Abstract
We studied a panel of mutant viruses containing wild-type and chimeric capsid HPV16 and HPV18 proteins. The mutant capsid protein expression, genome amplification, and episomal maintenance were comparable with the wild-type virus. However, the chimeric viruses varied in their titers from wild-type. We show that the intertypical mutant chimeric capsid viruses, that L2 affects the structure of L1 and that L1 affects the structure of L2 in the virion. These effects were measured using a panel of conformation-dependent neutralizing L1 MAbs and a L2 capsid surface peptide derived neutralizing antibody. These data suggest that variation of one capsid gene not only affects its own structure and antigenicity, but also affects the structure and antigenicity of the other capsid protein. Implications of our data suggest for the continued effectiveness of a vaccine, variation in both capsid proteins need to be considered and not just the protein the vaccine is directed against.
INTRODUCTION
Human papillomavirus (HPV) is the primary etiologic agent of cervical cancer. The viral capsid is approximately 50 to 55 nm in diameter, and has an icosahedral symmetry of T = 7 (de Villiers et al., 2004). The viral particle contains 360 copies of the major capsid protein L1, organized into 72 pentameric capsomeres. The atomic structure of a small, T=1 HPV16 L1 virus-like particle (VLPs) has been determined (Chen et al., 2000). Because VLPs, pseudoviruses (PsV) and quasiviruses (QV) are structurally similar to native virus, they have been used as surrogates for native virus in investigating the viral life cycle, structure, and host immunity. Recombinant-derived particles formed in monolayer cultures lack differentiation-dependent temporally correct and regulated capsid protein interactions. Therefore, these particles may or may not be accurate surrogates for the native virus. The organotypic (raft) tissue culture system is still the only in vitro method proven to reproducibly mimic epithelial differentiation to the extent that the full HPV life cycle can be studied and wherein infectious virions can be obtained from stratifying tissue in a differentiation-dependent culture system (Meyers et al., 1992; Ozbun, 2002a; Ozbun, 2002b; Song et al., 2010).
Recent publications have demonstrated that the native virus replicated in stratifying and differentiating host tissue differs in significant aspects from particles made using recombinant particles (Conway et al., 2009a; Conway et al., 2009b). For example, when the two N-terminal conserved HPV16 L2 cysteines were mutated in PsV or QV, the particles produced were non infectious (Campos and Ozbun, 2009; Gambhira et al., 2009). Moreover, when the same mutations were tested in native viral particles produced in stratifying and differentiating human epithelium, not only were the mutant viruses infectious but their titers were dramatically increased in some cases (Conway et al., 2009a). Additionally, the maturation time required for monolayer-culture derived PsV HPV16 is 24 h (Buck et al., 2005), but 20 days is required for maturation of differentiation-dependent grown native virus (Conway et al., 2009b). These brief examples suggest that the genetics and biochemistry of viral synthesis differ in recombinant particles formed within undifferentiating monolayer culture versus native virus formed within differentiating host epithelia.
The HPV capsid genes have high sequence homology. A recent manuscript described a series of intra- and/or inter-species cross-reactive epitopes suggesting that cross-reactivity only loosely follows phylogenetic relationships that are based on capsid gene sequence homology (Rizk et al., 2008). An important question is whether or not the sequence homology equates to similar requirements for virion assembly and maturation. One way to test this is to use HPV mutant constructs containing chimeric capsid genes. Recently, to test the relationship between sequence homology and virion morphogenesis we constructed a panel of mutant viruses containing wild-type and chimeric HPV16 and HPV18 capsid proteins (Chen et al., 2010). While aspects of their life cycles such as protein expression, genome amplification, genome episomal maintenance did not appear to be affected by the chimeric capsid proteins, the chimeric viruses showed variation in their viral titers. Due to the reduction of titers of some chimeric viruses, we hypothesized that the capsid proteins could mutually affect each other's structure-function in the viral particle, therefore affecting infectivity.
Using VLPs, type-specific, conformation-dependent neutralizing antibodies have been generated for neutralization and capsid structural studies (Bishop et al., 2007; Christensen et al., 2001; Christensen et al., 1996a; Culp et al., 2007; Rizk et al., 2008). Most L1 conformation-dependent MAbs are able to bind L1 VLPs as well as L1/L2 VLPs and virions. These data led to the idea that L2 has little impact on the conformation of L1 within the VLPs and virions. Studying the inhibition of PsV infection with a panel of polyclonal antibodies raised from HPV16 L2 peptides, potential neutralizing L2 sequences exposed on the capsid surface have been mapped (Kawana et al., 2001; Pastrana et al., 2005). To test our hypothesis that L1 and L2 can mutually affect each other's structure, we used a panel of conformation-dependent neutralizing L1 MAbs and a L2 capsid surface peptide derived neutralizing MAb and tested their abilities to neutralize infection by HPV18/HPV16 chimeric capsid protein mutant viruses. We observed with our intertypical chimeric capsid mutant viruses that L2 can affect the structure of L1 and that L1 can affect the structure of L2 in the native virus.
MATERIALS AND METHODS
Chimeric HPV Genomes and organotypic raft cultures
Chimeric mutant HPV genomes were described previously (Chen et al., 2010). Organotypic raft cultures were grown as previously described (Meyers et al., 2002).
Preparation of virus stocks
Virus stocks of each HPV chimera were prepared by peeling the epithelial tissue away from the collagen of three organotypic rafts. The peeled epithelial tissues were homogenized in 0.6 ml of ice-cold 1 M NaCl/0.05 M Na-Phosphate Buffer with a 7.5 ml homogenizer. The homogenizer was washed twice with 200 μl 1 M NaCl/0.05 M Na-Phosphate Buffer, pH 8. The homogenized viral solution was centrifuged at 10.5k for 10 min at 4 °C and the supernatant transferred to a 1.8 ml Nalgene cryovial. Each virus stock was made from three raft culture tissues and all stocks were stored at −20 °C.
HPV infection and Neutralization Assays
The infectivity measurement of chimeric HPV used the Limited Dilution RT-PCR titering assay as described previously (Alam et al., 2008; Conway et al., 2009a; Conway et al., 2009b; McLaughlin-Drubin, Christensen, and Meyers, 2004; McLaughlin-Drubin and Meyers, 2004; McLaughlin-Drubin and Meyers, 2005; McLaughlin-Drubin et al., 2003; Meyers et al., 2002; Meyers, Mayer, and Ozbun, 1997) using HaCaT cells, an immortalized human keratinocyte cell line (kindly provided by Norbert Fusenig, German Cancer Research Center). The virus titer was determined to be the last dilution at which the viral E1^E4 spliced transcript could be detected.
Neutralization assays were performed by infecting HaCaT cells with a 1:20 diluted viral sample that had been preincubated with either conformation-dependent neutralizing L1 monoclonal antibodies (MAbs) or the L2 surface peptide derived MAb diluted 1:20 in HaCaT culture medium as previously described (Chen et al., 2010; Conway et al., 2009a; Conway et al., 2009b; McLaughlin-Drubin, Christensen, and Meyers, 2004; McLaughlin-Drubin and Meyers, 2005; McLaughlin-Drubin et al., 2003). HPV18 and HPV16 L1 conformation-dependent, neutralizing MAbs H18.J4, H18.K2, H16.V5, H16.7E, H16.9A and H16.J4 were used (Bishop et al., 2007; Christensen et al., 2001; Christensen et al., 1996a; Culp et al., 2007; Rizk et al., 2008). In addition, a HPV16 L2-reactive monoclonal antibody RG-1 was used (Gambhira et al., 2007a; Pastrana et al., 2005). Following preincubation, HaCaT cells were incubated with the virus–antibody mixture for 2 days. Total RNA was then extracted and RT-PCR was performed as described (Chen et al., 2010; Conway et al., 2009b).
RESULTS
Using the HPV18 genome as the backbone, we recently showed that genomes containing the intertypical exchange of HPV18 L1 with the HPV16 L1 efficiently replicated and produced infectious virus, however, genomes containing an intertypical exchange of HPV18 L2 for the HPV16 L2 failed to produce detectable infectious virus (Chen et al., 2010). Expanding on this observation we studied a panel of eight chimeric constructs of individual capsid proteins. Seven of these mutants produced detectable infectious virus, but titers varied as much as 200-fold between the mutants. Importantly, one mutant did not produce detectable infectious virus (Chen et al., 2010). From these studies we identified a type-specific domain at the N-terminus of the L1 capsid protein of HPV18, which interferes with its ability to cooperate with the HPV16 L2 protein to form infectious viral particles. Deletion of this domain led to the cooperation of the HPV18 L1 protein and HPV 16 L2 protein as measured by the production of infectious progeny. However, this N-terminal sequence deletion of HPV18 L1 induced a change in the conformational structure as determined by the loss of neutralization with HPV18 L1 conformation-dependent neutralizing antibody (Chen et al., 2010). Based on the observed results, we hypothesized that the differences we see in the viral titers of the eight chimeric capsid mutants was related to changes in structure imposed by one capsid protein on the other capsid protein.
Changes in L1 structure imposed by chimeric L2 proteins as determined by neutralizing MAbs
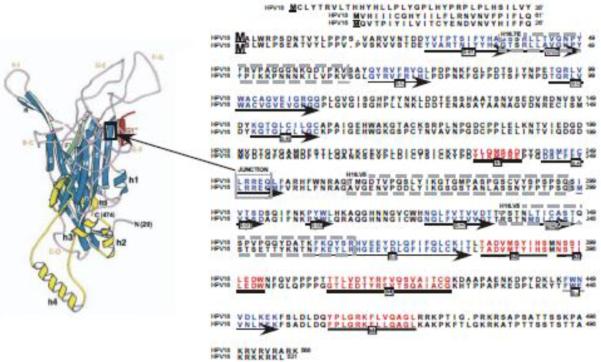
To test our hypothesis that L2 structural alterations can affect the conformation of L1, viral stocks were prepared by growing organotypic cultures of human keratinocyte cell lines infected with wild-type HPV16, HPV18, and with each of the eight chimeric mutant viruses (Fig. 1) (Chen et al., 2010). The chimeric viruses were made using HPV18 as the backbone and exchanging half of L2 or L1 with the corresponding half from HPV16 (Fig. 1) (Chen et al., 2010). A homologous sequence in the middle of the L2 and L1 proteins of HPV18 and HPV16 was used for the chimeric junction, allowing for the maintenance of sequence and structure around the junction sites. For L1 this region is in the C-terminal portion of beta sheet F (Fig. 2). Determining the structure of L2 has been difficult (Chen et al., 2010) therefore we chose a homologous sequence in the center of the protein to avoid reported functional domains (Becker et al., 2003; Bossis et al., 2005; Da Silva et al., 2001; Day and Schiller, 2006; Finnen et al., 2003; Florin et al., 2004; Florin et al., 2002; Giroglou et al., 2001; Graham, 2006; Heino et al., 2000; McMillan et al., 1999; Okun et al., 2001; Roden et al., 1994; Yang et al., 2003). Multiple stocks of each mutant virus were prepared, infectivity tested using a previously described Limited Dilution RT-PCR titering assay (Alam et al., 2008; Conway et al., 2009a; Conway et al., 2009b; McLaughlin-Drubin, Christensen, and Meyers, 2004; McLaughlin-Drubin and Meyers, 2004; McLaughlin-Drubin and Meyers, 2005; McLaughlin-Drubin et al., 2003; Meyers et al., 2002; Meyers, Mayer, and Ozbun, 1997), and reproducible titers were observed for independently derived stocks of each virus (Table 1).
Figure 1.
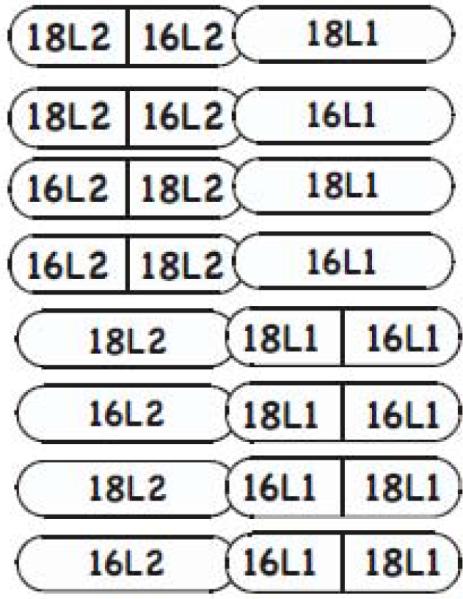
L2 and L1 half and half chimeric mutants (Chen et al., 2010).
Figure 2.
Position of the Chimeric Junction, HPV16 L1-Reactive Conformation-Dependent MAb Epitopes, Structural Motifs and Sequence Homology of HPV18 and HPV16 L1 Proteins. The L1 protein amino acids sequences for HPV18 and HPV16 are aligned. Sequence numbering begins with the `consensus' methionines used for producing VLPs, PsV, and QV (Chen et al., 2010). The extended N-terminal sequences present in native viruses are given prime numbers, 1'–61' for HPV18 and 1'–26' for HPV16. Structural motifs are marked underneath the appropriate sequences (Chen et al., 2000) by arrows for ß-sheets and bolden lines for helices. HPV16 L1-reactive conformation-dependent MAb epitopes (Christensen et al., 2001; Culp et al., 2007) are outlined by dashed line boxes. The chimeric junction, LRREQ amino acids 250–254 is outlined by a solid box.
TABLE 1.
| HPV CHIMERA | Titer Ave/SDa | H18.J4 | H18.K2 | H16.7E | H16.V5 |
|---|---|---|---|---|---|
| Wt HPV18 | 8,125/1157 | + b | + | − | − |
| Wt HPV16 | 10,000/0 | − c | − | + | + |
| HPV18-L2(18)L1(16) | 10,000/0 | − | − | + | + |
| HPV18-L2(16)L1(18) | 1/4.5 | + | + | − | − |
| HPV18-L2(18)L1(16/18) | 20/0 | − | − | + | − |
| HPV18-L2(18)L1(18/16) | 5,000/0 | + | − | − | + |
| HPV18-L2(16)L1(16/18) | 1,000/0 | − | − | + | − |
| HPV18-L2(16)L1(18/16) | 0/0 | NDd | ND | ND | ND |
| HPV18-L2(16/18)L1(18) | 100/0 | + | − | − | − |
| HPV18-L2(18/16)L1(18) | 20/45 | − | − | − | − |
| HPV18-L2(16/18)L1(16) | 100/0 | − | − | − | + |
| HPV18-L2(18/16)L1(16) | 73/46 | − | − | + | − |
Average titer/standard deviation of at least three separate virus stock preparations.
MAb was able to neutralize infection
MAb was not able to neutralize infection
Not done, no infectious virus
We predicted that the variations observed in the viral titers of raft tissues infected with one of the eight chimeric HPVs was due to a change in structure of the wild-type capsid protein induced by the chimeric capsid protein. To test our prediction we used conformation-dependent L1-reactive monoclonal antibodies (MAbs) in the Limited Dilution RT-PCR assays. To test whether the capsid structure had been affected, HPV16 and HPV18 L1-reactive conformation-dependent antibodies H16.7E, H16.V5, H18.J4, and H18.K2 were tested for their ability to neutralize infection by HPV18-L2(18)L1(16/18), HPV18-L2(18)L1(18/16), HPV18-L2(16)L1(16/18), HPV18-L2(16/18)L1(18), HPV18-L2(18/16)L1(18), HPV18-L2(16/18)L1(16), and HPV18-L2(18/16)L1(16) (Fig. 1). The conformation-dependent binding and neutralizing activity of all four MAbs has been previously characterized (Bishop et al., 2007; Chen et al., 2010; Christensen et al., 2001; Christensen et al., 1996b; Culp et al., 2007; Rizk et al., 2008). HPV16 L1 conformation-dependent neutralizing MAb H16.7E binds to the BC capsid loop (Christensen et al., 2001; Christensen et al., 1996b; Culp et al., 2007) and H16.V5 binds to the FG loop and to a lesser extent the HI loop (Christensen et al., 2001; Christensen et al., 1996b). In our chimeric virus the epitope for H16.7E would be present in the N-terminal half of HPV16 L1, whereas the epitope for H16.V5 would be present in the C-terminal half of HPV16 L1 (Fig. 2). In contrast to HPV16 L1-reactive MAbs, the binding regions for HPV18 L1-reactive MAbs H18.J4 and H18.K2 have not yet been mapped.
The titers for wild-type HPV18 and HPV16 were reproducibly high, measuring 7,500 and 10,000, respectively (Table 1). Type-specific, conformation-dependent MAbs H18.J4 and H18.K2 neutralized infection by wild-type HPV18. Likewise infection by wild-type HPV16 was neutralized by type-specific, conformation-dependent MAbs H16.7E and H16.V5 (Table 1 and Fig. 2). Therefore, all four MAbs were type-specific in their ability to neutralize infection. We then proceeded to perform neutralization assays on the viral stocks of each chimeric virus, with the exception of HPV18-L2(16)|L1(18/16), which never produced a stock of detectable infectious virus (Table 1). Of the other three chimeras that contained a wild-type L2 and a chimeric L1 protein, HPV18-L2(18)L1(16/18), HPV18-L2(18)L1(18/16), and HPV18-L2(16)L1(16/18) had average titers of 20, 5,000, and 1,000, respectively (Chen et al., 2010). As expected MAb H16.7E was able to neutralize HPV18-L2(18)L1(16/18) and HPV18-L2(16)L1(16/18) both of which contain the N-terminal half of the HPV16 L1 protein containing the H16.7E epitope. Both of these chimeric viruses contained the C-terminal half of the HPV18 L1 and as we expected, their infectivity was not neutralized by type-specific H16.V5 (Table 1). Conversely, the chimera HPV18-L2(18)L1(18/16) is just the opposite having only the C-terminal half of the HPV16 L1 protein. Since the type-specific H16.V5 has its epitope on the C-terminal half of L1, it could neutralize infection by this mutant virus (Table 1). As expected, H16.7E did not neutralize HPV18-L2(18)L1(18/16). Of these three chimeric viruses only infection by HPV18-L2(18)L1(18/16) was neutralized by MAb H18.J4. H18.J4 did not neutralize HPV18-L2(18)L1(16/18) and HPV18L2(16)L1(16/18) suggesting that the H18.J4 epitope lay on the N-terminal half of the HPV18 L1 protein. Interestingly, MAb H18.K2 had no effect on the infectivity of any of these three chimeric viruses. This would suggest that either the epitope was around the junction and subtle conformational differences were enough to prevent its binding, or that the L1 protein differed enough in its overall structure as a result of the chimeric structure to prevent binding.
Next we evaluated the neutralization activities of H16.7E, H16.V5, H18.J4, and H18.K2 on infectivity by the mutant viruses containing a wild-type L1 protein and a chimeric L2 protein. All the neutralization assays were done using a 1:20 dilution of the viral stock preincubated with a type-specific MAb and infectivity measured using the Limited Dilution RT-PCR titering assay. This means that a much lower amount of infectious chimeric virus was used per neutralization than used with the wild type virus, so it would be expected that the MAbs would neutralize the chimeric stocks. Based on their ability to neutralize infection by the high titer wild-type HPV16 and HPV18 virus stocks, we expected that the type-specific L1-reactive conformation-dependent MAbs would neutralize infection of viruses containing wild-type L1 proteins. H16.7E and H16.V5 were incapable of neutralizing infection by viruses containing a wild-type HPV18 L1. Similarly H18.J4 and HPV18.K2 were incapable of neutralizing infection by viruses containing a wild-type HPV16 L1 (Table 1).
When H16.7E and H16.V5 were tested with HPV18-L2(16/18)L1(16) only H16.V5 was able to neutralize infection. However, when H16.7E and H16.V5 were tested with HPV18-L2(18/16)L1(16) only H16.7E was able to neutralize infection. This was unexpected since both viruses have a wild-type HPV16 L1 protein. These results suggest that the chimeric L2 impacted the structure of L1 inducing a loss of function in the neutralization assay. When H18.J4 and H18.K2 were tested with HPV18-L2(16/18)L1(18) and HPV18-L2(18/16)L1(18) a similar result was observed, except that H18.J4 was only able to neutralize infection by HPV18-L2(16/18)L1(18) and neither HPV18-L2(16/18)L1(18) or HPV18-L2(18/16)L1(18) were neutralized by H18.K2 (Table 1). Again this suggests that the chimeric L2 impacted the structure of L1 inducing a loss of function phenotype in the infectivity neutralization assay.
To further support this observation, we used two additional conformation-dependent HPV16 L1-reactive MAbs, H16.J4 and H16.9A. These two MAbs were made using L1-only VLPs (Culp et al., 2007). When these MAbs were initially characterized it was found that while they bound efficiently to L1 only VLPs, binding was significantly decreased when VLPs containing L1 and L2 were used (Culp et al., 2007). In line with this observation neither H16.J4 nor H16.9A was able to neutralize infection by wild-type HPV16 (Table 2). Surprisingly, when tested on infectivity by HPV18-L2(16/18)L1(16) and HPV18-L2(18/16)L1(16), both MAbs H16.J4 and H16.9A neutralized infection. Again this suggests that the chimeric L2 protein impacts the structure of the wild-type L1 protein, but in this case inducing a gain of function phenotype in the infectivity neutralization assay.
TABLE 2.
MAb was not able to neutralize infection
MAb was able to neutralize infection
Changes in L2 structure imposed by chimeric L1 proteins as determined by a neutralizing MAb
After observing that a chimeric L2 protein could impact the structure of a wild-type L1 protein in HPV infectious particles we next were interested in testing whether a chimeric L1 protein could change the structure of the L2 protein within the virus. For these assays, we used the L2-reactive MAb RG-1 (Gambhira et al., 2007a; Pastrana et al., 2005). RG-1 was raised against the 17 to 36 amino acid peptide of HPV16 L2. This region shows high sequence homology among high-risk HPVs (Pastrana et al., 2005). RG-1 has also been shown to cross-react with HPV types other than HPV16 (Gambhira et al., 2007b; Kawana et al., 1999). Infectivity of both wild-type HPV16 and HPV18 were neutralized by RG-1 (Table 3). Infectivity of all four mutant viruses containing a chimeric L2 protein, HPV18-L2(16/18)L1(18), HPV18-L2(18/16)L1(18), HPV18-L2(16/18)L1(16), and HPV18-L2(18/16)L1(16) were neutralized by RG-1 (Table 3). This was expected since RG-1 could neutralize both wild-type HPV16 and HPV18 infection. However, when RG-1 was tested for its ability to neutralize infection of the mutant viruses containing a wild-type L2 protein and a chimeric L1 the results were mixed. RG-1 was effective in neutralizing infection by HPV18-L2(18)L1(16/18) and HPV18-L2(18)L1(18/16), but not infection by HPV18-L2(16)L1(16/18) (Table 3). Similar to what was observed with L1-reactive MAbs the results obtained with the L2-reactive MAb suggests that chimeric L1 impacts the structure of wild-type L2 in the native viral particle. The mutant HPV18-L2(16)L1(18/16) never produced infectious stocks of virus so it could not be tested in the neutralization assays.
TABLE 3.
| HPV CHIMERA | RG-1 |
|---|---|
| Wt HPV18 | + a |
| Wt HPV16 | + |
| HPV18-L2(18)L1(16/18) | − b |
| HPV18-L2(18)L1(18/16) | − |
| HPV18-L2(16)L1(16/18) | + |
| HPV18-L2(16)L1(18/16) | NDc |
| HPV18-L2(16/18)L1(18) | + |
| HPV18-L2(18/16)L1(18) | + |
| HPV18-L2(16/18)L1(16) | + |
| HPV18-L2(18/16)L1(16) | + |
MAb was able to neutralize infection
MAb was not able to neutralize infection
Not done, no infectious virus
DISCUSSION
VLPs comprised of full-length HPV16 L1 have not been crystallized. A deletion of ten amino acids from the `consensus' methionine was necessary to assemble VLPs that were competent for crystallographic analysis, resulting in a T=1 icosahedral lattice made from 12 L1 pentamers, opposed to the native T=7 icosahedral lattice made from 72 L1 pentamers (Chen et al., 2000). This structure was quite an accomplishment and allowed the field to make further hypotheses regarding the structure of HPV. However, the small size of the T=1 particle and lack of disulfide bonds suggests that the model may not be representative of all detailed temporal interactions that occur between and within the L1 pentamers of native virions during morphogenesis in stratifying and differentiating epithelial tissue. The recently published high resolution bovine papillomavirus type 1 (BPV-1) cryo-electron microscopy (cryo-EM) structure does provide more insight into intra- and inter-pentameric L1 interactions, but provides no information on interactions with the L2 capsid protein (Wolf et al., 2010).
HPV L1 pentamers are thought to form a network of intra- and interpentameric disulfide bonds to stabilize the capsid (Buck et al., 2005; Fligge et al., 2001; Kondo et al., 2007; Sapp et al., 1998). The cryo-EM structure of BPV-1 has a interpentameric L1 disulfide bond between two cysteine residues that are highly conserved among papillomaviruses, including HPVs (Wolf et al., 2010). BPV-1 also contains a intrapentameric disulfide bond between two cysteine residues that are not present in HPVs (Wolf et al., 2010).
L1 is sufficient to produce monolayer culture-derived particles; however, L2 has been shown to affect the final structure of the virion, in addition to enhancing infectivity and DNA encapsidation (Holmgren et al., 2005; Kirnbauer et al., 1992; Kondo et al., 2007). Papillomavirus capsids also contain an unknown amount of the minor capsid protein L2. Early cryo-EM studies of native BPV1 virus suggested that one L2 protein occludes the center of each pentavalent capsomere, totaling 12 L2 proteins per virion (Trus et al., 1997). This L1:L2 ratio of 30:1 was supported by SDS-PAGE analyses of L1/L2 VLPs (Bossis et al., 2005; Hagensee, Yaegashi, and Galloway, 1993; Kirnbauer et al., 1993; Volpers et al., 1994), however SDS-PAGE analysis of native HPV11 virions, biochemical analyses of HPV11 L1/L2 proteins, and cryo-EM images of HPV16 PsV suggest that papillomavirus particles can contain 36 and as much as 72 L2 proteins per particle (Buck et al., 2008; Doorbar and Gallimore, 1987; Finnen et al., 2003). However, recent high-resolution cryo-EM image reconstructions of native BPV failed to detect L2 protein density, suggesting that L2 signal may be averaged out of the reconstruction and that it is not maximally packaged into native virions or may be too mobile to visualize (Buck et al., 2008; Wolf et al., 2010).
Also, within the virion is a histone-associated, circular viral genome, which has been shown to alter the final structure of the virion (Fligge et al., 2001; Sapp et al., 1998). In addition to L2 and viral genomes it is also probable that unknown viral and/or cellular proteins/factors such as molecular chaperones and karyopherins may exist which influence the final structure of native virions (Bird et al., 2008; Chromy, Pipas, and Garcea, 2003). Our laboratory has recently demonstrated the presence of two different species of the HPV16 L1 protein in native virions that is not present in quasivirus particles (Conway et al., 2009b). The effect if any of having two species of the L1 protein on viral structure is unknown at this time, however it appears that native virion with both species of L1 localize to the gradient fractions exhibiting the highest concentration of mature infectious virions. In addition, virions isolated from these fractions exhibit a greater structural stability when compared to `mature' QV (Conway et al., 2009b).
L2 appears to interact with L1 capsomeres and not with preformed L1 VLPs, suggesting co-assembly of L1 and L2 into a virus particle (Finnen et al., 2003). Since L2 enhances assembly of L1 capsomeres in the absence of disulfide bonding, hydrophobic interactions between L2 and L1 are likely to initiate during early assembly events (Ishii et al., 2005). During differentiation co-assembly would require a regulated process mediated by interactions between the two proteins. Future investigations likely will discover other viral and/or cellular factors that are necessary for the proper assembly of virions associated with regulation of redox states within the cell, proper cellular localization and initial interaction of capsid proteins, the correct formation and regulation of disulfide bonds, and regulation of capsid protein expression. In vitro, the only system that can mimic all these interactions and mechanisms is the human epithelial organotypic culture allowing for complete stratification and differentiation of the natural host tissue.
Our laboratory's previous studies showed that while the HPV16 L1 protein could cooperate with the HPV18 L2 protein to produce infectious virus, the HPV18 L1 protein could not cooperate with the HPV16 L2 protein thereby failing to produce infectious virus. A region at the N-terminus of the HPV18 L1 protein was shown to be responsible for the inability to cooperate with the HPV16 L2 protein. Removal of this N-terminal region allowed for HPV18 L1 and HPV16 L2 proteins produce wild-type levels of infectious virus and these chimeric viruses exhibit subtle structural changes as measured by MAb neutralization activity (Chen et al., 2010). A panel of mutant viruses containing wild-type and chimeric HPV18 and HPV16 capsid proteins exhibited variation in their viral titers, while measurements of different aspects of their life cycles did not provide an answer to the variation in titers (Chen et al., 2010). We believed that due to the chimeric nature of one of the capsid proteins, subtle changes in the structure of the other capsid protein had occurred. Another possibility for the L2 chimerics is that they are able to occlude the binding sites for the L1 MAbs, however we feel this possibility is less likely. We measured the presence of these changes by their effect on the neutralization activity of MAbs. Our results strongly suggest that the structure of one capsid protein affected the structure of the other capsid protein.
In our study we hypothesized that the capsid proteins could mutually affect each other's structure in the viral particle and therefore affect titers. Our results supported this hypothesis showing not only a loss of neutralization (loss of function) but also gain of neutralization (gain of function) affected by the presence of a mutant L2 capsid protein. Similarly, the presence of a mutant L1 protein in the assembled viral particles affected the ability of a L2-reactive MAb to neutralize infection. The MAbs used in this study probably all react with surface exposed epitopes on the capsid. It is possible that a disturbance in structure equivalent to what is present in the chimeric viruses might be too defective to maintain themselves in a vaccinated population. Continued monitoring vaccinated populations for viral infection is warranted to identify increases in infection by variants and non vaccine types, but also to identify waning of the vaccinated host to block infection. Our data suggests that it is not only mutations in the particular vaccine directed capsid protein L1 or L2 that may alter the effectiveness of the vaccine, but mutations in the capsid protein that is not part of the vaccine could also alter effectiveness.
ACKNOWLEDGEMENTS
We thank Lynn Budgeon for histological expertise; and the members of the Meyers' laboratory for their critical reading of the manuscript and many helpful suggestions. This study was supported by a PHS grant from the National Institute of Allergy and Infectious Disease (R01AI57988).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alam S, Conway MJ, Chen HS, Meyers C. The cigarette smoke carcinogen benzo[a]pyrene enhances human papillomavirus synthesis. J Virol. 2008;82(2):1053–8. doi: 10.1128/JVI.01813-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird G, O'Donnell M, Moroianu J, Garcea RL. Possible role for cellular karyopherins in regulating polyomavirus and papillomavirus capsid assembly. J Virol. 2008;82(20):9848–57. doi: 10.1128/JVI.01221-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop B, Dasgupta J, Klein M, Garcea R, Christensen N, Zhao R, Chen X. Crystal structures of four types of human papillomavirus L1 capsid proteins: understanding the specificity of neutralizing monoclonal antibodies. Journal of Biological Chemistry. 2007;282(43):31803–31811. doi: 10.1074/jbc.M706380200. [DOI] [PubMed] [Google Scholar]
- Bossis I, Roden RB, Gambhira R, Yang R, Tagaya M, Howley PM, Meneses PI. Interaction of tSNARE syntaxin 18 with the papillomavirus minor capsid protein mediates infection. J Virol. 2005;79(11):6723–31. doi: 10.1128/JVI.79.11.6723-6731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck CB, Cheng N, Thompson CD, Lowy DR, Steven AC, Schiller JT, Trus BL. Arrangement of L2 within the papillomavirus capsid. J Virol. 2008;82(11):5190–7. doi: 10.1128/JVI.02726-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck CB, Thompson CD, Pang YY, Lowy DR, Schiller JT. Maturation of papillomavirus capsids. J Virol. 2005;79(5):2839–46. doi: 10.1128/JVI.79.5.2839-2846.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos SK, Ozbun MA. Two highly conserved cysteine residues in HPV16 L2 form an intramolecular disulfide bond and are critical for infectivity in human keratinocytes. PLoS One. 2009;4(2):e4463. doi: 10.1371/journal.pone.0004463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HS, Bromberg-White J, Conway MJ, Alam S, Meyers C. Study of infectious virus production from HPV18/16 capsid chimeras. Virology. 2010;405(2):289–99. doi: 10.1016/j.virol.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XS, Garcea RL, Goldberg I, Casini G, Harrison SC. Structure of small virus-like particles assembled from the L1 protein of human papillomavirus 16. Mol Cell. 2000;5(3):557–67. doi: 10.1016/s1097-2765(00)80449-9. [DOI] [PubMed] [Google Scholar]
- Christensen ND, Cladel NM, Reed CA, Budgeon LR, Embers ME, Skulsky DM, McClements WL, Ludmerer SW, Jansen KU. Hybrid papillomavirus L1 molecules assemble into virus-like particles that reconstitute conformational epitopes and induce neutralizing antibodies to distinct HPV types. Virology. 2001;291(2):324–34. doi: 10.1006/viro.2001.1220. [DOI] [PubMed] [Google Scholar]
- Christensen ND, Dillner J, Eklund C, Carter JJ, Wipf GC, Reed CA, Cladel NM, Galloway DA. Surface conformational and linear epitopes on HPV-16 and HPV-18 L1 virus-like particles as defined by monoclonal antibodies. Virology. 1996a;223(1):174–84. doi: 10.1006/viro.1996.0466. [DOI] [PubMed] [Google Scholar]
- Christensen ND, Dillner J, Eklund C, Carter JJ, Wipf GC, Reed CA, Cladel NM, Galloway DA. Surface conformational and linear epitopes on HPV-16 and HPV-18 L1 virus-like particles as defined by monoclonal antibodies. Virology. 1996b;223:174–184. doi: 10.1006/viro.1996.0466. [DOI] [PubMed] [Google Scholar]
- Chromy LR, Pipas JM, Garcea RL. Chaperone-mediated in vitro assembly of Polyomavirus capsids. Proc Natl Acad Sci U S A. 2003;100(18):10477–82. doi: 10.1073/pnas.1832245100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway MJ, Alam S, Christensen ND, Meyers C. Overlapping and independent structural roles for human papillomavirus type 16 L2 conserved cysteines. Virology. 2009a;393(2):295–303. doi: 10.1016/j.virol.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway MJ, Alam S, Ryndock EJ, Cruz L, Christensen ND, Roden RB, Meyers C. Tissue-spanning redox gradient-dependent assembly of native human papillomavirus type 16 virions. J Virol. 2009b;83(20):10515–26. doi: 10.1128/JVI.00731-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culp TD, Spatz CM, Reed CA, Christensen ND. Binding and neutralization efficiencies of monoclonal antibodies, Fab fragments, and scFv specific for L1 epitopes on the capsid of infectious HPV particles. Virology. 2007;361(2):435–46. doi: 10.1016/j.virol.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. Classification of papillomaviruses. Virology. 2004;324(1):17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Doorbar J, Gallimore PH. Identification of proteins encoded by the L1 and L2 open reading frames of human papillomavirus 1a. J Virol. 1987;61(9):2793–9. doi: 10.1128/jvi.61.9.2793-2799.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnen RL, Erickson KD, Chen XS, Garcea RL. Interactions between papillomavirus L1 and L2 capsid proteins. J Virol. 2003;77(8):4818–26. doi: 10.1128/JVI.77.8.4818-4826.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fligge C, Schafer F, Selinka HC, Sapp C, Sapp M. DNA-induced structural changes in the papillomavirus capsid. J Virol. 2001;75(16):7727–31. doi: 10.1128/JVI.75.16.7727-7731.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambhira R, Jagu S, Karanam B, Day PM, Roden R. Role of L2 cysteines in papillomavirus infection and neutralization. Virol J. 2009;6:176. doi: 10.1186/1743-422X-6-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambhira R, Jagu S, Karanam B, Gravitt PE, Culp TD, Christensen ND, Roden RB. Protection of Rabbits against Challenge with Rabbit Papillomaviruses by Immunization with the N Terminus of Human Papillomavirus Type 16 Minor Capsid Antigen L2. J Virol. 2007a;81(21):11585–92. doi: 10.1128/JVI.01577-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambhira R, Karanam B, Jagu S, Roberts JN, Buck CB, Bossis I, Alphs H, Culp T, Christensen ND, Roden RB. A protective and broadly cross-neutralizing epitope of Human Papillomavirus L2. J Virol. 2007b doi: 10.1128/JVI.00936-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagensee ME, Yaegashi N, Galloway DA. Self-assembly of human papillomavirus type 1 capsids by expression of the L1 protein alone or by coexpression of the L1 and L2 capsid proteins. J Virol. 1993;67(1):315–22. doi: 10.1128/jvi.67.1.315-322.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren SC, Patterson NA, Ozbun MA, Lambert PF. The minor capsid protein L2 contributes to two steps in the human papillomavirus type 31 life cycle. J Virol. 2005;79(7):3938–48. doi: 10.1128/JVI.79.7.3938-3948.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii Y, Ozaki S, Tanaka K, Kanda T. Human papillomavirus 16 minor capsid protein L2 helps capsomeres assemble independently of intercapsomeric disulfide bonding. Virus Genes. 2005;31(3):321–8. doi: 10.1007/s11262-005-3250-3. [DOI] [PubMed] [Google Scholar]
- Kawana K, Yoshikawa H, Taketani Y, Yoshiike K, Kanda T. Common neutralization epitope in minor capsid protein L2 of human papillomavirus types 16 and 6. J Virol. 1999;73(7):6188–90. doi: 10.1128/jvi.73.7.6188-6190.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawana Y, Kawana K, Yoshikawa H, Taketani Y, Yoshiike K, Kanda T. Human papillomavirus type 16 minor capsid protein l2 N-terminal region containing a common neutralization epitope binds to the cell surface and enters the cytoplasm. J Virol. 2001;75(5):2331–6. doi: 10.1128/JVI.75.5.2331-2336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirnbauer R, Booy F, Cheng N, Lowy DR, Schiller JT. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc Natl Acad Sci U S A. 1992;89(24):12180–4. doi: 10.1073/pnas.89.24.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirnbauer R, Taub J, Greenstone H, Roden R, Durst M, Gissmann L, Lowy DR, Schiller JT. Efficient self-assembly of human papillomavirus type 16 L1 and L1–L2 into virus-like particles. J Virol. 1993;67(12):6929–36. doi: 10.1128/jvi.67.12.6929-6936.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo K, Ishii Y, Ochi H, Matsumoto T, Yoshikawa H, Kanda T. Neutralization of HPV16, 18, 31, and 58 pseudovirions with antisera induced by immunizing rabbits with synthetic peptides representing segments of the HPV16 minor capsid protein L2 surface region. Virology. 2007;358(2):266–72. doi: 10.1016/j.virol.2006.08.037. [DOI] [PubMed] [Google Scholar]
- McLaughlin-Drubin ME, Christensen ND, Meyers C. Propagation, infection, and neutralization of authentic HPV16 virus. Virology. 2004;322(2):213–9. doi: 10.1016/j.virol.2004.02.011. [DOI] [PubMed] [Google Scholar]
- McLaughlin-Drubin ME, Meyers C. Evidence for the coexistence of two genital HPV types within the same host cell in vitro. Virology. 2004;321(2):173–80. doi: 10.1016/j.virol.2003.12.019. [DOI] [PubMed] [Google Scholar]
- McLaughlin-Drubin ME, Meyers C. Propagation of infectious, high-risk HPV in organotypic “raft” culture. Methods Mol Med. 2005;119:171–86. doi: 10.1385/1-59259-982-6:171. [DOI] [PubMed] [Google Scholar]
- McLaughlin-Drubin ME, Wilson S, Mullikin B, Suzich J, Meyers C. Human papillomavirus type 45 propagation, infection, and neutralization. Virology. 2003;312(1):1–7. doi: 10.1016/s0042-6822(03)00312-x. [DOI] [PubMed] [Google Scholar]
- Meyers C, Bromberg-White JL, Zhang J, Kaupas ME, Bryan JT, Lowe RS, Jansen KU. Infectious virions produced from a human papillomavirus type 18/16 genomic DNA chimera. J Virol. 2002;76(10):4723–33. doi: 10.1128/JVI.76.10.4723-4733.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers C, Frattini MG, Hudson JB, Laimins LA. Biosynthesis of human papillomavirus from a continuous cell line upon epithelial differentiation. Science. 1992;257(5072):971–3. doi: 10.1126/science.1323879. [DOI] [PubMed] [Google Scholar]
- Meyers C, Mayer TJ, Ozbun MA. Synthesis of infectious human papillomavirus type 18 in differentiating epithelium transfected with viral DNA. J Virol. 1997;71(10):7381–6. doi: 10.1128/jvi.71.10.7381-7386.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbun MA. Human papillomavirus type 31b infection of human keratinocytes and the onset of early transcription. J Virol. 2002a;76(22):11291–300. doi: 10.1128/JVI.76.22.11291-11300.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbun MA. Infectious human papillomavirus type 31b: purification and infection of an immortalized human keratinocyte cell line. J Gen Virol. 2002b;83(Pt 11):2753–63. doi: 10.1099/0022-1317-83-11-2753. [DOI] [PubMed] [Google Scholar]
- Pastrana DV, Gambhira R, Buck CB, Pang YY, Thompson CD, Culp TD, Christensen ND, Lowy DR, Schiller JT, Roden RB. Cross-neutralization of cutaneous and mucosal Papillomavirus types with anti-sera to the amino terminus of L2. Virology. 2005;337(2):365–72. doi: 10.1016/j.virol.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Rizk RZ, Christensen ND, Michael KM, Muller M, Sehr P, Waterboer T, Pawlita M. Reactivity pattern of 92 monoclonal antibodies with 15 human papillomavirus types. J Gen Virol. 2008;89(Pt 1):117–29. doi: 10.1099/vir.0.83145-0. [DOI] [PubMed] [Google Scholar]
- Sapp M, Fligge C, Petzak I, Harris JR, Streeck RE. Papillomavirus assembly requires trimerization of the major capsid protein by disulfides between two highly conserved cysteines. J Virol. 1998;72(7):6186–9. doi: 10.1128/jvi.72.7.6186-6189.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Moseley PL, Lowe SL, Ozbun MA. Inducible heat shock protein 70 enhances HPV31 viral genome replication and virion production during the differentiation-dependent life cycle in human keratinocytes. Virus Res. 2010;147(1):113–22. doi: 10.1016/j.virusres.2009.10.019. [DOI] [PubMed] [Google Scholar]
- Trus BL, Roden RB, Greenstone HL, Vrhel M, Schiller JT, Booy FP. Novel structural features of bovine papillomavirus capsid revealed by a three-dimensional reconstruction to 9 A resolution. Nat Struct Biol. 1997;4(5):413–20. doi: 10.1038/nsb0597-413. [DOI] [PubMed] [Google Scholar]
- Volpers C, Schirmacher P, Streeck RE, Sapp M. Assembly of the major and the minor capsid protein of human papillomavirus type 33 into virus-like particles and tubular structures in insect cells. Virology. 1994;200(2):504–12. doi: 10.1006/viro.1994.1213. [DOI] [PubMed] [Google Scholar]
- Wolf M, Garcea RL, Grigorieff N, Harrison SC. Subunit interactions in bovine papillomavirus. Proc Natl Acad Sci U S A. 2010;107(14):6298–303. doi: 10.1073/pnas.0914604107. [DOI] [PMC free article] [PubMed] [Google Scholar]