Abstract
The history of tomato (Solanum lycopersicum L.) improvement includes genetic bottlenecks, wild species introgressions, and divergence into distinct market classes. This history makes tomato an excellent model to investigate the effects of selection on genome variation. A combination of linkage mapping in two F2 populations and physical mapping with emerging genome sequence data was used to position 434 PCR-based markers including SNPs. Three-hundred-and-forty markers were used to genotype 102 tomato lines representing wild species, landraces, vintage cultivars, and contemporary (fresh market and processing) varieties. Principal component analysis confirmed genetic divergence between market classes of cultivated tomato (P <0.0001). A genome-wide survey indicated that linkage disequilibrium (LD) decays over 6–8 cM when all cultivated tomatoes, including vintage and contemporary, were considered together. Within contemporary processing varieties, LD decayed over 6–14 cM, and decay was over 3–16 cM within fresh market varieties. Significant inter-chromosomal (gametic phase) LD was detected in both fresh market and processing varieties between chromosomes 2 and 3, and 2 and 4, but in distinct chromosomal locations for each market class. Additional LD was detected between chromosomes 3 and 4, 3 and 11, and 4 and 6 in fresh market varieties and chromosomes 3 and 12 in processing varieties. These results suggest that breeding practices for market specialization in tomato have led to a genetic divergence between fresh market and processing types.
Keywords: Breeding, domestication, gametic phase, inter-chromosomal, selection
Introduction
The process of domestication and breeding has led to dramatic changes in the reproduction and morphology of crop species. The selection of individuals with favourable characteristics such as non-shattering seed pods, loss of germination inhibition in seeds, increased size of fruit, and compact plant habit has converted feral plants into forms amenable to cultivation (Tanksley and McCouch, 1997; Gepts, 2004; Doebley et al., 2006). These alterations in phenotype were the direct result of genetic changes underlying traits of interest to humans.
The effect of domestication and breeding on the genes and genomes of crop plants can be assessed using a range of approaches including linkage mapping and map-based cloning. As an alternative to analysis in controlled crosses, association mapping in unstructured and complex populations is now being applied to crops (Remington et al., 2001; Breseghello and Sorrells, 2006; Casa et al., 2008). In addition, the increased efficiency and accessibility of sequencing permits the application of advances in molecular evolution theory to detect the effects of artificial selection on genes and gene systems. Population level studies have been used to identify pathways (Whitt et al., 2002) and genes (Wang et al., 1999; Clark et al., 2004; Yamasaki et al., 2005) that were under selection during domestication and improvement. These studies are guided by observing signatures of selection in sequence data, including a reduction in diversity in cultivated germplasm relative to wild relatives, a reduction in diversity relative to control genes (neutral genes), and an excess of rare variants due to new mutations (Doebley, 2004). In addition to their value for identifying genes that were fixed during domestication, these approaches have the potential to identify the genes that explain existing phenotypic variation within breeding programmes.
Tomato (Solanum lycopersicum L.) has been a model for studying genes that distinguish domestic and wild plants. Mapping in wide crosses and the cloning of genes that affect specific traits has produced substantial insight into disease resistance, plant and fruit development, and specific biochemical pathways (Martin et al., 1993; Jones et al., 1994; Pnueli et al., 1998; Frary et al., 2000; Spassova et al., 2001). In species like tomato, fruit morphology is one of the major traits selected, and cultivated forms exhibit far greater phenotypic variation than their wild progenitors (Tanksley, 2004; Paran and van der Knaap, 2007). It is unlikely that allelic variation present in wild ancestors will explain all of the morphological changes that separate landraces, vintage cultivars or modern crop varieties from their wild relatives. For example, mutations of fruit shape genes (e.g. ovate and sun) have led to a high level of phenotypic variation (Liu et al., 2002; Xiao et al., 2008). In the case of ovate, this variation is found in wild progenitors (Tanksley, 2004) while sun originated as a gain-of-function mutation post-domestication (Xiao et al., 2008). Plant breeding balances the competing goals of introducing new variation, and selecting for specific alleles. Selection for the optimum alleles creates two problems. First, heritability declines as genetic variation declines. Thus, breeding progress will be limited as alleles are fixed throughout the genome. Second, fixation of favourable alleles at some loci may inadvertently fix undesirable genes that are linked. For example, linkage group 6 of cultivated sunflower (Helianthus annuus L.) contains several domestication-related loci, some of which provide positive effects, while others provide antagonistic effects relative to desired traits (Burke et al., 2005). Reintroduction of genetic diversity through wide crosses has been practised in cultivated tomato for nearly a century (Williams and St Clair, 1993; Sim et al., 2009). Practices that seek to introduce new variation may have negative consequences, such as the introduction of less favourable alleles and a restriction of recombination in some genomic regions. Introgression has been effective at introducing disease resistance not found in cultivated material (Francis et al., 2001; Kabelka et al., 2002), but has had mixed results with respect to fruit quality (Kabelka et al., 2004). A five-fold reduction in recombination has been documented in the region around the root-knot nematode resistance gene (Mi), which was introgressed from the wild species S. peruvianum (Messeguer et al., 1991). Thus, introgression of a trait may also lead to the inheritance of large linkage blocks associated with that trait. A major goal of marker-assisted breeding programmes is to be able to select for favourable combinations of genes, across genomes and within chromosomes (Frisch et al., 1999). Accomplishing this goal and balancing the competing demands of increasing genetic diversity while selecting desirable alleles will benefit from a description of genetic variation across the genome of breeding populations.
Several strategies have been employed to develop molecular resources for genome-wide analyses within tomato breeding germplasm. Although tomato was one of the first crops to have a saturated genetic linkage map (Tanksley et al., 1992), the nearly exclusive focus on wide crosses has left a paucity of genetic tools for investigating diversity within cultivated lineages. High-throughput markers remain a limited resource, since many markers selected based on polymorphisms in wide crosses are not polymorphic within cultivated germplasm (Jimenez-Gomez and Maloof, 2009). To overcome this limitation, several projects have identified genetic differences including simple sequence repeats (SSRs), insertion/deletion (indel), and single nucleotide polymorphisms (SNPs) among tomato varieties. Analysis of databases developed through large-scale sequencing of tomato ESTs resulted in the identification of approximately 609 potential simple sequence repeats (SSRs; Frary et al., 2005). Of these, 127 were mapped in the cultivated×wild (S. lycopersicum×S. pennellii) reference population, and 61 were polymorphic within cultivated tomato (Frary et al., 2005). Parallel strategies to develop high-throughput markers include in silico mining of SNPs from EST databases (Yang et al., 2004; Labate and Baldo, 2005), oligo-based microarray hybridization (Sim et al., 2009), and sequencing introns of conserved orthologous set (COS) genes (Van Deynze et al., 2007; Labate et al., 2009b). Since many of the SNPs from these studies have been validated in genotyping assays and show polymorphism within cultivated tomato, these marker resources provide an opportunity to assess cultivated germplasm genetically.
In order to organize these resources for the analysis of cultivated populations, a genetic map was developed based on 434 markers. Allele-specific primer extension (ASPE; Lee et al., 2004) markers were created based on previously identified SNPs and these were combined with existing framework RFLP markers, PCR-based SSR markers, and indel markers to develop an integrated linkage map based on two populations. This linkage map was combined with emerging sequence data for the tomato genome to organize markers relative to the tomato physical map. These markers have been used to genotype a collection of 93 S. lycopersicum accessions and nine wild species accessions. The resulting data were used to assess the extent of inter- and intra-chromosomal linkage disequilibrium (LD) in cultivated tomato. Given the history of tomato breeding, which includes introgression from wild species and breeding for distinct market specialization, we expected to identify differences in the pattern and distribution of genetic variation within the genomes of cultivated tomatoes representing different market classes. Specifically, the hypothesis that selection for market differentiation left a signature that could be detected through the analysis of genome-wide patterns of SNP variation was tested.
Materials and methods
Plant material
A set of 102 tomato accessions was assembled, including nine representatives of wild species, five Latin American cultivars, two unimproved breeding lines, 21 vintage cultivars, two greenhouse varieties, 24 fresh market varieties, and 39 processing varieties (Table 1; see Supplementary Table S1 at JXB online). The Latin American cultivars represent early domesticates while the vintage cultivars represent early tomato improvement. Fresh market and processing germplasm are varieties that are adapted to specific market niches and represent improvements made through contemporary plant breeding. These entries were selected from public breeding programmes that release commercially relevant parents and hybrids. Several processing lines were donated directly by seed companies. In addition, selected inbred lines were obtained through self-pollination of commercial hybrids followed by single-seed-descent selection to obtain inbred lines. These selections represent a sample of the alleles present in commercial hybrids, although they do not recreate the parents themselves. Also included were the parents of several important recombinant inbred and inbred backcross populations (Doganlar et al., 2002; Kabelka et al., 2002; Graham et al., 2004; Yang et al., 2005; Robbins et al., 2009). The collection also contained parents of populations utilized by the tomato research community such as segmental substitution lines (M82 and LA716; Eshed and Zamir, 1995) and a mutation library (Menda et al., 2004). Although a few wild tomato species were included in the collection, the focus was on cultivated materials so that the information gained may be directly applicable to tomato breeding programmes.
Table 1.
Number of markers polymorphic within different classes of tomato
| No. of entriesa | indel | SNP | SSR | Total | |
| Processing | 39 | 27 (22)b | 104 (64) | 39 (27) | 170 (113) |
| Fresh market | 24 | 22 (16) | 101 (62) | 38 (26) | 161 (104) |
| Vintage cultivars | 21 | 22 (16) | 51 (34) | 33 (22) | 106 (72) |
| Latin American cultivars | 5 | 18 (13) | 57 (38) | 42 (28) | 117 (79) |
| All S. lycopersicumc | 93 | 44 (34) | 154 (96) | 52 (37) | 250 (167) |
| Wild species | 9 | 63 (52) | 167 (117) | 65 (50) | 295 (219) |
| All entries | 102 | 70 (57) | 205 (135) | 65 (50) | 340 (242) |
The number of entries within each class.
The number in parentheses indicates the number of polymorphic markers with known genomic location either by either linkage or physical mapping.
All S. lycopersicum represents cultivated tomato and includes processing, fresh market, vintage, Latin American, and greenhouse cultivars as well as unimproved breeding lines.
The germplasm collection also contained the parents of two F2 mapping populations utilized to develop genetic linkage maps. The mapping population derived from Sun1642 (S. lycopersicum) and LA1589 (S. pimpinellifolium) consists of 100 F2 individuals (van der Knaap and Tanksley, 2001). The second mapping population consists of 200 F2 plants from a cross between Yellow Stuffer and LA1589 (van der Knaap and Tanksley, 2003).
Molecular marker genotyping
Markers used in this study are from various sources and are described in Supplementary Tables S2–S5 at JXB online. Framework markers (RFLP and SSR) used in map construction were from SGN (http://solgenomics.net). Additional SSRs with the prefix ‘TOM’ (Suliman-Pollatschek et al., 2002) were utilized (see Supplementary Table S5 at JXB online). Markers with the prefix ‘LEOH’ were developed based on SNPs or indels in EST sequences [Yang et al., 2004 (LEOH1-LEOH51), Francis et al., 2005 (LEOH100-LEOH365); see Supplementary Tables S2–S4 at JXB online]. Markers with the prefix ‘SL’ were developed based on SNPs and indels identified by Van Deynze et al. (2007; see Supplementary Tables S2 and S4 at JXB online). These ‘SL’ marker names correspond to the primers that amplify the locus followed by a number referring to the position of the polymorphism within the locus according to Van Deynze et al. (2007). The ‘SL’ markers spanning indels contain the suffix ‘i’ while all others are based on SNPs.
Genotyping was performed on two platforms, one for size polymorphisms (SSR, indel, and CAPS; see Supplementary Tables S3–S5 at JXB online) and a second for SNPs detected by an allele-specific primer extension (ASPE) assay (Lee et al., 2004) on the Luminex 200 system (Luminex Corporation, Austin, TX; see Supplementary Table S2 at JXB online). For markers based on indels, primers flanking the indel were designed using Primer3 (Rozen and Skaletsky, 2000). Size polymorphisms were detected using polyacrylamide gels on the Li-Cor-IR2 4200 system (Li-Cor Biosciences, Lincoln, NE) or agarose gels. To detect SNPs by ASPE (see Supplementary Table S2 at JXB online), allele specific primers were designed for each allele using Primo SNP 3.4 (Chang Bioscience; www.changbioscience.com/primo/primosnp.html) or BatchPrimer3 (You et al., 2008). SNP markers were then scored using the Luminex 200 system.
In order to determine marker genotypes, genomic DNA was isolated following the modified CTAB method described by Kabelka et al. (2002) and subjected to PCR. Conditions for PCR reactions were 10 mM TRIS-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 50 μM of each dNTP, 0.1 μM of each forward and reverse primers, 20 ng of template DNA, and 1 unit of Taq DNA polymerase in a total volume of 10–20 μl. To visualize PCR fragments on the Li-Cor system, an additional 0.1 nM of IRD 700 or 800 dye-labelled M-13 forward primer (Li-Cor Biosciences, Lincoln, NE) was added to the PCR reaction and one of the forward or reverse primers contained the M13 sequence as a tail on the 5' end. PCR amplification was performed following Sim et al. (2009) at a suitable annealing temperature between 45 °C and 60 °C (see Supplementary Tables S2–S5 at JXB online). Markers detected as a cleaved amplified polymorphic sequence (CAPS) were digested after PCR following Yang et al. (2004). For the ASPE assay, the locus was amplified using the primers and PCR conditions developed by Van Deynze et al. (2007). The PCR products were ethanol precipitated then rehydrated in 8 μl ddH2O. After this purification, 4 μl were used as a template in 10–15 μl ASPE reactions that included 1.25 mM MgCl2, 5 μM each of dATP, dGTP, and dTTP, 5 μM biotin-14-dCTP (Invitrogen Corporation, Carlsbad, CA), 25 nm of each ASPE primer, and 1 U of Platinum GenoType Tsp DNA Polymerase (Invitrogen Corporation, Carlsbad, CA) in 1× supplied buffer. Cycling conditions for the ASPE reactions were 2 min at 96 °C followed by 30 cycles of 30 s at 94 °C, 1 min at 55 °C, and 2 min at 74 °C.
Mapping markers
Linkage maps were developed for the Yellow Stuffer×LA1589 and Sun1642×LA1589 populations separately, then the two maps were combined chromosome by chromosome into an integrated map (Table 2; see Supplementary Table S6 at JXB online; Fig. 1) using Joinmap 3.0 (Van Ooijen and Voorrips, 2001). For all map construction, the thresholds for parameters within JoinMap were 1.00 for LOD, 0.4 for REC, 5.0 for jump, and 1 for ripple while employing the Kosambi mapping function.
Table 2.
Number of markers and size in cM of each chromosome in two linkage maps (Sun1642×LA1589 and Yellow Stuffer×LA1589) and the integrated map
| Chromosome | Framework markers | SNP and indel markers | Total markers | PCR-based markers | Markers with segregation distortion | Average cM between markers | Largest gap (cM) | Genome coverage (%)a | Total cM |
| Sun1642×LA1589 | |||||||||
| 1 | 25 | 16 | 41 | 29 | 6 | 3.4 | 13.4 | 100.0 | 135.0 |
| 2 | 17 | 13 | 30 | 20 | 6 | 1.9 | 9.6 | 99.2 | 55.7 |
| 3 | 15 | 18 | 33 | 22 | 5 | 3.4 | 11.3 | 78.4 | 108.7 |
| 4 | 13 | 12 | 25 | 17 | 0 | 4.8 | 21.4 | 86.3 | 114.1 |
| 5 | 11 | 7 | 18 | 10 | 2 | 5.5 | 19.6 | 94.1 | 94.3 |
| 6 | 8 | 9 | 17 | 10 | 8 | 5.2 | 12.7 | 100.0 | 83.2 |
| 7 | 10 | 6 | 16 | 9 | 5 | 6.1 | 17.3 | 98.3 | 91.6 |
| 8 | 12 | 7 | 19 | 11 | 0 | 4.6 | 11.4 | 97.1 | 82.4 |
| 9 | 12 | 5 | 17 | 9 | 0 | 5.2 | 17.1 | 87.7 | 83.3 |
| 10 | 12 | 11 | 23 | 15 | 4 | 4.2 | 11.8 | 100.0 | 93.1 |
| 11 | 9 | 6 | 15 | 8 | 9 | 7.1 | 14.7 | 100.0 | 99.5 |
| 12 | 9 | 9 | 18 | 11 | 10 | 5.0 | 11.9 | 87.1 | 85.8 |
| Total | 153 | 119 | 272 | 171 | 55 | 4.3 | 21.4 | 93.8 | 1126.7 |
| Yellow Stuffer×LA1589 | |||||||||
| 1 | 10 | 6 | 16 | 6 | 3 | 7.7 | 15.5 | 95.2 | 115.8 |
| 2 | 10 | 7 | 17 | 7 | 8 | 5.8 | 10.9 | 99.3 | 93.4 |
| 3 | 9 | 10 | 19 | 10 | 2 | 5.4 | 12.8 | 79.9 | 97.5 |
| 4 | 6 | 4 | 10 | 4 | 1 | 12.8 | 26.1 | 74.4 | 114.8 |
| 5 | 8 | 7 | 15 | 7 | 0 | 6.3 | 30.8 | 86.3 | 88.3 |
| 6 | 7 | 3 | 10 | 3 | 0 | 9.0 | 21.7 | 61.5 | 80.9 |
| 7 | 4 | 3 | 7 | 3 | 7 | 8.8 | 23.5 | 41.3 | 52.6 |
| 8 | 8 | 5 | 13 | 5 | 1 | 5.5 | 19.7 | 51.7 | 66.3 |
| 9 | 7 | 4 | 11 | 4 | 4 | 9.5 | 19.5 | 96.6 | 95.3 |
| 10 | 6 | 4 | 10 | 4 | 0 | 9.8 | 33.2 | 47.9 | 88.4 |
| 11 | 7 | 6 | 13 | 6 | 9 | 7.1 | 17.6 | 71.7 | 84.9 |
| 12 | 8 | 1 | 9 | 1 | 0 | 11.7 | 20.7 | 29.3 | 93.5 |
| Total | 90 | 60 | 150 | 60 | 35 | 7.8 | 33.2 | 71.9 | 1071.6 |
| Integrated | |||||||||
| 1 | 26 | 22 | 48 | 35 | – | 2.9 | 14.7 | 100.0 | 137.2 |
| 2 | 19 | 20 | 39 | 27 | – | 2.8 | 11.4 | 98.7 | 105.2 |
| 3 | 15 | 29 | 44 | 33 | – | 2.4 | 9.1 | 96.4 | 99.2 |
| 4 | 13 | 16 | 29 | 21 | – | 4.0 | 16.9 | 100.0 | 107.0 |
| 5 | 12 | 14 | 26 | 17 | – | 3.5 | 15.9 | 95.0 | 86.6 |
| 6 | 8 | 12 | 20 | 13 | – | 5.0 | 12.4 | 91.2 | 89.5 |
| 7 | 11 | 9 | 20 | 12 | – | 4.5 | 16.4 | 100.0 | 84.9 |
| 8 | 12 | 12 | 24 | 16 | – | 3.6 | 12.7 | 100.0 | 82.7 |
| 9 | 12 | 9 | 21 | 13 | – | 5.0 | 18.4 | 89.7 | 100.1 |
| 10 | 12 | 15 | 27 | 19 | – | 3.4 | 12.6 | 100.0 | 81.8 |
| 11 | 9 | 12 | 21 | 14 | – | 4.4 | 12.2 | 100.0 | 88.5 |
| 12 | 9 | 10 | 19 | 12 | – | 4.9 | 17.0 | 78.6 | 88.0 |
| Total | 158 | 180 | 338 | 232 | – | 3.6 | 18.4 | 96.0 | 1150.8 |
Percentage of the genome within 10 cM of at least one PCR-based (SSR, SNP, or indel) marker.
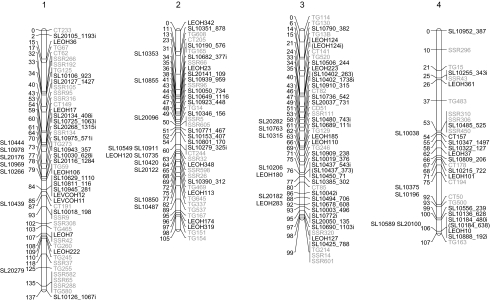
Fig. 1.
Integrated linkage map based on two F2 populations. Genetically mapped markers are on the right of the linkage groups while physically mapped markers are on the left. Marker names in grey text are framework markers used to merge the two F2 maps. Markers with the prefix ‘LEOH’ were previously developed [Yang et al., 2004 (LEOH1-LEOH51), Francis et al., 2005 (LEOH100-LEOH365)]. Markers with the prefix ‘SL’ were developed based on SNPs or indels (contains the suffix ‘i’) identified by Van Deynze et al. (2007). Markers in parentheses indicate that, although both a SNP and indel were created from the same locus, only one of the markers was mapped. Physically mapped markers are placed relative to the chromosome in their approximate position based on the framework markers. Markers on the far right of chromosome 9 were mapped using segmental substitution lines (Eshed and Zamir, 1995). Vertical lines indicate approximate boundaries of introgressions based on framework markers with the name of the segmental substitution line in vertical text to the left of the line. (Continued on page 6).
Several strategies were employed during the construction of each map to increase reliability. Segregation distortion was tested for each marker within JoinMap and the effect of skewed markers was investigated by comparing the map with and without the marker. If any marker noticeably expanded the map and had a relatively high mean χ2 contribution, the marker was removed from the map. Maps were first created with no order restraints and then compared with the Tomato-EXPEN 2000 map (SGN; http://solgenomics.net) by visually inspecting the order of the framework markers on each chromosome. For chromosome 4 where notable differences were detected, mapping was repeated using a fixed order of six framework markers (TG15, TG483, CT157, CT178, CT50, and TG163) based on the Tomato-EXPEN 2000 map. The order of these framework markers in the EXPEN 2000 map represents a robust order since this order is supported by several other genetic maps: EXPEN 1992 (Tanksley et al., 1992), EXPEN 2000 (Fulton et al., 2002), EXPIMP 2001 (Grandillo and Tanksley, 1996; Tanksley et al., 1996; Doganlar et al., 2002), EXPIMP 2008 (Gonzalo and van der Knaap, 2008), and EXHIR 1997 (Bernacchi and Tanksley, 1997). In addition, the position of TG163 is well established relative to the physical map. It was therefore decided to use a fixed order of framework markers based on these multiple maps and physical information from a BAC map. This new map was accepted only if the χ2 value decreased or increased reasonably. After the maps were constructed, genome coverage was calculated as the percentage of the genome that was within 10 cM of at least one PCR-based (SSR, SNP, or indel) marker.
The approximate position of markers that showed no segregation in either of the two mapping populations was identified based on the Tomato physical map (SGN; http://solgenomics.net). Tomato sequences with verified polymorphisms from ESTs (Yang et al., 2004; Francis et al., 2005) and conserved orthologous set (COS) introns (Van Deynze et al., 2007) were aligned with tomato genome sequence from the Tomato BAC sequences database (03-01-09; SGN; http://solgenomics.net) using BLASTN with the BLOSUM62 substitution matrix and a minimum expectation value (e-value) of 1e−10. The resulting hits were subjected to a two-step filtering process to identify highly probable marker–BAC alignments. Any BAC with >98% identity and >90% coverage of the query sequence was considered to contain the query locus. Because many BACs were in several stages of sequencing when these analyses were conducted (SGN; http://solgenomics.net), the remaining putative hits with >250 bp alignments were manually inspected to determine if the query sequence aligned to the edge of one of the unordered fragments of an unfinished BAC. In such instances, the BAC was considered to contain the query if the two sequences shared >98% identity. The BAC chromosome designation and data from the overgo analysis (bulk download SGN FTP site; http://solgenomics.net/bulk/input.pl?mode=ftp), were used to determine if each BAC containing a marker had a known chromosomal position on the tomato physical map, thereby indirectly placing the marker on the physical map.
Principal component analysis
Genotypic data from the germplasm collection was converted into allele frequencies based on their occurrence in the genome (0, 0.5, and 1) and analysed using the SAS PRINCOMP procedure (Version 9.1 for Windows, SAS Institute, Cary, NC). This approach allows for incorporation of SSR data that may be multi-allelic into the analysis. The eigenvalues of the first three principal components were extracted for each variety, and an analysis of variance (ANOVA) was performed using the General Linear Models procedure in order to test whether the market classes were significantly different.
Linkage disequilibrium analysis
Marker genotypes were used to measure the extent of LD within cultivated tomatoes (processing, fresh market, and vintage cultivars combined) as well as processing and fresh market cultivars separately. All other entries (greenhouse varieties, unimproved breeding lines, Latin American cultivars, and wild species) lacked sufficient representatives (<10 entries for each class) and were eliminated from the analysis. Only markers that were both placed on the integrated linkage map and polymorphic within cultivated tomato were used for LD analysis. Both the GGT 2.0 (van Berloo, 2008) and TASSEL (Bradbury et al., 2007) software were used to calculate pair-wise r2 values between 114 markers distributed throughout the genome. P values for each r2 estimate were calculated using 1000 permutations in TASSEL.
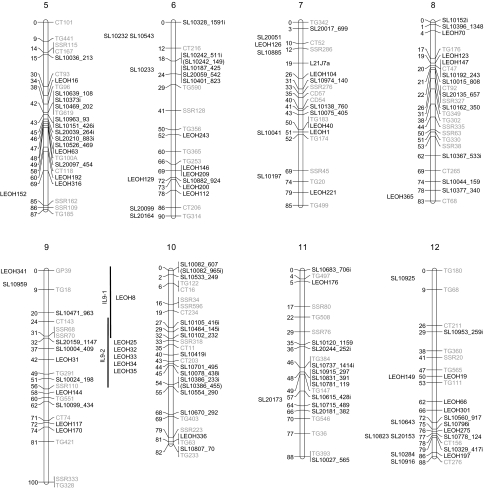
The decay of LD over genetic distance was investigated by plotting pair-wise r2 values against the distance (cM) between markers on the same chromosome (Fig. 3). A smooth line was fit to the data using second-degree locally weighted scatterplot smoothing (LOESS; Breseghello and Sorrells, 2006) as implemented in SAS. To describe the relationship between LD decay and genetic distance, two methods of establishing baseline r2 values were investigated. Critical values of r2 were based on a fixed value of 0.1 (Nordborg et al., 2002; Palaisa et al., 2003; Remington et al., 2001) and from the parametric 95th percentile of the distribution of the unlinked markers (Breseghello and Sorrells, 2006). The relationship between these baseline r2 values and genetic distance was determined using the LOESS curve and a 1 cM moving means approach. For the LOESS estimation of LD decay, genetic distance was estimated as the point where the LOESS curve first crosses the baseline r2 value. For the moving means approach, the distance between linked markers was used to divide marker pairs into bins of 1 cM. Markers separated by 0–0.9 cM were placed in the first bin, marker distances from 1–1.9 were in the second bin, etc. The mean of the r2 values within each bin was calculated and LD decay was estimated as the first bin where the baseline r2 value was lower than the bin mean.
Fig. 3.
Plots of linkage disequilibrium (LD) values (r2) against genetic distance (cM) between pairs of markers in multiple classes of cultivated tomato. All possible pair-wise combinations of markers on the same chromosome were plotted to visualize LD decay within chromosomes over the entire genome. The r2 values were calculated separately for processing and fresh market cultivars (B and C, respectively) as well as processing, fresh market, and vintage cultivar classes combined (A). Curves were fit for each plot by second-degree LOESS. The horizontal dotted lines indicate the baseline r2 values based on the 95th percentile of the distribution of unlinked r2 values (black) and the fixed r2 value of 0.1 (grey).
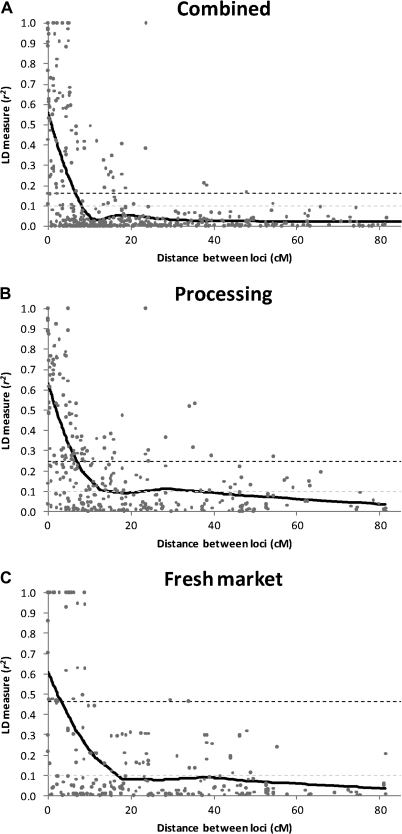
To visualize LD throughout the genome, heat maps were produced based on pair-wise r2 estimates and their P values for all marker pairs (Fig. 4). These heat maps were used to identify variation in disequilibrium between tomato classes at specific genomic locations. Differences were tested by comparing r2 estimates of marker pairs in the region using a paired t test in SAS. Only marker pairs with r2 estimates in both classes were included in the comparison.
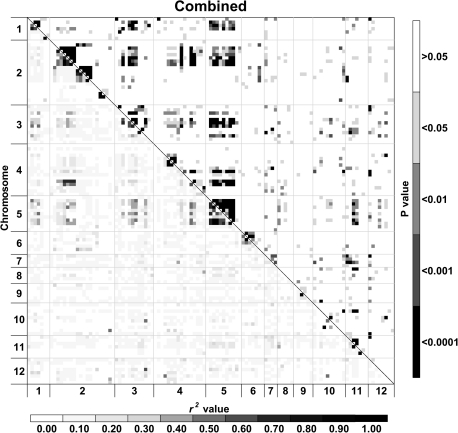
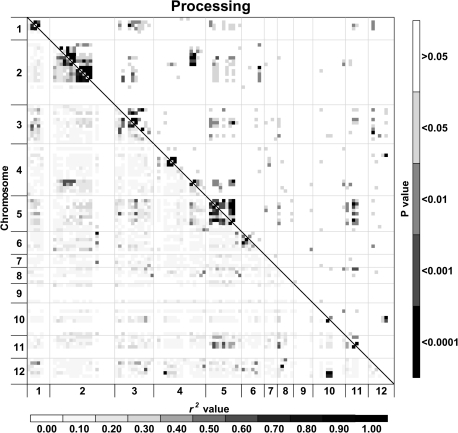
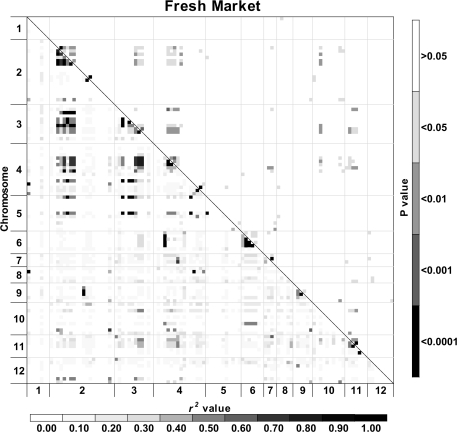
Fig. 4.
Heat maps of linkage disequilibrium (LD) values (r2) throughout the tomato genome. Markers were ordered on the x and y axes based on genomic location so that each cell of the heat map represents a single marker pair. The r2 values for each marker pair are on the bottom half of the heat map and are represented by shades of grey from 0.0 (white) increasing in darkness in equal increments of 0.1 to 1.0 (black). The P-values of each r2 estimate are on the top half of the heat map and are represented by shades of grey from non-significant (P >0.05; white) to highly significant (P <0.0001; black). The combined analysis includes processing, fresh market, and vintage cultivars.
Results
A germplasm collection representing currently relevant and historical tomato germplasm was genotyped with 340 indel, SNP, and SSR markers (see Supplementary Tables S2–S5 at JXB online). Markers had been pre-selected based on their potential for polymorphism within cultivated tomato. The majority of the markers were polymorphic within our collection of cultivated tomato varieties (74%) while over 85% were polymorphic within wild species. Fifty per cent of the markers were polymorphic within processing and 47% were polymorphic within fresh market germplasm (Table 1).
Genotypic information from the germplasm collection was utilized to identify markers that could be mapped in either of two F2 populations. For the Sun1642×LA1589 population, a total of 153 framework (SSR and RFLP) and 119 SNP and indel markers were mapped (Table 2). The order of the framework markers generally matched that of the Tomato-EXPEN 2000 map (SGN; http://solgenomics.net) without using a fixed marker order for all chromosomes except for chromosome 4. Using a fixed order of TG15, TG483, CT157, CT178, CT50, and TG163 derived from Tomato-EXPEN 2000 reduced the χ2 value from 123.7 to 25.5 and increased the map length from 56.7 cM to 114.1 cM. The total length of the Sun1642×LA1589 map was 1127 cM with an average of 4.3 cM between markers and the largest gap of 21.4 cM on chromosome 4. Segregation distortion was detected on chromosomes 6, 7, 11, and 12 (Table 2), with distorted markers adjacently located and skewed in the direction of the same parental allele indicating biased transmission. Ninety-four per cent of the genome was within 10 cM of at least one SSR, SNP or indel marker.
The Yellow Stuffer×LA1589 population map contains 90 framework markers with 60 new SNP and indel markers (Table 2). As with the Sun1642×LA1589 population, chromosome 4 was the only chromosome where a fixed marker order was employed. Using a fixed order increased the χ2 value from 36.7 to 124.8 and increased the map length from 97.4 cM to 114.8 cM. The Yellow Stuffer×LA1589 map had an average of 7.8 cM between markers, the largest gap of 33.2 cM on chromosome 10, and a total length of 1072 cM. Twenty-three per cent of the markers did not fit expected segregation ratios with the highest distortion on chromosomes 2, 7, 9, and 11 and distortion patterns indicating biased transmission. In this map, 72% of the genome was within 10 cM of a SSR, SNP or indel marker.
Eighty-five framework markers common to both maps allowed the creation of an integrated map with 338 markers including 180 new SNPs and indels (Table 2; Fig. 1). The average distance between markers was 3.6 cM with the largest gap on chromosome 9 of 18.4 cM. The total map length was 1151 cM with 96% of the genome within 10 cM of a PCR-based marker.
Emerging sequence data from the BAC-by-BAC international genome sequencing project were also used to identify the location of makers (see Supplementary Table S7 at JXB online). The sequence of 415 marker loci with verified polymorphisms was used as a BLAST query against the tomato genome sequence and 136 (33%) loci met the threshold for association with a BAC (see Materials and methods). The SGN data provided a chromosome assignment for 129 loci (31%), 60 (14%) of which had a precise location on the physical map. Forty-nine of the loci with a known chromosome from physical mapping were also placed on the genetic linkage map, allowing the two mapping methods to be compared. Out of these 49 loci, the chromosome designation of 48 (98%) matched. For the loci that were not placed on the linkage map, physical mapping provided the chromosome designation of 80 loci, 35 of which had a physical map position (see Supplementary Table S7 at JXB online). These loci were placed next to our integrated linkage map relative to the framework markers (Fig. 1). In addition, 18 polymorphic loci, whose physical position was previously determined (Van Deynze et al., 2007), were integrated into the map. Thus, 53 additional loci were added to the map based on physical position.
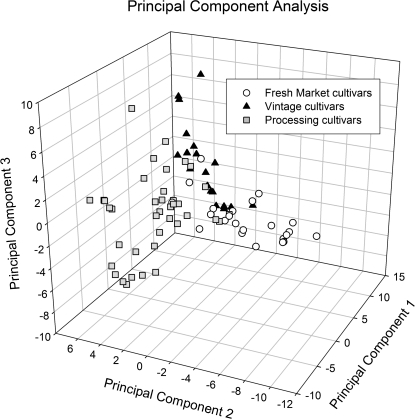
Principal Components Analysis (PCA) was used to visualize and test relationships between market classes within the collection of varieties. When processing, fresh market and vintage varieties were analysed together, the first three principal components explained 21.8% of the total variation and clear clusters emerged (Fig. 2). The hypothesis that market classes were distinct was tested by performing an analysis of variance (ANOVA) based on PCA. Both PC1 and PC2 were significantly different (P <0.0001). Mean separations demonstrated that all three classes were separated along PC1. For PC2, contemporary fresh market varieties were significantly different from contemporary processing and vintage varieties, but the latter two were not significantly different.
Fig. 2.
Graph of the first three Principal Components based on marker frequencies. Fresh market (circles), processing (squares), and vintage (triangles) cultivar groups are indicated by unique symbols.
Analysis of LD was performed for a data set consisting of contemporary and vintage varieties and separately for the two contemporary market classes. A difference was observed in both the decay of LD over genetic distance and the amount of inter-chromosomal LD between the three analyses. Based on the LOESS curves, the rate of LD decay was more pronounced for the combined entries followed by processing and then fresh market germplasm. The LOESS curves also indicate that LD decays over multiple centimorgans. The baseline r2 values of 0.160 (combined), 0.248 (processing), and 0.464 (fresh market) estimated by the 95th percentile method correspond to 6.9, 6.9, and 3.0 cM on the LOESS curves, respectively (Fig. 3; Table 3). By contrast, a fixed baseline r2 value of 0.1 equates to 8.0 (combined), 14.2 (processing), and 16.1 (fresh market) cM on the LOESS curve. Using a 1 cM moving means method, the 95th percentile baseline r2 values correspond to the 6 (combined), 6 (processing), and 2 (fresh market) cM bins, while the fixed baseline fell in bins 6, 9, and 10, respectively. In general, using a fixed r2 baseline provided larger decay estimates than the 95th percentile method. The difference in estimates between methods was especially large in fresh market varieties and probably reflects the distribution associated with unlinked loci. The baseline r2 values estimated by the 95th percentile method are based on the unlinked loci, and larger baseline estimates for fresh market cultivars reflect a high level of LD between markers on different chromosomes (inter-chromosomal LD) in this group. The patterns of LD can also be visualized across the genome from the diagonal of the heat maps (Fig. 4). Processing and fresh market germplasm share a similar degree of LD on chromosomes 3, 4, and 11. Processing cultivars have greater LD on chromosomes 1, 2, and 5, while LD is higher on chromosomes 6 and 9 for fresh market cultivars.
Table 3.
Summary of genome-wide linkage disequilibrium analysis
| Market class | No. marker pairsa |
r2 estimatesb |
Linkage disequilibrium decay (cM)e |
||||||
| Median | St. Dev. | 95th percentilec | P <0.01d | LOESSf |
Moving meansg |
||||
| 95th percentile method | Fixed r2 (0.1) method | 95th percentile method | Fixed r2 (0.1) method | ||||||
| Combinedh | 5248 | 0.011 | 0.102 | 0.160 | 8.1% | 6.6 | 8.0 | 6 | 6 |
| Processing | 3294 | 0.037 | 0.131 | 0.248 | 5.5% | 6.9 | 14.2 | 6 | 9 |
| Fresh market | 2622 | 0.031 | 0.187 | 0.464 | 2.0% | 3.0 | 16.1 | 2 | 10 |
The number of marker pairs includes only markers polymorphic within each market class.
Linkage disequilibrium was estimated as r2 values for all possible marker pairs using TASSEL (Bradbury et al., 2007) and GGT (van Berloo, 2008) software.
The 95th percentile of the distribution of r2 values for the unlinked markers. This value is the baseline r2 to estimate LD decay.
Percentage of r2 estimates with P value <0.01. P values of r2 estimates were calculated from 1000 permutations using TASSEL software (Bradbury et al., 2007).
Linkage disequilibrium decay was estimated over genetic distance by the relationship of a baseline r2 estimate to linked marker pairs using two methods, LOESS and 1 cM moving means. The baseline r2 value was either fixed at 0.1 or estimated using the 95th percentile of the unlinked markers. Values for r2 that exceed the baseline are considered to be in linkage disequilibrium.
For the LOESS estimation of LD decay, genetic distance was estimated as the point where the LOESS curve first crosses the baseline r2 value.
For the means estimation of LD decay, the r2 values of linked markers were grouped into bins of 1 cM based on the distance between markers. LD decay was estimated as the first bin where the baseline r2 value was lower than the bin mean.
The combined analysis includes processing, fresh market, and vintage cultivars.
The heat maps also reveal patterns of LD between markers on different chromosomes in the combined, processing, and fresh market groups, suggesting that inter-chromosomal LD is present within cultivated germplasm. Separating the market classes removed some of the observed inter-chromosomal LD, though residual patterns remain. Values of inter-chromosomal r2 tend to be higher in the fresh market germplasm, though statistically significant inter-chromosomal LD was detected for both market classes (Fig. 4). The location of the inter-chromosomal disequilibrium differs between these two classes (Fig. 4; Table 4). Pair-wise t tests of r2 values indicate that processing lines have significant disequilibrium between chromosomes 2 and 3, 2 and 4, and 3 and 12 (Table 4). Fresh market varieties have significant disequilibrium between chromosomes 2 and 3, 2 and 4, 3 and 4, 3 and 11, and 4 and 6. The regions of chromosomes 2, 3, and 4 that are in disequilibrium differ for the market classes with shifts on chromosome 2 and 4 being particularly important in distinguishing patterns (Table 4).
Table 4.
Comparison of inter-chromosomal linkage disequilibrium between processing and fresh market tomato germplasm
| Chromosomea | Positionb | Chromosomea | Positionb | No.c | Processing |
Fresh Market |
P-valuee | ||
| Mean r2d | St. Dev. | Mean r2d | St. Dev. | ||||||
| 2 | 36.3–47.3 | 3 | 71.2–87.9 | 33 | 0.0648 | 0.0682 | 0.5776 | 0.2813 | <0.0001 |
| 2 | 47.3–51.6 | 3 | 71.2–76.7 | 10 | 0.2094 | 0.0287 | 0.0203 | 0.0167 | <0.0001 |
| 2 | 36.3–45.2 | 4 | 100.0–105.7 | 10 | 0.2278 | 0.1610 | 0.0569 | 0.0525 | 0.0372 |
| 2 | 36.3–47.3 | 4 | 53.2–61.7 | 30 | 0.0294 | 0.0249 | 0.4362 | 0.2324 | <0.0001 |
| 3 | 76.7–87.9 | 4 | 53.2–61.7 | 17 | 0.0506 | 0.0407 | 0.4837 | 0.2346 | <0.0001 |
| 3 | 76.7–87.9 | 11 | 46.4–48.5 | 8 | 0.0581 | 0.0777 | 0.3346 | 0.1080 | 0.0009 |
| 3 | 52.5–94.9 | 12 | 49.7–65.8 | 13 | 0.1596 | 0.1496 | 0.0257 | 0.0196 | 0.0012 |
| 4 | 53.2–68.5 | 11 | 46.4–48.5 | 18 | 0.0249 | 0.0228 | 0.2358 | 0.1306 | <0.0001 |
Chromosomes being compared.
Genetic map position (cM) within the specified chromosomes. The position is derived from the integrated linkage map (Fig. 1).
Number of marker pairs in the comparison. Only marker pairs with r2 estimates in both classes were included.
Mean r2 values of all marker pairs between the two chromosomal regions.
P value of a paired t test of the mean r2 estimates of processing versus fresh market entries.
Discussion
In order to develop resources for the evaluation of genetic variation within cultivated tomato further, 434 markers were integrated based on a combination of linkage mapping in F2 populations and physical mapping relative to emerging sequence data. Three-hundred-and-forty markers, including 226 that were mapped based on linkage and/or physical location were used to genotype a collection of tomato lines representing wild species, landraces, vintage cultivars, and contemporary varieties. The markers differentiated the collection into market classes and >70% were polymorphic within cultivated tomatoes. These mapping and genotypic data are presented in Supplementary Tables S2–S7 at JXB online and are also available on the Tomato Mapping Resource Database under the sections Polymorphic Marker Search and Search Marker (http://www.tomatomap.net).
Our linkage map was generally consistent with the Tomato-EXPEN 2000 map. The integrated map is 21% shorter than the 1460.5 total cM of the Tomato-EXPEN 2000 map. This discrepancy may simply be due to the characteristics of the mapping populations (e.g. mapping parents of different species) or the general expansion of linkage maps with the addition of more markers. Our integrated map length is comparable with the Tomato-EXPIMP2001 (1275 total cM) and Tomato-EXPIMP2008 (1228 total cM) maps which have the same S. pimpinellifolium parent and fewer markers (145 and 181, respectively).
Segregation distortion was detected on chromosomes 6, 7, 11, and 12 for the Sun1642×LA1589 population, and chromosomes 2, 7, 9, and 11 for the Yellow Stuffer×LA1589 population. Segregation distortion is commonly observed in wide crosses of tomato and other species as the consequence of linkage between loci that operate in pre- and post-zygotic phases of reproduction (Zamir and Tadmor, 1986; Chetelat et al., 1989, 2000). The implications of distorted segregation on the map were tested by removing markers and repeating the mapping process. For the reported markers, segregation distortion does not significantly alter the map.
The use of BLAST to anchor markers to publicly available genome sequence data from the International Tomato Genome Sequencing Project (http://solgenomics.net/about/tomato_sequencing.pl) resulted in a physical association for 33% of our markers. At the time of our analysis, the sequencing effort was estimated to be 41% complete, suggesting that >80% of our markers will eventually be represented in BAC sequence. A high level of agreement (98%) was observed between markers that were mapped both physically and genetically. Thus, using the tomato genome sequence provides a robust method to identify the genomic location of unmapped loci. This approach will become the preferred method to map markers with the completion of a robust integrated tomato genome sequence in the near future.
Knowledge of the extent and structure of LD is important to assess the usefulness of association mapping strategies (Rostoks et al., 2006). The decay of LD over physical or genetic distance determines the depth of resolution as well as the density of markers needed for association analysis (Yu and Buckler, 2006). LD decay was estimated at 6–8 cM across all varieties, 6-14 cM within processing varieties, and 3–16 cM within fresh market varieties with the range dependent on the methods used to estimate threshold values and decay. The large range in fresh market estimates illustrates the difference between the methods used to establish a critical r2 value. Rather than selecting an arbitrary fixed value, the 95th percentile method relies on unlinked markers. As such, the estimate is influenced by inter-chromosomal LD and takes into account properties of the entries measured that may lead to population structure (Breseghello and Sorrells, 2006). Thus, estimates based on this method are more reflective of the sample. Our LD decay estimates are consistent with previous studies. In commercial European greenhouse varieties LD decayed over 15–20 cM (van Berloo et al., 2008). Labate et al. (2009a) found that intra-locus LD was high with a plateau at r2=0.6 over 1000 bp in 31 tomato landraces. Since LD decays over centimorgans in cultivated tomato, association mapping is theoretically feasible with a small number of markers.
Although our results suggest that marker numbers may be favourable for association mapping in cultivated tomato, the extent of inter-chromosomal LD between unlinked markers is likely to confound association analyses. For example, linkage disequilibrium between two genomic locations in a tomato mapping population resulted in a significant, but spurious marker–trait association that was not confirmed in subsequent populations (Robbins et al., 2009). Significant inter-chromosomal LD was identified within cultivated tomato that differed between fresh market and processing tomatoes. In a previous study, different patterns of inter-chromosomal LD were identified between cherry and beef-round tomatoes (van Berloo et al., 2008). The majority of chromosome pairs with disequilibrium differed from those we detected, suggesting that inter-chromosomal LD is population dependent and should be determined for each population of interest. In a separate study among tomato landraces, 19% of inter-locus marker pairs showed significant LD while only 10% of these were located on the same chromosome (Labate et al., 2009a). These results suggest that inter-chromosomal LD will complicate association analyses in cultivated tomato.
Linkage disequilibrium is caused by many factors including recombination rate, drift, mating system, selection, effective population size, and population structure (reviewed by Rafalski and Morgante, 2004). It appears that, in tomato, genetic bottlenecks, introgressions from wild species, and intense selection for market specialization have established haplotype blocks with disequilibrium over long physical distances. Such haplotype blocks have been identified in the genome of humans (Patil et al., 2001), mice (Wiltshire et al., 2003), dogs (Lindblad-Toh et al., 2005), rice (Tang et al., 2006; Li et al., 2009), and maize (Gore et al., 2009). It is hypothesized that, in tomato, some of the observed inter-chromosomal disequilibrium was produced by selection for the desired combinations of characters. The differences observed in LD patterns between fresh market and processing market types suggest that plant breeders may have selected for separate combinations of genes during the development of ideotypes for specialized markets.
Tomato has gone through several genetic bottlenecks during domestication, its introduction into Europe from Latin America, and its introduction into North America from Europe and the Caribbean (Rick, 1976; Miller and Tanskley, 1990; Labate et al., 2007). Early tomato improvement depended largely on mutation, spontaneous outcrossing, and recombination of available genetic variation to provide variability for selection (Rick, 1976). It was not until the 1920s that breeding programmes were established for tomato cultivar development (Stevens and Rick, 1986). Since then, the application of genetic principles and the continued innovation of breeding practices accelerated the pace of tomato improvement (Rick, 1976). High selection pressure for desired phenotypes in a limited germplasm pool, coupled with the high degree of self-pollination and multiple bottlenecks within the cultivated species have contributed to the narrow genetic base of tomato (Rick, 1976; Miller and Tanskley, 1990; Park et al., 2004). To overcome this challenge, breeding practices dating back to the 1930s have utilized wild tomato species for the introgression of new genetic variation, especially for disease resistance. At the same time, these practices reduced recombination in linkage blocks associated with introgressed segments (MacArthur and Butler, 1938; Alexander, 1949; Miller and Tanskley, 1990; Williams and St Clair, 1993; Park et al., 2004; Sim et al., 2009). Efforts to develop tomatoes specifically for mechanical harvest began in the late 1940s and by the mid 1960s, acceptable varieties were available (Rasmussen, 1968). The emphasis in breeding processing tomatoes suitable for mechanical harvest caused a divergence between fresh market and processing types. Results from this study support the hypothesis that breeding for market specialization is a major driving force for genetic differentiation between fresh market and processing varieties.
Our mapping and genetic data will provide a resource for researchers interested in using molecular markers for tomato improvement. Different patterns of LD between fresh market and processing varieties highlight how breeding practices have altered the genomes of market classes within cultivated tomato germplasm. The extent of inter-chromosomal LD in contemporary varieties leads us to hypothesize that market specialization has preserved certain favourable combinations of alleles. Breeders may choose to preserve these combinations, while also accessing and testing the affect of variation derived from different market classes. Extensive inter-chromosomal LD also suggests that association mapping should be conducted with caution to avoid detection of spurious marker–trait linkage.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Table S1. Description of 102 tomato accessions used in this study.
Supplementary Table S2. SNP markers in this study detected by allele specific primer extension (ASPE) assay.
Supplementary Table S3. SNPs detected as CAPS markers in this study.
Supplementary Table S4. Indel markers used in this study.
Supplementary Table S5. SSR markers used in this study.
Supplementary Table S6. Marker locations on the Sun1642×LA1589, Yellow Stuffer×LA1589, and integrated maps.
Supplementary Table S7. Location of markers placed on the tomato physical map compared to the integrated linkage map.
Acknowledgments
We would like to thank Tea Meulia and Jody Whittier of the OARDC Molecular and Cellular Imaging Center for support of the Luminex genotyping. We also acknowledge support of the National Research Initiative (NRI) Plant Genome Program of USDA's Cooperative State Research, Education and Extension Service for a Post-doctoral grant 2007-35300-18316 to MR, the USDA/NRI Plant Genome grant 2004-35300-14651 to DF, AV, and EV, the UDSA/NIFA/AFRI Plant Genome Solanaceae Coordinated Agricultural Project grant, and the Ohio Plant Biotechnology Consortium Competitive Grant 2007-025 to DF.
References
- Alexander LJ. Ohio W-R Globe, a new wilt-resistant glasshouse tomato variety. Research Bulletin. 1949;689 [Google Scholar]
- Bernacchi D, Tanksley SD. An interspecific backcross of Lycopersicon esculentum× L. hirsutum: linkage analysis and a QTL study of sexual compatibility factors and floral traits. Genetics. 1997;147:861–877. doi: 10.1093/genetics/147.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- Breseghello F, Sorrells ME. Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics. 2006;172:1165–1177. doi: 10.1534/genetics.105.044586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke JM, Knapp SJ, Rieseberg LH. Genetic consequences of selection during the evolution of cultivated sunflower. Genetics. 2005;171:1933–1940. doi: 10.1534/genetics.104.039057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casa AM, Pressoir G, Brown PJ, Mitchell SE, Rooney WL, Tuinstra MR, Franks CD, Kresovich S. Community resources and strategies for association mapping in sorghum. Crop Science. 2008;48:30–40. [Google Scholar]
- Chetelat RT, Rick CM, DeVerna JW. Isozyme analysis, chromosome pairing, and fertility of Lycopersicon esculentum× Solanum lycopersicoides diploid backcross hybrids. Genome. 1989;32:783–790. [Google Scholar]
- Chetelat RT, Meglic V, Cisneros P. A genetic map of tomato based on BC1 Lycopersicon esculentum× Solanum lycopersicoides reveals overall synteny but suppressed recombination between these homeologous genomes. Genetics. 2000;154:857–867. doi: 10.1093/genetics/154.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RM, Linton E, Messing J, Doebley JF. Pattern of diversity in the genomic region near the maize domestication gene tb1. Proceedings of the National Academy of Sciences, USA. 2004;101:700–707. doi: 10.1073/pnas.2237049100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J. The genetics of maize evolution. Annual Review of Genetics. 2004;38:37–59. doi: 10.1146/annurev.genet.38.072902.092425. [DOI] [PubMed] [Google Scholar]
- Doebley JF, Gaut BS, Smith BD. The molecular genetics of crop domestication. Cell. 2006;127:1309–1321. doi: 10.1016/j.cell.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Doganlar S, Frary A, Ku H, Tanksley S. Mapping quantitative trait loci in inbred backcross lines of Lycopersicon pimpinellifolium (LA1589) Genome. 2002;45:1189–1202. doi: 10.1139/g02-091. [DOI] [PubMed] [Google Scholar]
- Eshed Y, Zamir D. An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics. 1995;141:1147–1162. doi: 10.1093/genetics/141.3.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis DM, Kabelka E, Bell J, Franchino B, Clair DS. Resistance to bacterial canker in tomato (Lycopersicon hirsutum LA407) and its progeny derived from crosses to L. esculentum. Plant Disease. 2001;85:1171–1176. doi: 10.1094/PDIS.2001.85.11.1171. [DOI] [PubMed] [Google Scholar]
- Francis DM, Yang WC, van der Knaap E, Hogenhout S, Darrigues A. 2005. DNA-microarray detection of molecular markers for S. lycopersicum× S. lycopersicum crosses. 25–28 September, 2nd Solanaceae Genome Workshop, Ischia, Italy. [Google Scholar]
- Frary A, Xu YM, Liu JP, Mitchell S, Tedeschi E, Tanksley S. Development of a set of PCR-based anchor markers encompassing the tomato genome and evaluation of their usefulness for genetics and breeding experiments. Theoretical and Applied Genetics. 2005;111:291–312. doi: 10.1007/s00122-005-2023-7. [DOI] [PubMed] [Google Scholar]
- Frary A, Nesbitt TC, Frary A, Grandillo S, van der Knaap E, Cong B, Liu J, Meller J, Elber R, Alpert KB, Tanksley SD. fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science. 2000;289:85–88. doi: 10.1126/science.289.5476.85. [DOI] [PubMed] [Google Scholar]
- Frisch M, Bohn M, Melchinger AE. Comparison of selection strategies for marker-assisted backcrossing of a gene. Crop Science. 1999;39:1295–1301. [Google Scholar]
- Fulton TM, van der Hoeven R, Eannetta NT, Tanksley SD. Identification, analysis and utilization of a conserved ortholog set (COS) markers for comparative genomics in higher plants. The Plant Cell. 2002;14:1457–1467. doi: 10.1105/tpc.010479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gepts P. Crop domestication as a long-term selection experiment. Plant Breeding Reviews. 2004;24:1–44. [Google Scholar]
- Gonzalo MJ, van der Knaap E. A comparative analysis into the genetic bases of morphology in tomato varieties exhibiting elongated fruit shape. Theoretical and Applied Genetics. 2008;116:647–656. doi: 10.1007/s00122-007-0698-7. [DOI] [PubMed] [Google Scholar]
- Gore MA, Chia J, Elshire RJ, et al. A first-generation haplotype map of maize. Science. 2009;326:1115–1117. doi: 10.1126/science.1177837. [DOI] [PubMed] [Google Scholar]
- Graham EB, Frary A, Kang JJ, Jones CM, Gardner RG. A recombinant inbred line mapping population derived from a Lycopersicon esculentum× L. pimpinellifolium cross. Tomato Genetics Cooperative Report. 2004;54:22–25. [Google Scholar]
- Grandillo S, Tanksley SD. QTL analysis of horticultural traits differentiating the cultivated tomato from the closely related species Lycopersicon pimpinellifolium. Theoretical and Applied Genetics. 1996;92:935–951. doi: 10.1007/BF00224033. [DOI] [PubMed] [Google Scholar]
- Jiménez-Gómez J, Maloof J. Sequence diversity in three tomato species: SNPs, markers, and molecular evolution. BMC Plant Biology. 2009;9:85. doi: 10.1186/1471-2229-9-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DA, Thomas CM, Hammondkosack KE, Balintkurti PJ, Jones JDG. Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science. 1994;266:789–793. doi: 10.1126/science.7973631. [DOI] [PubMed] [Google Scholar]
- Kabelka E, Franchino B, Francis DM. Two Loci from Lycopersicon hirsutum LA407 Confer Resistance to Strains of Clavibacter michiganensis subsp. michiganensis. Phytopathology. 2002;92:504–510. doi: 10.1094/PHYTO.2002.92.5.504. [DOI] [PubMed] [Google Scholar]
- Kabelka E, Yang WC, Francis DM. Improved tomato fruit color within an inbred backcross line derived from Lycopersicon esculentum and L. hirsutum involves the interaction of loci. Journal of the American Society for Horticultural Science. 2004;129:250–257. [Google Scholar]
- Labate JA, Baldo AM. Tomato SNP discovery by EST mining and resequencing. Molecular Breeding. 2005;16:343–349. [Google Scholar]
- Labate JA, Robertson LD, Baldo AM. Multilocus sequence data reveal extensive departures from equilibrium in domesticated tomato (Solanum lycopersicum L.) Heredity. 2009a;103:257–267. doi: 10.1038/hdy.2009.58. [DOI] [PubMed] [Google Scholar]
- Labate JA, Robertson LD, Wu FN, Tanksley SD, Baldo AM. EST, COSII, and arbitrary gene markers give similar estimates of nucleotide diversity in cultivated tomato (Solanum lycopersicum L.) Theoretical and Applied Genetics. 2009b;118:1005–1014. doi: 10.1007/s00122-008-0957-2. [DOI] [PubMed] [Google Scholar]
- Labate JA, Grandillo S, Fulton T, et al. Tomato. In: Kole C, editor. Vegetables. Heidelberg: Springer-Verlag; 2007. pp. 1–125. [Google Scholar]
- Lee SH, Walker DR, Cregan PB, Boerma HR. Comparison of four flow cytometric SNP detection assays and their use in plant improvement. Theoretical and Applied Genetics. 2004;110:167–174. doi: 10.1007/s00122-004-1827-1. [DOI] [PubMed] [Google Scholar]
- Li XR, Tan LB, Zhu ZF, Huang HY, Liu Y, Hu SN, Sun CQ. Patterns of nucleotide diversity in wild and cultivated rice. Plant Systematics and Evolution. 2009;281:97–106. [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Liu J, Van Eck J, Cong B, Tanksley SD. A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proceedings of the National Academy of Sciences, USA. 2002;99:13302–13306. doi: 10.1073/pnas.162485999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur JW, Butler L. Size inheritance and geometric growth processes in the tomato fruit. Genetics. 1938;23:253–268. doi: 10.1093/genetics/23.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GB, Brommonschenkel SH, Chunwongse J, Frary A, Ganal MW, Spivey R, Wu T, Earle ED, Tanksley SD. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science. 1993;262:1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- Menda N, Semel Y, Peled D, Eshed Y, Zamir D. In silico screening of a saturated mutation library of tomato. The Plant Journal. 2004;38:861–872. doi: 10.1111/j.1365-313X.2004.02088.x. [DOI] [PubMed] [Google Scholar]
- Messeguer R, Ganal M, Devicente MC, Young ND, Bolkan H, Tanksley SD. High-resolution RFLP map around the root-knot nematode resistance gene (Mi) in tomato. Theoretical and Applied Genetics. 1991;82:529–536. doi: 10.1007/BF00226787. [DOI] [PubMed] [Google Scholar]
- Miller JC, Tanksley SD. RFLP analysis of phylogenetic relationships and genetic variation in the genus Lycopersicon. Theoretical and Applied Genetics. 1990;80:437–448. doi: 10.1007/BF00226743. [DOI] [PubMed] [Google Scholar]
- Nordborg M, Borevitz JO, Bergelson J, et al. The extent of linkage disequilibrium in Arabidopsis thaliana. Nature Genetics. 2002;30:190–193. doi: 10.1038/ng813. [DOI] [PubMed] [Google Scholar]
- Palaisa KA, Morgante M, Williams M, Rafalski A. Contrasting effects of selection on sequence diversity and linkage disequilibrium at two phytoene synthase loci. The Plant Cell. 2003;15:1795–1806. doi: 10.1105/tpc.012526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paran I, van der Knaap E. Genetic and molecular regulation of fruit and plant domestication traits in tomato and pepper. Journal of Experimental Botany. 2007;58:3841–3852. doi: 10.1093/jxb/erm257. [DOI] [PubMed] [Google Scholar]
- Park YH, West MAL, St Clair DA. Evaluation of AFLPs for germplasm fingerprinting and assessment of genetic diversity in cultivars of tomato (Lycopersicon esculentum L.) Genome. 2004;47:510–518. doi: 10.1139/g04-004. [DOI] [PubMed] [Google Scholar]
- Patil N, Berno AJ, Hinds DA, et al. Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science. 2001;294:1719–1723. doi: 10.1126/science.1065573. [DOI] [PubMed] [Google Scholar]
- Pnueli L, CarmelGoren L, Hareven D, Gutfinger T, Alvarez J, Ganal M, Zamir D, Lifschitz E. The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development. 1998;125:1979–1989. doi: 10.1242/dev.125.11.1979. [DOI] [PubMed] [Google Scholar]
- Rafalski A, Morgante M. Corn and humans: recombination and linkage disequilibrium in two genomes of similar size. Trends in Genetics. 2004;20:103–111. doi: 10.1016/j.tig.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Rasmussen WD. Advances in American agriculture: the mechanical tomato harvester as a case study. Technology and Culture. 1968;9:531–543. [Google Scholar]
- Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doeblay J, Kresovich S, Goodman MM, Buckler ES. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proceedings of the National Academy of Science, USA. 2001;98:11479–11484. doi: 10.1073/pnas.201394398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rick CM. Tomato, Lycopersicom esculentum(Solanaceae) In: Simmonds NW, editor. Evolution of crop plants. London: Longman Group; 1976. pp. 268–273. [Google Scholar]
- Robbins MD, Darrigues A, Sim S, Masud MAT, Francis DM. Characterization of hypersensitive resistance to bacterial spot race T3 (Xanthomonas perforans) from tomato accession PI 128216. Phytopathology. 2009;99:1037–1044. doi: 10.1094/PHYTO-99-9-1037. [DOI] [PubMed] [Google Scholar]
- Rostoks N, Ramsay L, MacKenzie K, et al. Recent history of artificial outcrossing facilitates whole-genome association mapping in elite inbred crop varieties. Proceedings of the National Academy of Sciences, USA. 2006;103:18656–18661. doi: 10.1073/pnas.0606133103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics methods and protocols: methods in molecular biology. Totowa: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Sim S, Robbins MD, Chilcott C, Zhu T, Francis DM. Oligonucleotide array discovery of polymorphisms in cultivated tomato (Solanum lycopersicum L.) reveals patterns of SNP variation associated with breeding. BMC Genomics. 2009;10 doi: 10.1186/1471-2164-10-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassova MI, Prins TW, Folkertsma RT, Klein-Lankhorst RM, Hille J, Goldbach RW, Prins M. The tomato gene Sw5 is a member of the coiled coil, nucleotide binding, leucine-rich repeat class of plant resistance genes and confers resistance to TSWV in tobacco. Molecular Breeding. 2001;7:151–161. [Google Scholar]
- Stevens MA, Rick CM. Genetics and breeding. In: Athernon JG, Rudich J, editors. The tomato crop. A scientific basis for iImprovement. London, England: Chapman and Hall; 1986. pp. 35–109. [Google Scholar]
- Suliman-Pollatschek S, Kashkush K, Shats H, Hillel J, Lavi U. Generation and mapping of AFLP, SSRs and SNPs in Lycopersicon esculentum. Cellular and Molecular Biology Letters. 2002;7:583–597. [PubMed] [Google Scholar]
- Tang T, Lu J, Huang JZ, He JH, McCouch SR, Shen Y, Kai Z, Purugganan MD, Shi SH, Wu CI. Genomic variation in rice: genesis of highly polymorphic linkage blocks during domestication. PLoS Genetics. 2006;2:1824–1833. doi: 10.1371/journal.pgen.0020199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley SD. The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. The Plant Cell. 2004;16:S181–S189. doi: 10.1105/tpc.018119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley SD, Grandillo S, Fulton TM, Zamir D, Eshed Y, Petiard V, Lopez J, Beck-Bunn T. Advanced backcross QTL analysis in a cross between an elite processing line of tomato and its wild relative L. pimpinellifolium. Theoretical and Applied Genetics. 1996;92:213–224. doi: 10.1007/BF00223378. [DOI] [PubMed] [Google Scholar]
- Tanksley SD, McCouch SR. Seed banks and molecular maps: unlocking genetic potential from the wild. Science. 1997;277:1063–1066. doi: 10.1126/science.277.5329.1063. [DOI] [PubMed] [Google Scholar]
- Tanksley SD, Ganal MW, Prince JP, et al. High-density molecular linkage maps of the tomato and potato genomes. Genetics. 1992;132:1141–1160. doi: 10.1093/genetics/132.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berloo R. GGT 2.0: versatile software for visualization and analysis of genetic data. Journal of Heredity. 2008;99:232–236. doi: 10.1093/jhered/esm109. [DOI] [PubMed] [Google Scholar]
- van Berloo R, Zhu AG, Ursem R, Verbakel H, Gort G, van Eeuwijk FA. Diversity and linkage disequilibrium analysis within a selected set of cultivated tomatoes. Theoretical and Applied Genetics. 2008;117:89–101. doi: 10.1007/s00122-008-0755-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Knaap E, Tanksley SD. Identification and characterization of a novel locus controlling early fruit development in tomato. Theoretical and Applied Genetics. 2001;103:353–358. [Google Scholar]
- van der Knaap E, Tanksley SD. The making of a bell pepper-shaped tomato fruit: identification of loci controlling fruit morphology in Yellow Stuffer tomato. Theoretical and Applied Genetics. 2003;107:139–147. doi: 10.1007/s00122-003-1224-1. [DOI] [PubMed] [Google Scholar]
- Van Deynze A, Stoffel K, Buell CR, Kozik A, Liu J, van der Knaap E, Francis D. Diversity in conserved genes in tomato. BMC Genomics. 2007;8:465. doi: 10.1186/1471-2164-8-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooijen JW, Voorrips RE. In: JoinMap® 3.0, Software for the calculation of genetic linkage maps. Anonymous, editor. Wageningen, the Netherlands: Plant Research International; 2001. [Google Scholar]
- Wang RL, Stec A, Hey J, Lukens L, Doebley J. The limits of selection during maize domestication. Nature. 1999;398:236–239. doi: 10.1038/18435. [DOI] [PubMed] [Google Scholar]
- Whitt SR, Wilson LM, Tenaillon MI, Gaut BS, Buckler ES. Genetic diversity and selection in the maize starch pathway. Proceedings of the National Academy of Sciences, USA. 2002;99:12959–12962. doi: 10.1073/pnas.202476999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CE, St Clair DA. Phenetic relationships and levels of variability detected by restriction fragment length polymorphism and random amplified polymorphic DNA analysis of cultivated and wild accessions of Lycopersicon esculentum. Genome. 1993;36:619–630. doi: 10.1139/g93-083. [DOI] [PubMed] [Google Scholar]
- Wiltshire T, Pletcher MT, Batalov S, et al. Genome-wide single-nucleotide polymorphism analysis defines haplotype patterns in mouse. Proceedings of the National Academy of Sciences, USA. 2003;100:3380–3385. doi: 10.1073/pnas.0130101100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Jiang N, Schaffner E, Stockinger EJ, van der Knaap E. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science. 2008;319:1527–1530. doi: 10.1126/science.1153040. [DOI] [PubMed] [Google Scholar]
- Yamasaki M, Tenaillon MI, Bi IV, Schroeder SG, Sanchez-Villeda H, Doebley JF, Gaut BS, McMullen MD. A large-scale screen for artificial selection in maize identifies candidate agronomic loci for domestication and crop improvement. The Plant Cell. 2005;17:2859–2872. doi: 10.1105/tpc.105.037242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Bai XD, Kabelka E, Eaton C, Kamoun S, van der Knaap E, Francis D. Discovery of single nucleotide polymorphisms in Lycopersicon esculentum by computer aided analysis of expressed sequence tags. Molecular Breeding. 2004;14:21–34. [Google Scholar]
- Yang W, Sacks EJ, Ivey MLL, Miller SA, Francis DM. Resistance in Lycopersicon esculentum intraspeciflc crosses to race T1 strains of Xanthomonas campestris pv. vesicatoria causing bacterial spot of tomato. Phytopathology. 2005;95:519–527. doi: 10.1094/PHYTO-95-0519. [DOI] [PubMed] [Google Scholar]
- You F, Huo N, Gu Y, Luo M, Ma Y, Hane D, Lazo G, Dvorak J, Anderson O. BatchPrimer3: A high throughput web application for PCR and sequencing primer design. BMC Bioinformatics. 2008;9:253. doi: 10.1186/1471-2105-9-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JM, Buckler ES. Genetic association mapping and genome organization of maize. Current Opinion in Biotechnology. 2006;17:155–160. doi: 10.1016/j.copbio.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Zamir D, Tadmor Y. Unequal segregation of nuclear genes in plants. Botanical Gazette. 1986;147:355–358. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.