Abstract
Plant retinoblastoma-related (RBR) proteins are primarily considered as key regulators of G1/S phase transition, with functional roles in a variety of cellular events during plant growth and organ development. Polyclonal antibody against the C-terminal region of the Arabidopsis RBR1 protein also specifically recognizes the alfalfa 115 kDa MsRBR protein, as shown by the antigen competition assay. The MsRBR protein was detected in all cell cycle phases, with a moderate increase in samples representing G2/M cells. Antibody against the human phospho-pRb peptide (Ser807/811) cross-reacted with the same 115 kDa MsRBR protein and with the in vitro phosphorylated MsRBR protein C-terminal fragment. Phospho-MsRBR protein was low in G1 cells. Its amount increased upon entry into the S phase and remained high during the G2/M phases. Roscovitine treatment abolished the activity of alfalfa MsCDKA1;1 and MsCDKB2;1, and the phospho-MsRBR protein level was significantly decreased in the treated cells. Colchicine block increased the detected levels of both forms of MsRBR protein. Reduced levels of the MsRBR protein in cells at stationary phase or grown in hormone-free medium can be a sign of the division-dependent presence of plant RBR proteins. Immunolocalization of the phospho-MsRBR protein indicated spots of variable number and size in the labelled interphase nuclei and high signal intensity of nuclear granules in prophase. Structures similar to phospho-MsRBR proteins cannot be recognized in later mitotic phases. Based on the presented western blot and immunolocalization data, the possible involvement of RBR proteins in G2/M phase regulation in plant cells is discussed.
Keywords: Auxin, cell cycle, cell synchronization, colchicine, cyclin-dependent kinases, phosphorylation, prophase, retinoblastoma-related protein, roscovitine, immunolocalization
Introduction
In the last decades, substantial progress has been made in the discovery of genes and protein complexes regulating cell division in different organs of higher plants, as discussed in recent reviews (Berckmans and De Veylder, 2009; Nieuwland et al., 2009; Sabelli and Larkins, 2009; Boruc et al., 2010). From these studies one can conclude that several components and mechanisms are functional in plant cell cycle control that also play a role in other eukaryotic cells. In addition to this phylogenetic conservation, plant-specific regulatory elements are responsible for perception or mediation of developmental and environmental signals to modulate cell division activities during completion of the plant life cycle. One surprising example is the family of plant retinoblastoma-related (RBR) proteins sharing homology with the human tumour suppressor retinoblastoma (pRb) protein and operating in gametophyte and sporophyte development (Johnston et al., 2010) or in the maintenance of meristem integrity and function (Borghi et al., 2010).
Maize cDNA clones encoding structural and functional counterparts of the pRb protein from higher eukaryotes were first identified from endosperm and seedling libraries (Grafi et al., 1996; Xie et al., 1996; Ach et al., 1997). The list of cloned plant RBR genes is continuously expanding, and the phylogenetic tree reveals significant differences between dicot and monocot plant species as far as the number of RBR genes is concerned. While dicot plants only have a single gene, monocot cereal species carry at least two distinct genes with characteristic expression patterns (Lendvai et al., 2007; Miskolczi et al., 2007). Microarray analysis showed constitutive expression of the Arabidopsis RBR1 gene during cell cycle progression in synchronized cells (de Almeida et al., 2009).
Studies on the mammalian cell division cycle have revealed that pRb proteins act as ‘pocket domain’ proteins regulating G1 to S phase transition through phosphorylation-dependent interaction with the E2F family transcription factors, repressing or activating genes required for cell cycle progression. The Rb–E2F complexes are involved in several basic cellular events such as oncological transformation, apoptosis, or cell differentiation (reviewed by Korenjak and Brehm, 2005; Poznic, 2009).
Similarly, in plants, the RBR protein functions are controlled by phosphorylation and protein–protein interactions (reviewed by Gutierrez, 1998; Durfee et al., 2000). Like human pRB proteins, plant RBR proteins are composed of an N-terminal region, A and B domains in the pocket region, and a C-terminal domain. These proteins have several potential cyclin-dependent kinase (CDK) phosphorylation sites (Durfee et al., 2000; Boniotti and Gutierrez, 2001; Miskolczi et al., 2007). Divergent functions of CDK–cyclin complexes in plant cell cycle control are determined by specific binding between various members of the kinase and cyclin families (Dudits et al., 2007; Nieuwland et al., 2007). More than 150 CDK proteins from 41 species can be categorized into eight classes: CDKA–CDKG and the CDK-like kinases (CDKLs) by using the predicted cyclin-binding motifs as an essential criterion. Characteristic members of the CDKA class share the canonical PSTAIRE motif, and their interactions with D-type cyclins are required for the formation of active kinase complexes that can phosphorylate RBR proteins, as was shown for tobacco, Arabidopsis, and wheat (Nakagami et al., 1999; Boniotti and Gutierrez, 2001). These plant cyclins may have the LxCxE motif that mediates the binding of a variety of proteins to RBR proteins (Soni et al., 1995; Huntley et al., 1998). The NtRBR1 protein of tobacco was phosphorylated by the cyclin D3;3–CDKA complex and this kinase activity was detected in extracts from G1 and S phase cells of synchronized tobacco BY-2 culture (Nakagami et al., 2002). The recombinant C-terminal domain [glutathione S-transferase (GST)–ZmRBR-C] of the maize RBR protein could serve as a substrate for p13SUC1-bound kinase complex from synchronized wheat cells. This kinase activity remained high for several hours after release from the hydroxyurea (HU) block (Boniotti and Gutierrez, 2001). Kawamura et al. (2006) generated antibodies against the C-terminal region of the NtRBR1 protein and different phosphoserine peptides containing sequences from NtRBR1. The NtRBR1 protein was phosphorylated by both CDKA and CDKB immunoprecipitated from actively growing cells. Antibodies recognizing specific phosphoserines cross-reacted differentially with the NtRBR1 protein in various phases of the cell cycle. The recently described PsRBR1 protein from pea was found to be able to form a complex with D-type cyclin (Pissa; cyclin D3;1) containing the canonical pRb-binding LxCxE motif in the N-terminal region (Shimizu-Sato et al., 2008). The authors detected the phosphorylated forms of the PsRBR1 protein by immunoprecipitation after in vivo labelling with [32P]inorganic phosphate.
Since cellular structures undergo dynamic changes during progression through the consecutive phases of the cell division cycle, therefore the localization of regulatory proteins is a key determinant in functionality. Boruc et al. (2010) presented the spatiotemporal occurrence of 60 Arabidopsis cell cycle proteins fused to green fluorescent protein in Arabidopsis and tobacco cells. In this study the AtRBR1 protein was shown to be localized in the nucleus of interphase cells. So far there have been no reports on the localization of phospho-RBR proteins in plant cells.
According to the dominant view, RBR proteins are responsible for a major G1 checkpoint, blocking S phase entry and cell growth. In this work, the molecular tools for monitoring both the MsRBR and the phospho-MsRBR proteins in cultured Medicago cells are extended. Limited fluctuation in the MsRBR protein level during the whole cell cycle including G2/M phases is shown. Western blot analysis revealed a lower level of phospho-MsRBR protein in G1 cells as compared with S or G2/M cells. Localization of phospho-MsRBR protein in spots of interphase nuclei and in nuclear granules in prophase cells is a novel finding in plant RBR research. Taking together the presented immunodetection data, a functional role for plant RBR proteins in mitotic events is postulated. Plant hormones can directly influence plant cell division activity. Reduced amounts of MsRBR and phospho-MsRBR proteins in cells during the stationary phase of growth, or the lack of MsRBR protein accumulation in non-dividing cells cultured in hormone-free medium for a prolonged time suggest a link between the presence of RBR proteins and plant cell division activity.
Materials and methods
Plant cell cultures, cell synchronization, and hormone starvation experiments
Medicago sativa ssp. varia genotype A2 cell suspension culture was maintained by weekly subculturing in Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) supplemented with 2 mg l−1 2,4-dichlorophenoxyacetic acid (2,4-D) and 0.2 mg l−1 kinetin according to Bögre et al. (1988).
Synchronization of the cell cycle was started by a 1:4 dilution of a 7-day-old alfalfa suspension culture. After 3 d cells were treated with 10 mM HU (Sigma, St Louis, MO, USA) for 36 h. The cells were then washed three times with pre-conditioned MS medium (taken from an A2 suspension culture of the same age after subculture) and cultured further for synchronous growth in the original volume (Magyar et al., 1993). Second inhibitors, 100 μM roscovitine or 0.05% colchicine, were used to stop cells in the desired cell cycle phases by adding them to the medium, 2 h after washing the HU from the cells. Samples were collected at the indicated time points for protein extraction, cytology, and flow cytometric analysis.
The hormone starvation and re-addition experiment was started by extensive washing of a 7-day-old Medicago A2 cell culture with hormone-free MS medium. Subsequently the cell culture was divided and grown in either hormone-free MS medium or in the presence of 1 mg l−1 2,4-D with 0.2 mg l−1 kinetin for 5 d. Afterwards hormones were re-added to the medium lacking growth hormones (1 mg l−1 2,4-D with 0.2 mg l−1 kinetin). The parallel cell culture was grown in the presence of 1 mg or 2 mg of 2,4-D and samples were taken at the indicated time points.
Protein extraction and immunoblotting
Protein extracts from cells were prepared according to Magyar et al. (1997) in buffer containing 25 mM TRIS-HCl pH 7.6, 15 mM MgCl2, 15 mM EGTA, 75 mM NaCl, 60 mM β-glycerophosphate, 1 mM dithiothreitol (DTT), 0.1% NP-40, 0.1 mM Na3VO4, 1 mM NaF, 1 mM phenylmethylsulphonyl fluoride (PMSF), protease inhibitors (Complete, Roche, Mannheim, Germany), and PhosSTOP phosphatase inhibitor cocktail (Roche, Mannheim, Germany). A 100 μg aliquot or the indicated amount of cleared protein extracts were subjected to SDS–PAGE with the appropriate concentration of acrylamide from 6% to 12% and transferred to polyvinyldifluoride (PVDF; Millipore). The amount of RBR protein was determined by immunoblot analysis as described previously by Horvath et al. (2006). To test the cross-reaction of anti-AtRBR1 antibody with the MsRBR protein, 5 ng of purified recombinant His-tagged C-terminal fragment of the MsRBR protein was used in immunoblot assays. In order to test specific cross-reaction of anti-AtRBR1 antibody with the MsRBR1 protein, an antigen competition assay was performed. The antibody was pre-incubated with the purified (His)6-tagged C-terminal part of the MsRBR1 protein at 4 °C for 3 h prior to use in the immunoblotting assay. The immunoblotting experiment was run in duplicate; the first time with antibody pre-incubated with recombinant protein and the second time with a control antibody (not pre-incubated). All other parameters of the immunoblotting experiment remained constant throughout the experiment. Polyclonal antibody produced against a phosphopeptide corresponding to residues around Ser807/811 of human Rb protein [Phospho-Rb (Ser807/811) Antibody #9308, Cell Signaling Technology] was used according to the manufacturer's instructions to monitor the phospho-MsRBR protein level. To demonstrate cross-reaction of the antibody with phospho-MsRb protein in vitro, phosphorylation was carried out. The purified recombinant His-tagged C-terminal fragment of the MsRBR protein was phosphorylated by the CDK complex bound to p13SUC1 (Sigma). Samples were taken at the indicated time points. The reaction was monitored using an immunoblot (with anti-phospho-pRb antibody) and by detecting incorporated [32P]inorganic phosphate (using PhosphorImager SI, Molecular Dynamics). Anti-MedsaCDKA1;1 or anti-MedsaCDKB2;1 antibodies were used according to Magyar et al. (1997).
Immunoprecipitation and kinase activity assays
For the immunoprecipitation procedures, equal amounts of protein extracts in homogenization buffer (see above) were incubated with 2–4 μg of antibodies (anti-MsCDKA1;1 and anti-MsCDKB2;1). The kinase reaction was initiated by the addition of 30 μl of reaction mixture (50 mM TRIS-HCl, pH 7.6, 15 mM MgCl2, 5 mM EGTA, 1 mM DTT, 10 μM ATP) containing 2.5 μg of histone H1 or GST–MsRBR substrate and 0.25 MBq of [32P]ATP. Detailed descriptions of these methods have been given previously (Magyar et al., 1993).
In vitro phosphatase treatment
Samples from M. sativa ssp. varia genotype A2 suspension culture were harvested both 4 d (exponential growing phase) and 7 d (stationary phase) after subculturing. Protein extracts were prepared in buffer containing 20 mM HEPES pH 7.6 and EDTA-free complete protease inhibitor cocktail (Roche, Mannheim, Germany). A 50 μg aliquot of total protein was incubated in calf intestinal alkaline phosphatase (CIAP) buffer at 37 °C for 30 min in the presence or absence of 5 U of enzyme. The reactions were stopped by boiling in SDS loading buffer. Samples were separated on a 6% polyacrylamide gel, then blotted onto a PVDF membrane.
Flow cytometry
For flow cytometric analysis, 1 ml of cell culture was filtered with miracloth, then the cells were chopped with a sharp razor blade in Galbraith's buffer (45 mM MgCl2, 20 mM MOPS, 30 mM sodium citrate, 0.1% Triton X-100, pH 7.0) in 6 cm plastic Petri dishes on ice (Galbraith et al., 1983). Nuclei (in 1 ml of buffer) were filtered into 1.5 ml microfuge tubes through 20 μm sieves. Nuclei were stained with propidium iodide (4 μg ml−1).
Nuclei (10×103) were used for flow cytometric determination of the relative DNA content with a FACSCalibur flow cytometer from Becton Dickinson (Franklin Lakes, NJ, USA). Cell cycle phase analysis was carried out by the ModFit® software.
Immunolocalization and confocal laser scanning microscopy
A 2-day-old suspension culture of Medicago was fixed for 15 min in 4% (w/v) formaldehyde solution in phosphate-buffered saline (PBS) with 0.1% Triton X-100. Fixed cells were washed twice for 5 min with PBS and once for 5 min with 0.5% MES (2-N-morpholinoethanesulphonic acid) at pH 5.8. Cell walls were partially digested for 35 min with chromatographically purified lyophilized enzymes from Worthington Biochemical Corporation (Lakewood, NJ, USA). The enzyme mixture was 1% cellulase and 0.5% pectinase in 0.5% MES, pH 5.8. After washing with PBS (3×5 min), cells were settled on poly-L-lysine-coated coverslips. Excess solution was removed and cells were permeabilized for 20 min with 0.5% Triton X-100 in PBS to allow antibody penetration, followed by 2×5 min washes with PBS and 1×20 min with PBS+ [PBS with 5% (v/v) fish gelatin]. Cells were incubated for 1.5 h at 37 °C (or overnight at 4 °C) with primary antibodies in PBS+. Chicken anti-retinoblastoma antibody and rabbit anti-phospho-Rb antibodies were used at a 1:200 and 1:100 dilution, respectively. Following 3×5 min washes with PBS+, cells were incubated for 1 h at 37 °C with anti-chicken fluorescein isothiocyanate (FITC)-conjugated and anti-rabbit AlexaFluor 488-conjugated antibodies (Invitrogen) diluted 1:300 in PBS+. Cells were then washed 3×5 min with PBS containing 100 ng ml−1 DNA staining dye DAPI (4',6-diamidino-2-phenylindole; Invitrogen) and mounted with Fluoromount-G anti-fade mounting solution (Southern Biotech). Confocal laser scanning microscopy was performed using an Olympus Fluoview FV1000 confocal laser scanning microscope (Olympus Life Science Europa GmbH, Hamburg, Germany). The microscope configuration is as follows: objective lenses, UPLSAPO ×20 (dry, NA:0.75), UPLFLN ×40 (oil, NA:1.3), and UPLSAPO ×60 (oil, NA:1.35); sampling speed, 4 μs pixel−1; line averaging, 2×; scanning mode, sequential unidirectional; excitation, 405 nm (DAPI) and 488 nm (FITC and AlexaFluor 488); laser transmissivity, <1% and 5% were used for DAPI and Alexa Fluor 488, respectively; main dichroic beamsplitter, DM405/488; intermediate dichroic beamsplitter, SDM 490; DAPI signal was detected between 425 nm and 475 nm, FITC and Alexa Fluor 488 were detected between 500 nm and 600 nm, and pseudocoloured red and green, respectively. Differential interference contrast (DIC) images were captured with a 488 nm laser line. Composite images were prepared using the ‘import image sequence’ and ‘make montage’ functions of the ImageJ software (National Institutes of Health, USA, version 1.41).
Results
Immunodetection of MsRBR and phospho-MsRBR proteins in extracts from cultured alfalfa cells
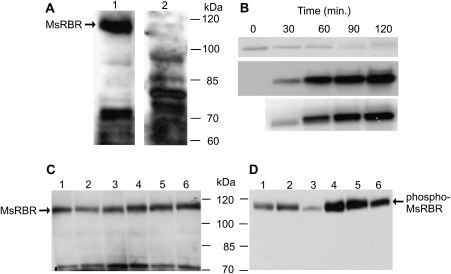
At present a polyclonal antibody is available that was produced against the recombinant AtRBR1 protein C-terminal fragment consisting of 236 amino acids from Arabidopsis (Horváth et al., 2006). The cross-reaction of this anti-AtRBR1 antibody with the His-tagged C-terminal fragment of the MsRBR protein produced in vitro was tested. This antibody recognized even a low amount (5 ng) of recombinant protein (data not shown), and a 115 kDa protein was detected as a major band in western blots of total protein extract (Fig. 1A, lane 1). The 115 kDa protein band corresponds to the predicted molecular mass (114.2 kDa) of the alfalfa RBR protein. The specificity of this cross-reaction was tested by antigen competition assay. Pre-incubation of this antibody with the purified (His)6-tagged C-terminal part of the MsRBR protein removed the cross-reacting antibody and resulted in the disappearance of the 115 kDa band (Fig. 1A, lane2). This finding supports the use of this antibody in the alfalfa experimental system. Since the amount of phosphorylated MsRBR protein has functional significance, the phosphoprotein was monitored by immunoblotting with antibody produced against the phosphorylated pRb peptide containing Ser807/811 (Cell Signaling Technology). In the alfalfa MsRBR protein Ser931 corresponds to the SPL motif. In order to test the specificity of this peptide antibody, the recombinant (His)6-tagged C-terminal fragment of the MsRBR protein was phosphorylated using CDK complex bound to GST–p13SUC1 beads. (Fig. 1B, middle panel). The phosphorylation reaction was also monitored by detection of 32P incorporation (Fig. 1B, lower panel). The two parameters are in good agreement with each other. Using whole-cell extracts, both antibodies produced against either AtRBR1 protein or phospho-pRb peptide recognized the same 115 kDa size protein (Fig. 1C, D). Here the sensitivity of phospho-MsRBR protein to CIAP treatment is also demonstrated. Dephosphorylation reduced the amount of detected protein cross-reacting with the anti-phospho-pRb antibody (Fig. 1D, lanes 3 and 6). The results of these western blots provide additional information on the accumulation of MsRBR and phospho-MsRBR proteins in cells at different phases of the growing period of the A2 suspension culture. The frequency of dividing cells harvested at 4 d after subculturing is high, because the suspension culture is in the exponential phase. As shown in Fig. 1C, D, lanes 4–6 these cells contain higher amounts of both forms of MsRBR proteins than do cells harvested at 7 d after subculturing representing the stationary phase of growth (Fig. 1C, D, lanes 1–3). In particular, the elevated levels of phospho-MsRBR protein in cells in a state of active division are indicated by this experiment. Careful tests using the above antibody set have opened up a way to monitor different forms of MsRBR protein in synchronized or hormone-treated alfalfa cells.
Fig. 1.
Specificity test of antibodies used for detection of the Medicago retinoblastoma-related protein (MsRBR) and its phosphorylated form (phospho-MsRBR) in cells at stationary or exponential growing phase. (A) Antigen competition assay for anti-AtRBR1 antibody: 1, western blot of total protein extract with anti-AtRBR1 antibody detected the MsRBR1 protein (115 kDa); 2, immunoblot of total protein extract with anti-AtRBR1 antibody pre-incubated with the purified (His)6-tagged C-terminal part of the MsRBR1 protein failed to detect the MsRBR protein (115 kDa). (B) Functionality test of anti-human pRb phosphopeptide antibody by western blot of the in vitro phosphorylated recombinant C-terminal fragment of MsRBR protein after incubation with p13SUC1-bound kinase complex. Upper panel, Ponceau S-stained filter used for immunoblot assay; middle panel, immunoblot with antibody produced against the phosphopeptide corresponding to residues around Ser807/811 of human pRb; lower panel, detection of incorporated [32P]inorganic phosphate by Phosphor Imager SI (Molecular Dynamics). (C) Alfalfa cells at exponential phase have an increased amount of the MsRBR protein in comparison with cells at stationary phase. Lanes 1–3, protein extracts from 7-day-old cultures (stationary phase); lanes 4–6, protein extracts from 4-day-old cultures (exponential phase); 1, 4, control cultures; 2, 5, protein extracts treated with phosphatase buffer; 3, 6, protein extracts with calf intestinal alkaline phosphatase (CIAP). (D) Western blots with the anti-human pRb phosphopeptide antibody detected reduced amounts of phospho-MsRBR protein after phosphatase treatment and showed significantly higher amounts of phospho-MsRBR protein in cells at exponential growing phase. Lanes 1–3, protein extracts from 7-day-old cultures (stationary phase); lanes 4–6, protein extracts from 4-day-old cultures (exponential phase); 1, 4, control cultures; 2, 5, protein extracts treated with phosphatase buffer; 3, 6, protein extracts with CIAP.
Medicago CDKA- and CDKB-type complexes phosphorylate the recombinant MsRBR protein
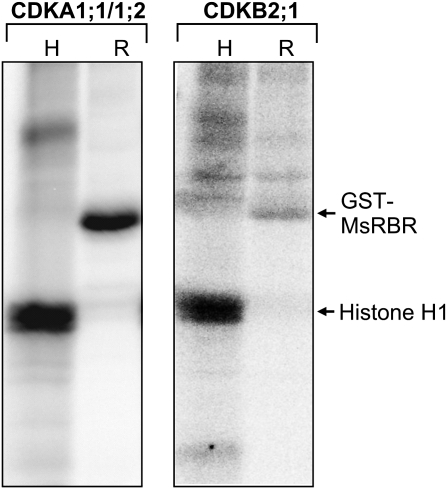
Based on the number of CDK phosphorylation sites and cell cycle phase-dependent activities of different plant CDKs, the phosphorylation of the MsRBR protein C-terminal fragment was tested by immunoprecipitated MedsaCDKA1;1/1;2 and MedsaCDKB2;1 complexes in vitro (Fig. 2). Both the A- and B-type kinases phosphorylated the histone H1 and the C-terminal fragment of the MsRBR protein. Previously it had been demonstrated that the phosphorylation function of MedsaCDKA1;1 is dependent on the presence of a cyclin partner (MedsaCYCD3,1) (Dudits et al., 2007). This cyclin variant has the characteristic LxCxE pRb-binding motif. On the other hand the mitosis-specific MedsaCDKB2;1 also interacted with the MedtrCYCD1;1 cyclin, having a similar pRb-binding motif, in yeast two-hybrid assays (Mészáros et al., 2000).
Fig. 2.
The MsRBR protein can serve as substrate for both the PSTAIRE (CDKA1;1/1/2) and the mitotic PPTALRE (CDKB2;1) kinases. Immunoprecipitated cyclin kinase complexes phosphorylate the histone H1 protein (H) and the MsRBR protein C-terminal fragment fused to GST (R).
Depletion of growth hormones causes a significant reduction of the level of RBR protein in cultured alfalfa cells
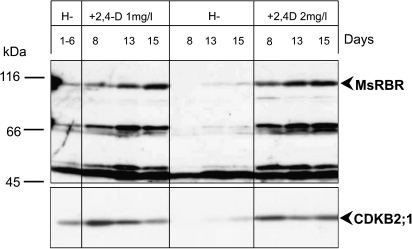
The Medicago A2 cell line is routinely cultured in medium supplemented with 2 mg l−1 2,4-D and 0.2 mg l−1 kinetin. To test hormonal responses the A2 cells were cultured in medium lacking growth hormones for 6 d and subsequently grown either in hormone-free medium or in the presence of 1 mg l−1 or 2 mg l−1 2,4-D in combination with 0.2 mg l−1 kinetin. As indicated by western blot analysis, low but detectable amounts of the MsRBR protein were characteristic for cells cultured in hormone-free medium (H–) for 6 d (Fig. 3). In these cells the G2/M phase-specific MedsaCDKB2;1 protein was also present. Re-addition of 1 mg l−1 or 2 mg l−1 2,4-D with 0.2 mg l−1 kinetin resulted in a gradual increase in the full-size MsRBR protein, reaching maximum level after 2 weeks. This hormonal treatment also stimulated the synthesis of the mitotic kinase protein. During the continuation of the hormone-free culture period alfalfa cells essentially reduced accumulation of the MsRBR protein. Based on the significant decrease in the mitotic kinase protein as a marker for cell division, it can be concluded that these alfalfa cells have decreased proliferation function. In this experiment, the western blot detected a 50 kDa protein that is present under the prolonged hormone-free culture period. Without knowing its nature it may be assumed to be an indicator of the living state of cells in this treatment combination. The very low amount of the full size MsRBR protein in these cells grown in hormone-free medium supports the conclusion that the presence of the RBR protein is linked to division activity in in vitro cultured alfalfa cells.
Fig. 3.
Growth hormone-dependent presence of the MsRBR protein in cultured alfalfa cells. Western blot of total protein extracts isolated from cells grown in hormone-free medium or 2,4-dichlorophenoxy acetic acid (2,4-D)- and kinetin-supplemented medium for the indicated time periods. The anti-AtRBR1 antibody and anti-CDKB2;1 antibody detected a significant reduction in the cross-reacting proteins in cells exposed to hormone-free conditions. The low amounts of the mitotic kinase protein indicate reduced division activities in these cells. The presence of a 50 kDa protein in cells from the hormone-free treatment combination reflects the living state of the analysed cells.
Cell cycle phase-dependent variation in amounts of MsRBR and phospho-MsRBR proteins
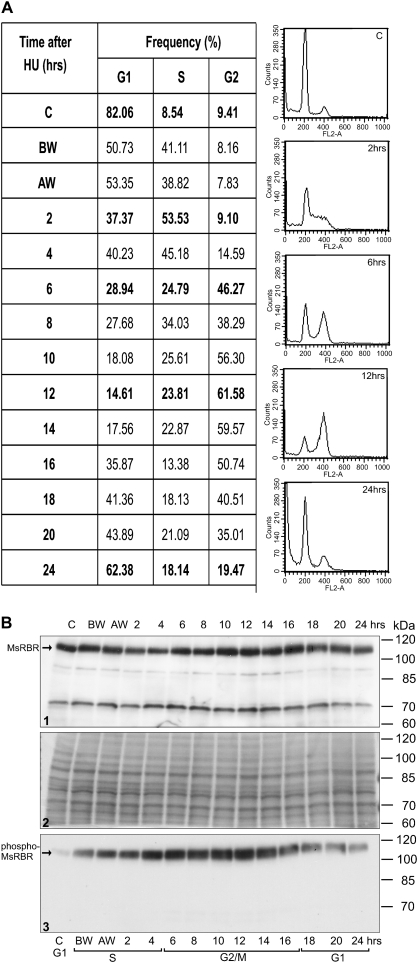
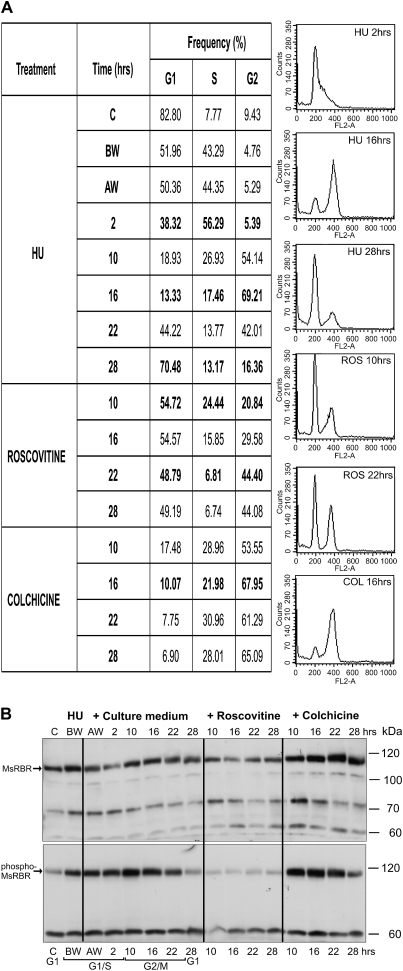
Actively dividing Medicago cells grown in suspension culture can be synchronized with HU treatment. As shown by representative profiles and frequency data from flow cytometry in Fig. 4A the control culture represents G1 cells (82%). Alfalfa cells were subjected to 36 h HU treatment and, after removing the inhibitor, cells progress through the S phase with the highest frequency at 2–4 h (53% and 45%). The maximum number of G2/M cells could be detected in samples from cultures collected after 10–16 h (56, 61, 59, and 50%). The frequency of G1 cells increased after 16 h, indicating the initiation of a new cycle. A western blot of total protein extracts detected the 115 kDa full size MsRBR protein in all samples (Fig. 4B, lane 1). The amounts of cross-reacting MsRBR protein showed limited variation during cell cycle progression. Interestingly the G2/M cells had slightly elevated levels of this protein. After 16 h the blot showed a gradual decrease of the MsRBR protein level, which is in good agreement with the increasing frequency of G1 cells. Western blotting of the same protein samples with anti-phospho-pRb antibody indicated a low protein phosphorylation level in the control culture with 82% G1 cells (Fig. 4B, lane 3). The protein level was elevated in samples with S phase cells (53% and 45%). The protein cross-reacting with antibodies against the phospho-pRb peptide was found to be high in protein extracts isolated from cell populations representing a significant frequency (56, 61, and 59%) of G2/M phase cells. Subsequently the phospho-MsRBR protein level was reduced in samples containing more G1 cells. In this experiment both antibodies cross-reacted with the full size MsRBR protein of 115 kDa. Both forms of the MsRBR protein are present in all cell cycle phases, and the phosphorylated MsRBR protein exhibits higher variation. The differences between samples from G1 and G2/M cells are significant.
Fig. 4.
Variation in the amounts of the MsRBR and the phospho-MsRBR proteins in alfalfa cells during cell cycle progression after synchronization with 10 mM hydroxyurea (HU) treatment. (A) Frequency data of G1, S, and G2 cells at various time points after removal of the inhibitor (HU) with characteristic DNA histograms from flow cytometry. (B) Western blot of protein extracts by anti-AtRBR1 antibody (1) and anti-human phospho-pRb peptide antibody (3). Ponceau S-stained filter with 50 μg protein samples (2). C, control, non-synchronized culture; BW, before washing out HU; AW, after washing out HU.
Roscovitine reduced whereas colchicine increased both forms of MsRBR protein
In order to dissect the G2/M phases in the analysis of levels of the MsRBR protein and its phosphorylated form, double synchronization based on HU treatment followed by roscovitine or colchicine inhibition of cell cycle progression was applied. Roscovitine is a specific inhibitor of CDKs and it can block cell cycle progression in either late G1 phase or late G2 phase, as was shown in BY-2 tobacco cells (Planchais et al., 1997). Colchicine is a well-known inhibitor of mitotic chromosome segregation by interacting with tubulin (Bhattacharyya et al., 2008). In the present study alfalfa cells were exposed to HU (10 mM) for 36 h. After the release of the HU block the cells were grown in culture medium for 2 h and subsequently treated with either roscovitine (100 μM) or colchicine (0.05%). The reference HU-treated cells were further grown in culture medium. First the cell cycle parameters were identified by flow cytometry after these treatment combinations (Fig. 5A). The maximum number of S phase cells (43, 44, and 56%) was found in samples from the time points of washing out HU and 2 h after the release of the HU block. Cultures from 10, 16, and 22 h contain an increased percentage of cells in G2/M phase with increased frequency (54, 69, and 42%). G1 cells form the majority in samples from 28 h. After roscovitine treatment G1 phase cells could be detected at 54% and later at 48% frequency. In the colchicine-treated cultures G2/M cells represented the majority (61% and 67%) of all cell types.
Fig. 5.
Roscovitine reduces and colchicine increases the amounts of MsRBR and phospho-MsRBR proteins in alfalfa cells exposed to double synchronization. (A) Frequency data of G1, S, and G2 cells at various time points after removal of the inhibitor [hydroxyurea (HU) at 10 mM] and treatment with roscovitine or colchicine for the indicated time periods. Characteristic DNA histograms from flow cytometry are also shown. (B) Western blot of protein extracts with anti-AtRBR1 antibody (upper) and anti-human pRb phosphopeptide antibody (low). C, control, non-synchronized culture; BW, before washing out HU; AW, after washing out HU.
Figure 5B (upper panel) presents the variation in levels of MsRBR protein as detected by western blot of protein extracts from cells synchronized by the above treatments. Using the control non-treated G1 cells as reference, increased MsRBR levels could be observed in cells grown in the presence of HU (before washing). After release from the HU block (after washing) a clear reduction was recognized in samples with a significant number of cells in mid to late S phase. The minimum amount was found in the sample from 2 h with 56% S phase cells. Compared with these time points, increased amounts of MsRBR protein were detected in samples from 10–16 h and 22 h, with a high representation of G2/M cells. Later the cross-reacting protein levels decreased when the frequency of G1 cells increased in the cultures. These data show similar trends in cell cycle phase-dependent changes as in the previous experiment (Fig. 4B, lane 1). As far as MsRBR protein amounts are concerned, the subsequent treatments of the synchronized cells resulted in different responses. The inhibitor roscovitine lowered the MsRBR protein levels, whereas the colchicine block enhanced the accumulation of this protein in cells enriched for G2/M phases. Using the same protein samples, the phospho-MsRBR levels were monitored in cell populations with various frequencies of cells in different division phases. As shown previously, G1 cells have relatively low amounts of the cross-reacting protein (Fig. 5B, lower panel). The roscovitine treatment significantly reduced the amount of the phosphorylated form of MsRBR protein. In contrast, the presence of colchicine caused accumulation of phospho-MsRBR protein. The lower molecular weight bands may represent unspecific cross-reactions since they show different patterns when compared with the full-size MsRBR protein.
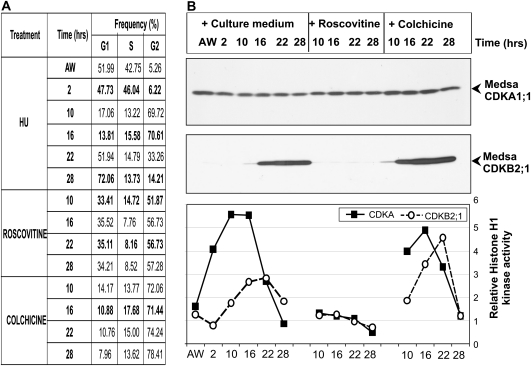
To characterize kinase markers in cultures exposed to the double synchronization treatment, an additional experiment was carried out. As shown in Fig. 6A, the distribution of cells in various phases of the cell cycle was similar to that in the previous experiment (see Fig. 5A). Cell samples enriched in different cell cycle stages were used for analysis of CDK activities and quantification of these kinase proteins (Fig. 6B). The MedsaCDKA1;1 protein was constantly present in cells released from HU inhibition as well as in those treated with either roscovitine or colchicine. Western blot of the same protein samples failed to detect the mitotic MedsaCDKB2;1 in the roscovitine-treated cells. In the presence of this inhibitor, the histone H1 phosphorylation function of both CDKs was blocked. After HU treatment these CDKs presented different activity profiles, with a maximum histone H1 phosphorylation at 10 h and 16 h in the case of CDKA and 16 h and 22 h for CDKB. In the colchicine-treated cells the activity of the mitotic MedsaCDKB2;1 was increased to a maximum in samples from 22 h.
Fig. 6.
Changes in amounts of CDKA1;1 and CDKB2;1 proteins and their activities in alfalfa cells after double synchronization based on 10 mM hydroxyurea (HU) treatment combined with roscovitine (100 μM) or colchicine (0.05%) treatment. (A) Frequency data of G1, S, and G2 cells at various time points after removal of the inhibitor (HU) subsequently grown in normal culture medium and treated with either roscovitine or colchicine. (B) Top, western blot of protein extracts with anti-CDKA1;1 antibody; middle, western blot of protein extracts with anti-CDKB2;1 antibody; bottom, histone H1 phosphorylation activities of CDKA1;1 and CDKB2;1 in the synchronized cells. AW, after washing out HU.
These sets of experiments confirmed differences in cell cycle-dependent changes in the amounts of MsRBR and phospho-MsRBR proteins. Roscovitine treatment of cells caused only limited reduction in MsRBR protein levels, whereas the phosphorylated form was significantly lowered possibly because of the lack of CDK activities. In contrast, the colchicine-treated cells found in G2 phase at high frequency (>70%) showed elevated levels of both protein forms. Accumulation of MsRBR proteins after DNA replication may be linked to the block of mitotic progression.
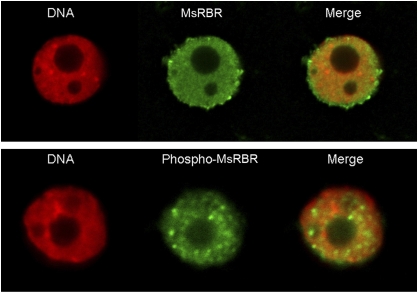
Immunolocalization of the phospho-MsRBR protein in cultured alfalfa cells
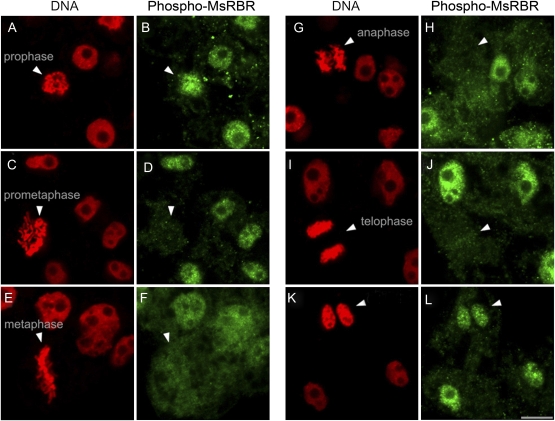
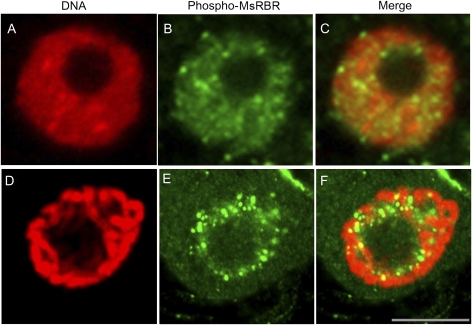
In the present study it was not possible to obtain clear immunolocalization signals by using antibody produced in hens against the C-terminus of the AtRBR1 protein in immunofluorescence microscopy of cultured alfalfa cells. Non-uniform and inconsistent cytoplasmic and cell wall staining of cells may be due to enhanced recognition of certain non-specific epitopes detected by this antibody during the immunolocalization procedure based on the formaldehyde fixation. During sample preparation, nuclei were released from some cells. These nuclei showed homogenous staining with the anti-AtRBR protein antibody. In contrast, the anti-phospho-pRb peptide antibody recognized nuclear spots of varying sizes, indicating a concentrated presence of the phosphorylated form in these structures (Fig. 7). These spots were randomly scattered in the nuclei. Similar structures were detected in nuclei of intact alfalfa cells by using this antibody. Figures 8 and 9 show the localization of the phospho-MsRBR protein in interphase and mitotic cells from asynchronous suspension cultures. The series of immunofluorescence images shown in Fig. 8 present cells with interphase nuclei together with cells at various mitotic phases. The nuclei of interphase cells were stained intensively with anti-phospho-pRb peptide antibodies and these signals were concentrated into spots as observed with isolated nuclei (Fig. 8B–L). Analysis of mitotic cells revealed labelled granules in prophase cells (Fig. 8B). These structures detected in prophase cells cannot be recognized in later mitotic stages such as prometaphase, metaphase, anaphase, and telophase (Fig. 8D–L). Images in Fig. 9 provide a more detailed insight into both interphase nuclei with immunoreactive structures (Fig. 9B) and prophase cells with positive nuclear granules (Fig. 9E). In this cell the condensation of chromosomes is well advanced and the merged pictures indicate an interchromosomal position of nuclear granules. The presented immunolocalization data strengthen the previous western blot results indicating the presence of phospho-MsRBR proteins not only in interphase cells before and during S phase but also in G2/M phases of the cell cycle. An unexpected finding is that this form of MsRBR protein is localized in spots or granules possibly representing protein complexes with unknown composition and function. Additional studies are needed to characterize these structures.
Fig. 7.
Differences in immunofluorescence labelling of MsRBR and phospho-MsRBR proteins in isolated nuclei released from alfalfa cells indicate different nuclear organization for the two forms of MsRBR protein. Upper panel: diffused, uniform staining of nucleoplasm (green) after immunolocalization with anti-AtRBR1 antibody. Lower panel: granular labelling of the nucleus (green) after immunolocalization with anti-human pRb phosphopeptide antibody. DNA stained with DAPI and pseudocoloured red.
Fig. 8.
Organization of the phospho-MsRBR protein into granular structures is restricted to mitotic prophase. According to the distribution of the phospho-MsRBR protein these characteristic granules cannot be detected in later mitotic stages. Immunodetection of phospho-MsRBR protein (green panels) is shown at various phases of the cell division cycle in formaldehyde-fixed suspension-cultured cells of alfalfa. Arrowheads indicate various mitotic phases (A, B, prophase; C, D, prometaphase; E, F, metaphase; G, H, anaphase; I, J, telophase) or early G1 cells (K, L). Note the significantly reduced labelling intensities at metaphase, anaphase, and telophase stages of mitosis. This figure also presents interphase nuclei (B, D, F, H, J, and L) with concentration of labelling signals in nuclear spots. Localization of phospho-MsRBR protein is shown in the green panel. DNA is stained with DAPI and pseudocoloured red (red panels). Bar=10 μm
Fig. 9.
Concentration of the phospho-MsRBR protein (green panel) into nuclear spots in interphase cells (A–C) and granules in prophase cells (D–I) after formaldehyde fixation. Prophase cells with advanced chromosome condensation have a considerable number of phospho-MsRBR protein-positive granules with an interchromosomal position. DNA is stained with DAPI (bar=10 μm) and pseudocoloured red (red panel).
Discussion
The retinoblastoma pathway has emerged as an important regulatory component in the cell division cycle of higher plants just like it is in other eukaryotes. Studies on plant RBR proteins discovered a wide variety of additional roles such as cell fate determination and influencing chromatin dynamics or genomic imprinting (de Jager et al., 2005; Park et al., 2005; Sablowski, 2007; Costa and Gutierrez-Marcos, 2008; Paz Sanchez et al., 2008; Chen et al., 2009; Sabelli and Larkins, 2009). For a deeper understanding of the molecular basis of these divergent functions of plant RBR proteins it is essential to know the cell cycle phase-dependent changes in levels and post-translational modifications of these proteins. In these studies, immunodetection techniques were used to monitor the MsRBR and the phospho-MsRBR proteins in samples from synchronized alfalfa cells. Antibodies against the C-terminal fragment of AtRBR1 protein specifically recognized the 115 kDa MsRBR protein as shown by the antigen competition assay (Fig. 1A). In total protein extracts from cultured alfalfa cells this polyclonal antibody detected smaller cross-reacting bands, indicating a wider spectrum of unspecific interactions for the antibody tested. The higher signal intensity in lane 2 (Fig. 1A) might originate from the concentration of antibody during the pre-incubation procedure. The present observation that antibodies generated either against the C-terminal fragment of the Arabidopsis AtRBR1 protein or the phosphopeptide from human pRb protein cross-reacted with the same 115 kDa alfalfa protein with the predicted molecular size provides a sufficient basis to use these antibodies in alfalfa cell systems (Fig. 1C, D). According to the sequence data, the C-terminal regions of the Arabidopsis and alfalfa RBR proteins share a significant degree of amino acid sequence identity (74%). The homologous regions cover a substantial portion of these fragments including the NVYVSPLRGS motif that may correspond to the NIYISPLK-S motif of the human pRb protein which has a conserved 807/811serine (S) phosphorylation site. In attempts to monitor the phosphorylated form of the MsRBR protein by using anti-phosphopeptide antibody, it has to be emphasized that this methodology detects a single phosphorylation site whereas post-translational modifications can occur at additional residues with different functional consequences. Therefore, the methodology used here is not sufficient for determining the ratio between the MsRBR and the phospho-MsRBR proteins. The functionality and specificity of this anti-phosphopeptide antibody was demonstrated by detection of the in vitro phosphorylated form of the Medicago RBR protein C-terminal fragment by western blot (Fig. 1B). The pRb proteins from mammalian cells show phosphorylation-dependent migration patterns, and the hyperphosphorylated pRb protein is usually detected as a more slowly migrating band (Farkas et al., 2002). As a result of the protein separation protocol employed (electrophoresis in denaturing polyacrylamide gels) the MsRBR protein band was diffuse and a clear shift between the two protein forms could not be recognized. Similarly, no band shift was reported in the case of the NtRBR1 and the phospho-NtRBR1 proteins (Kawamura et al., 2006). After inorganic phosphate incorporation into pea axillary buds the anti-PsRBR antibody immunoprecipitated higher and lower molecular mass forms of the phosphorylated PsRBR1 that were sensitive to λ-phosphatase treatment (Shimizu-Sato et al., 2008). In the present study the intensity of the band corresponding to the phospho-MsRBR (115 kDa) protein was reduced by phosphatase treatment, as an indication of correct recognition of the phosphorylated form by the anti-phosphopeptide antibody used (Fig. 1D, lanes 3 and 6).
It was demonstrated previously that Arabidopsis, tobacco, wheat, and maize RBR proteins can serve as substrates for CDK–cyclin complexes (Boniotti and Gutierrez, 2001). The in vitro phosphorylation reactions revealed that PSTAIRE-type CDKs and D-type cyclins are involved in this post-translational modification. Nakagami et al. (2002) reported tobacco NtRBR1 phosphorylation by NictaCycD3;3-associated kinase during a very short period in the middle of the G1 phase whereas p13SUC1-bound kinases were active in phosphorylation of NtRBR1 during late G1 phase to late S phase. These authors also showed that different types of tobacco CDKs (CDKA and CDKB) can phosphorylate the 110 kDa NtRBR protein in vitro (Kawamura et al., 2006). The present study supports the potential role of non-PSTAIRE kinases; namely the mitotic Cdc2F–MedsaCDKB2;1 complex is also capable of phosphorylating the C-terminal region of the alfalfa MsRBR protein as is shown in the case of the MedsaCDKA1;1 PSTAIRE kinase (Fig.2). The activity of MedsaCDKB2;1 is linked to cells in G2 phase and mitosis (see Magyar et al., 1997 and Fig. 6B). The MedsaCDKA1;1 protein is detectable during the whole cell cycle, with increased activities in cells accumulated in G1–S and early G2 phase after HU block (Fig. 6B). The histone H1 phosphorylation function of both CDKs was sensitive to roscovitine treatment. It is an interesting finding that the MedsaCDKB2;1 protein was undetectable in treated cells. That is a selective effect of this inhibitor in comparison with the response of MedsaCDKA1;1. The lack of B-type kinase protein accumulation can originate from blocking an unknown component required for either expression of the MedsaCDKB2;1 gene or synthesis and stability of this kinase protein. One potential candidate can be the A-type CDK since this kinase is active earlier in G1–S phases and its function is known to be inhibited by roscovitine. Detection of this unexpected effect of roscovitine may initiate further experiments.
Analysis of the MsRBR protein in cells from cultures at different growing phases revealed an increase in both forms of this protein during exponential growth (Fig. 1C, D, lanes 4–6). Differences between cells in the stationary and exponential phase of the culture period were more pronounced in the case of the phospho-MsRBR protein. Linking the presence of the MsRBR proteins to the proliferative state was also supported by the hormone starvation experiment (Fig. 3). In this experiment quantification of the mitotic kinase (CDKB2;1) protein was used as a marker for division activity that was induced already during the first 2 d by re-addition of hormones after a hormone-free growing period of 6 d. The amounts of the MsRBR protein(115 kDa) increased gradually and reached a maximum after 15 d of growth in a complete culture medium. Based on the observed changes, close correlation between these two proteins is not expected since the MsRBR protein slightly changes between cell cycle phases, whereas the mitotic kinase CDKB;2,1 protein is exclusively found in G2/M phase cells (Figs 4B, 6B). When alfalfa cells were cultured in hormone-free medium for a further 8–15 d, both the MsRBR protein and the mitotic kinase CDKB;2,1 proteins were reduced to the detection threshold in protein extracts; therefore, it was concluded that the prolonged withdrawal of synthetic auxin and kinetin stopped the division of A2 alfalfa cells and the non-dividing state of these cells was reflected by the very low level of the MsRBR protein. The cell division-dependent presence of plant RBR proteins could also be seen in the Arabidopsis experimental system. Only the non-phosphorylated AtRBR1 protein was detected in stationary phase MM2d Arabidopsis cells. Transfer into fresh medium resulted in the detection of increasing amounts of AtRBR1 protein and after 8 h its phosphorylated form was recognized by western blots (Hirano et al., 2008). The role of RBRs in dividing plant cells can also be deduced from experiments by these authors where MM2d cells were grown in low sucrose medium. AtRBR1 was depleted to undetectable levels within 12 h after transfer of cells into the medium lacking sucrose. In interpreting these findings as an indication for a link between the dividing state of cells and the RBR protein it should be emphasized that the present and cited studies were carried out with plant cells cultured in vitro. The functional characteristics of plant RBRs can be essentially different in vivo. Wyrzykowska et al. (2006) showed growth inhibition and initiation of differentiation by transient overexpression of the AtRBR1 gene. In tobacco plants the induced RBR protein synthesis caused a complete and rapid cessation of cell division. These observations highlight an essential divergence of RBR protein functions in plant organs such as shoot apical meristem and in hormone-stimulated, fast cycling cultured cells.
Despite the above limitation, cultured plant cells at present provide an optimal experimental system for efficient synchronization of the division cycle to analyse phase-specific events. In the present study the A2 cell line of M. sativa that is well adapted to in vitro conditions was used and its short cycle time allowed the enrichment of cell populations in various phases by using specific inhibitors such as HU, roscovitine, or colchicine. Western blotting of total protein extracts from alfalfa cells with either antibody against the C-terminal fragment of AtRBR1 protein or anti-human phospho-pRb peptide antibody detected the expected size of RBR protein (115 kDa) and additional cross-reacting bands of various sizes and quantities (Figs 1, 4B, 5B). These signals are considered to be the results of non-specific interactions or degradation; therefore, the present analysis is restricted to the full-size MsRBR protein and its phosphorylated form. Western blot analysis of protein samples detected the MsRBR protein during the whole division cycle with limited changes. The observed increase in MsRBR protein levels in samples with a higher frequency of G2/M phase cells was shown more clearly by the colchicine-blocked cells, although a cell cycle phase-independent action of this inhibitor on the RBR protein accumulation cannot be excluded. During HU+roscovitine treatment the MsRBR protein level was slightly lower in comparison with the HU-treated cells. The sequentially applied HU and colchicine combination resulted in a significantly elevated MsRBR protein level compared with the single HU treatment (Fig. 5B). Cell cycle phase dependence could be more clearly recognized in alterations of phospho-MsRBR protein levels (Figs 4B, 5B). The reduced level of the phosphorylated form in G1 cells can be safely concluded from the presented data. Comparing amounts of cross-reacting MsRBR and phospho-MsRBR proteins in Fig. 4B and Fig. 5B reveals that G1 cells primarily have the hypophosphorylated form. Hirano et al. (2008) demonstrated by pull-down assay that the non-phosphorylated AtRBR1 protein could bind to E2Fa protein. These findings are in agreement with the general model of G1/S phase transition control where the hypophosphorylated Rb proteins inhibit E2F functions in the activation of S phase-specific genes (Poznic, 2009). The release of this inhibition is expected to occur when the RBR proteins are phosphorylated by CDK complexes. The role of CDKs in phosphorylation of RBR protein was shown by different studies on plant systems. The present work demonstrates that in alfalfa cells the MedsaCDKA1;1 and the mitosis-specific MedsaCDKB1,2 can be responsible for this modification of the MsRBR protein (Fig. 2).
Based on the widely accepted model emphasizing the regulatory role of the Rb proteins in G1/S phase transition, experimental works on plant RBR proteins have essentially focused on this checkpoint. Functional roles for plant RBR proteins in the G2/M phases have not been proposed so far. The present western blot studies detected significant amounts of both MsRBR and phospho-MsRBR proteins in cells representing G2/M phases after chemical synchronization (Figs 4B, 5B). The presence of these proteins after S phase was more clearly demonstrated by western blot of proteins from the colchicine-treated cells. Occurrence of the phosphorylated form coincided with CDK activities including the PSTAIRE CDKA1;1 and the mitosis-specific CDKB2;1. In accordance with western blot data the immunolocalization studies also revealed the presence of phospho-MsRBR protein as granular structures in interphase nuclei and prophase cells (Figs 7–9). These figures present several interphase cells with nuclei having intense labelling concentrated into spots of various sizes. According to the results from labelling with anti-AtRBR1 antibody, a homogenous distribution of signals could be seen in the released nuclei. This observation is in agreement with the localization data presented by Boruc et al. (2010). Although the functional significance of these differences in the organization of MsRBR and phospho-MsRBR proteins has yet to be elucidated, one possible hypothesis is that only the phosphorylated form participates in protein complexes with the capability to form the detected nuclear structures. This question clearly is worthy of further investigation by using additional approaches. As shown by the present immunolocalization experiments, the phospho-MsRBR protein is detectable as granules in prophase cells with an advanced stage of chromosome condensation. Based on this observation it will be interesting to test a possible involvement of plant RBR proteins in building up chromosome structures. In Drosophila the Rb family proteins can have a function in the mitotic cell. The Rbf1 protein is required to target the condensin II subunit dCAPD3 to chromosomes and thus promotes chromosome condensation and faithful transmission (Longworth et al., 2008). These authors showed that the overexpression of the Drosophila Rbf1 gene resulted in hypercondensation of mitotic chromosomes. Additional mitotic functions of pRb proteins have been widely demonstrated in mammalian cell systems. Loss of pRb protein functions may induce cell cycle deregulation, leading to a malignant phenotype (Giacinti and Giordano, 2006). The pRb protein has been shown to regulate the expression of the mitotic checkpoint protein Mad2, which promotes chromosome instability when misregulated in mammalian cells (Hernando et al., 2004; Sotillo et al., 2007). Zheng et al. (2000) demonstrated that the pRb protein can interact with hsHec1p carrying the LxCxE motif during G2/M phases. This protein is required for proper chromosome segregation. The serine/threonine Aurora kinases have been shown to play a critical role in mitosis. Nair et al. (2009) demonstrated that phosphorylation of the pRb protein at Ser780 by Aurora B influences the polyploid cell formation caused by inhibition of this kinase. As shown by Karantza et al. (1993), cells overexpressing the pRb proteins accumulated uniformly in the G2 phase, whereas cells expressing endogenous levels of the pRb protein were found throughout the whole cell cycle. These results suggest that the pRb protein may interact with some protein of the cell cycle-regulatory machinery during the G2 phase. The present immunodetection study based on both western blot analysis and immunolocalization with immunofluorescence microscopy provides substantial support for considering plant RBR proteins to be present in G2/M cells as regulators of the cell cycle during post-DNA synthetic events.
Acknowledgments
This work was supported by a grant from OTKA (The Hungarian Scientific Research Grant), grant number: NK-69227. EÁ was supported by the János Bolyai Research Fellowship of the Hungarian Academy of Sciences. The authors thank Mátyás Cserháti and Erzsébet Fejes for critical reading, and Klára Godó for valuable assistance in the preparation of this manuscript.
References
- Ach RA, Durfee T, Miller AB, Taranto P, Hanley-Bowdoin L, Zambrski P, Gruissem W. RRB1 and RRB2 encode maize retinoblastoma-related proteins that interact with a plant D-type cyclin and geminivirus replication protein. Molecular and Cellular Biology. 1997;17:5077–5086. doi: 10.1128/mcb.17.9.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berckmans B, De Veylder L. Transcriptional control of the cell cycle. Current Opinion in Plant Biology. 2009;12:599–605. doi: 10.1016/j.pbi.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya B, Panda D, Gupta S, Banerjee M. Anti-mitotic activity of colchicine and the structural basis for its interaction with tubulin. Medicinal Research Reviews. 2008;28:155–183. doi: 10.1002/med.20097. [DOI] [PubMed] [Google Scholar]
- Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- Bögre L, Oláh Z, Dudits D. Ca2-dependent protein kinase from alfalfa (Medicago varia): partial purification and autophosphorylation. Plant Science. 1988;58:135–144. [Google Scholar]
- Boniotti MB, Gutierrez C. A cell-cycle-regulated kinase activity phosphorylates plant retinoblastoma protein and contains, in Arabidopsis, a CDKA/cyclin D complex. The Plant Journal. 2001;28:341–350. doi: 10.1046/j.1365-313x.2001.01160.x. [DOI] [PubMed] [Google Scholar]
- Borghi L, Gutzat R, Fütterer J, Laizet Y, Hennig L, Gruissem W. Arabidopsis RETINOBLASTOMA-RELATED is required for stem cell maintenance, cell differentiation, and lateral organ production. The Plant Cell. 2010;22:1792–1811. doi: 10.1105/tpc.110.074591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boruc J, Mylle E, Duda M, De Clercq R, Rombauts S, Geelen D, Hilson P, Inzé D, Van Damme D, Russinova E. Systematic localization of the Arabidopsis core cell cycle proteins reveals novel cell division complexes. Plant Physiology. 2010;152:553–565. doi: 10.1104/pp.109.148643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Hafidh S, Poh SH, Twell D, Berger F. Proliferation and cell fate establishment during Arabidopsis male gametogenesis depends on the retinoblastoma protein. Proceedings of the National Academy of Sciences, USA. 2009;106:7257–7262. doi: 10.1073/pnas.0810992106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa LM, Gutierrez-Marcos JF. Retinoblastoma makes its mark on imprinting in plants. PLoS Biology. 2008;6:1631–1633. doi: 10.1371/journal.pbio.0060212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida J, Engler J, De Veylder L, et al. Systematic analysis of cell-cycle gene expression during Arabidopsis development. The Plant Journal. 2009;59:645–660. doi: 10.1111/j.1365-313X.2009.03893.x. [DOI] [PubMed] [Google Scholar]
- de Jager SM, Maughan S, Dewitte W, Scofield S, Murray JA. The developmental context of cell-cycle control in plants. Seminars in Cell and Developmental Biology. 2005;16:385–396. doi: 10.1016/j.semcdb.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Dudits D, Cserháti M, Miskolczi P, Horváth VG. The growing family of plant cyclin-dependent kinases with multiple functions in cellular and developmental regulation. In: Inzé D, editor. Cell cycle control and plant development. Oxfore: Blackwell Publishing; 2007. pp. 1–30. [Google Scholar]
- Durfee T, Feiler HS, Gruissem W. Retinoblastoma-related proteins in plants: homologues or orthologues of their metazoan counterparts? Plant Molecular Biology. 2000;43:635–642. doi: 10.1023/a:1006426808185. [DOI] [PubMed] [Google Scholar]
- Farkas T, Hansen K, Holm K, Lukas J, Bartek J. Distinct phosphorylation events regulate p130- and p107-mediated repression of E2F-4. Journal of Biological Chemistry. 2002;277:26741–26752. doi: 10.1074/jbc.M200381200. [DOI] [PubMed] [Google Scholar]
- Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science. 1993;220:1049–1051. doi: 10.1126/science.220.4601.1049. [DOI] [PubMed] [Google Scholar]
- Giacinti C, Giordano A. RB and cell cycle progression. Oncogene. 2006;25:5220–5227. doi: 10.1038/sj.onc.1209615. [DOI] [PubMed] [Google Scholar]
- Grafi G, Burnett RJ, Helentjaris T, Larkins BA, DeCaprio JA, Sellers WR, Kaelin WG., Jr. A maize cDNA encoding a member of the retinoblastoma protein family: involvement in endoreduplication. Proceedings of the National Academy of Sciences, USA. 1996;93:8962–8967. doi: 10.1073/pnas.93.17.8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez C. The retinoblastoma pathway in plant cell cycle and development. Current Opinion in Plant Biology. 1998;1:492–497. doi: 10.1016/s1369-5266(98)80041-1. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Hernando E, Nahle Z, Juan G, et al. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 2004;430:797–802. doi: 10.1038/nature02820. [DOI] [PubMed] [Google Scholar]
- Hirano H, Harashima H, Shinmyo A, Sekine M. Arabidopsis retinoblastoma-related protein1 is involved in G1 phase cell cycle arrest caused by sucrose starvation. Plant Molecular Biology. 2008;66:259–275. doi: 10.1007/s11103-007-9268-2. [DOI] [PubMed] [Google Scholar]
- Horváth BM, Magyar Z, Zhang Y, Hamburger AW, Bakó L, Visser RG, Bachem CW, Bögre L. EBP1 regulates organ size through cell growth and proliferation in plants. EMBO Journal. 2006;25:4909–4920. doi: 10.1038/sj.emboj.7601362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley R, Healy S, Freeman D, et al. The maize retinoblastoma protein homologue ZmRb-1 is regulated during leaf development and displays conserved interactions with G1/S regulators and plant cyclin D (CycD) proteins. Plant Molecular Biology. 1998;37:155–169. doi: 10.1023/a:1005902226256. [DOI] [PubMed] [Google Scholar]
- Johnston AJ, Kirioukhova O, Barrell PJ, Rutten T, Moore JM, Baskar R, Grossniklaus U, Gruissem W. Dosage-sensitive function of RETINOBLASTOMA RELATED and convergent epigenetic control are required during the Arabidopsis life cycle. PLoS Genetics. 2010 doi: 10.1371/journal.pgen.1000988. 6, e1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karantza V, Maroo A, Fay D, Sedivy JM. Overproduction of Rb protein after the G1/S boundary causes G2 arrest. Molecular Cell Biology. 1993;13:6640–6652. doi: 10.1128/mcb.13.11.6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura K, Kato K, Shinmyo A, Sekine M. Tobacco retinoblastoma-related protein is phosphorylated by different types of cyclin-dependent kinases during the cell cycle. Plant Biotechnology. 2006;23:467–473. [Google Scholar]
- Korenjak M, Brehm A. E2F–Rb complexes regulating transcription of genes important for differentiation and development. Current Opinion in Genetics and Development. 2005;15:520–527. doi: 10.1016/j.gde.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Lendvai A, Pettkó-Szandtner A, Csordás-Tóth E, Miskolczi P, Horváth GV, Györgyey J, Dudits D. Dicot and monocot plants differ in retinoblastoma-related protein subfamilies. Journal of Experimental Botany. 2007;58:1663–75. doi: 10.1093/jxb/erm022. [DOI] [PubMed] [Google Scholar]
- Longworth MS, Herr A, Ji JY, Dyson NJ. RBF1 promotes chromatin condensation through a conserved interaction with the Condensin II protein dCAP-D3. Genes and Development. 2008;22:1011–1024. doi: 10.1101/gad.1631508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magyar Z, Bakó L, Bögre L, Dedeoglu D, Kapros T, Dudits D. Active cdc2 genes and cell cycle phase-specific cdc2-related kinase complexes in hormone-stimulated alfalfa cells. The Plant Journal. 1993;4:151–161. [Google Scholar]
- Magyar Z, Mészáros T, Miskolczi P, et al. Cell cycle phase specificity of putative cyclin-dependent kinase variants in synchronized alfalfa cells. The Plant Cell. 1997;9:223–235. doi: 10.1105/tpc.9.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mészáros T, Miskolczi P, Ayaydin F, Pettkó-Szandtner A, Peres A, Magyar Z, Horváth GV, Bakó L, Fehér A, Dudits D. Multiple cyclin-dependent kinase complexes and phosphatases control G2/M progression in alfalfa cells. Plant Molecular Biology. 2000;43:595–605. doi: 10.1023/a:1006412413671. [DOI] [PubMed] [Google Scholar]
- Miskolczi P, Lendvai Á Horváth GV, Pettkó-Szandtner A, Dudits D. Conserved functions of retinoblastoma proteins: from purple retina to green plant cells. Plant Science. 2007;172:671–683. [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Plant Physiology. 1962;15:473–497. [Google Scholar]
- Nair JS, Ho AL, Tse AN, Coward J, Cheema H, Ambrosini G, Keen N, Schwartz GK. Aurora B kinase regulates the postmitotic endoreduplication checkpoint via phosphorylation of the retinoblastoma protein at serine 780. Molecular Biology of the Cell. 2009;20:2218–2228. doi: 10.1091/mbc.E08-08-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H, Kawamura K, Sugisaka K, Sekine M, Shinmyo A. Phosphorylation of retinoblastoma-related protein by the cyclin D/cyclin-dependent kinase complex is activated at the G1/S-phase transition in tobacco. The Plant Cell. 2002;14:1847–1857. doi: 10.1105/tpc.002550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H, Sekine M, Murakami H, Shinmyo A. Tobacco retinoblastoma-related protein phosphorylated by a distinct cyclin-dependent kinase complex with Cdc2/cyclin D. in vitro. The Plant Journal. 1999;18:243–252. doi: 10.1046/j.1365-313x.1999.00449.x. [DOI] [PubMed] [Google Scholar]
- Nieuwland J, Menges M, Murray JAH. The plant cyclins. In: Inzé D, editor. Cell cycle control and plant development. Oxford: Blackwell Publishing; 2007. pp. 31–62. [Google Scholar]
- Nieuwland J, Scofield S, Murray JA. Control of division and differentiation of plant stem cells and their derivatives. Seminars in Cell and Developmental Biology. 2009;20:1134–1142. doi: 10.1016/j.semcdb.2009.09.011. [DOI] [PubMed] [Google Scholar]
- Park JA, Ahn JW, Kim YK, Kim J, Kim SJ, Kim JK, Kim WT, Pai HS. Retinoblastoma protein regulates cell proliferation, differentiation, and endoreduplication in plants. The Plant Journal. 2005;42:153–163. doi: 10.1111/j.1365-313X.2005.02361.x. [DOI] [PubMed] [Google Scholar]
- Paz Sanchez M, Caro E, Desvoyes B, Ramirez-Parra E, Gutierrez C. Chromatin dynamics during the plant cell cycle. Seminars in Cell and Developmental Biology. 2008;19:537–546. doi: 10.1016/j.semcdb.2008.07.014. [DOI] [PubMed] [Google Scholar]
- Planchais S, Glab N, Tréhin C, Perennes C, Bureau JM, Meijer L, Bergounioux C. Roscovitine, a novel cyclin-dependent kinase inhibitor, characterizes restriction point and G2/M transition in tobacco BY-2 cell suspension. The Plant Journal. 1997;12:191–202. doi: 10.1046/j.1365-313x.1997.12010191.x. [DOI] [PubMed] [Google Scholar]
- Poznic M. Retinoblastoma protein: a central processing unit. Journal of Biosciences. 2009;34:305–312. doi: 10.1007/s12038-009-0034-2. [DOI] [PubMed] [Google Scholar]
- Sabelli PA, Larkins BA. The contribution of cell cycle regulation to endosperm development. Sexual Plant Reproduction. 2009;22:207–219. doi: 10.1007/s00497-009-0105-4. [DOI] [PubMed] [Google Scholar]
- Sablowski R. The dynamic plant stem cell niches. Current Opinion in Plant Biology. 2007;10:639–644. doi: 10.1016/j.pbi.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Shimizu-Sato S, Ike Y, Mori H. PsRBR1 encodes a pea retinoblastoma-related protein that is phosphorylated in axillary buds during dormancy-to-growth transition. Plant Molecular Biology. 2008;66:125–135. doi: 10.1007/s11103-007-9257-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni R, Carmichael JP, Shah ZH, Murray JAH. A family of cyclin D homologs from plants differentially controlled by growth regulators and containing the conserved retinoblastoma protein interaction motif. The Plant Cell. 1995;7:85–103. doi: 10.1105/tpc.7.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo R, Hernando E, Diaz-Rodriguez E, Teruya-Feldstein J, Cordon-Cardo C, Lowe SW, Benezra R. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrzykowska J, Schorderet M, Pien S, Gruissem W, Fleming AJ. Induction of differentiation in the shoot apical meristem by transient overexpression of a retinoblastoma-related protein. Plant Physiology. 2006;141:1338–1348. doi: 10.1104/pp.106.083022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Sanz-Burgos AP, Hannon GJ, Gutierrez C. Plant cells contain a novel member of the retinoblastoma family of growth regulatory proteins. EMBO Journal. 1996;15:4900–4908. [PMC free article] [PubMed] [Google Scholar]
- Zheng L, ChenY Riley DJ, Chen PL, Lee WH. Retinoblastoma protein enhances the fidelity of chromosome segregation mediated by hsHec1p. Molecular and Cellular Biology. 2000;20:3529–3537. doi: 10.1128/mcb.20.10.3529-3537.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]