Abstract
An indica pyramiding line, DK151, and its recurrent parent, IR64, were evaluated under drought stress and non-stress conditions for three consecutive seasons. DK151 showed significantly improved tolerance to drought. The DNA methylation changes in DK151 and IR64 under drought stress and subsequent recovery were assessed using methylation-sensitive amplified polymorphism analysis. Our results indicate that drought-induced genome-wide DNA methylation changes accounted for ∼12.1% of the total site-specific methylation differences in the rice genome. This drought-induced DNA methylation pattern showed three interesting properties. The most important one was its genotypic specificity reflected by large differences in the detected DNA methylation/demethylation sites between DK151 and IR64, which result from introgressed genomic fragments in DK151. Second, most drought-induced methylation/demethylation sites were of two major types distinguished by their reversibility, including 70% of the sites at which drought-induced epigenetic changes were reversed to their original status after recovery, and 29% of sites at which the drought-induced DNA demethylation/methylation changes remain even after recovery. Third, the drought-induced DNA methylation alteration showed a significant level of developmental and tissue specificity. Together, these properties are expected to have contributed greatly to rice response and adaptation to drought stress. Thus, induced epigenetic changes in rice genome can be considered as a very important regulatory mechanism for rice plants to adapt to drought and possibly other environmental stresses.
Keywords: DNA methylation, drought tolerance, MSAP, rice
Introduction
Plants are constantly challenged by environmental (both abiotic and biotic) perturbations, and thus have developed remarkable capabilities to modulate their physiological and developmental machinery through genome-wide gene expression changes in response to these environmental perturbations (Zhou et al., 2007). Recent evidence indicates that epigenetic mechanisms, such as DNA methylation and histone modification, play a crucial role in regulating gene expression in plant responses to environmental stress (Razin and Cedar, 1992; Cullis, 2005; Boyko et al., 2007; Boyko and Kovalchuk, 2008). For example, environmental stimuli such as salinity and water stress can cause demethylation at coding regions of certain genes and subsequently activate their expression (Choi and Sano, 2007). Also, specific gene expression patterns under epigenetic control are reversible and may show transgenerational inheritance (Bender, 2004; Long et al., 2006; Zhao et al., 2007).
DNA methylation exists in virtually all organisms. In eukaryotes, DNA methylation frequently occurs at the 5-position of cytosine, yielding 5-methylcytosine (5mC). Under normal conditions, the ratio of methylated to total cytosines varies from 20% to 30% in plants (Finnegan et al., 1998), and methylcytosine usually occurs in CpG, CpNpG and CpHpH (H=A, T, C) sequences (Cao and Jacobsen, 2002; Zhang et al., 2006). It was also reported that DNA sequence polymorphisms might cause methylation differences and there are numerous cytosine methylation polymorphisms between different plant genotypes (Hua et al., 2005; Lu et al., 2005; Ruiz et al., 2005; Akimoto et al., 2007). Previous studies indicated that the transposon-rich heterochromatic regions in Arabidopsis are often heavily methylated (Lippman et al., 2004; Zhang et al., 2006). Genome-wide high-resolution mapping of DNA methylation revealed that over one-third of expressed genes in Arabidopsis show methylation within transcribed regions, while only ∼5% of genes showed methylation within their promoter regions and expression of these promoter methylated genes tend to show a greater degree of tissue specificity (Zilberman et al., 2006). Thus, DNA methylation within genes is a common feature of eukaryotic genomes (Tran et al., 2005).
Drought stress is the most important constraint limiting rice production in most rain-fed systems worldwide. Rice varieties differ greatly in their tolerance to drought. Genetically, drought tolerance of rice is a complex trait under polygenic control, and involves complex morpho-physiological mechanisms (Li and Xu, 2007). At the molecular level, drought can induce genome-wide changes in gene expression in rice (Zhou et al., 2007). Epigenetic mechanisms are involved in this type of stress-induced genome-wide differential gene expression. For instance, the mutant allele (met1) at the tobacco DNA methyltransferase 1 locus was reportedly able to remove methylation at some genomic regions, resulting in specific expression of 31 stress response-related genes (Wada et al., 2004). However, little is known about the general pattern of DNA methylation linked with rice responses to drought, and its relationship with drought tolerance in rice.
We describe here the DNA methylation patterns of a drought-tolerant (DT) rice line and its drought-sensitive parent under drought and non-stress conditions. The differences between the two lines in their spatial and temporal patterns of DNA methylation revealed a possible role of this epigenetic mechanism in rice adaptation to drought stress.
Materials and methods
Plant materials and genotyping
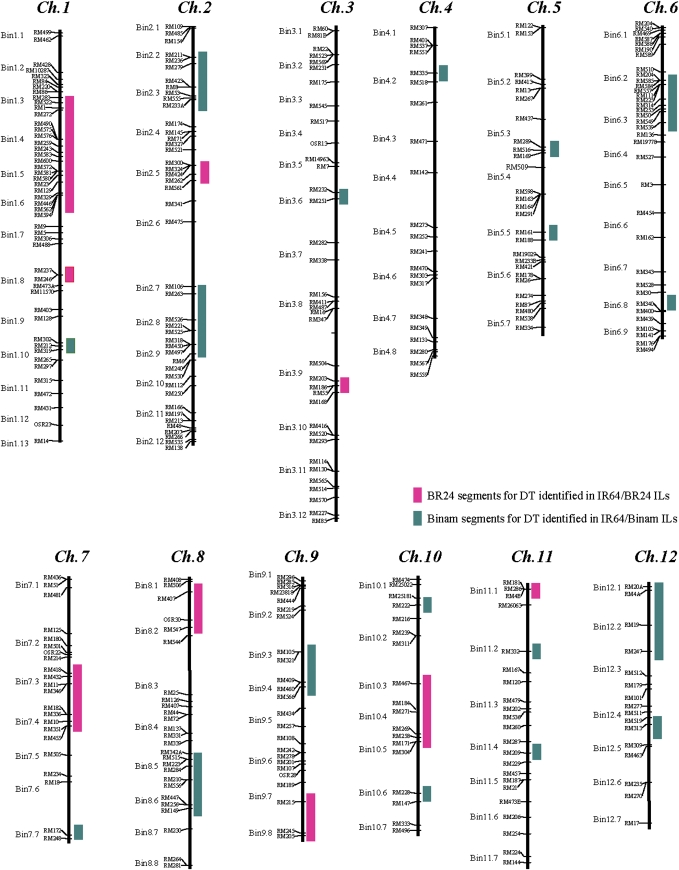
Two rice lines, DK151 and IR64, were used in this experiment. IR64 is a drought-sensitive rice variety developed at the International Rice Research Institute (IRRI, Manila, Philippines) and has been widely grown in many Asian countries for more than a decade. DK151 is a DT F7 line derived from a cross between two DT IR64 introgression lines, DGI 187 and DGI 74 (Fig. S1; Supplementary data available at JXB online). An extensive study using 625 simple sequence repeat markers across the rice genome (http://www.gramene.org/markers/), shows that DK151 differs from IR64 at 27 genomic segments from two donors, BR24 (an indica upland landrace from Bangladesh) and Binam (a japonica landrace from Iran) (Fig. 1).
Fig. 1.
Genetic composition of DK151 at 27 genomic regions (loci) introgressed and pyramided from two different donors, BR24 and Binam, in the IR64 genetic background. Ch., chromosome.
Phenotyping experiments and data analysis
DK151 and IR64 were evaluated in replicated experiments under drought stress and irrigated conditions consecutively in the 2004 dry season (DS), 2004 wet season (WS), and 2005 DS at the IRRI experimental farm. In the 2004 DS, seeds of both lines were sown in the seedling nursery on 15 December 2003 and 25-d-old seedlings of each line were transplanted into a three-row plot with 45 plants per plot at a spacing of 15×25 cm between rows and plants within each plot and three replications for each line. Two treatments were used. For drought stress treatment, water was drained, and irrigation was held at 55 d after transplanting at the peak tillering stage until maturity (terminal drought). In the irrigated control, the field was irrigated at weekly intervals and a constant water layer was maintained in the field until 2 weeks before harvesting. In the 2004 WS, the same experimental design was used to evaluate the yield and components of DK151 and IR64 in a replicated field experiment under the drought and control conditions at the IRRI experimental farm. Seeds of DK151 and IR64 were sown on 22 June 2004, and 21-d-old seedlings of each line were separately transplanted into a rain-fed upland field (stress treatment) and a lowland field (control) on 13 July. In the upland field, no irrigation was provided, whereas in the lowland field, a constant water layer was maintained by regular irrigation until maturity. In the 2005 DS, the same experimental design was used as for the 2004 DS except that only two replications were used under the irrigated control, while three replications were used in the drought treatment. Seeds were sown on 15 January 2005 and transplanted into the field on 4 February. The field management for the irrigated filed was similar to that of the 2004 DS. The following traits were measured in one or more experiments: plant height (in cm from soil surface to the tip of the tallest panicle per plant); heading date (in days from sowing to heading); and panicle number per plant, which were measured in the field; grain yield [tonnes/hectare (t/ha)]; filled grain number per panicle; spikelet fertility (%); and thousand grain weight (GW, in g), which were measured on 10 plants sampled at maturity from the middle row of each plot. Analysis of variance and t tests were used to compare the differences between DK151 and IR64 for all measured traits using the SAS program GLM (SAS, 1999).
Analysis of DNA methylation-sensitive amplified polymorphisms (MSAPs)
Seeds of DK151 and IR64 were sterilized in 0.1% NaClO (v/v) and then germinated at 26 °C in the incubator for 48 h. The germinated seeds were placed in PVC tubes filled with Turface commercial potting mix (Applied Industrial Materials, Corp., Buffalo Grove, IL, USA) with three tubes per line in the IRRI greenhouse. The tubes were watered with alternate applications of half-strength nutrient solution (Yoshida et al., 1976) and distilled water. For the drought stress treatments, plants were stressed by removing the hole plug of each tube and slowly draining the solution at the tillering, booting, and heading stages. The stress was maintained until leaves of the treated plants rolled completely and their leaf relative water content reached 70–75%. Then, the stressed plants were recovered by rewatering. Leaf and root tissues were collected from the drought-stressed, well-watered, and recovered plants at the tillering stage. At the booting and heading stages, only leaf tissues were sampled for the three treatments. Three replicates were prepared from each sample for analysing DNA methylation. After collection, samples were snap frozen in liquid nitrogen and kept at –80 °C freezer for total DNA extraction.
Genomic DNA of both DK151 and IR64 samples collected from above treatments was isolated using DNeasy plant mini Kit (Qiagen 69103; Qiagen, Hilden, Germany) following the product instructions, and MSAP analyses of the samples were performed as described previously (Xiong et al., 1999) with minor modifications. Briefly, double enzyme combinations, EcoRI/MspI and EcoRI/HpaII, were used to digest the DNA samples using the designed adapters, primary and secondary PCR primers (Table S1). Double enzyme digestion and ligation were performed in one step with a 25-μl reaction volume including 300 ng genomic DNA, 1×T4 ligase buffer (with 1 mmol ATP), 1×YANG†/TANGO buffer, 3 U of EcoRI and 3 U of HpaII/MspI, 1.5 of U T4 ligase, 5 pmol of EcoRI adapter, and 50 pmol of HpaII–MspI adapter. This reaction mixture was incubated at 37 °C for 8 h, and then stored at 4 °C. The resultant products were diluted 20-fold and used as templates in the following pre-amplification. Then, two consecutive PCRs were used to selectively amplify the EcoRI–HpaII and EcoRI–MspI DNA fragments. The total volume of pre-amplification was 20 μl, containing 2 μl of the diluted mixture mentioned above, 1×PCR buffer, 2 μl of 10 mM dNTP, 10 μmol EcoRI (E1), and H/M primer (HM1) (Table S1), 0.5 U of Taq DNA polymerase. The PCR was performed for 30 cycles consisting of 30 s at 94 °C, 1 min annealing at 56 °C, and 1 min extension at 72 °C. After checking the quality of the pre-amplified amplicons by agarose gel electrophoresis, the amplicons were diluted 20-fold and used for the second selective amplification with the same primers but containing two selection nucleotides at the 3' end. The selective PCR amplification profile followed the protocol described by Zhong et al. (2009). The final amplicons were denatured, separated on 6% denaturing polyacrylamide gels, and visualized by silver staining.
A set of 26 randomly selected differentially amplified fragments were isolated, re-amplified, and purified with the Wizard SV gel and PCR clean-up system (Promega, Madison, WI, USA). The purified DNA fragments were cloned with T-vector (Takara, Dalian, China) for sequencing. The sequences obtained were analysed by NCBI BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and EMBL BLAST (http://www.ebi.ac.uk/Tools/blast/).
Results
Genotypic and phenotypic differences between DK151 and IR64
Table 1 shows the results of the phenotyping experiments. Under the severe drought stress of the 2004 DS and the 2004 WS where IR64 suffered yield losses of, respectively, 96.4% and 98.1%, DK151 yielded 3.1 and 17.1 times as much as IR64, indicating its good level of DT. The better DT of DK151 was associated with 7–8 d of earlier heading and significantly improved fertility and grain filling under drought. However, under the irrigated control conditions, DK151 suffered significant yield penalty by 17.3% in the 2004 WS. Under the mild stress of the 2005 DS when IR64 suffered 30.2% yield loss, DK151 yielded twice as much as IR64, though its yield was 23.2% lower than IR64 under the irrigated control conditions. Interestingly, DK151 produced significantly higher yield under stress than irrigated control in the 2005 DS, indicating that it apparently became more adapted to the mild rain-fed conditions.
Table 1.
The mean performances of DK151 and IR64 for grain yield and related traits under drought stress and non-stress conditions evaluated in three consecutive seasons
| Season | Genotype | Treatment | Yield (t/ha)a | HD (d) | PH (cm) | PN | GN | SF (%) | GW (g) |
| 2004 DS | DK151 | Stress | 0.39* | 83.7** | 56.0 | 13.7 | – | 43.1* | – |
| IR64 | Stress | 0.12** | 91.1* | 54.8 | 14.0 | – | 37.8** | – | |
| DK151 | Control | 3.59 | 76.3** | 78.6 | 21.7 | – | 79.3** | – | |
| IR64 | Control | 3.58 | 81.1* | 78.6 | 20.3 | – | 86.6* | – | |
| 2004 WS | DK151 | Stress | 1.61* | 74.0** | 82.2* | 12.3 | 145.0* | 69.4* | 25.9* |
| IR64 | Stress | 0.09** | 81.7* | 77.6** | 11.5 | 47.9** | 27.2** | 17.2** | |
| DK151 | Control | 3.99** | 74.7** | 93.5 | 17.2 | 135.7 | 83.9 | 27.6* | |
| IR64 | Control | 4.83* | 80.1* | 93.5 | 19.0 | 136.0 | 84.1 | 25.0** | |
| 2005 DS | DK151 | Stress | 5.97* | – | – | – | 178.7* | 76.6* | 25.7* |
| IR64 | Stress | 2.98** | – | – | – | 48.8** | 62.1** | 20.1** | |
| DK151 | Control | 3.23** | – | – | – | 121.5** | 85.6 | 27.9* | |
| IR64 | Control | 4.21* | – | – | – | 143.1* | 88.5 | 25.0** |
HD, PH, PN, GN, SF, and GW are heading date, plant height, panicle number per plant, filled grain number per panicle, spikelet fertility, and 1000 grain weight, respectively.
* or ** after the mean trait values of DK151 and IR64 indicate statistically significant difference at P < 0.01.
General properties of DNA methylation patterns in rice
Using 45–65 pairs of primer combinations, 1180–1211 fragments were amplified in each leaf or root sample of DK151 and IR64 (Table S2). According to the presence or absence of the bands from specific isoschizomer digestions (Li et al., 2002, 2009), the amplified DNA fragments could be divided into four types: type I represents the band presence for both enzyme combinations; type II is the band presence only for EcoRI/HpaII; type III is the band presence for EcoRI/MspI; and type IV represents the band absence for both enzyme combinations. Here, type II represents cases of semi-methylated bands while types III and IV represent situations of full methylation.
Tables 2 and 3 show some general cytosine methylation patterns in DK151 and IR64 under well-watered, drought-stressed, and subsequent recovery conditions. When measured by the total number and percentage of the methylated bands (types II+III+IV), under well-watered conditions, the overall level of DNA methylation ranged from 16.1% (174 bands) in roots of IR64 at the tillering stage to 24.6% (289 bands) in leaves of DK151 at the booting stage. When measured by the number and percentage of total fully methylated bands at three developmental stages, there was a general pattern of control>stress<recovery, in both varieties and tissues. At the tillering stage, the total DNA methylation level in leaves and roots of both lines decreased considerably under drought stress, and bounced back slightly after subsequent rewatering. However, where specific methylation types are concerned, drought stress specifically induced more than doubled type II (hemi-methylated) bands at the expense of type IV (fully methylated) bands in both DK151 and IR64, as compared with the control and rewatered conditions. More methylated DNA bands were detected in leaves of both lines at the booting and heading stages than at the tillering stage, and the overall methylation levels in leaves of both lines were relatively stable under well-watered, drought stress, and subsequent recovery conditions (Table 3). The overall methylation level in DK151 was consistently higher than IR64 by 1–2% in both leaves and roots under all three water conditions.
Table 2.
DNA methylation changes in leaves and roots of DK151 and IR64 at the tillering stage under three water conditions
| Samples | Leaves |
Roots |
||||||||||
| DK151 |
IR64 |
DK151 |
IR64 |
|||||||||
| MSAP band typea | Control | Stress | Recovery | Control | Stress | Recovery | Control | Stress | Recovery | Control | Stress | Recovery |
| I | 943 | 961 | 973 | 847 | 871 | 882 | 1012 | 1017 | 1033 | 906 | 934 | 931 |
| II | 38 | 100 | 42 | 39 | 92 | 41 | 31 | 80 | 23 | 23 | 66 | 29 |
| III | 119 | 112 | 97 | 86 | 69 | 60 | 42 | 62 | 44 | 23 | 29 | 26 |
| IV | 111 | 38 | 99 | 108 | 48 | 97 | 126 | 52 | 111 | 128 | 51 | 94 |
| Total amplified bands | 1211 | 1211 | 1211 | 1080 | 1080 | 1080 | 1211 | 1211 | 1211 | 1080 | 1080 | 1080 |
| Total methylated bands | 268 | 250 | 238 | 233 | 209 | 198 | 199 | 194 | 178 | 174 | 146 | 149 |
| MSAP (%) | 22.13 | 20.64 | 19.65 | 21.57 | 19.35 | 18.33 | 16.43 | 16.02 | 14.70 | 16.11 | 13.52 | 13.80 |
| Fully methylated bands | 230 | 150 | 196 | 194 | 117 | 157 | 168 | 114 | 155 | 151 | 80 | 120 |
| Fully methylated ratio (%) | 19.0 | 12.4 | 16.2 | 18.0 | 10.8 | 14.5 | 13.9 | 9.4 | 12.8 | 14.0 | 7.4 | 11.1 |
Type II are hemi-methylated bands and types III+IV are fully methylated bands. Total methylated bands, II+III+IV.
Table 3.
DNA methylation changes in leaf of DK151 and IR64 at booting and heading stages
| Growth stage | Booting |
Heading |
||||||||||
| Genotype | DK151 |
IR64 |
DK151 |
IR64 |
||||||||
| MSAP band type | Control | Stress | Recovery | Control | Stress | Recovery | Control | Stress | Recovery | Control | Stress | Recovery |
| I | 888 | 886 | 889 | 899 | 901 | 899 | 891 | 901 | 888 | 898 | 899 | 898 |
| II | 79 | 83 | 82 | 78 | 79 | 80 | 81 | 84 | 82 | 78 | 82 | 76 |
| III | 166 | 167 | 164 | 166 | 164 | 166 | 166 | 164 | 166 | 166 | 165 | 166 |
| IV | 44 | 41 | 42 | 13 | 12 | 11 | 39 | 28 | 41 | 14 | 10 | 16 |
| Total amplified bands | 1177 | 1177 | 1177 | 1156 | 1156 | 1156 | 1177 | 1177 | 1177 | 1156 | 1156 | 1156 |
| Total methylated bandsa | 289 | 291 | 288 | 257 | 255 | 257 | 286 | 276 | 289 | 258 | 257 | 258 |
| MSAP (%) | 24.55 | 24.72 | 24.47 | 22.23 | 22.06 | 22.23 | 24.3 | 23.45 | 24.55 | 22.32 | 22.23 | 22.32 |
| Fully methylated bandsb | 210 | 208 | 206 | 179 | 176 | 177 | 205 | 192 | 207 | 180 | 175 | 182 |
| Full methylated ratio (%) | 17.84 | 17.67 | 17.5 | 15.48 | 15.22 | 15.31 | 17.42 | 16.31 | 17.59 | 15.57 | 15.14 | 15.74 |
Type II are hemi-methylated bands and types III+IV are fully methylated bands. Total methylated bands = II+III+IV.
Genotypic, tissue, and developmental differences in DNA methylation pattern under different water treatments
More detailed comparisons revealed some interesting results regarding the tissue and developmental patterns of DNA methylation/demethylation and their genotypic differences (Table 4, Fig. 2). First, at the tillering stage, drought resulted in significantly more cytosine demethylation than cytosine methylation in both leaf and root tissues with an average of 330 demethylated (class a+b+c) bands compared with 95 methylated (class d+e+f) bands, plus 100 class g bands that were unchanged under drought but changed after recovery. In the leaf tissue, the overall levels of drought-induced DNA methylation and demethylation were much higher at the tillering stage (162 and 37) than at the booting (12 and 8) and heading (37 and 24) stages.
Table 4.
Summary of DNA methylation pattern changes of DK151 and IR64 under three water conditions
| Band classa | Tillering stage |
Booting stage |
Heading stage |
|||||||||||||||
| Leaves |
Roots |
DK151 |
IR64 |
Leaves |
Leaves |
|||||||||||||
| DK151 | IR64 | Comm.b | DK151 | IR64 | Comm. | Leaf | Root | Comm. | Leaf | Root | Comm. | DK151 | IR64 | Comm. | DK151 | IR64 | Comm. | |
| a | 79 | 75 | 31 | 88 | 75 | 43 | 79 | 88 | 71 | 75 | 75 | 56 | 6 | 8 | 2 | 31 | 8 | 3 |
| b | 23 | 26 | 10 | 23 | 27 | 10 | 23 | 23 | 11 | 26 | 27 | 12 | 0 | 0 | 0 | 0 | 1 | 0 |
| c | 0 | 0 | 0 | 2 | 7 | 1 | 0 | 2 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| (a+b+c) | 102 | 101 | 41 | 113 | 109 | 54 | 102 | 113 | 82 | 101 | 109 | 68 | 6 | 8 | 2 | 31 | 9 | 3 |
| d | 11 | 12 | 6 | 20 | 8 | 5 | 11 | 20 | 10 | 12 | 8 | 2 | 3 | 5 | 2 | 18 | 2 | 1 |
| e | 11 | 10 | 2 | 21 | 13 | 5 | 11 | 21 | 10 | 10 | 13 | 13 | 2 | 0 | 0 | 3 | 2 | 0 |
| f | 0 | 1 | 0 | 4 | 4 | 2 | 0 | 4 | 0 | 1 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| (d+e+f) | 22 | 23 | 8 | 45 | 25 | 12 | 22 | 45 | 20 | 23 | 25 | 16 | 5 | 5 | 2 | 21 | 4 | 1 |
| g | 37 | 31 | 13 | 28 | 26 | 9 | 37 | 28 | 28 | 31 | 26 | 26 | 7 | 3 | 1 | 1 | 1 | 0 |
| h | 1048 | 922 | 876 | 1020 | 918 | 865 | 1048 | 1020 | 964 | 922 | 918 | 858 | 1159 | 1140 | 1105 | 1122 | 1142 | 1077 |
| i | 2 | 3 | 0 | 5 | 2 | 0 | 2 | 5 | 0 | 3 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| Total | 1211 | 1080 | 938 | 1211 | 1080 | 940 | 1211 | 1211 | 1105 | 1080 | 1080 | 968 | 1177 | 1156 | 1110 | 1177 | 1156 | 1081 |
a: demethylated by drought, but remethylated after recovery; b: demethylated by drought, and remaining hypomethylated after recovery; c: demethylated by drought, but remethylated in a different pattern after recovery; d: methylated by drought, but demethylated after recovery; e: methylated by drought, and remaining methylated after recovery; f: methylated by drought, but demethylated in a different pattern after recovery; g: DNA methylation pattern remained unchanged under drought, but changed after recovery; h: DNA methylation pattern was unchanged under all three conditions; i: others.
Comm., the number of common bands shared by DK151 and IR64 or two different tissues of the same genotype.
Fig. 2.
Comparisons of rice genotypic, tissue, and developmental specificities in drought-induced DNA demethylation (a+b+c)/methylation (d+e+f) patterns under three water conditions: (A) between rice genotypes in different tissues at the tillering stage; (B) between leaves and roots in different genotypes at the tillering stage; and (C) between the booting and heading stages in different rice genotypes under non-stress conditions. Here, the ‘class’ refers to the classification of DNA methylation/demethylation bands defined in Table 4.
Second, under non-stress conditions, the differences in the detected DNA methylation bands were small between different genotypes (22.9%), between leaves and roots (15.9%), and between the booting and heading stages (4.5%) (Table 4, class h). However, drought-induced DNA methylation showed much greater genotypic, tissue, and developmental differences. Under drought at the tillering stage, the commonly demethylated bands shared by DK151 and IR64 accounted for only 25.3% of the total detected demethylated bands in leaves and 32.1% in roots. This number was 21.6% in leaves and 20.7% in roots (Fig. 2A, B). Similarly, DK151 and IR64 had 50% and 8.1% commonly demethylated bands, and shared only 37.5% and 4.2% commonly methylated bands at the booting and heading stages, respectively, under drought conditions (Fig. 2C). In other words, drought-induced methylation and demethylation sites were largely different in the genome of DK151 from that of IR64. The maximum genotypic difference between DK151 and IR64 was found in drought-stressed roots at the tillering stage, where DK151 had 80% more methylation sites than IR64 (Fig. 2B) and in leaves at the heading stage when DK151 had 3.44 times more demethylation sites and 5.25 times more methylation sites than IR64, respectively Fig. 2C.
Third, drought-induced DNA demethylation bands consisted of approximately two-thirds of class a (demethylated by drought, but reversed after recovery) and one-third of class b (demethylated by drought, and remaining hypomethylated after recovery) with few class c bands detected (demethylated by drought, but remethylated in different patterns after recovery) (Table 4). DK151 had a slightly greater portion (79.6%) of class a bands than IR64 (74.8%). On the other hand, drought-induced DNA methylation consisted of approximately equal portions of class d (methylated by drought, but demethylated by recovery) and class e (methylated by drought, and remaining methylated after recovery) with few bands of class f (methylated by drought, but demethylated in different patterns after recovery) detected.
BLAST results of the differentially methylated DNA sequences
A random set of 26 drought-induced polymorphic DNA methylation bands detected above were cloned and sequenced. The sequences of the cloned bands have an average size of 200 bp, ranging from 90 to 297 bp (Table 5) and were found to be distributed widely on the rice genome except for chromosomes 2, 8, and 9, indicating a genome-wide alteration in DNA methylation/demethylation induced by drought.
Table 5.
BLAST results of a randomly selected set of 26 polymorphic methylated DNA fragments
| MSAP fragment |
Accession No. | Nuclear/protein identity (%) | E value | Sequence homology | |||
| Name | Primer combination | Size (bp) | Chr. | ||||
| M1 | E01/HM37 | 130 | 6 | GenBank:AY785763.1 | 96 | 6.E-42 | Putative polynucleotide adenylyltransferase |
| M2 | E09/HM310 | 233 | 4 | EMBL:CA766808 | 98 | 6.E-66 | IRRI Drought Stress Panicle Library Oryza sativa, cDNA clone |
| GenBank:CI445285.1 | 98 | 2.E-66 | Callus UVB-irradiated callus, 24 h after treatment, cDNA clone | ||||
| M3 | E09/HM311 | 189 | EMBL:AG876068 | O. sativa indica group genomic DNA, BAC end sequence | |||
| M4 | E10/HM311 | 256 | EMBL:AG876068 | 80 | 8.E-46 | O. sativa indica group genomic DNA, BAC end sequence | |
| M5 | E02/HM39 | 157 | 5 | GenBank:AC105768.2 | 95 | 2.E-07 | OJ1122_B08, complete sequence |
| M6 | E10/HM316 | 160 | 1 | GenBank:AP003372.2 | 98 | 4.E-34 | Hypothetical protein |
| M7 | E07/HM313 | 243 | 1 | GenBank:AB254027.1 | 98 | 6.E-53 | atp6 gene for ATPase subunit 6 and ORF79 gene |
| M8 | E08/HM314 | 157 | 4 | GenBank:CK058985.1 | 99 | 1.E-53 | PA64s panicle fertile cDNA |
| M9 | E09/HM312 | 120 | 12 | GenBank:AL731881.4 | 90 | 2.E-23 | Genomic DNA, chromosome 12 (hypothetical protein) |
| M10 | E04/HM37 | 183 | 7 | GenBank:AP005830.4 | 93 | 4.E-74 | Hypothetical protein |
| M11 | E06/HM39 | 90 | 4 | GenBank:AK288604.1 | 90 | 5.E-10 | O. sativa japonica group cDNA |
| EMBL:M22826 | 90 | 2.E-09 | Ribosomal protein L22 (rpL22) gene | ||||
| M12 | E07/HM38 | 272 | 10 | Swiss-Prot:Q10LD3 | 90 | 8.E-23 | Retrotransposon protein, putative, Ty3-gypsy |
| GenBank:AB014740.1 | 93 | 1.E-75 | gypsy-type retrotransposon RIRE8A DNA | ||||
| M13 | E08/HM39 | 297 | 5 | GenBank:BK000929.1 | 96 | 7.E-112 | O. sativa transposon Rim2-M344 |
| Swiss-Prot :Q94I15 | 96 | 6.E-40 | Putative retroelement | ||||
| M14 | E04/HM36 | 200 | 9 | GenBank:CI659730.1 | 94 | 9.E-51 | Leaf of seedling γ-irradiated (4 min), cDNA clone |
| Swiss-Prot :Q9AYB3 | 82 | 2.E-10 | Putative uncharacterized protein | ||||
| M15 | E06/HM33 | 165 | GenBank:AE017283.1 | 98 | 8.E-69 | Aspartyl aminopeptidase | |
| M16 | E10/HM316 | 190 | GenBank:CP000284.1 | 81 | 3.E-18 | N-6 DNA methylase flagellatus | |
| GenBank:AM039952.1 | 73 | 7.E-13 | Type I site-specific deoxyribonuclease | ||||
| M17 | E05/HM312 | 222 | 3 | GenBank:AC136284.1 | 78 | 5.E-35 | Genomic sequence for O. sativa |
| GenBank:CI437782.1 | 72 | 7.E-21 | O. sativa callus UVB-iradiated callus, immediately after treatment | ||||
| GenBank:CI050422.1 | 86 | 1.E-17 | Cold-treated cDNA clone | ||||
| GenBank:CI413574.1 | 86 | 1.E-17 | 100 ppm ZnSO4 for 1 week, cDNA clone | ||||
| GenBank:CI083616.1 | 86 | 1.E-17 | ABA: abscisic acid-treated callus cDNA clone | ||||
| M18 | E05/HM38 | 167 | 10 | GenBank:AC069145 | 100 | 1.E-34 | Genomic sequence |
| M19 | E7/HM310 | 155 | 1 | GenBank:AP003453.3 | 96 | 1.E-53 | O. sativa genomic DNA |
| EMBL:AY873625 | 90 | 4.E-40 | Transposon insertional mutants | ||||
| M20 | E7/HM310 | 143 | 7 | EMBL:EE590765 | 93 | 9.E-63 | Rice, mixture of leaf, root, panicle, cDNA |
| EMBL:EU155081 | 89 | 2.E-38 | Retrotransposon Tos17 | ||||
| M21 | E09/HM32 | 214 | 5 | Swiss-Prot :Q2QMZ1 | 96 | 3.E-25 | HAT family dimerization domain-containing protein, O. sativa |
| M22 | E09/HM32 | 132 | 5 | GenBank:AK289009.1 | 92 | 9.E-35 | O. sativa cDNA, clone: J090089C11 |
| Swiss-Prot :Q2QTE7 | 93 | 5.E-08 | Retrotransposon protein, putative, Ty3-gypsy subclass | ||||
| Swiss-Prot :Q9AYB7 | 93 | 8.E-08 | Similar to Sorghum bicolor 22 kDa akafirincluster | ||||
| M23 | E10/HM32 | 200 | 4 | GenBank:AJ440220.1 | 82 | 1.E-09 | O. sativa a9 gene for plasma membrane H+-ATPase |
| GenBank:CA766881.2 | 77 | 2.E-02 | Drought Stress Panicle Library Indica, cDNA clone | ||||
| M24 | E02/HM31 | 149 | 12 | GenBank:AL713950.4 | 83 | 9.E-23 | BAC OJ1004_F11 |
| GenBank:CB635907.1 | 80 | 6.E-07 | cDNA clone OSIIEb16M12 | ||||
| M25 | E02/HM31 | 90 | GenBank:AK289070.1 | 89 | 4.E-16 | cDNA, clone: J090094F22 | |
| Swiss-Prot :Q5H9W5 | 80 | 3.E-02 | B1168G10.5 protein, O. sativa | ||||
| M26 | E03/HM33 | 209 | 6 | GenBank:AF443596.1 | 93 | 1.E-03 | Zea mays enhancer of zeste-like protein 1 (mez1) mRNA |
| EMBL:EG710286 | 100 | 9.E-06 | Rice young panicle cDNA clone | ||||
Chr., chromosome.
Based on the BLAST results (Table 5), five of the cloned fragments were homologous to genes encoding polynucleotide adenylyltransferase, ribosomal protein, aspartyl aminopeptidase, zeste-like protein 1, and type I site-specific deoxyribonuclease; whereas seven were highly homologous to the cDNA sequences responding to abiotic stresses (drought, cold, ABA, ZnSO4, UV exposure, and γ-ray irradiation); and four were related to transposons/retrotransposons. These results indicate that the drought-induced methylation/demethylation bands detected involved genes of a wide range of functions, including those related to stress responsiveness.
Discussion
Plants are known to respond to environmental stresses by adjusting their physiological and developmental machinery by differentially regulating genome-wide gene expression (López-Maury et al., 2008). In this regard, epigenetic mechanisms such as DNA methylation/demethylation are expected to play a key role (Lu et al., 2008; Angers et al., 2010). Indeed, we found that drought was able to induce genome-wide changes in DNA methylation status and these changes, when averaged across different genotypes, tissues, and developmental stages, accounted for ∼12.1% of the total site-specific methylation differences in the rice genome as detected by the MSAP analysis. In particular, drought tended to reduce the overall DNA methylation levels in leaves and roots of both rice lines at the tillering stage. Our results are consistent with previous reports that showed that environmental factors such as cold, heavy metals, aluminum toxicity, and salt tend to cause demethylation of genomic DNA (Lizal and Relichova, 2001; Alina et al., 2004; Choi and Sano, 2007; Zhong et al., 2009). Furthermore, we observed three interesting properties of drought-induced DNA methylation changes in rice, i.e. its genotypic, tissue, and developmental specificities, which appear to shed some light on the possible roles of the epigenetic mechanisms in rice adaptation to drought stress.
Our results indicate that the genotypic specificity of epigenetic mechanisms such as DNA methylation/demethylation plays a very important role in regulating rice responses and thus adaptation to drought stress. In this study, although the overall levels of DNA methylation/demethylation in DT DK151 and drought-sensitive IR64 were similar, they shared a very small portion of commonly methylated and demethylated fragments detected by the MSAP technology. While this result is similar to the reported global methylation pattern of genomic DNA from different rice varieties (Takata et al., 2005), largely different sets of genes were expectedly differentially expressed in DK151 and IR64 under drought because the large differences in their drought-induced methylation/demethylation sites are expected to cause differential gene expression in the detected methylated/demethylated sites between the two lines. Consistent with this expectation, dramatic differences have been observed between rice genotypes that differ greatly in their DT (Fu et al., 2007). Apparently, the large differences in DNA methylation/demethylation patterns and drought tolerance between DK151 and IR64 result from the introgressed genomic fragments from two donors, Binam and BR24 (Fig. 1). Additional efforts are being made to identify DT candidate genes/pathways that differentiate DK151 and IR64 by linking the differentially expressed genes with the introgression segments and by bioinformatic analyses.
We found that reversibility was another important property of loci or genomic regions that had gone through drought-induced epigenetic changes. Two major types of drought-induced methylation changes were identified in this study, including 61.8% of class a sites plus 8.2% of class d sites at which drought-induced epigenetic changes were reversed to their original status after recovery, plus 19.6% of class b and 9.4% of class e sites at which the drought-induced DNA demethylation/methylation changes remain even after recovery (Table 4). Although this reversibility of DNA epigenetic processes is reportedly affected by complex gene–environment interactions (Ramchandani et al., 1999), and hypothesized to result from active demethylation or from passive loss of methylation (Zhu et al., 2007; Zhang et al., 2010), it remains unclear what molecular mechanism(s) are actually involved in the stress-induced epigenetic changes and subsequent recovery, and if and how they are involved in the expression and transmission behaviour of the regions (loci) involved.
Finally, we observed that drought-induced DNA methylation/demethylation alteration showed a significant level of developmental and tissue specificity. For example, the overall cytosine methylation level induced by drought dropped much more significantly at the tillering stage than at the booting and heading stages. Furthermore, a lower level of DNA methylation was observed in roots than in leaves at the same developmental stage in both lines, indicating unique biological functions of rice roots and leaves in response to drought stress. While consistent with previous reports on tissue-dependent DNA methylation pattern and its possible role in regulating tissue-specific gene expression (Aceituno et al., 2008; Lu et al., 2008), our results suggest that the developmental and tissue specificity of epigenetic changes in the rice genome could be a very important regulatory mechanism for rice plants in adapting to adverse environments, though how these developmental and tissue-specific epigenetic changes are controlled at the molecular level remains to be elucidated.
Conclusions
In conclusion, our results indicate that drought could induce genome-wide changes in DNA methylation/demethylation, accounting for ∼12.1% of total site-specific methylation differences in the rice genome. This drought-induced DNA methylation pattern in rice showed three interesting properties. The most important one was its genotypic specificity, as reflected by large differences in the detected DNA methylation/demethylation sites between DT DK151 and drought-sensitive IR64, which result from a small number of introgressed genomic fragments in DK151. Second, most drought-induced methylation/demethylation sites were of two major types distinguished by their reversibility, including 70% of methylation/demethylation sites at which drought-induced epigenetic changes were reversed to their original status after recovery, and 29% of sites at which the drought-induced DNA demethylation/methylation changes remain even after recovery. Third, the drought-induced DNA methylation/demethylation alteration showed a significant level of developmental and tissue specificity with the overall DNA methylation level induced by drought dropping much more significantly at the tillering stage than at the booting and heading stages. Together, these properties are expected to have contributed greatly to rice responses and adaptation to drought stress through regulating genome-wide gene expression.
Supplementary data
Supplementary Table 1 lists the adapter and primer sequences.
Supplementary Table 2 shows the alteration of DNA methylation pattern of DK151 and IR64 under three water conditions.
Supplementary Fig. 1 shows the backcross and intercross breeding procedures for developing drought-tolerant pyramiding line DK151.
Acknowledgments
Financial support from the National 863 Project of China (#2007AA10Z191), CAAS/ICS core funding, the Ministry of Agriculture of China ‘948’ project (#2006-G51), the CGIAR Generation Challenge Program project (#12), the Rockefeller Foundation project (#2005 FS029), and the Bill and Melinda Gates Foundation project (OPP51587) to ZKL. WSW was also supported by a scholarship from the National Science Foundation of China.
Glossary
Abbreviations
- DS
dry season
- DT
drough-tolerant
- MSAP
methylation-sensitive amplified polymorphisms
- WS
wet season
References
- Aceituno FF, Nick M, Seung YR, Rodrigo AG. The rules of gene expression in plants: organ identity and gene body methylation are key factors for regulation of gene expression in. Arabidopsis thaliana. BMC Genomics. 2008;9:438. doi: 10.1186/1471-2164-9-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimoto K, Katakami H, Kim HJ, Ogawa E, Sano CM, Wada Y, Sano H. Epigenetic inheritance in rice plants. Annals of Botany. 2007;100:205–217. doi: 10.1093/aob/mcm110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alina R, Sgorbati S, Santagostino A, Labra M, Ghiani A, Citterio S. Specific hypomethylation of DNA is induced by heavy metals in white clover and industrial hemp. Physiologia Plantarum. 2004;121:472–480. [Google Scholar]
- Angers B, Castonguay E, Massicotte R. Environmentally induced phenotypes and DNA methylation: how to deal with unpredictable conditions until the next generation and after. Molecular Ecology. 2010;19:1283–1295. doi: 10.1111/j.1365-294X.2010.04580.x. [DOI] [PubMed] [Google Scholar]
- Bender J. DNA methylation and epigenetics. Annual Review of Plant Biology. 2004;55:41–68. doi: 10.1146/annurev.arplant.55.031903.141641. [DOI] [PubMed] [Google Scholar]
- Boyko A, Kathiria P, Zemp FJ, Yao Y, Pogribny I, Kovalchuk I. Transgenerational changes in the genome stability and methylation in pathogen-infected plants: (virus-induced plant genome instability) Nucleic Acids Research. 2007;35:1714–1725. doi: 10.1093/nar/gkm029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko A, Kovalchuk I. Epigenetic control of plant stress response. Environmental and Molecular Mutagenesis. 2008;49:61–72. doi: 10.1002/em.20347. [DOI] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proceedings of the National Academy of Sciences, USA. 2002;99:16491–16498. doi: 10.1073/pnas.162371599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi CS, Sano H. Abiotic-stress induces demethylation and transcriptional activation of a gene encoding a glycerophosphodiesterase-like protein in tobacco plants. Molecular Genetics and Genomics. 2007;277:589–600. doi: 10.1007/s00438-007-0209-1. [DOI] [PubMed] [Google Scholar]
- Cullis CA. Mechanisms and control of rapid genomic changes in flax. Annals of Botany. 2005;95:201–206. doi: 10.1093/aob/mci013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Genger RK, Peacock WJ, Dennis ES. DNA methylation in plants. Annual Review of Plant Physiology and Plant Molecular Biology. 1998;49:223–247. doi: 10.1146/annurev.arplant.49.1.223. [DOI] [PubMed] [Google Scholar]
- Fu BY, Xiong JH, Zhu LH, Zhao XQ, Xu HX, Gao YM, Li YS, Xu JL, Li ZK. Identification of functional candidate genes for drought tolerance in rice. Molecular Genetics and Genomics. 2007;278:599–609. doi: 10.1007/s00438-007-0276-3. [DOI] [PubMed] [Google Scholar]
- Hua Y, Chen XF, Xiong JH, Zhang YP, Zhu YG. Isolation and analysis of differentially methylated fragment CIDM7 in rice induced by cold stress. Hereditas. 2005;27:595–600. [PubMed] [Google Scholar]
- Li XL, Lin ZX, Nie YC, Guo XP, Zhang XL. Methylation-sensitive amplification polymorphism of epigenetics changes in cotton under salt stress. Acta Agronomica Sinica. 2009;35:588–596. [Google Scholar]
- Li XQ, Xu ML, Korban SS. DNA methylation profiles differ between field- and in vitro-grown leaves of apple. Journal of Plant Physiology. 2002;159:1229–1234. [Google Scholar]
- Li ZK, Xu JL. Breeding for drought and salt tolerant rice (Oryza sativa L.): progress and perspectives. In: Jenks MA, Hasegawa PM, Jain SM, editors. Advances in molecular breeding towards salinity and drought tolerance. Dordrecht: Springer; 2007. pp. 531–564. [Google Scholar]
- Lippman Z, Gendrel AV, Black M, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- Lizal P, Relichova J. The effect of day length, vernalization and DNA demethylation on the flowering time in. Arabidopsis thaliana. Physiologia Plantarum. 2001;113:121–127. [Google Scholar]
- Long LK, Lin XY, Zhai JZ, Kou HP, Yang W, Liu B. Heritable alteration in DNA methylation pattern occurred specifically at mobile elements in rice plants following hydrostatic pressurization. Biochemical and Biophysical Research Communications. 2006;340:369–376. doi: 10.1016/j.bbrc.2005.12.015. [DOI] [PubMed] [Google Scholar]
- López-Maury L, Marguerat S, Bähler J. Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat Rev Genet. 2008;9:583–93. doi: 10.1038/nrg2398. [DOI] [PubMed] [Google Scholar]
- Lu GY, Wu XM, Chen BY, Gao GZ, Xu K, Li XZ. DNA methylation analysis by MSAP during seed germination in rape. Chinese Science Bulletin. 2005;24:2750–2756. [Google Scholar]
- Lu YL, Rong TZ, Cao MJ. Analysis of DNA methylation in different maize tissues. Journal of Genetics and Genomics. 2008;35:41–48. doi: 10.1016/S1673-8527(08)60006-5. [DOI] [PubMed] [Google Scholar]
- Ramchandani S, Bhattacharya SK, Cervoni N, Szyf M. DNA methylation is a reversible biological signal. Proceedings of the National Academy of Sciences, USA. 1999;96:6107–6012. doi: 10.1073/pnas.96.11.6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A, Cedar H. DNA methylation—biochemistry and biological significance. Journal of Chromatography. 1992;581:31–40. [Google Scholar]
- Ruiz GL, Cervera MT, Martine ZJM. DNA methylation increases throughout Arabidopsis development. Planta. 2005;222:301–306. doi: 10.1007/s00425-005-1524-6. [DOI] [PubMed] [Google Scholar]
- SAS Institute. SAS/Stat User's Guide, Version 8.2. Cary, NC,: SAS Institute, USA; 1999. [Google Scholar]
- Takata M, Yuji K, Yoshio S. DNA methylation polymorphisms in rice and wild rice strains: detection of epigenetic markers. Breeding Sciences. 2005;55:57–63. [Google Scholar]
- Tran RK, Henikoff JG, Zilberman D, Ditt RF, Jacobsen SE, Henikoff S. DNA methylation profiling identifies CG methylation clusters in Arabidopsis genes. Current Biology. 2005;15:154–159. doi: 10.1016/j.cub.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Wada Y, Miyamoto K, Kusano T, Sano H. Association between up-regulation of stress-responsive genes and hypomethylation of genomic DNA in tobacco plants. Molecular Genetics and Genomics. 2004;271:658–666. doi: 10.1007/s00438-004-1018-4. [DOI] [PubMed] [Google Scholar]
- Xiong LZ, Xu CG, Maroof S, Zhang QF. Patterns of cytosine methylation in an elite rice hybrid and its parental lines detected by a methylation sensitive amplification polymorphism technique. Molecular and General Genetics. 1999;261:439–446. doi: 10.1007/s004380050986. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Forna DA, Cock JH, Gomez KA. Laboratory manual for physiological studies of rice. Los Baños, Philippines: International Rice Research Institute. 1976 [Google Scholar]
- Zhang MS, Kimatu JN, Xu KZ, Liu B. DNA cytosine methylation in plant development. Journal of Genetics and Genomics. 2010;37:1–12. doi: 10.1016/S1673-8527(09)60020-5. [DOI] [PubMed] [Google Scholar]
- Zhang XY, Yazaki J, Sundaresan A, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Zhao XX, Chai Y, Liu B. Epigenetic inheritance and variation of DNA methylation level and pattern in maize intra-specific hybrids. Plant Sciences. 2007;172:930–938. [Google Scholar]
- Zhong L, Xu YH, Wang JB. DNA-methylation changes induced by salt stress in wheat. Triticum aestivum. African Journal of Biotechnology. 2009;8:6201–6207. [Google Scholar]
- Zhou J, Wang X, Jiao Y, et al. Global genome expression analysis of rice in response to drought and high-salinity in shoot, flag leaf, and panicle. Plant Molecular Biology. 2007;63:591–608. doi: 10.1007/s11103-006-9111-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Kapoor A, Sridhar VV, Agius F, Zhu JK. The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Current Biology. 2007;17:54–59. doi: 10.1016/j.cub.2006.10.059. [DOI] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an inter dependence between methylation and transcription. Nature Genetics. 2006;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.