Abstract
The primary inflorescence stem of Arabidopsis thaliana is rich in lignified cell walls, in both vascular bundles and interfascicular fibres. Previous gene expression studies demonstrated a correlation between expression of phenylpropanoid biosynthetic genes and a subset of genes encoding ATP-binding cassette (ABC) transporters, especially in the ABCB/multi-drug resistance/P-glycoprotein (ABCB/MDR/PGP) and ABCG/pleiotropic drug resistance (ABCG/PDR) subfamilies. The objective of this study was to characterize these ABC transporters in terms of their gene expression and their function in development of lignified cells. Based on in silico analyses, four ABC transporters were selected for detailed investigation: ABCB11/MDR8, ABCB14/MDR12, ABCB15/MDR13, and ABCG33/PDR5. Promoter::glucuronidase reporter assays for each gene indicated that promoters of ABCB11, ABCB14, ABCB15, and ABCG33 transporters are active in the vascular tissues of primary stem, and in some cases in interfascicular tissues as well. Homozygous T-DNA insertion mutant lines showed no apparent irregular xylem phenotype or alterations in interfascicular fibre lignification or morphology in comparison with wild type. However, in abcb14-1 mutants, stem vascular morphology was slightly disorganized, with decreased phloem area in the vascular bundle and decreased xylem vessel lumen diameter. In addition, abcb14-1 mutants showed both decreased polar auxin transport through whole stems and altered auxin distribution in the procambium. It is proposed that both ABCB14 and ABCB15 promote auxin transport since inflorescence stems in both mutants showed a reduction in polar auxin transport, which was not observed for any of the ABCG subfamily mutants tested. In the case of ABCB14, the reduction in auxin transport is correlated with a mild disruption of vascular development in the inflorescence stem.
Keywords: Arabidopsis thaliana, ATP-binding cassette transporter, auxin, cis-element, lignin, monolignol, polar auxin transporter, vascular bundle
Introduction
Lignified secondary cell walls are essential for plants, allowing water conduction in the vasculature and providing mechanical strength to the plant body. Lignin is the major strengthening component in the secondary cell wall and consists of a complex polymer of monolignols, which are produced by the phenylpropanoid pathway, and deposited into the thickened secondary polysaccharide wall. Intensive research using genetic and biochemical approaches has revealed Arabidopsis thaliana phenylpropanoid pathway genes that are involved in developmental monolignol biosynthesis (Boerjan et al., 2003; Van Holme et al., 2008). However, the mechanisms of monolignol export from the cytoplasm to the cell wall are not clear. In developing pine tracheids, autoradiographic evidence implicated either membrane transporters or diffusion as routes for monolignol export rather than Golgi-mediated vesicle export (Kaneda et al., 2008).
The inflorescence stem of Arabidopsis has been used to study the lignification process (Turner et al., 2001), due to the abundance of lignin-rich cell types such as tracheary elements in the primary xylem tissue, and interfascicular fibres (IFFs) between the vascular bundles (Lev-Yadun, 1997; Altamura et al., 2001; Lev-Yadun et al., 2005). Moreover, the vascular tissues and IFFs differentiate at predictable positions in the stem, with fibre differentiation following vascular bundle differentiation towards the bottom of the stem. Using whole transcriptome profiling, Ehlting et al. (2005) determined which genes, including those encoding phenylpropanoid pathway enzymes, are differentially expressed along the axis of bolting stems in Arabidopsis. Based on co-expression analysis with phenylpropanoid genes, multiple ATP-binding cassette (ABC) transporter genes were identified whose gene expression was correlated with lignification.
The ABC transporter superfamily is one of the largest transporter protein families in plants (Rea, 2007; Verrier et al., 2008). More than 120 genes have been annotated in the Arabidopsis genome based on specific nucleotide binding motifs and multiple transmembrane domains (Sanchez-Fernandez et al., 2001; Verrier et al., 2008). Plant ABC transporters have diverse transport substrates including fatty acids, cuticular lipids, auxin, heavy metals, xenobiotics, and secondary metabolites. Multiple members of the ABCB/PGP/MDR subfamily are involved in transport of auxin (Geisler and Murphy, 2006), one of the most important hormones for plant differentiation and response to environmental signals. Polar auxin transport and accumulation are important factors determining vascular differentiation and are critical for the development of cells with secondary cell walls (Woodward and Bartel, 2005; Demura and Fukuda, 2007).
In this study, ABC transporter candidate genes whose expression is correlated with phenylpropanoid biosynthetic gene expression in the Arabidopsis inflorescence stem (Ehlting et al., 2005), were examined. The top four candidate ABC transporter genes were expressed in the developing stem vasculature, indicating that they may function, directly or indirectly, in vascular/fibre differentiation, secondary cell wall deposition, or lignification. Multiple alleles of mutant lines for all four genes were tested with regard to xylem and interfascicular fibre phenotypes related to lignin and auxin transport. Mutants with lesions in ABCB candidate genes, e.g. abcb14-1 and abcb15, showed reduced auxin transport in the stem and one mutant, abcb14-1, also had altered vascular bundle organization. However, none of the mutant lines displayed phenotypic changes in lignification patterns, possibly due to redundant functions of the many co-expressed ABC transporters.
Materials and methods
Plant materials and growth conditions
A. thaliana (ecotype Col-0) was used as the wild-type ecotype. Wild-type and transgenic seeds were surface-sterilized with 3% H2O2 in 50% ethanol. Sterile seeds were plated on A. thaliana (AT) medium (Haughn and Somerville, 1986) containing 0.9% agar in plastic Petri dishes and kept at 4 °C in the dark for 2 d. Seedlings were grown at 21 °C under continuous light for 7 d. Plants were transferred to soil (MetroMix#3; SunGro, USA) and grown at 21 °C under long-day conditions (16 h light/ 8 h dark) or 24 h light in a Conviron E7/2 growth chamber (Winnipeg, Canada).
T-DNA insertion mutant lines for candidate ABC transporters (Supplementary Table S1 available at JXB online) were obtained from the Arabidopsis Biological Resource Center (ABRC, The Ohio State University, USA). The presence of the T-DNA insertion in the gene of interest was confirmed with PCR, using a combination of gene-specific primers, and T-DNA-specific primers (LBa1 and LBb1) described in Supplementary Table S2 (at JXB online). RNAi lines for ABCB14 (N213339–N213349) were obtained from the Nottingham Arabidopsis Stock Center (Nottingham, UK; http://www.agrikola.org) and the insertion validated by PCR using RNAi vector-specific primers. Homozygous lines of abcb15 T-DNA mutant lines (SALK_034562 and SALK_036871) were the kind gift of Dr Edgar Spalding (University of Wisconsin, Madison, WI, USA). Mutant lines were screened for reduced mRNA levels using reverse transcription-PCR (RT-PCR) (Supplementary Fig. S1 at JXB online). Total RNA from T-DNA insertion mutants and RNAi mutants was extracted using TRIzol reagent (Invitrogen), then used as a template for cDNA synthesis with oligo(dT) primers. The cDNA obtained was used for semi-quantitative RT-PCR with target gene-specific primers (Supplementary Table S2 at JXB online).
In silico promoter cis-element analysis
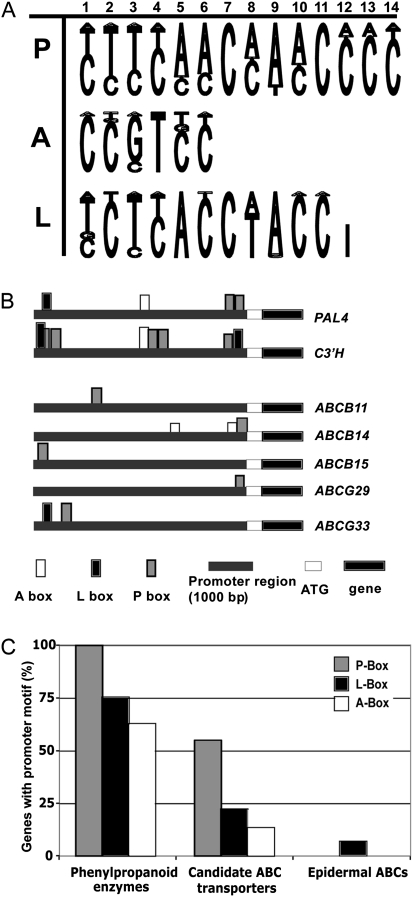
Based on earlier studies of phenylpropanoid biosynthesis pathway-specific cis-elements (Lois et al., 1989; da Costa e Silva et al., 1993; Mizutani et al., 1997), 1000 bp upstream promoter regions of candidate ABC transporter genes were examined using ‘MotifViz’ (http://biowulf.bu.edu/MotifViz). Upstream promoter region sequences were obtained from the ‘The Arabidopsis Information Resource’ (TAIR) database (http://Arabidopsis.org) and analysed using consensus matrices for three weighting elements: A box, P box, and L box (Fig. 1A). The consensus matrices were established from published putative cis-acting elements common to phenylpropanoid gene promoters, using the similarity to the previously established in vivo DNA footprint of the parsley phenylalanine ammonia lyase promoter (Petroselinum crispum PcPAL1; Lois et al., 1989) (see Fig. 1 and Supplementary Table S3 at JXB online).
Fig. 1.
In silico promoter analysis of candidate ABC transporters whose expression is correlated with lignin biosynthesis pathway enzyme genes. (A) Sequence logos visualizing the consensus of AC-rich P, A, and L boxes. (B) The presence of P, A, and L boxes, motifs commonly found upstream from genes encoding phenylpropanoid biosynthetic enzymes, as illustrated for phenylalanine ammonia-lyase (PAL) and coumaryl-ester-3′-hydroxylase (C3′H) promoters, mapped upstream of the translational start codon for each candidate ABC transporter gene. The coding region is shown in black. (C) The proportion of genes that contain a P, L, or A motif within each gene group: eight phenylpropanoid biosynthetic enzyme genes; nine stem ABC transporters, which positively correlated with phenylpropanoid gene expression; and, as a negative control, five epidermally expressed ABC transporters of the Arabidopsis stem.
Generation and transgenic plants
To generate promoter::β-glucuronidase (GUS) fusion lines of the selected ABC transporter genes, a 1-kb fragment upstream of the ABCG33/PDR5 (At2g37280), ABCB14/MDR12 (At1g28010), ABCB11/MDR8 (At1g02520), and ABCB15/MDR13 (At3g28345) genes were amplified by PCR. Upstream fragments were amplified from purified genomic wild-type Col-0 DNA using proofreading DNA polymerase, Pwo (Roche, Germany) and primers described in Supplementary Table S2 at JXB online. PCR products were digested with the respective restriction enzymes, cloned into pUC10, and validated by sequencing. After BamHI digestion, inserts were purified and subcloned into the pCAMBIA1281Z binary vector. The constructs (pCAMBIA-ABCG33promoter::GUS, pCAMBIA-ABCB11promoter::GUS, pCAMBIA-ABCB14promoter::GUS, and pCAMBIA-ABCB15promoter::GUS) were introduced into Agrobacterium tumefaciens strain LB4404. Arabidopsis Colombia ecotype was transformed with these constructs by the floral dip method (Clough and Bent, 1998) or spraying (Chung et al., 2000). Transgenic T1 plants were selected on AT medium plates that contained 30 μg/ml hygromycin.
For in situ GUS staining, inflorescence stems and seedlings of T2 plants were first incubated in 90% acetone for 30 min under vacuum. Then, tissues were rinsed three times with buffer A [50 mM Na-phosphate buffer pH 7.0, 500 nM Fe(CN), 0.1% Triton X-100] and incubated with X-Gluc (1 mM 5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid) in buffer A at 37 °C overnight. Stained tissues were cleared with 50% ethanol. Arabidopsis tissues with GUS staining were transferred to 100% acetone; then the inflorescences were infiltrated in Spurr's resin through a graded resin series using a low-temperature microwave (PELCO 3450 Laboratory Microwave, USA). The samples, under vacuum, were microwaved for 3 min each in a graded resin series at 28 °C and polymerized at 70 °C. The polymerized samples were sliced into 1- to 2-μm sections. The sections were viewed with a Leica DMR compound microscope (Leica, Wetzlar, Germany) and images were captured with a Q-Cam camera (Q-Imaging, Burnaby, BC, Canada).
For histochemical staining of pABCB14::GUS expression in stems, longitudinally dissected stems from young transgenic and wild-type plants were incubated in 50 μM C12-fluorescein di-β-D-glucuronide (ImaGene Green, Molecular Probes I-2908, Invitrogen; www.invitrogen.com) at room temperature for 3 h. The dissected surface of the stem was observed using fluorescence microscopy to detect the yellow–green emission of fluorescent GUS products.
Light microscopy and lignin histochemistry
Sections of Arabidopsis inflorescence stems at defined developmental stages were hand-cut with a razor and mounted on glass slides. Adjacent sections were stained with toluidine blue for structural reference or phloroglucinol for lignin. The sections were observed using a Leica DMR microscope with fluorescent source.
Polar auxin transport assays
Basipetal auxin transport in inflorescence stems was measured using [3H]indole acetic acid ([3H]IAA; GE Healthcare catalogue number TRK781), using the protocol of Okada et al. (1991). When Arabidopsis inflorescence stems were 10–15 cm long, the top 3 cm of the stems were harvested and all floral parts were removed. The apical 2–3 mm of the stems were submerged in 30 μl of radiolabelled IAA solution (5 mM MES buffer containing 1.45 μM IAA, including 100 nM [3H]IAA, with 1% sucrose, pH 5.5) for 24 h at room temperature in darkness. The basal 1.5-cm region of stems, which had been upended in the vial, were carefully removed, ensuring that they never contacted the radioactive solution, and were placed into a vial with 3 ml of scintillation cocktail (Sigma) for 24 h. For controls, 10 μM 1-N-naphthylphthalamic acid (NPA) was added to the radiolabelled IAA solution or stems were incubated with the basal, rather than the apical, portion of the stem (upright) in the radiolabelled solution. 3H content was measured by scintillation counting (Beckman LS600IC) for 2 min. Between 5 and 10 stems per genotype were used in each experiment in two to five replicates. Where assumptions of normalcy were met, polar auxin transport in mutant or RNAi lines was compared with wild type using Student's t-test.
pDR5::GUS in abcb14-1 and wild-type genotypic backgrounds
Plants containing the pDR5::GUS construct (Ulmasov et al., 1997) were crossed with the abcb14-1 mutant or wild-type (Col-0) line. The presence of the pDR5::GUS construct and the T-DNA insertions in the mutants were validated by PCR using gene-specific primers and T-DNA-specific primers, as described above. Excised stems were treated with 1.45 μM IAA for 12 h and subsequently stained with X-Gluc as described above. For high-resolution analysis of auxin distribution in wild-type and abcb14 RNAi plants, crosses between homozygous abcb14 RNAi and homozygous pDR5rev::3XVENUS-N7 plants (Heisler et al., 2005) were made to generate double heterozygous plants. The resulting F1 plants were assayed for abundance of ABCB14 transcripts (Supplementary Fig. S1 at JXB online) and VENUS fluorescence to confirm the genotypes. Wild-type plants heterozygous for the pDR5rev::3XVENUS-N7 element were used as controls. Inflorescence stem segments from 10- to 15-cm high inflorescences were excised and immediately longitudinally bisected and mounted in 10% glycerol. These samples were imaged using a Quorum Wave FX spinning-disc scan head on a Leica DMI6000 microscope using a Hamamatsu ImagEM CCD camera. VENUS fluorescence was excited using a 491 laser with a 528- to 566-nm filter combination and a 30- to 35-μm thick image stack was acquired at 1-μm intervals. Image analysis was processed using Volocity (Improvision) software and the fluorescence index of any image was calculated as follows: the volume, in μm3, of each fluorescent nucleus was multiplied by the average greyscale value of that nucleus (e.g. 505 μm3×230=1.1×105) and these values were summed to produce the fluorescence index value for that field of view. After confirmation of the normal distribution of the data, statistical analysis of these values was performed using Student's t-test.
Phylogenetic reconstruction of the Arabidopsis ABCB/MDR/PGP subfamily
ABCB/MDR protein sequences from Arabidopsis were retrieved from TAIR (v9; www.Arabidopsis.org) and aligned using Dialign2 (Morgenstern et al., 2006). Only amino acid positions with >40% diagonal similarity were retained for further analyses. The corresponding alignment is given in Supplementary Fig. S4 (at JXB online). Maximum parsimony analyses employing a heuristic search were performed using ‘PAUP’ (v4.0b8; Swofford, 2003). Bootstrap analyses were performed with 100 replicates.
Results
Selection of Arabidopsis stem ABC transporter genes correlated with lignification.
Selecting candidate ABC transporter genes that are correlated with vascular and interfascicular fibre maturation was possible by observing the changes in gene expression level observed along the inflorescence stem (Ehlting et al., 2005). This Arabidopsis full genome microarray data set showed that 14 ABC transporter genes (Table 1) were differentially expressed along the axis of developing primary stems. Of these, nine displayed expression profiles that were positively correlated with the expression of known phenylpropanoid biosynthetic genes. Some positively correlated genes, such as ABCB14/MDR12, ABCG29/PDR1, and ABCG33/PDR5, were more highly expressed in the shoot apex, a region of the stem where vascular bundle development is pronounced but interfascicular fibres have not yet begun to lignify. Others showed increasing expression towards the base of the stem where interfascicular fibres are lignifying, in a pattern closely matching that of phenylpropanoid biosynthetic genes (ABCB11/MDR8, ABCB15/MDR13, ABCG22/WBC23, ABCG36/PDR8/PEN3, Table 1).
Table 1.
ABC transporter genes whose expression is correlated with phenylpropanoid biosynthetic genes in Arabidopsis inflorescence stems according to Ehlting et al. (2005)
| MIPS locus | ABC transporter nomenclaturea | Synonymsb | Correlation with phenylpropanoid gene expression | Pattern of expression in stem | |
| 1 | At3g28345 | ABCB15 | MDR13/PGP15 | positive | High from 3 cm to base |
| 2 | At1g02520 | ABCB11 | MDR8/PGP11 | positive | High from 3 cm to base |
| 3 | At5g06530 | ABCG22 | WBC23 | positive | Increasing towards base |
| 4 | At1g59870 | ABCG36 | PDR8/PEN3 | positive | Increasing towards base |
| 5 | At5g61690 | ABCA11 | ATH15 | positive | High in 7- to 9-cm segment |
| 6 | At3g16340 | ABCG29 | PDR1 | positive | High at 3- to 5-cm segment |
| 7 | At1g28010 | ABCB14 | MDR12/PGP14 | positive | Stem apex only |
| 8 | At2g37280 | ABCG33 | PDR5 | positive | Stem apex only |
| 9 | At1g66950 | ABCG39 | PDR13 | positive | Expressed in lower base |
| 10 | At4g15230 | ABCG30 | PDR2 | negative | Negative in 3- to 5-cm stem |
| 11 | At5g61730 | ABCA9 | ATH11 | negative | Weakly expressed in base |
| 12 | At4g25960 | ABCB2 | MDR2/ PGP2 | negative | Negative at base |
| 13 | At2g26910 | ABCG32 | PDR4 | negative | Weakly expressed in base |
| 14 | At2g13610 | ABCG5 | WBC5 | negative | Negative at base |
Operon ‘longmer’ microarrays were used to compare gene expression in Arabidopsis stem segments over a developmental gradient from stem apex to base, with reference to the 0- to 2-cm apical region prior to lignification.
Plant ABC proteins updated nomenclature by Verrier et al. (2008).
Arabidopsis ABC protein system by (Sanchez-Fernandez et al. 2001), Martinoia et al. (2002).
Genes encoding phenylpropanoid biosynthetic enzymes contain short conserved cis-acting elements in their promoter region (reviewed by Rogers and Campbell, 2004). If these elements are present in selected ABC transporter gene promoters, there is an increased probability that these ABC transporter genes would be co-regulated with lignin biosynthesis pathway genes and could be involved in development of lignified cells. The 1000 bp upstream from each candidate ABC transporter gene were tested for the presence of three putative cis-acting elements: A box (CCGTCC), P box (CTTCAACCAACCCC), and L box (TCTCACCTACC) motifs (Fig. 1A), using ‘MotifViz’ (http://biowulf.bu.edu/MotifViz) with A, P, and L box nucleotide matrices (Supplementary Table S3 at JXB online). Eight genes encoding monolignol biosynthesis pathway enzymes all contained the P box motif and four out of the eight biosynthetic genes contained all three elements (examples are shown in Fig. 1B). Five out of nine candidate ABC transporters whose gene expression was correlated with lignification also contained the P box in their upstream sequence (Fig. 1B). While phenylpropanoid gene promoters were likely to contain also L and A motifs, ABC transporter gene promoters contained these elements in some, but not all, cases (Fig. 1C). As a negative control, the promoter regions of ABC transporter genes, whose expression is higher in the epidermis than the stem as a whole (Suh et al., 2005), were also examined for the occurrence of these cis-elements. Epidermal ABC transporters’ promoters contained no P or A motifs and only rarely L boxes (Fig. 1C). Among stem ABC candidates, ABCG36/PDR8/PEN3, ABCG22/WBC23, ABCG39/PDR13, and ABCA9/ATH11 lacked any of these motifs. The motifs present in the remaining candidate ABC transporters’ promoters support the hypothesis that these genes may be involved in lignification.
One of the problems with microarray data sets is that often the extracted mRNA arises from multiple tissues and cell types. Cross-referencing multiple data sets is one way to narrow gene expression to a specific cell type or developmental stage of interest, in this case, lignification. In order to eliminate candidates that are expressed in non-lignifying tissues, a second microarray dataset was used, describing epidermis-enriched gene expression in the inflorescence apex and base (Suh et al., 2005) where ABC transporters are known to be involved in lipid export to the cuticle (reviewed by Bird, 2008). Candidate ABC transporter genes whose expression was correlated with phenylpropanoid biosynthetic genes but whose epidermal expression makes them unlikely candidates for developmental lignification-related processes in secondary cell walls were thus eliminated (Supplementary Table S4 at JXB online). Both ABCG41/PDR13 and ABCG39/PDR8/PEN3, which had been identified in a forward genetic screen for powdery mildew resistance (Stein et al., 2006), fell into this category.
Candidate ABC transporters were examined for predicted subcellular targeting information. If a transporter were involved in monolignol export, it would be predicted to be located at the plasma membrane (PM), requiring it to be trafficked via the endoplasmic reticulum (ER) and Golgi during its biosynthesis. N-terminal hydrophobic signal sequences predict N-terminal targeting to the ER and endomembrane system, while an internal hydrophobic sequence of amino acids can act as a ‘start transfer’ signal to target a membrane protein with a cytoplasmic N-terminus to the ER. Several of the ABC transporter candidates have been detected in the PM by either proteomics (ABCB11/MDR8; Dunkley et al., 2006) or green fluorescent protein fusion localization (ABCG39/PDR8/ PEN3; Stein et al., 2006). Using Aramemnon, a compendium of web-based targeting prediction programs (http://aramemnon.botanik.uni-koeln.de/), six ABC transporters are predicted to have ‘Other’ as a protein destination (Supplementary Table S4 at JXB online). Thus, they could have internal start transfer sequences and cytoplasmic N-terminal sequences, the targeting prediction expected for the protein architecture of both the ABCB/MDR and ABCG/PDR subfamilies (Verrier et al., 2008). In some cases, sequences predicted targeting of ABC transporters to the chloroplasts (ABCG29/PDR1) (Koo and Ohlrogge, 2002) or the targeting predictions were unclear (ABCA11/ATH), thus these candidates were not investigated further.
After predicted targeting information was considered, the remaining four candidate ABC transporters, summarized in Table 2, were further investigated by promoter activity assays and analysis of T-DNA insertion mutants.
Table 2.
Candidate ABC transporter genes have higher stem than epidermal gene expression, are predicted to target by internal signal sequences (‘other’ than nucleus, ER, mitochondrial or chloroplast) and contain cis-element motifs typical of lignin biosynthetic genes
| MIPS locus | ABC transporter nomenclaturea | Synonymsb | Epidermal expression patternc | Targeting predictiond | P/L/A boxe | |
| 1 | At1g02520 | ABCB11 | MDR8 | ND | Other | P |
| 2 | At1g28010 | ABCB14 | MDR12 | Stem>Epi | Other | AP |
| 3 | At3g28345 | ABCB15 | MDR13 | Stem>Epi | Other | P |
| 4 | At2g37280 | ABCG33 | PDR5 | Stem>Epi | Other | LP |
Data from stem epidermal peel with reference to whole stems by Suh et al. (2005). ND, no data.
Subcellular targeting predictions using Aramemnon.
Presence of P/L, and A nucleotide elements in the 1000 bp upstream of candidate genes was determined using MotifViz.
Promoter::GUS assays indicate that ABCB11, ABCB14, ABCB15, ABCG33 have vascular expression patterns.
The previously described microarray gene expression data indicated that the candidate ABC transporter genes have differential developmental expression along the inflorescence stem. To increase spatial resolution of expression, promoter::GUS constructs were generated to distinguish whether these promoters are active in the tissues of the vasculature or interfascicular regions.
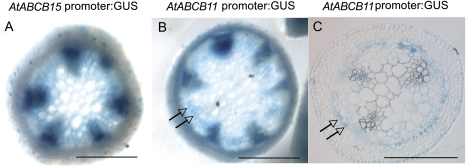
Transgenic plants expressing either the upstream sequences of ABCB11 or ABCB15 fused to GUS showed chromogenic substrate staining in overlapping patterns in both the apical (Fig. 2) and basal (Fig. 3) regions of inflorescence stems. Hand-cut sections of the top 2 cm of the inflorescence stems showed GUS activity in the vascular bundles, with some activity in the pith including the interfascicular regions (Fig. 2A, B). In resin sections of plants expressing pABCB11::GUS, the strong promoter activity found in vascular bundles could be resolved into differentiating xylem tracheary elements, parenchyma cells, and phloem of the young stem (Fig. 2C). In addition, indigo GUS substrate was observed in fibre precursor cells, i.e. parenchymous cells between vascular bundles.
Fig. 2.
Promoter::GUS activity assays for ABC transporter genes ABCB11 and ABCB15 in apical stem segments. (A) Hand-cut section of pABCB15::GUS transgenic stem segments, 3 cm from the shoot apical meristem, showing strong promoter activity in vascular bundles and interfascicular regions. (B) pABCB11::GUS plants at the same developmental stage with similar strong vascular bundle and interfascicular promoter activity. (C) Resin section of stems from pABCB11::GUS transgenic plants with GUS activity staining in interfascicular fibre precursors, xylem parenchyma, tracheary elements, and phloem. Arrows indicate interfascicular region. Scale bar 250 μm.
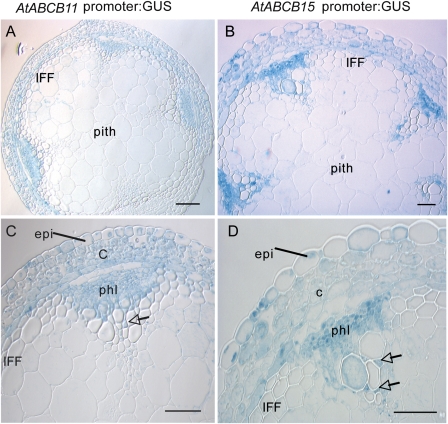
Fig. 3.
Promoter:: GUS activity assays for pABCB11 and pABCB15 in mature stem segments, within 1 cm of rosette. (A) and (C) Resin section of stems from pABCB11::GUS transgenic plants with GUS activity staining in vascular bundles, both in phloem (phl) and xylem parenchyma (arrow), cortex (c), and epidermis (epi). (B) and (D) Resin section of stems from pABCB15::GUS transgenic plants with GUS activity staining in vascular bundles, both phloem and xylem, as well as cortex. There is no activity in the mature interfascicular fibres (IFF). Scale bar 50 μm.
In the more mature base of the stem, both pABCB15::GUS and pABCB11::GUS activity was strong in the phloem, as well as in xylem parenchyma (Fig. 3A, B). As the stem matured, there was a subtle shift in the pattern of both genes’ promoter activity: the vascular bundles were still stained but there was additional GUS activity in the photosynthetic cortex and the epidermis, including guard cells (Fig. 3C, D). At this stage of development, the majority of the interfascicular fibres are mature around the stem periphery (Altamura et al., 2001) and promoter activity in vascular bundles was stronger than interfascicular fibre expression. The strong expression in the cortex and epidermis, combined with continuous expression in the phloem (Fig. 3C, D), could explain the pattern of gene expression as determined by microarrays, which indicated a steady increase in intensity towards the base of the stem.
As with the ABCB subfamily members described above, the upstream sequence of ABCG33/PDR5 fused to GUS produced reporter activity in the vascular bundles, in both xylem and phloem, and also in differentiating fibres/interfascicular parenchyma cells of the young stem (Supplementary Fig. S2 at JXB online). Promoter activity of pABCG33::GUS decreased in intensity towards the base of the stem, as predicted by the microarray data. In the stem base, the epidermis and the phloem contained GUS product, with weak staining in pith and cortex cells.
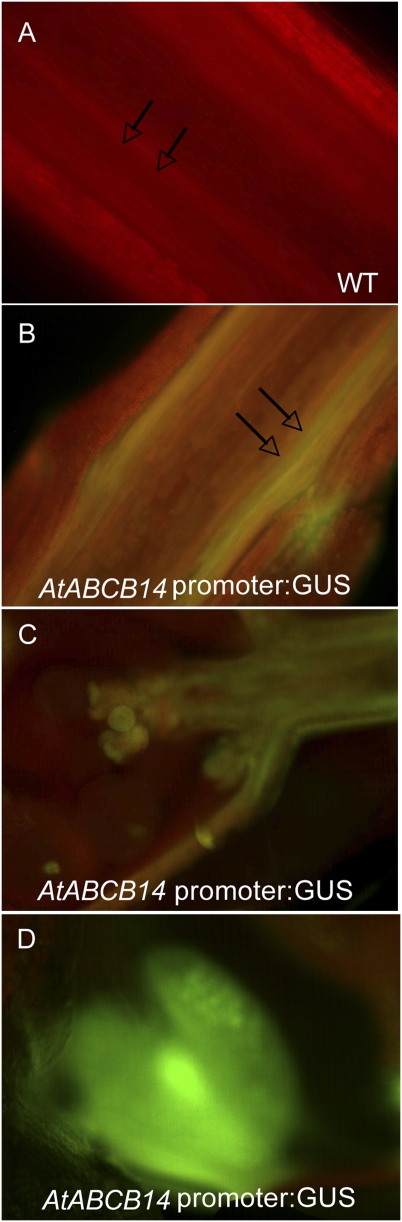
The upstream sequence that produced the weakest GUS activity was that of ABCB14. The conventional chromogenic substrate did not produce detectable signal in transgenic plants. Using a more sensitive fluorogenic substrate, ABCB14 promoter activity was observed in the procambium, in vascular bundles in the elongation region of the stem apex, and in developing pollen/anthers (Fig. 4).
Fig. 4.
Promoter:: GUS activity assays for ABCB14::GUS, with activity detected by fluorogenic substrate ImaGene Green. (A) Wild-type control, apical 1 cm of inflorescence stems longitudinal section lacking yellow–green fluorescence and displaying only red chlorophyll autofluorescence. (B) pABCB14::GUS transgenic plants stained in the vascular bundles of the apical 1 cm of stem. (C) Apex of stem with clusters of floral buds and procambium showed yellow–green fluorescence from GUS activity. (D) Anther of pABCB14::GUS transgenic plants with green fluorescence.
When whole seedlings were stained, GUS activity was observed in developing vascular cylinders above the differentiation zone in seedling roots of pABCB15::GUS, pABCB11::GUS, and pABCG33::GUS (Supplementary Fig. S3 at JXB online).
Reverse genetics of candidate ABC transporter genes in Arabidopsis.
For functional analysis of the selected ABC transporter genes, T-DNA insertion mutant lines were identified from the SIGnal database (http://signal.salk.edu) and obtained from the ABRC (Supplementary Table S1 at JXB online). Lines were selected based on the position of the insertion in the gene, in an exon or promoter, to increase the likelihood of disrupting function. All lines were tested with RT-PCR to assess the presence or absence of transcript (Supplementary Fig. S1 at JXB online).
Based on the expression patterns and correlations between phenylpropanoid biosynthetic gene expression and the candidate ABC transporters, it was predicted that the loss of function of a candidate ABC transporter gene might lead to abnormal vascular tissue development. Transverse hand-cut sections of inflorescence stems were cut at various developmental stages and stained with toluidine blue or phloroglucinol. Vascular bundle morphology was compared between wild type and mutant at early stages of development, i.e. 3 cm from the top of a 10-cm stem, and in mature vascular bundles at the base of the stem. There were no changes in lignification or significant differences from wild-type vasculature in the mutant lines (abcb11, Salk_094249; abcb14-1, Salk_016005; abcb14-2, Salk_026876; abcb15-1, WiscDsLox501E11; abcb15-2, Sail1187-C04; abcg29, Salk_113825; abcg33, Salk_002380), with the exception of the abcb14-1 T-DNA insertion line.
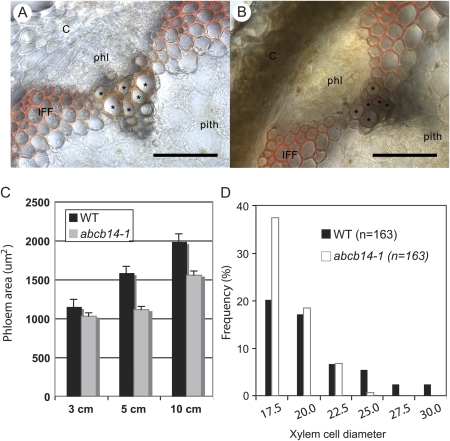
The abcb14-1 mutant stem had slightly altered vascular bundles, with smaller diameter vessels and the phloem occupying a smaller cross-sectional area than in wild-type stems (Fig. 5). In transverse sections along the developmental axis of the stem, the phloem area of abcb14-1 plants was significantly smaller than wild-type phloem throughout the stem (Fig. 5C). In addition, the vessel elements of the abcb14-1 metaxylem in vascular bundles were reduced in diameter compared with wild-type xylem (Fig. 5A compared with B). To quantify this reduction, xylem cell diameters from the largest lignified cells in mutant and wild type were binned into size classes. The abcb14-1 xylem cells tended to fall in the smallest size class and only wild type were in the largest size classes (Fig. 5D). This abcb14-1 Salk line was the same mutant line studied by Lee et al. (2008) and, similar to their data, no transcript was detected in this line (Supplementary Fig. S1 at JXB online). Unlike their RT-PCR, transcript was detected in abcb14-2 (Supplementary Fig. S1 at JXB online), so this allele was not used for further characterization.
Fig. 5.
Altered vasculature phenotype in the inflorescence stems of abcb14-1 mutant. The vascular bundles of (A) wild-type (Col-0) stem and (B) abcb14-1 stems show xylem (asterisks) with smaller vessel diameters in the mutants, demonstrated with phloroglucinol staining of lignified cells. Scale bars 50 μm. C, cortex; IFF, interfascicular fibre; phl, phloem. (C) The phloem areas of abcb14-1 stems are significantly smaller than the phloem areas in wild-type stems, measured by length and width of cross-sectioned phloem tissue. The sample size was 28–36 in wild-type, and 97–110 in abcb14-1 mutant stem. Means ± standard error. (D) The frequency distribution of metaxylem vessel diameters in wild-type (Col-0) and abcb14-1 mutant stem. The sample size was 163 cells, binned into size classes. More abcb14-1 xylem cells are in the smallest size class and only wild type are in the largest size classes.
None of the ABC transporter mutants studied had changes in interfascicular fibre development, morphology, or lignification. Unlike abcb1/pgp1 and abcb19/pgp19, the inflorescence stem development of all the mutants tested was identical to wild type except for abcb14-1, which showed slow stem development. As reported by Lee et al. (2008) knockout lines of abcb14-1 had more rosette leaves at the time of bolting and the bolting was delayed compared with the wild-type stem. At the end of the growth, however, the final stem height was the same as wild type, as found in pgp19 and pgp1 mutants (Noh et al., 2001; Geisler et al., 2005).
Polar auxin transport
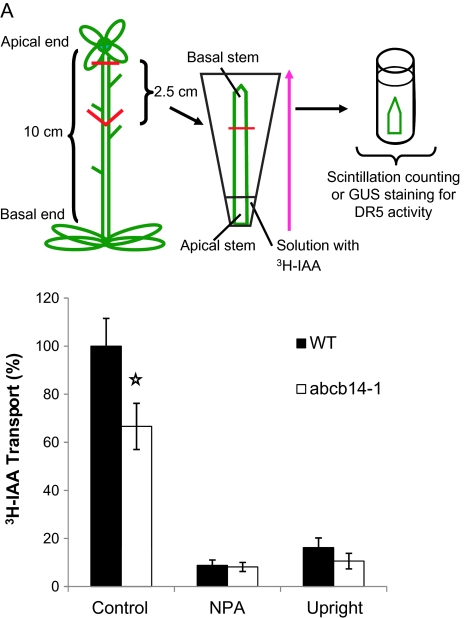
There is extensive evidence linking auxin to vascular tissue development (Mattsson et al., 1999; Woodward and Bartel, 2005; Turner et al., 2007), which led us to perform experiments to test whether auxin transport is altered in the abcb14-1 mutant plants. The morphological changes in the stem in the mutant pointed to changes in cell differentiation in the vasculature. In addition, ABCB14 belongs to the same ABC transporter subfamily as ABCB1/MDR1/PGP1 (At2g36910) and ABCB19/AtMDR11/AtPGP19 (At3g28860), which have both been identified as polar auxin transporters (Geisler and Murphy, 2006). Therefore, polar auxin transport assays were performed using inflorescence stems of wild-type Arabidopsis and abcb14-1 mutants (Fig. 6). The apical 2.5 cm of the stem was inverted in a [3H]IAA solution for 24 h to allow auxin transport. The amount of [3H]IAA that was transported to the distal portion of the stem was measured by excising the basal 1.5 cm of stem, which had never been in contact with the radiolabel, and immersing it in scintillation fluid. abcb14-1 mutants had significantly reduced polar auxin transport that was about half of the wild-type transport levels (Fig. 6B) (Student's t-test, P<0.05). The polarity of the auxin transport was confirmed by placing the stems upright, rather than inverting the apical end of the stem into the [3H]IAA, which resulted in transport being diminished to 20% of the wild-type control (Fig. 6B). Similarly, when the auxin transport inhibitor NPA was included with the [3H]IAA, then transport was strongly reduced, suggesting a transporter-mediated, rather than a bulk flow, mechanism.
Fig. 6.
Polar auxin transport decreases in inflorescence stems of abcb14-1 mutants. (A) Schematic diagram of polar auxin transport assay for inflorescence stem. The apical 2.5 cm of stem was dissected, inverted into [3H]IAA (centre) and incubated for 24 h. The uppermost stem segment was removed and radioactivity measured. A pink arrow indicates directional IAA transport from the apical side of the stem. (B) The amount of transported IAA in abcb14-1 inflorescence stems was significantly reduced compared with wild type. Mean ± standard error, Students t-test (P<0.05). Controls were treated with polar auxin inhibitor NPA or placed with the basal end of the stem in [3H]IAA and apical zone facing up (upright) to show that auxin transport was polar.
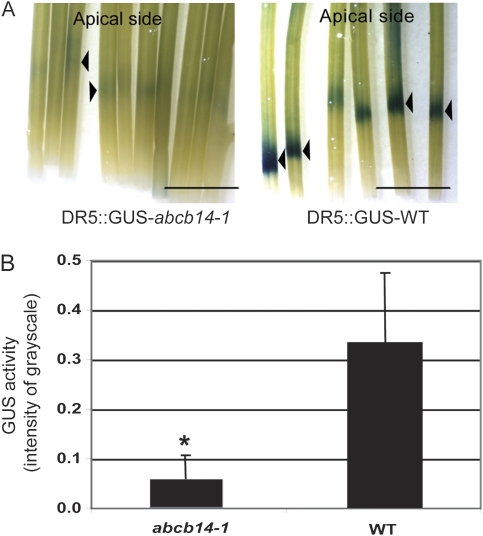
In order to observe altered auxin distribution in the abcb14-1 mutants, plants carrying the synthetic auxin-responsive promoter DR5 fused to GUS (Ulmasov et al., 1997) were crossed into both abcb14-1 and wild type. When inflorescence stems were tested for GUS activity, there were no auxin distribution differences detected in abcb14-1 mutants compared with wild type, even in the apical procambial region where the altered vasculature of the mutant phenotype was presumably determined. However, when exogenous IAA was applied in the polar auxin transport assay, and the stems tested for GUS activity, pDR5::GUS-wild-type stems showed a strong blue product in a band pattern near the cut end of the stem, while pDR5::GUS abcb14-1 stems only had a faint blue band (Fig. 7A). This could be quantified from digital images as pixel greyscale densities, which demonstrated that pDR5::GUS abcb14-1 stems translocated significantly less product than pDR5::GUS-wild-type controls (P<0.05 using Student's t-test; Fig. 7B).
Fig. 7.
Decreased polar auxin transport in inflorescence stems of abcb14-1 mutants detected with auxin response reporter pDR5::GUS. (A) Polar auxin transport was detected by induction of pDR5::GUS reporter activity in wild-type (wild type) and abcb14-1 stems. (B) Quantification of GUS activity by measuring substrate intensity in the basal stem segment suggests that less auxin was transported in the mutant background. Bar 5 mm, mean±SD. Three independent experiments were performed. An asterisk indicates significant differences from wild type by Student's t-test (P<0.01, n=40).
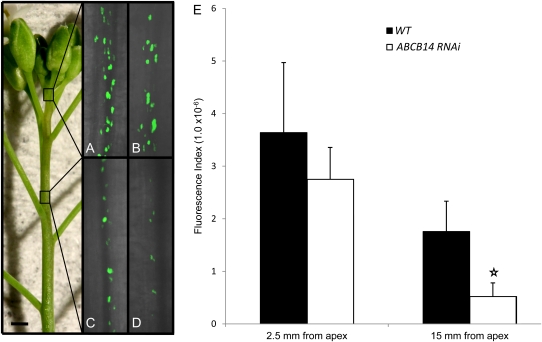
The pDR5::GUS construct has been extensively used as a reliable indicator of auxin accumulation in plants and gave results that were consistent with radiolabelled bulk polar auxin transport assays. However, it was not sensitive enough to detect subtle changes in auxin distribution in the procambium of the stem. A more sensitive pDR5rev promoter containing nine tandem repeats of the auxin-responsive element (TGTCTC) driving the expression of either green fluorescent protein or yellow fluorescent protein (VENUS) have been used in a number of high-impact studies to visualize discrete auxin gradients (Benkova et al., 2003; Friml et al., 2003; Heisler et al., 2005). To assess whether abcb14 loss of function influenced auxin distribution, the fluorescence of the nuclear localized VENUS protein driven by the auxin-responsive promoter pDR5rev was examined in wild-type and ABCB14 RNAi plants. The ABCB14 RNAi plants were obtained from the Arabidopsis Genomic RNAi Knock-out Line Analysis resource (AGRIKOLA; www.agrikola.org) and several lines were confirmed to have reduced levels or absent ABCB14 transcripts (Supplementary Fig. S1 at JXB online). Since ABCB14 expression is localized to the apical region of the inflorescence stem, this region of the inflorescence stem was examined. Near the shoot apical meristem, the procambial cells of the developing vascular strands showed strong fluorescent nuclei, indicating the presence of auxin at this developmental stage (Fig. 8 A, B). As vascular development proceeded, the number and intensity of fluorescent nuclei decreased (15 mm from the apical meristem) in the wild-type and ABCB14 RNAi stems and the fluorescent signal dropped closer to the apex in the ABCB14 RNAi stems (Fig. 8C, D). The expression of the nuclear localized VENUS protein could be quantified using a fluorescence index that accounts for both the intensity of nuclear fluorescence and volume of fluorescence. At all developmental stages, there was less fluorescence in the ABCB14 RNAi plants (Fig. 8E), with statistically significant differences emerging between wild-type and ABCB14 RNAi plants at 15 mm from the apex. This observation is in agreement with the reduced polar auxin transport observed in abcb14-1 mutants and supports our hypothesis that ABCB14 is required for auxin flow during early stages of inflorescence development.
Fig. 8.
Decreased auxin reporter (pDR5rev:VENUS) fluorescence in vascular strands of apical inflorescence stem segments from abcb14 RNAi plants. Representative images of nuclear localized pDR5rev::3XVENUS fluorescence in vascular strands from wild-type (A, C) and abcb14 RNAi (B, D) inflorescence stems at ∼2.5 mm (A, B) and 15 mm (C, D) from the inflorescence apex. (E) Quantification of values generated from average brightness and total volume of fluorescence from wild-type and abcb14 RNAi plants imaged under identical conditions. White star indicates a significant difference between images derived from wild type compared with abcb14 RNAi imaged at 15 mm from the apex (Student's t-test, P<0.05). Error bars represent the standard error and size bar is 2 mm.
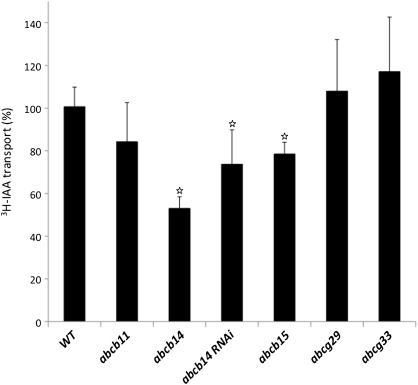
Since ABCB14 RNAi plants were used instead of abcb14-1 mutants in the pDR5rev::VENUS experiments, the polar IAA transport phenotype in ABCB14 RNAi plants was assessed and it was found that these plant lines also had significantly reduced polar IAA transport with similar reductions to the abcb14-1 mutant (Student's t-test, P<0.05; Fig. 9). Since several of the other candidate ABC transporters were from the ABCB subfamily, mutant lines (abcb11, abcb14, abcb15, abcg33, as well as abcg29 and abcg33) were tested for altered polar auxin transport (Fig. 9). Mutants in every candidate member of the ABCB subfamily tested had some level of reduced polar auxin transport, with reductions ranging from 20% to 50%. In abcb11, polar IAA transport was reduced to 80% of wild-type levels but was not significantly different after statistical analysis due to the high variance observed between different experiments. For the abcb15 mutants, polar IAA transport was reduced to 75% of wild-type levels (Student's t-test, P<0.05; Fig. 9). In contrast to the abcb subfamily mutants, there were no lines of abcg subfamily mutants with altered polar IAA transport. Polar IAA transport in abcg29 and abcg33 did not differ from wild type (Student's t-test, P>0.05; Fig. 9). These results suggest that the function of several of the ABCB transporters in the stem, at least ABCB14 and ABCB15, could be related to polar auxin transport.
Fig. 9.
Polar auxin transport of inflorescence stems in ABC transporter mutants. Average polar auxin transport in mutant stems in the ABCB subfamily was reduced, while ABCG subfamily mutants were similar to wild type. For this analysis, two to five independent experiments were repeated, resulting in 8–43 stems examined. White stars indicate that the differences between the mutant and wild type were considered significant (P<0.05) using Student's t-test. Error bars represent the standard error of percentages obtained in different replicate experiments.
Discussion
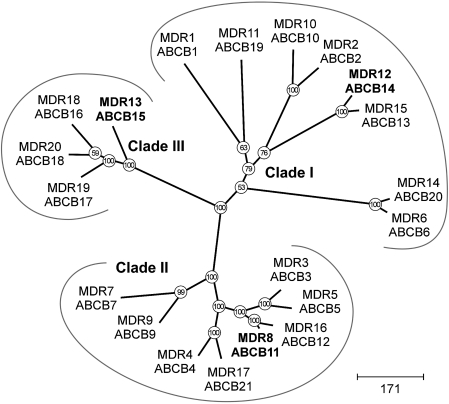
Members of the Arabidopsis ABCB subfamily of ABC transporters have been shown to function in polar auxin transport (Noh et al., 2001; Geisler and Murphy, 2006; Mvarec et al., 2008) and here it was shown that plants with mutations in additional ABCB subfamily members are characterized by reduced stem polar auxin transport. This suggests that this set of ABCB subfamily genes, whose expression was correlated with lignification in stems, plays a role in auxin transport. Phylogenetic analysis indicates that the ABCB subfamily can be subdivided into three clades (Fig. 10, Supplementary Fig. S4 at JXB online; Geisler and Murphy, 2006). Our candidate genes came from each of the clades: (i) ABCB14 from clade I whose prototype is the auxin exporter ABCB1; (ii) ABCB11 from clade II whose prototype is ABCB4 (a putative auxin importer); (iii) ABCB15 from clade III whose prototype was originally annotated as AtPGP8 (Geisler and Murphy, 2006) although that was more recently considered a pseudogene by Verrier et al. (2008). Mutants of abcb14-1 had the strongest polar auxin transport defect and this gene is a member of clade I, which also contains the best-characterized auxin-export ABC transporters, namely ABCB1/MDR1/PGP1 and ABCB19/PGP19 (Noh et al., 2001; Geisler et al., 2005; Yang and Murphy, 2009). The other mutants analysed in the present study are members of the phylogenetically divergent clades II and III. This may indicate that auxin transport function is not well correlated with primary sequence. For example, when ABCB2 was heterologously expressed in Schizosaccharomyces pombe, it did not show auxin transport activity and it is a member of clade I, which contains the most auxin transporters (Yang and Murphy, 2009).
Fig. 10.
Phylogenetic reconstruction of the Arabidopsis ABCB/MDR/PGP subfamily of ABC transporters. Arabidopsis ABCB subfamily form three clades, including clade I, which contains auxin transporters ABCB1/ABCB19. Proteins analysed in this study are highlighted in bold type. Arabidopsis protein sequences were aligned using ‘Dialign2’ and positions with at least 40% diagonal similarity were used for maximum parsimony analysis using ‘PAUP’. Bootstrap values (from 100 replicates) are given at each node.
The diversity of the ABCB transporter subfamily gene expression appears to contribute to net auxin transport in stems. It is likely that ABCB19/PGP19 is the key ABC transporter contributing to auxin transport in the Arabidopsis stem, based on the original mutant phenotype described by Noh et al. (2001), where their mdr1 mutant, now called abcb19/pgp19, had stem polar auxin transport levels at ∼40% of wild type. This hypothesis is supported by the evidence that ABCB1 and ABCB19 bind NPA (Murphy et al., 2002; Kim et al., 2010) and that the residual auxin transport in the abcb14 mutant was NPA sensitive. ABCB1/PGP1 is also expressed in the stem and the reduction in polar auxin transport in the double abcb1 abcb19 mutant phenotype suggests that these ABCBs act additively. The contributions to polar auxin transport made by ABCB14, and to a smaller extent by ABCB15, could represent either redundant or specialized functions in the stem depending on cell-specific gene expression.
In addition to the auxin transport phenotype, abcb14-1 mutants displayed subtle changes in the vascular tissue organization. The size of the phloem area was smaller than wild-type phloem and metaxylem vessels had smaller diameters than wild type. Loss of ABCB14 during procambial development could produce changes in local auxin gradients affecting vascular development generally and/or radial cell expansion of cells during procambium differentiation. The results obtained from examining the expression of the auxin reporter pDR5rev::VENUS in ABCB14 RNAi plants support this interpretation. Current models propose that pin-formed (PIN) auxin transporters, with their polar subcellular localization, export auxin along cell files, while ABCB transporters both stabilize PINs and act independently to export auxin diffusely (Mravec et al., 2008; Robert and Friml, 2009). Since it is the PINs, rather than ABCBs that produce polarity of transport, the abcb phenotype could be explained by a secondary effect on PIN proteins in the absence of ABCBs. It is clear from recent studies (Blakeslee et al., 2007; Titapiwatanakun et al., 2009), that there can be direct interactions between PINs and ABCB proteins in membrane microdomains as well as indirect, tissue-specific functional interactions.
The polar auxin transport phenotype observed in abcb14-1 mutants could be due to either changes in the vasculature, creating a smaller ‘canal’ through which auxin could flow, or changes in transport by ABCBs/PINs. However, since abcb15 also had decreased polar auxin transport phenotypes, without changes in vasculature, the decreased polar auxin transport observed was probably not due to the altered architecture of the vasculature.
A recent paper tested the abcb14-1 mutant for defects in guard cell responses to altered carbon dioxide levels and guard cell import of malate from the apoplast to control osmotic pressure (Lee et al., 2008). These authors found that ABCB14 was highly expressed in guard cells but they did not detect promoter activity in the stem procambium. Both their and our studies showed pABCB14 expression in pollen. Our GUS assay results did not show a guard cell pattern for ABCB14 expression, which might reflect that in our study, 1000 bp upstream of ABCB14 were fused to the GUS reporter instead of 2000 bp, as in Lee et al. (2008). In the upstream region used by Lee et al. (2008), there is a small open reading frame, encoding a predicted protein of 61 amino acids that is 1225 bases upstream of the ATG of the ABCB14 gene. For this reason, and to be consistent with the 1000 bp used for the in silico promoter analysis, a smaller upstream segment was used.
It is difficult to reconcile a role for the same gene product, ABCB14, in both import of malate in the guard cells and auxin transport in the stem. However, in two plant lines with reduced ABCB14 transcripts, i.e. abcb14 null mutants and ABCB14 RNAi lines, polar auxin transport in the stem is decreased. The auxin distribution in the procambial region of the inflorescence stem was altered in the ABCB14 RNAi lines and the abcb14-1 mutants had disrupted vascular development. Thus while stem auxin transport in addition to guard cell malate transport may be a surprising function, several lines of data support that it is a real function. If ABCB14 can translocate both anions, then perhaps it is importing auxin, similar to ABCB4 (Terasaka et al., 2005; Yang and Murphy, 2009).
In this study, it was hypothesized that ABC transporters were exporting monolignols during lignification, based on their coordinate expression with phenylpropanoid metabolic genes in a microarray co-expression analysis (Ehlting et al., 2005). Testing this hypothesis has proved challenging on several fronts: first, there was an abundance of ABC transporters identified in the expression profiling experiment, thus the list of candidate genes had to be narrowed using a combination of in silico prediction tools. On the next level, the promoter::GUS assays suggested that ABC transporter candidates were expressed in overlapping fashions, in both lignifying tissues such as interfascicular fibres and xylem, and non-lignifying tissues like phloem. The abcb and abcg mutants appeared phenotypically wild type with regard to lignification, a finding that was not unexpected when the overlapping expression patterns, and the possibility of redundancy, are considered. Only the generation of plant lines with multiple gene reductions, ideally in specific cell types and developmental stages, will be able to overcome this redundancy.
Although a role in monolignol export cannot be concluded or excluded, it is clear that auxin transport is disrupted in the abcb mutants. This makes it difficult to test a possible additional role in monolignol transport, since auxin is required for normal vascular development and lignification is a late event in this very same differentiation process. In addition, ABC transporters can be induced by xenobiotics or some secondary metabolites (Jasinski et al., 2001). Thus blocking transport might lead to a build-up of transport substrate, resulting in the induction of other ABC transporters for detoxification. These exported compounds might still be incorporated into the secondary cell wall, resulting in a wild-type appearance of the mutants. When lignin biosynthetic genes are altered, lignin monomer composition is altered, e.g. increased H-lignin at the expense of S- and G-lignin in C3'H down-regulated plants (Franke et al., 2002; Coleman et al., 2008), suggesting that the export and polymerization processes are somewhat flexible.
Alternatives to ABC transporter-mediated monolignol export are outlined in a review of secondary metabolism transport (Yazaki, 2006). A large family of transporters, called ‘Multi-drug and toxin exporter’ (MATE) could use the proton gradient energy to export monolignols, although there is no experimental evidence to evaluate this hypothesis. Overall, GUS staining in vasculature confirmed that the selected ABC transporters were being expressed in temporal association with lignifying cells, indicating that the microarray data sets were accurate. However, their expression was not restricted to lignifying cells suggesting that they may play a variety of functions. The overlapping gene expression patterns in vascular tissues indicate that the developing xylem tracheary elements, their surrounding parenchyma cells, and the phloem expresses multiple ABC transporters including ABCB11, ABCB14, ABCB15, and ABCG33.
ABC transporters fulfill highly divergent biochemical and physiological functions in organisms from all kingdoms. This is highlighted by the large size of the encoding gene family, most of which still await functional characterization. Testing hypotheses regarding biochemical functions of individual ABC transporters is a challenging task and the role of ABC transporters in monolignol export remains a plausible yet unproven hypothesis.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1 shows the transcriptional level by RT-PCR in ABC transporter candidate gene mutants.
Supplementary Fig. S2 shows the gene expression profile of candidate ABC transporter gene pABCG33::GUS in stem vasculature.
Supplementary Fig. S3 shows the GUS expression profiles of candidate ABC transporter genes in seedling root tissue.
Supplementary Fig. S4 shows protein alignment for phylogenetic reconstruction.
Supplementary Table S1 lists the T-DNA insertion lines and RNAi and their polymorphism sites.
Supplementary Table S2 lists primers used in this study.
Supplementary Table S3 is a nucleotide matrix of the P, A, and L boxes.
Supplementary Table S4 gives locus information and gene names used in this study for ABC transporter candidate genes.
Acknowledgments
The help of the staff of the UBC Bioimaging Facility is gratefully acknowledged. pDR5::GUS construct was a gift to Tamara Western from Elizabeth Schultz at the University of Lethbridge. The authors also thank Elliot Meyerowitz for making the pDR5rev::3XVENUS-N7 line available. This study was funded by the Canadian Natural Sciences and Engineering Research Council Green Crop Research Network project 2a and a Discovery grant to ALS.
Glossary
Abbreviations
- ABC
ATP-binding cassette
- AT
Arabidopsis thaliana
- ER
endoplasmic reticulum
- GUS
β-glucuronidase
- IAA
indole acetic acid
- IFF
interfascicular fibre
- MDR
multi-drug resistance
- NPA
1-N-naphthylphthalamic acid
- PGP
P-glycoprotein
- PM
plasma membrane
- RT-PCR
reverse transcription-polymerase chain reaction
- TAIR
The Arabidopsis Information Resource
References
- Altamura M, Possenti M, Matteucci A, Baima S, Ruberti I, Morelli G. Development of the vascular system in the inflorescence stem of Arabidopsis. New Phytologist. 2001;151:381–389. [Google Scholar]
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- Bird D. The role of ABC transporters in cuticular lipid secretion. Plant Science. 2008;174:563–569. [Google Scholar]
- Blakeslee JJ, Bandyopadhyay A, Lee AR, et al. Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. The Plant Cell. 2007;19:131–147. doi: 10.1105/tpc.106.040782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annual Review of Plant Biology. 2003;54:519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- Chung MH, Chen M-K, Pan S- M. Floral spray transformation can efficiently generate Arabidopsis transgenic plants. Transgenic Research. 2000;9:471–476. doi: 10.1023/a:1026522104478. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Coleman HD, Park JY, Nair R, Chapple C, Mansfield SD. RNAi-mediated suppression of p-coumaroyl-CoA 3′ hydroxylase in hybrid poplar impacts on lignin deposition and soluble secondary metabolism. Proceedings of the National Academy of Sciences, USA. 2008;105:4501–4506. doi: 10.1073/pnas.0706537105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa e Silva O, Klein L, Schmelzer E, Trezzini GF, Hahlbrock K. BPF-1, a pathogen-induced DNA-binding protein involved in the plant defense response. The Plant Journal. 1993;4:125–135. doi: 10.1046/j.1365-313x.1993.04010125.x. [DOI] [PubMed] [Google Scholar]
- Demura T, Fukuda H. Transcriptional regulation in wood formation. Trends in Plant Science. 2007;12:65–70. doi: 10.1016/j.tplants.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Dunkley TP, Hester S, Shadforth IP, Runions J, et al. Mapping the Arabidopsis organelle proteome. Proceedings of the National Academy of Sciences, USA. 2006;103:6518–6523. doi: 10.1073/pnas.0506958103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlting J, Mattheus N, Aeschliman DS, et al. Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. The Plant Journal. 2005;42:618–640. doi: 10.1111/j.1365-313X.2005.02403.x. [DOI] [PubMed] [Google Scholar]
- Franke R, Humphreys JM, Hemm MR, Denault JW, Ruegger MO, Cusumano JC, Chapple C. The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. The Plant Journal. 2002;30:33–45. doi: 10.1046/j.1365-313x.2002.01266.x. [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jurgens G. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature. 2003;426:147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- Geisler M, Blakeslee JJ, Bouchard R, et al. Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. The Plant Journal. 2005;44:179–194. doi: 10.1111/j.1365-313X.2005.02519.x. [DOI] [PubMed] [Google Scholar]
- Geisler M, Murphy AS. The ABC of auxin transport: the role of P-glycoproteins in plant development. FEBS Letters. 2006;580:1094–1102. doi: 10.1016/j.febslet.2005.11.054. [DOI] [PubMed] [Google Scholar]
- Haughn GW, Somerville C. Sulfonylurea-resistant mutants of Arabidopsis thaliana. Molecular and General Genetics. 1986;204:430–434. [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Current Biology. 2005;15:1899–1911. doi: 10.1016/j.cub.2005.09.052. [DOI] [PubMed] [Google Scholar]
- Jasinski M, Stukkens Y, Degand H, Purnelle B, Marchand-Brynaert J, Boutry M. A plant plasma membrane ATP binding cassette-type transporter is involved in antifungal terpenoid secretion. The Plant Cell. 2001;13:1095–1107. [PMC free article] [PubMed] [Google Scholar]
- Kaneda M, Rensing KH, Wong JCT, Banno B, Mansfield SD, Samuels AL. Tracking monolignols during wood development in lodgepole pine. Plant Physiology. 2008;147:1–11. doi: 10.1104/pp.108.121533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-Y, Henrichs S, Bailly A, Vincenzetti V, Sovero V, Mancuso S, Pollman S, Kim D, Geisler M, Nam H-G. Identification of an ABCB/P-glycoprotein-specific inhibitor of auxin transport by chemical genomics. Journal of Biological Chemistry. 2010;285:23309–23317. doi: 10.1074/jbc.M110.105981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo AJK, Ohlrogge JB. The predicted candidates of Arabidopsis inner envelope membrane proteins and their expression profiles. Plant Physiology. 2002;130:1–14. doi: 10.1104/pp.008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Choi Y, Burla B, Kim YY, Jeon B, Maeshima M, Yoo JY, Martinoia E, Lee Y. The ABC transporter AtABCB14 is a malate importer and modulates stomatal response to CO2. Nature Cell Biology. 2008;10:1217–1223. doi: 10.1038/ncb1782. [DOI] [PubMed] [Google Scholar]
- Lev-Yadun S. Fibres and fibre-sclereids in wild-type Arabidopsis thaliana. Annals of Botany. 1997;80:125–129. [Google Scholar]
- Lev-Yadun S, Wyatt SE, Flaishman MA. The inflorescence stem fibers of Arabidopsis thaliana Revoluta (ifl1) mutant. Journal of Plant Growth Regulation. 2005;23:301–306. [Google Scholar]
- Lois R, Dietrich A, Hahlbrock K, Schulz W. CG-1, a parsley light-induced DNA-binding protein. EMBO Journal. 1989;8:1641–1648. doi: 10.1002/j.1460-2075.1989.tb03554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinoia E, Klein M, Geisler M, Bovet L, Forestier C, Kolukisaoglu U, Muller-Rober B, Schulz B. Multifunctionality of plant ABC transporters: more than just detoxifiers. Planta. 2002;214:345–355. doi: 10.1007/s004250100661. [DOI] [PubMed] [Google Scholar]
- Mattsson J, Sung ZR, Berluth T. Responses of plant vascular systems to auxin transport inhibition. Development. 1999;126:2979–2991. doi: 10.1242/dev.126.13.2979. [DOI] [PubMed] [Google Scholar]
- Mizutani M, Ohta D, Sato R. Isolation of a cDNA and a genomic clone encoding cinnamate 4-hydroxylase from Arabidopsis and its expression manner in planta. Plant Physiology. 1997;113:755–763. doi: 10.1104/pp.113.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern B, Prohaska SJ, Pöhler D, Stadler PF. Multiple sequence alignment with user-defined anchor points. Algorithms for Molecular Biology. 2006;1:6–18. doi: 10.1186/1748-7188-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mravec J, Kubes M, Bielach A, Gaykova V, Petrasek J, Skupa P, Chand S, Benkova E, Zazimalova E, Friml J. Interaction of PIN and PGP transport mechanisms in auxin distribution-dependent development. Development. 2008;135:3345–3354. doi: 10.1242/dev.021071. [DOI] [PubMed] [Google Scholar]
- Murphy A, Hoogner KR, Peer WA, Taiz L. Identification, purification, and molecular cloning of N-1-naphthylphthalmic acid-binding plasma membrane-associated aminopeptidases from Arabidopsis. Plant Physiology. 2002;128:935–950. doi: 10.1104/pp.010519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh B, Murphy A, Spalding EP. Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. The Plant Cell. 2001;13:2441–2454. doi: 10.1105/tpc.010350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Ueda J, Komaki MK, Bell CJ, Shimura Y. Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. The Plant Cell. 1991;3:677–684. doi: 10.1105/tpc.3.7.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea PA. Plant ATP-binding cassette transporters. Annual Review of Plant Biology. 2007;58:347–375. doi: 10.1146/annurev.arplant.57.032905.105406. [DOI] [PubMed] [Google Scholar]
- Robert HS, Friml J. Auxin and other signals on the move in plants. Nature Chemical Biology. 2009;5:325–332. doi: 10.1038/nchembio.170. [DOI] [PubMed] [Google Scholar]
- Rogers LA, Campbell MM. The genetic control of lignin deposition during plant growth and development. New Phytologist. 2004;164:17–30. doi: 10.1111/j.1469-8137.2004.01143.x. [DOI] [PubMed] [Google Scholar]
- Sanchez-Fernandez R, Emyr Davies TG. The Arabidopsis thaliana ABC protein superfamily, a complete inventory. Journal of Biological Chemistry. 2001;276:30231–30244. doi: 10.1074/jbc.M103104200. [DOI] [PubMed] [Google Scholar]
- Stein M, Dittgen J, Sanchez-Rodrıguez C, Hou B-H, Molina A, Schulze-Lefert P, Lipka V, Somerville S. Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. The Plant Cell. 2006;18:731–746. doi: 10.1105/tpc.105.038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh MC, Samuels AL, Jetter R, Kunst L, Pollard M, Ohlrogge J, Beisson F. Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiology. 2005;139:1649–1665. doi: 10.1104/pp.105.070805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. Sunderland, MA, USA: Sinauer Associates; 2003. PAUP*: phylogenetic analysis using parsimony (*and other methods). Version 4. [Google Scholar]
- Terasaka K, Blakeslee JJ, Titapiwatanakun B, et al. PGP4, an ATP binding cassette P-glycoprotein, catalyzes auxin transport in Arabidopsis thaliana roots. The Plant Cell. 2005;17:2922–2939. doi: 10.1105/tpc.105.035816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titapiwatanakun B, Blakeslee JJ, Bandyopadhyay A, et al. ABCB19/PGP19 stabilizes PIN1 in membrane microdomains in Arabidopsis. The Plant Journal. 2009;57:27–44. doi: 10.1111/j.1365-313X.2008.03668.x. [DOI] [PubMed] [Google Scholar]
- Turner S, Taylor N, Jones L. Mutations of the secondary cell wall. Plant Molecular Biology. 2001;47:209–219. [PubMed] [Google Scholar]
- Turner S, Gallois P, Brown D. Tracheary element differentiation. Annual Review of Plant Biology. 2007;58:407–433. doi: 10.1146/annurev.arplant.57.032905.105236. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. ARF1, a transcription factor that binds to auxin response elements. Science. 1997;276:1865–1868. doi: 10.1126/science.276.5320.1865. [DOI] [PubMed] [Google Scholar]
- Van Holme R, Morreel K, Ralph J, Boerjan W. Lignin engineering. Current Opinion in Plant Biology. 2008;11:278–285. doi: 10.1016/j.pbi.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Verrier P, Bird D, Burla B, et al. Plant ABC proteins – a unified nomenclature and updated inventory. Trends in Plant Science. 2008;13:151–159. doi: 10.1016/j.tplants.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. Auxin: regulation, action, interaction. Annals of Botany. 2005;95:707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Murphy AS. Functional expression and characterization of Arabidopsis ABCB. AUX1 and PIN auxin transporters in Schizosaccharomyces pombe. The Plant Journal. 2009;59:179–191. doi: 10.1111/j.1365-313X.2009.03856.x. [DOI] [PubMed] [Google Scholar]
- Yazaki K. ABC transporters involved in the transport of plant secondary metabolites. FEBS Letters. 2006;580:1183–1191. doi: 10.1016/j.febslet.2005.12.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.