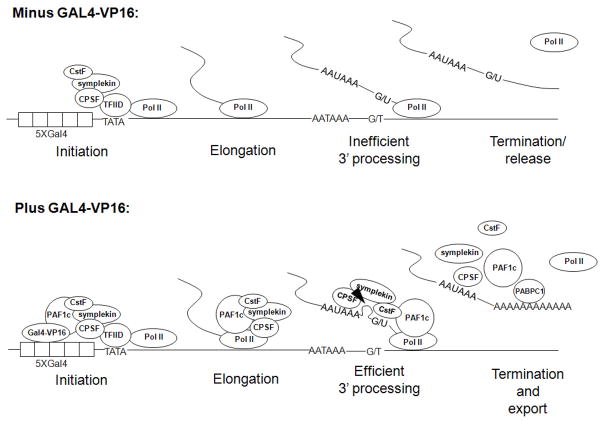
The immobilized template assay did however provide insight into how GAL4-VP16 might function to enhance transcription-coupled polyadenylation. In addition to providing direct evidence that GAL4-VP16 recruits PAF1c to the template, the assay revealed lower levels of template-associated symplekin, like RNAP II, after initiation of GAL4-VP16-activated transcription. As illustrated in the model shown in Figure 7, we suggest that this reflects a change in symplekin such that it travels with RNAP II to facilitate 3′ end formation. Although further work will be required to confirm this, it is consistent with the behavior of RNAP II on immobilized templates when transcription is initiated (Yudkovsky et al. 2000; Uhlmann et al. 2007; Ahn et al. 2009) and with the ability of symplekin to associate with RNAP II (Xiang et al. 2010). Furthermore, we suggest that this involves PAF1c, which is recruited to the promoter, likely by the direct interaction between its Paf1 subunit and GAL4-VP16, and also associates with symplekin. Interestingly, recent studies have established an important role for symplekin specifically in transcription-coupled polyadenylation (Xiang et al. 2010). Symplekin was shown to bind to and stimulate the RNAP II CTD phosphatase Ssu72, and this interaction, and Ssu72 catalytic activity, was found to be necessary for transcription-coupled (but not uncoupled) polyadenylation in vitro. Significantly, these studies also analyzed GAL4-VP16 activated transcription-processing, and it will be interesting to determine whether PAF1c might also function in these interactions.
Figure 7. Model for GAL4-VP16 induced activation of transcription-coupled polyadenylation.
In the absence of GAL4-VP16 (top), certain polyadenylation factors are recruited to the template, such as CPSF by interaction with TFIID. However, under these conditions the factors are not properly integrated into elongation complexes, and subsequent polyadenylation is inefficient. In the presence of GAL4-VP-16 (bottom), PAF1c is recruited to the promoter region, by direct interaction with GAL4-VP16. PAF1c then facilitates proper coordination of poly(A) factors with elongation complexes, which enhances the efficiency of 3′ end formation. Symplekin may play an especially important role in this process (see text). After poly(A) tail formation, PABPC1 is recruited to the poly(A) tail by PAF1c to facilitate mRNA export to the cytoplasm.
PAF1c is known to play a number of roles in the transcription process. Initial studies in yeast provided evidence that the complex could function as an elongation factor (Shi et al. 1996; Costa and Arndt 2000; Krogan et al. 2002; Mueller and Jaehning 2002; Pokholok et al. 2002; Squazzo et al. 2002), and more recent reports have provided evidence that mammalian PAF1c is in fact an elongation factor (Chen et al. 2009; Kim et al. 2010). However, recruitment to the promoter seems to be required for PAF1c to function in elongation, as Chen et al. (2009) observed PAF1c-mediated stimulation of elongation with DNA templates containing the adenovirus MLP promoter, but not with oligo(dC) tailed templates. PAF1c can also likely function in ways that do not involve elongation. For example, PAF1c (Cdc73) has been shown to associate with the promoter region of the CCND1 gene and inhibit its expression by recruiting the histone methylase SUV39H1 to induce repressive H3K9 methylation (Woodard et al. 2005; Yang et al. 2010). Recruitment of PAF1c to the c-Myc gene promoter was also shown to lead to repression of its expression (Lin et al. 2008). Additionally, microarray analysis of Cdc73-depleted HeLa cells identified several hundred genes that were either positively or negatively affected (Rozenblatt-Rosen et al. 2009). The roles of PAF1c in gene expression are thus complex, and presumably determined at least in part by how and if PAF1c is recruited to specific promoters. This suggests that gene-specific transcription factors (TFs) may be important for establishing PAF1c target genes, and our demonstration of a direct interaction between the PAF1c subunit Paf1 and the VP16 activation domain, and that GAL4-VP16 can recruit PAF1c to a DNA template, indicates that this may involve direct interactions between certain TFs and PAF1c.
Our experiments showed that when PAF1c subunits were depleted from 293T cells by siRNA knockdown, not only was 3′ end processing of a reporter transcript affected, but the ratio of mature mRNA in the cytoplasm versus the nucleus was also significantly reduced. This suggests that PAF1c plays a role in mRNA nuclear export in addition to its role in 3′ end processing. In this regard, immunopurified PAF1c associates with the cytoplasmic poly(A) binding protein PABPC1 (our unpublished data), which is thought to function in mRNA nuclear export in yeast and mammals (Brune et al. 2005; Hosoda et al. 2006). PAF1c depletion might cause inefficient PABPC1 recruitment to poly(A) tails and defective nuclear export (Fig. 7). Moreover, in humans the PAF1c subunit Leo1 interacts with Ranbp2, a nuclear pore complex protein (Forler et al. 2004; Rozenblatt-Rosen et al. 2005). Recently, Farber et al. (2010) reported that Cdc73 is required for proper histone mRNA 3′ processing and export to the cytoplasm. Together, these results support the view that PAF1c may have yet an additional function, to facilitate nuclear export of the mRNAs of target genes.
Utilization of alternative polyadenylation (AP) sites is emerging as a widespread mechanism for control of gene expression during differentiation and disease. Several recent studies have provided evidence that rapidly proliferating cells tend to use proximal poly(A) sites, while non-proliferating or slowly growing cells favor distal sites (Sandberg et al. 2008; Ji et al. 2009; Mayr et al. 2009). Little is known about the molecular mechanisms responsible for these changes in poly(A) site choice. We showed previously that changes in the levels of CstF-64 influence AP in the IgM heavy chain pre-mRNA (Takagaki et al. 1996; Takagaki and Manley 1998), and it may be that changes in the levels or activity of polyadenylation factors provides one mechanism for regulating AP. We suggest that the increased polyadenylation efficiency brought about by transcriptional activators as described here may provide another, such that the increased efficiency would favor proximal poly(A) sites. Indeed, a recent bioinformatics study provides support for this idea: Up-regulated genes tend to use promoter-proximal poly(A) sites, while down-regulated ones prefer distal poly(A) sites (B. Tian, personal communication). Additionally, PAF complex mutants in yeast are known to alter poly(A) site utilization for certain genes (Penheiter et al. 2005), while in humans siRNA-mediated depletion of Cdc73 was found to reduce expression and extend transcription of a target gene, Ints6, potentially favoring utilization of downstream poly(A) sites (Rozenblatt-Rosen et al. 2009). Therefore, it will be of considerable interest to determine if the increase in 3′ processing efficiency brought about by GAL4-VP16, mediated by PAF1c, that we described here has the potential to influence utilization of alternative polyadenylation sites.
EXPERIMENTAL PROCEDURES
DNA templates
The DNA template used for transcription-coupled polyadenylation assays contains GAL4 binding sites upstream of E4 core promoter and SVL or L3 poly(A) site downstream. The DNA template for reporter assay (pGL3G5E4) was constructed by inserting the region containing five tandem GAL4 binding sites and the E4 core promoter (G5E4) into the pGL3 plasmid (Promega). pG5E4SVL was made by deleting the luciferase gene from pGL3G5E4. pG3G5E4L3 was constructed by inserting the G5E4 region into pG3L3 (Takagaki et al. 1988). Mutant templates were made by PCR-based mutagenesis.
In vitro transcription-coupled polyadenylation assay
Transcription-coupled polyadenylation was carried out at 30°C for 1h in reaction mixtures (20μl) containing 10μl nuclear extract (Dignam et al. 1983), 100ng of Gal4-VP16, 12mM HEPES (pH 7.9), 500ng of the DNA templates (pG5E4SVL or pG3G5E4L3), 0.5mM each of ATP, GTP and CTP, 15μM cold UTP, 10μCi of [α-32P]UTP, 4mM MgCl2, 20mM creatine phosphate (di-tris), 2.4% PEG8000, 12% glycerol, 60mM KCl, 0.12mM EDTA, 0.12mM DTT and 0.3mM PMSF. The reaction was stopped by adding proteinase K. RNA products were separated into nonpolyadenylated and polyadenylated fractions by oligo(dT) selection, thereafter nonpolyadenylated and polyadenylated fractions were analyzed on 5% denaturing gel. Radioactivity was detected using a phosphorimager and quantified by NIH ImageJ (http://rsb.info.nih.gov/ij/index.html).
RNase protection assays
RNA probes were prepared by T7 transcription with pG3SVL (Takagaki et al. 1988) linearized with SalI. RNase protection was carried out according to methods described previously (Gilman 2001).
Antibodies
Antibodies for PAF1c subunits (Cdc73, Paf1, Leo1 and Ctr9) and anti-Symplekin were from Bethyl Laboratories. Anti-CPSF100 and anti-CstF64 antibodies were generated in our lab (Takagaki et al. 1990). Anti-His and anti-Pol II (N20) were purchased from Santa Cruz. Anti-actin and anti-GST were obtained from Sigma and GenScript, respectively.
Recombinant proteins
Recombinant His-tagged GAL4-VP16, GAL4-p53 and Paf1 were prepared by bacterial overexpression and purified with Talon resin (Clontech). Purification of PAF1c from 293T cells was carried out as follows. pIRES-puro vector containing Flag-tagged Paf1 sequence was transfected into 293T cells. Two weeks after selection with puromycin, positive clones were screened and expanded to larger cell culture. NE was prepared, incubated overnight with 200μl of M2 agarose beads (Sigma) equilibrated with buffer C (Dignam et al. 1983), and PAF1c was eluted with 400 μg/ml Flag peptide (Sigma) as described (Zhu et al. 2005a).
siRNA experiments
293T cells were transfected with siRNA targeting GFP (5′-gcgugaucuucaccgacaa-3′) or PAF1c subunits Cdc73 (5′-cagcgaucuacucaagucaaa-3′), Paf1 (5′-aagcagcagtttaccgaggaa-3′) and Ctr9 (5′-gcacguauagauggcaauu-3′) by using Lipofectamine 2000 (Invitrogen). 48h after transfection, cells were splitted to a new plate and second transfection was done with the same siRNA. NE was prepared from each transfectant 96h after the first transfection (Lee and Green 1990). For reporter assays, reporter plasmid pGL3G5E4 and GAL4-VP16 expression plasmid were transfected 48h after siRNA transfection. Total RNAs were harvested 24h after transfection of the reporter plasmid and cells fractionated into cytoplasmic and nuclear fractions (Wang et al. 2006). 3′ end cleavage was analyzed by RNase protection assays.
GST-pulldown assays
NE (100 μl) was diluted with buffer D (400 μl) and NP-40 was added to 0.1%. Then, 5 μg of purified GST or GST-VP16 was incubated overnight with the diluted NE and Glutathione agarose beads (20 μl) which is equilibrated with buffer D (Dignam et al. 1983). After extensive washing with buffer D without NP-40, bound proteins were eluted by boiling and subjected to western analysis.
Far-western analysis
Purified PAF1c was resolved on 8% SDS-PAGE and transferred to a nitrocellulose membrane. After denaturating-renaturation using guanidine hydrochloride, the membrane was rinsed with PBST buffer containing 0.05% Tween and blocked overnight with the same buffer with 5% nonfat milk. The membrane was then probed with 2 μg of GST or GST-VP16 in binding buffer (20mM Tris (pH 7.9), 100mM NaCl, 0.1% Tween, 10% glycerol, 1mM MgCl2, 0.1mM ZnSO4, 1mM PMSF), and then subjected to western analysis using anti-GST antibodies.
Immobilized template assay
Immobilized template assays were performed according to a procedure described previously (Johnson et al. 2004). NE was incubated with the immobilized template under the same conditions as for transcription-coupled polyadenylation assays, except that PEG was omitted from reaction mixtures. After extensive washing with the same buffer, proteins bound to the templates were analyzed by western blotting. Protein bands were detected and quantitated using LI-COR Odyssey.
Supplementary Material
Acknowledgments
We thank Nishta Rao for preparing HeLa nuclear extract. We are grateful to Dr. Jerry Workman and Dr. Russ Carstens for providing 5SG5E4 plasmid and pIRES-puro3 plasmid, respectively. We also thank Dr. Bin Tian for communicating results prior to publication. This study was supported by NIH grant R01 GM28983 to JLM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahn SH, Keogh MC, Buratowski S. Ctk1 promotes dissociation of basal transcription factors from elongating RNA polymerase II. EMBO J. 2009;28:205–212. doi: 10.1038/emboj.2008.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D. The mRNA assembly line: transcription and processing machines in the same factory. Curr Opin Cell Biol. 2002;14:336–342. doi: 10.1016/s0955-0674(02)00333-2. [DOI] [PubMed] [Google Scholar]
- Brune C, Munchel SE, Fischer N, Podtelejnikov AV, Weis K. Yeast poly(A)-binding protein Pab1 shuttles between the nucleus and the cytoplasm and functions in mRNA export. RNA. 2005;11:517–531. doi: 10.1261/rna.7291205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo O, Manley JL. The transcriptional coactivator PC4/Sub1 has multiple functions in RNA polymerase II transcription. EMBO J. 2005;24:1009–1020. doi: 10.1038/sj.emboj.7600575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Yamaguchi Y, Tsugeno Y, Yamamoto J, Yamada T, Nakamura M, Hisatake K, Handa H. DSIF, the Paf1 complex, and Tat-SF1 have nonredundant, cooperative roles in RNA polymerase II elongation. Genes Dev. 2009;23:2765–2777. doi: 10.1101/gad.1834709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa PJ, Arndt KM. Synthetic lethal interactions suggest a role for the Saccharomyces cerevisiae Rtf1 protein in transcription elongation. Genetics. 2000;156:535–547. doi: 10.1093/genetics/156.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousens DJ, Greaves R, Goding CR, O’Hare P. The C-terminal 79 amino acids of the herpes simplex virus regulatory protein, Vmw65, efficiently activate transcription in yeast and mammalian cells in chimeric DNA-binding proteins. EMBO J. 1989;8:2337–2342. doi: 10.1002/j.1460-2075.1989.tb08361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantonel JC, Murthy KG, Manley JL, Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber LJ, Kort EJ, Wang P, Chen J, Teh BT. The tumor suppressor parafibromin is required for posttranscriptional processing of histone mRNA. Mol Carcinog. 2010;49:215–223. doi: 10.1002/mc.20591. [DOI] [PubMed] [Google Scholar]
- Forler D, Rabut G, Ciccarelli FD, Herold A, Kocher T, Niggeweg R, Bork P, Ellenberg J, Izaurralde E. RanBP2/Nup358 provides a major binding site for NXF1-p15 dimers at the nuclear pore complex and functions in nuclear mRNA export. Mol Cell Biol. 2004;24:1155–1167. doi: 10.1128/MCB.24.3.1155-1167.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman M. Ribonuclease protection assay. Curr Protoc Mol Biol. 2001;Chapter 4(Unit 4):7. doi: 10.1002/0471142727.mb0407s24. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- Hirose Y, Tacke R, Manley JL. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev. 1999;13:1234–1239. doi: 10.1101/gad.13.10.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoda N, Lejeune F, Maquat LE. Evidence that poly(A) binding protein C1 binds nuclear pre-mRNA poly(A) tails. Mol Cell Biol. 2006;26:3085–3097. doi: 10.1128/MCB.26.8.3085-3097.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci U S A. 2009;106:7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KM, Wang J, Smallwood A, Carey M. The immobilized template assay for measuring cooperativity in eukaryotic transcription complex assembly. Methods Enzymol. 2004;380:207–219. doi: 10.1016/S0076-6879(04)80010-7. [DOI] [PubMed] [Google Scholar]
- Kaneko S, Rozenblatt-Rosen O, Meyerson M, Manley JL. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3′ processing and transcription termination. Genes Dev. 2007;21:1779–1789. doi: 10.1101/gad.1565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guermah M, Roeder RG. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell. 2010;140:491–503. doi: 10.1016/j.cell.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, Greenblatt JF, Shilatifard A. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell. 2003a;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol. 2002;22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, Shilatifard A, Buratowski S, Greenblatt J. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2003b;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KA, Green MR. Small-scale preparation of extracts from radiolabeled cells efficient in pre-mRNA splicing. Methods Enzymol. 1990;181:20–30. doi: 10.1016/0076-6879(90)81108-7. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol Cell. 2002;9:1101–1111. doi: 10.1016/s1097-2765(02)00518-x. [DOI] [PubMed] [Google Scholar]
- Lin L, Zhang JH, Panicker LM, Simonds WF. The parafibromin tumor suppressor protein inhibits cell proliferation by repression of the c-myc proto-oncogene. Proc Natl Acad Sci U S A. 2008;105:17420–17425. doi: 10.1073/pnas.0710725105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley DL. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997a;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997b;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- Mueller CL, Jaehning JA. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol Cell Biol. 2002;22:1971–1980. doi: 10.1128/MCB.22.7.1971-1980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller CL, Porter SE, Hoffman MG, Jaehning JA. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol Cell. 2004;14:447–456. doi: 10.1016/s1097-2765(04)00257-6. [DOI] [PubMed] [Google Scholar]
- Muntean AG, Tan J, Sitwala K, Huang Y, Bronstein J, Connelly JA, Basrur V, Elenitoba-Johnson KS, Hess JL. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell. 2010;17:609–621. doi: 10.1016/j.ccr.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, Dubois G, Mazo A, Croce CM, Canaani E. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10:1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- Nordick K, Hoffman MG, Betz JL, Jaehning JA. Direct interactions between the Paf1 complex and a cleavage and polyadenylation factor are revealed by dissociation of Paf1 from RNA polymerase II. Eukaryot Cell. 2008;7:1158–1167. doi: 10.1128/EC.00434-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazin MJ, Hermann JW, Kadonaga JT. Promoter structure and transcriptional activation with chromatin templates assembled in vitro. A single Gal4-VP16 dimer binds to chromatin or to DNA with comparable affinity. J Biol Chem. 1998;273:34653–34660. doi: 10.1074/jbc.273.51.34653. [DOI] [PubMed] [Google Scholar]
- Penheiter KL, Washburn TM, Porter SE, Hoffman MG, Jaehning JA. A posttranscriptional role for the yeast Paf1-RNA polymerase II complex is revealed by identification of primary targets. Mol Cell. 2005;20:213–223. doi: 10.1016/j.molcel.2005.08.023. [DOI] [PubMed] [Google Scholar]
- Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Hannett NM, Young RA. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol Cell. 2002;9:799–809. doi: 10.1016/s1097-2765(02)00502-6. [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- Rosonina E, Bakowski MA, McCracken S, Blencowe BJ. Transcriptional activators control splicing and 3′-end cleavage levels. J Biol Chem. 2003;278:43034–43040. doi: 10.1074/jbc.M307289200. [DOI] [PubMed] [Google Scholar]
- Rosonina E, Ip JY, Calarco JA, Bakowski MA, Emili A, McCracken S, Tucker P, Ingles CJ, Blencowe BJ. Role for PSF in mediating transcriptional activator-dependent stimulation of pre-mRNA processing in vivo. Mol Cell Biol. 2005;25:6734–6746. doi: 10.1128/MCB.25.15.6734-6746.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Hughes CM, Nannepaga SJ, Shanmugam KS, Copeland TD, Guszczynski T, Resau JH, Meyerson M. The parafibromin tumor suppressor protein is part of a human Paf1 complex. Mol Cell Biol. 2005;25:612–620. doi: 10.1128/MCB.25.2.612-620.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Nagaike T, Francis JM, Kaneko S, Glatt KA, Hughes CM, LaFramboise T, Manley JL, Meyerson M. The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3′ mRNA processing factors. Proc Natl Acad Sci U S A. 2009;106:755–760. doi: 10.1073/pnas.0812023106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Finkelstein A, Wolf AJ, Wade PA, Burton ZF, Jaehning JA. Paf1p, an RNA polymerase II-associated factor in Saccharomyces cerevisiae, may have both positive and negative roles in transcription. Mol Cell Biol. 1996;16:669–676. doi: 10.1128/mcb.16.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squazzo SL, Costa PJ, Lindstrom DL, Kumer KE, Simic R, Jennings JL, Link AJ, Arndt KM, Hartzog GA. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 2002;21:1764–1774. doi: 10.1093/emboj/21.7.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Ryner LC, Manley JL. Separation and characterization of a poly(A) polymerase and a cleavage/specificity factor required for pre-mRNA polyadenylation. Cell. 1988;52:731–742. doi: 10.1016/0092-8674(88)90411-4. [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL, MacDonald CC, Wilusz J, Shenk T. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes Dev. 1990;4:2112–2120. doi: 10.1101/gad.4.12a.2112. [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Seipelt RL, Peterson ML, Manley JL. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87:941–952. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol Cell. 1998;2:761–771. doi: 10.1016/s1097-2765(00)80291-9. [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol. 2000;20:1515–1525. doi: 10.1128/mcb.20.5.1515-1525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triezenberg SJ, Kingsbury RC, McKnight SL. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 1988;2:718–729. doi: 10.1101/gad.2.6.718. [DOI] [PubMed] [Google Scholar]
- Uhlmann T, Boeing S, Lehmbacher M, Meisterernst M. The VP16 activation domain establishes an active mediator lacking CDK8 in vivo. J Biol Chem. 2007;282:2163–2173. doi: 10.1074/jbc.M608451200. [DOI] [PubMed] [Google Scholar]
- Venkataraman K, Brown KM, Gilmartin GM. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 2005;19:1315–1327. doi: 10.1101/gad.1298605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhu W, Levy DE. Nuclear and cytoplasmic mRNA quantification by SYBR green based real-time RT-PCR. Methods. 2006;39:356–362. doi: 10.1016/j.ymeth.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Woodard GE, Lin L, Zhang JH, Agarwal SK, Marx SJ, Simonds WF. Parafibromin, product of the hyperparathyroidism-jaw tumor syndrome gene HRPT2, regulates cyclin D1/PRAD1 expression. Oncogene. 2005;24:1272–1276. doi: 10.1038/sj.onc.1208274. [DOI] [PubMed] [Google Scholar]
- Xiang K, Nagaike T, Xiang S, Kilic T, Beh MM, Manley JL, Tong L. Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature. 2010 doi: 10.1038/nature09391. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YJ, Han JW, Youn HD, Cho EJ. The tumor suppressor, parafibromin, mediates histone H3 K9 methylation for cyclin D1 repression. Nucleic Acids Res. 2010;38:382–390. doi: 10.1093/nar/gkp991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yart A, Gstaiger M, Wirbelauer C, Pecnik M, Anastasiou D, Hess D, Krek W. The HRPT2 tumor suppressor gene product parafibromin associates with human PAF1 and RNA polymerase II. Mol Cell Biol. 2005;25:5052–5060. doi: 10.1128/MCB.25.12.5052-5060.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudkovsky N, Ranish JA, Hahn S. A transcription reinitiation intermediate that is stabilized by activator. Nature. 2000;408:225–229. doi: 10.1038/35041603. [DOI] [PubMed] [Google Scholar]
- Zhu B, Mandal SS, Pham AD, Zheng Y, Erdjument-Bromage H, Batra SK, Tempst P, Reinberg D. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev. 2005a;19:1668–1673. doi: 10.1101/gad.1292105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell. 2005b;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.