Abstract
The brittle culm (bc) mutants of Gramineae plants having brittle skeletal structures are valuable materials for studying secondary cell walls. In contrast to other recessive bc mutants, rice Bc6 is a semi-dominant bc mutant with easily breakable plant bodies. In this study, the Bc6 gene was cloned by positional cloning. Bc6 encodes a cellulose synthase catalytic subunit, OsCesA9, and has a missense mutation in its highly conserved region. In culms of the Bc6 mutant, the proportion of cellulose was reduced by 38%, while that of hemicellulose was increased by 34%. Introduction of the semi-dominant Bc6 mutant gene into wild-type rice significantly reduced the percentage of cellulose, causing brittle phenotypes. Transmission electron microscopy analysis revealed that Bc6 mutation reduced the cell wall thickness of sclerenchymal cells in culms. In rice expressing a reporter construct, BC6 promoter activity was detected in the culms, nodes, and flowers, and was localized primarily in xylem tissues. This expression pattern was highly similar to that of BC1, which encodes a COBRA-like protein involved in cellulose synthesis in secondary cell walls in rice. These results indicate that BC6 is a secondary cell wall-specific CesA that plays an important role in proper deposition of cellulose in the secondary cell walls.
Keywords: Brittle culm, cellulose synthesis, CesA protein, dominant-negative form, hemicellulose, rice, secondary cell wall, transgenic plant
Introduction
Secondary cell walls function as skeletal frameworks and furnish the plant body with mechanical strength. Studies on Arabidopsis thaliana mutants have identified several components involved in the formation of secondary cell walls. The secondary cell wall-specific cellulose synthase catalytic subunits (CesAs), AtCesA8, AtCesA7, and AtCesA4, were first identified from their respective mutants irregular xylem 1 (irx1), irx3, and irx5, which show collapsed morphology in the xylem (Turner and Somerville, 1997; Taylor et al., 1999, 2000, 2003). A series of studies on Arabidopsis fragile fiber (fra) mutants with reduced inflorescence stem mechanical strength revealed that in addition to CesA proteins, the formation of secondary cell walls requires katanin, kinesin-like protein, and phosphatidylinositol phosphatase (Burk et al., 2001; Zhong et al., 2002, 2003, 2004). Decreased cellulose content is common to all these mutants, suggesting that proper synthesis and accumulation of cellulose microfibrils are central events in the formation of secondary cell walls in higher plants.
CesA was first cloned in cotton fibre as a plant homologue of bacterial cellulose synthase (Pear et al., 1996). In general, CesA proteins have eight transmembrane domains, a large cytoplasmic region between the second and third transmembrane domains, and a relatively small N-terminal cytoplasmic domain. To date, 10 CesA genes have been identified in the genome of Arabidopsis and nine in that of rice (Oryza sativa). These genes may be categorized into two groups according to their involvement in cellulose synthesis in primary or secondary cell walls. In Arabidopsis, the expression patterns of secondary cell wall-specific CesA genes encoding AtCesA4, AtCesA7, and AtCesA8 are highly correlated. These CesAs are known to form a cellulose synthase complex (CSC) (Taylor et al., 2003; Brown et al., 2005; Atanassov et al., 2009). However, the specific role of each CesA component in the overall cellulose synthesis pathway remains obscure.
Some mutations in the CesA genes confer resistance to herbicide and pathogens. The isoxaben resistant 1 (ixr1) and ixr2 mutants of Arabidopsis have missense mutations in the conserved C-terminal regions of AtCesA3 and AtCesA6, respectively (Scheible et al., 2001; Desprez et al., 2002). The Arabidopsis cev1 mutant with a mutation in the second cytoplasmic domain of AtCesA3 shows constitutive expression of the jasmonate-responsive genes, vegetative storage protein 1, protodermal factor 1.2, thionin 2.1/pathogenesis-related protein 13, and basic chitinase B/pathogenesis-related protein 3, and this confers greater resistance to powdery mildew diseases (Ellis and Turner, 2001; Ellis et al., 2002). In addition, altered secondary cell wall integrity due to defects in secondary cell wall-specific CesAs results in enhanced resistance to soil bacteria through activation of the abscisic acid (ABA) pathway independent of signalling by salicylic acid, ethylene, or jasmonate (Hernández-Blanco et al., 2007).
The brittle culm (bc) mutants of Gramineae plants exhibit reduced mechanical strength of the plant body, especially in culms (Kokubo et al., 1989, 1991; Ching et al., 2006; Sindhu et al., 2007). In rice, nine bc mutants (bc1, bc2, bc3, bc4, bc5, bc6, bc7, bc10, and bc11) have been found to date, and some of them were used as classic genome markers. These bc mutants are valuable materials for understanding the mechanism of secondary cell wall formation. Rice BC1 and maize (Zea mays) Brittle stalk 2 encode COBRA-like glycosylphosphatidylinositol-anchored proteins (Li et al., 2003; Ching et al., 2006). OsDRP2B encoding a plant classical dynamin has been identified as the causative gene for the rice bc3 mutant (Hirano et al., 2010). Rice bc7 and bc11 have mutations in OsCesA4, a secondary cell wall-specific CesA protein whose sequence is highly similar to AtCesA8/IRX1 (Yan et al., 2007). Furthermore, on the basis of sequence similarity, OsCesA4, OsCesA7, and OsCesA9 have been proposed to correspond to AtCesA8, AtCesA4, and AtCesA7, respectively (Tanaka et al., 2003).
With respect to its genetic semi-dominance, rice Bc6 is unique among bc mutants of Gramineae plants. Here, it is reported that rice Bc6 has a missense mutation in the OsCesA9 gene. On the basis of cell wall properties of the Bc6 mutant and the expression pattern of the BC6 gene, it is proposed that OsCesA9 is a secondary cell wall-specific CesA of rice and that the mechanism for cellulose synthesis in secondary cell walls is highly conserved between rice and Arabidopsis.
Materials and methods
Plant materials
Seeds of the Bc6 mutant (RGS number 420) and IR68 were provided by Dr Khush of the International Rice Research Institute (IRRI, Laguna, Philippines). Seeds of Taichung 65 (T65) was distributed from National Genetic Institute (Mishima, Japan). A japonica cultivar, T65, was mainly used to represent wild-type control. Rice plants were grown under field conditions at the National Institute of Agrobiological Sciences, Tsukuba, Ibaraki, Japan.
Analysis of cell wall polysaccharides
Fractionation and quantification of cell wall polysaccharides were performed as described (Aohara et al., 2009). Plant tissues were homogenized to a fine powder using a mortar and pestle in liquid nitrogen. The homogenates were washed twice with water, heated in 80% (v/v) ethanol at 100 °C for 15 min to inactivate endogenous cell wall enzymes, and then treated with α-amylase (100 U, Type VII-A from porcine pancreas, Sigma-Aldrich, St Louis MO, USA) in 50 mM 3-morpholinopropanesulphonic acid-NaOH buffer (pH 6.5) at 37 °C for 4 h. After removal of solubilized starch by centrifugation at 1500 g, the cell wall materials were sequentially extracted at 100 °C for 10 min with water, 50 mM EDTA (pH 6.8) (pectin fraction), and 17.5% (w/v) NaOH containing 0.04% NaBH4 (hemicellulose fraction). The residual precipitate was washed with water, ethanol, and diethyl ether, and collected as the cellulose fraction. Hemicellulose was neutralized with acetic acid, dialysed against water at 4 °C for 1 d, and lyophilized. The sugar content in each fraction was measured by the phenol–sulphuric acid method (Dubois et al., 1956) using glucose as the standard.
Klason lignin content was determined using cell walls prepared from culms of Bc6 mutants and T65 according to the method reported by Kirk and Obst (1998).
Determination of sugar composition
Hemicellulose was hydrolysed in 72% (v/v) H2SO4 at 4 °C for 1 h, followed by addition of 8 vols of water and hydrolysis at 100 °C for 4 h. After neutralization with solid BaCO3, the content of each monosaccharide was determined by high-performance anion-exchange chromatography (HPAEC) using a Dionex DX-500 liquid chromatograph equipped with a CarboPac PA-1 column and a pulsed amperometric detector (PAD) (Dionex, Sunnyvale, CA, USA) as described previously (Ishikawa et al., 2000).
Histology
The uppermost internode was fixed in an FAA solution (water/ethanol/acetic acid/formaldehyde=45:45:5:5, v/v), dehydrated through a graded ethanol/t-butyl alcohol series (0–100% t-butyl alcohol), embedded in paraffin, and sectioned with a microtome (Leica, RM2125RT, Leica Microsystems, Wetzlar, Germany) at a thickness of 10 μm. The sections were washed in a xylene/ethanol series (0–50% ethanol, v/v), stained with 1% (w/v) Safranin O (Waldeck, Münster, Germany) for 24 h, washed with an ethanol/water series (50–95% ethanol, v/v), stained with 0.5% (w/v) Fast Green FCF (Wako, Tokyo, Japan) for 45 s, and washed with 95% and 100% ethanol (v/v). To detect lignin in cell walls, hand-cut sections of culm were stained with 2% (w/v) phloroglucinol and then treated with 18% (w/v) HCl. The sections were observed under a light microscope (Eclipse E400, Nikon, Tokyo, Japan).
Transmission electron microscopy
Tissues were fixed in 50 mM phosphate buffer (pH 7.0) containing 2% (v/v) glutaraldehyde. After washing with the phosphate buffer, tissues were post-fixed with 2% (w/v) OsO4 dissolved in the phosphate buffer. Tissues were dehydrated through a graded series of acetone and gradually infiltrated with Spurr's resin, which was polymerized by incubation at 70 °C overnight. Ultra-thin, 90–100 nm sections were cut with a diamond knife on a Sorvall MT-IIB ultramicrotome (Thermo Electron Corporation, Asheville, NC, USA) and stained with 2% (w/v) uranyl acetate for 15 min followed by lead citrate for 5 min. The sections were observed with a Hitachi H-7500 electron microscope (Hitachi Science Systems, Ibaraki, Japan) at an acceleration voltage of 100 kV.
Positional cloning
For positional cloning of the BC6 gene, the Bc6 mutant was crossed with a japonica cultivar, Toride 1, and the resulting F2 population was analysed for their brittle phenotype and genotype. Together with DNA markers reported previously (Yamamoto and Sasaki, 1997), the co-dominant DNA markers designed on the basis of genomes of a japonica cultivar, Nipponbare (http://rgp.dna.affrc.go.jp/), and an indica cultivar, 93-11 (http://btn.genomics.org.cn:8080/rice/) were used (Table 1). Genomic DNA was extracted from leaves of F2 plants as described (Aohara et al., 2009). PCR was performed with Phusion DNA polymerase (Finnzymes, Espoo, Finland) under the following conditions: 10 s denaturation at 98 °C, 20 s annealing at 60 °C, and 30 s extension at 72 °C, 35 cycles.
Table 1.
DNA markers used for positional cloning
| Marker | Forward primer | Reverse primer | Length (bp)a | Type |
| 5838-EcoRI | 5′-GTCCCACATGTCAACAGAGC-3′ | 5′-CACACAATCCAGGAGAAGGC-3′ | 264 | CAPS (EcoRI)b |
| E61522 | 5′-GCGTTTGAGGAAGTTACCAC-3′ | 5′-TTTAGTCGAGGCAGAGATCC-3′ | 347 | CAPS (EcoRI) |
| 5579-54kb | 5′-TGGTATGGTCTGTGATTGGG-3′ | 5′-TAGAGTGAAAAGGCTGAGGC-3′ | 349 | SNPc |
| 5579-121kb | 5′-CCCGGTACACACAACACACC-3′ | 5′-TCCCACTTTGCACTCCCTGG-3′ | 303 | CAPS (EcoRI) |
| 5420-101kb | 5′-TGATCCCCTCAATCTGGCAG-3′ | 5′-AAGTAGCAGGTCCATCGAAC-3′ | 348 | Indeld |
| 5568-19kb | 5′-TGAGCTAGCGATGTGGCTGG-3′ | 5′-CTAGCTACACTACACACGGC-3′ | 379 | CAPS (EcoRI) |
The length of the fragment amplified by PCR from genomic DNA of Toride 1 is shown.
Cleaved amplified polymorphic sequence. The restriction enzyme used is shown in parentheses.
Single nucleotide polymorphism.
Marker with a difference amplified fragment length due to an insertion or deletion event.
The genomic fragment of Bc6 was amplified by PCR using a set of specific primers, gBc6-F1 (5′-GAAGCTTTCTAGAAGTCCCGCCAAACC-3′) and gBc6-R1 (5′-GCGGACGCATCTCACACAACTGAGGCC-3′). The nucleotide sequence was determined with an ABI genetic analyzer (PRISM 3100, ABI, Foster City, CA, USA), and compared with that of 93-11. Bc6 cDNA was amplified by reverse transcription-PCR (RT-PCR) using a set of primers, Bc6cDNA-F (5′-GGCTCTAGAGCGATCGATCGCCCTTCC-3′) and Bc6cDNA-R (5′-ACCTCTAGAGCTGCAATCTGAATATATTCC-3′), and sequenced.
Introduction of the Bc6 gene
A genomic fragment containing the mutant Bc6 gene (9374 bp), including 4.8 kbp of upstream sequence and 492 bp downstream, was digested with HindIII and subcloned into the binary vector pBIGRZ (Akiyama et al., 1997) to yield the mutant construct, gBc6/pBIGRZ. The wild-type BC6 gene construct, gBC6/pBIGRZ, was generated by changing the mutated nucleotide at position 7112 from G to wild-type A by PCR mutagenesis with primers, PM-F1 (5′-GCAGTTCCCGCAGAGGTTCGACGGC-3′) and PM-R1 (5′-ATCGACGTCCACGACCGATACGCC-3′). These constructs were introduced into the wild-type plant, T65, by an Agrobacterium (Rhizobium radiobacter)-mediated method using strain EHA101 (Hood et al., 1986; Toki, 1997). The T2 generation of transgenic plant was used for cell wall analysis.
Promoter:GUS assay
PromoterBC6:β-glucuronidase (pBC6:GUS) activity was assayed according to the method of Kosugi et al. (1991). The 4.8 kbp promoter region of BC6 was amplified by PCR using specific primers, gBc6-F1 and pBC6-R1 (5′-GAGGATCCATGGCCGCGCAACAACGGCCGG-3′), and fused with the GUS gene in the pBIGRZ vector, yielding pBC6:GUS. The nucleotide sequence of the construct was confirmed before transformation into the wild-type plant, T65, as described above. Culms, leaves, nodes, and seedlings of the transgenic plants harbouring the pBC6:GUS gene were cut into pieces, fixed in 5% (w/v) agar, and hand-sectioned. Sections from the transgenic plants were stained with a solution containing 0.5 mM 5-bromo-4-chloro-3-indolyl-β-D-glucuronide, 0.5 mM potassium ferrocyanide, 0.5 mM potassium ferricyanide, and 50 mM phosphate buffer (pH 7.4) at 37 °C for 24 h, and observed under a microscope (Eclipse E400).
Quantitative analysis of BC1 and BC6 mRNAs
Relative amounts of BC1, BC3, BC6, OsCesA4, and OsCesA7 mRNA were estimated by quantitative RT-PCR. Single-stranded cDNA was synthesized from total RNA of the tissues or organs using oligo(dT)12–18 primer. The following specific primers were designed using the Primer3 program (http://frodo.wi.mit.edu/): for BC1 (Os03g0416200), BC1-RTP-F1 (5′-CGCATGAACTACACCCAGTG-3′) and BC1-RTP-R1 (5′-TCCATGAGCAGGTCGTTGTA-3′); for BC3 (Os02g0738900), BC3-RTP-F1 (5′-GGCCGAAACGATGAGATTTA-3′) and BC3-RTP-R1 (5′- AACATCAGCAGCTTGCATTG-3′); for BC6 (Os09g0422500), BC6-RTP-F1 (5′-TTAGCACGTTTGCGAGTTTG-3′) and BC6-RTP-R1 (5′-GAACTCGTCGTCCTCGTCTC-3′); for OsCesA4 (Os01g0750300), OsCesA4-RTP-F1 (5′-CTAATGCGACGAAGACGATG-3′) and OsCesA4-RTP-R1 (5′-GATTTAACGGTGCCCTCTCA-3′); for OsCesA7 (Os10g0467800), OsCesA7-RTP-F1 (5′-TCCATCTTCTCCCTCGTCTG-3′) and OsCesA7-RTP-R1 (5′-GAATCATCCATCCGGTCATC-3′); and for ACTIN1 (Os03g0718100), ACT1-RTP-F1 (5′-TTCCTACATCGCCCTGGACT-3′) and ACT1-RTP-R1 (5′-AGCCTTGGCAATCCACATCT-3′). The PCR was performed with a SYBR Premix Ex Taq kit (Takara Bio Inc., Otsu, Japan) under the following conditions: 10 s denaturing at 95 °C, 30 s annealing at 60 °C, and 20 s amplification at 72 C, 40 cycles. The PCR products were detected with Opticon 2 (Bio-Rad, Hercules, CA, USA), and the mRNA amounts relative to ACTIN1 mRNA were calculated.
Results
Reduced cellulose content in the Bc6 mutant
The rice Bc6 mutant was generated from an indica cultivar, IR68, by treatment with ethyl methanesulphonate (Singh et al., 1994). As with rice bc1 and bc7 mutants (Li et al., 2003; Yan et al., 2007), culms and leaves of Bc6 mutants were easily broken when bent. The brittle phenotype was also observed in Bc6/BC6 heterozygotes, suggesting that Bc6 is a dominant mutation as reported previously (Sanchez and Khush, 2000; Singh et al., 1994). No pleiotropic phenotypes in the appearance of Bc6 were observed, such as dwarfism or withering that are observed for rice bc3 and bc11 mutants (Iwata and Omura, 1989; Zhang et al., 2009; Hirano et al., 2010). Because the genetic background of IR68 and Bc6 could not be confirmed to date, homozygous Bc6 mutant plants were compared with the wild-type japonica cultivar, T65.
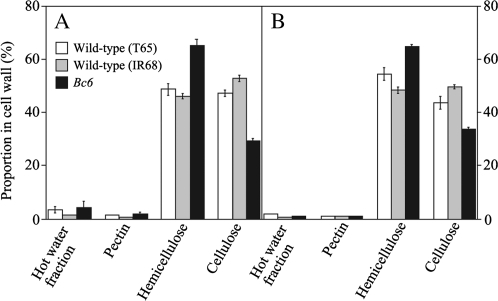
To characterize cell wall defects in the Bc6 mutant, cell wall polysaccharides were extracted, and the amount in different fractions (hot water, pectin, hemicellulose, and cellulose) were compared with those in wild-type plants. As shown in Fig. 1A, culms of Bc6 exhibited a 38% decrease in the proportion of cellulose in cell wall polysaccharides compared with that of T65 (Fig. 1). The value corresponds to a 31% decrease in the cellulose content based on weight compared with that of the wild-type plant, T65 (here, the content means the cellulose amount per fresh weight). The decrease in the cellulose content was comparable with that of bc1 (∼30% of the content based on weight) (Li et al., 2003). Conversely, the proportion of hemicellulose in the cell wall polysaccharides was increased by 34% compared with that of T65, which corresponds to a 48% increase in hemicellulose content based on weight. The increased hemicellulose content is probably a compensation reaction of rice with reduced cellulose content. Indeed, a similar increase in the hemicellulose content has also been observed for the bc1 mutant (Li et al., 2003). A decrease in the cellulose proportion and an increase in the hemicellulose proportion were also observed in the matured leaves of Bc6 mutants in samples taken at the same time as the culms, but the alterations in the components were rather mild compared with those of the culms (Fig. 1B). Essentially the same results were also obtained in a comparison of Bc6 mutants with the parental line, IR68 (Fig. 1).
Fig. 1.
Proportions of cell wall fractions. The amounts of cell wall fractions extracted from culms (A) and leaves (B) were measured in plants sampled 2 weeks after heading. Open, shaded, and filled bars indicate data for wild-type T65, IR68, and the Bc6 mutant, respectively. Values shown are averages of three plants, and the bars represent standard errors.
Sugar composition analysis by HPAEC-PAD was used to examine whether the proportion of particular polysaccharides was increased by the Bc6 mutation. In both Bc6 and wild-type plants, the hemicellulose mainly consisted of xylose, glucose, and L-arabinose (Table 2). These results indicate that Bc6 mutation did not affect the sugar composition of hemicellulose in culms; therefore Bc6 mutation does not seem to cause the accumulation of particular hemicellulosic polysaccharides in the cell walls.
Table 2.
Sugar composition of hemicellulose
| Sugar | Composition (mol %) |
||
| T65 | IR68 | Bc6 | |
| L-Arabinose | 8.5 | 13.7 | 9.3 |
| L-Fucose | 0.0 | 0.1 | 0.0 |
| Galactose | 1.3 | 4.0 | 1.5 |
| Glucose | 18.4 | 10.4 | 15.3 |
| Mannose | 0.0 | 0.0 | 0.0 |
| L-Rhamnose | 0.3 | 0.7 | 0.3 |
| Xylose | 69.6 | 69.8 | 72.0 |
| Galacturonic acid | 1.2 | 0.5 | 1.2 |
| Glucuronic acid | 0.7 | 0.9 | 0.4 |
Tissue organization of Bc6
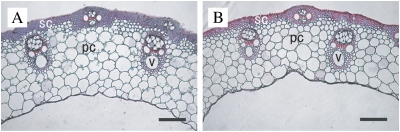
To address the effects of Bc6 mutation on tissue morphology, transverse sections of Bc6 culms stained with Safranin O and Fast Green FCF were observed microscopically. Bc6 mutants exhibited neither the altered xylem tissue organization nor the incomplete cell wall found in Arabidopsis irx mutants (Turner and Sommerville, 1997) (Fig. 2A, B).
Fig. 2.
Comparison of tissue morphology in culms. Transverse sections of the uppermost culms of T65 (A) and the Bc6 mutant (B) were double-stained with Safranin O and Fast Green FCF. Scale bars indicate 100 μm. pc, parenchymal cell; sc, sclerenchymal cell; v, vascular bundle.
To examine lignin accumulation in the tissue, transverse sections were treated with phloroglucinol to stain lignin-rich cell walls selectively. Although phloroglucinol staining appeared slightly stronger in Bc6 mutants than in T65 (see Supplementary Fig. S1 available at JXB online), quantitative analysis by the Klason method (Kirk and Obst, 1988) did not demonstrate a significant increase in lignin content in culms of Bc6 (Bc6, 2.23±0.65 mg g−1 fresh weight; T65, 2.42±0.20 mg g−1 fresh weight). Other cell wall components such as cellulose and hemicellulose may influence the staining.
Cloning of the Bc6 gene
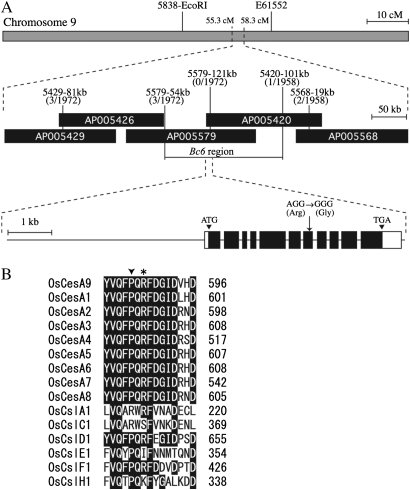
Positional cloning of the Bc6 causative gene was performed using the F2 population generated by crossing Bc6 with the japonica cultivar, Toride 1. To determine the genotype of F2 plants, several DNA markers were designed on the basis of the genomes of the japonica cultivar, Nipponbare, and the indica cultivar, 93-11 (Table 1). The BC6 gene was mapped between 5838-EcoRI and E61552 markers on chromosome 9 (Fig. 3A), consistent with its location on chromosome 9 in the classical linkage map (Sanchez and Khush, 2000). By using ∼1000 F2 plants, the BC6 locus was further narrowed to a 170 kb region covered by bacterial artificial chromosome (BAC) clones AP005579 and AP005420 (Fig. 3A). This region included the Os09g0422500 gene encoding OsCesA9, which at 79% identity shares the highest sequence similarity with AtCesA7 among 10 Arabidopsis CesAs. Sequence analysis of OsCesA9 identified a missense mutation that substitutes a highly conserved arginine residue with glycine (R588G, accession no. AB527075; Fig. 3A). The R588 residue is located in the middle region of the second cytoplasmic domain, but is not part of a QXXRW motif (791–795 in OsCesA9): the motif was shown to be required for catalytic activity of chitin synthase in yeast (Nagahashi et al., 1995; Somerville 2006). In Arabidopsis, fra5, a semi-dominant P557T mutation of AtCesA7, causes decreased cellulose content in fibre cells, perturbing cell wall thickening. Importantly, P557 of AtCesA7 corresponds to P586 of OsCesA9, a site quite near the R588 of the OsCesA9 residue mutated in Bc6 (Fig. 3B). Taken together, these results suggest that this region of CesA is essential for proper cellulose synthesis in secondary cell walls of both Arabidopsis and rice. Supporting this hypothesis, the region was found to be highly conserved among all CesAs of rice (Fig. 3B).
Fig. 3.
Cloning of BC6. (A) Map-based cloning using an F2 population of ∼1000 plants. The BC6 locus was narrowed to a 170 kb region of chromosome 9, flanked by markers 5579-54kb and 5420-101kb. BAC clones AP005579 and AP005420 encompass the OsCesA9 gene. The Bc6 mutant had the missense mutation, R588G, in the OsCesA9 gene. Black and white boxes indicate coding and untranslated regions, respectively. (B) Alignment of amino acid sequences of OsCesAs and OsCsls. The sequence of the highly conserved region of the second cytoplasmic domain of OsCesA9 was aligned with the corresponding regions of other OsCesAs and OsCsls by using the ClustalW program. Residues conserved among all OsCesA members are shown in black. The asterisk indicates the residue mutated in Bc6. An arrowhead indicates the residue corresponding to P557 of AtCesA7, which is substituted with threonine by semi-dominant fra5 mutation. Accession numbers for the sequences of OsCesAs and OsCsls are as follows: OsCesA1, Os05g0176100; OsCesA2, Os03g0808100; OsCesA3, Os07g0424400; OsCesA5, Os03g0837100; OsCesA6, Os07g0252400; OsCesA8, Os07g0208500; OsCslA1, Os02g0192500; OsCslC1, Os01g0766900; OsCslD1, Os10g0578200; OsCslE1, Os09g0478100; OsCslF1, Os07g0553000; OsCslH1, Os10g0341700.
Introduction of the Bc6 gene into the wild-type plant
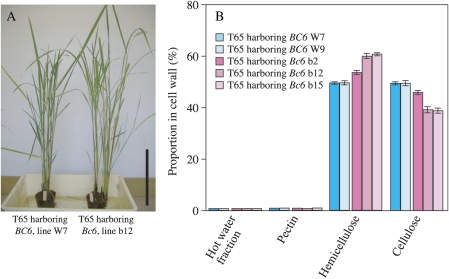
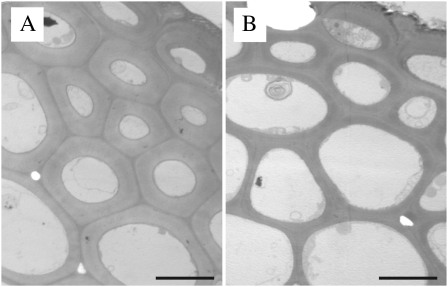
To confirm that the missense mutation in OsCesA9 is responsible for the brittle phenotype and decreased cellulose content of Bc6, the mutant Bc6 gene was introduced into the wild-type plant, T65. As a control, the missense R588G mutation was corrected by PCR, and this wild-type BC6 gene was introduced into T65. Transgenic plants harbouring mutant Bc6 (lines b12 and b15) had the brittle phenotype in culms and leaves, but did not show any morphological alterations such as dwarfism as observed for Bc6 mutants (Fig. 4A). Along with the brittle phenotype, a transgenic plant harbouring mutant Bc6 (line b12) also showed an apparently reduced proportion of cellulose in the cell walls (21% decrease in proportion), which corresponds to a 24% decrease in cellulose content based on weight compared with that harbouring the wild-type gene, BC6 (line W7, Fig. 4B). These facts confirm that Bc6 mutation is due to the R588G substitution. Introduction of the mutant Bc6 gene appeared to reduce the thickness of secondary cell walls in epidermal and sclerenchymal cells (Fig. 5). These results suggested that Bc6 protein with the R588G mutation perturbs cellulose synthesis in secondary cell walls by acting as a dominant-negative form. The cellulose deficiency in these transgenic plants was milder than that in Bc6 mutants (31% decrease based on weight) (Figs 1A, 4B), presumably because of the presence of wild-type BC6 protein in the transgenic plants. Furthermore, the decrease in the proportion of cellulose of one transgenic line harbouring the mutant Bc6 (line b2) was milder than that of other lines (lines b12 and b15) (Fig. 4B). Hence, this mutant gene behaved as a dose-responsive semi-dominant rather than fully dominant form in this experiment.
Fig. 4.
Introduction of the semi-dominant Bc6 gene into wild-type plants. Introduction of the genes, including the 4.8 kbp upstream region, into T65 yielded 15 lines of transgenic plants harbouring the mutant Bc6 gene and seven lines harbouring the wild-type BC6 gene. (A) Appearance of representative lines of wild-type BC6 (W7, left) and mutant Bc6 (b12, right) transgenic plants. The scale bar indicates 30 cm. (B) Proportions of cell wall fractions in culms of transgenic plants harbouring wild-type BC6 (lines W7 and W9, blue bars) and the mutant Bc6 (lines b2, b12, and b15, pink bars). Values shown are averages of three plants, and the bars represent standard errors.
Fig. 5.
Cell wall structure in transgenic plants. Transmission electron microscopic images of sclerenchymal cells in culms of transgenic plants harbouring the wild-type BC6 (line W7) (A) or mutant Bc6 gene (line b12) (B). Scale bars indicate 5 μm.
Expression pattern of BC6
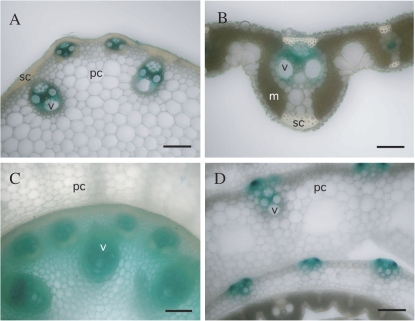
The pattern of expression of BC6 was assessed by analysing T65 plants transformed with pBC6:GUS, a binary vector in which the 4.8 kbp region upstream of the BC6 gene was fused with the GUS reporter gene. BC6 promoter activity was detected in leaves, culms, and nodes, with relatively strong expression in culm vascular bundles 2 weeks after heading (Fig. 6A–C). Promoter activity was also observed in young tissues such as developing leaves (Fig. 6D). Although Bc6 mutation appeared to reduce cell wall thickness in sclerenchymal cells (Fig. 5), promoter activity was not detected in developed sclerenchymal cells (Fig. 6A, B). It is possible that the reporter gene activity did not completely mirror the expression of the BC6 gene product, OsCesA9 protein. These patterns of BC6 promoter-driven gene expression were similar to the expression pattern of BC1 demonstrated by in situ hybridization (Li et al., 2003).
Fig. 6.
Pattern of BC6 promoter-driven GUS activity. The transverse sections of a culm (A), a mature leaf (B), a node (C), and a young leaf (D) in pBC6:GUS rice plants were GUS stained and observed under an optical microscope. Identical expression patterns were observed for three independent pBC6:GUS lines. Scale bars indicate 100 μm in A, C, and D, and 250 μm in B. m, mesophyll cell; pc, parenchymal cell; sc, sclerenchymal cell; v, vascular bundle.
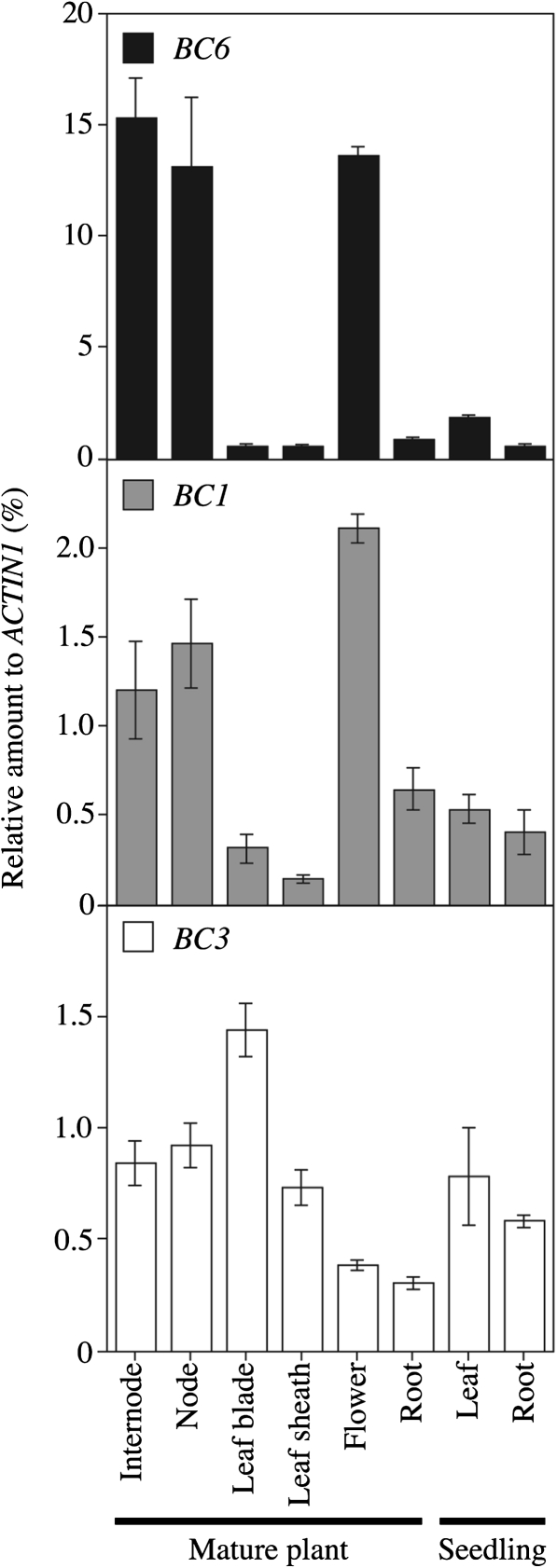
In Arabidopsis, the COBL4 gene is co-expressed with the secondary cell wall-specific CesA genes and is presumed to play a role in the cellulose synthesis of secondary cell walls, although the precise molecular functions of COBRA-like proteins remain unclear (Schindelman et al., 2001; Roudier et al., 2005). To examine the relationship between the secondary cell wall-specific CesA and COBRA-like proteins in rice, BC6 and BC1 mRNAs were quantitated in several tissues. Consistent with the results of the pBC6:GUS analysis, relatively high levels of BC6 mRNA were detected in culms and nodes (Fig. 7A). The level of Bc6 mRNA in roots was relatively low. Indeed, the brittle phenotype in roots was not clear compared with those in culms and leaves (data not shown). Despite the apparent brittle phenotype in the leaves of Bc6 mutants, the level was low in both leaf blades and sheaths. Importantly, both BC6 and BC1 were highly expressed in culms, nodes, and flowers (Fig. 7A, B). These results indicated that BC6 and BC1 are co-expressed during development of secondary cell walls. On the other hand, the expression of BC6 was not related to that of BC3, suggesting that BC6 and BC3 are differently regulated.
Fig. 7.
Comparison between BC6, BC1, and BC3 expression patterns. The amounts of BC6, BC1, and BC3 mRNA relative to that of ACTIN1 mRNA were determined by quantitative RT-PCR. The template cDNA was prepared using total RNA extracted from two wild-type plants. Values are averages of three technical replicates, and the bars represent standard errors.
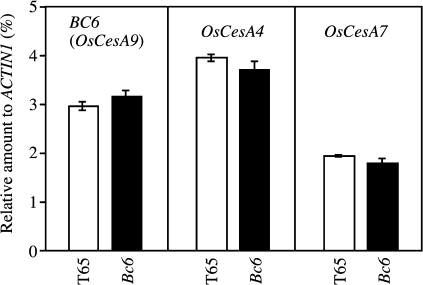
The effect of the mutation on the expression of Bc6 was also examined in developing leaf blades. Bc6 mutants showed accumulation of BC6 mRNA comparable with T65 (Fig. 8). Furthermore, Bc6 mutation barely influenced the mRNA levels of other CesA genes, OsCesA4 and OsCesA7, which are expected to participate in cellulose synthesis in secondary cell walls, together with BC6 (OsCesA9).
Fig. 8.
Expression levels of BC6, OsCesA4, and OsCesA9 in the Bc6 mutant. Relative amounts of BC6, OsCesA4, and OsCesA9 mRNA in developing leaf blades of Bc6 mutants were compared with those in T65. Values shown are averages of three plants, and the bars represent standard errors.
Discussion
Brittle phenotypes caused by mutation of OsCesA9
The bc loci have been used for genetic analysis of rice for >70 years, but several causative genes are yet to be cloned. The present study demonstrates that the BC6 gene encodes OsCesA9, a secondary cell wall-specific CesA. Insertional disruption of OsCesA9 by the endogenous retrotransposon Tos17 was previously reported (Tanaka et al., 2003). While ND2395 and NF1011 mutations in that study were recessive, the Bc6 missense mutation, R588G, in OsCesA9 reported here is semi-dominant. The phenotypes of Bc6 were milder than those of ND2395 and NF1011, i.e. Bc6 caused a 31% decrease in cellulose content relative to T65, while ND2395 and NF1011 showed a 91% and 46% decrease, respectively, compared with the wild-type plant. Furthermore, Bc6 did not affect plant growth, whereas ND2395 and NF1011 plants exhibited dwarfism, small leaves, and thin culms. These differences suggest that the transposon insertions in OsCesA9 affect the formation of primary cell walls, while the Bc6 missense mutation, R588G, does not. Indeed, a link between secondary cell wall integrity and primary cell wall deposition and remodelling has also been reported in Arabidopsis. The AtCesA7 mutation, murus10, results in altered structure of pectins and xyloglucan, leading to dwarfism (Bosca et al., 2006). In the case of the OsCesA4 gene, bc7 mutation comprising deletions in exon 10 and intron 10 does not affect growth and development, but the bc11 missense mutation, G858R, causes severe dwarfism (Yan et al., 2007; Zhang et al., 2009). The severity of the phenotype probably depends on both mutation type and site.
Dominant-negative form of CesA
The semi-dominant brittle phenotype of Bc6 was found to be caused by a missense mutation, R588G, in the second cytoplasmic domain of OsCesA9. Importantly, this mutated residue was located near to P586, and this site corresponds to P557 of AtCesA7 that is altered in the semi-dominant fra5 mutant of Arabidopsis. This region, which is distinct from the putative catalytic motif, QXXRW, is highly conserved among the CesA genes of higher plants (Fig. 3). Such conservation is not seen in the corresponding region in cellulose synthase-like proteins (Csls) catalysing related biosynthetic reactions of polysaccharides such as the β-1,4-mannan, β-1,3:1,4-glucan, and the β-1,4-glucan backbone of xyloglucan (Liepman et al., 2005; Burton et al., 2006; Cocuron et al, 2007). These facts suggest the possibility that the region has a function other than the catalysis of β-1,4-glucan synthesis. In the study on the Arabidopsis fra5 mutant, Zhong et al. (2003) suggested that the missense P557T mutation of AtCesA7 affects the interaction between CesA proteins or between CesA and other cellular components. In the present study, introduction of the semi-dominant Bc6 mutant gene into T65 caused decreased cellulose content and brittle phenotype. It is possible that the presence of the mutated CesA protein interferes with proper formation of functional CSC. Supporting this hypothesis, the phenotypes of the transgenic plants were milder than those of Bc6 homozygous mutants, possibly because of the presence of wild-type BC6 protein. Involvement of the second cytoplasmic domain in the formation of CSC has not yet been demonstrated, whereas the interaction of CesAs through their N-terminal RING-finger-like motifs, depending on the redox state, has been demonstrated (Kurek et al., 2002). It remains unknown, however, how the CesA dimers formed through interaction of RING-finger-like motifs assemble to form the rosette structure that includes perhaps 36 CesAs. An epitope tagging analysis of the interaction between secondary cell wall-specific AtCesAs detected higher order CesA oligomerization beyond dimerization (Atanassov et al., 2009). The conserved region of the second cytoplasmic domain of OsCesA9, including R588 and P586, could be a site for a protein–protein interaction to form such higher order oligomerization.
Cellulose synthesis of secondary cell walls in rice
Quantitative analysis of gene expression revealed correlated expression of BC6 and BC1 in rice. BC6 and BC1 represent rice counterparts of AtCesA7/IRX3 and AtCOBL4/IRX6, respectively. AtCesA7/IRX3 and AtCOBL4/IRX6 are co-expressed in tissues during secondary cell wall development, and loss-of-function mutation of either of these genes results in diminished cellulose content and loss of mechanical strength of the plant body (Brown et al., 2005). In addition, disruptions of OsCesA4 and OsCesA7, the respective rice orthologues of AtCesA8/IRX1 and AtCesA4/IRX5, cause the brittle phenotype and decreased cellulose content in rice (Tanaka et al., 2003; Yan et al., 2007; Zhang et al., 2009). The shared requirement for these components in secondary cell wall cellulose synthesis suggests that the mechanism of cellulose synthesis is highly conserved between Arabidopsis and rice.
On the other hand, Gramineae plants, including rice, generally show resistance to isoxaben, a strong inhibitor of cellulose synthesis in dicotyledonous plants, including Arabidopsis (Desprez et al., 2002). The differential sensitivity to isoxaben suggests that the mechanism of cellulose synthesis differs at least partially between rice and Arabidopsis. Future analyses of rice cell wall mutants, including bc mutants, should help to clarify the differences in cellulose synthesis between Gramineae and dicotyledonous plants.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Lignin staining of sclerenchymal cells. Sclerenchymal cells of T65 (A) and the Bc6 mutant (B) were stained with phloroglucinol-HCl. Scale bars indicate 50 μm. pc, parenchymal cell; sc, sclerenchymal cell; v, vascular bundle.
Acknowledgments
We are grateful to Dr Khush of IRRI (Laguna, Philippines) for providing the Bc6 mutant (RGS number 420) and IR68. This research was supported in part by a Grant-in-aid for Scientific Research to TK (no. 22770030) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Glossary
Abbreviations
- At
Arabidopsis thaliana
- BAC
bacterial artificial chromosome
- bc
brittle culm
- CesA
cellulose synthase catalytic subunit
- CSC
cellulose synthase complex
- Csl
cellulose synthase-like protein
- fra
fragile fiber
- HPAEC-PAD
high-performance anion-exchange chromatography with pulsed amperometric detection
- irx
irregular xylem
- ixr
isoxaben-resistant
- Os
Oryza sativa
- pBC6:GUS
PromoterBC6:β-glucuronidase
- T65
Taichung 65
References
- Akiyama K, Nakamura S, Suzuki T, Wisniewska I, Sasaki N, Kawasaki S. Development of a system of rice transformation with long genome inserts for their functional analysis for positional cloning. Plant and Cell Physiology. 1997;38:s94. [Google Scholar]
- Aohara T, Kotake T, Kaneko Y, Takatsuji H, Tsumuraya Y, Kawasaki S. Rice BRITTLE CULM 5 (BRITTLE NODE) is involved in secondary cell wall formation in the sclerenchyma tissue of nodes. Plant and Cell Physiology. 2009;50:1886–1897. doi: 10.1093/pcp/pcp133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanassov, Pittman JK, Turner SR. Elucidating the mechanism of assembly and subunit interaction of the cellulose synthase complex of Arabidopsis secondary cell walls. Journal of Biological Chemistry. 2009;284:3833–3841. doi: 10.1074/jbc.M807456200. [DOI] [PubMed] [Google Scholar]
- Bosca S, Barton CJ, Taylor NG, Ryden P, Neumetzler L, Pauly M, Roberts K, Seifert GJ. Interactions between MUR10/CesA7-dependent secondary cellulose biosynthesis and primary cell wall structure. Plant Physiology. 2006;142:1353–1363. doi: 10.1104/pp.106.087700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DM, Zeef LAH, Ellis J, Goodacre R, Turner SR. Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. The Plant Cell. 2005;17:2281–2295. doi: 10.1105/tpc.105.031542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk DH, Liu B, Zhong R, Morrison WH, Ye Z-H. A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. The Plant Cell. 2001;13:807–827. [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Wilson SM, Hrmova M, Harvey AJ, Shirley NJ, Medhurst A, Stone BA, Newbigin EJ, Bacic A, Fincher GB. Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-β-d-glucans. Science. 2006;311:1940–1942. doi: 10.1126/science.1122975. [DOI] [PubMed] [Google Scholar]
- Ching A, Dhugga KS, Appenzeller L, Meeley R, Bourett TM, Howard RJ, Rafalski A. Brittle stalk 2 encodes a putative glycosylphosphatidylinositol-anchored protein that affects mechanical strength of maize tissues by altering the composition and structure of secondary cell walls. Planta. 2006;224:1174–1184. doi: 10.1007/s00425-006-0299-8. [DOI] [PubMed] [Google Scholar]
- Cocuron J-C, Lerouxel O, Drakakaki G, Alonso AP, Liepman AH, Keegstra K, Raikhel N, Wilkerson CG. A gene from the cellulose synthase-like C family encodes a β-1,4 glucan synthase. Proceedings of the National Academy of Sciences, USA. 2007;104:8550–8555. doi: 10.1073/pnas.0703133104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T, Vernhettes S, Fagard M, Refrégier G, Desnos T, Aletti E, Py N, Pelletier S, Höfte H. Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant Physiology. 2002;128:482–490. doi: 10.1104/pp.010822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. Colorimetric method for determination of sugars and related substances. Analytical Chemistry. 1956;28:350–356. [Google Scholar]
- Ellis C, Karafyllidis I, Wasternack C, Turner JG. The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. The Plant Cell. 2002;14:1557–1566. doi: 10.1105/tpc.002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C, Turner JG. The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. The Plant Cell. 2001;13:1025–1033. doi: 10.1105/tpc.13.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Blanco C, Feng DX, Hu J, et al. Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. The Plant Cell. 2007;19:890–903. doi: 10.1105/tpc.106.048058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K, Kotake T, Kamihara K, Tsuna K, Aohara T, Kaneko Y, Takatsuji H, Tsumuraya Y, Kawasaki S. Rice BRITTLE CULM 3 (BC3) encodes a classical dynamin OsDRP2B essential for proper secondary cell wall synthesis. Planta. 2010;232:95–108. doi: 10.1007/s00425-010-1145-6. [DOI] [PubMed] [Google Scholar]
- Hood EE, Helmer GL, Fraley RT, Chilton M- D. The hypervirulence of Agrobacterium tumefaciens A281 is encoded in a region of pTiBo542 outside of T-DNA. Journal of Bacteriology. 1986;168:1291–1301. doi: 10.1128/jb.168.3.1291-1301.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa M, Kuroyama H, Takeuchi Y, Tsumuraya Y. Characterization of pectin methyltransferase from soybean hypocotyls. Planta. 2000;210:782–791. doi: 10.1007/s004250050680. [DOI] [PubMed] [Google Scholar]
- Iwata N, Omura T. Linkage studies in rice (Oryza sativa L). On some mutants derived from chronic gamma irradiation. Journal of the Faculty of Agriculture, Kyushu University. 1977;21:117–127. [Google Scholar]
- Kirk TK, Obst JR. Lignin determination. Methods in Enzymology. 1988;161:89–101. [Google Scholar]
- Kokubo A, Kuraishi S, Sakurai N. Culm strength of barley: correlation among maximum bending stress, cell wall dimensions, and cellulose content. Plant Physiology. 1989;91:876–882. doi: 10.1104/pp.91.3.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokubo A, Sakurai N, Kuraishi S, Takeda K. Culm brittleness of barley (Hordeum vulgare L.) mutants is caused by smaller number of cellulose molecules in cell wall. Plant Physiology. 1991;97:509–514. doi: 10.1104/pp.97.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Suzuka I, Ohashi Y, Murakami T, Arai Y. Upstream sequences of rice proliferating cell nuclear antigen (PCNA) gene mediate expression of PCNA–GUS chimeric gene in meristems of transgenic tobacco plants. Nucleic Acids Research. 1991;19:1571–1576. doi: 10.1093/nar/19.7.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurek I, Kawagoe Y, Jacob-Wilk D, Doblin M, Delmer D. Dimerization of cotton fiber cellulose synthase catalytic subunits occurs via oxidation of the zinc-binding domains. Proceedings of the National Academy of Sciences, USA. 2002;20:11109–11114. doi: 10.1073/pnas.162077099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Qian Q, Zhou Y, Yan M, Sun L, Zhang M, Fu Z, Wang Y, Han B, Pang X, Chen M, Li J. BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. The Plant Cell. 2003;15:2020–2031. doi: 10.1105/tpc.011775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman AH, Wilkerson CG, Keegstra K. Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proceedings of the National Academy of Sciences, USA. 2005;102:2221–2226. doi: 10.1073/pnas.0409179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahashi S, Sudoh M, Ono N, Sawada R, Yamaguchi E, Uchida Y, Mio T, Takagi M, Arisawa M, Yamada-Okabe H. Characterization of chitin synthase 2 of Saccharomyces cerevisiae: implication of two highly conserved domains as possible catalytic sites. Journal of Biological Chemistry. 1995;270:13961–13967. doi: 10.1074/jbc.270.23.13961. [DOI] [PubMed] [Google Scholar]
- Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM. Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proceedings of the National Academy of Sciences, USA. 1996;93:12637–12642. doi: 10.1073/pnas.93.22.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier F, Fernandez AG, Fujita M, et al. COBRA, an Arabidopsis extracellular glycosyl-phosphatidyl inositol-anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. The Plant Cell. 2005;17:1749–1763. doi: 10.1105/tpc.105.031732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez AC, Khush GS. Chromosomal localization of five mutant genes in rice, Oryza sativa, using primary trisomics. Plant Breeding. 2000;119:84–86. [Google Scholar]
- Scheible W-R, Eshed R, Richmond T, Delmer D, Somerville C. Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis Ixr mutants. Proceedings of the National Academy of Sciences, USA. 2001;98:10079–10084. doi: 10.1073/pnas.191361598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelman G, Morikami A, Jung J, Baskin TI, Carpita NC, Derbyshire P, McCann MC, Benfey PN. COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in. Arabidopsis. Genes and Development. 2001;15:1115–1127. doi: 10.1101/gad.879101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindhu A, Langewisch T, Olek A, Multani DS, McCann MC, Vermerris W, Carpita NC, Johal G. Maize brittle stalk2 encodes a COBRA-like protein expressed in early organ development but required for tissue flexibility at maturity. Plant Physiology. 2007;145:1444–1459. doi: 10.1104/pp.107.102582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K, Multani DS, Khush GS. A new brittle culm mutant in rice. Rice Genetic Newsletter. 1994;11:91–92. [Google Scholar]
- Somerville C. Cellulose synthesis in higher plants. Annual Review of Cell and Developmental Biology. 2006;22:53–78. doi: 10.1146/annurev.cellbio.22.022206.160206. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Murata K, Yamazaki M, Onosato K, Miyao A, Hirochika H. Three distinct rice cellulose synthase catalytic subunit genes required for cellulose synthesis in the secondary wall. Plant Physiology. 2003;133:73–83. doi: 10.1104/pp.103.022442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR. Interactions among three distinct CesA proteins essential for cellulose synthesis. Proceedings of the National Academy of Sciences, USA. 2003;100:1450–1455. doi: 10.1073/pnas.0337628100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Laurie S, Turner SR. Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. The Plant Cell. 2000;12:2529–2539. doi: 10.1105/tpc.12.12.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Scheible W-R, Cutler S, Somerville CR, Turner SR. The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. The Plant Cell. 1999;11:769–779. doi: 10.1105/tpc.11.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toki S. Rapid and efficient Agrobacterium-mediated transformation in rice. Plant Molecular Biology Reporter. 1997;15:16–21. [Google Scholar]
- Turner SR, Somerville CR. Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. The Plant Cell. 1997;9:689–701. doi: 10.1105/tpc.9.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Sasaki T. Large-scale EST sequencing in rice. Plant Molecular Biology. 1997;35:135–144. [PubMed] [Google Scholar]
- Yan C, Yan S, Zeng X, Zhang Z, Gu M. Fine mapping and isolation of Bc7(t), allelic to OsCesA4. Journal of Genetics and Genomics. 2007;34:1019–1027. doi: 10.1016/S1673-8527(07)60115-5. [DOI] [PubMed] [Google Scholar]
- Zhang B, Deng L, Qian Q, Xiong G, Zeng D, Li R, Guo L, Li J, Zhou Y. A missense mutation in the transmembrane domain of CESA4 affects protein abundance in the plasma membrane and results in abnormal cell wall biosynthesis in rice. Plant Molecular Biology. 2009;71:509–524. doi: 10.1007/s11103-009-9536-4. [DOI] [PubMed] [Google Scholar]
- Zhong R, Burk DH, Morrison WH III, Ye Z- H. A kinesin-like protein is essential for oriented deposition of cellulose microfibrils and cell wall strength. The Plant Cell. 2002;14:3101–3117. doi: 10.1105/tpc.005801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Burk DH, Morrison WH III, Ye Z- H. FRAGILE FIBER3, an Arabidopsis gene encoding a type II inositol polyphosphate 5-phosphatase, is required for secondary wall synthesis and actin organization in fiber cells. The Plant Cell. 2004;16:3242–3259. doi: 10.1105/tpc.104.027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Morrison WH III, Freshour GD, Hahn MG, Ye Z- H. Expression of a mutant form of cellulose synthase AtCesA7 causes dominant negative effect on cellulose biosynthesis. Plant Physiology. 2003;132:786–795. doi: 10.1104/pp.102.019331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.