Abstract
Most fruit trees in the Rosaceae exhibit self-incompatibility, which is controlled by the pistil S gene, encoding a ribonuclease (S-RNase), and the pollen S gene at the S-locus. The pollen S in Prunus is an F-box protein gene (SLF/SFB) located near the S-RNase, but it has not been identified in Pyrus and Malus. In the Japanese pear, various F-box protein genes (PpSFBB-α–γ) linked to the S-RNase are proposed as the pollen S candidate. Two bacterial artificial chromosome (BAC) contigs around the S-RNase genes of Japanese pear were constructed, and 649 kb around S4-RNase and 378 kb around S2-RNase were sequenced. Six and 10 pollen-specific F-box protein genes (designated as PpSFBB4-u1–u4, 4-d1–d2 and PpSFBB2-u1–u5, 2-d1–d5, respectively) were found, but PpSFBB4-α–γ and PpSFBB2-γ were absent. The PpSFBB4 genes showed 66.2–93.1% amino acid identity with the PpSFBB2 genes, which indicated clustering of related polymorphic F-box protein genes between haplotypes near the S-RNase of the Japanese pear. Phylogenetic analysis classified 36 F-box protein genes of Pyrus and Malus into two major groups (I and II), and also generated gene pairs of PpSFBB genes and PpSFBB/Malus F-box protein genes. Group I consisted of gene pairs with 76.3–94.9% identity, while group II consisted of gene pairs with higher identities (>92%) than group I. This grouping suggests that less polymorphic PpSFBB genes in group II are non-S pollen genes and that the pollen S candidates are included in the group I PpSFBB genes.
Keywords: BAC contig, F-box protein, pollen S gene, Pyrus pyrifolia, self-incompatibility, S-locus, S-RNase
Introduction
Self-incompatibility (SI) is a genetic system that prevents self-fertilization in flowering plants by the recognition and rejection of self-pollen (de Nettancourt, 2001). In the Rosaceae, Solanaceae, and Plantaginaceae families, SI is classified as gametophytic SI (GSI), and is controlled by a single S-locus with multiple S-haplotypes. Each S-haplotype contains two genetically linked genes, the pistil S gene and the pollen S gene, which determine the S-haplotype specificity of the pistil and pollen, respectively (McCubbin and Kao, 2000). The pistil S encodes a ribonuclease known as S-RNase (McClure et al., 1989; Ishimizu et al., 1996; Xue et al., 1996). The RNase activity of S-RNases is essential for rejection of self-pollen, and the degradation of rRNA by S-RNases inside the self-pollen tube results in inhibition of pollen growth (McClure et al., 1990; Huang et al., 1994). Thus, it is thought that the self S-RNase inhibits growth of the self-pollen tube via degradation of pollen rRNAs. On the other hand, the identity and function of the pollen S remained unknown for a long time. Recently, F-box protein genes were identified as the pollen S genes by sequence analyses of cosmid and bacterial artificial chromosome (BAC) contigs around S-RNase in Prunus species of the Rosaceae, in Petunia inflata of the Solanaceae, and in Antirrhinum hispanicum of the Plantaginaceae. These F-box protein genes were termed SLF (S-locus F-box) or SFB (S-haplotype-specific F-box protein) (Lai et al., 2002; Entani et al., 2003; Ushijima et al., 2003; Sijacic et al., 2004). Transformation experiments in P. inflata and analyses of pollen-part self-compatible (SC) mutants in Prunus species provided evidence that SLF/SFB genes are the pollen S genes (Sijacic et al., 2004; Ushijima et al., 2004; Sonneveld et al., 2005; Hauck et al., 2006; Tsukamoto et al., 2006; Vilanova et al., 2006). Generally, F-box proteins function as one of the four major subunits (CUL1, SKP1, RBX1, and F-box) that make up the SCF complex, which regulates protein stability through the ubiquitin–proteasome system (Lechner et al., 2006). The model for S-RNase degradation proposes that the non-self-interaction between S-RNase and SLF/SFB leads to S-RNase ubiquitylation and degradation by the 26S proteasome (McClure and Franklin-Tong, 2006).
In Rosaceae, the pollen S has been identified only in Prunus (almond, apricots, and cherry), but not in Pyrus (pear) and Malus (apple). The Rosaceae comprises three subfamilies: Rosoideae, Dryadoideae, and Spiraeoideae. Prunus, Pyrus, and Malus are all included in Spiraeoideae (Potter et al., 2007). Therefore, it is likely that the pollen S genes in Pyrus and Malus are also F-box protein genes. Recently, S-locus-linked and pollen-specific polymorphic F-box protein genes were isolated from apple (Malus×domestica) and Japanese pear (Pyrus pyrifolia), and these have been proposed as good candidates for the pollen S genes. Cheng et al. (2006) cloned two S-locus-linked F-box protein genes (MdSLF1 and MdSLF2) from apple by reverse transcription-PCR (RT-PCR) with degenerate primers designed from the conserved SLF/SFB sequences. Sassa et al. (2007) found several pollen-specific polymorphic F-box protein genes termed SFBB (S locus F-box brothers) in BAC contig sequences around apple S-RNase genes. These SFBB genes include MdSFBB3-α and MdSFBB3-β around S3-RNase, and MdSFBB9-α and MdSFBB9-β around S9-RNase. Using RT-PCR, they also cloned various PpSFBB genes (PpSFBB-α, PpSFBB-β, and PpSFBB-γ) that are linked to S-RNase genes of the Japanese pear; PpSFBB4-α, PpSFBB4-β, and PpSFBB4-γ are linked to S4-RNase, and PpSFBB5-α, PpSFBB5-β, and PpSFBB5-γ are linked to S5-RNase. PpSFBB-γ genes that are linked to another eight S-RNase genes have been cloned. They show high amino acid sequence identities (97.5–99.7%) among the 10 S-haplotypes (Kakui et al., 2007). However, it is not clear whether PpSFBB genes are located near the S-RNase, like MdSFBB genes, or whether they are the pollen S genes. To identify the pollen S genes in the Japanese pear, a previously constructed BAC library from an S4 homozygote was used and a BAC contig of ∼570 kb around S4-RNase was assembled. Sequence analysis of the 240 kb spanning 51 kb upstream to 189 kb downstream of S4-RNase revealed a pollen-specific F-box protein gene (S4F-box0; S4-haplotype F-box protein gene) that differed from PpSFBB4-α–γ. S4F-box0 is located 127 kb downstream of S4-RNase (Okada et al., 2008). The SC cultivar ‘Osa Nijisseiki’ (S2S4sm) is a natural mutant derived from ‘Nijisseiki’ (S2S4). The S4-haplotype of ‘Osa Nijisseiki’ lacks the pistil S function but retains the pollen S function, and is termed the S4sm-haplotype, where ‘sm’ stands for ‘stylar-part mutant’ (Sato, 1993). The S4sm-haplotype has a 236 kb deletion, which includes S4-RNase and S4F-box0, suggesting that the pollen S4 allele is located outside of the region spanning 48 kb upstream to 188 kb downstream of S4-RNase—that is, outside the region that is deleted in the S4sm-haplotype (Okada et al., 2008).
In this study, the sequence outside of the deleted region in S4sm was analysed, and the 649 kb region from 290 kb upstream to 359 kb downstream of S4-RNase was determined; six PpSFBB4 genes were found. To evaluate the S-haplotype polymorphism of PpSFBB4 genes, a BAC library was constructed from the Japanese pear cultivar ‘Choujuuro’ (S2S3) to assemble a BAC contig around S2-RNase. A 378 kb region from 166 kb upstream to 212 kb downstream of S2-RNase was sequenced, and 10 PpSFBB2 genes were found. Relationships among 36 F-box protein genes of Pyrus and Malus were analysed by comparing their amino acid sequences and by phylogenetic clustering.
Materials and methods
Plant materials
One cultivar and three S homozygotes of the Japanese pear were used: ‘Choujuuro’ (S2S3), and S2, S3, and S4 homozygotes. The S2 and S3 homozygotes were selected from bud-selfed progeny of ‘Choujuuro’ (S2S3) (Terai et al., 1999). The S4 homozygote was segregated from bud-selfed progeny of ‘Nijisseiki’ (S2S4) (Okada et al., 2008). The leaves, mature pollen, and pistils were frozen in liquid nitrogen, and stored at –80 °C until use.
Construction and characterization of an S2S3 BAC library
An S2S3 BAC library was constructed and characterized according to the method of Okada et al. (2008). High molecular weight DNA was isolated from leaf tissue (3 g) of ‘Choujuuro’ (S2S3), partially digested with HindIII, and size-selected twice by pulsed field gel electrophoresis (PFGE). In the first size selection, an agarose slice containing DNA fragments of 60–210 kb was excised and embedded into a new 1% SeaPlaque GTG agarose gel (Cambrex, http://www.cambrex.com/). In the second size selection, two size fractions (145–185 kb and 185–205 kb) were recovered by digestion of agarose slices with β-agarase I. DNA from each fraction was separately ligated into HindIII-digested CopyControl pCC1BAC Cloning-Ready vector (EPICENTRE, http://www.epibio.com/) and transformed into Escherichia coli strain TransforMax EPI300 (EPICENTRE). Equal numbers of transformed cells were picked from each fraction and a total of 61 440 colonies were pooled in 64 individual 96-well plates with 12 columns and eight rows (10 colonies per well) and stored at –80 °C. The BAC plasmid was extracted from the randomly chosen BAC clones by the standard alkaline lysis method, digested with NotI, and separated by PFGE. Insert size was estimated by comparison with a PFGE lambda ladder (New England Biolabs, http://www.neb.com/).
Chromosome walking
Chromosome walking in the region around S-RNase was performed by PCR screening of the S2S3 BAC library and the previously constructed S4S4 BAC library (Okada et al., 2008). The PCR screening was performed in three consecutive steps as described by Okada et al. (2008). Chromosome walking around the S2-RNase was initiated by PCR screening of the S2S3 BAC library with an S-RNase-specific primer pair, ‘FTQQYQ’ and ‘anti-IIWPNV’ (Ishimizu et al., 1999).
BAC plasmids were isolated from positive BAC clones using the Plasmid Midi Kit (Qiagen, http://www1.qiagen.com/). Both ends of the BACs (∼600 bp) of the positive clones were sequenced using T7 and RP vector primers, and a primer pair was designed from each BAC-end sequence (Supplementary Table S1 available at JXB online). For chromosome walking, non-repetitive primer pairs were selected from the BAC-end primer pairs located at the outer ends of the contig by PCR amplification of plate pool templates, which were prepared by mixing all 960 BAC clones in each plate. Furthermore, S2-specific primer pairs were identified from among the non-repetitive primer pairs by PCR, using genomic DNA of the S2 and S3 homozygotes as templates. These S2-specific primer pairs were used for PCR screening of the BAC library (Supplementary Table S1). For PCR, genomic DNA was isolated from leaves of the S2 and S3 homozygotes by the modified cetyltrimethylammonium bromide (CTAB) method (Castillo et al., 2001).
To estimate insert sizes and compare restriction patterns, BAC plasmids were digested with NotI and subjected to PFGE. Based on overlapping of the BAC clones, their insert sizes, and restriction patterns, the physical distance was calculated to construct a BAC contig.
BAC subcloning
The plasmids of BAC clones were completely digested with HindIII or EcoRI, separated on a 0.7% agarose gel, and purified from the gels using GENECLEAN Kit III (Qbiogene, http://www.qbiogene.com/). Each fragment was ligated into pBluescriptII SK (+) and transformed into E. coli strain TOP10F’ (Invitrogen, http://www.invitrogen.com/). Inserts from subclones that were smaller than 7 kb were sequenced by primer walking, and those that were larger than 7 kb were sequenced after subcloning using other restriction enzymes. A primer was designed from each insert-end sequence. Using these primers, the regions outside of the subclones in the BAC plasmids were sequenced. The sequences from subclones and the outside sequences were assembled to construct contigs for each BAC clone. Gap regions, for which no sequence data were obtained, were amplified from BAC plasmids by PCR with the Expand High Fidelity PCR system (Roche Diagnostics, http://www.roche-diagnostics.jp/), and directly sequenced.
Nucleotide and amino acid sequence analysis
Nucleotide sequences were determined with the BigDye Terminator v3.1 Cycle Sequencing Kit using an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, http://www3.appliedbiosystems.com/). The sequence data were analysed using GENETYX-MAC Ver. 13 and ATGC Ver. 4 software packages (Genetyx, http://www.sdc.co.jp/genetyx/). Protein-coding sequences were predicted using the GENSCAN program (Burge and Karlin, 1997). Homology searches were carried out using the BLASTX program (Altschul et al., 1997). Deduced amino acid sequences were analysed by Pfam (http://pfam.janelia.org/) to search for protein motifs. Amino acid sequences were aligned using ClustalW (http://clustalw.ddbj.nig.ac.jp/top-j.html) and manually optimized. A phylogenetic tree was constructed by the Neighbor–Joining method (Saitou and Nei, 1987). A Harr plot analysis was performed using GENETYX-MAC Ver. 13 software.
RT-PCR
Total RNAs extracted from pollen, pistils, and leaves were subjected to first-strand cDNA synthesis using ReverTra Ace α (TOYOBO, http://www.toyobo.co.jp/). Using the Expand High Fidelity PCR system (Roche Diagnostics), PCR was then carried out with gene-specific primer pairs (Supplementary Table S2 at JXB online). PCR products were separated by electrophoresis on 1.5% or 0.8% agarose gels and visualized by ethidium bromide staining. The PCR products were purified from the gels, and sequenced to confirm gene specificity.
Sequence data
The 649 kb and 378 kb sequences around S4- and S2-RNase genes have been deposited in the EMBL/GenBank Data Libraries under accession nos AB545981 and AB545982, respectively.
Results
Sequence analysis of 649 kb around the S4-RNase gene
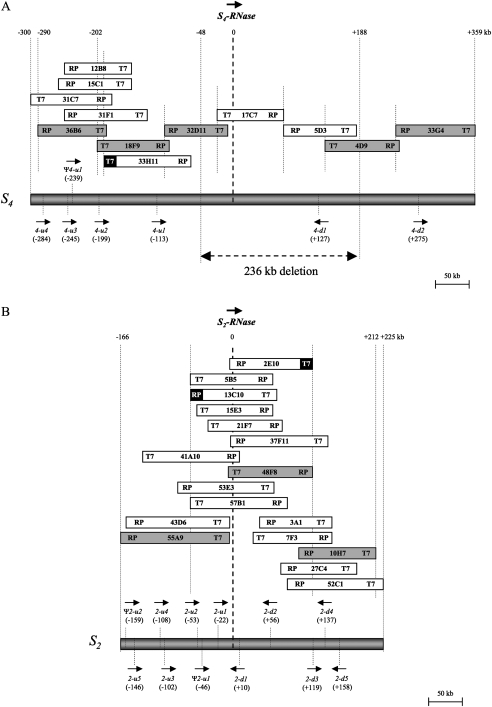
Previously, an S4 BAC contig spanning 202 kb upstream (18F9 T7-end) to 359 kb downstream (33G4 T7-end) of S4-RNase was constructed. In addition, the complete sequences of two BAC clones (17C7 and 5D3) and the partial sequences of two other BAC clones (32D11 and 4D9) were analysed to determine a 240 001 bp sequence spanning 51 kb upstream to 189 kb downstream of S4-RNase (Okada et al., 2008). In this study, 32D11 and 4D9 were sequenced in their entirety. To extend the S4 BAC contig upstream, chromosome walking was resumed using a non-repetitive BAC-end primer pair (33H11-T7). PCR screening of the S4S4 BAC library yielded five BAC clones: 12B8, 15C1, 31C7, 31F1, and 36B6. As a result, chromosome walking from S4-RNase produced a set of overlapping BAC clones (36B6, 18F9, 32D11, 17C7, 5D3, 4D9, and 33G4) covering ∼649 kb, spanning 290 kb upstream to 359 kb downstream of S4-RNase (Fig. 1A). Three BAC clones (36B6, 18F9, and 33G4) were subcloned and completely sequenced. The sequence assembly of the seven BAC clones (36B6, 18F9, 32D11, 17C7, 5D3, 4D9, and 33G4) yielded a 648 516 bp sequence spanning 290 kb upstream to 359 kb downstream of the S4-RNase.
Fig. 1.
Construction of BAC contigs, schematic genomic structures, and locations of (pseudo) F-box protein genes around S4-RNase (A) and S2-RNase (B) of the Japanese pear. Names, T7-, and RP-ends of BAC clones are shown in boxes. Black ends represent BAC-ends used for chromosome walking. Hatched boxes indicate BAC clones chosen for complete sequencing. Schematic genomic structures of S4- and S2-haplotypes are shown below the BAC contigs. The directions of transcription of S-RNase and PpSFBB genes are represented by arrows. Physical distances from S-RNase are indicated in parentheses. A double-headed arrow indicates the 236 kb deleted region in the S4sm-haplotype.
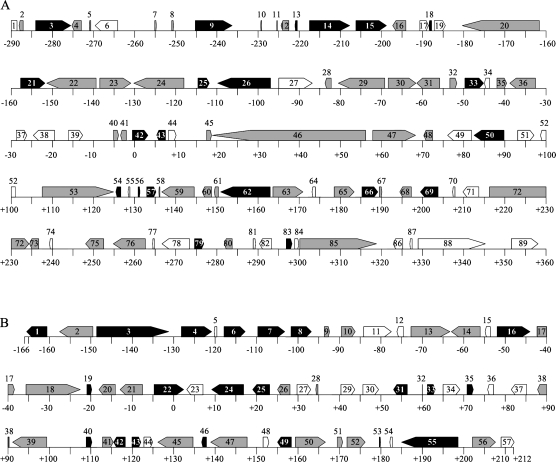
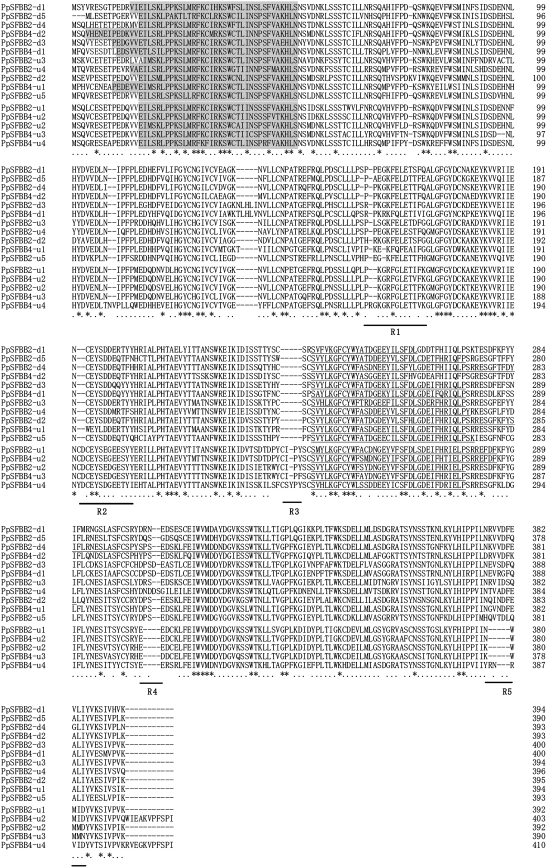
Analysis using GENSCAN software predicted 89 open reading frames (ORFs) in the S4 BAC 649 kb contig sequence (Fig. 2A). Among the 89 ORFs, 34 (ORF33–ORF66) were included in the 236 kb deleted region of the S4sm-haplotype that spans 48 kb upstream to 188 kb downstream of S4-RNase (Fig. 2A; Okada et al., 2008). The other 55 ORFs (ORF1–ORF32 and ORF67–ORF89) were located outside the deleted region. A BLASTX search of the 89 ORFs yielded 61 ORFs with significant similarity (E-value <e-4) to sequences of known proteins in the database (Table 1). Of the 89 ORFs, 40 showed similarity to a (retro) transposon, and 10 were similar to a hypothetical or predicted protein. ORF14 was similar to a zinc knuckle family protein, ORF43 to a zinc finger, ORF50 to a chromosome-associated kinesin KIF4A, and ORF62 to an unknown protein. ORF42 and ORF54 corresponded to S4-RNase and S4F-box0, respectively. Five ORFs (ORF3, ORF9, ORF15, ORF25, and ORF79) were located outside the deleted region, and showed similarity to MdSFBBs and PpSFBB9-γ. Using GENETYX-MAC Ver. 13 software, the predicted ORFs were reanalysed to determine the precise ORFs from the start (ATG) to the stop codon. ORF3, ORF9, ORF15, ORF25, and ORF79 encoded 410, 390, 403, 394, and 393 amino acid residues, respectively. A Pfam motif search predicted that these five proteins had an F-box domain at the N-terminus and an FBA_1 domain in the centre (Fig. 3). These ORFs showed pairwise deduced amino acid sequence identities ranging from 62.9% to 94.4% when compared with reported PpSFBB, MdSFBB, and MdSLF genes (Supplementary Table S3 at JXB online). They represent F-box protein genes that differ from PpSFBB4-α–γ, which are linked to S4-RNase (Sassa et al., 2007). Thus, these five ORFs were assigned as new PpSFBB4 genes. ORF3, ORF9, ORF15, and ORF25 were located ∼284, ∼245, ∼199, and ∼113 kb upstream of S4-RNase. ORF54 (S4F-box0) and ORF79 were located ∼127 kb and ∼275 kb downstream of S4-RNase. These PpSFBB4 genes upstream and downstream of S4-RNase were named PpSFBB4-u and PpSFBB4-d, respectively, and lower case numbers were assigned to the PpSFBB4-u and PpSFBB4-d located close to S4-RNase; therefore, ORF25, ORF15, ORF9, and ORF3 were designated as PpSFBB4-u1, PpSFBB4-u2, PpSFBB4-u3, and PpSFBB4-u4, and ORF54 (S4F-box0) and ORF79 were (re)named as PpSFBB4-d1 and PpSFBB4-d2, respectively. These PpSFBB4 genes around S4-RNase shared the same transcriptional orientation, except for PpSFBB4-d1 (Fig. 1A). Using ATGC Ver. 4 software, the S4 649 kb BAC contig sequence was searched for SFBB-like sequences. The analysis revealed a pseudogene (ΨPpSFBB4-u1) encoding a truncated F-box protein at ∼239 kb upstream of S4-RNase (Fig. 1A). PpSFBB4-u1–u4 and PpSFBB4-d1–d2 shared 67.2–86.2% amino acid sequence identities with each other (Table 3).
Fig. 2.
ORF maps of the region around S4-RNase (A) and S2-RNase (B). Arrowheads indicate the location and transcriptional direction of genes predicted by GENSCAN software. Open arrowheads indicate genes showing no significant homology to proteins in databases. Grey arrowheads represent transposable elements. Black arrowheads indicate non-transposon-like genes. The 649 kb and 378 kb sequences around S4-RNase and S2-RNase have been deposited with the EMBL/GenBank Data Libraries under accession nos AB545981 and AB545982, respectively.
Table 1.
Open reading frames (ORFs) predicted by GENSCAN in the 649 kb region around S4-RNase
| ORFs | Homologous protein | Species | Amino acid identity | Score (bits) | E-valuea | Accession no. | |
| ORF1 | None | ||||||
| ORF2 | Transposon protein | Oryza sativa | 64/122 (52%) | 134 | 9e-30 | DP000011 | |
| ORF3 | (SFBB4-u4) | PpSFBB9-γ | Pyrus pyrifolia | 291/378 (76%) | 566 | 4e-159 | AB297939 |
| ORF4 | Putative retroelement pol polyprotein | Arabidopsis thaliana | 24/36 (66%) | 50.8 | 4e-05 | AC006920 | |
| ORF5 | GAG-POL precursor | Vitis vinifera | 34/124 (27%) | 61.2 | 3e-08 | AB111100 | |
| ORF6 | None | ||||||
| ORF7 | Retrotransposon protein | Oryza sativa | 45/107 (42%) | 79.7 | 9e-14 | DP000009 | |
| ORF8 | Retrotransposon protein | Oryza sativa | 45/158 (28%) | 82.4 | 1e-14 | DP000009 | |
| ORF9 | (SFBB4-u3) | MdSFBB9-β | Malus×domestica | 350/390 (89%) | 726 | 0.0 | AB270792 |
| ORF10 | Retrotransposon protein | Oryza sativa | 31/78 (39%) | 54.3 | 6e-08 | DP000011 | |
| ORF11 | None | ||||||
| ORF12 | Retrotransposon protein | Oryza sativa | 36/82 (43%) | 81.6 | 6e-14 | DP000009 | |
| ORF13 | Hypothetical protein | Vitis vinifera | 59/181 (32%) | 71.6 | 3e-11 | AM472051 | |
| ORF14 | Zinc knuckle family protein | Oryza sativa | 60/182 (32%) | 100 | 4e-19 | DP000010 | |
| ORF15 | (SFBB4-u2) | MdSFBB9-α | Malus×domestica | 300/315 (95%) | 590 | 4e-166 | AB270792 |
| ORF16 | Retrotransposon protein | Oryza sativa | 71/165 (43%) | 124 | 7e-27 | DP000009 | |
| ORF17 | None | ||||||
| ORF18 | Predicted protein | Populus trichocarpa | 44/134 (32%) | 69.7 | 2e-10 | DS017968 | |
| ORF19 | None | ||||||
| ORF20 | Retrotransposon protein | Oryza sativa | 31/81 (38%) | 60.1 | 1e-09 | DP000086 | |
| ORF21 | Hypothetical protein | Vitis vinifera | 43/132 (32%) | 70.5 | 1e-10 | AM426737 | |
| ORF22 | Retrotransposon protein | Beta vulgaris | 61/173 (35%) | 82.0 | 3e-13 | EF101866 | |
| ORF23 | Retrotransposon gag protein | Asparagus officinalis | 418/767 (54%) | 772 | 0.0 | AC183435 | |
| ORF24 | Retrotransposon gag protein | Oryza sativa | 487/1032 (47%) | 921 | 0.0 | AC120534 | |
| ORF25 | (SFBB4-u1) | MdSFBB3-β | Malus×domestica | 306/394 (77%) | 628 | 4e-178 | AB270796 |
| ORF26 | Hypothetical protein | Vitis vinifera | 28/42 (66%) | 64.3 | 4e-08 | AM455744 | |
| ORF27 | None | ||||||
| ORF28 | Reverse transcriptase | Vigna radiata | 23/37 (62%) | 48.5 | 2e-04 | AY684634 | |
| ORF29 | Retrotransposon protein | Oryza sativa | 521/962 (54%) | 1028 | 0.0 | DP000011 | |
| ORF30 | Retrotransposon gag protein | Asparagus officinalis | 48/129 (37%) | 91.3 | 2e-16 | AC183436 | |
| ORF31 | Retrotransposon gag protein | Asparagus officinalis | 827/1636 (50%) | 1568 | 0.0 | AC183435 | |
| ORF32 | Integrase | Populus trichocarpa | 48/149 (32%) | 84.0 | 3e-14 | DQ536160 | |
| ORF33 | Predicted protein | Populus trichocarpa | 34/92 (36%) | 62.4 | 3e-08 | EQ134071 | |
| ORF34 | None | ||||||
| ORF35 | Retrotransposon gag protein | Asparagus officinalis | 95/194 (48%) | 172 | 4e-41 | AC183435 | |
| ORF36 | Retrotransposon gag protein | Asparagus officinalis | 117/405 (28%) | 141 | 2e-31 | AC183435 | |
| ORF37 | None | ||||||
| ORF38 | None | ||||||
| ORF39 | None | ||||||
| ORF40 | Transposon protein Pong subclass | Zea mays | 31/120 (25%) | 60.1 | 3e-07 | EU964924 | |
| ORF41 | Transposon protein Pong subclass | Zea mays | 185/380 (48%) | 355 | 8e-96 | EU962682 | |
| ORF42 | (S4-RNase) | S4-RNase | Pyrus pyrifolia | 49/50 (98%) | 115 | 2e-24 | AB014072 |
| ORF43 | Zinc finger | Medicago truncatula | 23/63 (36%) | 47.8 | 6e-04 | AC148290 | |
| ORF44 | None | ||||||
| ORF45 | Retrotransposon gag protein | Asparagus officinalis | 65/291 (22%) | 59.7 | 5e-07 | AC183435 | |
| ORF46 | Retroelement pol polyprotein-like | Arabidopsis thaliana | 667/1331 (50%) | 1272 | 0.0 | AB024037 | |
| ORF47 | Retrotransposon gag protein | Asparagus officinalis | 793/1644 (48%) | 1496 | 0.0 | AC183435 | |
| ORF48 | Retrotransposon protein | Oryza sativa | 352/728 (48%) | 673 | 0.0 | DP000011 | |
| ORF49 | None | ||||||
| ORF50 | Chromosome-associated kinesin KIF4A | Ricinus communis | 37/96 (38%) | 60.5 | 2e-07 | EQ974117 | |
| ORF51 | None | ||||||
| ORF52 | None | ||||||
| ORF53 | Retrotransposon protein | Beta vulgaris | 101/201 (50%) | 191 | 7e-46 | EF101866 | |
| ORF54 | (SFBB4-d1) | S4F-box0 | Pyrus pyrifolia | 400/400 (100%) | 834 | 0.0 | AB308360 |
| ORF55 | None | ||||||
| ORF56 | Hypothetical protein | Vitis vinifera | 45/154 (29%) | 72.0 | 2e-11 | AM429787 | |
| ORF57 | Hypothetical protein | Vitis vinifera | 50/135 (37%) | 81.3 | 2e-13 | AM467140 | |
| ORF58 | RNase H family protein | Asparagus officinalis | 35/63 (55%) | 78.6 | 2e-18 | AC183436 | |
| ORF59 | Retrotransposon protein | Oryza sativa | 67/121 (55%) | 137 | 1e-30 | DP000009 | |
| ORF60 | Retrotransposon protein | Oryza sativa | 127/297 (42%) | 234 | 2e-59 | DP000009 | |
| ORF61 | Retrotransposon gag protein | Asparagus officinalis | 37/129 (28%) | 74.7 | 1e-11 | AC183435 | |
| ORF62 | Unknown protein | Arabidopsis thaliana | 51/74 (68%) | 102 | 6e-20 | AK117191 | |
| ORF63 | Retrotransposon gag protein | Asparagus officinalis | 119/299 (39%) | 195 | 3e-47 | AC183435 | |
| ORF64 | None | ||||||
| ORF65 | Retrotransposon protein | Beta vulgaris | 35/44 (79%) | 76.3 | 2e-18 | EF101866 | |
| ORF66 | Hypothetical protein | Vitis vinifera | 39/110 (35%) | 55.1 | 5e-06 | AM489256 | |
| ORF67 | RNase H family protein | Asparagus officinalis | 58/121 (47%) | 115 | 2e-24 | AC183436 | |
| ORF68 | Retrotransposon gag protein | Asparagus officinalis | 229/692 (33%) | 341 | 2e-91 | AC183435 | |
| ORF69 | Hypothetical protein | Vitis vinifera | 68/227 (29%) | 84.7 | 3e-14 | AM451669 | |
| ORF70 | None | ||||||
| ORF71 | None | ||||||
| ORF72 | Retrotransposon gag protein | Asparagus officinalis | 714/1299 (54%) | 1440 | 0.0 | AC183435 | |
| ORF73 | Retrotransposon gag protein | Asparagus officinalis | 401/608 (65%) | 821 | 0.0 | AC183435 | |
| ORF74 | None | ||||||
| ORF75 | Integrase | Populus trichocarpa | 212/535 (39%) | 357 | 2e-96 | DQ536178 | |
| ORF76 | Retrotransposon gag protein | Asparagus officinalis | 167/506 (33%) | 223 | 1e-55 | AC183435 | |
| ORF77 | None | ||||||
| ORF78 | None | ||||||
| ORF79 | (SFBB4-d2) | MdSFBB3-β | Malus×domestica | 293/393 (74%) | 561 | 4e-158 | AB270796 |
| ORF80 | Retrotransposon protein | Oryza sativa | 175/487 (35%) | 225 | 9e-57 | DP000009 | |
| ORF81 | None | ||||||
| ORF82 | None | ||||||
| ORF83 | Hypothetical protein | Vitis vinifera | 27/46 (58%) | 56.2 | 1e-06 | AM482339 | |
| ORF84 | None | ||||||
| ORF85 | Retrotransposon protein | Oryza sativa | 242/432 (56%) | 471 | 4e-130 | DP000009 | |
| ORF86 | None | ||||||
| ORF87 | None | ||||||
| ORF88 | None | ||||||
| ORF89 | None |
a Significant similarity corresponds to an E-value <e −4.
Fig. 3.
Alignment of deduced amino acid sequences of PpSFBB4-u1–u4, 4-d1–d2 and PpSFBB2-u1–u5, 2-d1–d5. Amino acid sequences were aligned using ClustalW. Conserved sites and relatively conserved sites are marked with asterisks and dots, respectively. F-box domains and FBA_1 domains of F-box proteins are coloured and underlined, respectively. Accession numbers for the F-box protein genes are as follows: PpSFBB4-u1–u4, 4-d1–d2 (AB545981) and PpSFBB2-u1–u5, 2-d1–d5 (AB545982).
Table 3.
Pairwise amino acid sequence identities (%) of PpSFBB4 and PpSFBB2 genes
| PpSFBB4-u2 | PpSFBB4-u3 | PpSFBB4-u4 | PpSFBB4-d1 | PpSFBB4-d2 | PpSFBB4-α | PpSFBB4-β | PpSFBB4-γ | PpSFBB2-u1 | PpSFBB2-u2 | PpSFBB2-u3 | PpSFBB2-u4 | PpSFBB2-u5 | PpSFBB2-d1 | PpSFBB2-d2 | PpSFBB2-d3 | PpSFBB2-d4 | PpSFBB2-d5 | PpSFBB2-γ | |
| PpSFBB4-u1 | 71.4 | 70.6 | 72.7 | 73.1 | 71.8 | 69.6 | 71.0 | 66.3 | 72.3 | 73.6 | 73.3 | 71.2 | 74.0 | 72.8 | 76.3 | 72.6 | 76.3 | 71.5 | 66.3 |
| PpSFBB4-u2 | – | 86.2 | 73.7 | 67.3 | 69.7 | 83.2 | 67.0 | 70.8 | 92.1 | 87.8 | 71.5 | 66.5 | 68.5 | 70.3 | 72.4 | 72.0 | 75.1 | 66.2 | 70.3 |
| PpSFBB4-u3 | – | 70.3 | 67.2 | 69.5 | 80.5 | 67.2 | 67.4 | 84.9 | 93.1 | 70.1 | 66.9 | 68.8 | 70.8 | 70.6 | 72.4 | 73.7 | 67.1 | 66.9 | |
| PpSFBB4-u4 | – | 69.7 | 68.8 | 70.9 | 67.9 | 77.5 | 71.2 | 73.2 | 68.4 | 67.1 | 70.4 | 70.2 | 70.4 | 69.7 | 72.7 | 69.4 | 77.3 | ||
| PpSFBB4-d1 | – | 71.2 | 70.7 | 69.6 | 63.6 | 68.7 | 68.7 | 68.8 | 68.6 | 72.5 | 73.9 | 72.3 | 82.8 | 74.3 | 73.1 | 63.6 | |||
| PpSFBB4-d2 | – | 69.2 | 67.9 | 63.7 | 67.6 | 70.7 | 69.7 | 68.1 | 71.2 | 77.4 | 73.0 | 74.8 | 84.2 | 73.6 | 63.4 | ||||
| PpSFBB4-α | – | 68.7 | 68.2 | 81.9 | 81.6 | 70.2 | 68.7 | 70.7 | 69.4 | 70.5 | 72.5 | 73.0 | 69.2 | 67.7 | |||||
| PpSFBB4-β | – | 62.3 | 66.6 | 68.4 | 68.8 | 94.9 | 69.2 | 71.8 | 69.0 | 71.3 | 73.0 | 70.5 | 62.1 | ||||||
| PpSFBB4-γ | – | 68.2 | 70.0 | 64.9 | 62.5 | 65.5 | 65.6 | 65.1 | 65.1 | 69.2 | 65.5 | 99.0 | |||||||
| PpSFBB2-u1 | – | 86.0 | 69.7 | 66.3 | 69.7 | 70.5 | 72.0 | 70.5 | 73.3 | 67.9 | 67.7 | ||||||||
| PpSFBB2-u2 | – | 70.5 | 67.9 | 70.7 | 72.8 | 72.8 | 74.1 | 75.1 | 70.2 | 69.5 | |||||||||
| PpSFBB2-u3 | – | 69.5 | 70.7 | 74.1 | 71.1 | 72.3 | 73.8 | 71.0 | 64.9 | ||||||||||
| PpSFBB2-u4 | – | 68.4 | 70.8 | 69.0 | 70.9 | 72.5 | 70.0 | 63.0 | |||||||||||
| PpSFBB2-u5 | – | 71.0 | 74.0 | 72.8 | 74.3 | 71.8 | 65.5 | ||||||||||||
| PpSFBB2-d1 | – | 72.8 | 75.4 | 81.2 | 79.0 | 65.4 | |||||||||||||
| PpSFBB2-d2 | – | 74.9 | 77.8 | 73.6 | 64.3 | ||||||||||||||
| PpSFBB2-d3 | – | 77.1 | 73.6 | 65.1 | |||||||||||||||
| PpSFBB2-d4 | – | 78.7 | 69.4 | ||||||||||||||||
| PpSFBB2-d5 | – | 65.3 |
Values >90% are shown in bold.
To examine expression of PpSFBB4-u1–u4 and PpSFBB4-d1–d2, total RNA was extracted from pollen, pistils, and leaves of the S4 homozygote. RT-PCR analyses were conducted using gene-specific primer pairs (Supplementary Table S2 at JXB online). PpSFBB4-u1–u4 and PpSFBB4-d1–d2 were all specifically expressed in pollen, but not in pistils or leaves (Supplementary Fig. S1A). The PpSFBB4-d2-specific primer pair yielded fragments of 1373 bp and 1142 bp, which both were derived from the PpSFBB4-d2 transcript, because the forward primer annealed to the 5' untranslated region (UTR) and the coding region of PpSFBB4-d2 (Supplementary Table S2, Fig. S1A). Thus, in the 649 kb sequence around S4-RNase there were six F-box protein genes (PpSFBB4-u1–u4 and PpSFBB4-d1–d2) with pollen-specific expression. The three PpSFBB genes previously shown to be linked to the S4-RNase, PpSFBB4-α–γ, were not within the sequenced region.
Construction of a BAC contig around the S2-RNase gene
To analyse the sequence polymorphism of PpSFBB4-u1–u4 and PpSFBB4-d1–d2 in another haplotype, a BAC library was constructed from the Japanese pear cultivar ‘Choujuuro’ (S2S3). The BAC library consisted of two sublibraries derived from two DNA size fractions. One sublibrary, which was derived from the 145–185 kb size fraction, consisted of 30 720 clones with an average insert size of 111 kb. The other sublibrary, which was derived from the 185–205 kb size fraction, consisted of 30 720 clones with an average insert size of 127 kb. The average insert size of the whole BAC library was ∼119 kb. The haploid genome size of pear is estimated to be 496–536 Mb (Arumuganathan and Earle, 1991). Therefore, the BAC library represented ∼14-fold genome coverage, giving a >99% theoretical probability of recovering any single-copy DNA sequences in the genome.
To construct a BAC contig around S2-RNase, chromosome walking was initiated from S2-RNase. PCR screening of the BAC library of ‘Choujuuro’ with an S-RNase-specific primer pair yielded 10 BAC clones containing S2-RNase: 2E10, 5B5, 13C10, 15E3, 21F7, 37F11, 41A10, 48F8, 53E3, and 57B1. These BAC clones were aligned by PCR analysis with primer pairs designed from each BAC-end sequence, and a first contig was constructed based on the insert size and restriction pattern of the BAC plasmids (Fig. 1B).
For chromosome walking, two non-repetitive and S2-haplotype specific primer pairs, 13C10-RP and 2E10-T7, were selected from the BAC-end primer pairs located at the outer ends of the first contig (Supplementary Table S1 at JXB online). PCR screening of the BAC library with 13C10-RP yielded two BAC clones (43D6 and 55A9) upstream of S2-RNase. PCR screening of the BAC library with 2E10-T7 yielded five BAC clones (3A1, 7F3, 10H7, 27C4, and 52C1) downstream of S2-RNase. Finally, chromosome walking from S2-RNase yielded a total of 17 BAC clones. These were aligned to construct a BAC contig of ∼391 kb spanning 166 kb upstream to 225 kb downstream of S2-RNase (Fig. 1B).
Sequence analysis of 378 kb around the S2-RNase gene
To identify the genes around S2-RNase, three overlapping BAC clones, 55A9, 48F8, and 10H7, were subcloned and completely sequenced (Fig. 1B). Sequence assembly of the three BAC clones yielded a 378 419 bp sequence. Analysis using GENSCAN software predicted 57 ORFs in the 378 kb region (Fig. 2B). A BLASTX search of these ORFs yielded 41 ORFs with significant similarity (E-value <e-4) to sequences of known proteins in the database (Table 2). ORF22 corresponded to S2-RNase. Among the 57 ORFs, 20 were similar to (retro) transposons and four were similar to a hypothetical protein. ORF1 was similar to a serine-threonine protein kinase, ORF4 to the DNA glycosylase DEMETER, ORF6 to a DNA glycosylase, ORF40 and ORF42 to TIR-NBS-LRR-type disease resistance proteins, and ORF53 to a cyclin-like F-box. Ten ORFs (ORF3, ORF7, ORF8, ORF16, ORF19, ORF24, ORF31, ORF43, ORF46, and ORF49) showed high sequence similarity to MdSFBB genes, MdSLF genes, PpSFBB4-β, or S4F-box0. Using GENETYX-MAC Ver. 13 software, the predicted ORFs were reanalysed to determine the precise ORFs from the start (ATG) to the stop codon. ORF3, ORF7, ORF8, ORF16, ORF19, ORF24, ORF31, ORF43, ORF46, and ORF49 encoded 393, 396, 394, 392, 392, 394, 395, 400, 393, and 390 amino acid residues, respectively. A Pfam motif search predicted that these proteins had an F-box domain at the N-terminus and an FBA_1 domain in the centre (Fig. 3). When compared with reported PpSFBB, MdSFBB, and MdSLF genes, these ORFs showed pairwise deduced amino acid sequence identities ranging from 62.0% to 94.9% (Supplementary Table S3 at JXB online). The F-box protein genes differed from PpSFBB2-γ, which was reported to be linked to S2-RNase (Kakui et al., 2007). Thus, these ORFs were assigned as new PpSFBB2 genes. ORF3, ORF7, ORF8, ORF16, and ORF19 were located ∼146, ∼108, ∼102, ∼53, and ∼22 kb, respectively, upstream of S2-RNase. ORF24, ORF31, ORF43, ORF46, and ORF49 were located ∼10, ∼56, ∼119, ∼137, and ∼158 kb, respectively, downstream of S2-RNase. These new PpSFBB2 genes upstream and downstream of S2-RNase were named PpSFBB2-u and PpSFBB2-d5, respectively, and lower case numbers were assigned to the PpSFBB2-u and PpSFBB2-d located close to S2-RNase. Therefore, ORF19, ORF16, ORF8, ORF7, and ORF3 were designated as PpSFBB2-u1, PpSFBB2-u2, PpSFBB2-u3, PpSFBB2-u4, and PpSFBB2-u5, and ORF24, ORF31, ORF43, ORF46, and ORF49 were designated as PpSFBB2-d1, PpSFBB2-d2, PpSFBB2-d3, PpSFBB2-d4, and PpSFBB2-d5, respectively. These PpSFBB2 genes around S2-RNase shared variable transcriptional orientations (Fig. 1B). Using ATGC Ver. 4 software, the 378 kb S2 BAC contig sequence was searched for SFBB-like sequences. The analysis revealed two pseudogenes (ΨPpSFBB2-u1 and ΨPpSFBB2-u2) encoding truncated F-box proteins that were located ∼46 kb upstream and ∼159 kb upstream of S2-RNase, respectively (Fig. 1B). PpSFBB2-u1–u5 and PpSFBB2-d1–d5 shared 66.3–86.0% amino acid sequence identity with each other, and showed 66.2–93.1% identity with PpSFBB4-u1–u4 and PpSFBB4-d1–d2 (Table 3).
Table 2.
Open reading frames (ORFs) predicted by GENSCAN in the 378 kb region around S2-RNase
| ORFs | Homologous protein | Species | Amino acid identity | Score (bits) | E-valuea | Accession no. | |
| ORF1 | Serine-threonine protein kinase | Ricinus communis | 262/411 (63%) | 506 | 2e-141 | EQ974075 | |
| ORF2 | Retrotransposon protein | Oryza sativa | 189/479 (39%) | 355 | 1e-95 | DP000011 | |
| ORF3 | (SFBB2-u5) | S2-locus F-box | Malus×domestica | 280/312 (89%) | 602 | 9e-170 | DQ422811 |
| ORF4 | DNA glycosylase DEMETER | Arabidopsis thaliana | 351/1051 (33%) | 385 | 1e-110 | DQ335243 | |
| ORF5 | None | ||||||
| ORF6 | DNA glycosylase | Populus trichocarpa | 196/291 (67%) | 365 | 4e-99 | CM000346 | |
| ORF7 | (SFBB2-u4) | PpSFBB4-β | Pyrus pyrifolia | 376/396 (94%) | 752 | 0.0 | AB270798 |
| ORF8 | (SFBB2-u3) | S1-locus F-box | Malus×domestica | 369/394 (93%) | 747 | 0.0 | DQ422810 |
| ORF9 | Retrotransposon protein | Oryza sativa | 78/248 (31%) | 104 | 2e-20 | DP000010 | |
| ORF10 | Retrotransposon protein | Oryza sativa | 132/267 (49%) | 249 | 8e-64 | DP000010 | |
| ORF11 | None | ||||||
| ORF12 | None | ||||||
| ORF13 | Retrotransposon protein | Oryza sativa | 38/82 (46%) | 83.6 | 3e-14 | DP000086 | |
| ORF14 | GAG-POL precursor | Vitis vinifera | 65/208 (31%) | 105 | 2e-20 | AB111100 | |
| ORF15 | None | ||||||
| ORF16 | (SFBB2-u2) | MdSFBB9-β | Malus×domestica | 369/391 (94%) | 768 | 0.0 | AB270792 |
| ORF17 | Retrotransposon protein | Oryza sativa | 332/820 (40%) | 570 | 4e-160 | DP000011 | |
| ORF18 | Retrotransposon protein | Oryza sativa | 96/180 (53%) | 184 | 5e-44 | DP000011 | |
| ORF19 | (SFBB2-u1) | MdSFBB9-α | Malus×domestica | 361/392 (92%) | 694 | 0.0 | AB270792 |
| ORF20 | Retrotransposon protein | Oryza sativa | 269/693 (38%) | 466 | 8e-129 | DP000009 | |
| ORF21 | Retrotransposon protein | Beta vulgaris | 41/57 (71%) | 89.0 | 9e-16 | EF101866 | |
| ORF22 | (S2-RNase) | S2-RNase | Pyrus pyrifolia | 191/191 (100%) | 410 | 5e-112 | AB014073 |
| ORF23 | None | ||||||
| ORF24 | (SFBB2-d1) | MdSFBB3-α | Malus×domestica | 366/394 (92%) | 735 | 0.0 | AB270795 |
| ORF25 | Hypothetical protein | Vitis vinifera | 22/36 (61%) | 46.2 | 3e-09 | AM426737 | |
| ORF26 | Retrotransposon protein | Oryza sativa | 34/68 (50%) | 80.9 | 4e-13 | DP000009 | |
| ORF27 | None | ||||||
| ORF28 | Retrotransposon protein | Beta vulgaris | 25/38 (65%) | 54.3 | 4e-06 | EF101866 | |
| ORF29 | None | ||||||
| ORF30 | None | ||||||
| ORF31 | (SFBB2-d2) | MdSFBB3-β | Malus×domestica | 304/394 (77%) | 637 | 0.0 | AB270796 |
| ORF32 | None | ||||||
| ORF33 | Hypothetical protein | Vitis vinifera | 69/195 (35%) | 88.2 | 9e-16 | AM483001 | |
| ORF34 | None | ||||||
| ORF35 | Hypothetical protein | Vitis vinifera | 49/180 (27%) | 60.8 | 2e-07 | AM423348 | |
| ORF36 | None | ||||||
| ORF37 | None | ||||||
| ORF38 | Retroelement pol polyprotein-like | Arabidopsis thaliana | 151/243 (62%) | 238 | 1e-60 | AB024037 | |
| ORF39 | Retrotransposon gag protein | Asparagus officinalis | 72/152 (47%) | 146 | 5e-33 | AC183435 | |
| ORF40 | TIR-NBS-LRR-type disease resistance protein | Populus trichocarpa | 109/213 (51%) | 204 | 3e-51 | DQ513203 | |
| ORF41 | LTR retrotransposon like protein | Arabidopsis thaliana | 148/283 (52%) | 283 | 4e-74 | AL022140 | |
| ORF42 | TIR-NBS-LRR-type disease resistance protein | Populus trichocarpa | 66/90 (73%) | 137 | 3e-31 | DQ513203 | |
| ORF43 | (SFBB2-d3) | S4F-box0 | Pyrus pyrifolia | 331/400 (82%) | 681 | 0.0 | AB308360 |
| ORF44 | None | ||||||
| ORF45 | Retrotransposon protein | Oryza sativa | 145/328 (44%) | 249 | 2e-63 | DP000011 | |
| ORF46 | (SFBB2-d4) | MdSFBB3-β | Malus×domestica | 366/392 (93%) | 772 | 0.0 | AB270796 |
| ORF47 | Putative retroelement polyprotein | Arabidopsis thaliana | 388/919 (42%) | 652 | 0.0 | AC018460 | |
| ORF48 | None | ||||||
| ORF49 | (SFBB2-d5) | MdSFBB3-β | Malus×domestica | 309/388 (79%) | 647 | 0.0 | AB270796 |
| ORF50 | Retrotransposon protein | Oryza sativa | 178/526 (33%) | 233 | 1e-58 | DP000010 | |
| ORF51 | Polyprotein 1 | Petunia vein clearing virus | 67/284 (23%) | 66.2 | 7e-09 | AY228106 | |
| ORF52 | Retrotransposon protein | Oryza sativa | 145/350 (41%) | 255 | 2e-65 | DP000011 | |
| ORF53 | Cyclin-like F-box | Medicago truncatula | 41/89 (46%) | 88.2 | 2e-16 | AC150889 | |
| ORF54 | None | ||||||
| ORF55 | Hypothetical protein | Prunus persica | 42/91 (46%) | 92.8 | 6e-17 | DQ863257 | |
| ORF56 | Retrotransposon protein | Beta vulgaris | 64/86 (74%) | 139 | 7e-31 | EF101866 | |
| ORF57 | None |
a Significant similarity corresponds to an E-value <e −4.
Total RNA was extracted from pollen, pistils, and leaves of the S2 homozygote to examine the expression of PpSFBB2-u1–u5 and SFBB2-d1–d5. RT-PCR analyses were conducted using gene-specific primer pairs (Supplementary Table S2 at JXB online). PpSFBB2-u1–u5 and PpSFBB2-d1–d5 were all specifically expressed in pollen, but not in pistils or leaves (Supplementary Fig. S1B). Thus, in the 378 kb sequence around S2-RNase there were 10 F-box protein genes (PpSFBB2-u1–u5 and PpSFBB2-d1–d5) with pollen-specific expression. The PpSFBB2-γ gene, previously shown to be linked to the S2-RNase, was not within the sequenced region.
Comparison of deduced amino acid sequences between the PpSFBB4 and PpSFBB2 genes
The pairwise deduced amino acid sequence identities of nine PpSFBB4 genes (PpSFBB4-u1–u4, PpSFBB4-d1–d2, and PpSFBB4-α–γ) and 11 PpSFBB2 genes (PpSFBB2-u1–u5, PpSFBB2-d1–d5, and PpSFBB2-γ) were compared within and between haplotypes (Table 3). Sequence identity among the PpSFBB4 genes ranged from 62.3% to 86.2%, and among PpSFBB2 genes ranged from 63.0% to 86.0%. The PpSFBB4 and PpSFBB2 genes showed 62.1–99.0% identity between haplotypes. Identities of >90% were found between PpSFBB4-u2 and PpSFBB2-u1 (92.1%), PpSFBB4-u3 and PpSFBB2-u2 (93.1%), PpSFBB4-β and PpSFBB2-u4 (94.9%), and PpSFBB4-γ and PpSFBB2-γ (99.0%). Identities ranging from 80% to 90% were found between PpSFBB4-u2 and PpSFBB2-u2 (87.8%), PpSFBB4-u3 and PpSFBB2-u1 (84.9%), PpSFBB4-d1 and PpSFBB2-d3 (82.8%), PpSFBB4-d2 and PpSFBB2-d4 (84.2%), PpSFBB2-u1 and PpSFBB4-α (81.9%), and PpSFBB2-u2 and PpSFBB4-α (81.6%). The other 89 pairwise comparisons showed identities of <80%.
Phylogenetic analysis of the F-box protein genes of Pyrus and Malus
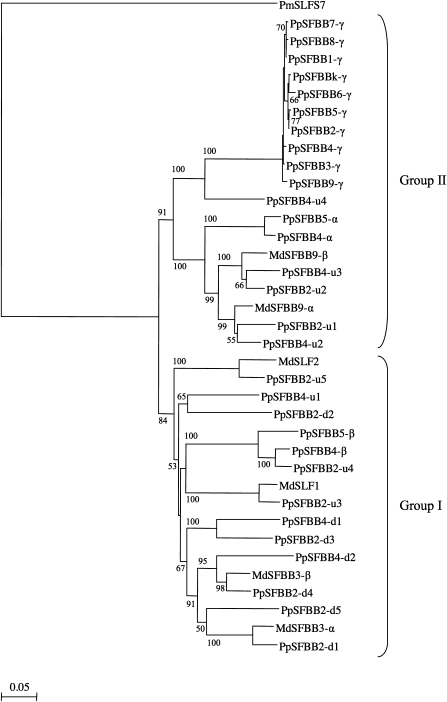
Most PpSFBB4 and PpSFBB2 genes cloned in this study shared the highest amino acid sequence identities with the F-box protein genes of Malus (MdSFBB and MdSLF genes), although PpSFBB4-u4 and PpSFB2-u4 showed the highest identities with PpSFBB3, 4, 9-γ (77.5%) and PpSFBB4-β (94.9%) derived from the same species, respectively (Supplementary Table S3 at JXB online). The deduced amino acid sequences of the 36 F-box protein genes of Pyrus and Malus were aligned with PmSLFS7 of P. mume using ClustalW, and a rooted phylogenetic tree was constructed by the Neighbor–Joining method with PmSLFS7 as an outgroup (Fig. 4). F-box protein genes of Pyrus and Malus did not form taxa-independent clusters, and several PpSFBB genes were positioned closest to MdSFBB and MdSLF genes. The F-box protein genes of Pyrus and Malus were grouped into two major groups: group I (84% bootstrap value) and group II (91% bootstrap value). Group I included PpSFBB4-u1, 4-d1–d2, PpSFBB2-u3–u5, 2-d1–d5, PpSFBB-β genes, MdSFBB3 genes, and MdSLF genes, while group II included PpSFBB4-u2–u4, PpSFBB2-u1–u2, PpSFBB-α genes, PpSFBB-γ genes, and MdSFBB9 genes. Comparing group I with group II, amino acid sequences were conserved in F-box domains, but were divergent in the five regions designated as R1, R2, R3, R4, and R5. In these regions, sequences and/or insertions/deletions (indels) were relatively conserved within each group (Fig. 3).
Fig. 4.
Phylogenetic analysis of the F-box protein genes of Pyrus and Malus, and Japanese apricot PmSLFS7. The phylogenetic tree was constructed using the Neighbor–Joining method. PmSLFS7 was used as an outgroup. Numbers besides the branches are bootstrap values >50%. The bar under the tree represents the number of amino acid substitutions per site.
Discussion
The results of a previous study suggested that the pollen S4 allele is distal to the region from 48 kb upstream to 188 kb downstream of S4-RNase (Okada et al., 2008). In this study, the BAC contig around S4-RNase was extended to 659 kb, and a 648 516 bp region spanning 290 kb upstream to 359 kb downstream of S4-RNase was sequenced. Sequence analysis of the 649 kb region predicted five new pollen-specific F-box protein genes (PpSFBB4-u1–u4 and PpSFBB4-d2). The 649 kb sequence around S4-RNase included six PpSFBB4 genes including PpSFBB4-d1 (S4F-box0), but not PpSFBB4-α–γ. In addition, a BAC library was constructed from ‘Choujuuro’ (S2S3), and a BAC contig of 391 kb around S2-RNase was assembled. Sequence analysis of a 378 419 bp region spanning 166 kb upstream to 212 kb downstream of S2-RNase predicted 10 new pollen-specific F-box protein genes (PpSFBB2-u1–u5, 2-d1–d5). The 378 kb sequence around S2-RNase included 10 PpSFBB2 genes, but not PpSFBB2-γ. The predicted products of PpSFBB4-u1–u4, 4-d1–d2, and PpSFBB2-u1–u5, 2-d1–d5 showed typical features of F-box proteins: an F-box domain at the N-terminus and an FBA_1 domain in the centre (Fig. 3). These results indicated that F-box protein genes with pollen-specific expression are clustered around the S-RNase of Japanese pear, and that PpSFBB4-α–γ and PpSFBB2-γ, which are linked to the S-RNase, were located outside the sequenced region.
Organization of the F-box protein gene cluster around the S-RNase gene of Japanese pear
Among PpSFBB4-u1–u4, 4-d1–d2 and PpSFBB2-u1–u5, 2-d1–d5, some genes may be located more distantly from S-locus regions. Entani et al. (2003) conducted pattern matching analysis of homologies (Harr plot analysis) for the sequences around PmS1- and PmS7-RNases of P. mume. Their results revealed that highly divergent S1- and S7-locus regions are surrounded by co-linear flanking regions, and that S1- and S7-locus regions are ∼27 kb and 15 kb long, respectively. Harr plot analysis of the 649 kb and 378 kb sequences around S4- and S2-RNases was conducted, and no co-linearity was found between these sequences (data not shown). This result suggests that both the 649 kb and 378 kb sequences are a part of the S-locus region, or that either sequence could contain both the S-locus region and its flanking region. Sequence analysis of the 649 kb and 378 kb regions predicted 40 and 20 transposon-like sequences around S4-RNase and S2-RNase, respectively (Tables 1, 2). The S-locus, which controls S-RNase-based GSI, contains many transposon-like sequences. For example, transposon-like sequences were found in three out of 12 ORFs in 72 kb of the P. dulcis Sc-haplotype (Ushijima et al., 2003), in four out of 11 ORFs in 64 kb of the A. hispanicum S2-haplotype (Lai et al., 2002), and in 31 out of 50 ORFs in 328 kb of the P. inflata S2-haplotype (Wang et al., 2004). These transposon-like sequences generate polymorphisms among S-haplotypes, and might contribute to suppression of recombination between S-RNase and SLF/SFB. In the sequenced regions around S4-RNase and S2-RNase, the non-co-linearity, the abundant (retro) transposon insertions, and the absence of PpSFBB4-α–γ and PpSFBB2-γ suggest that the 649 kb and 378 kb sequences around S4-RNase and S2-RNase are part of the S-locus region, and that the S-locus regions of the Japanese pear are probably larger than those of Prunus species.
The organization of the F-box protein gene clusters around the S4-RNase and S2-RNase was compared when S4-RNase and S2-RNase were fixed in the same transcriptional orientation (Fig. 1). PpSFBB4-u1 and PpSFBB4-d1 are located ∼113 kb upstream and ∼127 kb downstream of S4-RNase, whereas PpSFBB2-u1 and PpSFBB2-d1 are located close to S2-RNase (∼22 kb upstream and ∼10 kb downstream of S2-RNase, respectively). The average densities of F-box protein genes were one gene/108 kb around S4-RNase and one gene/38 kb around S2-RNase. Together, these results suggest that F-box protein genes are clustered in the region around S2-RNase more tightly than in the region around S4-RNase.
F-box protein genes, SLF/SFB and SLF-like genes (SLFL), were identified in cosmid and fosmid contigs around the S-RNase of Prunus species. SLF/SFB genes are the pollen S genes, but SLFL genes are probably not involved in SI recognition (Entani et al., 2003; Ushijima et al., 2003). SLF/SFB and SLFL1–SLFL3 cloned from the same haplotypes show low amino acid sequence identity with each other. For example, PmSLFS7 is 11.7–16.9% identical to PmSLFL1S7, PmSLFL2S7, and PmSLFL3S7, which share 26.9–45.3% identity with each other (Entani et al., 2003; Matsumoto et al., 2008). PdSFBc and PdSFBd are 18.7 and 20.2% identical to PdSLFc and PdSLFd (orthologuess of PmSLFL1 of P. mume), respectively (Ushijima et al., 2003). In contrast to Prunus species, PpSFBB4-u1–u4, 4–d1–d2 and PpSFBB2-u1–u5, 2-d1–d5 shared 67.2–86.2% and 66.3–86.0% identity within each haplotype, respectively (Table 3). This indicates that the region around an S-RNase of the Japanese pear comprises related F-box protein genes, which is different from the F-box protein gene organization around the S-RNases in Prunus, in which there are clusters of F-box protein genes that show low levels of identity to each other. The amino acid sequence identities between PpSFBB4-u1–u4, 4-d1–d2 and PpSFBB2-u1–u5, 2-d1–d5 ranged from 66.2% to 93.1% (Table 3), and were higher than those within each haplotype (66.3–86.2%). These similarities between haplotypes indicated that related polymorphic F-box protein genes between haplotypes were clustered in the regions around S4-RNase and S2-RNase.
Classification of PpSFBB genes based on phylogenetic analysis and sequence polymorphism
In Prunus species, F-box protein genes around S-RNase genes were grouped into two major classes, the SLF/SFB clade and the SLFL clade, by a phylogenetic analysis (Matsumoto et al., 2008). SLF/SFB genes show lower levels of allelic sequence identity (77.8–81.3% for PmSLF genes, 68.4–76.4% for PdSFB genes, and 75.1–81.1% for PavSFB genes, respectively) than SLFL genes (88.5–92.0% for PmSLFL1, 95.8–98.6% for PmSLFL2, and 95.1% for PdSLF) (Entani et al., 2003; Ushijima et al., 2003; Ikeda et al., 2004; Matsumoto et al., 2008). The sequence differences of the F-box protein genes among haplotypes implied that SLF/SFB genes with lower levels of identity were pollen S candidates, and that SLFL genes with high levels of identity were not (Entani et al., 2003; Ushijima et al., 2003).
The phylogenetic relationships and sequence differences of F-box protein genes of Pyrus and Malus would be useful for delineating pollen S candidates from PpSFBB4-u1–u4, 4-d1–d2 and PpSFBB2-u1–u5, 2-d1–d5. Phylogenetic analysis based on the deduced amino acid sequences of 36 F-box protein genes of Pyrus and Malus allowed them to be classified into two major groups, I and II (Fig. 4). PpSFBB4-u2–u4, PpSFBB2-u1–u2, PpSFBB-α genes, PpSFBB-γ genes and MdSFBB9 genes were classified into group II, and the other PpSFBB, MdSFBB3, and MdSLF genes were in group I. The phylogenetic analysis also generated a PpSFBB-γ subgroup and 10 gene pairs of PpSFBB genes and PpSFBB/Malus F-box protein genes. Sequence identities between the paired genes ranged from 76.3% to 96.4% (Table 3, Supplementary Table S3 at JXB online), which were higher than those among PpS2-, PpS4-, PpS5-, MdS1-, MdS2-, MdS3-, and MdS9-RNases (60.9–71.1%). Group I consisted of gene pairs with high levels of identity (>91%): MdSLF2/PpSFBB2-u5 (91.3% identity), PpSFBB4-β/PpSFBB2-u4 (94.9% identity), MdSLF1/PpSFBB2-u3 (93.9% identity), MdSFBB3-β/PpSFBB2-d4 (93.1% identity), MdSFBB3-α/PpSFBB2-d1 (92.9% identity); and gene pairs with low levels of identity: PpSFBB4-u1/PpSFBB2-d2 (76.3% identity) and PpSFBB4-d1/PpSFBB2-d3 (82.8% identity). Group II consisted of the PpSFBB-γ subgroup sharing 97.5–99.7% identity among 10 haplotypes (Kakui et al., 2007), and gene pairs with high levels of identities (>92%): PpSFBB5-α/PpSFBB4-α (96.4% identity, Sassa et al., 2007), PpSFBB4-u3/PpSFBB2-u2 (93.1% identity), and PpSFBB2-u1/PpSFBB4-u2 (92.1% identity). The gene pairs with low levels of identity were included in group I, not in group II, suggesting that pollen S candidates were included in the group I.
Therefore, the group I F-box protein genes from the region around S-RNase with low levels of sequence identity, PpSFBB4-u1/PpSFBB2-d2 and PpSFBB4-d1/PpSFBB2-d3, are expected to be pollen S candidates of Japanese pear. In a previous study, PpSFBB4-d1 (S4F-box0) was thought unlikely to be the pollen S4 allele, because it is found in the deleted region of the S4sm-haplotype (Okada et al., 2008). Interestingly, Saito et al. (2002) observed that S4sm pollen is rejected by pistils harbouring not only the S4-haplotype, but also the S1-haplotype. It seems, therefore, that S4sm pollen has a dual specificity for S4-RNase and S1-RNase, which have high amino acid identity (90.0%) (Ishimizu et al., 1998). This dual specificity is probably due to the loss of PpSFBB4-d1, and S4sm pollen might come to recognize S1-RNase. Therefore, PpSFBB4-d1 might also be a pollen S candidate. However, SLF genes of A. hispanicum and P. inflata share high levels of amino acid identity among haplotypes (97–99% and 88.4–89.7%, respectively; Zhou et al., 2003; Sijacic et al., 2004). There is no evidence for a co-evolutionary relationship between SLF/SFB and S-RNase in A. hispanicum and P. inflata, or in Prunus species, which implies that sequence polymorphism between haplotypes can no longer be considered a reliable diagnostic feature of pollen S genes, and functional analysis must be used to identify pollen S genes (Newbigin et al., 2008). Therefore, all PpSFBB4 genes and PpSFBB2 genes should be considered as pollen S gene candidates.
However, it is not a reasonable interpretation that all PpSFBB genes act in concert as the pollen S genes. Among several F-box protein genes around the S-RNases of Prunus species, A. hispanicum, and P. inflata, one F-box protein gene, SLF/SFB, functions as the pollen S gene; the other F-box protein genes, SLFL genes, are non-S pollen genes (Entani et al., 2003; Ushijima et al., 2003; Zhou et al., 2003; Wang et al., 2004; Hua et al., 2007). Therefore, several PpSFBB genes in a particular haplotype are probably not pollen S genes. The non-pollen S proteins of P. inflata, PiSLFLs, either fail to interact with S-RNase or interact much more weakly than PiSLF. When the deduced amino acid sequences of PiSLF and PiSLFLs were compared, three PiSLF-specific regions (SR1, SR2, and SR3) that confer on PiSLF its unique function in SI were revealed (Hua et al., 2007). Although the interactions of PpSFBBs with S-RNase have not yet been analysed, five regions (R1, R2, R3, R4, and R5) were identified where amino acid sequences were variable between the group I and II F-box proteins (Fig. 3). The sequence differences in these regions might account for different interactions with S-RNase between the group I and II F-box proteins. Therefore, there remains the possibility that the less polymorphic group II F-box protein genes are non-pollen S genes.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Expression of PpSFBB genes located around S4-RNase (A) and S2-RNase (B).
Table S1. Primer pairs used to construct BAC contigs around S-RNase.
Table S2. Gene-specific RT-PCR primer pairs.
Table S3. Pairwise amino acid sequence identities (%) of PpSFBB genes with previously reported PpSFBB, MdSFBB, and MdSLF genes.
Acknowledgments
This work was supported in part by Grants-in-Aid for Scientific Research (B) (nos 16380028; 19380020) and (C) (no. 17580027) from the Ministry of Education, Culture, Sports, Science and Technology, Japan. K.O. was supported by research fellowships of the Japan Society for the Promotion of Science for Young Scientists.
Glossary
Abbreviations
- GSI
gametophytic self-incompatibility
- PFGE
pulsed field gel electrophoresis
- SC
self-compatible
- SFB
S haplotype-specific F-box protein
- SFBB
S locus F-box brothers
- SI
self-incompatibility
- SLF
S locus F-box
References
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumuganathan K, Earle ED. Nuclear DNA content of some important plant species. Plant Molecular Biology Reporter. 1991;9:208–218. [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. Journal of Molecular Biology. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Castillo C, Takasaki T, Saito T, Yoshimura Y, Norioka S, Nakanishi T. Reconsideration of S-genotype assignments, and discovery of a new allele based on S-RNase PCR-RFLPs in Japanese pear cultivars. Breeding Science. 2001;51:5–11. [Google Scholar]
- Cheng JH, Han ZH, Xu XF, Li TZ. Isolation and identification of the pollen-expressed polymorphic F-box genes linked to the S-locus in apple (Malus×domestica) Sexual Plant Reproduction. 2006;19:175–183. [Google Scholar]
- de Nettancourt D. Incompatibility and incongruity in wild and cultivated plants. Berlin: Springer-Verlag; 2001. [Google Scholar]
- Entani T, Iwano M, Shiba H, Che FS, Isogai A, Takayama S. Comparative analysis of the self-incompatibility (S-) locus region of Prunus mume: identification of a pollen-expressed F-box gene with allelic diversity. Genes to Cells. 2003;8:203–213. doi: 10.1046/j.1365-2443.2003.00626.x. [DOI] [PubMed] [Google Scholar]
- Hauck NR, Ikeda K, Tao R, Iezzoni AF. The mutated S1-haplotype in sour cherry has an altered S-haplotype-specific F-box protein gene. Journal of Heredity. 2006;97:514–520. doi: 10.1093/jhered/esl029. [DOI] [PubMed] [Google Scholar]
- Hua ZH, Meng XY, Kao T- H. Comparison of Petunia inflata S-locus F-box protein (Pi SLF) with Pi SLF-like proteins reveals its unique function in S-RNase based self-incompatibility. The Plant Cell. 2007;19:3593–3609. doi: 10.1105/tpc.107.055426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Lee HS, Karunanandaa B, Kao TH. Ribonuclease activity of Petunia inflata S-proteins is essential for rejection of self-pollen. The Plant Cell. 1994;6:1021–1028. doi: 10.1105/tpc.6.7.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Igic B, Ushijima K, Yamane H, Hauck NR, Nakano R, Sassa H, Iezzoni AF, Kohn JR, Tao R. Primary structural features of the S-haplotype-specific F-box protein, SFB in Prunus. Sexual Plant Reproduction. 2004;16:235–243. [Google Scholar]
- Ishimizu T, Inoue K, Shimonaka M, Saito T, Terai O, Norioka S. PCR-based method for identifying the S-genotypes of Japanese pear cultivars. Theoretical and Applied Genetics. 1999;98:961–967. [Google Scholar]
- Ishimizu T, Sato Y, Saito T, Yoshimura Y, Norioka S, Nakanishi T, Sakiyama F. Identification and partial amino acid sequences of seven S-RNases associated with self-incompatibility of Japanese pear, Pyrus pyrifolia Nakai. Journal of Biochemistry. 1996;120:326–334. doi: 10.1093/oxfordjournals.jbchem.a021417. [DOI] [PubMed] [Google Scholar]
- Ishimizu T, Shinkawa T, Sakiyama F, Norioka S. Primary structural features of rosaceous S-RNases associated with gametophytic self-incompatibility. Plant Molecular Biology. 1998;37:931–941. doi: 10.1023/a:1006078500664. [DOI] [PubMed] [Google Scholar]
- Kakui H, Tsuzuki T, Koba T, Sassa H. Polymorphism of SFBB-gamma and its use for S genotyping in Japanese pear (Pyrus pyrifolia) Plant Cell Reports. 2007;26:1619–1625. doi: 10.1007/s00299-007-0386-8. [DOI] [PubMed] [Google Scholar]
- Lai Z, Ma WS, Han B, Liang LZ, Zhang YS, Hong GF, Xue YB. An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Molecular Biology. 2002;50:29–42. doi: 10.1023/a:1016050018779. [DOI] [PubMed] [Google Scholar]
- Lechner E, Achard P, Vansiri A, Potuschak T, Genschik P. F-box proteins everywhere. Current Opinion in Plant Biology. 2006;9:631–638. doi: 10.1016/j.pbi.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Matsumoto D, Yamane H, Tao R. Characterization of SLFL1, a pollen-expressed F-box gene located in the Prunus S locus. Sexual Plant Reproduction. 2008;21:113–121. [Google Scholar]
- McClure BA, Franklin-Tong V. Gametophytic self-incompatibility: understanding the cellular mechanisms involved in ‘self’ pollen tube inhibition. Planta. 2006;224:233–245. doi: 10.1007/s00425-006-0284-2. [DOI] [PubMed] [Google Scholar]
- McClure BA, Gray JE, Anderson MA, Clarke AE. Self-incompatibility in Nicotiana alata involves degradation of pollen ribosomal RNA. Nature. 1990;347:757–760. [Google Scholar]
- McClure BA, Haring V, Ebert PR, Anderson MA, Simpson RJ, Sakiyama F, Clarke AE. Style self-incompatibility gene products of Nicotiana alata are ribonucleases. Nature. 1989;342:955–957. doi: 10.1038/342955a0. [DOI] [PubMed] [Google Scholar]
- McCubbin AG, Kao TH. Molecular recognition and response in pollen and pistil interactions. Annual Review of Cell and Developmental Biology. 2000;16:333–364. doi: 10.1146/annurev.cellbio.16.1.333. [DOI] [PubMed] [Google Scholar]
- Newbigin E, Paape T, Kohn JR. RNase-based self-incompatibility: puzzled by pollen S. The Plant Cell. 2008;20:2286–2292. doi: 10.1105/tpc.108.060327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Tonaka N, Moriya Y, Norioka N, Sawamura Y, Matsumoto T, Nakanishi T, Takasaki-Yasuda T. Deletion of a 236 kb region around S4-RNase in a stylar-part mutant S4sm-haplotype of Japanese pear. Plant Molecular Biology. 2008;66:389–400. doi: 10.1007/s11103-007-9277-1. [DOI] [PubMed] [Google Scholar]
- Potter D, Eriksson T, Evans RC, et al. Phylogeny and classification of Rosaceae. Plant Systematics and Evolution. 2007;266:5–43. [Google Scholar]
- Saito T, Sato Y, Sawamura Y, Shoda M, Kotobuki K. Studies on breeding of self-compatibility in Japanese pear. 2. Characteristic of pollen of S4sm gene originated from ‘Osa-nijisseiki’. Journal of the Japanese Society for Horticultural Science 71. suppl. 2002;2):123. (in Japanese) [Google Scholar]
- Saitou N, Nei M. The neighbor–joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sassa H, Kakui H, Miyamoto M, Suzuki Y, Hanada T, Ushijima K, Kusaba M, Hirano H, Koba T. S locus F-box brothers: multiple and pollen-specific F-box genes with S haplotype-specific polymorphisms in apple and Japanese pear. Genetics. 2007;175:1869–1881. doi: 10.1534/genetics.106.068858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijacic P, Wang X, Skirpan AL, Wang Y, Dowd PE, McCubbin AG, Huang S, Kao TH. Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature. 2004;429:302–305. doi: 10.1038/nature02523. [DOI] [PubMed] [Google Scholar]
- Sato Y. Breeding of self-compatible Japanese pear. In: Hayashi T, Omura M, Scott NS, editors. Techniques on gene diagnosis and breeding in fruit trees. 1993. pp. 241–247. Tsukuba, Japan: FTRS (Fruit Tree Research Station) [Google Scholar]
- Sonneveld T, Tobutt KR, Vaughan SP, Robbins TP. Loss of pollen S function in two self-compatible selections of Prunus avium is associated with deletion/mutation of an S haplotype-specific F-box gene. The Plant Cell. 2005;17:37–51. doi: 10.1105/tpc.104.026963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terai O, Sato Y, Saito T, Abe K, Kotobuki K. Identification of homozygotes of self-incompatibility gene (S-gene), as useful tools to determine the S-genotype in Japanese pear, Pyrus pyrifolia Nakai. Bulletin of the National Institute of Fruit Tree Science. 1999;32:31–38. [Google Scholar]
- Tsukamoto T, Hauck NR, Tao R, Jiang N, Iezzoni AF. Molecular characterization of three non-functional S-haplotypes in sour cherry (Prunus cerasus) Plant Molecular Biology. 2006;62:371–383. doi: 10.1007/s11103-006-9026-x. [DOI] [PubMed] [Google Scholar]
- Ushijima K, Sassa H, Dandekar AM, Gradziel TM, Tao R, Hirano H. Structural and transcriptional analysis of the self-incompatibility locus of almond: identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. The Plant Cell. 2003;15:771–781. doi: 10.1105/tpc.009290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima K, Yamane H, Watari A, Kakehi E, Ikeda K, Hauck NR, Iezzoni AF, Tao RT. The S haplotype-specific F-box protein gene, SFB, is defective in self-compatible haplotypes of Prunus avium and P. mume. The Plant Journal. 2004;39:573–586. doi: 10.1111/j.1365-313X.2004.02154.x. [DOI] [PubMed] [Google Scholar]
- Vilanova S, Badenes ML, Burgos L, Martinez-Calvo J, Llacer G, Romero C. Self-compatibility of two apricot selections is associated with two pollen-part mutations of different nature. Plant Physiology. 2006;142:629–641. doi: 10.1104/pp.106.083865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tsukamoto T, Yi KW, Wang X, Huang SS, McCubbin AG, Kao TH. Chromosome walking in the Petunia inflata self-incompatibility (S) locus and gene identification in an 881 kb contig containing S2-RNase. Plant Molecular Biology. 2004;54:727–742. doi: 10.1023/B:PLAN.0000040901.98982.82. [DOI] [PubMed] [Google Scholar]
- Xue YB, Carpenter R, Dickinson HG, Coen ES. Origin of allelic diversity in antirrhinum S locus RNases. The Plant Cell. 1996;8:805–814. doi: 10.1105/tpc.8.5.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JL, Wang F, Ma WS, Zhang YS, Han B, Xue YB. Structural and transcriptional analysis of S-locus F-box genes in. Antirrhinum. Sexual Plant Reproduction. 2003;16:165–177. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.