Abstract
This manuscript describes the high-throughput analysis and isolation of bacterial cells encapsulated in agarose microparticles using fluorescence-activated cell sorting (FACS). Flow-focusing microfluidic systems were used to create monodisperse microparticles that were ~30μm in diameter. The dimensions of these particles made them compatible with flow cytometry and FACS, and the sensitivity of these techniques reduced the incubation time for cell replication before analyses were carried out. The small volume of the microparticles (~1–50 picoliters) minimized the quantity of reagents needed for bacterial studies. This platform made it possible to screen and isolate bacteria, and apply a combination of techniques to rapidly determine the target of biologically active small molecules. As a pilot study, Escherichia coli cells were encapsulated in agarose microparticles, incubated in the presence of varying concentrations of rifampicin, and analyzed using FACS. The minimum inhibitory concentration of rifampicin was determined, and spontaneous mutants that had developed resistance to the antibiotic were isolated via FACS and characterized by DNA sequencing. The β-subunit of RNA polymerase, RpoB, was confirmed as the target of rifampicin, and Q513L was the mutation most frequently observed. Using this approach, the time and quantity of antibiotics required for the isolation of mutants was reduced by 8- and 150-fold, respectively, compared to conventional microbiological techniques using nutrient agar plates. We envision that this technique will have an important impact on research in chemical biology, natural products chemistry, and the discovery and characterization of biologically active secondary metabolites.
INTRODUCTION
This paper describes a microfluidic technique for encapsulating, growing, and analyzing bacteria in agarose microparticles to rapidly screen and isolate cells that display phenotypes of interest. As a proof-of-principle, we used the bacterial RNA polymerase inhibitor rifampicin and identified spontaneous mutations in Escherichia coli (E. coli) that conveyed resistance to this antibiotic. We created agarose microparticles using microfluidic flow-focusing devices fabricated in poly(dimethylsiloxane) (PDMS). The microparticles are mechanically stable and have a user-defined diameter that ranges from 20 to 150 μm; these characteristics enable us to use flow cytometry (FC) and fluorescence-activated cell sorting (FACS) for the high-throughput analysis, selection, and isolation of encapsulated cells based on phenotypes.
Microbiologists frequently culture bacterial cells on agar plates to screen, select, and isolate monoclonal populations of cells. ‘Agar plates, toothpicks, and logic’ (1) played a central role in microbiology over the past century; however, approaches based on the growth of cells on agar nutrient media have significant drawbacks for culturing organisms that grow slowly or require environmental cues (2). The most common techniques for identifying targets of biologically active small molecules in bacteria involve large-scale screens to isolate mutants that have spontaneous resistance to the compounds, isolation and amplification of their genomic DNA, and sequencing to identify genetic changes responsible for the acquired resistance. The isolation of mutants by growing bacteria on agar typically requires large quantities of the chemical(s) of interest and multiple iterations of screening, which can make these assays expensive and slow. These requirements can be particularly problematic if the reagents are difficult to obtain and are only available in small quantities (e.g. structurally complex secondary metabolites and natural products that are not yet available through total syntheses) or when the exposure of users to hazardous compounds should be minimized. Furthermore, the time required for each cycle is constrained by the growth of an individual cell into a colony that is large enough to visualize and pick. This characteristic limits the throughput of assays for the rapid screening of biological and chemical targets.
To address these limitations, several different methods have been proposed for encapsulating bacterial cells in aqueous droplets (3–5), liquid plugs (6, 7), or hydrogels (8–13) for high-throughput analysis. The most widely used of these techniques encapsulate cells in agarose (2, 14–17) and have been has been used to study a variety of phenomena, including: (1) antibiotic susceptibility (18, 19); (2) bacterial uptake of small molecules by electroporation (20); (3) enrichment of slow-growing bacteria (21, 22); and (4) growth of ‘uncultured’ bacteria for phylogenetic analyses (2).
Despite their demonstrated applications, the methods currently available for fabricating agarose particles provide limited control over the diameter of particles and produce particles with a large coefficient of variance (e.g., ≥ 40%) (12, 23–26). The large variance in the diameter of these particles can be problematic when studying cells that are present at a low frequency in a heterogeneous population of cells. Large particles with diameters that are incompatible with FC or FACS are removed by filtration prior to analysis. Strains that are underrepresented in a mixed population of cells may be encapsulated in large particles and removed before they are analyzed, which biases the composition of the sample and poses a challenge for the detection and analysis of ‘rare’ cells in a mixed population.
To transcend the limitations of conventional techniques for encapsulating bacteria in agarose, we used PDMS flow-focusing microfluidic systems to form monodisperse microparticles (27, 28). The mechanism of drop formation in these microfluidic junctions is well understood and can be exploited to create monodisperse droplets and bubbles (29–31). We isolated single cells in microparticles and demonstrated the rapid determination of the minimum inhibitory concentration (MIC) of rifampicin to E. coli strain MG1655 and the screening and isolation of spontaneous mutants that are resistant to the antibiotic. This approach makes it possible to isolate strains of bacteria based on changes in genotype and phenotype and will be useful for evolving and engineering proteins in microbes (4).
RESULTS AND DISCUSSION
The flow-focusing microfluidic devices consisted of a cross-junction with three inputs and one outlet channel through which microparticles flowed out of the device and were collected. The dispersed phase liquid admixed with cells was pumped into the junction through an inlet channel where it met the continuous phase, mineral oil, which was flowing from two orthogonally oriented channels. This geometry caused the mineral oil streams to focus the dispersed phase into a thin stream that broke into droplets as it flowed through a constriction into the outlet channel (Figure 1). The geometry of the device and the physics of fluids at the micron-scale ensured the stable and reproducible breakup of the dispersed phase into uniform droplets (32).
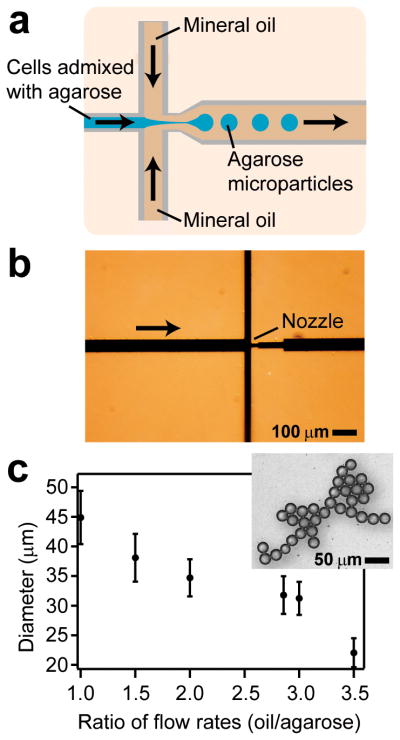
Figure 1.
Production of microparticles in PDMS flow-focusing microfluidic devices. a) A schematic diagram of the PDMS device depicting the direction of fluid flow and formation of agarose droplets suspended in mineral oil at the nozzle. A video (Movie S1) showing droplet formation inside the device is included in the Supplementary Information. b) An image of a PDMS microfluidic device showing the nozzle region of the device; the channels were filled with black ink to increase the contrast. The arrow shows the direction of flow of the agarose solution admixed with cells. c) A plot showing the relationship between the ratio of flow rates of mineral oil and agarose and the resulting diameter of microparticles that were formed. The error bars represent the coefficient of variance for each ratio tested. The image inset shows agarose microparticles produced using flow rates of 150 (oil) and 50 (agarose) μL h−1.
Characterization of microparticles
We measured the diameter of the agarose microparticles produced in the PDMS microfluidic devices at different ratios of flow rates of the mineral oil and solution of agarose. Although we fabricated microfluidic channels with a range of different dimensions, we present data from one set of devices with critical dimensions that are described in Supporting Information. As we increased the ratio of the flow rates of oil to agarose, the diameter of the microparticles decreased to ~20 μm (Figure 1C). The coefficient of variance for the microparticle diameter at each ratio of flow rates was 10% and indicated that the microparticles were moderately monodisperse. To analyze microparticles using FACS or FC, we fabricated colloids that were ~30 μm in diameter; we chose this diameter for the particles to accommodate a standard core size of 40 μm in most FC and FACS instruments. Microparticles with a diameter of 60–160 μm or larger can be created using other types of microfluidic devices (Supporting Information) or by fabricating channels/nozzles with dimensions that are larger than those described here.
Growth and analysis of encapsulated cells
To visualize cells in microparticles via microscopy, FC, and FACS, we used a strain of E. coli MG1655 with a plasmid encoding an ampicillin resistance and a lac operon for controlling the transcription of egfp (we refer to this strain as MG1655-placEGFP). Using the Poisson equation as a guide, we diluted a cell culture such that ~10% of a population of agarose microparticles would contain single cells and the majority of the remaining microparticles would be empty.
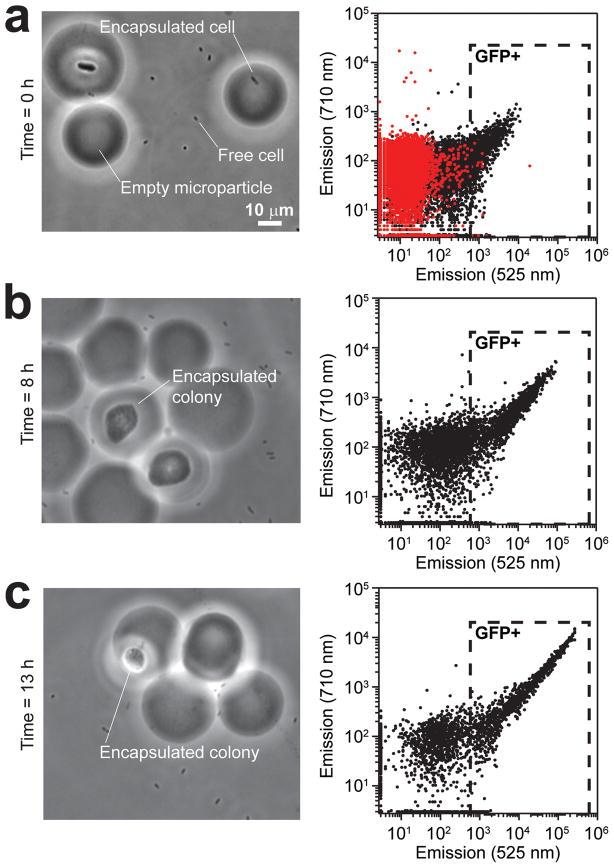
We gated the data in FC experiments and analyzed the fluorescence emission of the microparticles (Figure 2). To differentiate between the autofluorescence of empty agarose microparticles (red data points in Figure 2a) and the fluorescence emitted from EGFP-expressing cells encapsulated in microparticles (black data points), we excited the sample at λ=488 nm and compared the fluorescence emission at two wavelengths: 525 nm and 710 nm. The fluorescence emission of EGFP is high at λ=525 nm and low at λ=710 nm. Using this guideline, we gated the population of microparticles to determine the percentage containing fluorescent cells, and found that approximately 11% of the microparticles contained EGFP at the initial time point (Figure 2a). This percentage of fluorescent microparticles agreed well with our calculation using the Poisson distribution, indicating that most of the fluorescent microparticles contained single cells. When we imaged the microparticles using epifluorescence microscopy, we found that some microparticles contained more than one cell, which may have arisen from four experimental variables: (1) the Poisson distribution predicts that ~1% of the microparticles contain more than one cell; (2) aggregates of cells formed in the overnight culture were not dispersed completely in the agarose solution; (3) cells aggregated in the syringe during the formation of the microparticles in the microfluidic device; and (4) single cells encapsulated in microparticles replicated during sample collection from the microfluidic outlet channel. For example, a microparticle (top left) in image for the ‘time zero’ sample (Figure 2a) probably contained an aggregate of multiple cells.
Figure 2.
Growth of E. coli cells in agarose microparticles. Samples were collected, analyzed, and imaged at three time points: t = 0, 8, and 13 h. Images of microparticles are shown in the left column, corresponding to these time points. The right column contains plots of fluorescence from FC for each corresponding time point. The dotted rectangle shows the gate drawn for the GFP-positive population. Red dots in the scatter plot for the t = 0 h sample indicate background fluorescence emission from empty microparticles. Black dots indicate fluorescence signal from E. coli cells encapsulated in microparticles.
The incubation of microparticles containing single cells led to the formation of microcolonies within the agarose and demonstrated that the nutrients in the microparticles and gas exchange through the mineral oil were sufficient for cell growth. We have found that the growth rate of E. coli cells in agarose microparticles suspended in mineral oil was very similar to their growth on an agarose pad. The microencapsulated cells reproduced with a doubling time of 17 ± 3 min at 37 °C, which is comparable to the doubling time of 19 ± 5 min at 37 °C on an agarose pad. We observed that the doubling time increased after 1.5 h when cells were encapsulated in 36-μm diameter microparticles that were suspended in oil. However, the maximum time of growth in particles will depend on the medium in which they are suspended (oil vs. nutrient broth), the size of the particles, and the number of cells encapsulated per particle.
We also observed a time-dependent increase in the percentage of microparticles containing EGFP-expressing cells during the incubation of an unsorted collection of microparticles suspended in mineral oil at 37 °C without shaking (Figure 2). After 8 h of incubation in mineral oil, 24% of the microparticles were fluorescent (compared to 11% at 0 h), and after 13 h of incubation the number of fluorescent microparticles increased to 40%. We hypothesized that the increase in the number of fluorescent microparticles over time was due to the released cells from ruptured microparticles, which led to attachment of the freed cells to the surface of intact microparticles that were initially empty.
Cells formed microcolonies that were 5 – 20 μm in diameter within 30 μm (diameter) microparticles after 5 h of growth in mineral oil. It was difficult to determine the size of these colonies with precision using 2D optical microscopy due to the influence of the orientation of the colony/particle (Figure 2). A more reliable and sensitive approach was to use FACS to analyze the total fluorescence intensity of the encapsulated cells in the microparticles. For FACS, we transferred the colloids from mineral oil to an aqueous buffer (see Supporting Information). Some of the particles containing large colonies became mechanically unstable and released cells into the buffer during their transfer from the mineral oil. We suspected that these cells adsorbed to the surface of agarose microparticles and increased the number of fluorescent microparticles in our samples. To confirm this hypothesis we fabricated empty microparticles and admixed them with a suspension of MG1655-placEGFP cells. We immediately analyzed the sample by FC and found that ~40% of the empty microparticles were fluorescent.
The release of cells from microparticles can be an issue in some applications; however, it can be avoided by using short incubation times for cell growth and taking advantage of the sensitivity of the detectors in FC and FACS instruments. When microparticles were incubated for ≤ 5 hrs, we rarely observed cells released from microparticles. For applications that require larger microencapsulated communities, the optimization of the microparticle size, agarose concentration, and transfer of the microparticles from mineral oil to buffer will decrease colony heterogeneity that may arise from non-specific adsorption of cells to other microparticles.
Determination of the minimum inhibitory concentration
We used agarose microparticles to determine the MIC of rifampicin and compared the result to measurements made using techniques based on the macrodilution of batch cultures. In batch culture techniques, the MIC is the lowest concentration of antibiotic that inhibits visual growth of bacteria. These culture techniques require incubating cells for long periods of time (typically 12–18 h for E. coli, and > 24 h for slower-growing species) and consume significant amounts of the antibiotic (e.g. > 10 mg) as the cultures are typically grown in a dilution series and require several replicates at each concentration of compound. To decrease the amount of compound required for assay, the MIC can be determined using 96-well microtiter plates. However, performing assays in microtiter plates does not reduce the experimental time. In contrast, the microencapsulation technique we describe requires very small amounts of compounds (< 1 mg) and short incubation times (≤ 3 h), as FC makes it possible to quantitatively measure growth by measuring small changes in the fluorescence of microparticles. We anticipated that the determination of MICs using microfluidic encapsulation would provide a useful alternative to batch culture methods.
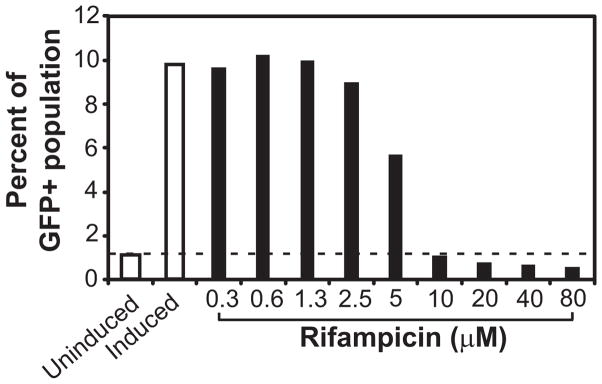
To measure an MIC of rifampicin against E. coli MG1655 we constructed a strain that contained a plasmid encoding an ampicillin resistance cassette and a tet operon for controlling the transcription of egfp (MG1655-ptetEGFP). We encapsulated single cells of MG1655-ptetEGFP in agarose microparticles and analyzed fluorescence using FC. To analyze FC data from microparticles, we included two controls without rifampicin: (1) uninduced; and (2) induced with anhydrotetracycline. Using these controls, we created a gate for the GFP-positive population of microparticles and used a percent population of the uninduced control that fell into the GFP-positive gate as a threshold to determine the concentration of rifampicin that completely inhibited the growth of bacteria. As we increased concentration of antibiotic the number of microparticles in the GFP-positive gate decreased (Figure 3). At 10 μM rifampicin, the percentage of the GFP-positive population approached the uninduced controls (1.1–1.2%), indicating that at this concentration, growth was inhibited. The MIC we determined using microparticles (10 μM) closely matches the value determined using a dilution technique (10–12.5 μM) (33, 34). The agar dilution method required 24–48 h of incubation after inoculating several plates of agar with cells and would have consumed at least 600 mL of solid media and 10 mg of antibiotic. By contrast, the microfluidic technique for determining the MIC of rifampicin reduced the total amount of reagents used by 60-fold and the experimental time by 16-fold.
Figure 3.
Determination of the minimum inhibitory concentration by encapsulating cells in agarose microparticles. The plot shows the percentage of a microparticle population that fell into the GFP-positive gate (GFP+) at various concentrations of the antibiotic (filled bars). Control samples (open bars) without antibiotic were either induced or uninduced to determine the lowest and highest percentages of cells in the GFP-positive gate. A dashed line was drawn to indicate the percent of GFP+ population from the uninduced control.
Screening and isolation of spontaneous mutants
The encapsulation of bacterial cells in microparticles combined with FACS may be a powerful tool for rapidly isolating strains of bacteria via changes in genotype and phenotype. To explore this application, we used this technique to isolate and characterize spontaneous mutants that are resistant to rifampicin. Although rifampicin is commercially available in large quantities, we used it as a surrogate of structurally complex natural products that are not readily available and for which screening and isolation of resistant mutants using conventional microbiological techniques may be challenging. The frequency of spontaneous mutants for resistance against small molecules in the rifamycin family is between 10−8–10−9 per bacterium per cell division (35, 36). Traditionally these mutants are selected on agar plates that contain growth media and an antibiotic at a concentration above its MIC. The isolation of spontaneous mutants using conventional approaches typically requires liters of solid media containing a high concentration of the target antibiotic (typically tens of milligrams per liter), and takes 2–4 days of incubation to obtain visible colonies on plates from which cells are isolated. A high-throughput approach using agarose microparticles with a picoliter volume combined with FACS should make it possible to reduce the time and amount of antibiotic for the isolation of mutants.
We encapsulated cells of strain MG1655-ptetEGFP in agarose microparticles and dosed them with rifampicin. As expected, most of the cells in microparticles did not express EGFP (99.8% of microparticles), indicating their sensitivity to rifampicin (35). A small sub-population of microparticles (0.2%; approximately 8 × 103 out of the 4 × 106 microparticles that we analyzed) was GFP-positive and was sorted into a tube containing liquid nutrient media. From these microparticles we isolated mutants, sequenced them, and identified an A1538T base-pair mutation in the rpoB ORF (Figure 4). This mutation results in a Q513L substitution, which has been described previously as conferring resistance to rifampicin (33, 34). Overall, the screen used a total of 65 μg of rifampicin and took 6 h to complete, compared to 15 mg of compound that would have been required for a 48 h screen using a traditional approach with agar plates (34).
Figure 4.
Spontaneous mutant screen of E. coli cells in agarose microparticles. a) We analyzed the microparticles using FC. By comparing the fluorescence emission at wavelengths of 525 nm and 710 nm, we differentiated a subpopulation with a positive EGFP signal for the induced control (the uppermost plot). The GFP-positive gate is drawn as a dashed box. As a negative control, we analyzed microparticles containing both inducer and rifampicin (the middle plot). The negative control had negligible emission in the GFP-positive gate since a monoculture of wildtype E. coli cells was sensitive to the antibiotic. By contrast, a small population of microparticles that contained cells from a pool of 480 independent cultures showed fluorescence emission in the GFP-positive gate despite the presence of rifampicin (the bottom plot). This GFP-positive subpopulation were isolated via FACS. b) Mutants that were isolated by FACS contained a mutation in the rpoB gene, resulting in a Q513L substitution. The panel shows the DNA sequence of the region, the corresponding amino acids (AA), and residue numbers. The mutant locus is indicated with a bold letter.
The microencapsulation technique has several advantages over conventional, Petri dish based approaches for isolating bacteria based on phenotypes. (1) The volume of the microparticles (e.g. several picoliters) minimizes the experimental time and consumption of reagents. Encapsulating cells in agarose microparticles makes it possible to grow the cells through only a few cycles of division and detect them by FACS. In contrast, a typical colony on an agar plate contains ~109 bacterial cells (37); a visible colony requires ~20 h of growth from a single cell that has a doubling time of 40 min. (2) FACS is a sensitive technique, which can detect low levels of fluorescence in a colloidal sample. This characteristic can be used to detect changes in fluorescence in individual encapsulated cells (38, 39). Using FACS for the analysis of cells expands the number of genotypes and phenotypes that can be simultaneously quantified in a high-throughput screen, as each can be coupled to a fluorescent reporter. Many FACS instruments can simultaneously measure up to 16 different fluorophores in a sample, which makes it possible to multiplex analyses. (3) The encapsulation and growth of cells in microparticles that are suspended in oil can protect the user from toxic reagents that are either incorporated into the colloids or are produced by cells. These compounds only diffuse out of the colloids if they are soluble in mineral oil. Replacing the mineral oil as the continuous phase with fluorous liquids, such as perfluorodecalin, will improve the insulation of the user from these compounds by reducing their solubility in the continuous phase fluid (8). This characteristic may be particularly useful in research with biosafety level two and three organisms and select chemicals. (4) The use of agarose as the hydrogel has several advantages, including biocompatibility, a gelling temperature that is close to the optimal growth temperatures for bacteria, relatively low cost, and availability in most biological laboratories. (5) PDMS devices are straightforward to fabricate and operate and users can easily manipulate the layout and dimensions of the systems using soft lithographic techniques (28). The devices described in this manuscript produce agarose microparticles at a frequency of >500 Hz. Droplet formation at frequencies of 103 – 104 Hz have been reported in the literature for similar microfluidic systems (29, 31), which will reduce the time required to encapsulate a large population of cells for analysis and selection. The devices in the manuscript produce monodisperse microparticles with a user-defined diameter, which makes it possible to analyze samples by FACS without first filtering out particles that are too large for the instrument. The ability to omit the filtration step avoids sample bias.
There are several characteristics of this approach that currently limit its application. Measuring the fluorescence emission of cells encapsulated in microparticles requires that cells produce fluorescent molecules or proteins. In many cases this criteria is not an issue and can be solved by genetic engineering. A fundamental limitation is that it may be difficult to rely on fluorescence detection when isolating and amplifying bacteria that have not been previously cultured or which are genetically intractable. A potential solution is to use forward light scattering for measuring cell growth in microparticles. In addition, long incubation periods can lead to release of cells from microparticles, which will be eliminated from analysis or which may adsorb on the surface of other colloids and contaminate them. The optimization of growth time and conditions can transcend this limitation. Finally, the small molecule we tested is an antibiotic and provides an obvious phenotype for screening as it prevents cell replication. The biological target of other small molecules may be more difficult to discern using this approach and may require the development of more sophisticated methods of analysis.
In addition to its application in the high-throughput screen for studying target proteins of biologically active small molecules in microorganisms, we anticipate that encapsulating and analyzing single bacterial cells in agarose microparticles will have a variety of applications in environmental microbiology and for studying the human microbiome. Since encapsulation of cells makes it possible to detect and analyze slow-growing and rare species in a mixed population by using FACS, bacterial strains that are isolated and amplified in microparticles can be sequenced to catalog microbes associated with a particular ecological niche. Manipulating the liquid in which the microparticles are dispersed and incubated provides control over the diffusion of secondary metabolites in and out of the colloids, which may promote growth of species that are otherwise difficult to culture using standard media (2, 40). Finally, this technique may find applications in areas that require phenotypic based screening of bacteria, including biosensing, biological engineering, and synthetic biology.
METHODS
Fabrication of microfluidic devices
We used photolithography and soft lithography to fabricate microfluidic devices. Details of the fabrication process can be found in the Supporting Information.
Fabrication and characterization of microparticles
We dissolved agarose in water or growth media at a final concentration of 2% v/v and used light mineral oil as the continuous phase for the emulsification of the agarose solution. To stabilize the pre-gelled agarose droplets and prevent coalescence, we added 4% (v/v) of the surfactant Span80 to the mineral oil. We filtered the agarose solutions and oil through 0.22 μm pore size syringe filters and loaded them into glass gas-tight syringes. We used positive displacement syringe pumps (Harvard Apparatus) to control the flow rate of fluids in the microfluidic devices. To prevent gelation of the agarose in the syringe and in the device during the production of microparticles, we heated the syringes on the pumps with heating tape to maintain a temperature of 40 °C; we heated the microfluidic device to 37 °C. Procedures for imaging microparticles and encapsulating cells are described in the Supporting Information.
Microbial cell culture and growth conditions
Details on bacterial strains and growth conditions can be found in the Supporting Information.
Using a Poisson distribution to encapsulate single cells
To guide the dilution of cells to encapsulate one cell per microparticle, we used the Poisson equation. Detailed description on the calculations can be found in the Supporting Information.
Flow cytometry and fluorescence-activated cell sorting
We used FACS to sort the population of microparticles containing EGFP-expressing cells into the wells of 96-well microplates, culture tubes, or onto LB agar plates(Figure S1). For these experiments, we used a SORP BD FACS Aria instrument at the Flow Cytometry Facility at the UW Carbone Cancer Center. Prior to analyzing the microparticles using FACS, we extracted the microparticles from the oil phase and transferred them into a solution of PBS or growth medium containing the appropriate antibiotics and small molecule inducers. To transfer the microparticles from oil to the buffer, we rinsed the microparticles with pure mineral oil seven times to remove the surfactant. For the first three rinse cycles, we spun the microparticles in microcentrifuge tubes at 100 x g for 3 min; we reduced the centrifugation time to 30 s for the subsequent four cycles. After rinsing with mineral oil, we carefully pipetted the microparticle-oil emulsion on top of 700 μL of growth medium or PBS in a microcentrifuge tube. The density of the microparticles was higher than the oil and the particles gradually moved into the aqueous solution. To prevent the possibility of clogging the FC or FACS, we filtered the aqueous suspension of microparticles through a Nylon filter with 60-μm diameter pores. This step was not necessary but was carried out at the request of the FACS facility. We acquired FC data by collecting >10,000 data points for each sample; for FACS, we used an 85 μm diameter nozzle tip at a liquid pressure of 45 psi.
Determination of the minimum inhibitory concentration
We determined the minimum inhibitory concentration (MIC) of rifampicin that prevents the growth of strain MG1655-ptetEGFP in liquid cultures (i.e. macrodilution method) (41) and in agarose microparticles. For the encapsulated cells in microparticles, we incubated the samples for 3 hr and induced the cells with anhydrotetracycline for 45 min prior to using FC. Details on MIC determination can be found in the Supporting Information.
Screen and isolation of spontaneous mutants
To increase the probability of isolating spontaneous mutants to rifampicin, we incubated 480 independent cultures of strain MG1655-placEGFP in 96-well microplates (200 μL per well). We treated encapsulated cells with the antibiotic for 3 h at 37 °C prior to FACS. We incubated sorted microparticles at 37°C for 17 h for release of cells, and subsequently plated the released cells on LB agar to obtain single colonies. Ten colonies were selected and their rpoB gene was amplified and sequenced. Details on this experiment can be found in the Supporting Information.
Supplementary Material
Acknowledgments
This manuscript is based upon work supported by the National Science Foundation under Grant No. DMR-0520527. ASU acknowledges the Japan Society for the Promotion of Science (JSPS) for support. YJE acknowledges a Genentech Graduate Fellowship and a William R. & Dorothy E. Sullivan Wisconsin Distinguished Graduate fellowship. MFC is funded by an NIH biotechnology training grant (grant 5T32GM08349). DBW acknowledges support from the Searle Scholar Program, DARPA, the Human Frontiers Science Program (RGY0069/2008-C), and USDA (WIS00974). We thank J. Weisshaar for vector pASK-IBA3plus.
Footnotes
Supporting Information Available: This material is available free via the Internet.
References
- 1.Shuman HA. Just toothpicks and logic: How some labs succeed at solving complex problems. Journal of Bacteriology. 2003;185:387–390. doi: 10.1128/JB.185.2.387-390.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zengler K, Toledo G, Rappe M, Elkins J, Mathur EJ, Short JM, Keller M. Cultivating the uncultured. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15681–15686. doi: 10.1073/pnas.252630999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huebner A, Bratton D, Whyte G, Yang M, Demello AJ, Abell C, Hollfelder F. Static microdroplet arrays: a microfluidic device for droplet trapping, incubation and release for enzymatic and cell-based assays. Lab Chip. 2009;9:692–698. doi: 10.1039/b813709a. [DOI] [PubMed] [Google Scholar]
- 4.Agresti JJ, Antipov E, Abate AR, Ahn K, Rowat AC, Baret JC, Marquez M, Klibanov AM, Griffiths AD, Weitz DA. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc Natl Acad Sci U S A. 2010;107:4004–4009. doi: 10.1073/pnas.0910781107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Granieri L, Baret JC, Griffiths AD, Merten CA. High-throughput screening of enzymes by retroviral display using droplet-based microfluidics. Chem Biol. 2010;17:229–235. doi: 10.1016/j.chembiol.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 6.Boedicker JQ, Li L, Kline TR, Ismagilov RF. Detecting bacteria and determining their susceptibility to antibiotics by stochastic confinement in nanoliter droplets using plug-based microfluidics. Lab on a Chip. 2008;8:1265–1272. doi: 10.1039/b804911d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu WS, Kim HJ, Lucchetta EM, Du WB, Ismagilov RF. Isolation, incubation, and parallel functional testing and identification by FISH of rare microbial single-copy cells from multi-species mixtures using the combination of chemistrode and stochastic confinement. Lab on a Chip. 2009;9:2153–2162. doi: 10.1039/b904958d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baret JC, Miller OJ, Taly V, Ryckelynck M, El-Harrak A, Frenz L, Rick C, Samuels ML, Hutchison JB, Agresti JJ, Link DR, Weitz DA, Griffiths AD. Fluorescence-activated droplet sorting (FADS): efficient microfluidic cell sorting based on enzymatic activity. Lab Chip. 2009;9:1850–1858. doi: 10.1039/b902504a. [DOI] [PubMed] [Google Scholar]
- 9.Morimoto Y, Tan WH, Tsuda Y, Takeuchi S. Monodisperse semi-permeable microcapsules for continuous observation of cells. Lab on a Chip. 2009;9:2217–2223. doi: 10.1039/b900035f. [DOI] [PubMed] [Google Scholar]
- 10.Panda P, Ali S, Lo E, Chung BG, Hatton TA, Khademhosseini A, Doyle PS. Stop-flow lithography to generate cell-laden microgel particles. Lab Chip. 2008;8:1056–1061. doi: 10.1039/b804234a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pereira T, Millar TJ, Chuck JA. Viability analysis of alginate encapsulated micro-organisms using fluorescent stains. Journal of Microencapsulation. 2005;22:787–792. doi: 10.1080/02652040500273829. [DOI] [PubMed] [Google Scholar]
- 12.Weaver JC, Williams GB, Klibanov A, Demain AL. GEL MICRODROPLETS - RAPID DETECTION AND ENUMERATION OF INDIVIDUAL MICROORGANISMS BY THEIR METABOLIC-ACTIVITY. Bio-Technology. 1988;6:1084–1089. [Google Scholar]
- 13.Workman VL, Dunnett SB, Kille P, Palmer DD. On-chip alginate microencapsulation of functional cells. Macromolecular Rapid Communications. 2008;29:165–170. [Google Scholar]
- 14.Bernath K, Hai MT, Mastrobattista E, Griffiths AD, Magdassi S, Tawfik DS. In vitro compartmentalization by double emulsions: sorting and gene enrichment by fluorescence activated cell sorting. Analytical Biochemistry. 2004;325:151–157. doi: 10.1016/j.ab.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 15.Katsuragi T, Tanaka S, Nagahiro S, Tani Y. Gel microdroplet technique leaving microorganisms alive for sorting by flow cytometry. Journal of Microbiological Methods. 2000;42:81–86. doi: 10.1016/s0167-7012(00)00179-2. [DOI] [PubMed] [Google Scholar]
- 16.Luo DW, Pullela SR, Marquez M, Cheng ZD. Cell encapsules with tunable transport and mechanical properties. Biomicrofluidics. 2007;1 doi: 10.1063/1.2757156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nir R, Yisraeli Y, Lamed R, Sahar E. FLOW-CYTOMETRY SORTING OF VIABLE BACTERIA AND YEASTS ACCORDING TO BETA-GALACTOSIDASE ACTIVITY. Applied and Environmental Microbiology. 1990;56:3861–3866. doi: 10.1128/aem.56.12.3861-3866.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akselband Y, Cabral C, Shapiro DS, McGrath P. Rapid mycobacteria drug susceptibility testing using Gel Microdrop (GMD) Growth Assay and flow cytometry. Journal of Microbiological Methods. 2005;62:181–197. doi: 10.1016/j.mimet.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 19.Ryan C, Nguyen BT, Sullivan SJ. RAPID ASSAY FOR MYCOBACTERIAL GROWTH AND ANTIBIOTIC SUSCEPTIBILITY USING GEL MICRODROP ENCAPSULATION. Journal of Clinical Microbiology. 1995;33:1720–1726. doi: 10.1128/jcm.33.7.1720-1726.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gift EA, Weaver JC. Simultaneous quantitative determination of electroporative molecular uptake and subsequent cell survival using gel microdrops and flow cytometry. Cytometry. 2000;39:243–249. [PubMed] [Google Scholar]
- 21.Akselband Y, Cabral C, Castor TP, Chikarmane HM, McGrath P. Enrichment of slow-growing marine microorganisms from mixed cultures using gel microdrop (GMD) growth assay and fluorescence-activated cell sorting. Journal of Experimental Marine Biology and Ecology. 2006;329:196–205. [Google Scholar]
- 22.Manome A, Zhang H, Tani Y, Katsuragi T, Kurane R, Tsuchida T. Application of gel microdroplet and flow cytometry techniques to selective enrichment of non-growing bacterial cells. Fems Microbiology Letters. 2001;197:29–33. doi: 10.1111/j.1574-6968.2001.tb10578.x. [DOI] [PubMed] [Google Scholar]
- 23.Nilsson K, Birnbaum S, Flygare S, Linse L, Schroder U, Jeppsson U, Larsson PO, Mosbach K, Brodelius P. A GENERAL-METHOD FOR THE IMMOBILIZATION OF CELLS WITH PRESERVED VIABILITY. European Journal of Applied Microbiology and Biotechnology. 1983;17:319–326. [Google Scholar]
- 24.Nir R, Lamed R, Gueta L, Sahar E. SINGLE-CELL ENTRAPMENT AND MICROCOLONY DEVELOPMENT WITHIN UNIFORM MICROSPHERES AMENABLE TO FLOW-CYTOMETRY. Applied and Environmental Microbiology. 1990;56:2870–2875. doi: 10.1128/aem.56.9.2870-2875.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams GB, Weaver JC, Demain AL. RAPID MICROBIAL DETECTION AND ENUMERATION USING GEL MICRODROPLETS AND COLORIMETRIC OR FLUORESCENCE INDICATOR SYSTEMS. Journal of Clinical Microbiology. 1990;28:1002–1008. doi: 10.1128/jcm.28.5.1002-1008.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tumarkin E, Kumacheva E. Microfluidic generation of microgels from synthetic and natural polymers. Chemical Society Reviews. 2009;38:2161–2168. doi: 10.1039/b809915b. [DOI] [PubMed] [Google Scholar]
- 27.McDonald JC, Duffy DC, Anderson JR, Chiu DT, Wu HK, Schueller OJA, Whitesides GM. Fabrication of microfluidic systems in poly(dimethylsiloxane) Electrophoresis. 2000;21:27–40. doi: 10.1002/(SICI)1522-2683(20000101)21:1<27::AID-ELPS27>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 28.Xia YN, Whitesides GM. Soft lithography. Angewandte Chemie-International Edition. 1998;37:551–575. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 29.Garstecki P, Fuerstman MJ, Stone HA, Whitesides GM. Formation of droplets and bubbles in a microfluidic T-junction - scaling and mechanism of break-up. Lab on a Chip. 2006;6:437–446. doi: 10.1039/b510841a. [DOI] [PubMed] [Google Scholar]
- 30.Hashimoto M, Shevkoplyas SS, Zasonska B, Szymborski T, Garstecki P, Whitesides GM. Formation of Bubbles and Droplets in Parallel, Coupled Flow-Focusing Geometries. Small. 2008;4:1795–1805. doi: 10.1002/smll.200800591. [DOI] [PubMed] [Google Scholar]
- 31.Xu S, Nie Z, Seo M, Lewis P, Kumacheva E, Stone HA, Garstecki P, Weibel DB, Gitlin I, Whitesides GM. Generation of monodisperse particles by using microfluidics: Control over size, shape, and composition (vol 44, pg 724, 2005) Angewandte Chemie-International Edition. 2005;44:3799–3799. doi: 10.1002/anie.200462226. [DOI] [PubMed] [Google Scholar]
- 32.Nie Z, Xu S, Seo M, Lewis PC, Kumacheva E. Polymer particles with various shapes and morphologies produced in continuous microfluidic reactors. J Am Chem Soc. 2005;127:8058–8063. doi: 10.1021/ja042494w. [DOI] [PubMed] [Google Scholar]
- 33.Jin DJ, Cashel M, Friedman DI, Nakamura Y, Walter WA, Gross CA. Effects of rifampicin resistant rpoB mutations on antitermination and interaction with nusA in Escherichia coli. J Mol Biol. 1988;204:247–261. doi: 10.1016/0022-2836(88)90573-6. [DOI] [PubMed] [Google Scholar]
- 34.Reynolds MG. Compensatory evolution in rifampin-resistant Escherichia coli. Genetics. 2000;156:1471–1481. doi: 10.1093/genetics/156.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Floss HG, Yu TW. Rifamycin-mode of action, resistance, and biosynthesis. Chemical Reviews. 2005;105:621–632. doi: 10.1021/cr030112j. [DOI] [PubMed] [Google Scholar]
- 36.Wehrli W, Staehelin M. Actions of the rifamycins. Bacteriol Rev. 1971;35:290–309. doi: 10.1128/br.35.3.290-309.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mashimo K, Nagata Y, Kawata M, Iwasaki H, Yamamoto K. Role of the RuvAB protein in avoiding spontaneous formation of deletion mutations in the Escherichia coli K-12 endogenous tonB gene. Biochem Biophys Res Commun. 2004;323:197–203. doi: 10.1016/j.bbrc.2004.08.078. [DOI] [PubMed] [Google Scholar]
- 38.Davey HM, Kell DB. Flow cytometry and cell sorting of heterogeneous microbial populations: the importance of single-cell analyses. Microbiol Rev. 1996;60:641–696. doi: 10.1128/mr.60.4.641-696.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Czechowska K, Johnson DR, van der Meer JR. Use of flow cytometric methods for single-cell analysis in environmental microbiology. Curr Opin Microbiol. 2008;11:205–212. doi: 10.1016/j.mib.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 40.Kaeberlein T, Lewis K, Epstein SS. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science. 2002;296:1127–1129. doi: 10.1126/science.1070633. [DOI] [PubMed] [Google Scholar]
- 41.Ferraro MJ Institute, C. a. L. S. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 5. Wayne; 2000. pp. 7–12. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.