Abstract
Macrophages and CD4+ lymphocytes are the major reservoirs for HIV-1 infection. CD63 is a tetraspanin transmembrane protein, which has been shown to play an essential role during HIV-1 replication in macrophages. In this study, we further confirm the requirement of CD63 in early HIV-1 replication events in both macrophages and a CD4+ cell line. Further analysis revealed that viral attachment and cell-cell fusion were unaffected by CD63 silencing. However, CD63-depleted macrophages showed a significant decrease in the initiation and completion of HIV-1 reverse transcription, affecting subsequent events of the HIV-1 life cycle. Integration of HIV-1 cDNA as well as the formation of 2-LTR circles was notably reduced. Reporter assays showed that CD63 down regulation reduced production of the early HIV protein Tat. In agreement, CD63 silencing also inhibited production of the late protein p24. These findings suggest that CD63 plays an early post-entry role prior to or at the reverse transcription step.
Keywords: HIV-1, Tetraspanin, CD63, siRNA, Macrophage, life cycle
INTRODUCTION
Macrophages and CD4+ T lymphocytes are major target cells for HIV-1 infection and replication, and play critical roles in multiple aspects of viral pathogenesis. Numerous host determinants have been identified that facilitate HIV-1 replication by performing essential steps of the viral life cycle from viral entry to egress (Bergamaschi et al., 2010). A recent large-scale siRNA screen revealed over 250 host factors that participate in a broad array of cellular functions and implicate new pathways in the HIV life cycle (Brass et al., 2008). HIV encodes only 15 proteins and thus must take advantage of multiple host cell functions for productive infection (Brass et al., 2008). HIV uses a CD4 receptor (Klatzmann et al., 1984), as well as CCR5 and CXCR4 co-receptors (Feng et al., 1996; Dragic et al., 1996) for entry into the host cells. Upon membrane fusion, the viral core containing the viral nucleocapsid along with HIV RNA, reverse transcriptase, integrase, protease, and viral accessory proteins is released into the cytoplasm. Uncoating involves viral proteins as well as a variety of host factors that promote dissociation of the matrix protein from the plasma membrane (Aiken, 2006). After uncoating, reverse transcriptase converts viral RNA into DNA which is then transported across the nucleus and integrates into host chromosome. HIV genes are transcribed using host enzymes, and viral mRNA is transported outside the nucleus, and is used as a blueprint for producing new HIV proteins and enzymes. Some studies have reported reverse transcription to be largely associated with cellular enzymes or cofactors in vitro (Hooker and Harrich, 2003; Warrilow et al., 2008, 2010). HIV integration and proviral transcription also require the use of host cell factors (Goodarzi et al., 1999; Greene, 1991). Following viral gene expression, the cellular GTPase Rab9 facilitates trafficking of viral proteins from the late endosome to the trans-Golgi (Murray et al., 2005). HIV assembly and release utilizes TSG101 and the Endosomal Sorting Complexes Required for Transport (ESCRTs), a network of cellular factors that sort proteins and facilitate HIV budding (von Schwedler et al., 2003; Morita et al., 2004).
Historically, all antiretroviral treatments targeted viral proteins. More recently, however, FDA-approved Maraviroc® (Selzentry) and Aplaviroc® (GlaxoSmithKline) are the first antiretroviral drugs that target a cellular gene CCR5 co-receptor, and have been demonstrated to be effective and shown to cause less resistance among HIV phenotypes than previous generations of drugs (Reeves and Peifer, 2005; Hughes et al., 2008). Like CCR5, CD63 is another cellular protein that may represent an important new therapeutic target for the development of antiretroviral drugs.
CD63 is a type II cellular membrane protein and belongs to the tetraspanin superfamily (Stipp et al., 2003; Hemler, 2005). It has been reported that CD63 incorporates into HIV-1 virions (Chertova et al., 2006; Pelchen-Matthews et al., 2003; Sato et al., 2008) and colocalizes with HIV-1 Env and Gag proteins in HIV-1 producing cells (Raposo et al., 2002; Pelchen-Mathews et al., 2003; Grigorov et al., 2006; Jolly et al., 2007; Perlman and Resh, 2006). In addition, CD63 has been shown to be associated with tetraspanin-enriched mircrodomains (TEMs) that act as the gateway for HIV-1 budding at the plasma membrane (Nydegger et al., 2006).
We have previously demonstrated a role for CD63 in HIV-1 infection events in macrophages and a CD4+ cell line. Our earlier studies showed that anti-CD63 monoclonal antibody treatment 30 min prior to and during infection markedly reduced HIV replication in macrophages (von Lindern et al., 2003). Inhibition was shown to occur during early infection, suggestive of involvement in virus entry or reverse transcription. Our more recent studies confirm the requirement of CD63 in HIV-1 replication by pretreatment with CD63-specific siRNA in both macrophages and a CD4+ cell line (Chen et al., 2008). In this study, we investigated the role of CD63 during sequential steps of the HIV-1 replication cycle including viral attachment, cell-cell fusion, reverse transcription, integration, and early and late viral protein expression in both human monocyte-derived macrophages (MDMs) and CD4+ U373-MAGI-CCR5 cells. These studies revealed that CD63-depleted macrophages showed a significant decrease in the initiation and completion of HIV-1 reverse transcription, affecting subsequent events of the HIV-1 life cycle. However, events prior to reverse transcription including viral attachment and cell-cell fusion were not affected. These findings further support a role for CD63 in an early, post-entry event of the HIV-1 replication cycle, particularly prior to or at the reverse transcription step.
RESULTS
CD63 mRNA down regulation by siRNA in human MDMs and U373-MAGI-CCR5 cells
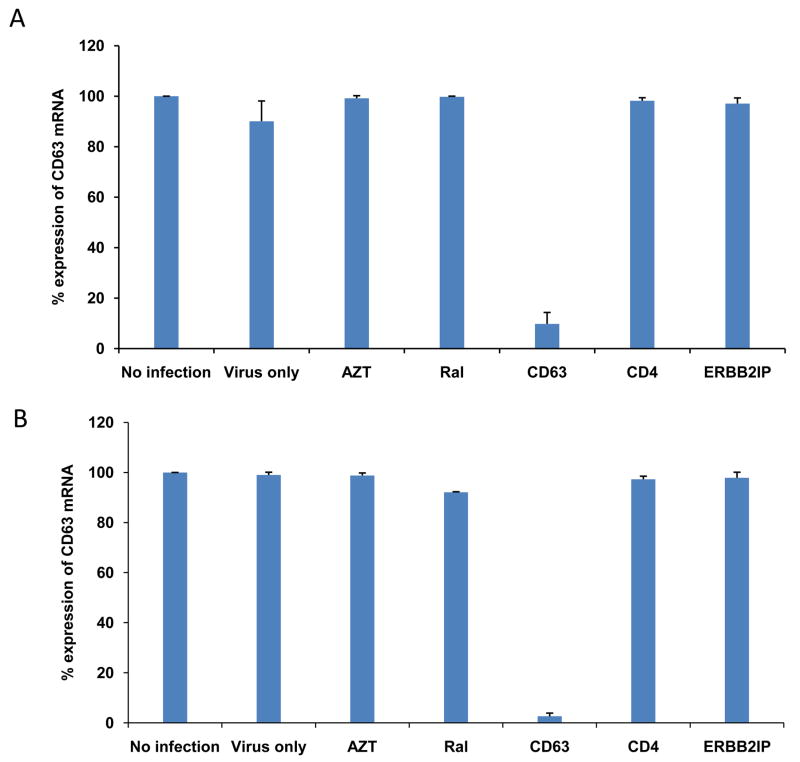
To confirm the down regulation of CD63 by siRNA in MDMs and U373-MAGI-CCR5 cells, mRNA was extracted and analyzed by quantitative reverse transcriptase PCR (qRT-PCR). CD63 mRNA expression was silenced by >90% or >95% following CD63 siRNA transfections of MDMs and U373-MAGI-CCR5 cells, respectively (Fig. 1A and B). CD63 mRNA was not significantly reduced by transfections of other siRNAs (CD4, ERBB2IP) or FDA approved HIV-1 inhibitors (AZT, RT inhibitor; Raltegravir, integrase inhibitor) used throughout this study indicating specificity for CD63 siRNA down regulation in both MDMs and U373-MAGI-CCR5 cells. ERBB2IP siRNA was used as a cellular target negative control, as our previous studies showed that silencing ERBB2IP does not inhibit HIV-1 (Murray et al., 2005). Western blot analysis revealed that CD63 protein expression was significantly reduced in CD63-siRNA transfectants compared to the cells transfected with control siRNAs or non-transfected cells (data not shown), consistent with our previous findings (Chen et al., 2008).
Figure 1. Confirmation of CD63 mRNA down regulation via real-time PCR.
(A) MDMs (5 × 105 cells/well) or (B) U373-MAGI-CCR5 cells (1 × 105 cells/well) were plated in 24-well plates and transfected with 50 nM siRNAs (CD63, CD4, and ERBB2IP). For controls, cells were treated with AZT (1 μM) or raltegravir (20 mM). Controls also included untreated cells or cells infected with HIV-1 SX (m.o.i.= 0.02) only. Total mRNA was isolated from each well using the Qiagen RNeasy kit 48 h after transfection. Quantitative RT-PCR was used to determine relative CD63 expression levels, normalizing to GAPDH expression as an internal control.
CD63 down regulation affects HIV-1 production in human MDMs and U373-MAGI-CCR5 cells
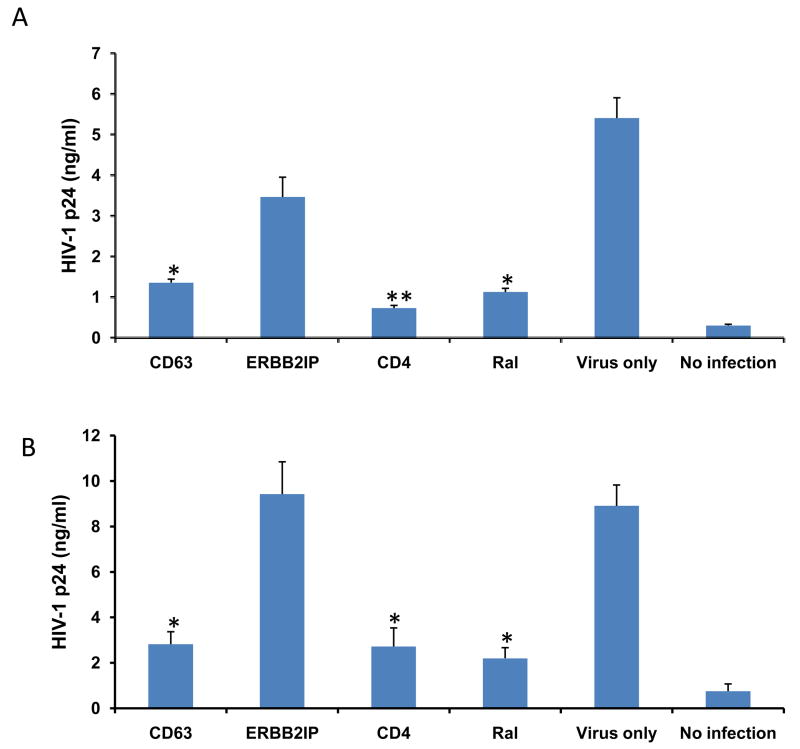
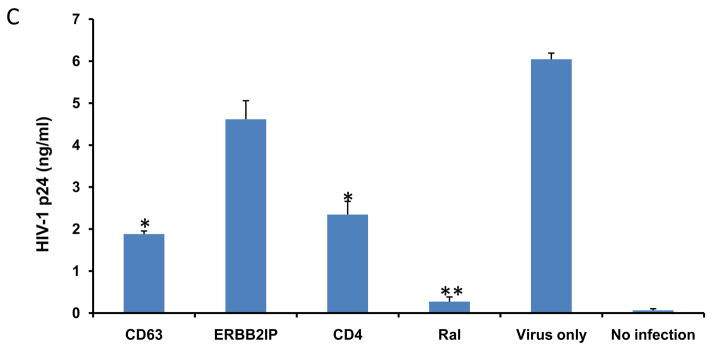
To gauge the effect of CD63 down regulation on viral production, MDMs and U373-MAGI-CCR5 cells were transfected with various specific siRNAs or ERBB2IP control siRNA, or were treated with the integrase inhibitor raltegravir, followed by HIV-1 SX infection 48 h post-transfection. HIV-1 SX virus production was assessed in culture supernatants by p24 ELISA at 7 days post-infection of MDMs (Fig. 2A) or at 5 days post-infection of U373-MAGI-CCR5 cells (Fig. 2B). As shown in Fig. 2A and B, virus production in both cell types was significantly reduced following CD63 or CD4 silencing, or raltegravir treatment compared to ERBB2IP siRNA transfected cells. (P<0.05). Similar results were also observed in MDMs lysates (Fig. 2C), where intracellular HIV-1 SX p24 production was dramatically reduced by silencing CD63, to the same extent as the CD4 siRNA control group.
Figure 2. Effects of CD63 silencing on HIV-1 replication in human MDMs and U373-MAGI-CCR5 cells.
(A) MDMs (5 × 105 cells/well) or (B) U373-MAGI-CCR5 cells (1 × 105 cells/well) were plated in triplicate in 24-well plates and transfected with 50 nM siRNAs (CD63, CD4, and ERBB2IP). Controls included cells treated with raltegravir (20 mM), untreated cells or cells treated with virus infection only. Forty eight hours post-transfection, cells were infected with HIV-1 SX (m.o.i. =0.02). Supernatants were harvested for p24 detection on day 5 post-infection for U373-MAGI-CCR5 cells, and on day 7 post-infection for MDMs using p24 Capture ELISA kit (ImmunoDiagnostics, Woburn, MA). (C) Cell lysates from MDMs on day 7 post-infection were also harvested for detection of intracellular p24. *P<0.05, **P<0.01, compared with ERBB2IP control group, respectively.
Anti-CD63 shows no inhibition of HIV-1 attachment to human MDMs
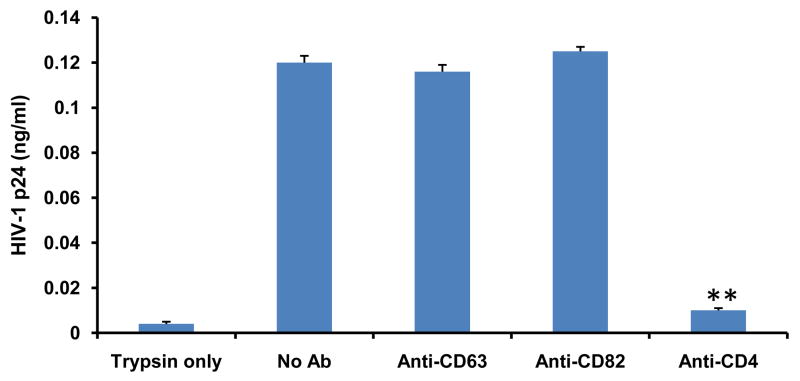
Because CD63 silencing affects HIV-1 replication, we looked systematically at the effect of CD63 inhibition at successive stages within the HIV-1 replication cycle. Since antibody-mediated neutralization of HIV-1 has been shown to inhibit virus-receptor binding, and interfere with events after binding (Ugolini et al., 1997), we investigated the effect of CD63 on HIV-1 attachment by virus-cell binding assay using anti-CD63. Here we showed soluble monoclonal antibodies to CD63 and control monoclonal antibody CD82 did not inhibit the adsorption of HIV-1 SX to MDMs (Fig. 3), however, soluble CD4 monoclonal antibody showed dramatic inhibition of virus binding compared to those of anti-CD63, anti-CD82 and virus only (P<0.01). In contrast, the anti-CD63 antibody showed no inhibition of HIV-1 binding to MDMs, suggesting that CD63 has no effect on viral attachment of MDMs.
Figure 3. Anti-CD63 does not inhibit HIV-1 attachment to human MDMs.
MDMs (1×103cells/well) were pretreated for 30 min at 37°C with Iscove’s modified Dulbecco’s medium (IMDM) alone, 25 μg/ml of anti-CD63 antibody (Invitrogen), 25 μg/ml of anti-CD82 antibody (Invitrogen), or 25 μg/ml of anti-CD4 antibody (BD Sciences) as a positive inhibition control. Thereafter, HIV-1 SX (m.o.i.= 0.02) was added, and incubated for 1 h at 4°C, and unbound virions were removed by washing with cold PBS containing 1% FBS. The cells were trypsinized, and the amount of cell-associated p24 in trypsin was determined with p24 Capture ELISA kit. Controls included cells treated with trypsin only and untreated (no Ab) cells. Experiments were performed in triplicate. **P<0.01, compared with anti-CD82 group.
CD63 has no effect on cell-cell fusion
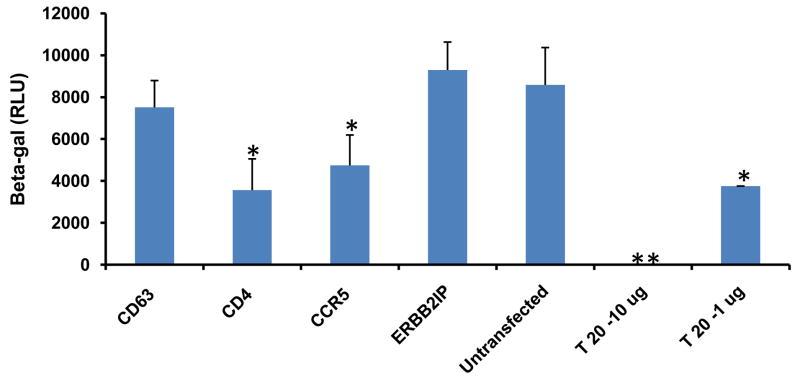
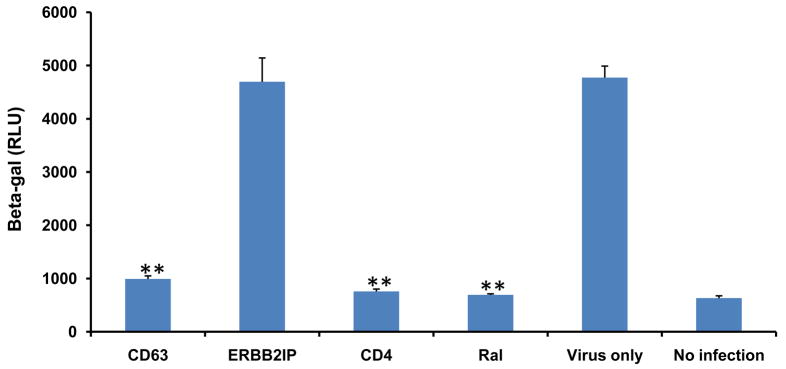
Since CD63 is not required for virus attachment (Fig. 3), the subsequent step after binding in the HIV life cycle is to assess virus entry. We therefore tested any potential role for CD63 in virus entry using cell-cell fusion assay by co-culturing HL2/3 cells, which express Tat and HIV receptor Env proteins gp41 and gp120, with U373-MAGI-CCR5 cells. HIV Env proteins on HL2/3 cells interact with primary receptor CD4 and co-receptor CCR5 on U373-MAGI-CCR5 cells, prompting cellular fusion via the same mechanism used by HIV; upon fusion, Tat activates β-gal expression. Decreased CD63 levels had no effect on β-gal activity, whereas decreased CD4 and CCR5 levels correlated with diminished β-gal activity, consistent with inhibition of viral fusion inhibitor drug T-20 (Fig. 4). Together, these data suggest that CD63 has no effect on viral entry of U373-MAGI-CCR5 cells.
Figure 4. CD63 silencing has no effect on cell-cell fusion.
U373-MAGI-CCR5 target cells (104cells/well) were transfected with CD63-, CD4-, or CCR5-specific siRNAs and ERBB2IP siRNA. For controls, fusion inhibitor T20 (1 μg/ml or 10 μg/ml) was used as well as fusion only conditions containing no siRNA. Forty-eight hours post-transfection U373-MAGI-CCR5 target cells were co-cultured with 2 × 104 of HL2/3 effector cells per well at 37°C for 12 h to allow fusion to occur. Fusion was monitored by assaying for Tat-dependent β-gal reporter gene activation stimulated by HIV-1 Tat from the HL2/3 cells. All experiments were performed in quadruplicate. *P<0.05, **P<0.01, compared with ERBB2IP group.
CD63 down regulation affects HIV-1 reverse transcription in human MDMs and U373-MAGI-CCR5 cells
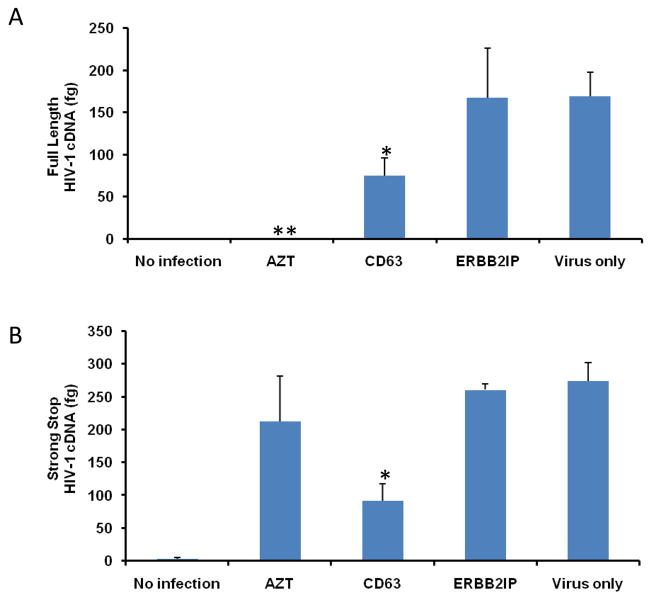
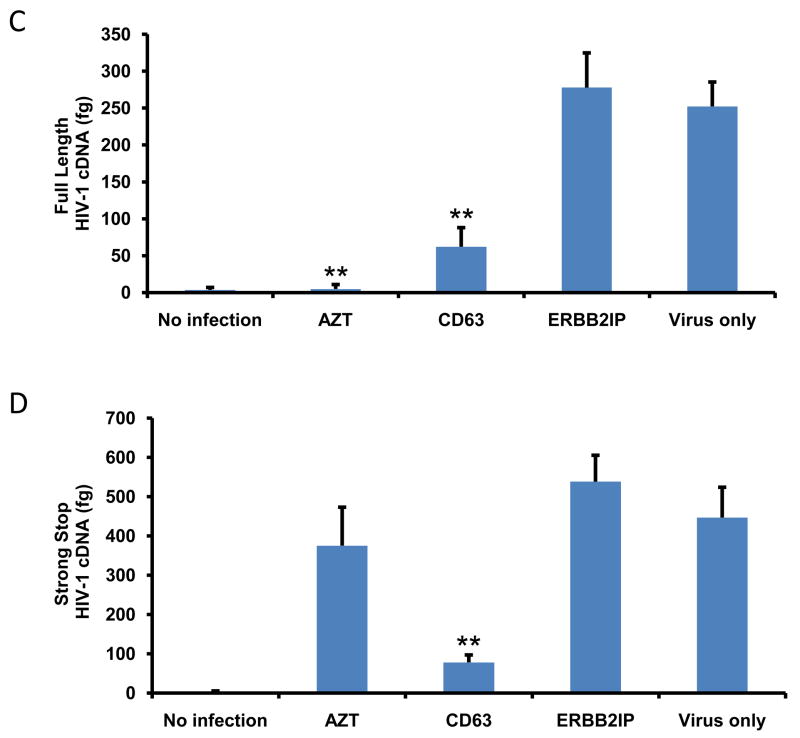
In order to clarify whether CD63 may be required for HIV-1 reverse transcription, small non-genomic DNA was isolated from control- and CD63 siRNA-transfected MDMs or U373-MAGI-CCR5 cells at 24 h post-infection with HIV-1 SX for quantification by real time PCR. As shown in Fig. 5A and C, CD63 silencing affected formation of full length HIV-1 cDNA, which decreased by approximately 55% in CD63-deficient MDMs and 75% in CD63-deficient U373-MAGI-CCR5 cells, compared to those transfected with control siRNA ERBB2IP or untransfected cells (P<0.05). In addition, we measured the formation of strong stop DNA, which is the first piece of DNA (147 bp) synthesized during the initiation of reverse transcription. HIV strong stop DNA formation (Fig. 5B and D) decreased by approximately 65% in CD63-deficient MDMs and 80% in CD63-deficient U373-MAGI-CCR5 cells, compared to those transfected with ERBB2IP or virus infection only (P<0.01). Inhibition of strong stop DNA formation is consistent with our earlier studies, in which MDMs were pretreated with anti-CD63, followed by HIV-1 infection (von Lindern et al., 2003). Full length viral DNA formation was not detected in cells treated with the reverse transcription inhibitor AZT (Fig. 5A and C), whereas viral strong stop formation was evident in cells treated with AZT (Fig. 5B and D). As expected, AZT does not inhibit strong stop formation, only full length HIV DNA (Arts and Wainberg, 1994). Collectively, these data indicate that CD63 serves a critical post-entry role early in the HIV life cycle either before or during reverse transcription strong stop DNA formation, at a step upstream of AZT. Sequential steps following reverse transcription in the HIV life cycle were assessed to determine if steps downstream of reverse transcription were impaired by CD63 silencing.
Figure 5. CD63 silencing reduces HIV-1 reverse transcription in human MDMs and U373-MAGI-CCR5 cells.
The effect of HIV-1 reverse transcription on the formation of full length cDNA (A and C) and the initiation of strong stop (B and D) were performed in MDMs (5 × 105 cells/well) and in U373-MAGI-CCR5 cells (1 × 105 cells/well) that were transfected with 50 nM siRNAs (CD63 and ERBB2IP). Forty-eight hours post-transfection, cells were infected with HIV-1 SX (m.o.i. = 0.02). For controls, cells were treated with reverse transcriptase inhibitor AZT (1 μM); cells infected with virus only; and untreated cells. Small non-genomic DNA was isolated from controls and CD63 siRNA-transfected MDMs and U373-MAGI-CCR5 cells at 24 h post infection. Primers M667/M661, along with probe MH603, was used to quantify full-length cDNA formation (A and C), and primers M667/AA55, along with probe MH603 was used to quantify strong stop cDNA formation (B and D) by real time PCR. *P<0.05, compared with ERBB2IP group.
Decreased integration of HIV-1 cDNA in both CD63 deficient human MDMs and U373-MAGI-CCR5 cells
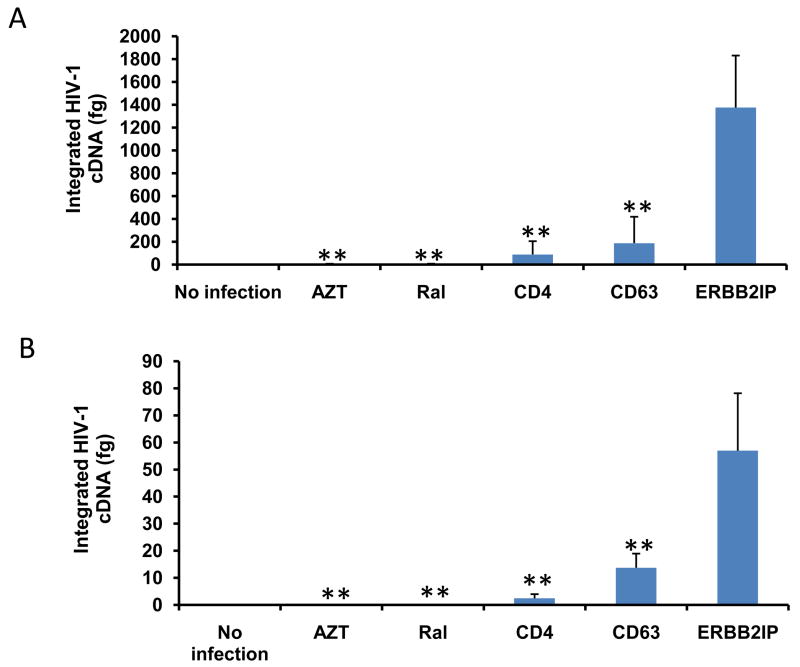
To address the effects of CD63 for proviral DNA integration, genomic DNA was isolated from cells post infection with HIV-1 SX. Quantitative real-time PCR was used to quantify differences in HIV-1 SX integration between control cells (ERBB2IP- or CD4-siRNA transfectants; AZT and raltegravir treated cells; virus infection only or cell only) and CD63-siRNA transfected cells. Down regulation of CD63 resulted in significantly reduced amounts of integrated HIV-1 cDNA in both MDMs (Fig. 6A) and U373-MAGI-CCR5 cells (Fig. 6B) (P<0.01).
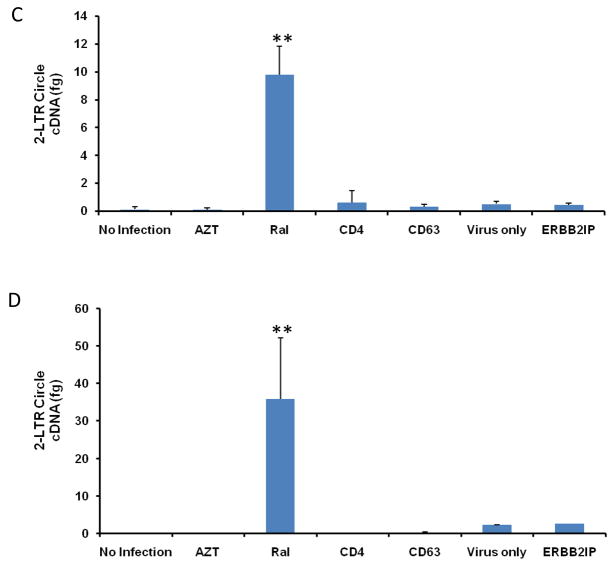
Figure 6. CD63 silencing decreases HIV-1 integration and 2-LTR circle formation in human MDMs and U373-MAGI-CCR5 cells.
(A) MDMs (5 × 105 cells/well) or (B) U373-MAGI-CCR5 cells (1 × 105 cells/well) were plated in 24-well plates and transfected with 50 nM siRNAs (CD63, CD4, and ERBB2IP). Controls include cells treated with raltegravir (20 mM) 24 h prior to infection, during infection and 48 h post- infection and untreated cells. Forty-eight hours post-transfection, cells were infected with HIV-1 SX (m.o.i. = 0.02). Genomic DNA was isolated from cells 5 days (MDMs) and 3 days (U373-MAGI-CCR5 cells) post-infection. Quantitative real-time PCR was used to quantify differences in HIV-1 SX integration between control cells and CD63-siRNA transfected cells using primers Alu-gag-LTR, and LTR probe (O’Doherty et al., 2002; Friedrich et al., 2010). Plasmid DNA was isolated from cells (C) 5 days (MDMs) and (D) 3 days (U373-MAGI-CCR5 cells) post HIV-1 SX infection. Quantitative real-time PCR was used to quantify differences between control cells and CD63-siRNA transfected cells using primers MH535/MH536 with probe MH603 to detect HIV 2-LTR circle formation (Butler et al., 2001; Friedrich et al., 2010). All experiments were performed in triplicate. *P<0.05, **P<0.01, compared with ERBB2IP group.
CD63 down-regulation does not induce HIV-1 2-LTR circle formation
To determine if CD63 expression is necessary immediately prior to viral integration, small non-genomic DNA was isolated from CD63-deficient cells after infection to quantify formation of 2-LTR circles. The cDNA circularizes to form 2-LTR circles if it does not integrate into the cellular genome (Bukrinsky et al., 1993; Farnet et al., 1991), and these 2-LTR circles can be quantified using real-time PCR. As shown in Fig. 6, down regulation of CD63 did not induce 2-LTR circle formation in either MDMs (Fig. 6C) or U373 cells (Fig. 6D). As expected, 2-LTR circles were dramatically formed in the cells treated with the integrase inhibitor raltegravir. These results strongly suggest that CD63 affects the HIV-1 replication cycle preceding integration.
Inhibition of HIV-1 Tat activity following CD63 down regulation
To determine if CD63 plays a role in early HIV-1 protein production, U373-MAGI-CCR5 cells were used in a β–galactosidase reporter gene assay to gauge the effect of CD63 silencing on Tat function. The LTR-HIV promoter controls expression of β–galactosidase induced by production of HIV Tat. U373-MAGI-CCR5 cells were transfected with various specific siRNAs or control siRNA, or were treated with raltegravir, followed by HIV-1 SX infection 48 h after transfection. Cells were cultured for 72 h, lysed, and then exposed to a luminescent β–gal substrate, and expressed as relative light units (RLU) (Fig. 7). In agreement, we also observed that Tat activity was dramatically inhibited to background levels in CD63 siRNA-transfectant to the same extent as siRNA specific for CD4 when compared to ERBB2IP control siRNA-transfectant or cells exposed to virus infection only (P<0.01). These data strongly indicate that CD63 supports HIV-1 replication at a step prior to gene expression.
Figure 7. CD63 silencing decreases early HIV-1 protein Tat activity in U373-MAGI-CCR5 cells.
U373-MAGI-CCR5 cells (1 × 105 cells/well) were plated in triplicate in 24-well plates and transfected with 50 nM siRNAs (CD63, CD4, and ERBB2IP). Controls included cells treated with raltegravir (20 mM), untreated cells or cells treated with virus infection only. Forty eight hours post-transfection, cells were infected with HIV-1 SX (m.o.i. = 0.02). Cell lysates were collected on day 3 post-infection, and Tat-driven β–galactosidase activity was analyzed using the Beta–Glo Assay System (Promega, Madison, WI), and was detected by Dynex MLX Luminometer. *P<0.05, **P<0.01, compared with ERBB2IP control group, respectively.
DISCUSSION
Extensive studies have revealed that HIV-1 exploits multiple host proteins during infection (Bergamaschi et al., 2010; Brass et al., 2008; Kumar et al., 2008; Bouazzaoui et al., 2006; Hamamoto et al., 2006; Stremlau et al., 2004; Krishnan and Zeichner, 2004). These proteins participate in a broad array of cellular functions and implicate different pathways in the viral life cycle. However, the mechanisms by which these proteins play relevant roles in HIV-1 pathogenesis are subjects of continued exploration.
In this study, we revealed the role of CD63 during sequential steps of the HIV-1 replication cycle which include viral attachment, viral entry (cell-cell fusion), reverse transcription, integration, early/late viral protein formation, and virion release from infected human MDMs and U373-MAGI-CCR5 cells. We found that CD63 was required for efficient HIV-1 reverse transcription and subsequent steps from integration to egress. However, entry mechanisms including cell membrane docking and cell-cell fusion life cycle steps that precede HIV-1 reverse transcription were unaffected. In all cases, the down regulation of CD63 via specific siRNA significantly reduced HIV-1 replication in both cell types, suggesting that CD63 is associated with a common mechanism in early events of the HIV-1 life cycle, specifically at a step preceding or at reverse transcription. Since our observation is an early post-entry step, it is possible that HIV-1 may be linked to an intracellular signaling event involving CD63. CD63 has been shown to induce signaling through tyrosine-dependent interaction of its carboxyterminal cytoplasmic tail with the AP-3 adaptor subunit (Rous et al., 2002). In addition, when HIV-1 gp120 binds to CD4 and CCR5 on the surface of macrophages, it has been shown to induce phosphorylation of receptor tyrosine kinases such as p56Lck, which in turn activates the Raf/MEK/ERK and PIP3 kinase pathways and intracellular calcium channels (Popik and Pitha, 1996), implying that an intracellular signaling route is likely to be important for the HIV-1 life cycle. However, it is not clear how HIV-1 is linked to CD63 signaling, which still remains to be explored.
Although recent studies (Ruiz-Mateos et al., 2008; Krementsov et al., 2009) showed that CD63 expression is not required for either the production or the infectivity of HIV-1, numerous studies have reported that CD63 incorporates into HIV-1 virions and have the potential to interfere with viral infection (Chertova et al., 2006; Pelchen-Matthews et al., 2003; Sato et al., 2008; Raposo et al., 2002; Grigorov et al., 2006; Jolly and Sattentau, 2007; Perlman and Resh, 2006; Nydegger et al., 2006). Our previous studies (von Lindern et al., 2003; Chen et al., 2008) demonstrated that CD63 plays an important supportive role in HIV-1 replication in macrophages. Although controversial data have been published in the last few years concerning effects of CD63 on HIV-1 replication in macrophages and other cell types, it is possible that this effect may be dependent on the cell type, viral strain, and virus concentration. Even though MDMs display a great heterogeneity in their capacities to replicate HIV-1, depending on the donors (up to a 3 log difference in viral production) (Bol et al., 2009; Eisert et al., 2001; Chang et al., 1996), we performed experiments in macrophages from several different donors as well a CD4+ cell line, infecting with HIV-1 R5 virus strain, and our results consistently present similar outcomes.
Understanding the virus-host interactions that lead to limited HIV replication in these cell types can be a powerful tool to identify molecular mechanisms of innate/intrinsic restriction of HIV replication. A key pharmacological strategy for treating individuals living with HIV has been to simultaneously target multiple virus-encoded enzymes required for replication to overcome emergence of drug resistance. Our laboratory has pursued an alternate strategy by identifying host factors, such as CD63, that are required for HIV-1 replication, as HIV-1 should be less likely to evolve resistance to drugs targeting cellular proteins.
In conclusion, we have revealed that CD63 plays an essential role in an early, post–entry event of the HIV-1 replication cycle, particularly prior to or at the reverse transcription step. Whether CD63 interacts directly with HIV or indirectly, by altering host dependent factors potentially required for reverse transcription, remains to be determined. In addition, a recent study (Lin et al., 2008) showed that expression of CD63 activates protein tyrosine kinase (PTK) and enhances the PTK-induced inhibition of ROMK channels. Thus, the role of CD63 in affecting the HIV-1 replication cycle induced by the stimulation of the PTK pathway is unknown and further research is needed to gain insight into the molecular signaling mechanisms, including CD63, PTK and viral proteins that possibly facilitate early events in the HIV replication cycle.
MATERIALS AND METHODS
Cells and culture conditions
U373-MAGI-CCR5 cells (obtained from NIH AIDS Research and Reference Reagent Program, and contributed by Drs. Michael Emerman and Adam Geballe), are a cell line derived from a glioblastoma that has been modified by stable transfection of LTR-β-galactosidase which is transactivated by HIV Tat protein during virus replication (Harrington and Geballe, 1993; Vodicka et al., 1997). U373-MAGI-CCR5 cells express CD4 and human chemokine receptor CCR5 on the cell surface, which allow infection by HIV R5 strains (Vodicka et al., 1997). U373-MAGI-CCR5 cells were propagated in Dulbecco’s modified Eagle’s (DMEM) (Gibco, CA) supplemented with 10% fetal bovine serum (Hyclone, IL), 0.2 mg/ml G418, 0.1 mg/ml hygromycin B, and 1.0 μg/ml puromycin. For infection experiments, U373-MAGI-CCR5 cells were maintained in 90% DMEM, 10% fetal bovine serum, and 1% penicillin/streptomycin (Sigma, MO).
HL2/3 cells (NIH AIDS Research and Reference Reagent Program, kindly contributed by Dr. Barbara K. Felber and Dr. George N. Pavlakis) were generated by cotransfection of HeLa cells with the plasmids pHXB2/3gpt, pSV2neo and selected in geneticin (G418). HL2/3 cells were selected on the basis of high-level production of Gag, Env, Tat, Rev, and Nef proteins. Co-cultivation with the CD4 producing cell line results in efficient cell fusion within 6–12 hours. Human embryonic kidney (HEK) 293T cells were purchased from Invitrogen. Both HL2/3 cells and 293T cells were maintained in DMEM supplemented with 10% fetal bovine serum, 1% non-essential amino acids (Sigma) and 1% penicillin/streptomycin.
MDMs were purified from healthy human PBMCs by adherence to plastic tissue culture dishes as described previously (O’Brien et al., 1990). Briefly, PBMCs were purified by Ficoll-Hypaque centrifugation from buffy coats of healthy HIV-negative blood donors prepared by the UTMB Blood Bank (Galveston, TX). MDMs were obtained by adherence for 6 days to plastic Petri dishes initially coated with human AB serum (Rich et al., 1992). During differentiation, macrophages were cultured in Iscove’s modified Dulbecco’s medium (IMDM) supplemented with 20% FCS; 1% L-glutamine and 1% Penicillin/Streptomycin.
Viruses
HIV-1 SX, which is a chimeric R5-tropic virus encoding the majority of the HIV-1 JRFL envelope protein in an HIV-1NL4–3 backbone, were purchased from the Virology Core Facility, Center for AIDS Research at Baylor College of Medicine, Houston, TX. In all experiments, MDMs and U373-MAGI-CCR5 cells were infected with HIV-1 SX (m.o.i. = 0.02).
siRNA transfections and infectivity assay
The CD63- and CD4-specific target siRNAs were designed and synthesized by Dharmacon (Lafayette, CO), ERBB2IP non-HIV specific siRNA served as a negative control in each experiment. The target sequences for siGENOME SMARTpool siRNA of CD63 includes D-017256 (01-GAGAUAAGGUGAUGUCAGA; 02-AAGGAGAACUAUUGUCUUA; 03-GGAUUAAUUUCAACGAGAA; 04-GAUGGAGAAUUACCCGAAA). The target sequences for siGENOME SMARTpool siRNA of CD4 includes D-005234 (01-GAACUGACCUGUACAGCUU; 02-AAUCAGGGCUCCUUCUUAA; 03-GAAGAAGAGCAUACAAUUC; 18-AUUACCAAGUGCCGGACUA); The target sequences for siGENOME SMARTpool siRNA of ERBB2IP includes D-031861 (01-UGAAACAGCUCACAUAUUU; 02-UGUGAAAUCUCAUAGCAUA; 03-CGAAGAGCCAAAUAUAAUA; 04-CCAAACGACCGACUUAUUC). siRNAs transfections were performed using oligofectamine (Invitrogen, CA) following the manufacturer’s instructions. In brief, MDMs adherent for 5 days were plated in 24-well plates (5 × 105 cells/well), and transfected with 50 nM siRNAs using oligofectamine in serum-free Opti-MEM I medium (Gibco) 24 h after plating. Cells were maintained in Iscove’s medium and 20% FCS after 4 h to terminate transfection. In order to measure down regulation of target genes, cells were lysed by cell lysis buffer (Promega, WI) and lysates were collected for Western blot analysis, or RNA was extracted for real time quantitative PCR analysis at 2 days post-transfection. U373-MAGI-CCR5 cells were transfected in 24-well plates (1 × 105 cells/well), using oligofectamine and 50 nM siRNAs (final concentration in supernatants). Subsequently, cells were fed with DMEM with 10% FCS after 4 h to terminate transfection, followed by detection of proteins or target genes down regulation as described above.
Forty eight hours post-transfection, MDMs or U373-MAGI-CCR5 cells were infected with HIV-1 SX (m.o.i. = 0.02). Culture supernatants or cell lysates were harvested for p24 detection on day 5 post-infection in U373-MAGI-CCR5 cells, and on day 7 post-infection in MDMs using p24 Capture ELISA kit (ImmunoDiagnostics, Woburn, MA).
In order to assess Tat protein expression, U373-MAGI-CCR5 cells were lysed on day 3 post-infection, and analyzed for β–galactosidase activity using the Beta–Glo Assay System (Promega) and a Dynex MLX Luminometer.
Virion attachment assay
The virion attachment assay was performed as previously described (Tanaka et al., 2003). Briefly, infections are performed in 96 well microtiter plates at a m.o.i. of 0.02. MDMs (103/well) were pretreated for 30 min at 37°C with medium only (no antibody), 25 μg/ml of anti-CD63 monoclonal antibody (Invitrogen, clone CLB-gran/12 CLB-180), 25 μg/ml of anti-CD82 monoclonal antibody (Invitrogen, clone C-16), or 25 μg/ml anti-CD4 monoclonal antibody (BD Sciences, clone L200) as a positive inhibition control. Thereafter, HIV-1 SX was added, which was continued in the presence of antibody, and incubated for 1 h at 4°C, and unbound virions were removed by washing with ice-cold PBS containing 1% FBS. For each condition, the cells were treated with 0.05% trypsin (Gibco) to cleave bound virus, and then the amount of cell-associated p24 in the trypsin was determined with p24 Capture ELISA kit.
Cell-cell fusion assay
U373-MAGI-CCR5 target cells, were plated in 96-well plates (104 cells/well) and cultured overnight, followed by transfection of CD63-, CD4-, CCR5-specific siRNAs and ERBB2IP control siRNA. The medium was removed 48 h post-transfection and 2 × 104 HL2/3 HeLa cells were added to each well in fresh medium. HL2/3 HeLa cells express Tat and HIV envelope proteins gp41 and gp120 that allow them to efficiently interact with CD4 and CCR5 on U373-MAGI-CCR5 target cells, prompting cellular fusion via the same mechanism used by HIV. The co-culture was incubated at 37°C for 12 h to allow fusion to occur; the fusion of U373-MAGI-CCR5 target cells and HL2/3 HeLa cells was monitored by assaying for Tat-dependent β-gal reporter gene activation stimulated by HIV-1 Tat from the HL2/3 HeLa cells. U373-MAGI-CCR5 and HL2/3 cells alone were used to determine background luminescence. To study the effect of each siRNAs on cell-cell fusion, fusion inhibitor, T-20, was used as positive control. The target cells were pre-treated with T-20 (10 μg/ml or 1 μg/ml) for 24 h followed by incubation with HL2/3 cells in the continued presence of the T-20. Subsequently, fusion was measured via luminometry expressed as β–galactosidase production.
Real time quantitative PCR
Primers and probes for real time PCR were custom ordered from Sigma-Aldrich. Full genomic DNA was isolated from MDMs or cell lines using the Qiagen Blood and Tissue Kit (Qiagen, Inc., CA). Small non-genomic DNA, such as reverse transcribed viral cDNA and 2-LTR circles were isolated using a Qiagen Miniprep kit. To detect strong stop of reverse transcription, primers M667 and AA55, along with probe MH603 (Butler et al., 2001) were used; to detect full length HIV DNA, primers M667 and M661 (Zack et al., 1990), along with probe MH603 were used; to identify 2-LTR circle formation, as described in our previous study (Friedrich et al., 2010), primers MH535 and MH536 were used with the MH603 probe (Butler et al., 2001). Total mRNA was isolated from siRNA-transfected cells and MDMs using RNeasy Mini Kits (Qiagen). For integration, a two-step PCR reaction was performed. For the initial PCR amplification, Alu forward and HIV-1 gag reverse primers were used (O’Doherty et al., 2002; Friedrich et al., 2010). The second step real-time PCR used the primers LTR forward, LTR reverse, and LTR probe (O’Doherty et al., 2002; Friedrich et al., 2010). CD63 specific primers and probe were purchased from Applied Biosytems (Carlsbad, CA). All reactions were performed using Applied Biosystems TaqMan Universal Master Mix and run using an Applied Biosystems 7500 Fast Real Time PCR system and 7500 Fast System Software. Silencing of target genes was determined by normalizing target gene expression to GAPDH expression (n=3).
Statistical analysis
The results are expressed as mean ± SD of at least four wells. Two-tailed, paired Student’s t-test was used to determine statistical significance. P value of <0.05*, and <0.01** were considered significant.
Acknowledgments
This work was supported by Public Health Service grant HL088999 from the National Heart, Lung, and Blood Institute. We thank the NIH AIDS Research and Reference Reagent Program for providing the U373-MAGI-CCR5 cells. We thank Edward Siwak, Ph.D., Associate Director of Virology Core Facility, Center for AIDS Research at Baylor College of Medicine, Houston, TX for providing HIV-1SX.. Also, we greatly appreciate Merck & CO., Inc. for generously providing raltegravir used in our studies, as well as Drs. Mark A. Endsley and William A. O’Brien for their excellent editorial suggestions.
Footnotes
We confirm that we have no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Aiken C. Viral and cellular factors that regulate HIV-1 uncoating. Curr Opin HIV AIDS. 2006;1:194–199. doi: 10.1097/01.COH.0000221591.11294.c1. [DOI] [PubMed] [Google Scholar]
- 2.Arts EJ, Wainberg MA. Preferential incorporation of nucleoside analogs after template switching during human immunodeficiency virus reverse transcription. Antimicrob Agents Chemother. 1994;38(5):1008–1016. doi: 10.1128/aac.38.5.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergamaschi A, Pancino G. Host hindrance to HIV-1 replication in monocytes and macrophages. Retrovirology. 2010;7:31. doi: 10.1186/1742-4690-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bol SM, van Remmerden Y, Sietzema JG, Kootstra NA, Schuitemaker H, van’t Wout AB. Donor variation in in vitro HIV-1 susceptibility of monocyte-derived macrophages. Virology. 2009;390:205–211. doi: 10.1016/j.virol.2009.05.027. [DOI] [PubMed] [Google Scholar]
- 5.Bouazzaoui A, Kreutz M, Eisert V, Dinauer N, Heinzelmann A, Hallenberger S, Strayle J, Walker R, Rubsamen-Waigmann H, Andreesen R, von Briesen H. Stimulated trans-acting factor of 50 kDa (Staf50) inhibits HIV-1 replication in human monocyte-derived macrophages. Virology. 2006;356:79–94. doi: 10.1016/j.virol.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 6.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319(5865):921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 7.Bukrinsky M, Sharova N, Stevenson M. Human immunodeficiency virus type 1 2-LTR circles reside in a nucleoprotein complex which is different from the preintegration complex. J Virol. 1993;67:6863–6865. doi: 10.1128/jvi.67.11.6863-6865.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butler SL, Hansen MS, Bushman FD. A quantitative assay for HIV DNA integration in vivo. Nat Med. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- 9.Chang J, Naif HM, Li S, Sullivan JS, Randle CM, Cunningham AL. Twin studies demonstrate a host cell genetic effect on productive human immunodeficiency virus infection of human monocytes and macrophages in vitro. J Virol. 1996;70:7792–7803. doi: 10.1128/jvi.70.11.7792-7803.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen H, Dziuba N, Friedrich B, von Lindern J, Murray JL, Rojo DR, Hodge TW, O’Brien WA, Ferguson MR. A critical role for CD63 in HIV replication and infection of macrophages and cell lines. Virology. 2008;379(2):191–196. doi: 10.1016/j.virol.2008.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chertova E, Chertov O, Coren LV, Roser JD, Trubey CM, Bess JW, Jr, Sowder RC, 2nd, Barsov E, Hood BL, Fisher RJ, Nagashima K, Conrads TP, Veenstra TD, Lifson JD, Ott DE. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J Virol. 2006;80(18):9039–9052. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, Cayanan C, Maddon PJ, Koup RA, Moore JP, Paxton WA. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381(6584):667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 13.Eisert V, Kreutz M, Becker K, Königs C, Alex U, Rübsamen-Waigmann H, Andreesen R, von Briesen H. Analysis of cellular factors influencing the replication of human immunodeficiency virus type I in human macrophages derived from blood of different healthy donors. Virology. 2001;286:31–44. doi: 10.1006/viro.2001.0940. [DOI] [PubMed] [Google Scholar]
- 14.Farnet CM, Haseltine WA. Circularization of human immunodeficiency virus type 1 DNA in vitro. J Virol. 1991;65:6942–6952. doi: 10.1128/jvi.65.12.6942-6952.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272(5263):872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 16.Friedrich B, Li G, Dziuba N, Ferguson MR. Quantitative PCR used to assess HIV-1 integration and 2-LTR circle formation in human macrophages, peripheral blood lymphocytes and a CD4+ cell line. Virol J. 2010;7:354. doi: 10.1186/1743-422X-7-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodarzi G, Pursley M, Felock P, Witmer M, Hazuda D, Brackmann K, Grandgenett D. Efficiency and fidelity of full-site integration reactions using recombinant simian immunodeficiency virus integrase. J Virol. 1999;73(10):8104–8111. doi: 10.1128/jvi.73.10.8104-8111.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greene WC. The molecular biology of human immunodeficiency virus type 1 infection. N Engl J Med. 1991;324(5):308–317. doi: 10.1056/NEJM199101313240506. [DOI] [PubMed] [Google Scholar]
- 19.Grigorov B, Arcanger F, Roingeard P, Darlix JL, Muriaux D. Assembly of infectious HIV-1 in human epithelial and T-lymphoblastic cell lines. J Mol Biol. 2006;359(4):848–862. doi: 10.1016/j.jmb.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 20.Hamamoto S, Nishitsuji H, Amagasa T, Kannagi M, Masuda T. Identification of a novel human immunodeficiency virus type 1 integrase interactor, Gemin2, that facilitates efficient viral cDNA synthesis in vivo. J Virol. 2006;80(12):5670–5677. doi: 10.1128/JVI.02471-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrington RD, Geballe AP. Cofactor requirement for human immunodeficiency virus type 1 entry into a CD4-expressing human cell line. J Virol. 1993;67:5939–5947. doi: 10.1128/jvi.67.10.5939-5947.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol. 2005;6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 23.Hooker CW, Harrich D. The first strand transfer reaction of HIV-1 reverse transcription is more efficient in infected cells than in cell-free natural endogenous reverse transcription reactions. J Clin Virol. 2003;26(2):229–238. doi: 10.1016/s1386-6532(02)00121-x. [DOI] [PubMed] [Google Scholar]
- 24.Hughes A, Barber T, Nelson M. New treatment options for HIV salvage patients: an overview of second generation PIs, NNRTIs, integrase inhibitors and CCR5 antagonists. J Infect. 2008;57(1):1–10. doi: 10.1016/j.jinf.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 25.Jolly C, Sattentau QJ. Human immunodeficiency virus type 1 assembly, budding, and cell-cell spread in T cells take place in tetraspanin-enriched plasma membrane domains. J Virol. 2007;81(15):7873–7884. doi: 10.1128/JVI.01845-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman JC, Montagnier L. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature. 1984;312(5996):767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 27.Krementsov DN, Weng J, Labele M, Roy NH, Thali M. Tetraspanins regulate cell-to-cell transmission of HIV-1. Retrovirol. 2009;6:64. doi: 10.1186/1742-4690-6-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krishnan V, Zeichner SL. Alterations in the expression of DEAD-box and other RNA binding proteins during HIV-1 replication. Retrovirology. 2004;1:42. doi: 10.1186/1742-4690-1-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar A, Zloza A, Moon RT, Watts J, Tenorio AR, Al-Harthi L. Active beta-catenin signaling is an inhibitory pathway for human immunodeficiency virus replication in peripheral blood mononuclear cells. J Virol. 2008;82(6):2813–2820. doi: 10.1128/JVI.02498-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin D, Kamsteeg EJ, Zhang Y, Jin Y, Sterling H, Yue P, Roos M, Duffield A, Spencer J, Caplan M, Wang WH. Expression of tetraspan protein CD63 activates protein-tyrosine kinase (PTK) and enhances the PTK-induced inhibition of ROMK channels. J Biol Chem. 2008;283(12):7674–7681. doi: 10.1074/jbc.M705574200. [DOI] [PubMed] [Google Scholar]
- 31.Morita E, Sundquist WI. Retrovirus budding. Annu Rev Cell Dev Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- 32.Murray JL, Mavrakis M, McDonald NJ, Yilla M, Sheng J, Bellini WJ, Zhao L, Le Doux JM, Shaw MW, Luo CC, Lippincott-Schwartz J, Sanchez A, Rubin DH, Hodge TW. Rab9 GTPase is required for replication of human immunodeficiency virus type 1, filoviruses, and measles virus. J Virol. 2005;79(18):11742–11751. doi: 10.1128/JVI.79.18.11742-11751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nydegger S, Khurana S, Krementsov DN, Foti M, Thali M. Mapping of tetraspanin-enriched microdomains that can function as gateways for HIV-1. J Cell Biol. 2006;173(5):795–807. doi: 10.1083/jcb.200508165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Brien WA, Koyanagi Y, Namazie A, Zhao JQ, Diagne A, Idler K, Zack JA, Chen IS. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature. 1990;348:69–73. doi: 10.1038/348069a0. [DOI] [PubMed] [Google Scholar]
- 35.O’Doherty U, Swiggard WJ, Jeyakumar D, McGain D, Malim MH. A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. J Virol. 2002;76:10942–10950. doi: 10.1128/JVI.76.21.10942-10950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pelchen-Matthews A, Kramer B, Marsh M. Infectious HIV-1 assembles in late endosomes in primary macrophages. J Cell Biol. 2003;162(3):443–455. doi: 10.1083/jcb.200304008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perlman M, Resh MD. Identification of an intracellular trafficking and assembly pathway for HIV-1 gag. Traffic. 2006;7(6):731–745. doi: 10.1111/j.1398-9219.2006.00428.x. [DOI] [PubMed] [Google Scholar]
- 38.Popik W, Pitha PM. Binding of human immunodeficiency virus type 1 to CD4 induces association of Lck and Raf-1 and activates Raf-1 by a Ras-independent pathway. Mol Cell Biol. 1996;16:6532–6541. doi: 10.1128/mcb.16.11.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raposo G, Moore M, Innes D, Leijendekker R, Leigh-Brown A, Benaroch P, Geuze H. Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic. 2002;3(10):718–729. doi: 10.1034/j.1600-0854.2002.31004.x. [DOI] [PubMed] [Google Scholar]
- 40.Reeves JD, Piefer AJ. Emerging drug targets for antiretroviral therapy. Drugs. 2005;65(13):1747–1766. doi: 10.2165/00003495-200565130-00002. [DOI] [PubMed] [Google Scholar]
- 41.Rich EA, Chen IS, Zack JA, Leonard ML, O’Brien WA. Increased susceptibility of differentiated mononuclear phagocytes to productive infection with human immunodeficiency virus-1 (HIV-1) J Clin Invest. 1992;89:176–183. doi: 10.1172/JCI115559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rous BA, Reaves BJ, Ihrke G, Briggs JA, Gray SR, Stephens DJ, Banting G, Luzio JP. Role of adaptor complex AP-3 in targeting wild- type and mutated CD63 to lysosomes. Mol Biol Cell. 2002;13(3):1071–1082. doi: 10.1091/mbc.01-08-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruiz-Mateos E, Pelchen-Matthews A, Deneka M, Marsh M. CD63 is not required for production of infectious human immunodeficiency virus type 1 in human macrophages. J Virol. 2008;82(10):4751–4761. doi: 10.1128/JVI.02320-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sato K, Aoki J, Misawa N, Daikoku E, Sano K, Tanaka Y, Koyanagi Y. Modulation of human immunodeficiency virus type 1 infectivity through incorporation of tetraspanin proteins. J Virol. 2008;82(2):1021–1033. doi: 10.1128/JVI.01044-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427(6977):848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 46.Stipp CS, Kolesnikova TV, Hemler ME. Functional domains in tetraspanin proteins. Trends Biochem Sci. 2003;28(2):106–112. doi: 10.1016/S0968-0004(02)00014-2. [DOI] [PubMed] [Google Scholar]
- 47.Tanaka M, Ueno T, Nakahara T, Sasaki K, Ishimoto A, Sakai H. Downregulation of CD4 is required for maintenance of viral infectivity of HIV-1. Virology. 2003;311(2):316–325. doi: 10.1016/s0042-6822(03)00126-0. [DOI] [PubMed] [Google Scholar]
- 48.Ugolini S, Mondor I, Parren PW, Burton DR, Tilley SA, Klasse PJ, Sattentau QJ. Inhibition of virus attachment to CD4+ target cells is a major mechanism of T cell line-adapted HIV-1 neutralization. J Exp Med. 1997;186(8):1287–1298. doi: 10.1084/jem.186.8.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vodicka MA, Goh WC, Wu LI, Rogel ME, Bartz SR, Schweickart VL, Raport CJ, Emerman M. Indicator cell lines for detection of primary strains of human and simian immunodeficiency viruses. Virology. 1997;233:193–198. doi: 10.1006/viro.1997.8606. [DOI] [PubMed] [Google Scholar]
- 50.von Lindern JJ, Rojo D, Grovit-Ferbas K, Yeramian C, Deng C, Herbein G, Ferguson MR, Pappas TC, Decker JM, Singh A, Collman RG, O’Brien WA. Potential role for CD63 in CCR5-mediated human immunodeficiency virus type 1 infection of macrophages. J Virol. 2003;77(6):3624–3633. doi: 10.1128/JVI.77.6.3624-3633.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.von Schwedler UK, Stuchell M, Müller B, Ward DM, Chung HY, Morita E, Wang HE, Davis T, He GP, Cimbora DM, Scott A, Kräusslich HG, Kaplan J, Morham SG, Sundquist WI. The protein network of HIV budding. Cell. 2003;114(6):701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 52.Warrilow D, Meredith L, Davis A, Burrell C, Li P, Harrich D. Cell factors stimulate human immunodeficiency virus type 1 reverse transcription in vitro. 2008;82(3):1425–1437. doi: 10.1128/JVI.01808-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Warrilow D, Warren K, Harrich D. Strand transfer and elongation of HIV-1 reverse transcription is facilitated by cell factors in vitro. PLoS One. 2010;5(10):e13229. doi: 10.1371/journal.pone.0013229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zack JA, Arrigo SJ, Weitsman SR, Go AS, Haislip A, Chen IS. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]