Abstract
The beneficial element silicon (Si) may affect radial oxygen loss (ROL) of rice roots depending on suberization of the exodermis and lignification of sclerenchyma. Thus, the effect of Si nutrition on the oxidation power of rice roots, suberization and lignification was examined. In addition, Si-induced alterations of the transcript levels of 265 genes related to suberin and lignin synthesis were studied by custom-made microarray and quantitative Real Time-PCR. Without Si supply, the oxidation zone of 12 cm long adventitious roots extended along the entire root length but with Si supply the oxidation zone was restricted to 5 cm behind the root tip. This pattern coincided with enhanced suberization of the exodermis and lignification of sclerenchyma by Si supply. Suberization of the exodermis started, with and without Si supply, at 4–5 cm and 8–9 cm distance from the root tip (drt), respectively. Si significantly increased transcript abundance of 12 genes, while two genes had a reduced transcript level. A gene coding for a leucine-rich repeat protein exhibited a 25-fold higher transcript level with Si nutrition. Physiological, histochemical, and molecular-biological data showing that Si has an active impact on rice root anatomy and gene transcription is presented here.
Keywords: Oryza sativa (rice); exodermis; leucine-rich repeat protein (LRR); lignin, radial oxygen loss (ROL); silicic acid, silicon; suberin
Introduction
Silicon (Si) is the second most abundant element in soils and nearly ubiquitously plant available. In soil solution, Si is present as silicic acid, Si(OH)4, at pH <9, at concentrations between 0.1 mM and 2.0 mM, which is in the same order of magnitude as potassium, calcium, and other major plant nutrients (Epstein, 1994; Bogdan and Schenk, 2008).
Although all soil-borne plants contain Si in their tissues with concentrations ranging from 0.1% up to 10% dry matter, Si is not considered as an essential element according to the definition by Arnon and Stout (1939). Albeit not essential, Si is a beneficial element because it supports the healthy development of many plant species, in particular, of graminaceae like rice. Si enhances growth and yield, improves mechanical strength and thus prevents lodging, and increases resistance to biotic and abiotic stresses like pests and diseases as well as to salinity, drought stress, and metal toxicity (Epstein, 1994, 1999; Ma and Yamaji, 2006).
The view of how Si affects plants has changed from a passive to a more active one, as the alleviatory impact of silicic acid on rice plants infected by the rice blast fungus Magnaporthe grisea has been attributed to the Si-induced cell wall fortification of rice leaves (Kim et al., 2002), while results from further studies suggested that Si enhanced phytoalexin and peroxidase transcript levels (Rodrigues et al., 2004, 2005) in infected rice leaves. In cucumber and wheat plants, Si also increased resistance to the fungal infection powdery mildew, and this is attributed to the enhanced production of phytoalexins in infected leaves (Fawe et al., 1998; Rémus-Borel et al., 2005). Moreover, Si nutrition increased lignin content and enhanced activities of peroxidase, polyphenol oxidase, and phenylalanine ammonia-lyase (PAL) in rice leaves exposed to rice blast (Cai et al., 2008). In maize plants, Si alleviated Al toxicity that was attributed to mediated phenol metabolism as Si treatment stimulated the release of phenolic compounds in roots of maize under Al stress (Kidd et al., 2001). Generally, the beneficial effects of Si are most obvious in plants encountering stress situations.
Rice is often grown in flooded soils under anaerobic and reducing conditions (Ponnamperuma, 1984). Rice, as well as other wetland species, has adapted to a low-oxygen environment by the internal aeration of root via the aerenchyma—a tissue containing gas-filled spaces, which provides a low-resistance pathway for the diffusion of oxygen within the root (Colmer, 2003a, 2006). To counteract the diffusion of oxygen from the root to the anaerobic rhizosphere, called radial oxygen loss (ROL), rice roots develop a barrier (Armstrong, 1979). This barrier against ROL is present in the basal parts of the root whereas there is no barrier to protect the apical zone of the root from ROL (Armstrong, 1971; Colmer, 2003b). In general, densely packed cells, suberin deposition, and lignification in the outer cell layers are thought to serve as barrier formation (Sorrell, 1994; Armstrong et al., 2000). In rice roots, the barrier against ROL is attributed to a suberized exodermis and lignified sclerenchyma cells (Kotula and Steudle, 2008). One of the factors controlling barrier formation was found to be the aeration of growth solution. Rice roots grown under anaerobic conditions contained higher amounts of suberin and lignin in the outer root parts forming a stronger barrier to ROL (Kotula et al., 2009a, b).
Lignin and suberin metabolism in plants share the phenylpropanoid pathway resulting in monolignols, which are secreted to the apoplast and polymerized to lignin (Boerjan et al., 2003). Suberin monomers are composed of fatty acid derivates, glycerol, and ferulic acid, with the latter being an intermediate of the phenylpropanoid pathway (Franke and Schreiber, 2007). The suberin monomers are released to the apoplast via an ATP-binding cassette (ABC) transporter and polymerized by class III peroxidases (POD) to suberin. Parts of this metabolic pathway were enhanced by Si supply in plants encountering stress (Kidd et al., 2001; Cai et al., 2008). As early as 1961, Okuda and Takahashi reported that the addition of silicon to nutrient solution increased the oxidation power of rice roots leading to an oxidation of Fe2+ and Mn2+ and subsequent precipitation on the root surface and hence to a reduced Fe and Mn uptake of the rice plant (Okuda and Takahashi, 1961).
To investigate the Si effect on rice roots further, experiments were conducted with rice plants grown in nutrient solution either with (+Si) or without silicic acid (–Si) to explore the oxidation power of the root, the development of the exodermis, endodermis, and the sclerenchyma, as well as the impact of Si on the transcription of candidate genes.
Materials and methods
Plant cultivation
Rice (Oryza sativa L. cv. Selenio) seeds were germinated in tap water for 7 d and then placed between two layers of filter paper standing in tap water for additional 7 d.
Each five seedlings were transferred to 5.0 l pots containing non-aerated nutrient solution (mM: 1.43 NH4NO3, 0.32 NaH2PO4.2H2O, 0.51 K2SO4, 1 CaCl2.2H2O, 1.6 MgSO4.7H2O; μM: 1.82 MnSO4, 0.03 (NH4)6Mo7O24, 9 H3BO3, 0.3 ZnSO4.7H2O, 0.15 CuSO4, and 35.81 Fe as sequestrene). The nutrient solution was changed every 7 d for the first 14 d, and then every 4 d. Plants were harvested after 28 d.
For the determination of oxidation power, histochemical analysis, and microarray experiments, plants were supplied with Si in the form of K2SiO3 at concentrations of 0 ppm Si (control) and 50 ppm Si (1.78 mM) and potassium in the control treatment was balanced with K2SO4 supply. For the evaluation of microarray results by quantitative Real Time-PCR (qRT-PCR), Si was applied as silica gel yielding a concentration in the range of 30–40 ppm Si. The pH-value of the nutrient solution was adjusted to 6.0 by the addition of 10% (v/v) H2SO4 and 0.75 M KOH. The plants were grown in a growth chamber (photoperiod, 14/10 h light/dark; temperature, 25/20 °C day/night; 75% relative humidity, and a light intensity of 220 μmol m−2 s−1).
The oxygen concentration in the nutrient solution was measured with an optical sensor using the Fibox 3 system (Presens, Regensburg, Germany). Oxygen concentration was reduced to 3–4 mg l−1 O2 within 24 h after renewing the nutrient solution and remained at this level. To determine root growth, adventitious roots were marked 1 cm behind the tip with a waterproof marker and root length growth was measured after 24 h.
Determination of silicon in plant material and nutrient solution
Plant matter was dried at 60 °C for 4 d and ground. Plant dry matter was digested overnight in a mixture of 1 M HCl and 2.3 M HF (1:2) (Novozamsky et al., 1984). After the addition of 3.2% boric acid, dye reagent (0.08 M sulphuric acid and 2% ammonium heptamolybdate), 3.3% tartaric acid, and 0.4% ascorbic acid, the silicon concentration in the digested plant material and nutrient solution was photometrically determined at 811 nm.
Visualization of oxidation power in FeS medium
To visualize the oxidation power, adventitious roots were embedded in semi-solid agar medium containing FeS. The medium was prepared by adding 0.8% agar to iron-free nutrient solution and subsequent heating to solubilize the agar. The solution was amended with 1.4 g l−1 FeSO4.7H2O and 0.32 g l−1 Na2S whereupon a black FeS precipitation developed (Trolldenier, 1988). Finally, the solution was buffered by addition of 0.5 g l−1 CaCO3 and adjusted to pH 6.0. Single adventitious root of 42-d-old plants was placed between two plastic plates (5×14 cm; 0.5 cm apart), which were sealed with plasticine, while the rest of the root system remained in nutrient solution. The media was poured between the plastic plates and the top was sealed with paraffin wax. The plates were covered with black foil and photographs of the plates were taken 24 h and 48 h after embedding in agar. For each treatment, four roots were investigated.
Histochemical examination of adventitious roots
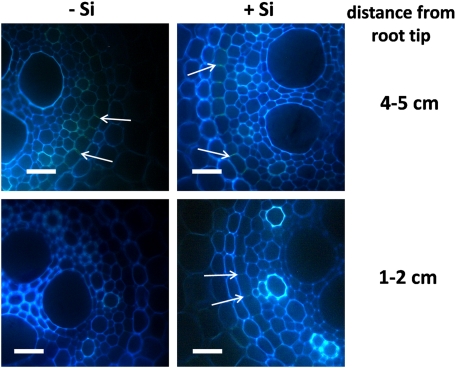
For quantification of the suberin layers and detection of lignification, free-hand cross-sections of adventitious rice roots of 42-d-old plants were taken at 1–2, 4–5, 8–9, and 12–13 cm distance from the root tip (drt) and stained with 0.1% (w/v) berberine hemi-sulphate for 60 min and with 0.5% (w/v) aniline blue for a further 30 min (Brundrett et al., 1988). Stained sections were mounted in 0.1% (w/v) FeCl3 in 50% (v/v) glycerine and viewed under an Axioskop fluorescence microscope (Zeiss, Jena, Germany) with UV illumination using excitation filter G 365, chromatic beam splitter FT 395, and barrier filter LP 420. Pictures were taken with the AxioCam MRc (Zeiss) and picture recording software (AxioVision Ac, Version 4.4, Zeiss). Under UV light, suberin exhibited a blue–white colour and lignin was yellowish-green.
Both –Si and +Si treatments were replicated four times with each replicate consisting of 10 plants. Per plant, one root without lateral roots was cut into four segments (1–2, 4–5, 8–9, and 12–13 cm drt), and from each segment one cross-section was taken. Under the microscope, 10 cells per cross-section were examined and the degree of suberization in the anticlinal cell walls was determined and allocated to one of four stages: 0% (stage I), 0–25% (stage II), 25–50% (stage III), and 50–100% (stage IV) suberization of the length of the anticlinal cell wall.
Collection of genes of interest
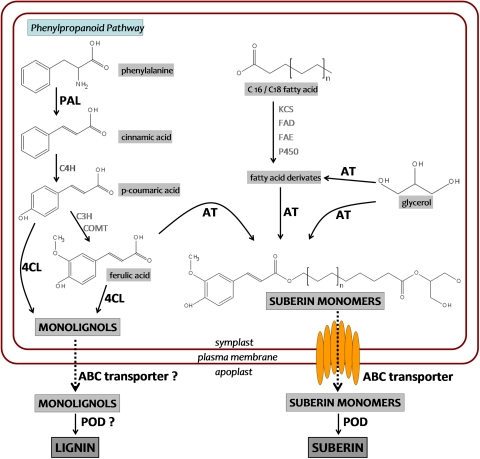
To study the effect of Si nutrition on the transcription of genes related to suberin and lignin synthesis, a list of candidate genes was created. A combined model of the suberin and lignin synthesis pathways was drawn containing 12 protein groups (Fig. 5). These proteins were used as search terms in the search for the putative function of genes in the Rice Genome Annotation Database of the Institute of Genomic Research (TIGR), http://www.tigr.org/tdb/e2k1/osa1. Resulting hits were gathered from the list of candidate genes.
Fig. 5.
Model of the suberin and lignin synthesis pathways. Abbreviations of the proteins are: 4CL, 4-coumarate-CoA ligase; ABC transporter, ATP binding cassette transporter; AT, acyltransferase; C3H, coumaroyl-CoA-3-hydroxylase; C4H, cinnamate 4-hydroxylase; COMT, caffeic acid O-methyltransferase; FAD, fatty acid desaturase; FAE, fatty acid elongase; KCS, β-ketoacyl-CoA synthase; PAL, phenylalanine ammonia-lyase; P450, cytochrome P450 monooxygenase; POD, peroxidase. Bold names indicate Si-induced changes in transcript abundance. (This figure is available in colour at JXB online.)
A hypothetical synthesis pathway for suberin in Arabidopsis (Franke and Schreiber, 2007) attributes key functions to acyltransferases (AT), ABC transporters, POD, and β-ketoacyl-CoA synthases (KCS). A query in the TIGR database for AT and ABC transporters resulted in 62 and 36 hits, respectively. For POD, there is a special database available at http://peroxibase.isb-sib.ch. A search here for genes that are expressed in rice roots resulted in 40 hits. Five genes were obtained from a list of rice KCS available at http://www.izmb.de/schreiber/elogases.shtml. Bernards et al. (2004) suggested that NADPH oxidases are involved in suberin synthesis, and a query in the TIGR database provided one rice gene with the corresponding annotation.
In the biosynthesis pathway of monolignols, the precursors of lignin, major functions are attributed to PAL, 4-coumarate-CoA ligases (4CL), and caffeic acid O-methyltransferases (COMT) (Goujon et al., 2003). A putative function search for rice genes with corresponding annotation in the TIGR database resulted in 8, 14, and 10 hits, respectively. Nine additional genes were gathered from a list of candidate genes for lignin synthesis in Arabidopsis (Goujon et al., 2003). Sequences of Arabidopsis genes were compared to the rice genome by a BLAST (Basic Local Alignment Search Tool) search at http://www.tigr.org/tdb/e2k1/osa1. The result with the highest score was chosen and the annotated function for the rice gene was adopted.
Moreover, a SSH-library with genes differentially expressed in suberized tissue compared to non-suberized tissue in Quercus suber provided additional candidate genes (Soler et al., 2007). Comparing gene sequences of the library with the rice genome by a BLAST search provided an additional 79 candidate genes. The BLAST search resulting in genes with unknown function annotation were not used as candidate genes. After deleting duplicate genes from the function search and the BLAST search, the list of candidate genes contained 265 rice genes.
Microarray analysis
Adventitious roots were harvested at 0–2 cm and 4–6 cm drt and frozen immediately in liquid nitrogen. For RNA isolation, roots were ground under liquid nitrogen and total RNA was isolated using the TRIsure® Reagent (Bioline, Luckenwalde, Germany) following the instructions of the manufacturer. For cDNA synthesis and hybridization with chips, RNA had to be concentrated. Therefore, the RNA of each of 10 biological replicates was pooled in equal amounts and precipitated by the addition of 0.1 vol. 5 M NaCl and 2 vols 96% ethanol. Pooled samples were stored overnight at –70 °C and then centrifuged for 10 min at 16 500 g. The pellet was washed with 500 μl 70% ethanol and centrifuged for 7 min at 16 500 g. The washed pellet was air-dried for 5 min and dissolved in H2O.
For each of the 265 candidate genes, a 50mer oligonucleotide with an aminolink-C6-modification at the 5'-end was designed and synthesized (Ocimum Biosolutions, Ijsselstein, Netherlands). To minimize cross-hybridization with non-target transcripts, the criteria used for oligonucleotide designing were a maximum overall similarity to all other genes in rice genome of 75% and the exclusion of complementary sequences with more than 15 contiguous bases (Kane et al., 2000). Synthetic 50mer oligonucleotides were spotted in triplicate on aldehyde-modified glass slides (VSS25, CEL Associates, Inc., Pearland, TX, USA) using an Affymetrix 417 arrayer. During reverse transcription 8 μg of purified total RNA was copied into fluorescein- (Fl) and biotin- (B) labelled cDNA. The Fl- and B-labelled cDNA of both samples were hybridized simultaneously in one experiment to the same array. After hybridization, the unbound and unspecifically fixed cDNA was removed from the array by three stringent washing steps with sodium chloride–sodium citrate buffer in decreasing concentrations. Specifically bound Fl- and B-labelled cDNAs were sequentially detected with a series of conjugate reporter molecules according to the tyramide signal amplification process, ultimately with Tyramide-Cy3 and Tyramide-Cy5. First, the DNA-chip was incubated with anti-Fl-horseradish peroxidase (HRP) antibody-enzyme conjugate, which specifically binds to the hybridized Fl-labelled cDNA probe. The enzyme portion of the conjugate, horseradish peroxidase, catalyses deposition of the Cy3-labelled tyramide amplification reagent. The reaction results in the immediate deposition of numerous Cy3 labels adjacent to immobilized HRP. Thereby, the amount of tyramide (Cy3) is greatly amplified relative to cDNA hapten (fluorescein). After inactivation of the residual HRP, the added streptavidin-HRP binds to biotin labelled cDNA and catalyses deposition of the Cy5-labelled tyramide amplification reagent. The array obtained through this process was subsequently scanned for the two distinct fluorescent dyes (Cy3 and Cy5), of the cDNA derived from the +Si roots and the –Si roots.
The scanning process of the hybridized chips with the Axon 4000B scanner (Axon Instruments, Foster City, CA, USA) included a 6-fold scan of each chip at different settings, changing both the photomultiplier tube and the laser power settings. The following primary analysis served as a quantification method and was performed with the software tool Gene Pix Pro 6.0™. Next, the secondary analysis was conducted using the data from the primary analysis. Therefore, data from different scans were first normalized by using the median of all background intensities, which were therefore checked for outliers. Three replicates for each gene were tested for outliers. Outliers amongst the gene replicates were eliminated according to the outlier test by Nalimov and the remaining data were averaged. The ratio of the two states for each individual gene was calculated. Finally, a t test (5% probability of error) was applied in order to detect differentially expressed genes between the groups of samples. The results of the microarray data are given as evidence for changes in transcript abundance. The microarray data have been submitted to the NCBI Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE23723 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE23723).
qRT-PCR
Total RNA (1 μg) and random hexamer primers were used to synthesize first-strand cDNA using the Revert Aid™ H Minus Kit (Fermentas, St Leon-Rot, Germany) following the manufacturer's instructions.
In qRT-PCR experiments, 80 ng cDNA was used as the template in a 25 μl reaction-mix containing 2.5 μl 10× buffer, 3.6 mM MgCl2, 0.2 mM dNTPs mix (Fermentas), 0.25 μl 1:1000 diluted SYBR-Green (Invitrogen, Carlsbad, CA, USA), 0.75 U HotStart-Taq-Polymerase (DNA cloning service, Hamburg, Germany), 0.25 μM forward and 0.25 μM reverse primers. The qRT-PCR runs were performed in the CFX96 cycler (Bio-Rad, München, Germany), using an initial 95 °C step for 10 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 30 s, and a final melting curve procedure with a stepwise increment of 1 °C ranging from 60 °C to 95 °C.
Prior to primer design, sequences of genes of interest were blasted against the rice genome at http://rice.plantbiology.msu.edu/ and http://blast.ncbi.nlm.nih.gov/ to obtain gene families or genes with similar sequences. For designing gene member-specific primers suitable for qRT-PCR assay, gene member-specific alignments were created with Vector NTI software (Invitrogen). Heterologous parts of the gene member sequence were used for primer design using Primer 3 (Rozen and Skaletsky, 2000). All primers had to fulfil internal quality standards, which included an amplicon size of 70–200 base pairs, a primer melting temperature of 59–61 °C, a primer size of 20–24 nucleotides, and a primer GC content of 45–65%. Moreover, primer efficiencies were tested with a 5-fold dilution series with 5 ng μl−1 cDNA as the highest concentration and only primer pairs with an efficiency of 90% or above were used for qRT-PCR. The sequences of forward and reverse primer pairs are shown in Table 1.
Table 1.
Sequences of primers used for qRT-PCR
| Gene ID | Forward primer (5′→3′) | Reverse primer (5′→3′) |
| LOC_Os01g22230 | CGGCCGTTGGATGCGAGTGATTT | GATCGACACGACACGACGACACT |
| LOC_Os01g24010 | CCTTGCTACAGCCGGCACCT | ACATGTTCGCGGGCACCCTG |
| LOC_Os01g42410 | GGACGCATTGAAAGAGAAGC | TTGGTCAGCAATATGGGACA |
| LOC_Os01g67540 | TGAGGCAACCGGGTGCATACCTTA | AAGCCACACGGCGCACTTTCTT |
| LOC_Os02g05630 | GTGGTGGTCGTCGTCTTCTT | CCCATAACTGAACTTGCCGT |
| LOC_Os02g06250 | TACCACTGTCGCCACCACTAACCA | AACGACGGCAACTCCGTGGAAA |
| LOC_Os02g41680 | TCACAAGCTCAAGCACCATC | CTCACCAAGCTTCTTGGCAT |
| LOC_Os03g08010 | TCAAGTTTGCTGAGCTGGTG | AAAACGACCAAGAGGAGGGT |
| LOC_Os05g20100 | GCGACGGAGCTGTTGGGCTT | GGCACCGGCTGCCATTCCTT |
| LOC_Os06g16350 | CAGCGCCATGGACAGCCACA | ACGGTGTCGGCCGTGGAGTA |
| LOC_Os06g22080 | GATCGCCAGGAACGTCGCCC | TGCCCTGTTGGGATCGAAGCAC |
| LOC_Os06g32990 | CGGCGAGCTTGTGCTGACCT | GAGTCCAAGGGTCCGGGCGT |
| LOC_Os06g39520 | TGTGAAGAGCGCTGGGCTGCTA | TCACCCCAGCACGGATAACGCT |
| LOC_Os07g44560 | TGCAGCACAACGACAACGGCAT | GCTGCAACAGCGACGAACGTCATA |
| LOC_Os08g02110 | TCCTGAATTGCCCGCCTTAGCTCT | TCACAAAGACGCGGCCACGAAA |
| LOC_Os09g32964 | CGCTCTTCTTCGAGGCGTTCAAGG | CTCCATGCTTCCCGGACTTGACTC |
| LOC_Os10g30610 | ATCATCTAATGAGGCACGGC | TCATTGTCTGGCTGCAGAAC |
| LOC_Os11g14050 | ATCAGGCACCATACCAAGCCAGC | TGGGAGGAATGCCGCCAGTGAAA |
The constitutively expressed eukaryotic elongation factor 1-alpha (eEF 1-α) (LOC_Os03g08010) was used as an endogenous control due to its stable transcript abundance in rice (Jain et al., 2006; Jain, 2009). For each target in qRT-PCR, three technical and three biological replicates were used. Relative quantity was calculated using the 2-ΔΔCT method (Livak and Schmittgen, 2001).
Results
The Si concentration of Si-treated plants was much higher than of –Si plants. Si concentration in the roots and shoots of +Si plants was 6–20 and 53–65 mg Si g−1 DW, while –Si plants contained only 0.6–1.7 and 1.9–2.6 mg Si g−1 DW in root and shoot, respectively.
Oxidation power
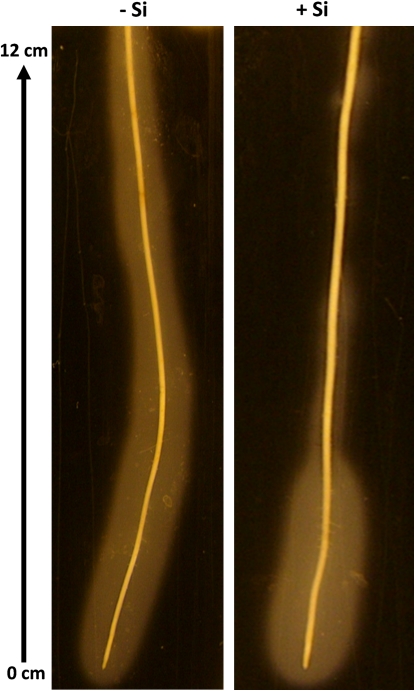
Adventitious roots of plants differentially supplied with silicic acid were embedded for 24 h in semi-solid agar medium containing FeS for the visualization of oxidation power. Roots of control (–Si) plants developed a bright zone (oxidized zone) in the surrounding medium over the whole length of the root from the tip to the basal parts (0–12 cm) (Fig. 1). By contrast, when plants were supplied with Si, bleaching of the medium was restricted to the first 5 cm, while the basal parts of the root did not brighten the medium. The oxidation pattern did not differ between the four replicates, and was the same after 48 h embedding in FeS-agar.
Fig. 1.
Oxidation power of adventitious rice roots as affected by silicic acid supply 24 h after embedding in FeS-agar medium. Plants were grown for 28 d in nutrient solution without Si or with 50 ppm Si. (This figure is available in colour at JXB online.)
Histochemical analysis of suberization and lignification
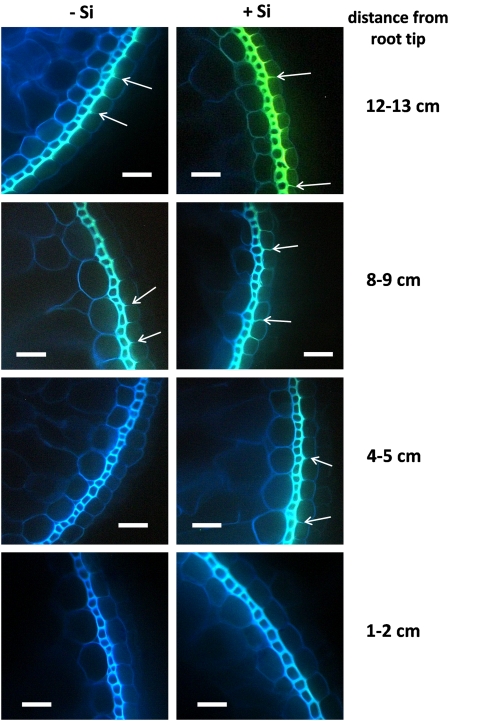
Suberization and lignification in adventitious roots were studied by staining root sections with berberine–aniline blue and examination under a fluorescence microscope. At 1–2 cm drt, no suberin and lignin depositions could be detected in the outer parts of –Si and +Si roots (Fig. 2). At 4–5 cm drt, suberin and lignin deposits were visible in the exodermis of +Si plants, but not in control plants. At 8–9 cm drt, suberin and lignin also occurred in –Si plants, but weaker than in the same section of +Si plants. With increasing distance from the root tip, suberization of the exodermis and lignification of sclerenchyma were enhanced in both treatments. The suberization of the anticlinal cell walls of the exodermis started from the proximal side, the sclerenchyma. The length of suberized cell walls was rated for quantification of suberization of the exodermis.
Fig. 2.
Influence of silicic acid supply on the suberization of the exodermal cell layer and lignification of the sclerenchyma of adventitious rice roots of plants grown in nutrient solution without Si or with 50 ppm Si for 28 d. Root sections were stained with berberine–aniline blue and examined under the fluorescence microscope. Suberized anticlinal cell walls are blue–white-coloured and indicated by arrows, whereas lignified sclerenchyma has a yellow colour. Scale bars=20 μm.
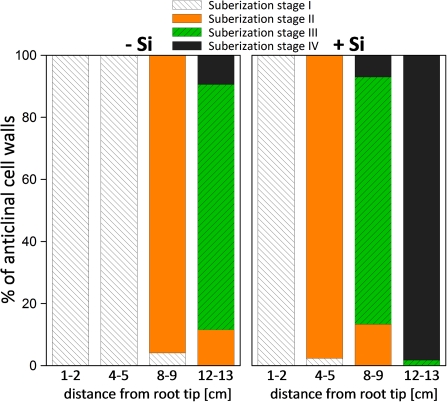
In –Si plants, the exodermal cell walls exhibited no suberin layers (stage I) at 1–2 cm and 4–5 cm drt (Fig. 3). At 8–9 cm drt, nearly all cell walls were suberized to 25% (stage II) of cell wall length, and in 12–13 cm drt, half of the anticlinal cell wall was suberized (stage III) for about 80% of the cells. By contrast, in plants supplied with silicic acid, suberization had already started at 4–5 cm drt and was more pronounced in the following sections. At 12–13 cm drt, nearly all the anticlinal exodermal cell walls were fully suberized (stage IV). In the inner part of the root, no suberin could be detected in root sections at 1–2 cm drt of –Si plants while in the same root sections of +Si plants, suberin in the endodermis was visible over the entire length of the anticlinal cell walls (Fig. 4). In the following root sections (4–5, 8–9, 12–13 cm drt) of both –Si and +Si plants, all anticlinal endodermal cell walls were fully suberized (data not shown).
Fig. 3.
Suberization of the anticlinal cell walls of the exodermis along the adventitious roots of rice plants as affected by silicic acid supply. Plants were grown in nutrient solution without or with 50 ppm Si for 28 d. The total number of evaluated cell walls per treatment and section=400. Suberization stages are: I=0%, II=0–25%, III=25–50%, IV=50–100% suberization of the length of the anticlinal cell wall. (This figure is available in colour at JXB online.)
Fig. 4.
Influence of silicic acid supply on the suberization of the endodermal cell layer of adventitious rice roots of plants grown in nutrient solution without Si or with 50 ppm Si for 28 d. Root sections were stained with berberine–aniline blue and examined under UV-light. Suberin is blue–white-coloured and indicated by arrows. Scale bars=20 μm.
Transcription analysis
Relative transcription levels of 265 genes in rice roots subjected to different Si treatments were investigated by using custom-made microarrays. In the root tip (0–2 cm drt), the transcript rate of eight genes was found to be significantly regulated by silicic acid supply (see Supplementary Table S1 at JXB online); transcript abundance of four genes decreased by a factor of 1.4–1.9 while transcripts of four genes were 1.4–2.5-fold up-regulated. Among the latter, two genes are coding for AT, one for an ABC-transporter and one for a POD.
In root segments at 4–6 cm drt, 19 gene transcripts showed significant regulation by Si treatment (see Supplementary Table S2 at JXB online); transcripts of four genes were down-regulated by a factor of 1.6–2.8, and transcripts of 15 genes were up-regulated from 1.5-fold to 2.9-fold. Among the genes with elevated transcript levels, four AT, four ABC-transporters, and two POD were found.
To gain independent biological replicates, further experiments were undertaken. RNA was isolated from root sections at 0–2 cm and 4–6 cm drt and used for cDNA synthesis. For confirmation of the microarray results, cDNA from –Si and +Si roots was used in qRT-PCR to detect Si-induced changes in transcript abundance of 19 genes. 12 genes exhibited significantly increased transcript abundance by silicic acid supply, whereas transcripts of two genes were less abundant. The transcript level of five genes was not altered by Si treatment (Table 2).
Table 2.
Relative quantity (RQ) of transcripts in root segments at 0–2 cm and 4–6 cm distance from the root tip (drt) as affected by silicic acid. Stars indicate significant regulation at α <0,05, t test, n=3.
| Gene ID | RQ +Si/–Si at 0–2 cm drt | RQ +Si/–Si at 4–6 cm drt | Gene annotation |
| LOC_Os02g41680 | 2.10* | 3.29* | Phenylalanine ammonia-lyase |
| LOC_Os01g67540 | 1.95* | 2.24* | 4-coumarate-CoA ligase |
| LOC_Os07g44560 | 0.69 | 2.91* | 4-coumarate-CoA ligase |
| LOC_Os05g20100 | 2.63* | 2.71* | Glycerol-3-phosphate acyltransferase |
| LOC_Os06g22080 | 2.05* | 2.00* | Diacylglycerol O-acyltransferase |
| LOC_Os01g24010 | 1.27 | 1.30 | ABC transporter |
| LOC_Os01g42410 | 1.56 | 1.19 | ABC transporter |
| LOC_Os10g30610 | 5.27* | 3.68* | ABC transporter |
| LOC_Os12g32814 | 1.74* | 1.57* | ABC transporter |
| LOC_Os01g22230 | 0.27* | 0.25* | Class III peroxidase |
| LOC_Os06g16350 | 4.08* | 7.11* | Class III peroxidase |
| LOC_Os06g32990 | 1.05 | 1.95* | Class III peroxidase |
| LOC_Os08g02110 | 3.40* | 3.03* | Class III peroxidase |
| LOC_Os09g32964 | 0.34* | 0.52* | Class III peroxidase |
| LOC_Os02g05630 | 2.27* | 1.55 | Protein phosphatase 2C |
| LOC_Os02g06250 | 0.38 | 1.16 | Phytosulphokine receptor |
| LOC_Os06g39520 | 1.13 | 1.10 | Myristoyl-acyl carrier protein thioesterase |
| LOC_Os09g23540 | 0.81 | 1.39 | Dehydrogenase |
| LOC_Os11g14050 | 11.00* | 25.40* | Leucine-rich repeat family protein |
The tendency of gene regulation by silicic acid supply as observed in the microarray analysis could be verified for 13 of the 19 genes in qRT-PCR analysis. The highest Si-induced change in transcript ratio was detected for a gene coding for a leucine-rich repeat family protein (LRR), which was over 25-fold more abundant at 4–6 cm drt of +Si plants than in control plants.
Discussion
Rice plants grown in nutrient solution with silicic acid for 28 d accumulated between 53 mg and 65 mg Si g−1 shoot DW, which was in the same range as reported for well-supplied rice plants (Epstein, 1994). Control plants grown without additional silicic acid contained only 1.9–2.6 mg Si g−1 shoot DW, which was 20–30 times less than in +Si plants.
Oxidation power, suberization, and lignification
Adventitious roots of –Si and +Si plants showed clearly different pattern of oxidation power in FeS-agar (Fig. 1). Roots of –Si plants bleached the medium over the whole length suggesting that the root along the entire axis released oxygen into the surrounding medium. With roots of +Si plants, discoloration of the agar was restricted to about 5 cm from the root tip while basal parts of the root did not brighten the medium. This indicated a clearly reduced ROL in older parts of roots supplied with silicic acid and hence a reduced oxidation power, which is in contrast to the literature (Okuda and Takahashi, 1964, cited in Lewin and Reimann, 1969). Okuda and Takahashi observed an increased deposition of iron and manganese oxides on the root surface of Si-supplied plants and concluded that Si promoted the oxidation power of rice roots. Unfortunately, the original publication was not accessible for detailed discussion.
Generally, the low oxidation power in basal parts of the root is caused by a barrier to radial oxygen loss (ROL) from aerenchyma to the rhizodermis, which is due to densely packed sclerenchyma cells with lignified secondary walls and a suberized exodermis with casparian bands (Kotula and Steudle, 2008). Silicic acid supply clearly enhanced the formation of casparian bands in the exodermis and endodermis (Figs 2, 4). Nearly all anticlinal cell walls of the exodermis were suberized for 50% of their length (stage III) at 8–9 cm drt in +Si plants, while control plants reached this stage only at 12–13 cm drt. At this stage, +Si plants had already fully developed casparian bands (stage IV) in the exodermis. Similarly, lignification of sclerenchyma was enhanced by silicic acid supply. Both the formation of casparian bands in the exodermis and the lignification of sclerenchyma cells reduce ROL, but the contribution of each process is unclear (Kotula et al., 2009a).
Suberin deposition in the exodermis was first observed in 4–5 cm drt and 8–9 cm drt for plants grown with and without Si, respectively, while endodermal suberin layers could be seen at 1–2 cm drt and 4–5 cm drt in +Si roots and –Si roots, respectively. This is comparable to the literature as, for adventitious roots of rice grown hydroponically without additional Si supply, initiation of exodermal casparian bands was found at 3 cm drt and well-developed casparian bands at 5 cm drt, while endodermal casparian bands were observed at 2 cm drt (Ranathunge et al., 2003). In onion (Allium cepa) roots, mature casparian bands in the endodermis were already observed at 0.8 cm drt (Ma and Peterson, 2003). For adventitious roots of corn (Zea mays) grown in aerated nutrient solution, casparian bands in the exodermis appeared first at 2–12 cm drt, with increasing drt in older roots, whereas casparian bands in the endodermis had already occurred at 1 cm drt, independent of root age. When the growth of corn roots was slowed down by the addition of polyethylene glycol, casparian bands developed within 1 cm drt (Perumalla and Peterson, 1986). Other environmental factors which reduce root growth such as salinity and drought stress were also shown to enhance the suberization of roots (North and Nobel, 1995; Gong et al., 2006; Schreiber et al., 2007). The aeration of the nutrient solution is also a factor controlling ROL of rice roots, as growth in stagnant deoxygenated 0.1% agar nutrient solution resulted in decreased ROL and elevated levels of suberin in the exodermis and lignin in the sclerenchyma compared with plants grown in aerated nutrient solution (Colmer, 2003b; Kotula et al., 2009a). Enhancement of barrier formation under anaerobic conditions was accompanied by root growth reduction (Kotula et al., 2009a). The nutrient solution contained no Si (Kotula et al., 2009a) or only 2.8 ppm (Colmer, 2003b), which was low compared with the Si concentration occurring in soil solution and in our experiments, and rapid depletion due to uptake by plants was probable since the nutrient solution was renewed every 7 d. By contrast, our experiments were conducted with high Si concentration under hypoxic conditions (3–4 mg l−1 O2). So it was shown that anaerobic conditions and Si nutrition separately induced a barrier against ROL. How perfect anaerobic conditions and constant high Si concentrations found under field conditions act together remains to be investigated.
Since, in our experiment, the root growth rate was unaffected by Si supply under hypoxic conditions (–Si: 1.05±0.05 cm d−1; +Si: 1.02±0.06 cm d−1) and no other environmental stresses were present, the alteration in suberin and lignin formation can be attributed to a direct effect of silicic acid.
Increased suberin content in the root may be useful not only to reduce ROL, but also to protect from biotic and abiotic stresses, since suberin was associated with partial resistance against fungal pathogens in roots (Thomas et al., 2007). Furthermore, stronger barriers to ROL were correlated with higher salt tolerance in rice (Krishnamurthy et al., 2009).
Gene regulation as affected by silicic acid
The biopolymer lignin is a complex mixture of phenolic compounds derived mainly from three hydroxycinnamyl alcohol monomers (monolignols), p-coumaryl, coniferyl, and sinapyl alcohols. The synthesis of all monolignols starts with phenylalanine, which is initially processed to cinnamic acid by PAL and to p-coumaric acid by cinnamate 4-hydroxylase (C4H) during the first steps of the phenylpropanoid pathway (Fig. 5). Thereafter, p-coumaric acid can be metabolized either by 4CL and further enzymes to the monolignol p-coumaryl alcohol or by coumaroyl-CoA-3-hydroxylase (C3H) and COMT to ferulic acid (Boerjan et al., 2003; Goujon et al., 2003). Ferulic acid can further be converted to monolignols by several enzymes including 4CL. The monolignols are transported to the apoplast, probably via ABC transporter, where POD are assumed to catalyse the formation of lignin by dehydrogenative polymerization of the monolignols (Boerjan et al., 2003). It was found that Si significantly increased the transcript abundance of PAL and 4CL, leading to enhanced synthesis of monolignols. Increased transcript abundance of ABC transporter and POD might facilitate transport to the apoplast and subsequent polymerization of monolignols, which is in line with the enhanced lignification.
Suberin is a biopolymer which consists of a polyaliphatic and a polyaromatic domain (Kolattukudy, 1984). Similar to lignin formation, suberin evolves from suberin monomers that polymerize in the apoplast under the catalytic force of POD (Fig. 5). The monomers are made up of ferulic acid, glycerol, and varying oxygenated fatty acid derivates, mainly ω-hydroxyacids and α,ω-dicarboxylic acids (Franke and Schreiber, 2007). The fatty acid modifications are attributed to KCS, fatty acid desaturases (FAD), fatty acid elongases (FAE), and cytochrome P450 monooxygenases (P450) (Franke et al., 2005; Franke and Schreiber, 2007). The monomers are assembled by AT in the symplast, whereupon ABC transporter release the monomers into the apoplast.
Si enhanced transcript abundance of AT, ABC transporters, and POD, indicating an increased formation of suberin, which coincides with the histochemical observations. None of the genes examined that are involved in fatty acid metabolism showed significant transcription changes as affected by Si, suggesting that fatty acids derivates exhibit no bottleneck in suberin synthesis.
The gene with the most highly elevated transcript level by Si supply was a leucine-rich repeat family protein (LRR). At 0–2 cm drt and at 4–6 cm drt, transcript rates were 11-fold and 25-fold higher in +Si roots than in –Si roots, respectively. The gene was selected from a SSH-library that contained genes differentially expressed in suberized tissue compared with non-suberized tissue in Quercus suber (Soler et al., 2007). LRR proteins belong to the receptor-like kinases (RLK), a major gene family with more than 1100 members in rice (Morillo and Tax, 2006). RLK are transmembrane proteins, which contain an extracellular domain that is linked via a transmembrane domain to a cytoplasmic serine/threonine protein kinase domain (Shiu et al., 2004). One of the motifs in the extracellular domain is the LRR-motif, and the LRR-RLKs form the biggest group of RLKs with 216 members in Arabidopsis, where the LRR-RLKs are best described (Diévart and Clark, 2003). LRR-RLKs are grouped into 13 subfamilies according to their LRR organization in the extracytoplasmic domain. Homologue Arabidopsis proteins to LOC_Os11g14050 are members of subfamily VIII-1. Since details about possible ligands for this LRR-group are not known, it would be interesting to further characterize this group and its involvement in suberin and lignin synthesis.
In the root tip, the transcript level of a gene coding for a protein phosphatase 2C was significantly enhanced. Protein phosphatases are regulating proteins, and the subgroup 2C is thought to be involved in stress signal transduction (Luan, 2003). Two genes had fewer transcript levels under +Si conditions, both genes coding for POD. This does not necessarily conflict with the other results due to the wide variability of the class III peroxidase group in terms of encoded genes and substrate specifities (Marjamaa et al., 2009). Hence, it is not unlikely that the POD exhibiting reduced transcript levels under +Si conditions might have functions different from those in suberin and lignin synthesis.
Conclusion
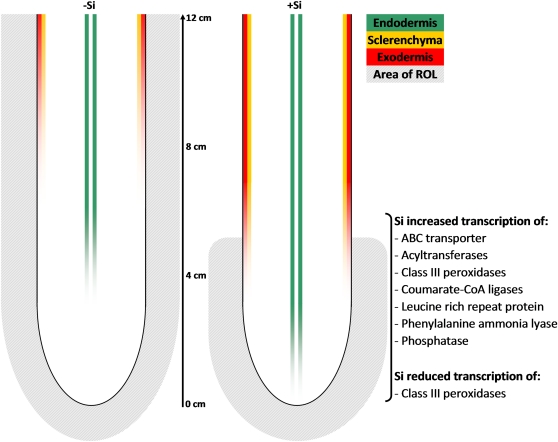
The effects of silicic acid on rice root anatomy and on the transcription of genes related to suberin and lignin synthesis are summarized in Fig. 6. Si nutrition of rice plants reduced the oxidation power of roots and enhanced the development of casparian bands in the exodermis and endodermis, as well as lignin depositions in the sclerenchyma. These changes are probably the reason for the reduced ROL and might be useful for the plants to grow in anaerobic soils and cope with unfavourable conditions. Increased suberization and lignification was accompanied by silicic acid triggered transcription of genes related to lignin and suberin meta-bolism. In addition, a high impact of silicic acid supply on transcript level of a LRR-RLK gene could be observed, highlighting the possibility that this regulating protein plays a central role either in perceiving a Si signal of an up-to-now unknown nature or in promoting suberin and lignin synthesis or in both.
Fig. 6.
Model of the rice root and transcript levels of genes as affected by silicic acid. Si supply reduced area of radial oxygen loss (ROL), and hence oxidation power, to a 5 cm distance from the root tip. This coincided with an increased suberization of exodermis and lignification of sclerenchyma by Si treatment. Also, Si supply enhanced the suberization of the endodermis. These changes indicated by Si supply were related to an increased transcription of genes associated with suberin and lignin synthesis. (This figure is available in colour at JXB online.)
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Table S1. Effect of silicic acid on relative quantity (RQ) of transcripts in root segments at 0–2 cm distance from the root tip as measured by microarray analysis.
Supplementary Table S2. Effect of silicic acid on relative quantity (RQ) of transcripts in root segments at 4–6 cm distance from the root tip as measured by microarray analysis.
Glossary
Abbreviations
- 4CL
4-coumarate-CoA ligase
- ABC
ATP binding cassette
- AT
acyltransferase
- B
biotin
- C3H
coumaroyl-CoA-3-hydroxylase
- C4H
cinnamate 4-hydroxylase
- COMT
caffeic acid O-methyltransferase
- drt
distance from root tip
- Fl
fluorescein
- FAD
fatty acid desaturase
- FAE
fatty acid elongase
- HRP
anti-Fl-horseradish peroxidase
- KCS
β-ketoacyl-CoA synthase
- LRR
leucine-rich repeat family protein
- P450
cytochrome P450 monooxygenase
- PAL
phenylalanine ammonia-lyase
- POD
class III peroxidase; RLK, receptor-like kinase
- ROL
radial oxygen loss
- TIGR
The Institute of Genomic Research
References
- Armstrong W. Radial oxygen losses from intact rice roots as affected by distance from the apex, respiration, and waterlogging. Physiologia Plantarum. 1971;25:192–197. [Google Scholar]
- Armstrong W. Aeration in higher plants. Advances in Botanical Research. 1979;7:225–332. [Google Scholar]
- Armstrong W, Cousins D, Armstrong J, Turner DW, Beckett PM. Oxygen distribution in wetland plant roots and permeability barriers to gas-exchange with the rhizosphere: a microelectrode and modelling study with Phragmites australis. Annals of Botany. 2000;86:687–703. [Google Scholar]
- Arnon DI, Stout PR. The essentiality of certain elements in minute quantity for plants with special reference to copper. Plant Physiology. 1939;14:371–375. doi: 10.1104/pp.14.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards MA, Summerhurst DK, Razem FA. Oxidases, peroxidases and hydrogen peroxide: the suberin connection. Phytochemistry Reviews. 2004;3:113–126. [Google Scholar]
- Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annual Review of Plant Physiology. 2003;54:519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- Bogdan K, Schenk MK. Arsenic in rice (Oryza sativa L.) related to dynamics of arsenic and silicic acid in paddy soils. Environmental Science and Technology. 2008;42:7885–7890. doi: 10.1021/es801194q. [DOI] [PubMed] [Google Scholar]
- Brundrett MC, Enstone DE, Peterson CA. A berberine–aniline blue fluorescent staining procedure for suberin, lignin, and callose in plant tissue. Protoplasma. 1988;1446:133–142. [Google Scholar]
- Cai K, Gao D, Luo S, Zeng R, Yang J, Zhu X. Physiological and cytological mechanisms of silicon-induced resistance in rice against blast disease. Physiologia Plantarum. 2008;134:324–333. doi: 10.1111/j.1399-3054.2008.01140.x. [DOI] [PubMed] [Google Scholar]
- Colmer TD. Long-distance transport of gases in plants: a perspective on internal aeration and radial oxygen loss from roots. Plant, Cell and Environment. 2003a;26:17–36. [Google Scholar]
- Colmer TD. Aerenchyma and an inducible barrier to radial oxygen loss facilitate root aeration in upland, paddy and deep-water rice (Oryza sativa L.) Annals of Botany. 2003b;91:301–309. doi: 10.1093/aob/mcf114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmer TD, Cox MCH, Voesenek LACJ. Root aeration in rice (Oryza sativa L.): evaluation of oxygen, carbon dioxide, and ethylene as possible regulators of root acclimatizations. New Phytologist. 2006;170:767–778. doi: 10.1111/j.1469-8137.2006.01725.x. [DOI] [PubMed] [Google Scholar]
- Diévart A, Clark SE. Using mutant alleles to determine the structure and function of leucine-rich repeat receptor-like kinases. Current Opinion in Plant Biology. 2003;6:507–516. doi: 10.1016/s1369-5266(03)00089-x. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein E. The anomaly of silicon in plant biology. Proceedings of the National Academy of Sciences, USA. 1994;91:11–17. doi: 10.1073/pnas.91.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein E. Silicon. Annual Review of Plant Physiology and Plant Molecular Biology. 1999;50:641–644. doi: 10.1146/annurev.arplant.50.1.641. [DOI] [PubMed] [Google Scholar]
- Fawe A, Abou-Zaid M, Menzies JG, Belanger RR. Silicon-mediated accumulation of flavonoid phytoalexins in cucumber. Phytopathology. 1998;88:396–401. doi: 10.1094/PHYTO.1998.88.5.396. [DOI] [PubMed] [Google Scholar]
- Franke R, Briesen I, Wojciechowski T, Faust A, Yephremov A, Nawrath C, Schreiber L. Apoplastic polyesters in Arabidopsis surface tissues: a typical suberin and a particular cutin. Phytochemistry. 2005;66:2643–2658. doi: 10.1016/j.phytochem.2005.09.027. [DOI] [PubMed] [Google Scholar]
- Franke R, Schreiber L. Suberin: a biopolyester forming apoplastic plant interfaces. Current Opinion in Plant Biology. 2007;10:252–259. doi: 10.1016/j.pbi.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Gong HJ, Randall DP, Flowers TJ. Silicon deposition in the root reduces sodium uptake in rice (Oryza sativa L.) seedlings by reducing bypass flow. Plant, Cell and Environment. 2006;29:1970–1979. doi: 10.1111/j.1365-3040.2006.01572.x. [DOI] [PubMed] [Google Scholar]
- Goujon T, Sibout R, Eudes A, MacKay J, Joulanin L. Genes involved in the biosynthesis of lignin precursors in Arabidopsis thaliana. Plant Physiology and Biochemistry. 2003;41:677–687. [Google Scholar]
- Jain M. Genome-wide identification of novel internal control genes for normalization of gene expression during various stages of development in rice. Plant Science. 2009;176:702–706. [Google Scholar]
- Jain M, Nijhawan A, Tyagi AK, Khurana JP. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochemical and Biophysical Research Communications. 2006;345:646–651. doi: 10.1016/j.bbrc.2006.04.140. [DOI] [PubMed] [Google Scholar]
- Kane MD, Jatkoe TA, Stumpf CR, Lu J, Thomas JD, Madore SJ. Assessment of the sensitivity and specificity of oligonucleotide (50mer) microarrays. Nucleic Acids Research. 2000;28:4552–4557. doi: 10.1093/nar/28.22.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd PS, Llugany M, Poschenrieder C, Gunse B, Barcelo J. The role of root exudates in aluminium resistance and silicon-induced amelioration of aluminium toxicity in three varieties of maize (Zea mays L.) Journal of Experimental Botany. 2001;359:1339–1352. [PubMed] [Google Scholar]
- Kim SG, Kim KW, Park EW, Choi D. Silicon-induced cell wall fortification of rice leaves: a possible cellular mechanism of enhanced host resistance to blast. Phytopathology. 2002;92:1095–1103. doi: 10.1094/PHYTO.2002.92.10.1095. [DOI] [PubMed] [Google Scholar]
- Kolattukudy PE. Biochemistry and function of cutin and suberin. Canadian Journal of Botany. 1984;62:2918–2933. [Google Scholar]
- Kotula L, Ranathunge K, Schreiber L, Steudle E. Functional and chemical comparison of apoplastic barriers to radial oxygen loss in roots of rice (Oryza sativa L.) grown in aerated or deoxygenated solution. Journal of Experimental Botany. 2009a;60:2155–2167. doi: 10.1093/jxb/erp089. [DOI] [PubMed] [Google Scholar]
- Kotula L, Ranathunge K, Steudle E. Apoplastic barriers effectively block oxygen permeability across outer cell layers of rice roots under deoxygenated conditions: roles of apoplastic pores and of respiration. New Phytologist. 2009b;184:909–917. doi: 10.1111/j.1469-8137.2009.03021.x. [DOI] [PubMed] [Google Scholar]
- Kotula L, Steudle E. Measurements of oxygen permeability coefficients of rice (Oryza sativa L.) roots using a new perfusion technique. Journal of Experimental Botany. 2008;60:567–580. doi: 10.1093/jxb/ern300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy P, Ranathunge K, Franke R, Prakash HS, Schreiber L, Mathew MK. The role of root apoplastic transport barriers in salt tolerance of rice (Oryza sativa L.) Planta. 2009;230:119–134. doi: 10.1007/s00425-009-0930-6. [DOI] [PubMed] [Google Scholar]
- Lewin J, Reimann BEF. Silicon and plant growth. Annual Review of Plant Physiology. 1969;20:289–304. [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luan S. Protein phosphatases in plants. Annual Review of Plant Biology. 2003;54:63–92. doi: 10.1146/annurev.arplant.54.031902.134743. [DOI] [PubMed] [Google Scholar]
- Ma JF, Yamaji N. Silicon uptake and accumulation in higher plants. Trends in Plant Science. 2006;11:392–397. doi: 10.1016/j.tplants.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Ma F, Peterson CA. Current insights into the development, structure, and chemistry of the endodermis and exodermis of roots. Canadian Journal of Botany. 2003;81:405–421. [Google Scholar]
- Marjamaa K, Kukkola EM, Fagerstedt KV. The role of xylem class III peroxidases in lignification. Journal of Experimental Botany. 2009;60:367–376. doi: 10.1093/jxb/ern278. [DOI] [PubMed] [Google Scholar]
- Morillo SA, Tax FE. Functional analysis of receptor-like kinases in monocots and dicots. Current Opinion in Plant Biology. 2006;9:460–469. doi: 10.1016/j.pbi.2006.07.009. [DOI] [PubMed] [Google Scholar]
- North GB, Nobel PS. Hydraulic conductivity of concentric root tissues of Agave deserti Engelm. under wet and drying conditions. New Phytologist. 1995;130:47–57. [Google Scholar]
- Novozamsky I, van Eck R, Houba VJG. A rapid determination of silicon in plant material. Communications in Soil Science and Plant Analysis. 1984;15:205–211. [Google Scholar]
- Okuda A, Takahashi E. Studies on the physiological role of silicon in crop plants. 4. Effect of silicon on the growth of barley, tomato, raddish, green onion, chinese cabbage and their nutrients uptake. Journal of the Science of Soil and Manure. 1961;32:623–626. [Google Scholar]
- Okuda A, Takahashi E. The role of silicon. In: Chandler RF, editor. The mineral nutrition of the rice plant. Baltimore, MD, USA: Johns Hopkins Press; 1964. pp. 132–146. [Google Scholar]
- Perumalla CJ, Peterson CA. Deposition of Casparian bands and suberin lamellae in the exodermis and endodermis of young corn and onion roots. Canadian Journal of Botany. 1986;64:1873–1878. [Google Scholar]
- Ponnamperuma FN. Effects of flooding on soils. In: Kozlowski TT, editor. Flooding and plant growth. New York, NY, USA: Academic Press; 1984. pp. 9–45. [Google Scholar]
- Ranathunge K, Steudle E, Lafitte R. Control of water uptake by rice (Oryza sativa L.): role of the outer part of the root. Planta. 2003;217:193–205. doi: 10.1007/s00425-003-0984-9. [DOI] [PubMed] [Google Scholar]
- Rémus-Borel W, Menzies JG, Bélanger RR. Silicon induces antifungical compounds in powdery mildew-infected wheat. Physiological and Molecular Plant Pathology. 2005;66:108–115. [Google Scholar]
- Rodrigues FÁ, Jurick WM, Datnoff LE, Jones JB, Rollins JA. Silicon influences cytological and molecular events in compatible and incompatible rice– Magnaporthe oryzae interactions. Physiological and Molecular Plant Pathology. 2005;66:144–159. [Google Scholar]
- Rodrigues FÁ, McNally DJ, Datnoff LE, Jones JB, Labbé C, Benhamou N, Menzies JG, Bélanger RR. Silicon enhances the accumulation of diterpenoid phytoalexins in rice: a potential mechanism for blast resistance. Phytopathology. 2004;94:177–183. doi: 10.1094/PHYTO.2004.94.2.177. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics methods and protocols: methods in molecular biology. Totowa, NJ, USA: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Schreiber L, Franke R, Hartmann K. Chemical composition of apoplastic transport barriers in roots: quantification of suberin depositions in endodermal and hypodermal root cell walls. In: Sattelmacher B, Horst WJ, editors. The apoplast of higher plants: compartment of storage, transport and reactions. The significance of the apoplast for the mineral nutrition of higher plants. Heidelberg, Germany: Springer; 2007. pp. 109–117. [Google Scholar]
- Shiu S-H, Karlowski WM, Pan R, Tzeng Y-H, Mayer KFX, Li W- H. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. The Plant Cell. 2004;16:1220–1234. doi: 10.1105/tpc.020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler M, Serra O, Molinas M, Huguet G, Fluch S, Figueras M. A genomic approach to suberin biosynthesis and cork differentiation. Plant Physiology. 2007;144:419–431. doi: 10.1104/pp.106.094227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrell BK. Airspace structure and mathematical modeling of oxygen diffusion, aeration and anoxia in Eleocharis sphacelata R. Br. roots. Australian Journal of Marine and Freshwater Research. 1994;45:1529–1541. [Google Scholar]
- Thomas R, Fang X, Ranathunge K, Anderson TR, Peterson CA, Bernards MA. Soybean root suberin:anatomical distribution, chemical composition and relation to partial resistance to Phytophthora sojae. Plant Physiology. 2007;144:299–311. doi: 10.1104/pp.106.091090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trolldenier G. Visulization of oxidation power of rice roots and of possible participation of bacteria in iron deposition. Zeitschrift für Pflanzenernährung und Bodenkunde. 1988;151:117–122. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.