Abstract
Chloroplastic thioredoxins f and m (TRX f and TRX m) mediate light regulation of carbon metabolism through the activation of Calvin cycle enzymes. The role of TRX f and m in the activation of Calvin cycle enzymes is best known among the TRX family. However, the discoveries of new potential targets extend the functions of chloroplastic TRXs to other processes in non-photosynthetic tissues. As occurs with numerous chloroplast proteins, their expression comes under light regulation. Here, the focus is on the light regulation of TRX f and TRX m in pea and Arabidopsis during the day/night cycle that is maintained during the subjective night. In pea (Pisum sativum), TRX f and TRX m1 expression is shown to be governed by a circadian oscillation exerted at both the transcriptional and protein levels. Binding shift assays indicate that this control probably involves the interaction of the CCA1 transcription factor and an evening element (EE) located in the PsTRX f and PsTRX m1 promoters. In Arabidopsis, among the multigene family of TRX f and TRX m, AtTRX f2 and AtTRX m2 mRNA showed similar circadian oscillatory regulation, suggesting that such regulation is conserved in plants. However, this oscillation was disrupted in plants overexpressing CCA1 (cca1-ox) or repressing CCA1 and LHY (cca1-lhy). The physiological role of the oscillatory regulation of chloroplastic TRX f and TRX m in plants during the day/night cycle is discussed.
Keywords: Arabidopsis mutants, CCA1, chloroplastic thioredoxins, circadian regulation, EE, Pisum sativum
Introduction
The ferredoxin/thioredoxin system constitutes a notable example of modulation of light-regulated Calvin cycle enzymes by chloroplastic thioredoxins (TRXs), a process that involves thiol–disulphide exchange. During photosynthesis, the reduced ferredoxin transfers its electron via a reductase to the TRX proteins, which in turn can reduce enzymes involved in carbon metabolism. Found in all organisms, TRXs are small proteins (14 kDa) that have a conserved redox active centre (WCXPC) involved in the reduction of a wide variety of proteins (Holmgren, 1985, 1989; Balmer et al., 2003, 2006a; Meyer et al., 2009). The genome of Arabidopsis thaliana contains four TRX m genes (AtTRX m1, m2, m3, and m4), two TRX f genes (AtTRX f1 and f2), one TRX x gene, two TRX y genes (AtTRX y1 and y2), and one TRX z gene (Arsova et al., 2010). In addition, the A. thaliana genome contains nine TRX h-type proteins presumably located in the cytosol, and two mitochondrial TRX o genes (Meyer et al., 2008). To date, in the pea genome, one TRX f (PsTRX f) and two TRX m (PsTRX m1 and m2) located in the chloroplast (López-Jaramillo et al., 1997; Pagano et al. 2000), one mitochondrial TRX o1 (Martí et al., 2009), and four cytosolic TRX h (Montrichard et al., 2003; Traverso et al., 2007, 2008) have been isolated. In the chloroplast, ferredoxin reduces the bridge formed between the two cysteines of TRX through ferredoxin-thioredoxin reductase (FTR) (Schürmann and Jacquot, 2000). In the cytosol and organelles, TRX h proteins are reduced by NADPH via NADP thioredoxin reductase (NTR).
Activation of Calvin cycle enzymes by TRX f and m is one of the best characterized processes, and the function of the proteins involved in carbon metabolism of the chloroplast has been well defined (Bassham and Krause, 1969; Jacquot et al., 1978; Crawford et al., 1979; López-Jaramillo et al., 1997; Collin et al., 2003, 2004). Among the Calvin cycle enzymes, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), fructose-1,6-bisphosphatase (FBPase), sedoheptulose-1,7-bisphosphatase (SBPase), phosphoribulokinase (PRK), and ADP-glucose pyrophosphorylase (AGPase) are activated by TRXs (Buchanan et al., 2002; Geigenberger et al., 2005). A chloroplast detoxifying protein, a subunit of ATP synthase, and a chlorophyll biosynthetic enzyme that belongs to the photosynthetic electron chain are also protein targets of the chloroplastic TRXs (Lemaire et al., 1999; Rey et al., 2005; Pérez-Ruiz et al., 2006; Ikegami et al., 2007; Kirchsteiger et al., 2009). Recently an additional mechanism of control has been proposed in which a Calvin cycle ternary complex including PRK, GAPDH, and a small protein, CP12, may represent a reservoir of inhibited enzymes that are reduced and dissociated by TRX f during the dark to light transition (Wedel et al., 1997; Howard et al., 2008; Marri et al., 2009). The activation of these enzymes at specific times of the day to avoid futile steps is crucial for metabolic processes, and this activation requirement suggests that TRX activation adapts the activities of the chloroplast to the availability of light energy and carbon status.
Light is the main environmental factor involved in the biosynthesis and regulation of chloroplast thioredoxin expression, and can affect many processes in which they are involved (Carrasco et al., 1992; Meyer et al., 2008). In addition, plants have endogenous biological clocks that enable them to organize their physiological, metabolic, and developmental processes to occur at optimal times (Dodd et al., 2005). The assimilation of carbon in the Calvin cycle is under the control of the light/dark cycle that regulates the activity of the enzymes to prevent futile steps and control the balance between sucrose and starch synthesis. It is therefore not surprising that the expression and function of the genes encoding proteins involved in sugar synthesis are under tight regulation by light and the biological clock. To date, transcripts of the genes of Neurospora crassa GAPDH, Chlamydomonas PRK, and Rubisco activase in several plant species have been shown to be under the control of the circadian clock (Martino-Catt and Ort, 1992; Pilgrim and McClung, 1993; Shinohara et al., 1998; Lemaire et al. 1999).
Work on circadian clock control in plant TRXs is limited to observations by Harmer et al. (2000) regarding the abundance of AtTRX f2 and AtTRX like5 mRNA (WCRKC THIOREDOXIN 1), which responds to oscillatory regulation. None of the well-known genes that code for enzymes activated by chloroplast TRXs has been found among the genes involved in carbon metabolism that function under the circadian clock. However, researchers have detected a cluster of genes that encode enzymes involved in starch mobilization that are under clock control, suggesting the presence of a circadian component in the regulation of carbohydrate metabolism (Jeannette and Prioul, 1994; Mérida et al., 1999). Interestingly, Chlamydomonas reinhardtii TRX m, TRX h, ferredoxin, PRK, and ferredoxin-NADP reductase (FNR) genes have been shown to be controlled by the circadian clock under continuous light conditions (Lemaire et al., 1999, 2002).
In this context, it is reasonable to conjecture that in addition to the light-dependent redox regulation exerted by chloroplastic TRXs to activate the proteins involved in photosynthesis and CO2 fixation, the expression of the TRX genes could be regulated by the circadian clock and anticipate dawn to develop their function. However, only a few studies examining the day/night cycle have focused on the transcriptional regulation of the TRX f and m genes under different light conditions, and no transcription factor controlling expression of these genes has yet been described. In this work, the effect of light to dark transition on pea TRX f and m1 gene expression as well as protein biosynthesis is characterized, and support for the hypothesis that these genes are regulated under the circadian clock is presented. The gel-shift assay findings document binding of the CCA1 recombinant transcription factor on a fragment that carries the evening element (EE) of the promoter regions of both chloroplastic PsTRX f and m1. In addition, the expression of Arabidopsis thaliana TRX f and m genes (AtTRX f1, AtTRX f2, AtTRX m1, AtTRX m2, AtTRX m3, and AtTRX m4) is analysed in Arabidopsis plants as well as in plants overexpressing CCA1, and in plants repressing CCA1 and LHY.
Materials and methods
Plant material and growth conditions
Pea (Pisum sativum var. Lincoln) leaves were used in protein, DNA, and RNA extraction and promoter isolation, as well as for northern blot, reverse transcription-PCR (RT-PCR), and western blotting experiments. Plants were grown in a green cabinet for 15 d, first in vermiculite and then in compost, with a 12 h/12 h photoperiod and light intensity of 80 μmol photons m−2 s−1 PAR, 23 °C temperature, and daily watering.
Arabidopsis thaliana ecotype Columbia seeds overexpressing CCA1 (cca1-ox) or repressing CCA1 and LHY (cca1-lhy) and transgenic seeds carrying constructions f444–β-glucuronidase (GUS; 444 bp), f126–GUS (126 bp), m1874–GUS (1874 bp), m200–GUS (200 bp), and m100–GUS (100 bp), described in a previous work (Barajas et al., 2007), were grown in MS medium supplemented with sucrose, agar, and kanamycin in a light cabinet under a 12 h light/12 h dark photoperiod, at 140 μmol photons m−2 s−1 PAR and 23 °C temperature.
For the circadian experiments with day/night cycles, the samples were collected every 4 h or 12 h over 48 h, and during the second night the light was kept on to simulate subjective night.
Extraction and gel analysis of RNA
Total RNA extracted from pea with Trizol (BD, Sparks, MD, USA) was used for northern blot analysis and reverse transcription. For northern blots, total pea leaf RNA (15 μg) was subjected to electrophoresis through agarose/formaldehyde gels and transferred onto Hybond-N+ membranes (Amersham Pharmacia Biotech). Hybridization was performed at 42 °C in a MOPS buffer (10× MOPS, 50% formamide, 0.5% SDS, 6× SSC, 5× Denhardt's solution, 50 μl of calf thymus) to random primed 3′-specific probes of PsCab, PsTRX f, and PsTRX m1 genes amplified by PCR (Kit of Readyprime, Amersham II Labelling System).
RT-PCR and semi-quantitative PCR analysis
For semi-quantitative RT-PCR, 2.5 μg or 5 μg of RNAs treated with DNase (DNase Turbo®-Invitrogen) were used for reverse transcription with retrotranscriptase RNase SuperScript-III® (Invitrogen, Carlsbad, CA, USA) and oligo(dT)12–15 primer, at 50 °C for 1 h. After selecting the dilutions, 1/50 of the reaction was taken for PCR using specific oligonucleotides (GUSI and II) for the GUS gene (Table 1). Amplified fragments of Pisum or Arabidopsis 18S were used as housekeeping genes to normalize the expression level.
Table 1.
Gene-specific oligonucleotides used for qPCR analysis
| Gene analysed | Oligosequence | |
| Pcab up | AtCAB | 5′-CTGCGGCATCAGAAGTCCTT-3′ |
| Pcab down | id | 5′-CCTTTGGCTTGGCAACAGTC-3′ |
| Pf1 up | AtTRXf1 | 5′-AAACAGCGAGGTCTGCTGCT-3′ |
| Pf1 down | id | 5′-TAACACCAGATTTCACATTACATACAAACA-3′ |
| Pf2 up | AtTRXf2 | 5′-TCCGTTATTCTCCGATTACATCTACC-3′ |
| Pf2 down | id | 5′-GAATTCGGGATCCGGCA-3′ |
| Pm1 up | AtTRXm1 | 5′-AATTCTAGGGTTTCCCGATTACG-3′ |
| Pm1 down | id | 5′-GAGTCCCATGTTGAATCGTTGA-3′ |
| Pm2 up | AtTRXm2 | 5′-TCTCCGGCTTCGTTGACC-3′ |
| Pm2 down | id | 5′-GAGCTTCACAGACGACGGCT-3′ |
| Pm3 up | AtTRXm3 | 5′-AGCGAAACCCCGGTGTTAG-3′ |
| Pm3 down | id | 5′-TTATCCTGTGGACCATCCGAC-3′ |
| Pm4 up | AtTRXm4 | 5′-AATCGCTCGCGGTGGAC-3′ |
| Pm4 down | id | 5′-GATTTGGTACTTCGACGGCG-3′ |
| 18S up | rRNA18s | 5′-AGTAAGCGCGAGTCATCAGCT-3′ |
| 18S down | id | 5′-CATTCAATCGGTAGGAGCGAC-3′ |
| Act up | rRNAAct | 5′-TGGTCGTACAACCGGTATTG-3′ |
| Act down | id | 5′-CAGTAAGGTCACGTCCAGCA-3′ |
| GUS I | GUS | 5′-AACGGGGAAACTCAGCAAGC-3′ |
| GUS II | id | 5′-TGTGAGCGTCGCAGAACATT-3′ |
| Probes | ||
| AtTRXf1 ATf1 | 5′ FAM-CCGGATGAACCAGTCTCTCATGTCTTTACC-Tamra3′ | |
| AtTRXf2 ATf2 | 5′ FAM-CCGGAGGATTTTCCCCCGTGAA-Tamra3′ | |
| AtTRXm1 ATm1 | 5′ FAM-CGAGGCGTTATCTGTGAAGCTCAGGACAC-Tamra3′ | |
| AtTRXm2 ATm2 | 5′ FAM-CGATTCATCAACCTAGGGTTTCTCGATTACGAA- Tamra3′ | |
| AtTRXm3 ATm3 | 5′ FAM- AGAATTCTACACGAGTTGGTGCGGTCCA-Tamra3′ | |
| AtTRXm4 ATm4 | 5′ FAM-AATCGCCTGTGAGGCTCAGGACACC-Tamra3′ | |
| AtCAB2 ATcab2 | 5′ TET-AAGCGGCCGTGTGACAATGAGGA-Tamra3′ | |
| rRNA18s P18S | 5′-FAM-CTGCCCTTTGTACACACCGCCCG-Tamra3′ |
Real-time quantitative RT-PCR analysis
For gene expression analysis by real-time PCR, samples of total RNA (500 ng) extracted from wild-type, cca1-ox, and cca1-lhy double-mutant rosettes using the Aurum™ Total RNA Mini kit (BioRad) were reverse transcribed with Multiscribe Reverse Transcriptase and random hexamer primers (Applied Biosystems, Foster City, CA, USA). For quantification, real-time PCRs were performed with an ABI PRISM 7700 Sequence Detection System (Applied Biosystems) and Taqman technology. The cDNA was amplified in a 2× TaqMan Master mix (Eurogentec S.A., University of Liège, Belgium) in a final volume of 25 μl. The PCR conditions were: 2 min at 50 °C; 10 min at 95 °C; and 40 cycles of 15 s at 95 °C, 1 min at 60 °C. The Ct value was determined using the instrument's software. Specific primers and probes of each gene were designed using Primer Express (Applied Biosystems) software, the corresponding pair of primers and probe being Pcab up/down, AtCab2 for AtCAB, Pf1 up/down, ATf1 for AtTRX f1, Pf2 up/down, ATf2 for AtTRX f2; Pm1 up/down, ATm1 for AtTRX m1, Pm2 up/down, ATm2 for AtTRX m2, Pm3 up/down, ATm3 for AtTRX m3, Pm4 up/down, ATm4 for AtTRX m4, and 18S up/down, P18S for rRNA 18S (Table 1). Relative quantification of gene expression was monitored after normalization by 18S rRNA expression as internal control, as fold variation over a calibrator using the 2–ΔΔCT method (Livak and Schmittgen, 2001).
Electrophoretic mobility shift assays (EMSAs)
The double-stranded oligonucleotides LumF and LumM containing the EE were obtained by annealing oligonucleotides LumFs (5′-AAGTGAAAAAAAAAAGAGATATTCGAAGGG-3′) and LumFa (5′-GATTCCCTTCGAATATCTCTTTTTTTTTTC-3′), and oligonucleotides LumMs (5′-TAAGTAGATATTGAAAGCAAGATTGAAAAAAATGTTG-3′) and LumMa (5′-AATCAAACATTTTTTTCAATCTTGCTTTCAATATCT-3'), respectively. A single mutation in the EE was introduced in the oligonucleotides LumFDs (5′-AAGTGAAAAAAAAAAGAGcTATTCGAAGGG-3′) and LumFDa (5′-GATTCCCTTCGAATAgCTCTTTTTTTTTTC-3′), and oligonucleotides LumMDs (5′-TAAGTAGcTATTGAAAGCAAGATTGAAAAAAATGTTG-3′) and LumMDa (5′-AATCAAACATTTTTTTCAATCTTGCTTTCAATAgCT-3′). The double-stranded mutated oligonucleotides were obtained by annealing. Fragments were labelled with [32P]dATP by fill-in reaction with Klenow. The radioactive probes were purified from 8% polyacrylamide gels. CCA1 protein was purified with plasmid pXCA-24, kindly provided by Elaine Tobin (UCLA, USA), which contains the full-length CCA1 cDNA fused to glutathione S-transferase (GST) cloned in pGEX-3X (Amersham Pharmacia Biotech) (Wang et al., 1997). Binding was carried out in 20 μl of 30 mM HEPES, pH 7.9, 125 mM KCl, 0.25 M dithiothreitol (DTT), 12% glycerol (Díaz et al., 2002). Mixes were incubated for 30 min on ice. In unspecific competition experiments, 0–0.5 μg of poly(dI–dC) was also included in the mixes. In specific competition experiments, the unlabelled double-stranded oligonucleotides (0.1–0.5 μg) were included and assayed under standard conditions. EMSAs were performed after adding 1/10 of loading buffer to the mixes in a 6% polyacrylamide pre-electrophoresed gel.
Results
Oscillation of pea TRX f and m1 mRNA in leaves during the day/night cycle
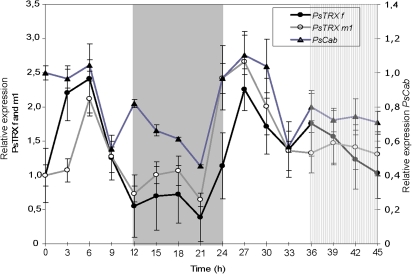
The levels of TRX f and TRX m1 mRNAs were measured during growth of pea seedlings in a photoperiod of 12 h light and 12 h darkness. To study the circadian regulation, the second dark period was replaced by a light period called subjective night. Figure 1 shows that PsTRX f transcripts peak after 6 h of illumination and then rapidly decline. The level remains almost undetectable until 21 h (night period) and starts to rise by the end of the same period, reaching a maximum of mRNA expression at 27 h (during the light period). A low transcript level is detected during the subjective night. PsTRX m1 transcripts also peak after 6 h of light of the first day and at 27 h of the second light phase (Fig. 1). The transcript levels steadily declined throughout the dark period and subjective night. This suggests that the oscillation of the PsTRX f and m1 mRNA level is likely to be under the regulation of the circadian clock. The decreases detected at 9 h and 33 h, a few hours before the end of the light period, anticipating the dark effect, reflect an oscillatory control and a circadian clock-related mechanism. The chlorophyll a/b-binding (CAB) genes are known to be light regulated and under the control of the circadian clock (Buetow et al., 1988). The P. sativum Cab-8 (PsCab8) gene is a member of the type-I Lhcb (chlorophyll a/b-binding protein) gene family and exhibits the most important rhythmic expression among them (Alexander et al., 1991), and a cDNA fragment (500 bp) was used as an Lhcb-specific probe. The PsCab8 gene shows an oscillatory expression pattern, which persists during subjective night (Fig. 1). During the daytime, the peaks appeared at 6 h and 27 h. The increase in mRNA content started before the end of the night phase, indicating anticipation of the expression before the light is switched on. According to previous reports (Millar and Kay, 1991; White et al, 1992), these results indicate that PsCab expression is under a circadian regulation, validating the experimental conditions.
Fig. 1.
Relative expression of PsCab, PsTRX f, and PsTRX m1 genes in leaves of pea during the day/night cycle. Plants were grown under a 12 h light/12 h night photoperiod, and leaf samples were collected at 3 h intervals over a period of 45 h. Total RNA was extracted and analysed by northern blotting (15 μg). Filters were hybridized with 3'-specific probes of PsCab, PsTRX f and PsTRX m1 genes, and loading differences were corrected by re-probing the filter with a cDNA fragment of the 18S rRNA. Clear areas in the graph indicate the day during the day/night periods, the grey area indicates the night, and the hatched area indicates the night period in which the light was kept on. The graph shows quantification of northern blot data normalized considering the lowest value in each case. Numbers on the ordinates indicate the real time in hours at which samples were collected for RNA isolation. (Fisher's exact test, P=0.01). (This figure is available in colour at JXB online.)
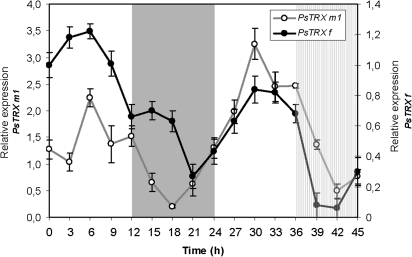
Oscillation of pea TRX f and TRX m1 proteins during the day/night cycle
Immunoblot analyses were performed with pea anti-TRX f and anti-TRX m antisera. Figure 2 shows that the amount of pea TRX f and m1 protein detected in leaves reaches maximum accumulation at 6 h and 30 h of the light periods, then drops to minimum levels during the night (21–18 h) and subjective night (42 h). The oscillation of pea TRX f and m proteins follows the same mRNA oscillation behaviour during the day/night cycle. Both TRX f and m mRNA and protein increases are seen to anticipate the light effect, indicating a circadian regulation.
Fig. 2.
Relative expression of PsTRX f (A) and PsTRX m1 (B) proteins determined after immunodetection by western blotting using specific antibodies against Pisum sativum TRX f and TRX m1 antibodies. Western blot analysis was performed in 50 μg of pea total protein extracts. Clear areas in the graph indicate the day during the day/night periods, the grey area indicates the night, and the hatched area indicates night period in which the light was kept on (hatched area). The graph is representative of two repetitions and shows relative quantification of bands from western blot that were normalized considering the lowest value in each case. Numbers on the ordinates indicate the real time in hour at which samples were collected for protein isolation.
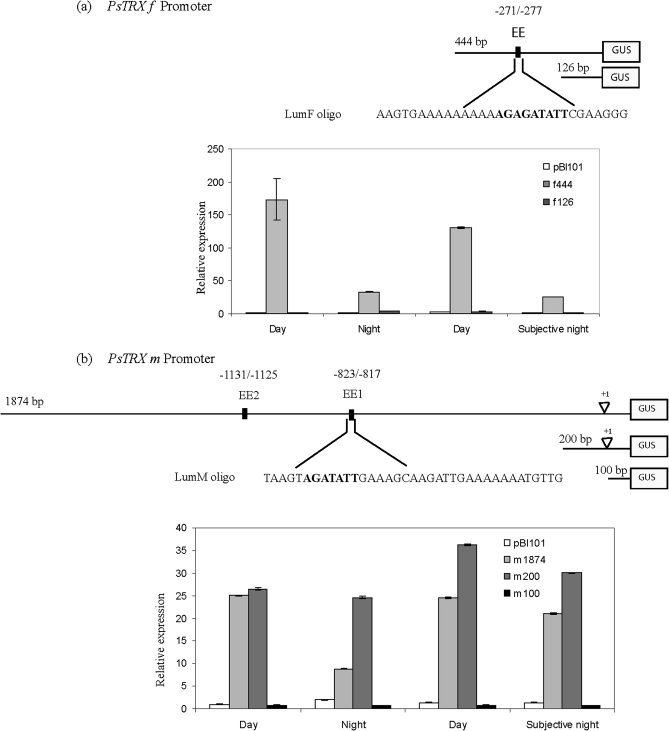
Oscillation of GUS mRNA levels under the control of the pea TRX f promoter in transgenic plants during the day/night cycle
To ascertain whether the circadian oscillation of PsTRX f and PsTRX m1 gene expression is governed by transcriptional control, a 444 bp fragment and a deleted fragment of the same region (126 bp) corresponding to the 5' upstream regions of PsTRX f, as well as a fragment of 1894 bp and two deleted fragments of 200 bp and 100 bp of PsTRX m1 promoter were isolated (Fig. 3). All were transcriptionally fused to the uidA (GUS) reporter gene via the pBI101 vector, and designated as: f444 and f126; and m1874, m200, and m100, respectively (Barajas et al., 2007). Arabidopsis plantlets carrying the different promoter constructs were used to determine the abundance of GUS transcripts. Figure 3a shows a peak of f444–GUS expression in the middle of light periods that decayed 6- to 7-fold during the night and the subjective night in comparison with the light periods. The f126–GUS, which lacks most of the light-dependent cis-elements, lost its GUS expression. These results suggest that the 444 bp 5' upstream region suffices to confer an oscillatory pattern of PsTRX f expression, and that the cis-elements necessary for the gene's transcription are located in the –444/–126 region.
Fig. 3.
Light/dark regulation of GUS mRNA expression under the PsTRX f and PsTRX m1 promoter. Transgenic plants carrying complete promoter constructs (f444–GUS or m1894–GUS) or truncated promoter constructs (f126–GUS, m200–GUS, or m100–GUS) were subjected to a 12 h/12 h photoperiod, the second night the light was kept on, and rosettes were collected at 12 h intervals over a 48 h period; mRNA levels of the GUS-encoding gene were determined in leaves by semi-quantitative RT-PCR as described in the Materials and methods. (a) EE site localization, LUMFs oligo sequence, and GUS expression under the PsTRX f promoter; (b) EEs site localizations, LUMMs oligo sequence, and GUS expression under the PsTRX m promoter. Samples were harvested for 48 h at midday and midnight from 20-day-old plants. Values are the average of three determinations of two cDNA preparations.
The m1874–GUS construct displayed a peak of expression during the mid-light periods that decayed 2.5 times in the night phase and 1.2 times during the subjective night. The m200–GUS construct shows a weaker oscillatory pattern of expression, suggesting that additional elements control the circadian process. Removing another 100 bp was sufficient to lose GUS expression.
CCA1 transcription factor binds specifically to EEs located in PsTRX f and m1 promoters
Analysis of the sequence of the proximal promoter of PsTRXs f in the –444/–126 region revealed an AATATCT sequence (–271 to –277 in an antisense orientation) similar to the core of a putative EE (Fig. 3a). EEs have been reported to be over-represented in a cluster of genes regulated by the circadian clock (Lam and Chua, 1989; Gilmartin et al., 1990; Harmer et al., 2000; Barajas et al., 2007). The EE sequence (AAA/TATATCT) is identical to the sequence CBS (CCA1-binding site) recognized in the TOC1 (Timing of Cab Expression 1) promoter (AAAATATCT) by the transcriptional repressors CCA1 (Circadian Clock Associated 1) and LHY (Late Elongated Hypocotyl) (Alabadí et al., 2001; Yanovsky and Kay, 2001). The homologous Myb-like transcription factors CCA1 and LHY together with TOC1 constitute a transcriptional feedback loop that is essential for a functional circadian clock in Arabidopsis.
As shown in Fig. 3b, the PsTRX m1 promoter showed two putative EE motifs in a sense orientation that differed by only one nucleotide from the core seven-nucleotide consensus sequence (EE1, –823 to –817; and EE2, –1131 to –1125; all numbered from the transcription start site).
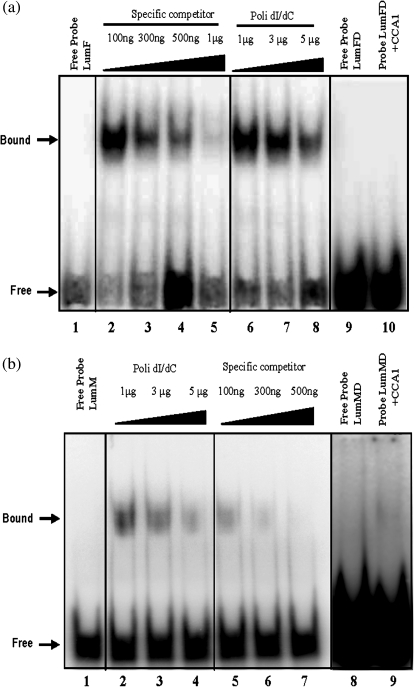
EMSAs were performed to determine whether a recombinant Arabidopsis CCA1 protein could bind the putative EE from the TRX f and TRX m upstream regions. Double-stranded probes of 30 bp (LumFs/LumFa) and 37 bp (LumMs/LumMa) (described in the Materials and methods) carrying the putative EE of the TRX f and TRX m regulatory regions, respectively, were constructed. As shown in Fig. 4a and b, the migration of both sequences was retarded in the presence of the recombinant Arabidopsis CCA1 protein. The shifted probes were stable with the addition of a large excess of the non-specific competitor poly(dI–dC) (Fig. 4a, lines 6–8; b, lines 2–4). However, when 300 ng of cold specific LumFs/LumFa or LumMs/LumMa competitors were added, the bandshift was fully displaced, suggesting a specific binding of CCA1 to the EE of PsTRX f and m1 promoters (Fig. 4a, b, specific competitor).
Fig. 4.
Functional properties of the pea EE motif present in PsTRX f (a) and PsTRX m1 (b) promoters. Electrophoretic mobility shift assays (EMSAs) of the recombinant Arabidopsis CCA1 with oligonucleotide probes derived from the PsTRX f and PsTRX m1 promoter. (a) The probe without recombinant CCA1 is designated Free Probe (lanes 1 and 9). Competition experiments were performed using increasing amounts either of the non-specific competitor [poly(dI–dC)] (lanes 6, 7, 8, and 10) or of the unlabelled probe (lanes 2, 3, 4, and 5). Increasing molar amounts of the unlabelled probe, 100, 300, and 500 ng, and 1 μg, are indicated by triangles. Lanes 1–8: a probe of 30 bp (LumF) derived from the PsTRX f promoters was 32P labelled 5′-AAGTGAAAAAAAAAAGAGATATTCGAAGGG-3′. Lanes 9 and 10: a substitution of the ATATT EE core by CTATT was made in the LumFD oligonucleotide and was 32P labelled 5′-AAGTGAAAAAAAAAAGAGcATATTCGAAGGG-3′. (b) The probe without recombinant CCA1 is designated Free Probe (lanes 1 and 8). Competition experiments were performed using increasing amounts either of the non-specific competitor [poly(dI–dC)] (lanes 2, 3, 4, and 9) or of the unlabelled probe (lanes 5, 6, and 7). Increasing molar amounts, 100, 300, and 500 ng, are indicated by triangles. Lanes 1–7: a probe of 37 bp (LumM) derived from the PsTRX m1 promoters was 32P labelled 5′-TAAGTAGATATTGAAAGCAAGATTGAAAAAAATGTTG-3′, Lanes 8 and 9: a substitution of the ATATT EE core by CTATT was made in the LumMD oligonucleotide and was 32P labelled 5′-TAAGTAGcTATTGAAAGCAAGATTGAAAAAAATGTTG-3′.
To identify the bases involved in the binding, EMSA was performed using new pairs of oligonucleotides where the ATATT core sequence was changed to cTATT. In Fig. 4a (lines 9 and 10) and b (lines 8 and 9), no binding was observed between the mutated variant and the CCA1 protein, suggesting that the first A base of the core sequence is essential to establish the complex. These results indicate that CCA1 binds in a specific manner to the EE sequence of the PsTRXs f and m1 promoters.
Oscillation of A. thaliana TRX f2 and m2 mRNA in rosettes of wild-type plants during the day/night cycle and day/subjective night
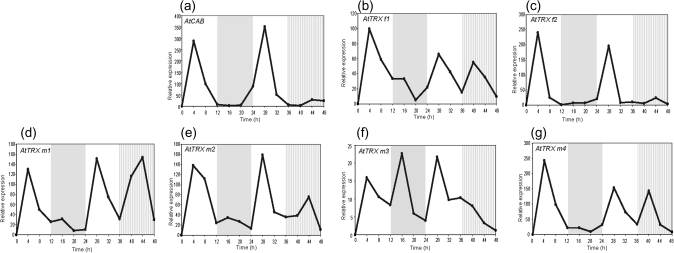
The in vitro interaction of CCA1 with the putative EE of the PsTRX f and PsTRX m1 upstream regions suggests CCA1 involvement in the circadian regulation of TRX f and TRX m1 in pea. To test this hypothesis, the circadian regulation of TRX f and TRX m was studied in the cca1 mutant background. Because such mutants are not yet available in pea, Arabidopsis and real-time PCR with specific oligonucleotides (Table 1) were used to establish the expression of the two TRX f [AtTRX f1 (locus At3g02730) and AtTRX f2 (locus At5g16400)], the four TRX m [AtTRX m1 (locus At1g03680), AtTRX m2 (locus At4g03520), AtTRX m3 (locus At2g15570), and AtTRX m4 (locus At3g15360)], and AtCab2 (locus At1g29920) genes during the diurnal cycle in Arabidopsis plantlets grown under the same oscillatory conditions as pea plantlets.
Figure 5c shows a peak of the AtTRX f2 transcripts after 4 h and 28 h illumination, corresponding to the light periods. Expression was almost undetectable during the first night and the subjective night, and a noteworthy oscillatory pattern was seen for AtTRX m2 (Fig. 5e), displaying two peaks of expression, at 4 h and 28 h of the corresponding mid-light periods. During the dark phase and subjective night the level of transcripts diminished 8-fold with respect to the maximum found in the light phase. These results suggest circadian control of the AtTRX f2 and AtTRX m2 genes. Although AtTRX f1, AtTRX m1, and AtTRX m4 are light induced, no oscillatory effect was observed over the photoperiod (Fig. 5b, d, g). These findings suggest that, among the TRX f and m genes of Arabidopsis, only AtTRX f2 and m2 are under circadian clock regulation.
Fig. 5.
Relative expression of AtTRXCAB (a), AtTRX f1 (b), AtTRX f2 (c), AtTRX m1 (d), AtTRX m2 (e), AtTRX m3 (f), and AtTRX m4 (g) genes in rosettes of Arabidopsis wild-type plants during the day/night cycle. Seedlings were grown under a 12 h light/12 h night photoperiod and rosette samples were collected at 4 h intervals over a period of 48 h; during the second night, the light was kept on. Total RNA was extracted and analysed by real-time PCR using specific oligonucleotides. Clear areas in the graph indicate the day during the day/night periods, the grey area indicates the night, and the hatched area indicates subjective night. The graph shows relative quantification of gene expression monitored after normalization by 18S rRNA expression as internal control, considering the lowest value in each case. Numbers on the ordinates indicate the real time in hours at which samples were collected for RNA isolation.
Figure 5a shows an oscillatory pattern of expression of the wild-type AtCab2 gene that is maintained throughout subjective night. Peaks of expression appeared at 4 h and 28 h of the light periods, similar to the case of PsCab8 transcripts. Expression was undetectable during the night phase and subjective night.
Disruption of Arabidopsis TRX f2 and m2 oscillatory expression in cca1-ox and cca1-lhy double-mutant plants
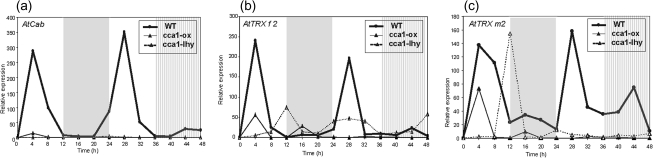
To determine whether CCA1 and LHY transcription factors are involved in the circadian oscillatory regulation of AtTRX f2 and m2 in Arabidopsis, the relative mRNA content of both genes was determined in plants that overexpress the CCA1 transcription factor (cca1-ox plants) and in plants that are defective in CCA1 and LHY transcription factors (cca1-lhy double-mutant) (Schaffer et al., 1998; Wang and Tobin, 1998; Mizoguchi et al. 2002). Figure 6a indicates that the peak of AtCab2 mRNA accumulation in cca1-ox plants was 12 times lower than that found in wild-type plants, and started to decrease rapidly after 4 h of illumination, the transcript level being very low during the rest of the photoperiod. Moreover, the expected peak of expression in the second light phase disappeared. In the cca1-lhy double-mutant line, the transcripts level was undetectable. Two small peaks of AtTRX f2 transcripts were detected in cca1-ox mutant plants during the first day (Fig. 6b). One peak at 4 h of the light phase coincided with the same peak found in wild-type plants, but with mRNA content 4.5 times lower; the second peak was detected at 16 h of the night phase. From midnight of the first day onward, expression became undetectable.
Fig. 6.
Relative expression of AtTRXCAB (a), AtTRX f2 (b), and AtTRX m2 (c) genes in rosettes of Arabidopsis wild-type plants, plants overexpressing CCA1 (cca1-ox), and plants repressing CCA1 and LHY (cca1-lhy) during the day/night cycle. Seeds were grown under a 12 h light/12 h night photoperiod and rosette samples were collected at 4 h intervals over a period of 48 h; during the second night, the light was kept on. Total RNA was extracted and analysed by real-time PCR using specific oligonucleotides. Clear areas in the graph indicate the day during the day/night periods, the grey area indicates the night, and the hatched area indicates subjective night. The graph shows the relative quantification of gene expression monitored after normalization by 18S rRNA expression as an internal control, considering the lowest value in each case. Numbers on the ordinates indicate the real time in hours at which samples were collected for RNA isolation.
In the cca1-lhy double-mutant line, AtTRX f2 transcripts displayed a small peak at the end of the light phase and a broad accumulation of mRNA from 24 h to 32 h of the second light period, declining during subjective night. Plants overexpressing CCA1 showed half the mRNA accumulation of AtTRX m2 detected in wild-type rosettes after 4 h of illumination, which decreased rapidly (Fig. 6c). In the cca1-lhy double-mutant line, AtTRX m2 transcripts were at least 10 times lower, and in the pattern of expression a single peak appeared at the end of the first light phase (Fig. 6c). Meanwhile, the transcripts decreased rapidly.
Altogether, these results reflect a loss of oscillatory regulation of AtCab2, AtTRX f2, and AtTRX m2 gene expression in both mutants, and strongly suggest that AtTRX f2 and m2 expression is controlled during the day/night cycle by CCA1 and LHY transcription factors through the circadian mechanism.
Discussion
In the chloroplast, the light-dependent ferredoxin/TRX system is one of the most studied with the ability to reduce and activate key enzymes of the Calvin cycle and proteins involved in different processes in plants (Buchanan, 1980; Balmer et al., 2006b). This system appears to be one of the best candidates to avoid uncontrolled steps of carbon metabolism during the day/night cycle by anticipating its reaction to environmental light changes through the control of the circadian clock (Bläsing et al., 2005; Yakir et al., 2007).
The database developed by Harmer and colleagues (2000) showed that A. thaliana chloroplast TRX f2 (AtTRX f2; At5g16400) and TRX-like 5 (At5g06690, WCRKC THIOREDOXIN 1) were governed by circadian regulation, peaking at 6 h and 28 h in the light periods. Lemaire et al. (1999) were the first to observe that the expression of Chlamydomonas TRX h, TRX m, ferredoxin, FTR, and PRK is under the control of the circadian oscillator. They suggested that TRX h might be an element in a light- and/or circadian-related transduction pathway. Research by Harmer et al. (2000) has shown that under circadian clock control are genes related to photosynthesis, carbon metabolism, sugar production and transport, as well as enzymes that participate in the regulation of sugar metabolism in higher plants (Pilgrim and McClung, 1993; McClung et al., 2000; Schaffer et al., 2001; Dodd et al., 2005; Lu et al., 2005). The up-regulation of these diverse genes near the end of the subjective day suggests that the clock plays an important role in allocating assimilated sugars to different pathways or to storage in the chloroplast.
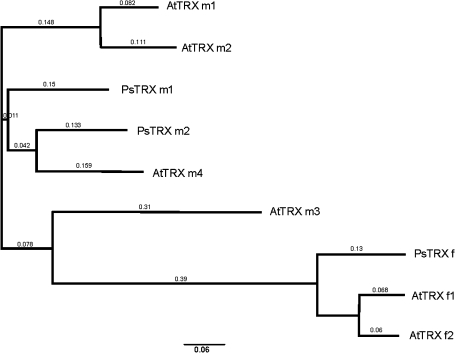
Sequence alignment and the construction of a phylogenetic tree revealed high homology between AtTRX f2 and PsTRX f, and between PsTRX m1 and AtTRX m2 (Fig. 7). Consistent with this was the finding that the orthologues PsTRX f and AtTRX f2 share a common mechanism that directs transcript content to oscillate in a circadian manner during the day/night cycle. The oscillation of PsTRX f protein content suggests regulation at the transcriptional level. Similar results were found for PsTRX m1 and AtTRX m2 mRNA accumulation, which displayed a circadian pattern of expression, and transcriptional regulation was corroborated by the oscillation of PsTRX m1 protein contents. None of the other chloroplastic A. thaliana TRXs (f or m) showed an oscillatory pattern of expression (Fig. 5). However, AtTRX f1, AtTRX m1, and AtTRX m4 mRNA expression appears to be directly induced by light; furthermore, AtTRX m3 does not display specific regulation under the day/night cycle. This TRX is highly expressed in flower petals during stage 15, whereas the other TRXs are in green tissues, cotyledons, rosettes, and leaves (Supplementary Fig. S1, Arabidopsis eFP Browser analysis, available at JXB online). These analyses provide proof of the regulation of PsTRX f, PsTRX m1, AtTRX f2, and AtTRX m2 mRNA levels by the circadian clock, as has been observed for other photosynthetic genes.
Fig. 7.
CLUSTALX phylogenetic tree for chloroplastic TRXs f and m from pea (Ps) and Arabidopsis (At). Accession numbers are: PsTRX f (CAA45098), PsTRX m1 (CAA53900), PsTRX m2 (CAC69854), AtTRX f1 (NP_186922), AtTRX f2 (NP_197144), AtTRX m1 (NP_849585), AtTRX m2 (NP_192261), AtTRX m3 (NP_179159), and AtTRX m4 (NP_188155).
Despite the challenges to understanding the circadian clock, it is well known that TOC1, CCA1, and LHY, the components of the core oscillator, comprise a negative feedback loop grounded in positive and negative transcriptional regulation (Schaffer et al., 1998; Wang and Tobin, 1998; Matsushika et al., 2000; Strayer et al., 2000). Light induces CCA1 and LHY expression and represses TOC1. CCA1 and LHY may directly activate clock-regulated genes that are expressed early in the day, while possibly repressing genes that are only expressed in the evening. CCA1 and LHY levels decrease during the day, a phenomenon that appears to weaken the repression of TOC1, whose levels increase towards the end of the day. Then TOC1 induces the expression of LHY and CCA1 (Wang and Tobin, 1998; Alabadí et al., 2001). CCA1 and LHY have partially redundant functions in the control of period length, but are required to sustain the intensity and circadian rhythmicity (Schaffer et al., 1998; Wang and Tobin, 1998: Green and Tobin, 1999; Alabadí et al., 2002; Mizoguchi et al., 2002).
In wild-type Arabidopsis plants, CCA1 peaks 1 h after dawn and disappears at the end of the day, and this phase oscillation precedes that of CAB gene expression by ∼3 h. Because the CAB gene encodes a chlorophyll a/b-binding protein (Lhcb), this sequence suggests that CCA1 acts as a positive regulator of Lhcb RNA and may be responsible for the circadian rhythm of Lhcb genes early in the morning (Wang et al., 1997; Green and Tobin, 1999). AtTRX f2 and m2 RNA peak 4 h after dawn, a phase relationship consistent with CCA1 driving the oscillation in their gene expression. The Arabidopsis microarray database (www.genevestigator.ethz.ch) corroborates the data presented here by showing that all TRX f and m genes are induced by light, yet only AtTRX f2 and m2 are circadian regulated.
It has been shown that overexpression of CCA1 and inhibition of CCA1 and LHY in Arabidopsis plants disrupts the circadian mRNA accumulation profile of AtTRX f2 and AtTRX m2 mRNA (Fig. 6, and Supplementary Fig. S2 at JXB online). Therefore, if CCA1 is the main factor driving the rhythm of TRX expression, TRX mRNA levels would be expected to be up-regulated in CCA1-overexpressing plants (cca1-ox). However, in this mutant, AtTRX f2 and AtTRX m2 mRNA are 4- and 2-fold lower, respectively, during the first light phase than in wild-type plants, and the circadian rhythm under continuous light is lost. Wang and Tobin (1998) have shown that the constitutive expression of CCA1 in Arabidopsis plants abolished the circadian rhythm in the expression of the Lhcb1 gene in continuous light. These results indicate that as with Lhcb1 RNA, the circadian rhythm of AtTRX f2 and m2 RNA is mediated by the oscillation of the CCA1 protein. Moreover, in plants with repressed CCA1 and LHY, the decrease in AtTRX f2 and AtTRX m2 mRNA is 2- and 18-fold lower than in control plants, respectively, and the first peak is shifted toward the night phase. Alabadí and colleagues (2002) showed that plants lacking CCA1 and with reduced LHY functioning (cca1-lhy-R mutant) were able to sustain oscillations in both constant light and constant darkness. However, as with AtTRX f2 and AtTRX m2, these plants continue to exhibit a degree of circadian function in light/dark cycles, indicating that the Arabidopsis circadian clock is not entirely dependent on CCA1 and LHY activities. In contrast to the earlier phase of activity seen in cca1-lhy-R plants compared with the wild type, the shifted peak in AtTRX f2 and AtTRX m2 mRNA suggests that the delays may reflect the need for an additional structural or energy requirement (Bvnning and Moser, 1973).
Analysis of the promoter regions of the pea chloroplastic f and m1 TRX genes identified the AATATCT sequence (Fig. 3) as similar to the EE, AAAATATCT, sequence described by Harmer et al. (2000). This element has been found to be over-represented in a cluster of 31 clock-regulated genes (Harmer et al., 2000), and the importance of CCA1 in the regulation of gene expression by light and the circadian clock has been observed in microarray experiments (Michael et al., 2008). The EE is known to be important for the rhythmic activity of several evening-phased promoters (Alabadi et al., 2001; Michael and McClung, 2002).
When the functional relevance of the EE was tested in f444–GUS and m1874–GUS, it was shown that this element confers circadian regulation with peak expression in light. The AtTRX f2 regulatory region displayed a circadian-regulated motif at position –987, while AtTRX m2 revealed several CCA1-binding sites at positions –178, –347, and –1868, and at 1003 in the antisense orientation, one circadian cis-motif (at position 528 in the antisense orientation) and two EEs (positions –1593, and 148 in the antisense orientation), suggesting that these motifs are important for oscillatory regulation. No functional EEs were detected in the promoter regions of the other TRXs, AtTRX f1, AtTRX m1, m3, or m4. However, among the putative regulatory elements of all Arabidopsis TRXs, the f and m promoters comprised several light-dependent elements such as GATA, GT1, and Ibox, which are conserved in the promoters of light-regulated genes such as LCHII type I Cab. Different bioinformatics analyses used to compare AtTRX f2 and m2 promoters did not reveal any relevant information that could help understand their circadian regulation, with the exception of the presence of circadian and EE cis-acting elements in the promoter sequences.
The present in vitro binding studies demonstrate that the CCA1 transcription factor binds in vitro directly to the EE of PsTRX f and m1, as has been shown in previous reports with other promoters (Alabadi et al., 2001; Farre et al., 2005). These results suggest that CCA1 is a regulator of TRX genes. The findings revealed that the A base located in the EE core is essential for binding to the CCA1 transcription factor, and therefore a single base pair difference suffices to specify the time of day when transcription occurs. Numerous studies have focused on motif element mutagenesis and mutant plants, and concluded that combinations of known and unknown motifs are necessary for the correct phasing of circadian genes (Matsushika et al., 2002; Mizogushi et al., 2002; Harmer and Kay, 2005).
The physiological relevance of the circadian regulation of chloroplast TRXs might be related to their redox regulation of light-dependent protein targets. The circadian clock appears as an additional regulatory mechanism that induces some TRX genes to anticipate the activation of protein involved in the regulation of photosynthesis, carbon fixation, and growth, and preventing the loss of balance between the supply and utilization of carbon (Smith and Stitt, 2007; Stitt et al., 2007). Extending the night leads to an acute carbon limitation and catabolism of protein, lipids, and other sources of carbon (Usadel et al., 2008). By comparing short- and long-period Arabidopsis mutants with wild-type plants, Dodd et al. (2005) showed that correct matching of the circadian clock period with that of the external light/dark cycle confers a substantial photosynthetic advantage, which in turn contributes to a higher chlorophyll content, and increased carbon fixation.
In conclusion, it was shown that PsTRX f, PsTRX m1, AtTRX f2, and AtTRX m2 mRNA are controlled by the circadian clock, and new evidence is provided that elements of the clock regulate the expression of genes involved in the redox regulation of proteins, probably through direct interaction of CCA1 or LHY transcription factors with an EE of the promoter region of these genes. PsTRX f and PsTRX m1 protein levels also oscillated during the day/night cycle, implying circadian regulation at the transcriptional level. However, AtTRX f2 and AtTRX m2 expression shows a circadian oscillation that is altered in lines that overexpress CCA1 as well as in those that repress CCA1 and LHY and have lower transcript levels. A complex mechanism entailing the interaction of proteins that are light and circadian clock dependent with other proteins induced by light only is envisaged. Thus at least two different but coordinated light-dependent pathways seem to be involved in the regulation of the expression of the genes analysed here.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Expression pattern of the TRX f1, f2, m1, m2, m3, and m4 isoforms of A. thaliana. Data were obtained from the ‘Arabidopsis eFP browser’ (http://bbc.botany.utoronto.ca/efp/cgibin/efpWeb.cgi).
Figure S2. Relative expression of AtTRXCAB (a), AtTRX f1 (b), AtTRX f2 (c), AtTRX m1 (d), AtTRX m2 (e), AtTRX m3 (f), and AtTRX m4 (g) genes in rosettes of Arabidopsis wild-type plants, plants overexpressing CCA1 (cca1-ox), and plants repressing CCA1 and LHY (cca1-lhy) during the day/night cycle. Seedlings were grown under a 12 h light/12 h night photoperiod and rosette samples were collected at 4 h intervals over a period of 48 h; during the second night, the light was kept on. Total RNA was extracted and analysed by real-time PCR using specific oligonucleotides. Clear areas in the graph indicate the day during the day/night periods, the grey area indicates the night, and the hatched area indicates subjective night. The graph shows the relative quantification of gene expression monitored after normalization by 18S rRNA expression as an internal control, considering the lowest value in each case. Numbers on the ordinates indicate the real time in hours at which samples were collected for RNA isolation.
Acknowledgments
The authors appreciate the assistance of Trinidad Moreno and Christèle Couderc for their technical support in this work. We gratefully acknowledge the provision of mutant lines cca1-ox and cca1-lhy by the laboratories of Dr Ángel Mérida (CSIC-Spain) and Dr George Coupland (Max Planck Institute, Germany). We thank K. Shashok for editing parts of the manuscript for clarity. This work was supported by Grants BFI2002-00401, BIO2005-00157, and BIO2006-2816 from the Ministerio de Educación y Ciencia (Spain), FEDER and Grant BIO154 from the Junta de Andalucía (Spain). JDB-L was supported by a pre-doctoral fellowship from the Junta de Andalucía, and ASR was supported by a post-doctoral I3P fellowship from the Consejo Superior de Investigaciones Científicas (Spain) and by a contract from the Junta de Andalucía.
References
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- Alabadí D, Yanovsky MJ, Más P, Harmer SL, Kay SA. Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Current Biology. 2002;12:757–761. doi: 10.1016/s0960-9822(02)00815-1. [DOI] [PubMed] [Google Scholar]
- Alexander L, Falconet D, Fristensky BW, White MJ, Watson JC, Roe BA, Thompson WF. Nucleotide sequence of Cab-8, a new type I gene encoding a chlorophyll a/b-binding protein of LHC II in Pisum. Plant Molecular Biology. 1991;17:523–526. doi: 10.1007/BF00040649. [DOI] [PubMed] [Google Scholar]
- Arsova B, Hoja U, Wimmelbacher M, Greiner E, Üstün S, Melzer M, Petersen K, Lein W, Börnke F. Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. The Plant Cell. 2010;22:1498–1515. doi: 10.1105/tpc.109.071001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer Y, Koller A, del Val G, Manieri W, Schürmann P, Buchanan BB. Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proceedings of the National Academy of Sciences, USA. 2003;100:370–375. doi: 10.1073/pnas.232703799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer Y, Vensel WH, Cai N, Manieri W, Schürmann P, Hurkman WJ, Buchanan BB. a. A complete ferredoxin/thioredoxin system regulates fundamental processes in amyloplasts. Proceedings of the National Academy of Sciences, USA. 2006;103:2988–2993. doi: 10.1073/pnas.0511040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer Y, Vensel WH, DuPont FM, Buchanan BB, Hurkman WJ. Proteome of amyloplasts isolated from developing wheat endosperm presents evidence of broad metabolic capability. Journal of Experimental Botany. 2006b;57:1591–1602. doi: 10.1093/jxb/erj156. [DOI] [PubMed] [Google Scholar]
- Barajas J, Serrato A, Olmedilla A, Chueca A, Sahrawy M. Localization in roots and flowers of pea chloroplast thioredoxin f and m proteins reveals new roles in non-photosynthetic organs. Plant Physiology. 2007;145:946–960. doi: 10.1104/pp.107.105593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham JA, Krause GH. Free energy changes and metabolic regulation in steady-state photosynthetic carbon reduction. Biochimica et Biophysica Acta. 1969;189:207–221. doi: 10.1016/0005-2728(69)90048-6. [DOI] [PubMed] [Google Scholar]
- Bläsing OE, Gibon Y, Gunther M, Hohne M, Morcuende R, Osuna D, Thimm O, Usadel B, Scheible WR, Stitt M. Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. The Plant Cell. 2005;17:3257–3281. doi: 10.1105/tpc.105.035261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan BB, Lara C, de la Torre A, Jacquot JP, Nishizawa AN. Thioredoxin and enzyme regulation. Federation Proceedings. 1980;39:1973. [Google Scholar]
- Buchanan BB, Schürmann P, Wolosiuk RA, Jacquot J. The ferredoxin/thioredoxin system: from discovery to molecular structures and beyond. Photosynthesis Research. 2002;73:215–222. doi: 10.1023/A:1020407432008. [DOI] [PubMed] [Google Scholar]
- Buetow DF, Chen H, Erdös G, Yi LSH. Regulation and expression of the multigene family coding light harvesting chlorophyll a/b-binding proteins of photosystem II. Photosynthesis Research. 1988;18:61–97. doi: 10.1007/BF00042980. [DOI] [PubMed] [Google Scholar]
- Bvnning E, Moser I. Light-induced phase shifts of circadian leaf movements of Phaseolus: comparison with the effects of potassium and of ethyl alcohol. Proceedings of the National Academy of Sciences, USA. 1973;12:3387–3389. doi: 10.1073/pnas.70.12.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco JL, Chueca A, Sahrawy M, Hermoso R, Lázaro JJ, López Gorgé J. Role of light in the in vivo and in vitro synthesis of spinach thioredoxin f. Physiologia Plantarum. 1992;84:236–242. [Google Scholar]
- Collin V, Issakidis-Bourget E, Marchand C, Hirasawa M, Lancelin JM, Knaff DB, Miginiac-Maslow M. The Arabidopsis plastidial thioredoxins: new functions and new insights into specificity. Journal of Biological Chemistry. 2003;278:23747–23752. doi: 10.1074/jbc.M302077200. [DOI] [PubMed] [Google Scholar]
- Collin V, Lamkemeyer P, Miginiac-Maslow M, Hirasawa M, Knaff DB, Dietz KJ, Issakidis-Bourguet E. Characterization of plastidial thioredoxins from Arabidopsis belonging to the new y-type1. Plant Physiology. 2004;136:4088–4095. doi: 10.1104/pp.104.052233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NA, Yee BC, Nishizawa AN, Buchanan BB. Occurrence of cytoplasmic f- and m-type thioredoxins in leaves. FEBS Letters. 1979;104:141–145. [Google Scholar]
- Díaz I, Vicente-Carbajosa J, Abraham Z, Martínez M, Isabel Almoneda I, Carbonero P. The GAMYB protein from barley interacts with the DOF transcription factor BPBF and activates endosperm-specific genes during seed development. The Plant Journal. 2002;29:453–464. doi: 10.1046/j.0960-7412.2001.01230.x. [DOI] [PubMed] [Google Scholar]
- Dodd AN, Salathia N, Hall A, Kevei E, Toth R, Nagy F, Hibberd JM, Millar AJ, Webb AAR. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- Farre EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA. Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Current Biology. 2005;15:47–54. doi: 10.1016/j.cub.2004.12.067. [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Kolbe A, Tiessen A. Redox regulation of carbon storage and partitioning in response to light and sugars. Journal of Experimental Botany. 2005;56:1469–1479. doi: 10.1093/jxb/eri178. [DOI] [PubMed] [Google Scholar]
- Gilmartin PM, Sarokin L, Memelink J, Chua NH. Molecular light switches for plant genes. The Plant Cell. 1990;2:369–378. doi: 10.1105/tpc.2.5.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RM, Tobin EM. Loss of the circadian clock associated protein 1 in Arabidopsis results in altered clock regulated gene expression. Proceedings of the National Academy of Sciences, USA. 1999;96:4176–4179. doi: 10.1073/pnas.96.7.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Hogenesc LB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- Harmer SL, Kay SA. Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. The Plant Cell. 2005;17:1926–1940. doi: 10.1105/tpc.105.033035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren A. Thioredoxin. Annual Review of Biochemistry. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- Holmgren A. Thioredoxin and glutaredoxin systems. Journal of Biological Chemistry. 1989;264:13963–13966. [PubMed] [Google Scholar]
- Howard TP, Metodiev M, Lloyd JC, Raines CA. Thioredoxin-mediated reversible dissociation of a stromal multiprotein complex in response to changes in light availability. Proceedings of the National Academy of Sciences, USA. 2008;105:4056–4061. doi: 10.1073/pnas.0710518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami A, Yoshimura N, Motohashi K, Takahashi S, Romano PG, Hisabori T, Takamiya K, Masuda T. The CHLI1 subunit of Arabidopsis thaliana magnesium chelatase is a target protein of the chloroplast thioredoxin. Journal of Biological Chemistry. 2007;282:19282–19291. doi: 10.1074/jbc.M703324200. [DOI] [PubMed] [Google Scholar]
- Jacquot JP, Vidal J, Gadal P, Schürmann P. Evidence for existence of several enzyme-specific thioredoxins in plants. FEBS Letters. 1978;96:243–246. [Google Scholar]
- Jeannette E, Prioul JL. Variation of ADP glucose pyrophosphorylase activity from maize leaf during day/night cycle. Plant and Cell Physiology. 1994;35:869–878. [Google Scholar]
- Kirchsteiger K, Pulido P, Gonzalez M, Cejudo FJ. NADPH thioredoxin reductase C controls the redox status of chloroplast 2-Cys peroxiredoxins in Arabidopsis thaliana. Molecular Plant. 2009;2:298–307. doi: 10.1093/mp/ssn082. [DOI] [PubMed] [Google Scholar]
- Lam E, Chua NH. ASF-2: a factor that binds to the cauliflower mosaic virus 35S promoter and a conserved GATA motif in Cab promoters. The Plant Cell. 1989;1:1147–1156. doi: 10.1105/tpc.1.12.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire SD, Miginiac-Malow M, Jacquot JP. Plant thioredoxins gene expression: control by light, circadian clock, and heavy metals. Methods in Enzymology. 2002;347:412–421. doi: 10.1016/s0076-6879(02)47041-3. [DOI] [PubMed] [Google Scholar]
- Lemaire SD, Stein M, Issakidis-Bourget E, Keryer E, Vanina Benoit, Pineau B, Gérard-Hirne C, Miginiac-Maslow M, Jacquot JP. The complex regulation of ferredoxin/thioredoxin-related genes by light and the circadian clock. Planta. 1999;209:221–229. doi: 10.1007/s004250050626. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- López-Jaramillo J, Chueca A, Jacquot JP, Hermoso R, Lázaro JJ, Sahrawy M, López-Gorgé J. High-yield expression of pea thioredoxin m and assessment of its efficiency in chloroplast fructose-1,6-bisphosphatase activation. Plant Physiology. 1997;114:1169–1175. doi: 10.1104/pp.114.4.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Gehan JP, Sharkey TD. Day length and circadian effects on starch degradation and maltose metabolism. Plant Physiology. 2005;138:2280–2291. doi: 10.1104/pp.105.061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marri L, Zaffagnini M, Collin V, Issakidis-Bourguet E, Lemaire SD, Pupillo P, Sparla F, Miginiac-Maslow M, Trost P. Prompt and easy activation by specific thioredoxins of Calvin cycle enzymes of Arabidopsis thaliana associated in the GAPDH/CP12/PRK supramolecular complex. Molecular Plant. 2009;2:259–269. doi: 10.1093/mp/ssn061. [DOI] [PubMed] [Google Scholar]
- Martí MC, Olmos E, Calvete JJ, Díaz I, Barranco-Medina S, Whelan J, Lázaro JJ, Sevilla F, Jiménez A. Mitochondrial and nuclear localization of a novel pea thioredoxin: identification of its mitochondrial target proteins. Plant Physiology. 2009;150:646–657. doi: 10.1104/pp.109.138073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino-Catt S, Ort DR. Low temperature interrupts circadian regulation of transcriptional activity in chilling-sensitive plants. Proceedings of the National Academy of Sciences, USA. 1992;89:3731–3735. doi: 10.1073/pnas.89.9.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushika A, Makino S, Kojima M, Mizuno T. Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: insight into the plant circadian clock. Plant and Cell Physiology. 2000;41:1002–1012. doi: 10.1093/pcp/pcd043. [DOI] [PubMed] [Google Scholar]
- Matsushika A, Makino S, Kojima M, Yamashino T, Mizuno T. The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: II. Characterization with CCA1-overexpressing plants. Plant and Cell Physiology. 2002;43:118–122. doi: 10.1093/pcp/pcf006. [DOI] [PubMed] [Google Scholar]
- McClung CR, Hsu M, Painter JE, Gagne JM, Karlsberg SD, Salome PA. Integrated temporal regulation of the phosphorespiratory pathway. Circadian regulation of two Arabidopsis genes encoding serine hydroxymethyltransferase. Plant Physiology. 2000;123:381–392. doi: 10.1104/pp.123.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mérida A, Rodriguez-Galan JM, Vincent C, Romero JM. Expression of the granule-bound starch synthase I (Waxy) gene from snapdragon is developmentally and circadian clock regulated. Plant and Cell Physiology. 1999;120:401–410. doi: 10.1104/pp.120.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer Y, Buchanan BB, Vignols F, Reichheld JP. Thioredoxins and glutaredoxins: unifying elements in redox biology. Annual Review of Genetics. 2009;43:335–367. doi: 10.1146/annurev-genet-102108-134201. [DOI] [PubMed] [Google Scholar]
- Meyer Y, Siala W, Bashandy T, Riondet C, Vignols C, Reichheld JP. Glutaredoxins and thioredoxins in plants. Biochimica et Biophysica Acta. 2008;1783:589–600. doi: 10.1016/j.bbamcr.2007.10.017. [DOI] [PubMed] [Google Scholar]
- Michael TP, McClung CR. Phase-specific circadian clock regulatory elements in Arabidopsis. Plant Physiology. 2002;130:627–638. doi: 10.1104/pp.004929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Mockler TC, Breton G, et al. Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genetics. 2008;4:0001–0015. doi: 10.1371/journal.pgen.0040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ, Kay SA. Circadian control of cab gene transcription and mRNA accumulation in Arabidopsis. The Plant Cell. 1991;3:541–550. doi: 10.1105/tpc.3.5.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song HR, Carré IA, Coupland G. LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Developmental Cell. 2002;2:629–641. doi: 10.1016/s1534-5807(02)00170-3. [DOI] [PubMed] [Google Scholar]
- Montrichard F, Renard M, Alkhalfioui F, Duval FD, Macherel D. Identification and differential expression of two thioredoxin h isoforms in germinating seeds from pea. Plant Physiology. 2003;132:1707–1715. doi: 10.1104/pp.102.019562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano EA, Chueca A, López-Gorge J. Expression of thioredoxins f and m and of their targets fructose-1,6-bisphosphatase and NADP-malate dehydrogenase, in pea plants grown under normal and light/temperature stress conditions. Journal of Experimental Botany. 2000;51:1299–1307. [PubMed] [Google Scholar]
- Pérez-Ruiz JM, Spínola MC, Kirchsteiger K, Moreno J, Sahrawy M, Cejudo FJ. Rice NTRC is a high-efficiency redox system for chloroplast protection against oxidative damage. The Plant Cell. 2006;18:2356–2368. doi: 10.1105/tpc.106.041541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilgrim ML, McClung CR. Differential involvement of the circadian clock in the expression of genes required for ribulose-1,5-bisphosphate carboxylase/oxygenase synthesis, assembly, and activation in Arabidopsis thaliana. Plant Physiology. 1993;103:553–564. doi: 10.1104/pp.103.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey P, Cuine S, Eymery F, Garin J, Court M, Jacquot J-P, Rouhier N, Broin M. Analysis of the proteins targeted by CDSP32, a plastidic thioredoxin participating in oxidative stress responses. The Plant Journal. 2005;41:31–42. doi: 10.1111/j.1365-313X.2004.02271.x. [DOI] [PubMed] [Google Scholar]
- Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, Wisman E. Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. The Plant Cell. 2001;13:113–123. doi: 10.1105/tpc.13.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, Coupland G. LATE ELONGATED HYPOCOTYL, an Arabidopsis gene encoding a MYB transcription factor, regulates circadian rhythmicity and photoperiodic responses. Cell. 1998;93:1219–1229. doi: 10.1016/s0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- Schürmann P, Jacquot JP. Plant thioredoxin systems revisited. Annual Review of Plant Physiology and Plant Molecular Biology. 2000;51:371–400. doi: 10.1146/annurev.arplant.51.1.371. [DOI] [PubMed] [Google Scholar]
- Shinohara ML, Loros J, Dunlap JC. Glyceraldehyde-3-phosphate dehydrogenase is regulated on a daily basis by the circadian clock. Journal of Biological Chemistry. 1998;273:446–452. doi: 10.1074/jbc.273.1.446. [DOI] [PubMed] [Google Scholar]
- Smith SM, Stitt M. Coordination of carbon supply and plant growth. Plant, Cell and Environment. 2007;30:1126–1149. doi: 10.1111/j.1365-3040.2007.01708.x. [DOI] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Más P, Panda S, Kreps JA, Kay SA. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science. 2000;289:768–771. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- Stitt M, Gibon Y, Lunn J, Piques M. Multilevel genomics analysis of carbon signalling during low carbon availability: coordination of the supply and of utilization carbon in a fluctuating environment. Functional Plant Biology. 2007;34:526–549. doi: 10.1071/FP06249. [DOI] [PubMed] [Google Scholar]
- Traverso JA, Vignols F, Cazalis R, Pulido A, Sahrawy M, Cejudo FJ, Meyer Y, Chueca A. PsTRXh1 and PsTRXh2 are both pea h-type thioredoxins with antagonistic behavior in redox imbalances. Plant Physiology. 2007;143:300–311. doi: 10.1104/pp.106.089524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traverso JA, Vignols F, Cazalis R, Serrato AJ, Pulido P, Sahrawy M, Meyer Y, Cejudo FJ, Chueca A. Immunocytochemical localization of Pisum sativum TRXs f and m in non-photosynthetic tissues. Journal of Experimental Botany. 2008;59:1267–1277. doi: 10.1093/jxb/ern037. [DOI] [PubMed] [Google Scholar]
- Usadel B, Bläsing OE, Gibon Y, Retzlaff K, Höhne M, Günther M, Stitt M. Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiology. 2008;146:1834–1861. doi: 10.1104/pp.107.115592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z-Y, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM. A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. The Plant Cell. 1997;9:491–507. doi: 10.1105/tpc.9.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z-Y, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93:1207–1217. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- Wedel N, Soll J, Paap BK. CP12 provides a new mode of light regulation of Calvin cycle activity in higher plants. Proceedings of the National Academy of Sciences, USA. 1997;94:10479–10484. doi: 10.1073/pnas.94.19.10479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MJ, Fristensky B, Falconet D, Childs LC, Watson JC, Alexander L, Roe BA, Thompson WF. Expression of the chlorophyll-a/b-protein multigene family in pea (Pisum sativum L.) Planta. 1992;188:190–198. doi: 10.1007/BF00216813. [DOI] [PubMed] [Google Scholar]
- Yakir E, Hilman D, Harir Y, Green R. Regulation of output from the plant circadian clock. FEBS Journal. 2007;274:335–345. doi: 10.1111/j.1742-4658.2006.05616.x. [DOI] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA. Signaling networks in the plant circadian system. Current Opinion in Plant Biology. 2001;4:429–435. doi: 10.1016/s1369-5266(00)00196-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.