Abstract
The discovery of the gene mutation responsible for Huntington's Disease (HD), huntingtin, in 1993 allowed for a better understanding of the pathology of the and enabled development of animal models. HD is caused by the expansion of a polyglutamine repeat region in the N-terminal of the huntingtin protein. Here we examine the behavioral, transcriptional, histopathological and anatomical characteristics of a knock-in HD mouse model with a 140 polyglutamine expansion in the huntingtin protein. This CAG 140 model contains a portion of the human exon 1 with 140 CAG repeats knocked into the mouse huntingtin gene. We have longitudinally examined the rearing behavior, accelerating rotarod, constant speed rotarod and gait for the heterozygote, homozygote and their non-transgenic littermates and have found a significant difference in the afflicted mice. However, while there were significant differences between the non-transgenic and the knock-in mice, these behaviors were not progressive. As in HD, we show that the CAG 140 mice also have a significant decrease in striatally enriched mRNA transcripts. In addition, striatal neuronal intranuclear inclusion density increases with age. Lastly these CAG 140 mice show slight cortical thinning compared to unaffected littermates, similarly to the cortical thinning recently reported in HD.
Keywords: Neuronal intranuclear inclusions, transcriptional analysis, Rotarod, Rearing behavior, Gait Analysis, Cortical Thinning
Introduction
Huntington's Disease (HD) is a progressive neurodegenerative autosomal dominant genetic disorder that afflicts approximately 3 to 8 in 100,000 individuals (Folstein, et al., 1987, Wexler, et al., 2004). In 1993, the gene that causes HD, huntingtin (htt), was discovered and sequenced (The Huntington's Disease Collaborative Research Group, 1993). HD is caused by an expanded polyglutamine tract which is encoded by the tri-nucleotide, CAG, in the first exon (The Huntington's Disease Collaborative Research Group, 1993). An individual with HD has more than 35 CAG repeats in the htt gene, normal repeat lengths of less than 35 do not result in the development of HD (The Huntington's Disease Collaborative Research Group, 1993). At this time, however, there is still little known regarding htt's native function or the exact mechanism of how the mutant htt protein causes HD.
Briefly, HD pathology mainly affects the medium spiny neurons in the striatum (Graveland, et al., 1985, Vonsattel, et al., 1985). In addition, progressive cortical thinning (Gomez-Anson, et al., 2009, Hobbs, et al., 2010, Hobbs, et al., 2009, Rosas, et al., 2005, Rosas, et al., 2008), striatal atrophy (Halliday, et al., 1998, Peinemann, et al., 2005, Ruocco, et al., 2006), and striatal specific transcriptional dysregulation (Albin, et al., 1991, Augood, et al., 1997, Hebb, et al., 2004, Hu, et al., 2004, Richfield, et al., 1995) are also observed in HD patients. Behaviorally, HD is characterized by a progression from mood swings and personality changes, to chorea, and dementia (Craufurd, et al., 2001, Lawrence, et al., 1998, Lawrence, et al., 1996, Naarding, et al., 2001, Thompson, et al., 2002). For a more comprehensive overview of the clinical symptoms, time course, and treatments for HD please see Phillips et al (Phillips, et al., 2008).
In 1996, the first transgenic mouse models of HD were developed by Mangiarini et al (Mangiarini, et al., 1996). The two transgenic lines, R6/1 and R6/2, have been among the most studied models in the HD field and consist of the 5' region of the human mutant htt inserted into the mouse genome. The R6/1 mice have approximately 116 repeats in the transgene while the R6/2 mice have approximately 144 repeats. The gene expression levels of the R6/1 and R6/2 in relation to the endogenous mouse htt (hdh) are 31% and 75% respectively. Phenotypically, these mice show resting tremors, clasping behavior when held by the tail, gait abnormalities and other motor skill deficits starting about 15 weeks in the R6/1 and 5 to 6 weeks in the R6/2 line (Carter, et al., 1999, Mangiarini, et al., 1996). A pathological hallmark of HD, neuronal intranuclear inclusions (NII) was first found in the R6 lines and has since been one of the most utilized histopathological markers in mouse models of HD as well as in humans (Becher, et al., 1998, Davies, et al., 1997, Gutekunst, et al., 1999, Morton, et al., 2000, Schilling, et al., 1999). Levels of striatally enriched transcripts, similar to that observed in humans, are also dysregulated in transgenic mice such as the R6 lines (Bibb, et al., 2000, Denovan-Wright and Robertson, 2000, Dowie, et al., 2009, Hebb, et al., 2004, Hebb, et al., 2008, Hu, et al., 2004, Kuhn, et al., 2007, Luthi-Carter, et al., 2000, McCaw, et al., 2004, van Dellen, et al., 2000)
Since the R6 and other HD models that are transgenic and have both the expanded 5' portion of the human mutant htt gene and the endogenous mouse hdh alleles, these models do not perfectly match the genetics of HD which is important especially when attempting to study allele specific knock-down of mutant htt (DiFiglia, et al., 2007, Drouet, et al., 2009, Harper, et al., 2005, Pfister, et al., 2009, Rodriguez-Lebron, et al., 2005). To more accurately model the genetic makeup of HD, various knock-in models have been developed. These models have replaced one of the hdh alleles with either a fused human/mouse mutant htt chimera (Menalled, et al., 2003, Menalled, et al., 2002, Wheeler, et al., 2002, Wheeler, et al., 2000) or simply expanded the CAG region of the mouse hdh gene itself (Lin, et al., 2001). The knock-in mouse models, like the transgenic mouse models, have been created with various lengths of expanded CAG region (Lin, et al., 2001, Menalled, et al., 2003, Menalled, et al., 2002, Wheeler, et al., 2002, Wheeler, et al., 2000). These knock-in mice tend to have a longer life span than the transgenic with a slower progression towards neuropathology seen in transgenic models consisting of truncated versions of htt (Hodgson, et al., 1999, Levine, et al., 1999, Menalled, et al., 2003, Menalled, et al., 2002, Schilling, et al., 1999, Slow, et al., 2003, Wheeler, et al., 2002, Wheeler, et al., 2000).
Menalled et al. (Menalled, et al., 2003) have developed a mouse knock-in model characterized by 140 CAG repeats. This CAG 140 model contains a small portion of the human gene, starting from 18 base pairs upstream of the CAG repeat region through 100 base pairs of first intron and was initially characterized by the Menalled group (Menalled, et al., 2003). The CAG 140 model was further characterized by Dorner and Hickey (Dorner, et al., 2007, Hickey, et al., 2008). To date, all characterizations of the CAG 140 mouse were restricted in the timing of the behavioral testing and the pathological aspects examined. Thus, here, we have undertaken a long-term longitudinal motor behavior study of the CAG 140 and a concomitant cross-sectional neuropathological study model to provide a comprehensive picture of the progression of disease in this mouse model. Additionally, the initial CAG 140 characterization studies were performed with homozygous mice (Dorner, et al., 2007, Hickey, et al., 2008, Menalled, et al., 2003). No study, to date, has directly compared the non-transgenic (nTG), heterozygote and the homozygote genotypes. To this end, we performed monthly rotarod, rearing, and gait behavioral analysis as well as a cross-sectional analysis of transcripts and NII levels at 3 month intervals for approximately 19 months in nTG, heterozygote and homozygote CAG 140 mice. The longitudinal and cross-sectional study reported here expands the characterization of the CAG 140 mouse model phenotype and provides a comprehensive set of dependent variables that can be used for future testing of therapeutics in the CAG 140 model.
Materials and Methods
Animals and tissue preparation
Ten week old CAG 140 knock-in mice on the C57BL6 background strain (Menalled, et al., 2003) were used for behavioral experiments (a kind gift of Scott Zeitlin). Briefly, Menalled et al. replaced a portion of the first exon of the mouse htt gene with the human equivalent (Menalled, et al., 2003). The knocked in region spans from 18 base pairs upstream of the CAG repeat region to the 100 base pairs into the first intron and introduces ~140 CAG repeats (Menalled, et al., 2003).
Genotype determination was performed by PCR analysis after isolation of genomic DNA from tail snips. PCR primers for the nTG gene are as follows: F- 5' ACGCATCCGCCTGTCAATTCTG 3' and R- 5' CTGAAACGACTTGAGCGACTC 3'. Primers for the knock-in gene are F- 5' GCCCGGCATTCTGCACGCTT 3' and R- 5'GAGTACGTGCTCGCTCGATG 3'. An initial 5 minute 94°C was followed by 36 cycles of 30 seconds at 94 °C, 30 seconds at 65 °C and 1 minute at 72 °C. Following the last cycle a final elongation step was performed at 72 °C for 7 minutes. PCR samples were run on a 2 % agarose gel and genotype was determined by either the presence of the nTG band at 534 bps and/or the knock-in band at 287 bps.
For all behavioral experiments, 8 nTG mice (6 males, 2 females), 13 heterozygous (8 males, 5 females), and 4 homozygous (2 males, 2 females) mice were tested. In addition a cross-sectional group of mice at 3, 6, 9, 12, 15 and 19/20 months of age were used for the in situ hybridization study as well as immunohistochemistry quantification of inclusion bodies and cortical thickness. The number of mice used in these cross-sectional studies varies and are indicated in the appropriate figure legends. No gender differences were observed in the cross-sectional studies.
Mice were euthanized with an overdose of pentobarbital (>150mg/kg) and the brains were removed and placed at −80°C until further processing was performed. All appropriate housing and handling procedures were followed in accordance with the Institutional Animal Care and Use Committees at the University of Florida.
Rearing Behavior
nTG, heterozygous, homozygous mice were individually placed in 1 L beakers in the dark and videotaped for 10 minutes. Testing was done at the end of the light cycle for the mice. The tapes were viewed by an individual who was ignorant of the mouse genotype to determine the number of times the mice reared up and touched the side of the beaker with their front paws. Rears that did not include a front paw touch were not counted. Average number of rears per minute was calculated. This task was performed once every two weeks initially and once a month when the mice were 4 months old.
Rotarod
The accelerating rotarod (Columbus Instruments, Columbus OH) started at 5 RPM and increased in speed at 0.3 RPM per second. The mice were allowed to stay on the apparatus until they fell off. The time that each mouse fell off was noted. The test was performed 4 consecutive times a day over 3 consecutive days. A minimum of a minute and half was allowed between each session. The maximum speed was then calculated from the latency to fall. We were unable to perform this test in the 15th month.
In addition, we measured the latency to fall off the rotarod at two different constant speeds. Starting at 10 RPM, the mice were allowed to continue to run on the rod until they fell or until 60 seconds elapsed. The test was performed again at 18 RPM. The test consisted of two consecutive 10 RPM-18 RPM switches over two consecutive days following the accelerating rotarod task described above. A minimum of a minute and half was allowed between each session. We were unable to perform this test in the 15th month.
Gait analysis
A gait apparatus with a runway of 50 centimeters by 10 cm wide with 10 cm high walls was used to test gait. Each mouse was initially allowed to be in a dark goal box for 5 minutes with a FrootLoop™. The mouse was then taken out and shown the corridor that was well lit overhead and on the side. Receipt paper was laid down in the corridor. The mouse was then placed back into the goal box for 1 minute, after which the front paws were painted blue and the rear paws orange with non-toxic children's finger paint. The mouse was then placed at the opposite end of the goal box and allowed to return to the goal box on its own volition. The receipt paper was then collected and the distance between the front and rear paws (Gait Width), and between the front footprints of the consecutive steps (Gait Length) were measured. In subsequent tests, mice were only allowed in the goal box for 1 minute before their feet were painted.
In Situ hybridization
Fresh-frozen mouse brain tissue was sectioned at 14μm on a cryostat. Approximately 25 slides with 5 sections per slide were processed for each mouse, and these slides were used for both immunohistochemisty (see immunohistochemisty section) and in situ hybridization. In situ hybridization was performed on the representative 14 μm thick coronal mouse brain sections (bregma +1.70 to −0.50) mounted on slides from using radiolabeled (33P) antisense gene-specific oligonucleotide probes. The probes used here are dopamine and cyclic AMP-regulated phosphoprotein with molecular weight 32 kDa (DARPP-32), pre-pro-Enkephalin (ppENK), Phosphodiesterase 10a (PDE10a), Phosphodiesterase 1b (PDE1b), Dopamine Receptor Type 2 (D2), Cannabinoid Receptor Type 1 (CB1), Neuronal Growth Factor I-A NGFi-A, dynamin and β-actin. The methods employed for in situ hybridization and quantification of the hybridization signal have been described previously in detail. (Denovan-Wright and Robertson, 2000, Hebb, et al., 2004, Hu, et al., 2004, Luthi-Carter, et al., 2000, Rodriguez-Lebron, et al., 2005)
Quantification of Transcripts
To determine the relative mRNA transcript levels, optical density (OD) was calculated using the Quantity One analysis software from Bio-Rad (Hercules, CA). Slide background was subtracted out and all values were normalized to the β-actin transcript levels. Heterozygote and homozygote percentage levels were calculated from nTG levels. Dynamin was determined using a rectangle shape outline.
Immunohistochemistry
Slides used for immunohistochemical staining were washed 3 times with PBS then fixed for 15 minutes with 4% paraformaldhyde. Three PBS washes were then performed to remove the paraformaldehyde, and then the samples were treated with a 3% (v/v) H202 and 10 % methanol solution for 10 minutes. Subsequently a blocking step was performed with 7.5% (v/v) Natural Horse Serum (NHS) and 0.1% Triton-X100 for 2 hours. Primary antibody solutions were made with mouse anti-htt mEM48 (MAB5374 from Millipore™ Billerica, MA at 1:2000), and mouse anti-NueN (MAB377 from Millipore™ Billerica, MA at 1:1000) with 1% NHS and 0.1% TritonX100. Primary was applied to the slides overnight at 4°C using Hybriwells™ (Grace Bio-Labs Bend, OR). After washing slides with PBS, they were incubated in biotinylated horse anti-mouse secondary with 1% NHS and 0.1% Triton-X100 for 2 hours. ABC solution from the Vectastain ABC kit (Vector Laboratories Burlingame, CA) was applied to prime the secondary antibodies on the tissue for an hour before NovaRed (Vector Laboratories, Burlingame, CA) visualization was performed.
Quantification of Inclusion Bodies
Using the program Stereo Investigator (MBF Bioscience, Williston, VT) striatal regions of the brain sections were outlined using the optical fractionator function. Using approximately 300μm separation between each stop in the setup parameters a representative number of inclusion bodies were counted in the section. Five sections were counted separated approximately by 350 μm. The average number of inclusions per stop was calculated. A minimum of three mice per group were used for this calculation. This sampling method yielded an estimate of NII density identically for each mouse. NII estimates were normalized for each mouse to the mean of the 6 month NII density in order to give an index of percentage increase over the sixth month time point.
Cortical Thickness
Measurements of cortical thickness were determined using the Quantity One program from Bio-Rad (Hercules, CA). The thickness of an average of 3 sections from the left and right hemispheres was determined from the dynamin in situ films.
Statistical Analysis
One-way or Two-Way analysis of variance (ANOVA) was performed where applicable. Two-way ANOVA was followed by Bonferoni/Dunn post-hoc tests. All longitudinal data were analyzed using repeated measures ANOVA. In these repeated measures analysis, no post hoc comparisons between groups at individual time points was performed due to the lack of a time X genotype interaction in all cases. Statistics and Graphs were generated in GraphPad Software (Prism, La Jolla, CA) or Statview 5.1 (SAS Institute, Inc. Cary, NC). P– values of <0.05 were accepted to be significant.
Results
Behavioral Abnormalities in HD mice
Rearings
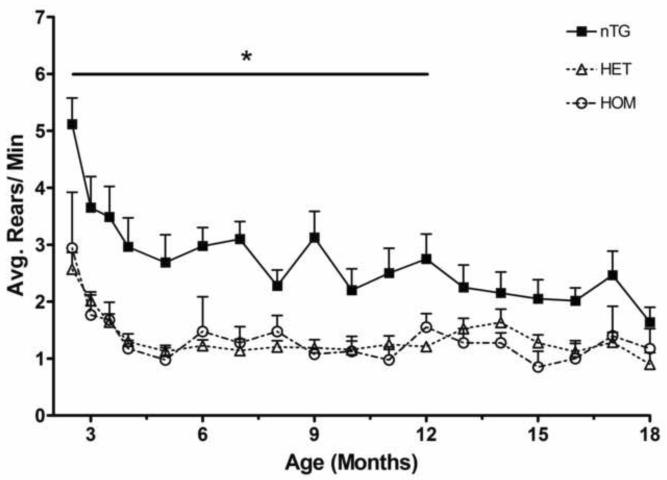
We placed individual mice in a 1 L beaker for 10 minutes in the dark and the number of rears was recorded. From the initial 2.5 month time point on, the nTG mice reared significantly more times than the knock-in mice. nTG mice averaged 2.74 rears per minute while both heterozygote and homozygote averaged 1.36 (Figure 1). There was no difference in rearing between heterozygote CAG 140 and homozygote CAG 140 mice. While there was a significant difference between the knock-in and the nTG mice, this difference was not progressive. There were some inconsistent gender differences prior to 9 months of age but these differences did not contribute significantly to the overall reduction of rearing behavior in CAG 140 mice (supplemental figure 1).
Figure 1.
Longitudinal rearing behavior of the CAG140 knock-in mouse model of Huntington 's disease shows a deficit in heterozygous (Het) and homozygous (Hom) mice. Mice were videotaped in a 1 L beaker for 10 minutes in the dark starting every two weeks for the first four testing periods then once a month thereafter. nTG mice reared significantly more than Het and Hom mice (ANOVA; p<0.0001). There was no significant difference between Het and Hom mice. [n: nTG= 8 (6 males, 2 females), Het= 13 (8 males, 5 females), Hom= 4 (2 males, 2 females)].
Rotarod
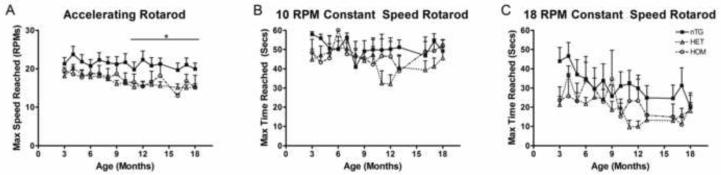
We used two different rotarod tests to determine if the CAG 140 mice exhibited a deficiency in motor skills. The accelerating rotarod started at a speed of 5 RPM (revolutions per minute) and increased in speed by 0.3 RPM each second (Figure 2A). nTG mice reached an average max speed of 21.3 RPM before falling, whereas the heterozygotes and homozygotes reached 16.8 and 17.2 RPM, respectively. The maximum speed that the nTG mice were able to perform was significantly higher than either the heterozygous or the homozygous CAG 140 mice. There was no significant difference between the maximum rotarod performance speeds of the heterozygous or homozygous CAG 140 mice.
Figure 2.
Latency to fall in the accelerating and constant speed rotarod. A) CAG 140 mice (Hets and Homs) displayed rotarod performance deficits beginning at 11 months of age (ANOVA; P<0.05). There was no difference between the performance of the Het and the Hom mice. B) 10 RPM constant speed rotarod testing revealed no differences between nTG and CAG 140 mice. C) All genotypes performed similarly in the 18 RPM constant speed rotarod test, although performance of this task was generally reduced with age (ANOVA; p<0.001). [n: nTG= 8 (6 males, 2 females), Het= 13 (8 males, 5 females), Hom= 4 (2 males, 2 females].
The constant speed rotarod performance was tested at two different speeds, 10 RPM and 18 RPM (Figure 2B and 2C). While the accelerating rotarod showed a difference between the genotypes, constant speed rotarod tests showed no difference between nTG and either the heterozygous CAG 140 or the homozygous CAG 140 mice. On the 10 RPM constant speed rotarod, the nTG, heterozygous and homozygous mice stayed on an average of 50.9 seconds, 43.7 seconds and 47.2 seconds respectively. The 18 RPM constant speed rotarod showed the nTG, heterozygous and homozygous stayed on an average of 31.8, 19.4 and 22.7 respectfully.
Gait Analysis
Like the rearings, gait analysis was performed once a month. The results for both stride length and stride width or ratios between the two measurements did not indicate a difference between any of the genotypes (data not shown).
Expression of the CAG 140 allele leads to transcriptional anomalies in the mouse brain
In situ hybridization of transcripts was performed on nTG, heterozygous and homozygous mice to ascertain the transcriptional changes associated with the expression of the CAG 140 allele of hdh. The striatally enriched transcripts examined here were DARPP-32, ppENK, PDE10a, PDE1b, D2 receptors, CB1 receptors, and NGFi-A, (also known as zif-268). β-actin was used to normalize the other transcripts. We examined 3 month old mice through 19/20 months at intervals of approximately 3 months. We examined the homozygous mice only out to 12 months.
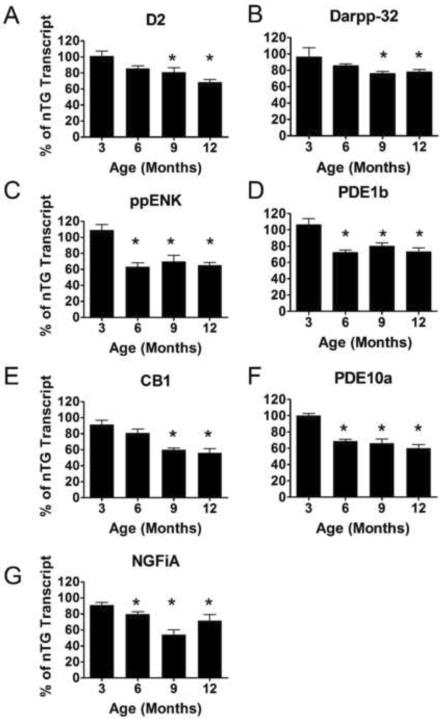
Transcriptional levels of heterozygote compared to nTG (Figure 4) started to decrease at 6 months in four transcripts (PDE1b, NGFiA, CB1 and ppENK). We saw a transcript reduction of approximately 20% when compared to nTG transcript levels at 9 months with the exception of ppENK and DARPP-32 which showed a 7 % increase and a 15 % decreases respectively. Transcripts continued to decrease over time and eventually by 19/20 months the transcripts were approximately reduced by 30% compared to controls. The transcript with the greatest reduction levels compared to nTG was PDE10a which decreased by 36%, while ppENK transcript levels decreased the least, by 14%, at the final time point.
Figure 4.
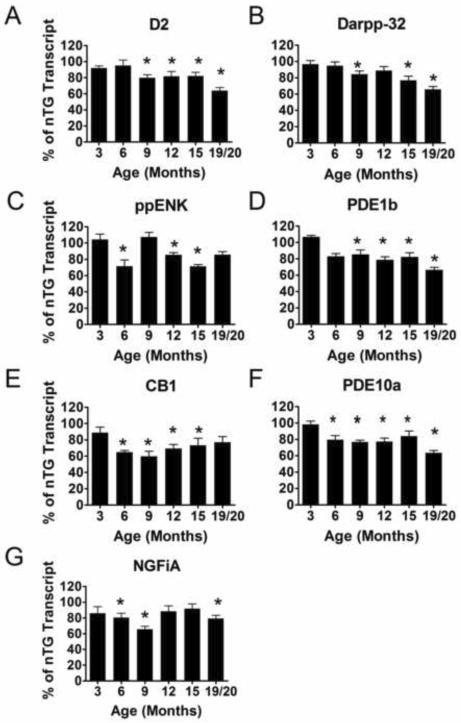
Striatal mRNA transcript in situ 33P hybridization of homozygote CAG 140 knock-in mouse model of Huntington's disease. The striatal area of the Hom mice was analyzed for optical density after mRNA transcript radioactive in situ hybridization in relation to the nTG mRNA transcripts after normalization to β-actin mRNA transcript optical density. An overall transcriptional down regulation was similar to that observed in the Het mice, in A) D2, B) DARPP-32, C) ppENK, D) PDE1b, E) CB1, F) PDE10a and G) NGFiA transcripts (ANOVA; p<0.001). All transcripts except D2 and DARPP-32 showed significant reduction starting at 6 months (C–G). Overall differences from Het and Hom mice are significant over age groups analyzed here (ANOVA; p<0.01). The asterisk represent significant differences between homozygote transcript levels and the nTG transcript levels at the corresponding age (p<0.05). [n: 3 months: nTG=6 (4males, 2 females), Hom=6 (3 males, 3 females), 6 months: nTG=7 (3 males, 4 females), Hom=11 (8 males, 3 females), 9 months: nTG=9 (7 males, 2 females), Hom=7 (4 males, 3 females), 12 months: nTG=3, Hom=8 (4 males, 4 females)].
Similar to heterozygous CAG 140 mice, homozygote transcript levels (Figure 5) began to decrease at 6 months. At 12 months, the average transcript reduction was 35% with the greatest reduction being CB1 at 56%, and the lowest reduction being DARPP-32 at 18% when compared to nTG mice transcript levels. For comparison, heterozygous mice at 12 months have an average reduction of 22% across the transcripts while homozygote at 12 months mice transcripts reduced by an average of 35% compared to nTG.
Figure 5.
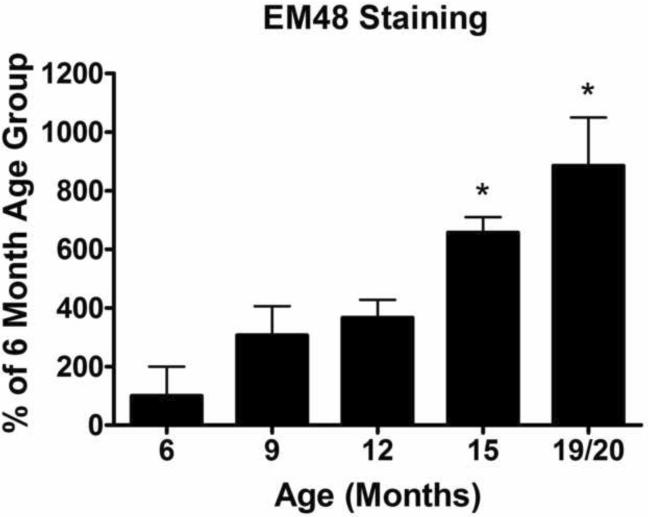
Quantification of Neuronal Intranuclear Inclusions (NIIs). NIIs were quantified using a sterologically based random sampling regime. NII density was estimated identically for each group and normalized to the 6 month age group. 6 months was chosen because NIIs were first detected at this age group. A significant increase in NIIs as the mice age was detected (One-Way ANOVA: p<0.0001). Starting at 15 months, there is a significant increase of NIIs over the 6 month baseline (p<0.01). [n: 6 months: Het= 4 (4 males), 9 months: Het= 5 (4 males, 1 females) 12 months: Het= 4 (2 males, 2 females), 15 months: Het= 3 (3 males), 18/19 months: Het= 5 (3 males, 2 females)].
Older CAG 140 mice exhibit an increase in NIIs
Sections from each genotype at the various ages were stained with EM48 (Millipore™ Billerica, MA) for semi-quantification of the NIIs. Figure 6 shows the percentage increase over the baseline levels of NIIs at 6 months. 6 months was the first time NIIs became evident in any of our mice and we used this time point as our normalization baseline. Similar to the heterozygous mice, the homozygote NIIs show an increase over time compared to the 6 month baseline but these differences did not reach statistical significance (Supplementary Figure 2). nTG mice, as expected, did not show significant inclusion staining at any time point (data not shown).
Figure 6.
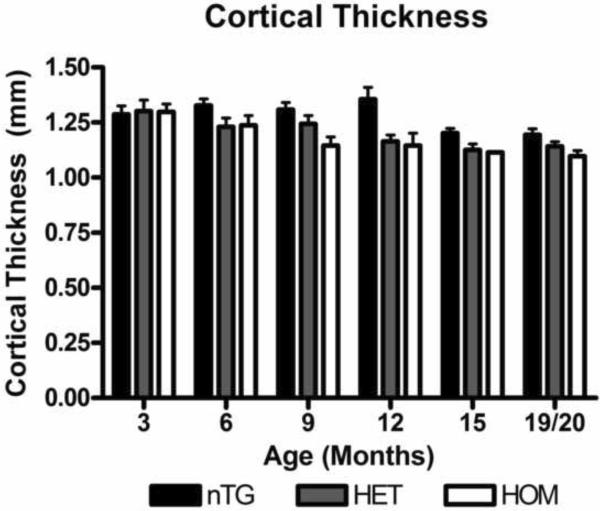
Cortical thickness of the CAG 140 knock-in mice decrease over time compared to the nTG mice. Using the dynamin in-situ film images of the mice examined previously, cortical thickness of the nTG, Het, and Hom mice were measured in millimeters using the Quantity One program. There is an overall significant difference between the nTG and the knock-in mice (ANOVA; p =0.0001). [n: 3 months: nTG=6 (4males, 2 females), Het =9 (5 males, 4 females), Hom=6 (3 males, 3 females), 6 months: nTG=7 (3 males, 4 females), Het= 5 (4 males, 1 females), Hom=11 (8 males, 3 females), 9 months: nTG=9 (7 males, 2 females), Het=6 (5 males, 1 females) Hom=7 (4 males, 3 females), 12 months: nTG=7 (4 males, 3 females), Het=11 (8 males, 3 females), Hom=8 (4 males, 4 females) 15 months: nTG=11 (4 males, 7 females), Het=12 (7 males, 5 females), Hom = 1 males, 19/20 months: nTG=12 (4 males, 8 females) Het=13(7 males, 6 females), Hom= 3 (2 males, 1 females)].
Significant increase in the percent of NIIs seen above baseline did not occur until 15 months for the heterozygous mice. At 19/20 months the heterozygous mice had 5.3 inclusions per 1mm2 equating to 8.8 times as many inclusions compared to the 6 month time point. There was no significant difference between heterozygote and homozygote inclusion counts. For an example of the homozygous NII stain see Supplementary Figure 2.
Cortical Thickness
Using the sections probed for dynamin from the in situ hybridization experiments, we measured the cortical thickness. Cortical thickness has been recently found to decrease in HD patients (Gomez-Anson, et al., 2009, Hobbs, et al., 2010, Hobbs, et al., 2009, Rosas, et al., 2005, Rosas, et al., 2008) and because dynamin transcript signal was found to be located primarily in the cortex, it provides a reasonable metric of cortical dimensions. When compared to the nTG mice, the cortices of the heterozygous and homozygous CAG 140 mice showed a significant difference across the genotypes (Figure 6, ANOVA; p=0.0001)
Using NeuN staining, the number of neurons in this region were counted and showed no significant decrease in the homozygous mice (supplementary Figure 2D). Thus, a neuronal population decline is unlikely to explain the loss of cortical thickness observed here. The region that was counted is outlined in supplementary figure 2D.
Discussion
Recently developed knock-in mouse models of HD have the advantage of recapitulating the human disease by having one or two copies of the mutant gene under the control of the endogenous promoter, as would be the case in the human HD. Here, we decided to characterize the CAG 140 knock-in model originally created by Menalled et al., (2003) to determine the fidelity of this model compared to the human disease with an eye towards eventually study siRNA knock-down technology in this model. When the present study started, there was very little information on the time course of the disease in the knock-in CAG 140 mouse model. The model was initially characterized by Menalled et al. (Menalled, et al., 2003) and more recently studied by Dorner and Hickey (Dorner, et al., 2007, Hickey, et al., 2008), but each of these characterizations examined only a few select time points and only examined homozygous mice.
In previous studies of CAG 140 mice, rotarod deficits were observed at 4 months, slight rearing differences seen at 6.5 months in a novel cage environment, and night time running wheel differences were detected at 4, 6 and 8 months (Hickey, et al., 2008). In these previous studies, while examining differences between nTG and the CAG 140 mice at selected time points, did not investigate both heterozygous and homozygous mice. In the present study, we examined the motor behavior over a longitudinal time course in one set of mice, a corresponding time-matched time course of striatal specific transcripts, and NIIs in a separate cross-sectional study. The present study also examined both the heterozygous and the homozygous CAG 140 mice, which, to date, have not been compared in a single study. The CAG 140 mice do not exhibit the shortened lifespan like the other more widely used models such as the R6/1, and thus allow for a more extended examination of the HD progression.
Behavioral Abnormalities
Behaviorally, the CAG 140 model exhibits slight and subtle overt differences compared nTG mice. There was no obvious gait, size or activity difference between the genotypes by simple observations. However, behavioral testing revealed a relatively mild pathological phenotype. Unlike other models, no clasping behavior was observed. As early as 2.5 months of age, we observed a significant difference in the rearing behavior of the HD mice (Figure 1). In HD patients, individuals have been reported to have slight motor, language and cognitive skill deficits that precede the major HD symptoms and could be an early indication of the disease (Feigin, et al., 2006, Gabrieli, et al., 1997, Ghilardi, et al., 2008, Robins Wahlin, et al., 2007) and it is not inconceivable that CAG 140 rearing behavior may be a manifestation of similar subtle early deficit in HD mice. While other models have shown gait abnormalities, no significant differences were seen in our study. Neither gait width and gait length nor the ratio between the two was significantly different from nTG (data not shown).
In HD, motor and cognitive skills progressively worsen as the individual ages (Harper, 1993, Josiassen, et al., 1983, Lawrence, et al., 1996). In the CAG 140 knock-in mouse model examined here, there was little to no progressive decline in the behavioral tasks utilized here. Rearing and rotarod behavioral tasks showed a large significant deficit when comparing nTG mice to the knock-in mice, but this deficit did not increase as the mice aged. In fact, for the rearing behavior, nTG mice approached the knock-in rearing numbers towards the end of this study indicating a possible age-related decline in normal mice. Gait analysis did not show a significant difference between nTG and knock-in mice. The lack of a progressive behavioral deficit in the CAG 140 mouse model is a drawback of the model but does not preclude the possibility that other behavioral paradigms might uncover a progressive deficit especially in light of the progressive neuropathological deficits uncovered in this study (see below).
Transcriptional Dysfunction
In humans, as well as other mouse models of HD, striatal specific transcript and protein levels are known to be progressively reduced. We examined striatal DARPP-32, ppENK, PDE10a, PDE1b, D2, CB1, and NGFi-A transcripts because of various reports that have shown these transcripts are altered in HD and in prominent HD mouse models (Albin, et al., 1991, Augood, et al., 1997, Bibb, et al., 2000, Chan, et al., 2002, Denovan-Wright and Robertson, 2000, Hebb, et al., 2004, Hu, et al., 2004, Luthi-Carter, et al., 2000, McCaw, et al., 2004, Menalled, et al., 2000, Richfield, et al., 1995). The transcripts examined here, are all enriched in the striatum and have a role in cell signaling to varying degrees (Herkenham, et al., 1991, Le Moine, et al., 1991, Le Moine, et al., 1990, Moratalla, et al., 1992, Reiner, et al., 1988, Scott, et al., 2005, Tsou, et al., 1998, Walaas and Greengard, 1984). Both D2 and CB1 receptors, when activated, help to the regulate the intracellular levels of cAMP (Bidaut-Russell and Howlett, 1991, Glass and Felder, 1997, Nishi, et al., 1997, Stoof and Kebabian, 1981, Stoof and Verheijden, 1986) which in turn regulates the phosphorylation of the striatal enriched protein DARRP-32 (Nishi, et al., 1997, Stoof and Kebabian, 1981, Stoof and Verheijden, 1986). PDE10a and PDE1b are striatal specific and have been shown to hydrolyze cAMP (Bender and Beavo, 2006, Fujishige, et al., 1999, Lakics, et al., 2010, Loughney, et al., 1999, Reed, et al., 1998). Studies regarding upstream binding sites of the gene that codes PDE10a revealed that there is a NGFiA binding site which suggests that NGFiA could help regulate the transcription of PDE10a (Hu, et al., 2004). The interconnections between these striatally enriched transcripts highlight their functional significance especially since mutant htt has been associated with decreases in these transcripts.
Indeed, just as seen in the transgenic R6 mouse model (Bibb, et al., 2000, Denovan-Wright and Robertson, 2000, Dowie, et al., 2009, Hebb, et al., 2004, Hebb, et al., 2008, Hu, et al., 2004, McCaw, et al., 2004, van Dellen, et al., 2000), and various other mouse models of HD (Chan, et al., 2002, Luthi-Carter, et al., 2000, Luthi-Carter, et al., 2002, Menalled, et al., 2000), there is an age-dependent decrease of striatal specific transcripts in the CAG 140 model. There is a decrease of the measured striatal mRNA levels in heterozygous CAG 140 mice starting at 6 months of age and a reduction of nearly 20% in most transcripts by 9 months of age. The trend for reduced striatal specific transcripts in CAG 140 mice continues as the mice age and at 19/20 months of age, an average reduction of 30% is observed. Homozygous mice already display a reduction of 20% by 6 months of age, and at 12 months of age, the homozygous mice show a reduction of approximately 35% which is an earlier onset of striatal transcriptional than heterozygous mice. The difference between the heterozygous and the homozygous striatal transcript levels seen at 12 months may indicate a gene dosing effect of the two expanded knock-in htt alleles present in the homozygous mice compared to the single copy in the heterozygotes. The decrease in relative mRNA of these various striatal transcripts in the CAG 140 mice corroborates what has been seen in the human HD as well as what has been seen in other mouse models of HD.
Neuronal Intranuclear Inclusions
Another well established pathological component in HD and HD models, in addition to transcript dysregulation, is the presence of the NIIs. In the present study, we have used an unbiased sampling method to determine NII density over the life-span of the CAG 140 mouse. We normalized the NII densities to the six month time point which was the first age where we could detect significant NIIs. Visualizing these data demonstrates an age related progression in CAG 140 mice (Figure 5).
Wild-type htt has been shown to associate with various transcription factors and that association is altered when mutant htt is present (Marcora, et al., 2003, Marcora and Kennedy, , Steffan, et al., 2000, Sugars, et al., 2004, Takano and Gusella, 2002, Zuccato, et al., 2007, Zuccato, et al., 2001). It would follow that the function and or availability of the transcription factors to perform their tasks would be altered if sequestered by mutant htt. Our observations support the hypothesis that transcription factor segregation due to mutant htt could in turn decrease mRNA levels (Figures 3 and 4). We see significant decrease of transcripts before a significant increase of NIIs in the CAG 140 mice. This may suggest sequestering of transcriptional factors takes place before the actual formation of detectable inclusion bodies (Schaffar, et al., 2004, Takahashi, et al., 2005, Yu, et al., 2002). While sequestering of transcription factors prior to the formation of NIIs may account for our early behavioral data, from our data we cannot conclude that this is occurring in the CAG 140 model.
Figure 3.
Striatal mRNA transcript in situ 33P hybridization of heterozygote CAG 140 knock-in mouse model of Huntington's disease. The striatal area of the Het mice was analyzed for optical density after mRNA transcript radioactive in situ hybridization in relation to the nTG mRNA transcripts after normalization to β-actin mRNA transcript optical density. An overall transcriptional down regulation was observed in A) D2, B) DARPP-32, C) ppENK, D) PDE1b, E) CB1, F) PDE10a and G) NGFiA (ANOVA; p<0.001) In D2, DARPP-32 and PDE1b (A, B and D) the transcriptional down regulation begins at 9 months and continues through the last age group. In CB1 and PDE10a transcripts (E and F), reductions begin earlier at 6 months continue to be significant through the course of the cross sectional analysis. ppENK and NGFiA (C and G) showed variable but over all transcriptional down regulation. Asterisk represent significant differences between heterozygote transcript levels and the nTG transcript levels at the corresponding age (p<0.05). [n: 3 months: nTG=6 (4 males, 2 females), Het =9 (5 males, 4 females), 6 months: nTG=7 (3 males, 4 females), Het= 5 (4 males, 1 females), 9 months: nTG=9 (7 males, 2 females), Het=6 (5 males, 1 females), 12 months: nTG=3(1 males, 2 females), Het=11 (8 males, 3 females), 15 months: nTG=11 (4 males, 7 females), Het=12 (7 males, 5 females), 19/20 months: nTG=12 (4 males, 8 females) Het=13 (7 males, 6 females)].
Cortical Thinning
Cortical thinning has recently been examined during the progression of HD. A number of studies have determined via functional magnetic resonance imaging (fMRI) that as the disease progresses in humans cortical areas begin to thin or atrophy (Gomez-Anson, et al., 2009, Hobbs, et al., 2009, Rosas, et al., 2008). The CAG 140 mouse model displays an overall significant difference between the genotypes (Figure 6). nTG mice also showed a significant decline in the cortical thickness over time. The nTG cortical thinning could be attributed to aging which has been seen in normal human aging (Fotenos, et al., 2005, Resnick, et al., 2003, Scahill, et al., 2003). Here, the knock-in mice might reach a threshold of cortical thinning and the nTG mice reach that point at a later stage. Examining neuron density at 9 to 12 months of the homozygous mice, did not reveal a consistent decrease in the number of neurons as visualized by NeuN staining (Supplementary Figure 1D). This lack of NeuN differences suggests that atrophy or loss of cell types other than neurons or significant atrophy of cortical neurons is potentially responsible for the observed cortical thinning. The implications of such cortical thinning in the CAG 140 mice cannot be determined from the present study. More precise learning behavioral tasks may provide a clue as to the effects of such cortical loss.
While a non-invasive method of determining transcript and/or NIIs in humans has not been developed, post-mortem studies have indicated that, in all likelihood, a progressive decrease of transcripts and a progressive increase of NIIIs occurs in HD. Cortical thinning may provide a real-time measurable progressive pathological marker for the CAG 140 mice. Additionally we have found a series of behavioral tests that show long-term, quantifiable, non-progressive deficits when compared to nTG. These tests along with other behavioral assays such as the open field anxiety test (Belzung, et al., 2001) and the object recognition test (Devito and Eichenbaum, 2010) could be used to ascertain the effectiveness of various treatments of HD.
In conclusion, the CAG 140 knock-in mouse model of HD has many aspects that are similar to the human disease. Not all of the observed pathological aspects observed in CAG 140 mimic the human disease perfectly but the histopathological characteristics do recapitulate many observations in the human disease and may provide good benchmarks for determining the efficacy of drugs and treatments. The progressive decreases in transcripts and cortical thinning as well as the increase in NIIs over time allow us to gauge treatments at various points in the disease. Thus, given the present and previously reported data (Dorner, et al., 2007, Hickey, et al., 2008, Menalled, et al., 2003), the best currently available functional outcome measures for the CAG 140 mouse model of HD are probably striatal transcripts, NIIs, and cortical thickness measurements.
Supplementary Material
Supplementary Figure 1 A) nTG mice's rearing behavior separated by gender and B) CAG 140 mice's rearing behavior separated by gender are shown here. Although it appears that male nTGs rear more than female nTGs prior to 9 months of age in (A), there was neither a significant main effect of gender (F(1,19) = 3.5, p = 0.07) nor was there a significant genotype × gender interaction (F(2,19) = 1.19, p > 0.3). [n: nTG= 8 (6 M, 2 F), Het= 13 (8 M, 5 F), Hom= 4 (2 M, 2 F)]. There were no gender differences detected in any behavioral or post mortem measurements throughout the rest of the experiment.
Supplementary Figure 2 A) Representative images of (i) nTG, (ii) Het, and (iii) Hom striatal area containing NIIs at 12 months. Arrows point to an example of NIIs. Scale bar represents 25μm. B) Representative image of cortical region where NeuN+ neurons were counted (black box) and measurement of cortical thickness (white arrow). Scale bar represents 250μm. Contrast levels were adjusted in (A and B) for better visualization in this figure. C) Hom progression of NIIs as a percent over the 6 month baseline. D) NeuN+ cells average per 1mm2. 9 month and 12 month old nTG and homozygote mice were stained with NeuN to determine the total cortical neurons in the given area seen in B). CC= Corpus Collosum.
Acknowledgments
This work was supported by NIH grant 1R01 NS48588-01 to RJM and EDW. ACR was supported by NIH training grant T32AG00196. The authors would like to thank Izzie Williams for technical support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albin RL, Qin Y, Young AB, Penney JB, Chesselet MF. Preproenkephalin messenger RNA-containing neurons in striatum of patients with symptomatic and presymptomatic Huntington's disease: an in situ hybridization study. Ann Neurol. 1991;30:542–549. doi: 10.1002/ana.410300406. [DOI] [PubMed] [Google Scholar]
- Augood SJ, Faull RL, Emson PC. Dopamine D1 and D2 receptor gene expression in the striatum in Huntington's disease. Ann Neurol. 1997;42:215–221. doi: 10.1002/ana.410420213. [DOI] [PubMed] [Google Scholar]
- Becher MW, Kotzuk JA, Sharp AH, Davies SW, Bates GP, Price DL, Ross CA. Intranuclear neuronal inclusions in Huntington's disease and dentatorubral and pallidoluysian atrophy: correlation between the density of inclusions and IT15 CAG triplet repeat length. Neurobiol Dis. 1998;4:387–397. doi: 10.1006/nbdi.1998.0168. [DOI] [PubMed] [Google Scholar]
- Belzung C, El Hage W, Moindrot N, Griebel G. Behavioral and neurochemical changes following predatory stress in mice. Neuropharmacology. 2001;41:400–408. doi: 10.1016/s0028-3908(01)00072-7. [DOI] [PubMed] [Google Scholar]
- Bender AT, Beavo JA. Cyclic nucleotide phosphodiesterases: molecular regulation to clinical use. Pharmacol Rev. 2006;58:488–520. doi: 10.1124/pr.58.3.5. [DOI] [PubMed] [Google Scholar]
- Bibb JA, Yan Z, Svenningsson P, Snyder GL, Pieribone VA, Horiuchi A, Nairn AC, Messer A, Greengard P. Severe deficiencies in dopamine signaling in presymptomatic Huntington's disease mice. Proc Natl Acad Sci U S A. 2000;97:6809–6814. doi: 10.1073/pnas.120166397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidaut-Russell M, Howlett AC. Cannabinoid receptor-regulated cyclic AMP accumulation in the rat striatum. J Neurochem. 1991;57:1769–1773. doi: 10.1111/j.1471-4159.1991.tb06379.x. [DOI] [PubMed] [Google Scholar]
- Carter RJ, Lione LA, Humby T, Mangiarini L, Mahal A, Bates GP, Dunnett SB, Morton AJ. Characterization of progressive motor deficits in mice transgenic for the human Huntington's disease mutation. J Neurosci. 1999;19:3248–3257. doi: 10.1523/JNEUROSCI.19-08-03248.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EY, Luthi-Carter R, Strand A, Solano SM, Hanson SA, DeJohn MM, Kooperberg C, Chase KO, DiFiglia M, Young AB, Leavitt BR, Cha JH, Aronin N, Hayden MR, Olson JM. Increased huntingtin protein length reduces the number of polyglutamine-induced gene expression changes in mouse models of Huntington's disease. Hum Mol Genet. 2002;11:1939–1951. doi: 10.1093/hmg/11.17.1939. [DOI] [PubMed] [Google Scholar]
- Craufurd D, Thompson JC, Snowden JS. Behavioral changes in Huntington Disease. Neuropsychiatry Neuropsychol Behav Neurol. 2001;14:219–226. [PubMed] [Google Scholar]
- Davies SW, Turmaine M, Cozens BA, DiFiglia M, Sharp AH, Ross CA, Scherzinger E, Wanker EE, Mangiarini L, Bates GP. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- Denovan-Wright EM, Robertson HA. Cannabinoid receptor messenger RNA levels decrease in a subset of neurons of the lateral striatum, cortex and hippocampus of transgenic Huntington's disease mice. Neuroscience. 2000;98:705–713. doi: 10.1016/s0306-4522(00)00157-3. [DOI] [PubMed] [Google Scholar]
- Devito LM, Eichenbaum H. Distinct contributions of the hippocampus and medial prefrontal cortex to the “what-where-when” components of episodic-like memory in mice. Behav Brain Res. 2010;215:318–325. doi: 10.1016/j.bbr.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFiglia M, Sena-Esteves M, Chase K, Sapp E, Pfister E, Sass M, Yoder J, Reeves P, Pandey RK, Rajeev KG, Manoharan M, Sah DW, Zamore PD, Aronin N. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc Natl Acad Sci U S A. 2007;104:17204–17209. doi: 10.1073/pnas.0708285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner JL, Miller BR, Barton SJ, Brock TJ, Rebec GV. Sex differences in behavior and striatal ascorbate release in the 140 CAG knock-in mouse model of Huntington's disease. Behav Brain Res. 2007;178:90–97. doi: 10.1016/j.bbr.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowie MJ, Bradshaw HB, Howard ML, Nicholson LF, Faull RL, Hannan AJ, Glass M. Altered CB1 receptor and endocannabinoid levels precede motor symptom onset in a transgenic mouse model of Huntington's disease. Neuroscience. 2009;163:456–465. doi: 10.1016/j.neuroscience.2009.06.014. [DOI] [PubMed] [Google Scholar]
- Drouet V, Perrin V, Hassig R, Dufour N, Auregan G, Alves S, Bonvento G, Brouillet E, Luthi-Carter R, Hantraye P, Deglon N. Sustained effects of nonallele-specific Huntingtin silencing. Ann Neurol. 2009;65:276–285. doi: 10.1002/ana.21569. [DOI] [PubMed] [Google Scholar]
- Feigin A, Ghilardi MF, Huang C, Ma Y, Carbon M, Guttman M, Paulsen JS, Ghez CP, Eidelberg D. Preclinical Huntington's disease: compensatory brain responses during learning. Ann Neurol. 2006;59:53–59. doi: 10.1002/ana.20684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folstein SE, Chase GA, Wahl WE, McDonnell AM, Folstein MF. Huntington disease in Maryland: clinical aspects of racial variation. Am J Hum Genet. 1987;41:168–179. [PMC free article] [PubMed] [Google Scholar]
- Fotenos AF, Snyder AZ, Girton LE, Morris JC, Buckner RL. Normative estimates of cross-sectional and longitudinal brain volume decline in aging and AD. Neurology. 2005;64:1032–1039. doi: 10.1212/01.WNL.0000154530.72969.11. [DOI] [PubMed] [Google Scholar]
- Fujishige K, Kotera J, Michibata H, Yuasa K, Takebayashi S, Okumura K, Omori K. Cloning and characterization of a novel human phosphodiesterase that hydrolyzes both cAMP and cGMP (PDE10A) J Biol Chem. 1999;274:18438–18445. doi: 10.1074/jbc.274.26.18438. [DOI] [PubMed] [Google Scholar]
- Gabrieli JD, Stebbins GT, Singh J, Willingham DB, Goetz CG. Intact mirror-tracing and impaired rotary-pursuit skill learning in patients with Huntington's disease: evidence for dissociable memory systems in skill learning. Neuropsychology. 1997;11:272–281. doi: 10.1037//0894-4105.11.2.272. [DOI] [PubMed] [Google Scholar]
- Ghilardi MF, Silvestri G, Feigin A, Mattis P, Zgaljardic D, Moisello C, Crupi D, Marinelli L, Dirocco A, Eidelberg D. Implicit and explicit aspects of sequence learning in pre-symptomatic Huntington's disease. Parkinsonism Relat Disord. 2008;14:457–464. doi: 10.1016/j.parkreldis.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass M, Felder CC. Concurrent stimulation of cannabinoid CB1 and dopamine D2 receptors augments cAMP accumulation in striatal neurons: evidence for a Gs linkage to the CB1 receptor. J Neurosci. 1997;17:5327–5333. doi: 10.1523/JNEUROSCI.17-14-05327.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Anson B, Alegret M, Munoz E, Monte GC, Alayrach E, Sanchez A, Boada M, Tolosa E. Prefrontal cortex volume reduction on MRI in preclinical Huntington's disease relates to visuomotor performance and CAG number. Parkinsonism Relat Disord. 2009;15:213–219. doi: 10.1016/j.parkreldis.2008.05.010. [DOI] [PubMed] [Google Scholar]
- Graveland GA, Williams RS, DiFiglia M. Evidence for degenerative and regenerative changes in neostriatal spiny neurons in Huntington's disease. Science. 1985;227:770–773. doi: 10.1126/science.3155875. [DOI] [PubMed] [Google Scholar]
- Gutekunst CA, Li SH, Yi H, Mulroy JS, Kuemmerle S, Jones R, Rye D, Ferrante RJ, Hersch SM, Li XJ. Nuclear and neuropil aggregates in Huntington's disease: relationship to neuropathology. J Neurosci. 1999;19:2522–2534. doi: 10.1523/JNEUROSCI.19-07-02522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday GM, McRitchie DA, Macdonald V, Double KL, Trent RJ, McCusker E. Regional specificity of brain atrophy in Huntington's disease. Exp Neurol. 1998;154:663–672. doi: 10.1006/exnr.1998.6919. [DOI] [PubMed] [Google Scholar]
- Harper PS. Clinical consequences of isolating the gene for Huntington's disease. Bmj. 1993;307:397–398. doi: 10.1136/bmj.307.6901.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper SQ, Staber PD, He X, Eliason SL, Martins IH, Mao Q, Yang L, Kotin RM, Paulson HL, Davidson BL. RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc Natl Acad Sci U S A. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebb AL, Robertson HA, Denovan-Wright EM. Striatal phosphodiesterase mRNA and protein levels are reduced in Huntington's disease transgenic mice prior to the onset of motor symptoms. Neuroscience. 2004;123:967–981. doi: 10.1016/j.neuroscience.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Hebb AL, Robertson HA, Denovan-Wright EM. Phosphodiesterase 10A inhibition is associated with locomotor and cognitive deficits and increased anxiety in mice. Eur Neuropsychopharmacol. 2008;18:339–363. doi: 10.1016/j.euroneuro.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Herkenham M, Lynn AB, de Costa BR, Richfield EK. Neuronal localization of cannabinoid receptors in the basal ganglia of the rat. Brain Res. 1991;547:267–274. doi: 10.1016/0006-8993(91)90970-7. [DOI] [PubMed] [Google Scholar]
- Hickey MA, Kosmalska A, Enayati J, Cohen R, Zeitlin S, Levine MS, Chesselet MF. Extensive early motor and non-motor behavioral deficits are followed by striatal neuronal loss in knock-in Huntington's disease mice. Neuroscience. 2008;157:280–295. doi: 10.1016/j.neuroscience.2008.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs NZ, Barnes J, Frost C, Henley SM, Wild EJ, Macdonald K, Barker RA, Scahill RI, Fox NC, Tabrizi SJ. Onset and progression of pathologic atrophy in Huntington disease: a longitudinal MR imaging study. AJNR Am J Neuroradiol. 2010;31:1036–1041. doi: 10.3174/ajnr.A2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs NZ, Henley SM, Ridgway G, Wild EJ, Barker R, Scahill RI, Barnes J, Fox NC, Tabrizi S. The Progression of Regional Atrophy in Premanifest and Early Huntington's Disease: A Longitudinal Voxel-Based Morphometry Study. J Neurol Neurosurg Psychiatry. 2009 doi: 10.1136/jnnp.2009.190702. [DOI] [PubMed] [Google Scholar]
- Hodgson JG, Agopyan N, Gutekunst CA, Leavitt BR, LePiane F, Singaraja R, Smith DJ, Bissada N, McCutcheon K, Nasir J, Jamot L, Li XJ, Stevens ME, Rosemond E, Roder JC, Phillips AG, Rubin EM, Hersch SM, Hayden MR. A YAC mouse model for Huntington's disease with full-length mutant huntingtin, cytoplasmic toxicity, and selective striatal neurodegeneration. Neuron. 1999;23:181–192. doi: 10.1016/s0896-6273(00)80764-3. [DOI] [PubMed] [Google Scholar]
- Hu H, McCaw EA, Hebb AL, Gomez GT, Denovan-Wright EM. Mutant huntingtin affects the rate of transcription of striatum-specific isoforms of phosphodiesterase 10A. Eur J Neurosci. 2004;20:3351–3363. doi: 10.1111/j.1460-9568.2004.03796.x. [DOI] [PubMed] [Google Scholar]
- Josiassen RC, Curry LM, Mancall EL. Development of neuropsychological deficits in Huntington's disease. Arch Neurol. 1983;40:791–796. doi: 10.1001/archneur.1983.04050120041005. [DOI] [PubMed] [Google Scholar]
- Kuhn A, Goldstein DR, Hodges A, Strand AD, Sengstag T, Kooperberg C, Becanovic K, Pouladi MA, Sathasivam K, Cha JH, Hannan AJ, Hayden MR, Leavitt BR, Dunnett SB, Ferrante RJ, Albin R, Shelbourne P, Delorenzi M, Augood SJ, Faull RL, Olson JM, Bates GP, Jones L, Luthi-Carter R. Mutant huntingtin's effects on striatal gene expression in mice recapitulate changes observed in human Huntington's disease brain and do not differ with mutant huntingtin length or wild-type huntingtin dosage. Hum Mol Genet. 2007;16:1845–1861. doi: 10.1093/hmg/ddm133. [DOI] [PubMed] [Google Scholar]
- Lakics V, Karran EH, Boess FG. Quantitative comparison of phosphodiesterase mRNA distribution in human brain and peripheral tissues. Neuropharmacology. 2010;59:367–374. doi: 10.1016/j.neuropharm.2010.05.004. [DOI] [PubMed] [Google Scholar]
- Lawrence AD, Hodges JR, Rosser AE, Kershaw A, ffrench-Constant C, Rubinsztein DC, Robbins TW, Sahakian BJ. Evidence for specific cognitive deficits in preclinical Huntington's disease. Brain. 1998;121(Pt 7):1329–1341. doi: 10.1093/brain/121.7.1329. [DOI] [PubMed] [Google Scholar]
- Lawrence AD, Sahakian BJ, Hodges JR, Rosser AE, Lange KW, Robbins TW. Executive and mnemonic functions in early Huntington's disease. Brain. 1996;119(Pt 5):1633–1645. doi: 10.1093/brain/119.5.1633. [DOI] [PubMed] [Google Scholar]
- Le Moine C, Normand E, Bloch B. Phenotypical characterization of the rat striatal neurons expressing the D1 dopamine receptor gene. Proc Natl Acad Sci U S A. 1991;88:4205–4209. doi: 10.1073/pnas.88.10.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Moine C, Normand E, Guitteny AF, Fouque B, Teoule R, Bloch B. Dopamine receptor gene expression by enkephalin neurons in rat forebrain. Proc Natl Acad Sci U S A. 1990;87:230–234. doi: 10.1073/pnas.87.1.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine MS, Klapstein GJ, Koppel A, Gruen E, Cepeda C, Vargas ME, Jokel ES, Carpenter EM, Zanjani H, Hurst RS, Efstratiadis A, Zeitlin S, and Chesselet MF. Enhanced sensitivity to N-methyl-D-aspartate receptor activation in transgenic and knockin mouse models of Huntington's disease. J Neurosci Res. 1999;58:515–532. [PubMed] [Google Scholar]
- Lin CH, Tallaksen-Greene S, Chien WM, Cearley JA, Jackson WS, Crouse AB, Ren S, Li XJ, Albin RL, Detloff PJ. Neurological abnormalities in a knock-in mouse model of Huntington's disease. Hum Mol Genet. 2001;10:137–144. doi: 10.1093/hmg/10.2.137. [DOI] [PubMed] [Google Scholar]
- Loughney K, Snyder PB, Uher L, Rosman GJ, Ferguson K, Florio VA. Isolation and characterization of PDE10A, a novel human 3', 5'-cyclic nucleotide phosphodiesterase. Gene. 1999;234:109–117. doi: 10.1016/s0378-1119(99)00171-7. [DOI] [PubMed] [Google Scholar]
- Luthi-Carter R, Strand A, Peters NL, Solano SM, Hollingsworth ZR, Menon AS, Frey AS, Spektor BS, Penney EB, Schilling G, Ross CA, Borchelt DR, Tapscott SJ, Young AB, Cha JH, Olson JM. Decreased expression of striatal signaling genes in a mouse model of Huntington's disease. Hum Mol Genet. 2000;9:1259–1271. doi: 10.1093/hmg/9.9.1259. [DOI] [PubMed] [Google Scholar]
- Luthi-Carter R, Strand AD, Hanson SA, Kooperberg C, Schilling G, La Spada AR, Merry DE, Young AB, Ross CA, Borchelt DR, Olson JM. Polyglutamine and transcription: gene expression changes shared by DRPLA and Huntington's disease mouse models reveal context-independent effects. Hum Mol Genet. 2002;11:1927–1937. doi: 10.1093/hmg/11.17.1927. [DOI] [PubMed] [Google Scholar]
- Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies SW, Bates GP. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- Marcora E, Gowan K, Lee JE. Stimulation of NeuroD activity by huntingtin and huntingtin-associated proteins HAP1 and MLK2. Proc Natl Acad Sci U S A. 2003;100:9578–9583. doi: 10.1073/pnas.1133382100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcora E, Kennedy MB. The Huntington's disease mutation impairs Huntingtin's role in the transport of NF-{kappa}B from the synapse to the nucleus. Hum Mol Genet. doi: 10.1093/hmg/ddq358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaw EA, Hu H, Gomez GT, Hebb AL, Kelly ME, Denovan-Wright EM. Structure, expression and regulation of the cannabinoid receptor gene (CB1) in Huntington's disease transgenic mice. Eur J Biochem. 2004;271:4909–4920. doi: 10.1111/j.1432-1033.2004.04460.x. [DOI] [PubMed] [Google Scholar]
- Menalled L, Zanjani H, MacKenzie L, Koppel A, Carpenter E, Zeitlin S, Chesselet MF. Decrease in striatal enkephalin mRNA in mouse models of Huntington's disease. Exp Neurol. 2000;162:328–342. doi: 10.1006/exnr.1999.7327. [DOI] [PubMed] [Google Scholar]
- Menalled LB, Sison JD, Dragatsis I, Zeitlin S, Chesselet MF. Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington's disease with 140 CAG repeats. J Comp Neurol. 2003;465:11–26. doi: 10.1002/cne.10776. [DOI] [PubMed] [Google Scholar]
- Menalled LB, Sison JD, Wu Y, Olivieri M, Li XJ, Li H, Zeitlin S, Chesselet MF. Early motor dysfunction and striosomal distribution of huntingtin microaggregates in Huntington's disease knock-in mice. J Neurosci. 2002;22:8266–8276. doi: 10.1523/JNEUROSCI.22-18-08266.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moratalla R, Robertson HA, Graybiel AM. Dynamic regulation of NGFI-A (zif268, egr1) gene expression in the striatum. J Neurosci. 1992;12:2609–2622. doi: 10.1523/JNEUROSCI.12-07-02609.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton AJ, Lagan MA, Skepper JN, Dunnett SB. Progressive formation of inclusions in the striatum and hippocampus of mice transgenic for the human Huntington's disease mutation. J Neurocytol. 2000;29:679–702. doi: 10.1023/a:1010887421592. [DOI] [PubMed] [Google Scholar]
- Naarding P, Kremer HP, Zitman FG. Huntington's disease: a review of the literature on prevalence and treatment of neuropsychiatric phenomena. Eur Psychiatry. 2001;16:439–445. doi: 10.1016/s0924-9338(01)00604-6. [DOI] [PubMed] [Google Scholar]
- Nishi A, Snyder GL, Greengard P. Bidirectional regulation of DARPP-32 phosphorylation by dopamine. J Neurosci. 1997;17:8147–8155. doi: 10.1523/JNEUROSCI.17-21-08147.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinemann A, Schuller S, Pohl C, Jahn T, Weindl A, Kassubek J. Executive dysfunction in early stages of Huntington's disease is associated with striatal and insular atrophy: a neuropsychological and voxel-based morphometric study. J Neurol Sci. 2005;239:11–19. doi: 10.1016/j.jns.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Pfister EL, Kennington L, Straubhaar J, Wagh S, Liu W, DiFiglia M, Landwehrmeyer B, Vonsattel JP, Zamore PD, Aronin N. Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington's disease patients. Curr Biol. 2009;19:774–778. doi: 10.1016/j.cub.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips W, Shannon KM, Barker RA. The current clinical management of Huntington's disease. Mov Disord. 2008;23:1491–1504. doi: 10.1002/mds.21971. [DOI] [PubMed] [Google Scholar]
- Reed TM, Browning JE, Blough RI, Vorhees CV, Repaske DR. Genomic structure and chromosome location of the murine PDE1B phosphodiesterase gene. Mamm Genome. 1998;9:571–576. doi: 10.1007/s003359900820. [DOI] [PubMed] [Google Scholar]
- Reiner A, Albin RL, Anderson KD, D'Amato CJ, Penney JB, Young AB. Differential loss of striatal projection neurons in Huntington disease. Proc Natl Acad Sci U S A. 1988;85:5733–5737. doi: 10.1073/pnas.85.15.5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick SM, Pham DL, Kraut MA, Zonderman AB, Davatzikos C. Longitudinal magnetic resonance imaging studies of older adults: a shrinking brain. J Neurosci. 2003;23:3295–3301. doi: 10.1523/JNEUROSCI.23-08-03295.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richfield EK, Maguire-Zeiss KA, Cox C, Gilmore J, Voorn P. Reduced expression of preproenkephalin in striatal neurons from Huntington's disease patients. Ann Neurol. 1995;37:335–343. doi: 10.1002/ana.410370309. [DOI] [PubMed] [Google Scholar]
- Robins Wahlin TB, Lundin A, Dear K. Early cognitive deficits in Swedish gene carriers of Huntington's disease. Neuropsychology. 2007;21:31–44. doi: 10.1037/0894-4105.21.1.31. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Lebron E, Denovan-Wright EM, Nash K, Lewin AS, Mandel RJ. Intrastriatal rAAV-mediated delivery of anti-huntingtin shRNAs induces partial reversal of disease progression in R6/1 Huntington's disease transgenic mice. Mol Ther. 2005;12:618–633. doi: 10.1016/j.ymthe.2005.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas HD, Hevelone ND, Zaleta AK, Greve DN, Salat DH, Fischl B. Regional cortical thinning in preclinical Huntington disease and its relationship to cognition. Neurology. 2005;65:745–747. doi: 10.1212/01.wnl.0000174432.87383.87. [DOI] [PubMed] [Google Scholar]
- Rosas HD, Salat DH, Lee SY, Zaleta AK, Pappu V, Fischl B, Greve D, Hevelone N, Hersch SM. Cerebral cortex and the clinical expression of Huntington's disease: complexity and heterogeneity. Brain. 2008;131:1057–1068. doi: 10.1093/brain/awn025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruocco HH, Lopes-Cendes I, Li LM, Santos-Silva M, Cendes F. Striatal and extrastriatal atrophy in Huntington's disease and its relationship with length of the CAG repeat. Braz J Med Biol Res. 2006;39:1129–1136. doi: 10.1590/s0100-879x2006000800016. [DOI] [PubMed] [Google Scholar]
- Scahill RI, Frost C, Jenkins R, Whitwell JL, Rossor MN, Fox NC. A longitudinal study of brain volume changes in normal aging using serial registered magnetic resonance imaging. Arch Neurol. 2003;60:989–994. doi: 10.1001/archneur.60.7.989. [DOI] [PubMed] [Google Scholar]
- Schaffar G, Breuer P, Boteva R, Behrends C, Tzvetkov N, Strippel N, Sakahira H, Siegers K, Hayer-Hartl M, Hartl FU. Cellular toxicity of polyglutamine expansion proteins: mechanism of transcription factor deactivation. Mol Cell. 2004;15:95–105. doi: 10.1016/j.molcel.2004.06.029. [DOI] [PubMed] [Google Scholar]
- Schilling G, Becher MW, Sharp AH, Jinnah HA, Duan K, Kotzuk JA, Slunt HH, Ratovitski T, Cooper JK, Jenkins NA, Copeland NG, Price DL, Ross CA, Borchelt DR. Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum Mol Genet. 1999;8:397–407. doi: 10.1093/hmg/8.3.397. [DOI] [PubMed] [Google Scholar]
- Scott L, Forssberg H, Aperia A, Diaz-Heijtz R. Locomotor effects of a D1R agonist are DARPP-32 dependent in adult but not weanling mice. Pediatr Res. 2005;58:779–783. doi: 10.1203/01.PDR.0000180553.23507.31. [DOI] [PubMed] [Google Scholar]
- Slow EJ, van Raamsdonk J, Rogers D, Coleman SH, Graham RK, Deng Y, Oh R, Bissada N, Hossain SM, Yang YZ, Li XJ, Simpson EM, Gutekunst CA, Leavitt BR, Hayden MR. Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum Mol Genet. 2003;12:1555–1567. doi: 10.1093/hmg/ddg169. [DOI] [PubMed] [Google Scholar]
- Steffan JS, Kazantsev A, Spasic-Boskovic O, Greenwald M, Zhu YZ, Gohler H, Wanker EE, Bates GP, Housman DE, Thompson LM. The Huntington's disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc Natl Acad Sci U S A. 2000;97:6763–6768. doi: 10.1073/pnas.100110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoof JC, Kebabian JW. Opposing roles for D-1 and D-2 dopamine receptors in efflux of cyclic AMP from rat neostriatum. Nature. 1981;294:366–368. doi: 10.1038/294366a0. [DOI] [PubMed] [Google Scholar]
- Stoof JC, Verheijden PF. D-2 receptor stimulation inhibits cyclic AMP formation brought about by D-1 receptor stimulation in rat neostriatum but not nucleus accumbens. Eur J Pharmacol. 1986;129:205–206. doi: 10.1016/0014-2999(86)90358-4. [DOI] [PubMed] [Google Scholar]
- Sugars KL, Brown R, Cook LJ, Swartz J, Rubinsztein DC. Decreased cAMP response element-mediated transcription: an early event in exon 1 and full-length cell models of Huntington's disease that contributes to polyglutamine pathogenesis. J Biol Chem. 2004;279:4988–4999. doi: 10.1074/jbc.M310226200. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Nozaki K, Tsuji S, Nishizawa M, Onodera O. Polyglutamine represses cAMP-responsive-element-mediated transcription without aggregate formation. Neuroreport. 2005;16:295–299. doi: 10.1097/00001756-200502280-00019. [DOI] [PubMed] [Google Scholar]
- Takano H, Gusella JF. The predominantly HEAT-like motif structure of huntingtin and its association and coincident nuclear entry with dorsal, an NF-kB/Rel/dorsal family transcription factor. BMC Neurosci. 2002;3:15. doi: 10.1186/1471-2202-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Huntington's Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Thompson JC, Snowden JS, Craufurd D, Neary D. Behavior in Huntington's disease: dissociating cognition-based and mood-based changes. J Neuropsychiatry Clin Neurosci. 2002;14:37–43. doi: 10.1176/jnp.14.1.37. [DOI] [PubMed] [Google Scholar]
- Tsou K, Brown S, Sanudo-Pena MC, Mackie K, Walker JM. Immunohistochemical distribution of cannabinoid CB1 receptors in the rat central nervous system. Neuroscience. 1998;83:393–411. doi: 10.1016/s0306-4522(97)00436-3. [DOI] [PubMed] [Google Scholar]
- van Dellen A, Welch J, Dixon RM, Cordery P, York D, Styles P, Blakemore C, Hannan AJ. N-Acetylaspartate and DARPP-32 levels decrease in the corpus striatum of Huntington's disease mice. Neuroreport. 2000;11:3751–3757. doi: 10.1097/00001756-200011270-00032. [DOI] [PubMed] [Google Scholar]
- Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED, Richardson EP., Jr. Neuropathological classification of Huntington's disease. J Neuropathol Exp Neurol. 1985;44:559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- Walaas SI, Greengard P. DARPP-32, a dopamine- and adenosine 3':5'-monophosphate-regulated phosphoprotein enriched in dopamine-innervated brain regions. I. Regional and cellular distribution in the rat brain. J Neurosci. 1984;4:84–98. doi: 10.1523/JNEUROSCI.04-01-00084.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler NS, Lorimer J, Porter J, Gomez F, Moskowitz C, Shackell E, Marder K, Penchaszadeh G, Roberts SA, Gayan J, Brocklebank D, Cherny SS, Cardon LR, Gray J, Dlouhy SR, Wiktorski S, Hodes ME, Conneally PM, Penney JB, Gusella J, Cha JH, Irizarry M, Rosas D, Hersch S, Hollingsworth Z, MacDonald M, Young AB, Andresen JM, Housman DE, De Young MM, Bonilla E, Stillings T, Negrette A, Snodgrass SR, Martinez-Jaurrieta MD, Ramos-Arroyo MA, Bickham J, Ramos JS, Marshall F, Shoulson I, Rey GJ, Feigin A, Arnheim N, Acevedo-Cruz A, Acosta L, Alvir J, Fischbeck K, Thompson LM, Young A, Dure L, O'Brien CJ, Paulsen J, Brickman A, Krch D, Peery S, Hogarth P, Higgins DS, Jr., Landwehrmeyer B. Venezuelan kindreds reveal that genetic and environmental factors modulate Huntington's disease age of onset. Proc Natl Acad Sci U S A. 2004;101:3498–3503. doi: 10.1073/pnas.0308679101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler VC, Gutekunst CA, Vrbanac V, Lebel LA, Schilling G, Hersch S, Friedlander RM, Gusella JF, Vonsattel JP, Borchelt DR, MacDonald ME. Early phenotypes that presage late-onset neurodegenerative disease allow testing of modifiers in Hdh CAG knock-in mice. Hum Mol Genet. 2002;11:633–640. doi: 10.1093/hmg/11.6.633. [DOI] [PubMed] [Google Scholar]
- Wheeler VC, White JK, Gutekunst CA, Vrbanac V, Weaver M, Li XJ, Li SH, Yi H, Vonsattel JP, Gusella JF, Hersch S, Auerbach W, Joyner AL, MacDonald ME. Long glutamine tracts cause nuclear localization of a novel form of huntingtin in medium spiny striatal neurons in HdhQ92 and HdhQ111 knock-in mice. Hum Mol Genet. 2000;9:503–513. doi: 10.1093/hmg/9.4.503. [DOI] [PubMed] [Google Scholar]
- Yu ZX, Li SH, Nguyen HP, Li XJ. Huntingtin inclusions do not deplete polyglutamine-containing transcription factors in HD mice. Hum Mol Genet. 2002;11:905–914. doi: 10.1093/hmg/11.8.905. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Belyaev N, Conforti P, Ooi L, Tartari M, Papadimou E, MacDonald M, Fossale E, Zeitlin S, Buckley N, Cattaneo E. Widespread disruption of repressor element-1 silencing transcription factor/neuron-restrictive silencer factor occupancy at its target genes in Huntington's disease. J Neurosci. 2007;27:6972–6983. doi: 10.1523/JNEUROSCI.4278-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato C, Ciammola A, Rigamonti D, Leavitt BR, Goffredo D, Conti L, MacDonald ME, Friedlander RM, Silani V, Hayden MR, Timmusk T, Sipione S, Cattaneo E. Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease. Science. 2001;293:493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 A) nTG mice's rearing behavior separated by gender and B) CAG 140 mice's rearing behavior separated by gender are shown here. Although it appears that male nTGs rear more than female nTGs prior to 9 months of age in (A), there was neither a significant main effect of gender (F(1,19) = 3.5, p = 0.07) nor was there a significant genotype × gender interaction (F(2,19) = 1.19, p > 0.3). [n: nTG= 8 (6 M, 2 F), Het= 13 (8 M, 5 F), Hom= 4 (2 M, 2 F)]. There were no gender differences detected in any behavioral or post mortem measurements throughout the rest of the experiment.
Supplementary Figure 2 A) Representative images of (i) nTG, (ii) Het, and (iii) Hom striatal area containing NIIs at 12 months. Arrows point to an example of NIIs. Scale bar represents 25μm. B) Representative image of cortical region where NeuN+ neurons were counted (black box) and measurement of cortical thickness (white arrow). Scale bar represents 250μm. Contrast levels were adjusted in (A and B) for better visualization in this figure. C) Hom progression of NIIs as a percent over the 6 month baseline. D) NeuN+ cells average per 1mm2. 9 month and 12 month old nTG and homozygote mice were stained with NeuN to determine the total cortical neurons in the given area seen in B). CC= Corpus Collosum.