Abstract
The evolution of multicellularity required the suppression of cancer. If every cell has some chance of becoming cancerous, large, long-lived organisms should have an increased risk of developing cancer compared to small, short-lived organisms. The lack of correlation between body size and cancer risk is known as Peto’s Paradox. Animals with 1,000 times more cells than humans do not exhibit an increased cancer risk, suggesting that natural mechanisms can suppress cancer 1,000 times more effectively than is done in human cells. Because cancer has proven difficult to cure, attention has turned to cancer prevention. In this review, like pharmaceutical companies mining natural products, we seek to understand how evolution has suppressed cancer to ultimately develop improved cancer prevention in humans.
The Evolutionary Theory of Cancer
Cancer is a consequence of multicellularity and a striking example of multi-level selection. The theory of cancer initiation and progression is deeply rooted in evolutionary and ecological concepts [1]. Cancer develops through somatic evolution, with genetic and epigenetic instability generating fitness variation among the cells of a body (Box 1). Selection at the level of organisms has led to the evolution of tumor suppressor mechanisms, such as cell cycle check points and apoptosis, which act as safe-guards to prevent somatic mutations from propagating in the cell population. Nonetheless, cancer occurs at astonishingly high rates and can be responsible for 20–46% of total deaths in multicellular animals ranging from mollusks to mammals [2].
Box 1. Somatic Evolution and the Development of Cancer.
Throughout an organism’s life, cells accumulate mutations caused by endogenous and exogenous damage, or errors in DNA synthesis, that is not properly repaired. In fact, somatic cells in tumors satisfy the three necessary and sufficient conditions for natural selection:
There must be variation within the population. A tumor is a heterogeneous population of cells with somatic genetic and epigenetic alterations.
The variation must be heritable. Genetic and epigenetic alterations (mutations) are inherited by both daughter cells when a cell divides.
There must be differential survival and reproduction (i.e. fitness). In some cases, the genetic and epigenetic mutations provide cells with survival and/or reproductive advantages over other cells.
The genetic and epigenetic changes in somatic cells can result in the six ‘hallmarks of cancer’, all of which provide a fitness advantage to somatic cells: (i) self sufficiency of growth-signals, (ii) insensitivity to anti-growth signals, (iii) evasion of apoptosis, (iv) sustained angiogenesis, (v) limitless replicative potential (stabilization of telomeres), and (vi) the ability to invade tissue and metastasize [69]. The somatic evolution taking place within mutant cell populations can result in cancer [1, 70]. Understanding this process through an evolutionary perspective is essential for knowing how a given treatment will affect the population dynamics and how we might be able to intervene in order to prevent the development of cancer all together.
Peto’s Paradox
The challenge of suppressing somatic evolution dramatically increases with larger bodies and longer lifespans. Because cancer develops through the accumulation of mutations, each proliferating cell is at risk of malignant transformation, assuming all proliferating cells have similar probabilities of mutation. Therefore, if an organism has more cells, i.e. more chances to initiate a tumor, the probability of getting cancer should increase. Similarly, if an organism has an extended lifespan, its cells have more time to accumulate mutations. Because the probability of carcinogenesis is an increasing function of age [3], an organism’s lifetime risk of cancer should also scale with its lifespan. It is well known that larger organisms generally have longer lifespans [4] which exacerbates this problem.
There appears to be no correlation between body size, longevity and cancer across species and the absence of such a relationship is referred to as Peto’s Paradox [2, 5]. Cancer rates across multicellular animals only vary by approximately two-fold even though the difference of size among mammals alone can be on the order of a million-fold [2]. Natural selection interacts with the life history of a species and should suppress cancer through the expected period of fertility of an organism. Therefore, given the relative age of an organism, we would expect cancer rates to be similar across species. The question of Peto’s Paradox is how has natural selection changed the biology of large, long-lived organisms to achieve this scaling.
The exact functional relationship between body size and expected cancer risk is unclear; however it is assumed to be an increasing function. In comparing laboratory rodents and humans, which differ in lifespan by a factor of 40 and size by three orders of magnitude, about 30% of both rodents and humans have cancer by the end of their life [6]. The general explanation for this is that large, long-lived animals are more resistant to carcinogenesis than small, short-lived animals [5, 7–9]; however, how they accomplish this resistance has yet to be established. Understanding this resistance can lead to new methods of cancer prevention in humans.
The Need and Potential for Cancer Prevention
Cancer has proven difficult to cure. Since former U.S. president Richard Nixon declared the “War on Cancer” almost 40 years ago, little progress has been made on reducing lifetime risk of cancer and increasing survival rates for patients with late stage diagnoses [10, 11]. The majority of cancer research focuses on treatment rather than prevention, and this often leads to the recurrence of tumors that are resistant to therapy. With 109–1012 cells in a tumor and perhaps 105 mutations [12–15], it appears that in many cases therapy selects for a resistant clone [1]. Increasingly, attention is turning to cancer prevention so as to avoid this scenario entirely.
A proven strategy in drug development has been to seek natural products that have been honed by millions of years of evolution to generate the desired effect [16]. The evolution of large multicellular organisms could hold the key to preventing cancer in humans. Peto’s Paradox suggests that large, long-lived animals such as the blue whale (Balaenoptera musculus) have evolved mechanisms capable of suppressing cancer 1,000 times better than humans. Research on how these large animals are suppressing cancer holds the promise of dramatic improvements in cancer prevention for humans.
Peto’s Paradox Appears to be Real
Cancer incidence records for wild and captive animals are not well documented for most species, making it difficult to directly compare incidence records of humans and other animals. However, it is still clear that cancer incidence does not scale with body size across species (Box 2). If blue whales got 1,000 times more cancer than humans, they would likely die before they were able to reproduce and the species would quickly go extinct [17]. The mere existence of whales suggests that is it possible to suppress cancer many-fold better than humans.
Box 2. All Whales Should Have Colorectal Cancer by Age 80.
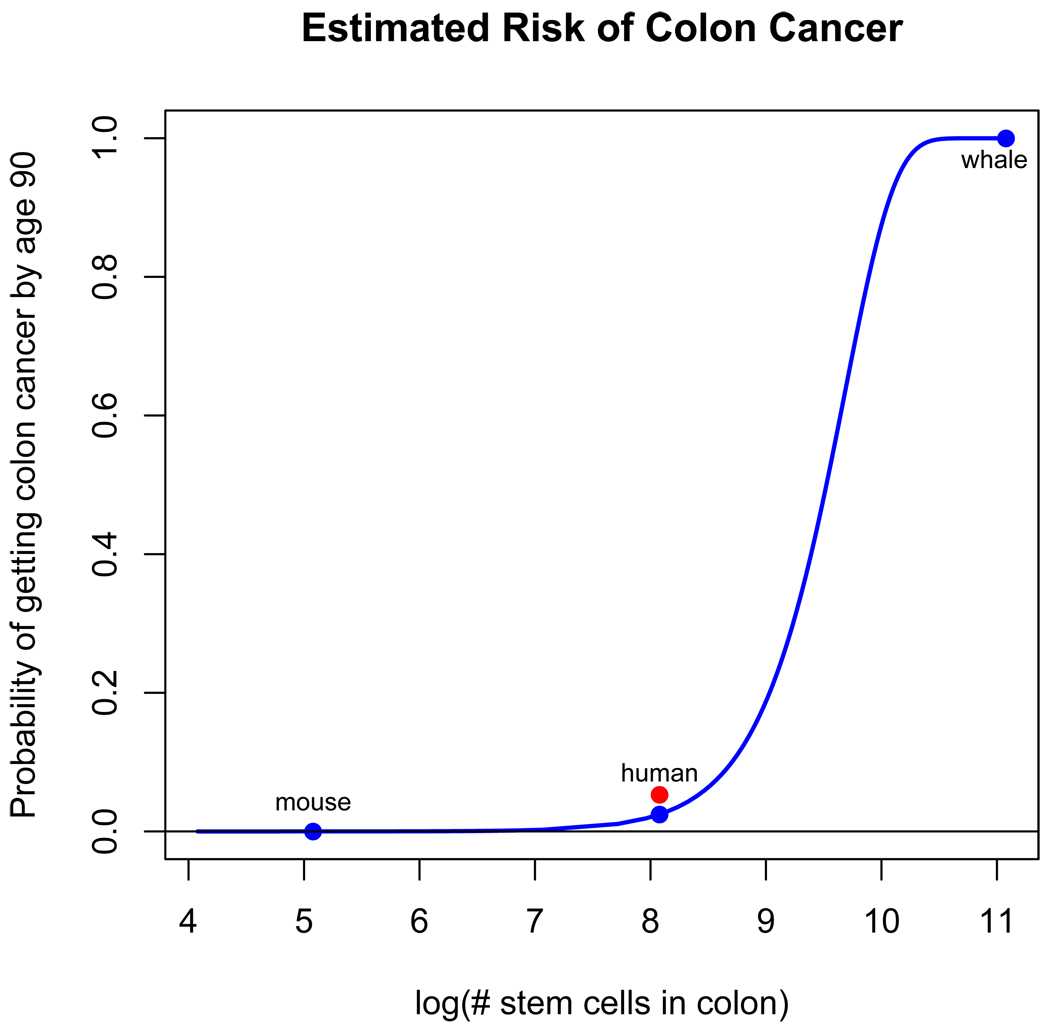
Calabrese and Shibata devised a simple mathematical equation to express the probability of a human developing colorectal cancer given their age [71]. Their equation produces results which closely match data from the Surveillance, Epidemiology and End Results (SEER) Program [72]. The probability of an individual developing colorectal cancer after a given number of cell divisions, which is proportional to age, is formulated in the equation,
where u is the mutation rate per gene per division, d is the number of stem cell divisions since birth, k is the number of rate limiting mutations required for cancer to occur, N is the number of effective stem cells per crypt and m is the number of crypts per colon.
The model also shows that the increased cancer risk observed in taller women in the SEER data set can be fit by simply increasing the parameter m to account for a larger colon [71]. Using the same rationale, we varied the parameter m from 1.5 × 103 to 1.5 × 1010 to see how the total number of stem cells in the colon changes the lifetime (90 year) risk of developing colorectal cancer (Figure I). We used the same values as Calabrese and Shibabta for all other parameters (Table S1) [71]. If we use the blue whale as an example of an animal that is 1,000 times the size of a human, where m could equal 1.5 × 1010 crypts, then this analysis reveals that over 50% of blue whales would have colorectal cancer by age 50 and all would have colorectal cancer by age 80. The chance of an individual person getting colorectal cancer by age 90 is only about 2.5% according to this model and just over 5% as reported by the American Cancer Society [10]. It is implausible that 100% of blue whales actually get colorectal cancer by age 80. Though we do not know how often blue whales are getting colorectal cancer, they have been reported to occasionally have other cancers [20, 21] and live over 100 years [35]. This model suggests that there is something fundamentally different in the initiation and progression of cancer in large, long-lived animals, like whales, compared to humans. Cancer rates for large, long-lived organisms could be made more similar to smaller animals by decreasing the mutation rate u, decreasing the rate of stem cell divisions which would decrease d, increasing the number of rate limiting mutations (k) needed to get cancer, or decreasing the number of proliferating stem cells per crypt (N).
Cancer death rates vary approximately two-fold across multicellular animals of drastically different sizes [2]. When wild mice are raised in protected laboratory conditions 46% die of cancer [18]. Cancer is also responsible for about 20% of dog deaths [19], roughly 25% of human deaths in the United States [10] and 18% of beluga whale deaths [20]. Rare cases of cancer are discovered in blue whales, giving no evidence of elevated cancer risk in these species [20, 21]. No matter the size or lifespan of the animal, cancer seems to account for approximately the same percentage of deaths.
Interestingly, within a species, size is associated with an increased cancer risk. In humans, 3–4mm above the average leg length results in an 80% higher risk of nonsmoking-related cancers [22]. Also, children with bone cancers tend to be taller and osteosarcomas occur in large dogs 200 times more frequently than small and medium breeds [23]. There has likely not been enough time to evolve additional mechanisms to protect them from this increased risk and counteract the extreme artificial selection for size. This suggests that animals which evolved to be larger as a species developed mechanisms to offset this increased cancer risk; while above average individuals do not possess additional defenses compared to smaller organisms within their species, and therefore fall victim to cancer with greater probability. This divergent trend within versus between species is an example of Simpson’s paradox [24].
Hypotheses to Resolve Peto’s Paradox
Limited research efforts have been focused on resolving Peto’s Paradox. However, there are many hypotheses that might explain how organisms could overcome the burden of cancer despite an increased number of cells and extended lifespan. Some have been previously proposed [2, 25–29] and others, to the best of our knowledge, are new in this review. Large bodies evolved independently along multiple lineages; therefore, we would not expect that all large, long-lived animals have evolved the same mechanism(s) to suppress cancer, unless the suppression stems from an innate characteristic common to all larger organisms. Differences in diet and carcinogenic exposures (including pathogens, which are only associated with 15% of human cancers [30]) are unlikely explanations because there are many-fold differences in size between organisms with similar environments (e.g., dolphins and whales) and similar diets (e.g., elephants and mice are both herbivores). Here we present some possible mechanisms that might have evolved to obliterate the expected correlation between body size, lifespan and cancer risk.
Tumor Suppression Mechanisms that might vary across Large, Long-lived Species
Lower Somatic Mutation Rates
If large animals have lower somatic mutation rates per cell generation, then more cell divisions would need to occur, compared to smaller animals, in order for a cell to acquire the necessary mutations to become malignant. Mutation rate is a function of the error rate and the rate at which these errors are repaired. This could be achieved through a number of mechanisms including better DNA damage detection and repair mechanisms. However, experimental data seem to suggest that mice and humans have comparable mutation rates [2], though better methods to measure somatic mutation rates in vivo are needed to explore this hypothesis.
Redundancy of Tumor Suppressor Genes
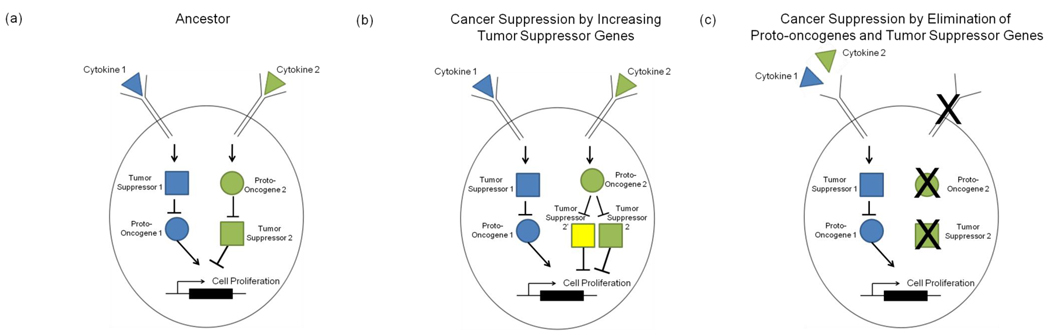
Added redundancy of tumor suppressor genes (TSGs) could also suppress cancer in large animals [2, 28] by requiring more mutations to occur to produce a malignant phenotype (Figure 1). Human cells require more mutations than mouse (Mus musculus) cells to create immortalized cultures [31]. Both the Rb and p53 pathways must be knocked out to immortalize human fibroblasts while mouse cells require only the p53 pathway to be inactivated [25]. Mice genetically engineered to have extra copies of Trp53 or Cdkn2A have increased tumor resistance [32, 33]. Interestingly, the current build of the elephant genome (Loxodonta africana) (Ensembl release 59) has 12 orthologs of the human gene TP53, in addition to one copy of the genes that encode p73 and p63. The human genome only has one of each of these genes (TP53, TP63, TP73) [34]. If these all function as tumor suppressors, it might explain how elephants can have such large bodies and long lifespans (70 years in the wild) [35] but not succumb to cancer any more so than smaller animals..
Figure 1. Alternative pathways to cancer hallmarks.
(a) Assume that the ancestor of a large, long-lived organism has two pathways initiated by cytokines (triangles) such that if either one is disrupted the result is a hallmark of cancer. We illustrate this concept with cell proliferation; however this could be replaced with any of the hallmarks. A large organism could decrease its risk of cancer by evolving redundant copies of tumor suppressor genes (squares) (b) or by removing proto-oncogenes (circles) and tumor suppressor genes to eliminate an entire pathway (c) so that there are fewer carcinogenic loci in the genome that are vulnerable to mutation. This option might be constrained by selective pressures on the remaining pathways to produce the adaptive phenotypes that had been encoded in the deleted pathway.
An opposing solution would be to eliminate some proto-oncogenes from the genomes of large, long-lived organisms. Having fewer proto-oncogenes, decreases the chance of getting an oncogenic mutation and therefore decreases the overall probability of a cell resulting in cancer. This is supported by an experiment demonstrating that Hras1 null mutant mice develop significantly fewer papillomas than wild type mice [36]. If there were fewer pathways that could generate the phenotypes necessary for cancer, there would be fewer vulnerabilities in the genome and a reduced likelihood of cancer (Figure 1). Of course proto-oncogenes are serving other functions, so eliminating them could be deleterious for other reasons.
Redundancy could also be in the form of expression. Many tumor suppressor genes are tissue specific [37]. Cells of larger species could have evolved expression patterns such that in any given cell more TSGs are expressed compared to smaller, shorter-lived animals, even though there might be the same number of TSGs in the genome. This hypothesis would predict that large animals would have more ubiquitously expressed TSGs than smaller species.
Lower selective advantage of mutant cells
A haploinsufficient gene in mice could be completely recessive in a larger animal, requiring mutations to occur on both alleles in order to gain a selective advantage over neighboring cells during carcinogenesis in the larger species [2]. This would decrease the possibility that mutations at this locus would contribute to progression towards cancer. This has been observed in a tissue-specific manner. The tumor suppressor Trp53 usually requires both alleles of the gene to be null in order to see a mutant phenotype; however, in some tissues, Trp53 is haploinsufficient and losing only one allele produces a phenotype in mice [37].
Different tissue architecture
Changes in tissue architecture could influence the frequency of cancers by altering the way cells are compartmentalized and/or the dynamics of the tissue [2]. Most tissues are comprised of small proliferative units, for example the crypts of the intestines. It has been proposed that this hierarchical structure is a crucial cancer prevention mechanism [38]. Since differentiating cells are evolutionary dead-ends, the effective population size of a somatic tissue probably depends mainly on the number and dynamics of stem cells (though a mutation which disrupts differentiation in a non-stem cell might also generate a carcinogenic cell lineage) [39]. Under a model of “serial differentiation” it is possible to increase the number of cells and the amount of cell turnover without increasing the number or proliferative activity of somatic stem cells, simply by adding non-stem stages [40]. Altering the number of stem cells, the crypt density or the dynamics of differentiation and division could enhance the tissue’s ability to prevent malignant transformation.
More efficient immune system
Immune system efficiency against virus-associated cancers might account for some differences observed in cancer rates within people [26], but this could apply to non-viral cancers and we propose immune surveillance might explain differences in cancer resistance across species of all sizes. Tumors are initially immunogenic. When mice are treated with carcinogens, tumorigenesis is delayed by immune system surveillance [41]. However, as the tumor co-evolves with the immune system, tumor variants that go undetected are selected (termed “immunoediting”) [42]. “Chronic antigenic stress” can result in exhaustion of the immune system leading to ineffective surveillance, similar to observations of chronic viral infections [42]. Large, long-lived organisms might have better immune surveillance for neoplastic cells than smaller organisms.
More sensitive or efficient apoptotic processes
The apoptotic propensity of cells might differ between large and small organisms. Cells from large bodies could be more sensitive to DNA damage or the activation of an oncogene and thus would be more apt to apoptose [26]. Support for this hypothesis comes from observations of human and mouse cell cultures. When human cells are irradiated, many die due to apoptosis triggered by DNA damage. A much higher percentage of mouse cells survive and continue dividing regardless of the gross DNA damage inflicted by the radiation [43]. Apoptosis due to DNA damage eliminates the damaged cell from the population instead of repairing the DNA and possibly propagating remaining mutations in the tissue. However, there is likely to be a trade-off between apoptosis preventing cancer, but causing senescence due to depletion of the stem cell pool [44].
Increased sensitivity to contact inhibition
Selfish cellular proliferation can also be suppressed by signals from the microenvironment [26]. For example, cell contact inhibition has been noted to differ between human, mouse and naked mole-rat (Heterocephalus glaber) cells. In culture, naked mole-rat cells stop dividing at much lower densities than human and mouse cells due to the early activation of the p16 pathway which results in hypersensitivity to contact inhibition [45]. Although naked mole-rats and mice are small animals, the former live significantly longer than the latter (28 years [46] versus 4 years [47]). In all 250 necropsies of naked mole-rats that died in captivity, none had died of cancer [48]. Hypersensitive contact inhibition might have evolved to suppress cancer so the naked mole-rat can live longer, though it has only been verified in vitro [45]. Signals for early cell senescence could be triggered in large, long-lived organisms to inhibit uncontrolled proliferation.
Shorter Telomeres
Telomere length appears to be a fundamental check on the proliferative capacity of cells [49]. Telomeres shorten with every cell cycle and when they become too short to protect the chromosomes’ ends, the cell senses those ends as DNA double strand breaks, usually leading to apoptosis [50, 51]. Even though stem cells express telomerase, which helps to rebuild telomeres, they generally do not express enough to prevent telomere shortening due to proliferation [51]. We hypothesize that large, long-lived animals might have shorter telomeres (or erode them faster) than smaller animals, limiting the number of times their cells can divide and reducing opportunities to accumulate carcinogenic mutations.
Characteristics of all large organisms that might act as tumor suppression mechanisms
Less Reactive Oxygen Species due to Lower Basal Metabolic Rate
A lower somatic mutation rate could also be a result of metabolism. Reactive oxygen species (ROS) are byproducts of metabolism and can cause DNA damage thought to contribute to aging and cancer [52–54]. The rate at which ROS are produced in a cell is a function of the basal metabolic rate (BMR) [55]. BMR per unit mass (mass-specific BMR) is proportional to (body mass)−1/4 [56] and has been shown to correlate with the amount of oxidative damage [57]. Knocking out oxidative repair genes, and therefore allowing for DNA damage from ROS to persist, results in increased tumor susceptibility in a variety of tissues suggesting that DNA damage caused by ROS plays a causal role in tumor formation [58]. Large animals should produce fewer ROS due to their lower BMR and consequently have less endogenous damage to their DNA and an overall lower somatic mutation rate [29].
The average BMR of women is 10% lower than that of men after adjusting for body mass, composition, activity and age [29] and women consistently have lower rates of cancer [10]. Naked mole-rats, for which spontaneous cancer has yet to be reported [48], have a mass-specific BMR that is much lower than expected given their size [35]. Caloric restriction inhibits cancers in animal models and one explanation for this is that the decrease in caloric intake lowers the metabolic rate, therefore producing less ROS and subjecting the DNA to less endogenous damage [59]. These observations could all be attributed to cells having less endogenous oxidative damage which effectively results in a lower somatic mutation rate and a reduced cancer risk.
Formation of hypertumors
Nagy et al. have proposed an alternative hypothesis to resolve Peto’s paradox [27]. Natural selection within a tumor might favor ‘cheater’ cells that take advantage of vasculature built by angiogenic cells. These ‘cheaters’ could grow and parasitize the primary tumor. This “hypertumor” would reduce the overall fitness of the tumor and might even cause the tumor to regress. Nagy et al. argue that lethal tumors must be drastically larger in larger animals, giving the hypertumor more time to evolve and force the parent tumor to become necrotic [27]. This model predicts that large animals would often carry macroscopic tumors which should be disproportionately more necrotic when compared to lethal tumors in smaller organisms [27], though this has yet to be verified experimentally. This hypothesis might be tested by serially passaging a cancer through mice, eventually generating enough cells that a hypertumor should evolve and the mean fitness of the tumor decrease.
Suggestions for the future
If the current understanding of cancer is correct, there must be something fundamentally different in large, long-lived organisms that enhances their suppression of carcinogenesis. These mechanisms have allowed for the evolution of large bodies and extended lifespans without increasing the burden of cancer. Most of the hypotheses that have been proposed have not been directly tested, and most related questions remain open (Box 3).
Box 3. Outstanding Questions for Peto’s Paradox.
What is the age-related incidence of cancer in most non-human (and non-experimental) animal species?
-
Which of the many suggested mechanisms are valid explanations for the lack of correlation between body size, longevity and cancer incidence? Compared to humans, do larger, long-lived organisms have:
Lower somatic mutation rates?
More copies of tumor suppressor genes?
Fewer proto-oncogenes?
Smaller selective advantages for somatic mutants?
Different tissue architecture (smaller proportion of stem cells or more quiescent stem cells)?
More efficient immune surveillance?
An apoptotic process highly sensitive to DNA damage?
Increased sensitivity to contact inhibition?
Shorter telomeres?
Less DNA damage due to fewer reactive oxygen species?
Is the presumed decrease in cancer incidence in lineages with lower than expected cancer incidence the result of several mechanisms that each contribute in a cumulative manner to decreasing the cancer risk of each cell, or rather of a single mechanism that has a drastic effect on a cell’s probability of becoming malignant?
Are such mechanisms shared among large, long-lived species or are they unique to each species?
Does the cancer protection come from some innate characteristic of large organisms (i.e. low mass specific basal metabolic rate)?
Can the cancer suppression mechanisms used by large, long-lived organisms be translated to humans as novel cancer preventive interventions?
Large bodies have evolved independently multiple times in the history of life, so each clade could have evolved different mechanism(s) to boost their tumor suppression abilities. An approach based on independent contrasts [60] of small and large species within each clade could prove fruitful for identifying cancer suppression mechanisms (Box 4).
Box 4. A Phylogenetic Approach to study Peto’s Paradox.
Large, long-lived organisms might have evolved to suppress cancer better than small animals by duplicating tumor suppressor genes [2, 28] or eliminating some proto-oncogenes from the genome. A simple linear regression cannot be used to study whether a correlation exists between body size and the copy number of cancer related genes because this assumes independence of each genome. In reality, the genomes share many traits in common due to evolutionary decent from a common ancestor. An independent contrast model [73] should be used to partition the variance among species into comparisons that are independent of their evolutionary relationships. This can be done by studying multiple clades, each composed of closely related species which have large variance in body size. Marine mammals belonging to the Order Cetacea are an ideal clade for this study since they range in size from small dolphins like the Commerson’s dolphin (∼50 kg) [35] to the largest mammal on Earth, the blue whale (over 100,000 kg) [35]. The split between dolphins and whales occurred only 25–30 million years ago [74]. Unfortunately, the genomes of these animals are not currently available. Studies should focus on clades that include animals larger than humans, as opposed to looking at differences among various sized rodents or between mouse and human, because the goal is to find a way of preventing cancer that is superior to endogenous tumor suppression mechanisms in humans.
Unfortunately we currently lack data necessary to make these analyses possible. Efforts should focus on sequencing genomes of large, long-lived species along with closely related small species, to determine if tumor suppressor genes duplicated, or oncogenes were deleted, during the evolution of a large body within a clade. Gene expression analyses of the same tissues might also reveal differential expression of cancer genes in large organisms. In addition there are standard assays that could be used in comparative analyses to test many of the hypotheses for resolving Peto’s paradox (Box 5), including measurements of DNA damage repair [61], telomere lengths [62], differentiation [63] and proliferation [64, 65], apoptosis [66], and reactive oxygen species [67].
Box 5. Suggestions for Future Experiments.
There are many experiments that could be done to test the hypotheses proposed to explain Peto’s Paradox; however, they are limited by currently available information and assays as well as the fact that large, long-lived animals cannot be easily genetically manipulated in a laboratory.
Lower somatic mutation rates: Mutation rates can be measured in elephant and whale cells in vitro; however with better assays and longitudinal tissue sampling, the in vivo somatic mutation rate could be estimated [75].
More copies of tumor suppressor genes or fewer proto-oncogenes: Copy number of cancer-associated genes can be studied using genomics to count the orthologs of known cancer genes using independent contrasts (Box 4). This is based on sequence information only, so functional studies would be necessary as follow-up.
Smaller selective advantages for somatic mutants: Fitness affects of mutations in cells of different animals might be estimated using in vitro cell competitions; however this is not a realistic environment and might not reflect the true fitness caused by the mutation in vivo. Modern genetically engineered organisms could be used to measure the fitness of isolated mutations in vivo [76, 77].
Different tissue architecture (smaller proportion of stem cells or more quiescent stem cells): The mitotic index could be measured for crypts in intestinal tissue samples from elephant and whale. Given reliable stem cell markers, stem cells could also be counted.
More efficient immune surveillance: It might be possible to measure the immune response to mutant proteins that vary from the endogenous sequence by different degrees.
An apoptotic process highly sensitive to DNA damage: Cells from animals like elephants and whales can be irradiated in vitro to quantify how many cells apoptose as a function of the amount of DNA damage.
Increased sensitivity to contact inhibition: Cells can be grown in vitro to determine how the density of the cultures when the cells stop growing compares to the density of cultured cells from smaller organisms, as was done with the naked mole-rat [45].
Shorter telomeres: Telomere lengths can be measured and compared across species by standard assays.
Less DNA damage due to fewer reactive oxygen species: New methods involving fluorgenic sensors for superoxide and hydroxyl radicals can detect ROS in cell cultures, tissues and in vivo [78].
The majority of cancer research is done on a very small subset of organisms which restricts our understanding of cancer to what we learn from those particular model systems. Furthermore, the qualities of model organisms that make them ideal to work with in laboratory conditions (short lifespan and small body) are the very things that make them poor models for cancer suppression [2]. The lack of functional data for non-model organisms is a major gap in the field. Function is often assumed from homology which is not necessarily correct. For example, TSGs in Drosophila are largely non-overlapping with human tumor suppressors [68]. Studies that aim at a better understanding of the evolution of cancer suppression mechanisms will have to expand the variety of organisms that are studied in the laboratory setting.
There are not many large, long-lived organisms that have been fully sequenced yet, so testing Peto’s Paradox by doing comparative genomic analyses is difficult with current data. We are also lacking robust epidemiological studies of cancer incidence in wildlife and captive populations. Captive populations will be useful for longitudinal studies and the predation-free environment will allow for better estimates of cancer rates. This will help researchers to better understand the nature of Peto’s Paradox.
Conclusion
There has been no observed correlation between body size, longevity and lifetime cancer risk [2]. Every additional cell and extra year of life should increase the probability of carcinogenesis. The fact that large, long-lived organisms are not over burdened by cancer suggests that they are more resistant to malignant transformation. Research focusing on what mechanisms have evolved to yield this cancer resistance will not only help explain Peto’s Paradox, but should also open new doors in the field of cancer prevention. Cancer treatments have not proven as effective as promised. If we can harness the cancer suppression mechanisms of large, long-lived organisms, then we could potentially eradicate cancer as a public health threat in humans. A pharmaceutical company’s initial step in developing a new class of drugs is to survey natural products to see if evolution has already invented a solution to their problem. We are proposing that cancer prevention research capitalize on the same strategy. People have been invested in cancer research for decades while evolution has been tuning cancer suppression mechanisms for over a billion years. It’s time to learn from the expert.
Supplementary Material
Figure I. Estimated probability of colorectal cancer by age 90 based on the number of cells in the colon.
The probability of getting colorectal cancer at a certain age was calculated with the equation p = 1-(1-(1-(1-u)d)k)Nm [71] where u is the mutation rate per gene per division, d is the number of stem cell divisions since birth, k is the number of rate limiting mutations required for cancer to occur, N is the number of effective stem cells per crypt and m is the number of crypts per colon [71]. Parameter values are listed in Table S1. This shows that assuming all other parameters are equal, larger animals should have a much greater lifetime risk of cancer when compared to smaller organisms. Blue dots for mouse, human and whale indicate the estimated risk of colon cancer occurring within 90 years of life given the approximate number of cells in a human colon, 1,000 times fewer cells to represent the mouse, and 1,000 times more cells to represent the whale. The estimate for 1,000 times smaller than a human (e.g. a mouse) is still barely above zero even after 90 years. In reality, a mouse only lives a maximum of 4 years [35], so based on this equation they should never get colorectal cancer. The red dot indicates the lifetime risk of colon cancer according to the American Cancer Society which is about 5.3% for men and women averaged together [10].
Acknowledgements
This work was supported in part by the US Department of Energy Computational Science Graduate Fellowship, DE-FG02-97ER25308, the Martha W. Rodgers Charitable Trust, a McLean Contributionship, the Landon AACR Innovator Award for Cancer Prevention, Research Scholar Grant #117209-RSG-09-163-01-CNE from the American Cancer Society and NIH grants R03 CA137811, P01 CA91955, P30 CA010815, R01 CA119224 and R01 CA140657.
Glossary
- Cancer
a disease defined by the uncontrolled growth of abnormal cells which have the ability to invade other tissues or spread to a new part of the body
- Malignancy
synonymous to cancer
- Malignant transformation
the process through which normal cells become cancerous
- Tumor suppressor gene (TSG)
a gene which increases the chance of progression to cancer when it is inactivated or deleted. These genes normally function to suppress the initiation of tumors, or prevent and repair damage to the genome
- Proto-oncogene
a gene which increases the chance of progression to cancer when it is over-expressed or inappropriately activated by mutation
- Metastasis
the spreading of cancerous cells from the initial tumor to a new location/tissue in the body
- Angiogenesis
the process of growing new blood vessels. The size of a tumor is limited by the diffusion distance of oxygen and glucose prior to angiogenesis
- Angiogenic cell
a cell producing factors to induce angiogenesis
- Late stage
cancer that has metastasized, requiring systemic therapy for treatment, rather than surgical excision
- Apoptosis
programmed cell death
- Rb
a tumor suppression protein that inhibits cell cycle progression until the cell is ready to continue to next phase of the cycle
- p53
a tumor suppressor protein that is involved in DNA repair, cell cycle regulation and apoptosis. In humans this is the product of the gene TP53 (GenBank Accession U94788) and the orthologous gene in mouse is Trp53 (GenBank Accession AY044188)
- p16 (CDKN2A)
a tumor suppressor protein encoded by the cyclin-dependent kinase inhibitor 2A gene which regulates the cell cycle. In mouse this is the product of gene Cdkn2A (GenBank Accession AF044335)
- p63
a tumor suppressor protein that is part of the p53 family and involved in cell differentiation. In human this is the product of the TP63 gene (GenBank Accession BC039815)
- p73
a tumor suppressor protein that is part of the p53 gene family and is involved in cell cycle regulation and apoptosis. In human this is the product of the TP73 gene (GenBank Accession BC117251)
- Hras1
(GenBank Accession AY373386) A proto-oncogene that codes for the Hras11 protein which promotes cell growth and division. It is frequently mutated in cancers
- Crypt
a well-like structure of epithelial cells. Stem cells remain at the base and as cells differentiate they move up the walls toward the top layer of the tissue. The surface of the intestine is made up of a sheet of crypts
- Haploinsufficient gene
A gene in a diploid organism that requires both alleles to be fully functional in order to exhibit a normal phenotype. A mutation in one allele will result in an abnormal phenotype
- Simpson’s Paradox
the observation that a statistical trend within groups opposes the trend between groups [24]. For example, two variables could be positively correlated within a species, but an interspecific comparison would reveal a negative correlation across groups
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Merlo L, et al. Cancer as an evolutionary and ecological process. Nature Reviews Cancer. 2006;6:924–935. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- 2.Leroi A, et al. Cancer selection. Nature reviews. Cancer. 2003;3:226–231. doi: 10.1038/nrc1016. [DOI] [PubMed] [Google Scholar]
- 3.Frank SA. Dynamics of cancer : incidence, inheritance, and evolution. Princeton University Press; 2007. [PubMed] [Google Scholar]
- 4.Speakman JR. Body size, energy metabolism and lifespan. J Exp Biol. 2005;208:1717–1730. doi: 10.1242/jeb.01556. [DOI] [PubMed] [Google Scholar]
- 5.Peto R, et al. Cancer and Aging in Mice and Men. Brit J Cancer. 1975;32:411–426. doi: 10.1038/bjc.1975.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rangarajan A, Weinberg RA. Opinion: Comparative biology of mouse versus human cells: modelling human cancer in mice. Nature Reviews Cancer. 2003;3:952–959. doi: 10.1038/nrc1235. [DOI] [PubMed] [Google Scholar]
- 7.Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- 8.Dawe CJ, et al. Neoplasms and related disorders of invertebrate and lower vertebrate animals; [proceedings. U.S. National Cancer Institute; 1969. for sale by the Supt. of Docs. [Google Scholar]
- 9.Graham J. Cancer Selection: The New Theory of Evolution. Aculeus Press; 1992. [Google Scholar]
- 10.American Cancer Society. Cancer Facts & Figures 2010. American Cancer Society; 2010. [Google Scholar]
- 11.Etzioni R, et al. The case for early detection. Nature Reviews Cancer. 2003;3:243–252. doi: 10.1038/nrc1041. [DOI] [PubMed] [Google Scholar]
- 12.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 14.Mardis ER, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bielas JH, et al. Human cancers express a mutator phenotype. Proc Natl Acad Sci U S A. 2006;103:18238–18242. doi: 10.1073/pnas.0607057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 17.Lichtenstein A. On evolutionary origin of cancer. Cancer Cell International. 2005;5:5. doi: 10.1186/1475-2867-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andervont HB, Dunn TB. Occurrence of tumors in wild house mice. J Natl Cancer Inst. 1962;28:1153–1163. [PubMed] [Google Scholar]
- 19.Morris J, Dobson J. Small Animal Oncology. Wiley-Blackwell; 2001. [Google Scholar]
- 20.Martineau D, et al. Cancer in wildlife, a case study: beluga from the St. Lawrence estuary, Québec, Canada. Environmental health perspectives. 2002;110:285–292. doi: 10.1289/ehp.02110285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newman SJ, Smith SA. Marine mammal neoplasia: a review. Vet Pathol. 2006;43:865–880. doi: 10.1354/vp.43-6-865. [DOI] [PubMed] [Google Scholar]
- 22.Albanes D. Height, early energy intake, and cancer. Evidence mounts for the relation of energy intake to adult malignancies. BMJ (Clinical research ed.) 1998;317:1331–1332. doi: 10.1136/bmj.317.7169.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altman AJ, Schwartz AD. Malignant diseases of infancy, childhood and adolescence. Major Probl Clin Pediatr. 1978;18:1–515. [PubMed] [Google Scholar]
- 24.Simpson EH. The Interpretation of Interaction in Contingency Tables. Journal of the Royal Statistical Society Ser. B. 1951:238–241. [Google Scholar]
- 25.Hahn W, Weinberg R. Rules for making human tumor cells. The New England journal of medicine. 2002;347:1593–1603. doi: 10.1056/NEJMra021902. [DOI] [PubMed] [Google Scholar]
- 26.Klein G. Toward a genetics of cancer resistance. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:859–863. doi: 10.1073/pnas.0811616106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagy JD, et al. Why don't all whales have cancer? A novel hypothesis resolving Peto's paradox. Integr Comp Biol. 2007;47:317–328. doi: 10.1093/icb/icm062. [DOI] [PubMed] [Google Scholar]
- 28.Nunney L. Lineage Selection and the Evolution of Multistage Carcinogenesis. Proceedings: Biological Sciences. 1999:266. doi: 10.1098/rspb.1999.0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Totter JR. Spontaneous cancer and its possible relationship to oxygen metabolism. Proc Natl Acad Sci U S A. 1980;77:1763–1767. doi: 10.1073/pnas.77.4.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.zur Hausen H. Viral Oncogenesis. JAIDS Journal of Acquired Immune Deficiency Syndromes. 1999;21:A7. [Google Scholar]
- 31.Rangarajan A, et al. Species- and cell type-specific requirements for cellular transformation. Cancer Cell. 2004;6:171–183. doi: 10.1016/j.ccr.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 32.García-Cao I, et al. "Super p53" mice exhibit enhanced DNA damage response, are tumor resistant and age normally. The EMBO journal. 2002;21:6225–6235. doi: 10.1093/emboj/cdf595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matheu A, et al. Increased gene dosage of Ink4a/Arf results in cancer resistance and normal aging. Genes & development. 2004;18:2736–2746. doi: 10.1101/gad.310304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belyi VA, et al. The origins and evolution of the p53 family of genes. Cold Spring Harb Perspect Biol. 2010;2 doi: 10.1101/cshperspect.a001198. a001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Magalhães JP, Costa J. A database of vertebrate longevity records and their relation to other life-history traits. Journal of evolutionary biology. 2009;22:1770–1774. doi: 10.1111/j.1420-9101.2009.01783.x. [DOI] [PubMed] [Google Scholar]
- 36.Ise K, et al. Targeted deletion of the H-ras gene decreases tumor formation in mouse skin carcinogenesis. Oncogene. 2000;19:2951–2956. doi: 10.1038/sj.onc.1203600. [DOI] [PubMed] [Google Scholar]
- 37.Payne S, Kemp C. Tumor suppressor genetics. Carcinogenesis. 2005;26:2031–2045. doi: 10.1093/carcin/bgi223. [DOI] [PubMed] [Google Scholar]
- 38.Gatenby R, et al. The evolutionary dynamics of cancer prevention. Nature Reviews Cancer. 2010;10:526–527. doi: 10.1038/nrc2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michor F. Chronic myeloid leukemia blast crisis arises from progenitors. Stem Cells. 2007;25:1114–1118. doi: 10.1634/stemcells.2006-0638. [DOI] [PubMed] [Google Scholar]
- 40.Pepper J, et al. Animal cell differentiation patterns suppress somatic evolution. PLoS computational biology. 2007;3:e250. doi: 10.1371/journal.pcbi.0030250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koebel C, et al. Adaptive immunity maintains occult cancer in an equilibrium state. Nature. 2007;450:903–907. doi: 10.1038/nature06309. [DOI] [PubMed] [Google Scholar]
- 42.Pawelec G, et al. Immunosenescence and cancer. Critical Reviews in Oncology/Hematology. 2010;75:165–172. doi: 10.1016/j.critrevonc.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 43.Humbert O, et al. Mismatch repair and differential sensitivity of mouse and human cells to methylating agents. Carcinogenesis. 1999;20:205–214. doi: 10.1093/carcin/20.2.205. [DOI] [PubMed] [Google Scholar]
- 44.Tyner SD, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 45.Seluanov A, et al. Hypersensitivity to contact inhibition provides a clue to cancer resistance of naked mole-rat. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19352–19357. doi: 10.1073/pnas.0905252106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buffenstein R, Jarvis JU. The naked mole rat--a new record for the oldest living rodent. Sci Aging Knowledge Environ. 2002;2002:pe7. doi: 10.1126/sageke.2002.21.pe7. [DOI] [PubMed] [Google Scholar]
- 47.Turturro A, et al. Growth curves and survival characteristics of the animals used in the Biomarkers of Aging Program. J Gerontol A Biol Sci Med Sci. 1999;54:B492–B501. doi: 10.1093/gerona/54.11.b492. [DOI] [PubMed] [Google Scholar]
- 48.Buffenstein R. The naked mole-rat: a new long-living model for human aging research. The journals of gerontology. Series A, Biological sciences and medical sciences. 2005;60:1369–1377. doi: 10.1093/gerona/60.11.1369. [DOI] [PubMed] [Google Scholar]
- 49.Monaghan P. Telomeres and life histories: the long and the short of it. Ann N Y Acad Sci. 2010;1206:130–142. doi: 10.1111/j.1749-6632.2010.05705.x. [DOI] [PubMed] [Google Scholar]
- 50.d"Adda di Fagagna F, et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 51.Shay JW, Wright WE. Telomeres and telomerase in normal and cancer stem cells. FEBS Lett. 2010;584:3819–3825. doi: 10.1016/j.febslet.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wiseman H, Halliwell B. Damage to DNA by reactive oxygen and nitrogen species: role in inflammatory disease and progression to cancer. Biochem J. 1996;313((Pt 1)):17–29. doi: 10.1042/bj3130017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sedelnikova OA, et al. Role of oxidatively induced DNA lesions in human pathogenesis. Mutat Res. 2010;704:152–159. doi: 10.1016/j.mrrev.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 55.Ku HH, et al. Relationship between mitochondrial superoxide and hydrogen peroxide production and longevity of mammalian species. Free Radic Biol Med. 1993;15:621–627. doi: 10.1016/0891-5849(93)90165-q. [DOI] [PubMed] [Google Scholar]
- 56.Savage VM, et al. Scaling of number, size, and metabolic rate of cells with body size in mammals. Proc Natl Acad Sci U S A. 2007;104:4718–4723. doi: 10.1073/pnas.0611235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adelman R, et al. Oxidative damage to DNA: relation to species metabolic rate and life span. Proc Natl Acad Sci U S A. 1988;85:2706–2708. doi: 10.1073/pnas.85.8.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xie Y, et al. Deficiencies in Mouse Myh and Ogg1 Result in Tumor Predisposition and G to T Mutations in Codon 12 of the K-Ras Oncogene in Lung Tumors. Cancer Research. 2004;64:3096–3102. doi: 10.1158/0008-5472.can-03-3834. [DOI] [PubMed] [Google Scholar]
- 59.Longo VD, Fontana L. Calorie restriction and cancer prevention: metabolic and molecular mechanisms. Trends Pharmacol Sci. 2010;31:89–98. doi: 10.1016/j.tips.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garland T, Jr, et al. Phylogenetic approaches in comparative physiology. J Exp Biol. 2005;208:3015–3035. doi: 10.1242/jeb.01745. [DOI] [PubMed] [Google Scholar]
- 61.Olive PL, Banath JP. The comet assay: a method to measure DNA damage in individual cells. Nat Protoc. 2006;1:23–29. doi: 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
- 62.Canela A, et al. Telomere length analysis. Methods Mol Biol. 2007;371:45–72. doi: 10.1007/978-1-59745-361-5_5. [DOI] [PubMed] [Google Scholar]
- 63.Li Y, et al. Differentiation of embryonic stem cells in adult bone marrow. J Genet Genomics. 2010;37:431–439. doi: 10.1016/S1673-8527(09)60062-X. [DOI] [PubMed] [Google Scholar]
- 64.Minor LK. Label-free cell-based functional assays. Comb Chem High Throughput Screen. 2008;11:573–580. doi: 10.2174/138620708785204072. [DOI] [PubMed] [Google Scholar]
- 65.Woosley JT. Measuring cell proliferation. Arch Pathol Lab Med. 1991;115:555–557. [PubMed] [Google Scholar]
- 66.Ribble D, et al. A simple technique for quantifying apoptosis in 96-well plates. BMC Biotechnol. 2005;5:12. doi: 10.1186/1472-6750-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Afanasev I. Detection of superoxide in cells, tissues and whole organisms. Front Biosci (Elite Ed) 2009;1:153–160. doi: 10.2741/E15. [DOI] [PubMed] [Google Scholar]
- 68.Pearson BJ, Sánchez Alvarado A. Regeneration, stem cells, and the evolution of tumor suppression. Cold Spring Harbor symposia on quantitative biology. 2008;73:565–572. doi: 10.1101/sqb.2008.73.045. [DOI] [PubMed] [Google Scholar]
- 69.Hanahan D. The Hallmarks of Cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 70.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 71.Calabrese P, Shibata D. A simple algebraic cancer equation: calculating how cancers may arise with normal mutation rates. BMC cancer. 2010;10:3. doi: 10.1186/1471-2407-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Surveillance, Epidemiology and End Results (SEER) Program: SEER*Stat Database: Incidence - SEER 11 Regs Public-Use. Sureveillance Research Program, Cancer Statistics Branch. 2001 http://seer.cancer.gov/
- 73.Felsenstein J. Inferring phylogenies. Sinauer Associates; 2003. [Google Scholar]
- 74.Murphy WJ, et al. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 2007;17:413–421. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Drummond AJ, et al. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics. 2002;161:1307–1320. doi: 10.1093/genetics/161.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang J, et al. Evidence for mutation showers. Proc Natl Acad Sci U S A. 2007;104:8403–8408. doi: 10.1073/pnas.0610902104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Frese KK, Tuveson DA. Maximizing mouse cancer models. Nat Rev Cancer. 2007;7:645–658. doi: 10.1038/nrc2192. [DOI] [PubMed] [Google Scholar]
- 78.Kundu K, et al. Hydrocyanines: a class of fluorescent sensors that can image reactive oxygen species in cell culture, tissue, and in vivo. Angew Chem Int Ed Engl. 2009;48:299–303. doi: 10.1002/anie.200804851. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.