Abstract
The response to vernalization and the expression of genes associated with responses to vernalization (VRNH1, VRNH2, and VRNH3) and photoperiod (PPDH1 and PPDH2) were analysed in four barley (Hordeum vulgare L.) lines: ‘Alexis’ (spring), ‘Plaisant’ (winter), SBCC058, and SBCC106 (Spanish inbred lines), grown under conditions of vernalization and short days (VSD) or no vernalization and long days (NVLD). The four genotypes differ in VRNH1. Their growth habits and responses to vernalization correlated with the level of expression of VRNH1 and the length of intron 1. ‘Alexis’ and ‘Plaisant’ behaved as expected. SBCC058 and SBCC106 showed an intermediate growth habit and flowered relatively late in the absence of vernalization. VRNH1 expression was induced by cold for all genotypes. Under VSD, VRNH1 expression was detected in the SBCC genotypes later than in ‘Alexis’ but earlier than in ‘Plaisant’. VRNH2 was repressed under short days while VRNH1 expression increased in parallel. VRNH3 was detected only in ‘Alexis’ under NVLD, whereas it was not expressed in plants with the active allele of VRNH2 (SBCC058 and ‘Plaisant’). Under VSD, PPDH2 was expressed in ‘Alexis’, SBCC058, and SBCC106, but it was only expressed weakly in ‘Alexis’ under NVLD. Further analysis of PPDH2 expression in two barley doubled haploid populations revealed that, under long days, HvFT3 and VRNH2 expression levels were related inversely. The timing of VRNH2 expression under a long photoperiod suggests that this gene might be involved in repression of PPDH2 and, indirectly, in the regulation of flowering time through an interaction with the day-length pathway.
Keywords: Barley, gene expression, landraces, Mediterranean conditions, photoperiod, vernalization
Introduction
The classic model of the genetic control of vernalization in barley (Takahashi and Yasuda, 1971) is based on three loci, Sh/sh, Sh2/sh2, and Sh3/sh3, among which epistatic relationships exist. Candidate genes for these three loci in barley have been proposed. HvBM5A (which corresponds to TmAP1 or WAP1 in wheat) was identified as a candidate for VRNH1, which is a synonym of Sh2 (Danyluk et al., 2003; Trevaskis et al., 2003; Yan et al., 2003). VRNH1 promotes the transition of the apex from the vegetative to the reproductive stage. The locus is always expressed at high basal levels in plants that have spring (dominant) alleles (Trevaskis et al., 2006). In winter varieties that are responsive to vernalization, VRNH1 expression is repressed until the plants are exposed to low temperatures (von Zitzewitz et al., 2005; Sasani et al., 2009). Allelic diversity at VRNH1 has been described, mostly in relation to deletions within the first intron (Fu et al., 2005; Cockram et al., 2007; Szűcs et al., 2007). These deletions presumably cause variation in the levels of VRNH1 expression in plants that have not been vernalized (Hemming et al., 2009) and hence lead to different flowering times (Trevaskis et al., 2003; von Zitzewitz et al., 2005).
A cluster of three genes, ZCCT-H, was identified as a candidate for VRNH2, which is synonymous with Sh (Yan et al., 2004). VRNH2 acts as a repressor of flowering and delays flowering in plants that have not been vernalized (Takahashi and Yasuda, 1971; Yan et al., 2004; Karsai et al., 2005). The allelic variation at the VRNH2 locus seems to be of the presence/absence type, although there is still debate over which of the three ZCCT-H genes is functionally responsible (Dubcovsky et al., 2005; Trevaskis et al., 2006; Szűcs et al., 2007). The spring VRNH2 allele is associated with a deletion of the three genes of the ZCCT-H cluster (Yan et al., 2004; Karsai et al., 2005; von Zitzewitz et al., 2005). Studies have found that in cereals VRN2 expression is repressed by short days and by a high level of VRN1 expression (Loukoianov et al., 2005; Trevaskis et al., 2006), which explains the long-known interaction between these two genes (Tranquilli and Dubcovsky, 2000).
HvFT1 is a candidate gene for VRNH3 (Sh3; Yan et al., 2006). It is homologous to the FLOWERING LOCUS T (FT) gene of Arabidopsis (Turck et al., 2008). In Arabidopsis, FT promotes flowering and is activated by long days (Corbesier et al., 2007). In cereals, FT also promotes flowering during long days (Yan et al., 2006; Faure et al., 2007; Hemming et al., 2008). In winter varieties, VRNH3 is only expressed after prolonged exposure to low temperatures (Yan et al., 2006; Hemming et al., 2008). The role of FT1 might extend beyond vernalization and it has been proposed to integrate the vernalization and day-length flowering pathways in cereals (Hemming et al., 2008; Distelfeld et al., 2009).
With regard to genes that are involved in responses to photoperiod, Laurie et al. (1994, 1995) identified two genes with large effects, PPDH1 and PPDH2. PPDH1 confers sensitivity to long photoperiod—that is, the dominant or sensitive allele induces earlier flowering with long days. Turner et al. (2005) identified HvPRR7, a pseudo-response regulator gene, as a candidate for PPDH1, and proposed a diagnostic single nucleotide polymorphism (SNP) that differentiated between alleles that conferred sensitivity and insensitivity to long photoperiod. The dominant PPDH1 allele might accelerate flowering by up-regulation of HvFT1 (Hemming et al., 2008), which is mediated by the activity of CONSTANS (Turner et al., 2005).
PPDH2 affects flowering under conditions with a short photoperiod (Laurie et al., 1995). Recently, HvFT3 has been identified as a candidate gene for PPDH2 (Faure et al., 2007; Kikuchi et al., 2009). Two alleles have been described: a dominant functional allele, which is frequently present in spring varieties, and a recessive non-functional allele, which is mostly present in winter varieties (Faure et al., 2007).
These five genes that are involved in the responses to vernalization and different day-lengths are the major players in the pathways that determine flowering time in barley and other cereals. These pathways, albeit not yet elucidated fully, are rich in interactions between the genes themselves and in responses to environmental cues (Greenup et al., 2009; Shimada et al., 2009; Higgins et al., 2010). The results of these interactions are complex phenotypic responses, which are aimed at the promotion of flowering when optimal environmental conditions are present. Hence, the genes involved in this system should be studied concurrently because their responses might depend on the allelic configurations of the other genes.
Materials and methods
Plant material
Four genotypes of barley (Hordeum vulgare L.) were chosen to assess differences in the expression of the five major genes involved in responses to temperature and day-length: SBCC058 and SBCC106 [inbred lines derived from landraces; belonging to the Spanish Barley Core Collection (SBCC); Igartua et al. (1998)], the French winter cultivar ‘Plaisant’ (‘Ager’בNymphe’), and the German spring cultivar ‘Alexis’ (Br.1622בTriumph’). The genotypes studied exhibit differences in the length of the first intron of the VRNH1 gene, as well as in some of the other major genes involved in the control of responses to vernalization and sensitivity to day-length (Table 1).
Table 1.
Genotypes for the genes associated with responses to vernalization and photoperiod in the cultivars and lines under study
| Cultivar or line | Vernalization and photoperiod genes | ||||
| VRNH1a | VRNH2b | VRNH3c | PPDH1d | PPDH2e | |
| ‘Plaisant’ | vrnh1 | VRNH2 | vrnH3 | PPDH1 | ppdH2 |
| SBCC106 | VRNH1-6 | VRNH2 | vrnH3 | PPDH1 | PPDH2 |
| SBCC058 | VRNH1-4 | VRNH2 | vrnH3 | PPDH1 | PPDH2 |
| ‘Alexis’ | VRNH1-3 | vrnH2 | vrnH3 | ppdH1 | PPDH2 |
| ‘Pané’ | VRNH1-4 | VRNH2 | vrnH3 | PPDH1 | PPDH2 |
| ‘Beka’ | VRNH1-1 | vrnH2 | vrnH3 | ppdH1 | PPDH2 |
| ‘Mogador’ | vrnh1 | VRNH2 | vrnH3 | ppdH1 | ppdH2 |
Alleles based on the size of intron 1, in accordance with Hemming et al. (2009).
Presence/absence of HvZCCT, in accordance with Karsai et al. (2005).
Alleles based on two SNPs in intron 1, as reported by Yan et al. (2006).
Alleles based on SNP22 of Turner et al. (2005).
Alleles based on amplification of a 431 bp product using primers FT3.1F (5'-ATCCATTGGTTGTGTGGCTCA-3') and FT3.2R (5'-ATCTGTCACCAACCTGCACA-3'), which amplify the entire region from exons 1 to 2 of the HvFT3 gene (‘Alexis’, SBCC058, SBCC106, ‘Pané’, and ‘Beka’). These primers give a null allele for ‘Plaisant’ and ‘Mogador’. The allele from ‘Plaisant’ (ppdH2) was amplified using the F4/R1 primers reported by Kikuchi et al. (2009). HvFT3 was localized on the long arm of chromosome 1H in the ‘Beka’בMogador’ mapping population (Supplementary Fig. S1 at JXB online), which matches the location of a QTL for response to a short photoperiod (Supplementary Fig. S2).
Doubled haploid (DH) lines from two different barley crosses [‘Alexis’בPané’ (Cuesta-Marcos et al., 2008a) and ‘Beka’בMogador’ (Cuesta-Marcos et al., 2008b), Table 1] were used to validate some of the results.
Plant growth conditions
The vernalization requirement of ‘Plaisant’, SBCC058, and ‘Alexis’ was evaluated at the Martonvásár (Hungary) phytotron, in accordance with the procedures described by Karsai et al. (2004). SBCC106 was not included in this experiment, but three landraces with the same genotype as SBCC106 in terms of VRNH1/VRNH2 were included. Vernalization was applied in 15 d increments, for a total of four treatments that ranged from no vernalization (0 d) to 45 d of vernalization at 3 °C, under a short-day regime (8 h light/16 h dark) and low light intensity (12±1 μmol m−2 s−1). After vernalization (or 14 d after germination for the samples not subjected to vernalization), the seedlings were transferred to a regime with long days (16 h light) and a high level of light intensity (340±22 μmol m−2 s−1) at 18 °C. For each plant, the number of days to heading, which corresponds to developmental phase 49 on the Zadoks scale (Zadoks et al., 1974), was recorded. The experiment was continued for a total of 150 d. Two plants were tested for each genotype and treatment.
For studies of gene expression, plants of ‘Plaisant’, SBCC058, SBCC106, and ‘Alexis’ were grown in pots in Zaragoza (Spain), in a sunlit glasshouse at 19±1 °C, with a 16 h light/8 h dark photoperiod. Ten days after sowing, when the plants had reached the two-leaf stage (stage 12 of the Zadoks scale), the pots were assigned to one of two groups of the same size and transferred to two growth chambers. Each group was exposed to a distinct experimental treatment. One was a vernalization treatment (VSD), for which the plants were grown at 7±1 °C under a short photoperiod (8 h light/16 h dark) and a low level of light intensity (12 μmol m−2 s−1). The second set of plants was grown under conditions of no vernalization and long days (NVLD) at 22±1 °C and a photoperiod of 16 h light/8 h dark with a high level of light intensity (220 μmol m−2 s−1). The intention was to include SBCC106 in the same experiment, but a seed identification error was detected and prevented the use of the results obtained. SBCC106 was later sown and grown for 10 d under the same conditions in the glasshouse until the two-leaf stage, together with SBCC058 and ‘Plaisant’, but in this case the plants were only subjected to the VSD treatment. Hereafter, the experiment that included ‘Alexis’, SBCC058, and ‘Plaisant’ will be referred to as Experiment 1 (or Exp1) and the later experiment that included SBCC058, SBCC106, and ‘Plaisant’ will be referred to as Experiment 2 (or Exp2). In Exp1, two samples were obtained per genotype for each sampling time, which resulted in two biological replicates. Each sample consisted of two plants that were harvested and pooled. In Exp2, three individual plants per sampling time and genotype were harvested, and were treated as three biological replicates. Harvesting took place on day 0 (just before transfer from the greenhouse to the growth chambers), and after 7, 14, 21, 28, and 35 d of each treatment. An additional sampling at day 42 was carried out in Exp2. In all the experiments, plants were harvested in the middle of the light period.
For the gene expression analysis of the DH lines (‘Alexis’בPané’ and ‘Beka’בMogador’), plants were grown in pots that contained soil in a sunlit glasshouse at a temperature of 19±1 °C, with long days (16 h light/8 h dark). Seven days after sowing, the pots were transferred to a growth chamber, where they were grown under conditions of NVLD. Plants were harvested after 10 d of treatment. At harvest, two samples were collected per genotype. Each sample consisted of the pooled leaf tissue of two plants per genotype, to reduce the effects of individual variation.
RT-PCR and real-time PCR analysis
RNA was extracted from 100 mg of tissue with TRIzol® Reagent (Invitrogen) and treated with DNase (DNase I Recombinant, RNase-free; Roche) to remove possible DNA contamination. An oligo(dT)20 primer (Invitrogen) was used to prime the synthesis of first-strand cDNA from 1 μl of RNA (2.25 μg of total RNA), using SuperScript III Reverse Transcriptase (Invitrogen) in accordance with the manufacturer's instructions. A single reverse transcription reaction was carried out for each RNA sample.
Primers for VRNH1, VRNH2, and Actin were designed in accordance with Trevaskis et al. (2006); primers for VRNH3 in accordance with Yan et al. (2006); and primers for PPDH1 in accordance with Hemming et al. (2008). For PPDH2, the forward primer was designed in accordance with Kikuchi et al. (2009) and the reverse primer in accordance with Faure et al. (2007). In all cases, the same primers were used for semi-quantitative PCR and quantitative real-time PCR (qRT-PCR). Each primer pair amplified cDNA-specific DNA products.
Semi-quantitative PCR
Semi-quantitative PCR was performed in a GeneAmp® PCR System 2700 (Applied Biosystems). Cycling conditions were 4 min at 94 °C, followed by cycles of 30 s denaturation at 94 °C, 30 s annealing at 55 °C, and 30 s elongation at 72 °C for Actin (30 cycles), VRNH1 (30 cycles), and VRNH2 (35 cycles). For PPDH1 (30 cycles) and PPDH2 (35 cycles), the annealing temperature was 57 °C, whereas for VRNH3 (35 cycles) the annealing temperature was set at 60 °C. The enzyme used was Platinum® Taq DNA Polymerase (Invitrogen), in accordance with the manufacturer's instructions. The PCR products were visualized on agarose gels.
Real-time PCR quantification
This was performed for samples obtained for each treatment at 0, 7, 21, and 35 d for Exp1. In Exp2, real-time PCR quantification was undertaken for groups of samples taken at 21 d and 35 d. Amplifications were carried out in 20 μl reactions that included 10 μl of SYBR Green Quantimix Easy SYG Kit (Biotools, Madrid, Spain), 0.3 μM of each primer, 4 mM MgCl2, and 4 μl of cDNA, which corresponded to ∼89 ng of total RNA.
Reactions were run on an ICycler iQ™ (BioRad). Cycling conditions were 6 min at 95 °C, followed by 40 cycles of 30 s at 95 °C, 30 s at 60 °C, and 50 s at 72 °C for VRNH1, VRNH2, VRNH3, Actin, and PPDH1. For PPDH2, the annealing temperature was 58 °C. This was followed by a melting curve program (55–95 °C), which involved incremental temperature increases of 0.5 °C with a hold for 10 s at each temperature. Fluorescence data were acquired during the 72 °C step and during the melting curve program. Three identical reactions (technical repeats) were performed per sample, for each cDNA–primer combination in each run. Actin expression levels were also quantified in the same run. Two biological repeats were carried out in Exp1 and three in Exp2. All experiments showed similar trends in separate biological repeats.
Expression levels were calculated using the ICycler iQ™ software package (BioRad). The expression of the genes at each time point was normalized to the expression of Actin. The amplification efficiencies of each primer set were calculated.
Sequencing of HvFT3 (PPDH2)
Polymorphisms in HvFT3 (PPDH2) were ascertained by sequencing. Primers were designed to amplify overlapping fragments on the basis of the sequence from the cultivar ‘Morex’ (AB476614; Supplementary Fig. S3 available at JXB online).
GenBank accession numbers for the HvFT3 nucleotide sequences described in this manuscript are as follows: ‘Alexis’, HM133570; ‘Beka’, HM133571; ‘Pané’, HM133572; and SBCC058, HM133573.
Statistical analysis
Statistical analysis of the differences in relative expression between genotypes and treatment times was carried out using the analysis of variance (ANOVA) procedure in SAS (SAS Institute, 1998). The variable used for the analysis was ΔCT (CT actin–CT target gene) for each treatment and genotype, at each sampling time. This variable was preferred over the more commonly used 2ΔCT because of the concerns expressed by Yuan et al. (2006) regarding its use for statistical analysis. These concerns were namely that the target variable for statistical analysis should be based directly on the CT value, because this parameter is influenced directly by the treatment, concentration, and the nature of the sample itself (in the present case, the different genotypes). The ANOVA model included biological replication, genotype, sampling time (0, 7, 21, and 35 d for Exp1; 21 d and 35 d for Exp2), and genotype-by-time interactions for each treatment (VSD and NVLD for Exp1, VSD only for Exp2) separately. Genotypes and treatments were considered as fixed factors. The variability due to biological repeats and their interaction with the other factors was used as the error term to test time and genotype, as well as their interaction. Each value included in the analysis was the average of three technical repeats, to protect against slight fluctuations in reading and small pipetting errors. Differences between genotypes at each sampling time were calculated for each gene using orthogonal contrasts between each pair of genotypes.
Results
Flowering time in response to vernalization
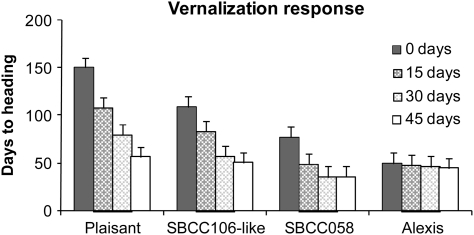
The lines studied differed in their responses to vernalization (Fig. 1). For all genotypes except ‘Alexis’, the length of time to flowering decreased as the period of exposure to low temperature increased. ‘Alexis’ was completely unaffected by exposure to the cold, regardless of the length of the cold period. The three SBCC106-like lines showed very consistent results. The period of cold treatment required for these three lines and SBCC058 to flower early was no more than 30 d. Without vernalization, SBCC058 flowered 28 d later than ‘Alexis’ and the genotypes similar to SBCC106 60 d later than ‘Alexis’, whereas ‘Plaisant’ did not reach this stage during the experimental period (150 d).
Fig. 1.
Days from sowing to flowering of four barley lines (‘Plaisant’, SBCC106-like, SBCC058, and ‘Alexis’; mean of two replications) after 0, 15, 30, or 45 d of vernalization (3 °C, 8 h light). Plants were grown in a phytotron at 18 °C with 16 h of light. Error bar (LSD 11.19 days, P=0.05).
Differences in gene expression
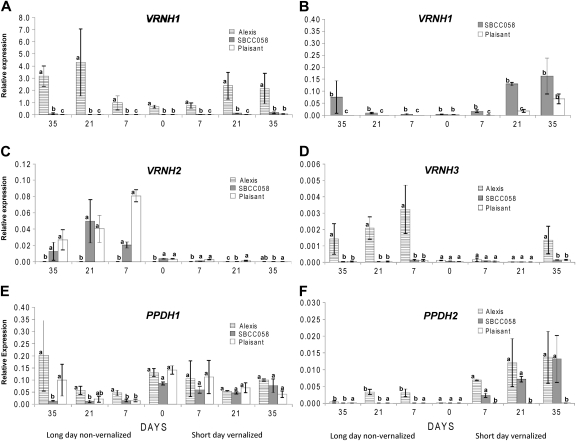
The expression patterns of three lines (‘Plaisant’, ‘Alexis’, and SBCC058) under VSD and NVLD treatments (Exp1) and that of SBCC106 under VSD conditions (Exp2) were analysed. Gene expression was assessed by qRT-PCR at every other sampling time (Figs 2, 4) and by semi-quantitative PCR at all sampling times (Figs 3, 5). For each gene, the number of cycles performed for the semi-quantitative PCR was set in accordance with the qRT-PCR results and corresponded to the point at which the differences in expression among genoytpes could best be differentiated. Differences among genotypes and sampling times were detected for VRNH1, VRNH2, VRNH3, and PPDH2 for the VSD treatment, and for all the genes for the NVLD treatment (Figs 2–5). Of the five genes studied, the level of expression of VRNH1 was the highest. The genes with the lowest expression levels were VRNH3 and PPDH2.
Fig. 2.
Relative expression levels of VRNH1 (A, B), VRNH2 (C), VRNH3 (D), PPDH1 (E), and PPDH2 (F) assayed by qRT-PCR in three barley lines, grown under conditions of vernalization and short days (VSD) or no vernalization and long days (NVLD). B shows enlarged graphs of VRNH1 expression for SBCC058 and ‘Plaisant’. The results shown are normalized with respect to the level of the housekeeping gene Actin for each genotype and treatment. Samples were taken from plants that were 10 d old (time 0) or after 7, 21, and 35 d of growth under each treatment. The variable of relative gene expression shown is 2ΔCT, where ΔCT is (CT Actin–CT target gene), for each genotype and treatment. Error bars represent the SEM. For the sampling times, bars with the same letter are not significantly different at P=0.05 according to orthogonal contrasts performed for an ANOVA that included all sampling times and genotypes per treatment.
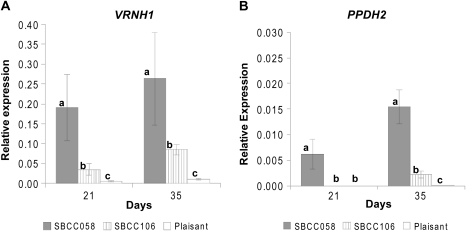
Fig. 4.
Relative expression levels of VRNH1 (A) and PPDH2 (B), assayed by qRT-PCR in three barley lines, grown under vernalization and short-day conditions (VSD). The results shown are normalized with respect to the housekeeping gene Actin for each genotype and treatment. Samples were taken after 21 d or 35 d of growth. The variable of relative gene expression shown is 2ΔCT, where ΔCT is (CT Actin–CT target gene), for each genotype and treatment. Error bars represent the SEM. For each sampling time, bars with the same letter are not significantly different at P=0.05 according to orthogonal contrasts performed for an ANOVA that included all sampling times and genotypes per treatment.
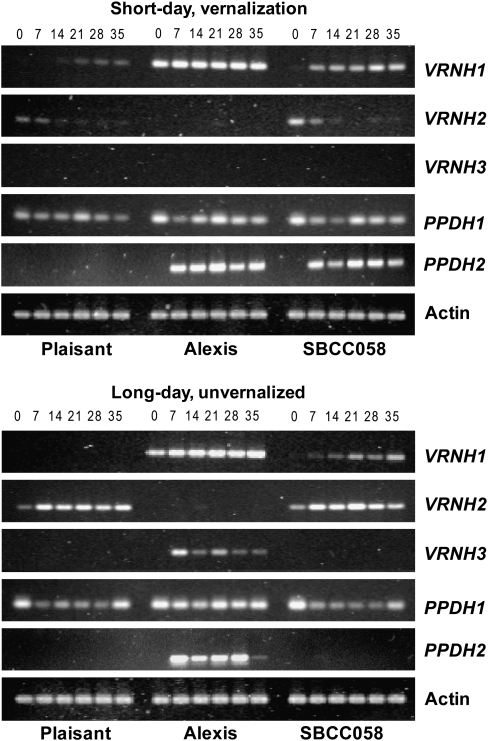
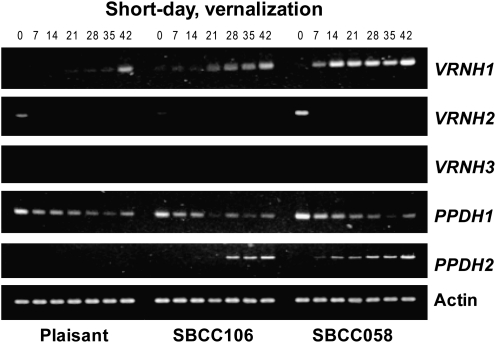
Fig. 3.
Semi-quantitative PCR for VRNH1 (30 cycles), VRNH2 (35 cycles), VRNH3 (35 cycles), PPDH1 (30 cycles), PPDH2 (35 cycles), and Actin (30 cycles) in three lines of barley vernalized under conditions of a short photoperiod (VSD), or grown without vernalization with long days (NVLD), over the course of 5 weeks (0, 7, 14, 21, 28, and 35 d).
Fig. 5.
Semi-quantitative PCR for VRNH1 (30 cycles), VRNH2 (35 cycles), VRNH3 (35 cycles), PPDH1 (30 cycles), PPDH2 (35 cycles), and Actin (30 cycles) in three lines of barley vernalized under conditions of a short photoperiod (VSD), in the course of 6 weeks (0, 7, 14, 21, 28, 35, and 42 d).
VRNH1 (HvBM5)
The expression of VRNH1 was much higher in ‘Alexis’ than in the other genotypes for both treatments (Figs 2A, 3). There was no expression of VRNH1 in SBCC058, SBCC106, or ‘Plaisant’ at day 0. Under the VSD treatment, VRNH1 expression increased gradually (Figs 2B–5), first in SBCC058 (around day 7) and then in ‘Plaisant’ (around day 35), with the level for SBCC106 being between the other two (Figs 4A, 5). Expression in ‘Plaisant’ remained significantly lower than that in SBCC106 and SBCC058 until day 35 (Fig. 4A).
Under the NVLD treatment, VRNH1 expression in SBCC058 also increased with time, but at a lower rate than for the VSD treatment (Figs 2B, 3). In contrast, VRNH1 expression in ‘Plaisant’ was undetectable with the NVLD treatment for the entire duration of the experiment (Figs 2B, 3). Expression of VRNH1 in SBCC058 was always significantly lower than that in ‘Alexis’ and significantly higher than that in ‘Plaisant’ for the later sampling times (days 21 and 35, Fig. 3).
VRNH2 (HvZCCTa, b)
Under the VSD treatment, VRNH2 expression in ‘Plaisant’, SBCC106, and SBCC058 was low and decreased with time, so that it had almost disappeared at 35 d (Figs 3, 5). The decrease in VRNH2 expression contrasted with the increase in VRNH1 expression in ‘Plaisant’, SBCC058 (Figs 2B, C, 3), and SBCC106 (Fig. 5). Under the NVLD treatment, VRNH2 expression for ‘Plaisant’ and SBCC058 increased after 7 d, was sustained, and then decreased slightly until the end of the experiment (Figs 2C, 3).
VRNH3 (HvFT1)
All genotypes carried the same recessive allele in VRNH3 (Table 1); however, differences in expression were detected. The expression of VRNH3 was very low under the VSD treatment for all genotypes in both experiments and for all sampling times. Indeed, no expression was apparent in the semi-quantitative PCR gels (Figs 3, 5) after 35 cycles. By qRT-PCR, VRNH3 expression could only be detected in ‘Alexis’ at the last sampling time (Fig. 2D). Under NVLD conditions, VRNH3 could already be detected in ‘Alexis’ after 7 d, whereas SBCC058 and ‘Plaisant’ exhibited hardly any expression of this gene (Figs 2D, 3).
PPDH1 (HvPRR7)
qRT-PCR did not detect any significant differences in PPDH1 expression among genotypes and sampling times for the VSD treatment (Fig. 2E), nor were there apparent differences among the four genotypes in the semi-quantitative assays (Figs 3, 5), even though ‘Alexis’ carries an allele different from the other three genotypes. In contrast, significant differences were observed among genotypes under the NVLD conditions (Fig. 2E). Overall, in Exp1, PPDH1 expression increased over time for ‘Alexis’ and ‘Plaisant’, but not for SBCC058. At day 35, ‘Alexis’ and ‘Plaisant’ exhibited significantly higher transcript levels than SBCC058 (Figs 2E, 3).
PPDH2 (HvFT3)
In the case of PPDH2, differences among genotypes and times were found for both treatments (Figs 2F–5). Differences can be explained partly by the presence of ppdH2 (the non-functional allele) in ‘Plaisant’, which caused the absence of transcripts in this variety. All the other lines analysed in Exp1 and Exp2 had the same functional allele (Table 1). Differences between sampling times stemmed mostly from the fact that the expression at day 0 was almost zero, as compared with later sampling times (in genotypes other than ‘Plaisant’). PPDH2 exhibited a higher level of expression under VSD conditions than under the NVLD treatment (Figs 2F–5). The levels of PPDH2 expression in ‘Alexis’ and SBCC058 were very similar, with transcripts being detected after just 7 d of VSD treatment and the levels then increasing slightly with time (Figs 2F, 3). In contrast, expression was detected later in SBCC106 in the VSD treatment in Exp2, at 21 d, as compared with ‘Alexis’ and SBCC058 (Figs 4B, 5). Moreover, PPDH2 expression in SBCC106 did not reach the level attained in SBCC058 at 35 d (Fig. 4B). For the NVLD treatment, PPDH2 expression was detected only in ‘Alexis’ (Figs 2F, 3). No expression was found in SBCC058 even though it carries the same allele as ‘Alexis’. After 35 d, when ‘Alexis’ had already flowered, the level of PPDH2 transcripts in ‘Alexis’ had decreased again.
To investigate the differences in the expression of PPDH2 among the genotypes that carried functional alleles (‘Alexis’ and SBCC058), two different experiments were carried out: (i) HvFT3 (PPDH2) was sequenced in several genotypes (including ‘Alexis’ and SBCC058) that have the functional allele of this gene; and (ii) the expression profile of PPDH2 in two different DH populations was analysed under NVLD conditions.
Sequencing of HvFT3 in ‘Alexis’ and SBCC058
The differences in expression of HvFT3 (PPDH2) between ‘Alexis’ and SBCC058 in Exp1 were apparently due to differences in regulation, because these lines both carry putatively functional alleles. To ensure that the difference in expression pattern was not due to sequence polymorphisms, which might produce functional changes, 1922 bp of the HvFT3 gene was sequenced. In addition to ‘Alexis’ and SBCC058, the gene from the Spanish cultivar ‘Pané’ (SBCC167) and from the French spring cultivar ‘Beka’ (SBCC169) were also sequenced. All of these carry the functional allele of HvFT3.
The sequences obtained for the four genotypes were the same and 99% identical to that of cultivar ‘Morex’ (AB476614) (Supplementary Fig. S3 at JXB online). The only observed polymorphism within the coding sequence, after comparison with ‘Morex’, was in exon 3. This SNP does not produce a change in the amino acid sequence. Therefore, the differences observed in the HvFT3 expression profiles of the studied lines were not caused by polymorphisms in the coding sequence of the gene.
Regulation of HvFT3 expression under conditions that do not typically induce its expression
The expression of HvFT3 detected in ‘Alexis’ under a long photoperiod was unexpected, because this gene has been thought to respond only to short days. Confirmation of the response of this gene to conditions that had been thought not to induce its expression (NVLD treatment) was sought in a different set of plant materials. The DH populations ‘Alexis’בPané’ (Cuesta-Marcos et al., 2008a) and ‘Beka’בMogador’ (Cuesta-Marcos et al., 2008b) segregate at VRNH1 and VRNH2. Therefore, they can be used to assess the possible effects of genes involved in the response to vernalization on HvFT3. In the first population, ‘Pané’ has all five alleles for the genes involved in responses to vernalization and photoperiod that are carried by SBCC058 (therefore, the population segregates for VRNH1, VRNH2, and PPDH1; Table 1). In the second population, ‘Beka’ carries the functional HvFT3 allele, whereas ‘Mogador’ has the non-functional allele, the same as ‘Plaisant’. Thus, this population segregates for VRNH1, VRNH2, and HvFT3 (Table 1), although only lines with the functional allele of HvFT3 were chosen for this experiment.
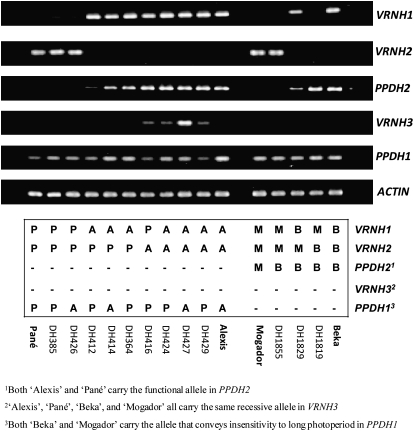
In ‘Pané’, HvFT3 was not transcribed, whereas ‘Alexis’ and ‘Beka’ showed high levels of HvFT3 expression (Fig. 6). Expression of HvFT3 was not detected in DH lines that carried the functional allele but showed high levels of expression of VRNH2 (DH lines 385 and 426). However, HvFT3 expression could be detected in some DH lines that did carry VRNH2, but in which the level of VRNH2 expression was apparently lower (DH lines 412 and 414). Indeed, lines without VRNH2 exhibited the highest level of expression of HvFT3 (DH lines 416, 424, 427, and 429). This suggests that VRNH2 might play a role in the down-regulation of HvFT3 (Fig. 6). There seemed to be no relationship between the genotype for PPDH1 and the expression of HvFT3. Finally, expression of VRNH3 was detected in only a few DH lines of the ‘Alexis’בPané’ population, namely those that carried the spring allele of VRNH1 from ‘Alexis’ or in which VRNH2 was absent (Fig. 6).
Fig. 6.
Semi-quantitative PCR for VRNH1 (30 cycles), VRNH2 (35 cycles), VRNH3 (35 cycles), PPDH1 (30 cycles), PPDH2 (35 cycles), and Actin (30 cycles) in DH lines of two barley mapping populations grown for 10 d without vernalization under conditions of a long photoperiod (NVLD). The genetic constitution of the parental lines was as follows: ‘Alexis’ (VRNH1-3, vrnH2, PPDH2, ppdH1), ‘Pané’ (VRNH1-4, VRNH2, PPDH2, PPDH1), ‘Beka’ (VRNH1-1, vrnH2, PPDH2, ppdH1), and ‘Mogador’ (vrnH1, VRNH2, ppdH2, ppdH1); VRNH1 alleles are coded in accordance with Hemming et al. (2009). P, ‘Pané’ allele; A, ‘Alexis’ allele; B, ‘Beka’ allele; M, ‘Mogador’ allele. 1Both ‘Alexis’ and ‘Pané’ carry the functional allele in PPDH2. 2‘Alexis’, ‘Pané’, ‘Beka’, and ‘Mogador’ all carry the same recessive allele in VRNH3. 3Both ‘Beka’ and ‘Mogador’ carry the allele that conveys insensitivity to a long photoperiod in PPDH1.
Discussion
Expression of VRNH1 is responsible for a gradation in the vernalization requirements of barley
‘Alexis’ and ‘Plaisant’ presented flowering behaviours, responses to vernalization, and expression patterns for flowering genes that were in accordance with expectations for typical varieties with spring and winter growth habits, respectively. Typically, spring cereal varieties do not require a cold period prior to heading, whereas in winter varieties it is an essential prerequisite for flowering (Roberts et al., 1988). However, SBCC106 and SBCC058 displayed intermediate responses. This situation resembles the gradation of vernalization requirements described by Takahashi and Yasuda (1971), which was associated with an allelic series of what is now known as VRNH1. The phenology of the four lines studied seems to be associated with the respective VRNH1 alleles. Polymorphism at VRNH1 has been described by several authors, and in some cases has been related to differences in function (Fu et al., 2005; Cockram et al., 2007; Szűcs et al., 2007).
The cultivar ‘Plaisant’, with a winter growth habit, carries an allele of VRNH1 with a full-length intron 1. This cultivar flowers very late in the absence of vernalization, and expression of VRNH1 is undetectable in plants that have not been vernalized, as reported for other winter cultivars by Trevaskis et al. (2003), von Zitzewitz et al. (2005), and Hemming et al. (2009). As expected (von Zitzewitz et al., 2005; Trevaskis et al., 2006), cold treatment induced the expression of VRNH1 and decreased the time to flowering.
SBCC106 and SBCC058, with an intermediate growth habit, carry alleles of VRNH1 with deletions of ∼0.5 kb (VRNH1-6 in Hemming et al., 2009) and ∼4 kb (VRNH1-4, in Hemming et al., 2009) in intron 1, respectively. These deletions have been reported previously: VRNH1-4 in cultivars ‘Albacete’ and ‘Calicuchima-sib’ (von Zitzewitz et al., 2005, Szűcs et al., 2007) and VRNH1-6 in cultivar ‘Express’ (Cockram et al., 2007). SBCC106 and SBCC058 flowered relatively late in the absence of vernalization, whereas cold treatment induced increased VRNH1 expression and decreased the time to flowering.
The small deletion (∼0.5 kb) in intron 1 that is carried by cultivar SBCC106 is sufficient to enable the detection of VRNH1 expression in plants that have not been vernalized and are grown under a short photoperiod (Hemming et al., 2009). The low level of VRNH1 expression associated with this allele appears to be sufficient to allow flowering in the absence of vernalization, after a long vegetative period, as already reported by Hemming et al. (2009).
‘Alexis’, a spring cultivar, carries a VRNH1 allele with a very large deletion in intron 1 (∼9 kb), which was first described in cultivar ‘Triumph’ (von Zitzewitz et al., 2005) and corresponds to allele VRNH1-3 reported by Hemming et al. (2009). This cultivar flowered early in the absence of vernalization and exhibited high levels of VRNH1 expression in plants that had not been vernalized. Cold treatment was found to induce an increase in VRNH1-3 expression levels, although flowering time did not change significantly, as reported by Hemming et al. (2009).
In general, the growth habits of these four barley lines and their responses to vernalization were correlated with their level of expression of VRNH1 and the size of the deletion in intron 1. The larger the deletion, the higher the levels of VRNH1 transcript in plants both with and without vernalization, and the earlier the plants tended to flower. This is consistent with data presented for several VRNH1 alleles by other researchers (Cockram et al., 2007; Szűcs et al., 2007; Hemming et al., 2009). However, this is the first report that shows increased expression of the VRNH1-6 allele in response to vernalization. In addition, the duration of the cold treatment required to trigger VRNH1 expression differed for the four alleles studied. ‘Alexis’, the spring cultivar, exhibited high expression from the very beginning, whereas SBCC058, SBCC106, and ‘Plaisant’, in this order, exhibited increasingly long lag periods until expression was detected.
Oliver et al. (2009) showed that, in barley, as in Arabidopsis, flowering induced by vernalization is associated with epigenetic changes at the VRNH1 gene that promote an active chromatin state. In this earlier study, two cultivars of barley were used, a winter type (‘Sonja’), with a full-length intron 1, and a spring type (‘Morex’), which carries a large deletion in the first intron of VRNH1 (VRNH1-1). It was suggested that regions of the first intron that are present in the winter cultivar could be important for the repression of VRNH1 before vernalization. A similar mechanism might also be responsible for the differing behaviour of the VRNH1 alleles studied here, which are characterized by differences at intron 1.
It has been proposed that genotypes that carry the VRNH1 allele found in SBCC058, even in the presence of VRNH2, should be classified agronomically as ‘spring’ varieties with a reduced requirement for vernalization (Cockram et al., 2007; Szűcs et al., 2007), whereas the SBCC106 allele confers a strict winter habit, with a requirement for full vernalization (Cockram et al., 2007). The results of the present study provide evidence that indicates that lines SBCC106 (VRNH1-6) and SBCC058 (VRNH1-4) exhibit patterns of expression of VRNH1 that are intermediate between those of the varieties with habits of winter and spring growth. The intermediate nature of the vernalization response of SBCC058 was confirmed recently using a different set of materials (Casao et al., 2010). In this previous study, the introgression of the SBCC058 VRNH1 allele into a winter-type background reduced but did not cancel the vernalization requirement of the winter-type cultivar.
Thus, as other researchers have suggested, different VRNH1 alleles are associated with different growth habits and flowering times. It is proposed that VRNH1 polymorphism can be used as the basis for the adaptation of cultivars to enable them to grow in particular regions. SBCC058 and SBCC106 are representative of the two main VRNH1/VRNH2 haplotypes found in a large class of Spanish barleys (Casas et al., 2008). In fact, out of the 159 landraces represented in the SBCC, 47 carry the VRNH1/VRNH2 haplotype of SBCC058 and 93 carry the same haplotype as SBCC106. This latter haplotype has been found at very low frequencies in European barley germplasm (5C+Z in Cockram et al., 2007). These VRNH1 alleles found in Spanish barleys could confer advantages that enable adaptation to the Mediterranean climate that predominates in the Iberian Peninsula, with winters that are milder than those in more northerly latitudes.
Expression analysis of vernalization and photoperiod genes
Expression analyses might help to explain the causes that underlie the variety of phenotypic responses that are observed with respect to vernalization. As far as we know, this is the first study that has examined the time course of expression of the five major genes that are associated with responses to vernalization and photoperiod in barley simultaneously.
The interactions among VRNH1, VRNH2, and VRNH3 form a feedback regulatory loop, which means that modification of the transcript levels of any one of these genes affects the transcript levels of the others (Distelfeld et al., 2009). This model predicts that, under the conditions that are prevalent during winter after sowing in autumn (low temperature and short photoperiod), VRNH2 is repressed (by a lack of long days) and VRNH1 is expressed increasingly as the number of cold days increases. This was confirmed by the results described herein for this kind of genotype (‘Plaisant’, SBCC106, and SBCC058). In general, the expression of VRNH2 was accompanied by almost complete absence of VRNH1 expression under a long photoperiod. These results agree with those reported by Yan et al. (2006). However, in SBCC058, simultaneous expression of these two genes under the conditions of NVLD was detected, but at levels that might indicate a rise in VRNH1 and the beginning of a decrease in VRNH2 expression.
VRNH3 expression was not detected in any of the lines under the VSD treatment because the conditions did not induce the expression of this gene, as expected (long days are required). Slight expression of VRNH3 could only be detected in ‘Alexis’ after 35 d (Fig. 2). A low level of expression of VRNH3 was also observed under short days (12 h light) by Kikuchi et al. (2009) in the spring cultivar ‘Morex’.
Under conditions with a long photoperiod, expression of VRNH3 was not detected in genotypes in which VRNH2 was present, as predicted by the feedback model and shown experimentally by Hemming et al. (2008). VRNH3 expression was detected only in ‘Alexis’, although its level was low. ‘Alexis’ has a genotype that conveys insensitivity to long photoperiod, and high levels of VRNH3 expression have only been reported in the literature for genotypes with an active PPDH1 allele (Turner et al., 2005; Faure et al., 2007; Hemming et al., 2008; Kikuchi et al., 2009).
Correlation between VRNH2 expression and HvFT3 repression
In the present study, HvFT3, the candidate gene for PPDH2 (Faure et al., 2007; Kikuchi et al., 2009), was also analysed. Under conditions with a short photoperiod, HvFT3 was expressed in all the genotypes that carried the active allele. There are numerous reports that describe the effect of this gene under conditions of short days (Laurie et al. 1995; Faure et al., 2007; Karsai et al., 2008; Kikuchi et al., 2009). HvFT3 was also expressed weakly under conditions of a long photoperiod in ‘Alexis’. This result was unexpected, but confirms similar observations by Faure et al. (2007) and Kikuchi et al. (2009) in the spring cultivars ‘Triumph’ and ‘Morex’, respectively. The lack of expression of HvFT3 in SBCC058 during the NVLD treatment must have been caused by a mechanism of repression that is absent in ‘Alexis’, because both genotypes share the same HvFT3 allele (Table 1) and no differences were detected between them in terms of the nucleotide sequence.
A possible role for HvFT3 in the determination of flowering time is also supported by a previous quantitative trait locus (QTL) analysis on the population ‘Beka’בMogador’ (Cuesta-Marcos et al., 2008b). It was found that HvFT3 (Supplementary Fig. S2 at JXB online) corresponded to a major QTL that affected flowering under short days in the field and in glasshouse experiments; this QTL was detected in earlier studies and was also found previously to be associated with HvFT3 (Laurie et al., 1995; Faure et al., 2007; Kikuchi et al., 2009). However, previously a QTL with a peak at HvTF3 was also found under conditions of long days in a glasshouse, although its influence on flowering time under these conditions was weaker than that during short days (Cuesta-Marcos et al., 2008b). This result seems to be consistent with the expression pattern of HvFT3 that was detected under conditions of long days in ‘Alexis’. Karsai et al. (2008) also found an effect of PPDH2 under conditions of a long photoperiod in the ‘Dicktoo’בMorex’ population.
By further analysis of HvFT3 expression in DH lines of two barley populations, it was found that, under conditions of long days, the expression levels of HvFT3 and VRNH2 were related inversely (Fig. 6). This suggests a possible role for VRNH2 in the down-regulation of HvFT3 expression and, indirectly, in the regulation of flowering time through an interaction with the pathway that affects responses to day-length. Expression of HvFT3 under conditions of a long photoperiod was detected in ‘Alexis’ and some DH lines, but not in SBCC058 or ‘Pané’. VRNH2 (absent in ‘Alexis’) was expressed in SBCC058 under these conditions. Therefore, VRNH2 expression might repress HvFT3 expression in SBCC058. Interaction of VRNH2 with the photoperiod pathway has already been described by Hemming et al. (2008). They reported an interaction between VRNH2, VRNH3, and PPDH1 under a long photoperiod, such that deletion of VRNH2 was associated with expression of VRNH3 and early flowering only when combined with the PPDH1 allele that conveys sensitivity to a long photoperiod. They concluded that VRNH2 counteracts the effects of PPDH1 to prevent flowering before vernalization. The present results illustrate that VRNH2 also offsets the effects of PPDH2 under conditions of a long photoperiod. In the absence of VRNH2, expression of PPDH2 and VRNH3 could be observed in plants with both alleles of PPDH1.
The interaction of VRNH2 with two of the representatives of the FT gene family suggests the possibility of similar mechanisms of action for both HvFT1 and HvFT3. This hypothesis would be consistent with the results of Kikuchi et al. (2009). In this previous study, it was hypothesized that HvFT3 functions indirectly to promote flowering and that its activity can be modulated by photoperiod signals, even with a short photoperiod. An important role for VRNH2 in the promotion to flowering has been proposed recently by Distelfeld and Dubcovsky (2010), although they acknowledge that its function has not been elucidated completely. In this study, it is suggested that VRNH2, a gene shown previously to act as a repressor of HvFT1, also appears to act as a repressor of HvFT3. By this mechanism, the pattern of HvFT3 expression with respect to day-length might be determined fully or partially by VRNH2: VRNH2 is not expressed under conditions of short days, therefore HvFT3 is expressed. However, VRNH2 is expressed under conditions of long days and represses the expression of HvFT3. In barley varieties in which the VRNH2 locus is deleted, HvFT3 is expressed under conditions of long days. However, HvFT3 is expressed at a lower level under long days than under short days, which suggests that a promoter of HvFT3 is activated more strongly under conditions of short days or there is an additional repressor activity under conditions of long days.
Supplementary data
Supplementary data are available at JXB online
Figure S1. Mapping of HvFT3 in the ‘Beka’בMogador’ population.
Figure S2. QTL analysis for traits related to flowering time in the ‘Beka’בMogador’ population.
Figure S3. Sequencing of HvFT3.
Acknowledgments
This study was funded by grants AGL2007-63625 and HH2008-0013 from the Spanish Ministry of Science and Technology and by the European Regional Development Fund. Germplasm from the SBCC is maintained with funding from project RFP2004-00015-00-00. MCC was supported by an I3P Predoctoral Fellowship from CSIC. Substantial help from one anonymous reviewer was greatly appreciated.
References
- Casao MC, Igartua E, Karsai I, Bhat PR, Cuadrado N, Gracia MP, Lasa JM, Casas AM. Introgression of an intermediate VRNH1 allele leads to reduced vernalization requirement without affecting freezing tolerance. Molecular Breeding. 2010 DOI 10.1007/s11032-010-9497-y (in press) [Google Scholar]
- Casas AM, Yahiaoui S, Ciudad FJ, et al. Vrn-H1 and Vrn-H2 allelic diversity in barley may explain specific adaptation to the Mediterranean environments. In: Molina Cano JL, Christou P, Graner A, editors. Cereal science and technology for feeding ten billion people: genomics era and beyond. Options Méditerranéennes. 2008. Serie A, No. 81. Zaragoza: CIHEAM/IRTA, 105–109. [Google Scholar]
- Cockram J, Chiapparino E, Taylor SA, Stamati K, Donini P, Laurie DA, O'Sullivan DM. Haplotype analysis of vernalization loci in European barley germplasm reveals novel VRN-H1 alleles and a predominant winter VRN-H1/VRN-H2 multi-locus haplotype. Theoretical and Applied Genetics. 2007;115:993–1001. doi: 10.1007/s00122-007-0626-x. [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- Cuesta-Marcos A, Casas AM, Yahiaoui S, Gracia MP, Lasa JM, Igartua E. Joint analysis for heading date QTL in small interconnected barley populations. Molecular Breeding. 2008a;21:383–399. [Google Scholar]
- Cuesta-Marcos A, Igartua E, Ciudad FJ, et al. Heading date QTL in a spring×winter barley cross evaluated in Mediterranean environments. Molecular Breeding. 2008b;21:455–471. [Google Scholar]
- Danyluk J, Kane NA, Breton G, Limin AE, Fowler DB, Sarhan F. TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiology. 2003;132:1849–1860. doi: 10.1104/pp.103.023523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distelfeld A, Li C, Dubcovsky J. Regulation of flowering in temperate cereals. Current Opinion in Plant Biology. 2009;12:178–184. doi: 10.1016/j.pbi.2008.12.010. [DOI] [PubMed] [Google Scholar]
- Distelfeld A, Dubcovsky J. Characterization of the maintained vegetative phase deletions from diploid wheat and their effect on VRN2 and FT transcript levels. Molecular Genetics and Genomics. 2010;283:223–232. doi: 10.1007/s00438-009-0510-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky J, Chen C, Yan L. Molecular characterization of the allelic variation at the VRN-H2 vernalization locus in barley. Molecular Breeding. 2005;15:395–407. [Google Scholar]
- Faure S, Higgins J, Turner A, Laurie DA. The FLOWERING LOCUS T-like gene family in barley. Hordeum vulgare. Genetics. 2007;176:599–609. doi: 10.1534/genetics.106.069500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D, Szűcs P, Yan L, Helguera M, Skinner J, Hayes P, Dubcovsky J. Large deletions within the first intron of the VRN-1 are associated with spring growth habit in barley and wheat. Molecular Genetics and Genomics. 2005;273:54–65. doi: 10.1007/s00438-004-1095-4. [DOI] [PubMed] [Google Scholar]
- Greenup A, Peacock WJ, Dennis ES, Trevaskis B. The molecular biology of seasonal flowering-responses in Arabidopsis and the cereals. Annals of Botany. 2009;103:1165–1172. doi: 10.1093/aob/mcp063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemming MN, Peacock WJ, Dennis ES, Trevaskis B. Low-temperature and daylength cues are integrated to regulate FLOWERING LOCUS T in barley. Plant Physiology. 2008;147:355–366. doi: 10.1104/pp.108.116418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemming MN, Fieg S, Peacock WJ, Dennis ES, Trevaskis B. Regions associated with repression of the barley Hordeum vulgare. VERNALIZATION1 gene are not required for cold induction. Molecular Genetics and Genomics. 2009;282:107–117. doi: 10.1007/s00438-009-0449-3. [DOI] [PubMed] [Google Scholar]
- Higgins JA, Bailey PC, Laurie DA. Comparative genomics of flowering time pathways using Brachypodium distachyon as a model for the temperate grasses. PLoS One. 2010;5:e10065. doi: 10.1371/journal.pone.0010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igartua E, Gracia MP, Lasa JM, Medina B, Molina-Cano JL, Montoya JL, Romagosa I. The Spanish barley core collection. Genetic Resources & Crop Evolution. 1998;45:475–481. [Google Scholar]
- Karsai I, Hayes PM, Kling J, Matus IA, Mészáros K, Láng L, Bedő Z, Sato K. Genetic variation in component traits of heading date in Hordeum vulgare subsp. spontaneum accessions characterized in controlled environments. Crop Science. 2004;44:1622–1632. [Google Scholar]
- Karsai I, Szűcs P, Kőszegi B, Hayes PM, Casas A, Bedő Z, Veisz O. Effects of photo and thermo cycles on flowering time in barley: a genetical phenomics approach. Journal of Experimental Botany. 2008;59:2707–2715. doi: 10.1093/jxb/ern131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsai I, Szűcs P, Mészáros K, Filichkina T, Hayes PM, Skinner JS, Láng L, Bedő Z. The VRN-H2 locus is a major determinant of flowering time in a facultative×winter growth habit barley Hordeum vulgare L. mapping population. Theoretical and Applied Genetics. 2005;110:1458–1466. doi: 10.1007/s00122-005-1979-7. [DOI] [PubMed] [Google Scholar]
- Kikuchi R, Kawahigashi H, Ando T, Tonooka T, Handa H. Molecular and functional characterization of PEBP genes in barley reveal the diversification of their roles in flowering. Plant Physiology. 2009;149:1341–1353. doi: 10.1104/pp.108.132134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie DA, Pratchett N, Bezant JH, Snape JW. Genetic analysis of a photoperiod response gene on the short arm of chromosome 2 (2H) of Hordeum vulgare (barley) Heredity. 1994;72:619–627. [Google Scholar]
- Laurie DA, Pratchett N, Bezant JH, Snape JW. RFLP mapping of five major genes and eight quantitative trait loci controlling flowering time in a winter×spring barley Hordeum vulgare L. cross. Genome. 1995;38:575–585. doi: 10.1139/g95-074. [DOI] [PubMed] [Google Scholar]
- Loukoianov A, Yan L, Blechl A, Sanchez A, Dubcovsky J. Regulation of VRN-1 vernalization genes in normal and transgenic polyploid wheat. Plant Physiology. 2005;138:2364–2373. doi: 10.1104/pp.105.064287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver SN, Finnegan EJ, Dennis ES, Peacock WJ, Trevaskis B. Vernalization-induced flowering in cereals is associated with changes in histone methylation at the VERNALIZATION1 gene. Proceedings of the National Academy of Sciences, USA. 2009;106:8386–8391. doi: 10.1073/pnas.0903566106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts EH, Summerfield RJ, Cooper JP, Ellis RH. Environmental-control of flowering in barley (Hordeum vulgare L). I. Photoperiod limits to long-day responses, photoperiod-insensitive phases and effects of low-temperature and short-day vernalization. Annals of Botany. 1988;62:127–144. [Google Scholar]
- SAS Institute. SAS/STATTM User's Guide Release 6.03 Edition. 1028. Cary, NC: SAS Institute Inc; 1998. [Google Scholar]
- Sasani S, Hemming MN, Oliver SN, et al. The influence of vernalization and daylength on expression of flowering-time genes in the shoot apex and leaves of barley Hordeum vulgare. Journal of Experimental Botany. 2009;60:2169–2178. doi: 10.1093/jxb/erp098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada S, Ogawa T, Kitagawa S, et al. A genetic network of flowering-time genes in wheat leaves, in which an APETALA1/FRUITFULL-like gene, VRN1, is upstream of FLOWERING LOCUS T. The Plant Journal. 2009;58:668–681. doi: 10.1111/j.1365-313X.2009.03806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szűcs P, Skinner JS, Karsai I, Cuesta-Marcos A, Haggard KG, Corey AE, Chen THH, Hayes PM. Validation of the VRN-H2/VRN-H1 epistatic model in barley reveals that intron length variation in VRN-H1 may account for a continuum of vernalization sensitivity. Molecular Genetics and Genomics. 2007;277:249–261. doi: 10.1007/s00438-006-0195-8. [DOI] [PubMed] [Google Scholar]
- Takahashi R, Yasuda S. Genetics of earliness and growth habit in barley. In: Nilan RA, editor. Barley genetics II. Washington State University Press; 1971. pp. 388–408. [Google Scholar]
- Tranquilli G, Dubcovsky J. Epistatic interaction between vernalization genes Vrn-Am1 and Vrn-Am2 in diploid wheat. Journal of Heredity. 2000;91:304–306. doi: 10.1093/jhered/91.4.304. [DOI] [PubMed] [Google Scholar]
- Trevaskis B, Bagnall DJ, Ellis MH, Peacock WJ, Dennis ES. MADS box genes control vernalization-induced flowering in cereals. Proceedings of the National Academy of Science, USA. 2003;100:13099–13104. doi: 10.1073/pnas.1635053100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B, Hemming MN, Peacock WJ, Dennis ES. HvVRN2 responds to daylength, whereas HvVRN1 is regulated by vernalization and developmental status. Plant Physiology. 2006;140:1397–1405. doi: 10.1104/pp.105.073486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annual Review of Plant Biology. 2008;59:573–594. doi: 10.1146/annurev.arplant.59.032607.092755. [DOI] [PubMed] [Google Scholar]
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science. 2005;310:1031–1034. doi: 10.1126/science.1117619. [DOI] [PubMed] [Google Scholar]
- von Zitzewitz J, Szűcs P, Dubcovsky J, Yan L, Pecchioni N, Francia E, Casas A, Chen THH, Hayes PM, Skinner JS. Molecular and structural characterization of barley vernalization genes. Plant Molecular Biology. 2005;59:449–467. doi: 10.1007/s11103-005-0351-2. [DOI] [PubMed] [Google Scholar]
- Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Dubcovsky J. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proceedings of the National Academy of Sciences, USA. 2006;103:19581–19586. doi: 10.1073/pnas.0607142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science. 2004;303:1640–1644. doi: 10.1126/science.1094305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J. Positional cloning of wheat vernalization gene VRN1. Proceedings of the National Academy of Sciences, USA. 2003;100:6263–6268. doi: 10.1073/pnas.0937399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan JS, Reed A, Chen F, Stewart CN., Jr. Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006;7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadoks JC, Chang TT, Konzak CF. A decimal code for the growth stages of cereals. Weed Research. 1974;14:415–421. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.