Abstract
Programmed cell death (PCD) has a key role in defence and development of all multicellular organisms. In plants, there is a large gap in our knowledge of the molecular machinery involved at the various stages of PCD, especially the early steps. Here, we identify kiss of death (KOD) encoding a 25-amino-acid peptide that activates a PCD pathway in Arabidopsis thaliana. Two mutant alleles of KOD exhibited a reduced PCD of the suspensor, a single file of cells that support embryo development, and a reduced PCD of root hairs after a 55°C heat shock. KOD expression was found to be inducible by biotic and abiotic stresses. Furthermore, KOD expression was sufficient to cause death in leaves or seedlings and to activate caspase-like activities. In addition, KOD-induced PCD required light in leaves and was repressed by the PCD-suppressor genes AtBax inhibitor 1 and p35. KOD expression resulted in depolarization of the mitochondrial membrane, placing KOD above mitochondria dysfunction, an early step in plant PCD. A KOD∷GFP fusion, however, localized in the cytosol of cells and not mitochondria.
Keywords: BAX inhibitor 1, caspase-like, embryogenesis, p35
Introduction
Programmed cell death (PCD) is fundamental to development and defence mechanisms in plants. Although animal PCD features such as cell shrinkage, chromatin condensation and DNA fragmentation, are observed in plant cells (Danon and Gallois, 1998), there are very few genes showing true homology with animal PCD genes (Hoeberichts and Woltering, 2003). For example, the proteases with caspase-like enzymatic activities in plants are not related to animal caspases (Bonneau et al, 2008). Therefore, even if ancestral mechanisms may persist in animal and plant PCD, each phylum seems to have evolved convergent features independently. Plant PCD linked to biotic and abiotic stress is probably the most well-characterized form of plant PCD, for example the hypersensitive response (HR) elicited by pathogen attacks (Heath, 2000). By contrast, less is known about the mechanisms of developmentally regulated PCD. Unravelling further PCD cascade components in plants is therefore key to the understanding of this mechanism and its evolution. There are several experimental systems that have been used to investigate developmental PCD in plants. Studying Zinnia cell cultures undergoing PCD to differentiate into tracheary elements has aided greatly in the elucidation of the order of events in the developmental PCD pathway. For example, in this system, mitochondrial membrane depolarization and release of cytochrome c precedes vacuole rupture in the PCD pathway (Yu et al, 2002). Additionally, it was found that vacuole rupture results in cessation of cytoplasmic streaming followed by nuclear degradation, the latter of which involved the DNase ZEN1 (Groover et al, 1997; Obara et al, 2001; Ito and Fukuda, 2002). Corroborating with the Zinnia system, the timing of events at the organelle level, such as vacuole rupture and nuclear degradation, have also been seen in the emerging PCD model Aponogeton madagascariensis (Lace plant) (Gunawardena et al, 2004); suggesting that these features are common events in plant developmental PCD. The terminally differentiated suspensor is an organ, which has also been used to study the regulation of developmental PCD. The developmental stages of embryo suspensors were observed nearly 90 years ago (Souèges, 1919), but still to date little is known about the regulation of PCD in the suspensor at a molecular level. In the suspensors of somatic embryos, hallmarks of PCD such as DNA laddering and cytoskeleton reorganization have been described (Bozhkov et al, 2005). Additionally, PCD of somatic suspensors in Norway spruce embryonic cell cultures was demonstrated to be dependent on a functional metacaspase facilitating nuclear degradation (Suarez et al, 2004) and activating a caspase-like protease (Bozhkov et al, 2003). Studies in Arabidopsis have helped identify genes involved with the developmental PCD pathway, for example the vacuolar processing enzyme δ is required for the PCD of two seed-coat cell layers in developing Arabidopsis seeds and presumably acts at the execution stage of the PCD pathway (Nakaune et al, 2005). In summary, work in the last 10 years using various experimental systems has shed light on some of the steps involved with the developmental PCD process at the organelle and molecular level. However, with regards to the molecular components, those as yet identified would appear mainly to act at the execution phase of PCD rather than at the decision/initiation phase. In particular, there are no cascade components demonstrated to be above mitochondrial dysfunction. Therefore, there is a gap in our knowledge of the molecular components involved at the early stage of the developmental PCD process. Here, we propose that the 25-amino-acid (aa) peptide kiss of death (KOD) is a pro-PCD component in Arabidopsis that acts during the initial stages of the PCD process in development. The existence of KOD suggests that small peptides may be important regulators of PCD that have so far been overlooked as a consequence of their small size.
Results
Identification of the KOD gene
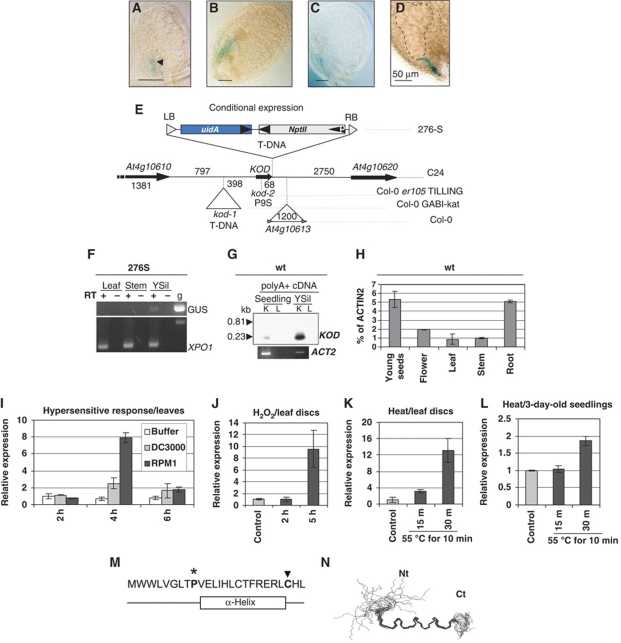
To identify marker genes of suspensor development in Arabidopsis, we generated a population of pΔGUS-promoter trap lines and screened for GUS activity during embryogenesis (Devic et al, 1995). The line 276S expressed GUS in the suspensor only, from the globular stage onwards (Figure 1A–D). When 276S was used as a marker line in a yoda mutant background, GUS expression was lost in the abnormal suspensors that did not undergo PCD (Lukowitz et al, 2004). This indicates that GUS expression in 276S was a marker of suspensor cell fate, possibly associated with PCD. The T-DNA in line 276S was found to be inserted in chromosome IV between the genes annotated At4g10610 and the LINE retrotransposon insertion annotated At4g10613 in Col-0 or between the genes At4g10610 and At4g10620 in C24, which does not contain the retrotransposon (Figure 1E). The T-DNA insertion site was found 9 bp downstream of a 75-bp open reading frame (ORF) corresponding to a deduced 25-aa peptide (Figure 1M) that we named KOD. Although this locus is not annotated in for example TAIR, the analysis software NetPlantGene attributes a high-coding probability to the ORF. The ORF is not part of the cDNA sequences of either the upstream or downstream genes in the T-DNA Express and TAIR databases. The transcript was, however, detected using real-time polymerase chain reaction (RT–PCR), with special care to control for the absence of genomic DNA as the ORF contains no intron. Consistent with the GUS histochemical pattern, the KOD∷GUS transcript fusion was detected only in siliques of line 276S using primer oexTi15 in KOD and oGUSj in the GUS gene (Figure 1F). The amplification signal was weak as expected for a gene expressed in an eight-cell organ. In wild type, the longest amplified fragment from the transcript extended up to 150 bp upstream the ATG of the 75-bp ORF; forward primers located further upstream, including in the transcript of the upstream gene At4g10610, did not yield any product (Figure 1G). Three-prime rapid amplification of cDNA ends (3′RACE) experiments and sequencing placed the polyA tail 30 bp downstream of the stop codon. Using quantitative RT (QRT)–PCR analysis, we found low-level expression in all tissues tested with highest expression at about 5% of ACTIN2 in dissected, white and green seeds and in roots (Figure 2H). Therefore, the promoter trap in 276S may have allowed detection of KOD∷GUS expression only in the suspensor, but lower levels of expression of the native gene occur in other tissues. Further, QRT–PCR analysis was carried out to analyse expression in conditions inducing PCD. The HR of plants resistant to microbial pathogens involves a form of PCD (Heath, 2000). KOD expression was induced eight-fold in leaves, 4 h after infiltration with 108 c.f.u./ml Pseudomonas syringae pv. tomato DC3000 expressing the resistance to P. syringae pv. maculicola 1 (RPM1) avirulence gene, which triggers the HR PCD in the Arabidopsis Col-0 ecotype (Figure 1I). This induction did not occur with the virulent strain DC3000, which does not induce HR PCD. The avrRpm1/RPM1 gene-for-gene interaction has been shown to cause local accumulation of H2O2 in Arabidopsis challenged by P. syringae pv. tomato (Grant et al, 2000). In addition, external application of H2O2 does induce PCD in leaf discs of Arabidopsis (He et al, 2008). In leaf discs challenged with 30 mM H2O2, KOD expression was induced nine-fold at the 5-h time point (Figure 1J). To investigate abiotic stress, we used heat shock. A 10-min heat shock at 55°C has been shown to trigger PCD in Arabidopsis cells (Reape et al, 2008). KOD expression was found to be induced 13-fold in heat-shocked leaf discs (Figure 1K) and two-fold in 3-day-old seedlings (Figure 1l) that were composed of two cotyledons and a main root of circa 5 mm. In both cases, induction occurred as early as 30 min. On a different note, the KOD peptide was synthesized to obtain its structure using nuclear magnetic resonance (NMR). NMR spectra showed that KOD adopts a definite α-helix (Figure 1N), starting at P9 (Supplementary Figure S1). The peptide is predicted to be amphipilic with two hydrophobic ridges of the α-helix possibly playing a role in protein or membrane interaction (Supplementary Figure S1). The biotic and abiotic stress induction of KOD combined with suspensor expression was suggestive of a possible involvement in PCD regulation.
Figure 1.
Identification and expression profile of KOD. (A–E) Trap line and KOD locus. GUS expression pattern in line 276S. Arrowhead shows the boundary between the suspensor and the embryo proper. Stages of embryo development (A) globular, (B) late globular, (C) heart, (D) torpedo. Bars=50 μm. (E) Locus and structure of the promoter trap T-DNA in 276S (ecotype C24). At4g10613: LINE retrotransposon in Col-0; kod-1: T-DNA position in the GABI-Kat line. P9S, non-synonymous substitution in kod-2 (TILLING). uidA, β-glucuronidase gene. DNA sizes are given in bp. (F–L) Expression analysis of KOD. (F) RT–PCR analysis on tissues from line 276S using primer oexTi15 in KOD and oGUSj in the GUS gene. ‘GUS’ indicates the transcriptional fusion between KOD and the GUS sequence. XPO1, a ubiquitous gene (At5g17020) as a control. g, genomic DNA; YSil, young silique. (G) RT–PCR on polyA+ cDNA of WT, blotted and probed with a radiolabelled KOD ORF. K, Primer pair (oexTi15; oSUPR1) amplifying the KOD transcript (0.23 kb); L, primer pair (oTi05; oSUPR1) with oTi05 outside the transcript (0.81 kb). Expected sizes are shown; ACT2, Actin2 control. (H) KOD expression in Col-0 wild-type tissues using Taqman QRT–PCR. Actin2 is used as the reference gene. (I) KOD expression using Taqman QRT–PCR in WT leaves following infiltration with 10 mM MgCl2 (Buffer), P. syringae pv. tomato DC3000 wild-type (virulent) or expressing AvrRPM1 (avirulent). 18S is used as the reference gene. Each sample was measured in triplicate and is shown relative to expression at 2 h post-inoculation (hpi). (J) KOD expression using Taqman QRT–PCR in WT leaf discs floated on SDW (control) or 30 mM H2O2 for 2 and 5 h. 18S is used as the reference gene. Each sample was measured in triplicate and is shown relative to expression in the control. (K) KOD expression using Taqman QRT–PCR in WT leaf discs floated on SDW at RT (control) or 15 and 30 min after heat shock at 55°C for 10 min. 18S is used as the reference gene. Each sample was measured in triplicate and is shown relative to expression of control. (L) KOD expression using Taqman QRT–PCR in WT 3-day-old seedlings floated on SDW at RT (control) or 15 and 30 min after heat shock at 55°C for 10 min. 18S is used as the reference gene. Each sample was measured in triplicate and is shown relative to expression of control. (M) Amino-acid sequence of KOD and position of α-helix, see Supplementary Figure S1. Star and arrow highlight 2 aa required for full functionality of KOD, see Figure 5C. (N) Overlay of 22 calculated structures from NMR spectra. Nt, N-terminus; Ct, C-terminus. Residues 9–21-fold into an α-helix, whereas the N-terminal region is disordered.
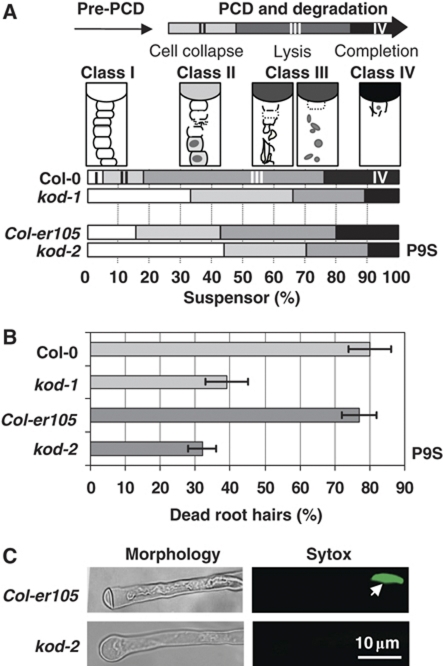
Figure 2.
Phenotype in Arabidopsis KOD mutant lines. Mutant lines are kod-1, T-DNA knockdown (GABI line); kod-2, P9S point mutation in Col-er105 background (TILLING line). (A) Stacked column chart with the percentage of suspensors in classes I–IV for 100 embryos of each genotype. Seeds containing embryos at the bent-cotyledon stage were clarified using Hoyers solution and observed under DIC. Each suspensor was assigned to one of the four stages (classes I–IV) of suspensor elimination described in Supplementary Figure S2. (B) Reduced root hair PCD in 3-day-old seedlings of the two mutant backgrounds. PCD was induced using 10 min at 55°C. Cell death was scored 6 h after treatment using plasma membrane retraction and sytox green positive nucleus as criteria. Untreated seedlings of all backgrounds had around 10% dead root hairs. Errors bars are 2 × s.e. of triplicates of five seedlings (>100 root hairs total) each. (C) Most common root hair morphology in wild-type Col-er105 (dead root hair) or kod-2 (live root hair). (Left panel) White light microscopy. (Right panel) Fluorescence microscopy of one root hair incubated with sytox green using FITC filter. White arrow points at a sytox-positive nucleus, indicative of a loss of plasma membrane permeability.
KOD modulates suspensor elimination during embryogenesis
To investigate the possible role of KOD in the PCD of the suspensor, we analysed two mutant alleles, kod-1 and kod-2. kod-1 is a GABI-kat allele (Col-0) that carries a T-DNA inserted in the promoter region, 398 bp upstream of the ORF. KOD transcription was strongly downregulated in seeds of kod-1 (Supplementary Figure S3A). kod-2 is a targeting induced local lesions in genomes (TILLING) allele (in Col-er105) with a proline to serine (P9S) point mutation. Proline often has the role of initiator at the N-terminus of α-helices (Kim and Kang, 1999). Therefore, the P9S mutation in kod-2 may affect KOD function by altering its folding. Phenotypic analysis of the suspensor was carried out in the kod-1 and kod-2 backgrounds with 100 embryos each being analysed using microscopy. No defect in the development of the embryo proper was found, but the occurrence of suspensor death varied between genetic backgrounds. We first scored the phenotypes of Col-0 embryos at the bent-cotyledon stage, the last stage at which the suspensor was visible. The suspensors were assigned to four different classes from intact (I) to absent (IV) (Figure 2A; Supplementary Figure S2). Only 5% of Col-0 suspensors were intact (class I); hence 95% of the suspensors had entered PCD. By contrast, 32% of kod-1 embryos showed intact suspensors, a six-fold increase in the number of suspensors failing to initiate PCD. Similarly, 44% of kod-2 embryos had intact suspensors, three times as many as in wild-type Col-er105. We concluded that KOD appears to have a pro-PCD effect, directly or indirectly, with P9 crucial for that function as the mutation correlated with a substantial reduction of suspensor cell death.
KOD modulates root hair PCD
Overall, seedlings and plants of mutant lines looked similar to the corresponding wild type. However, when 3-day-old seedlings were heat shocked, root hairs gave a reduced PCD phenotype in both mutant backgrounds. Ten-minute 55°C heat shocks have been shown to induce hallmarks of PCD in Arabidopsis cells including cell shrinkage that is absent from necrotic cells (McCabe and Leaver, 2000; Reape et al, 2008). Three-day-old seedlings with a root of Circa 5 mm were submitted to a 10 min heat shock at 55°C. After 6 h, 80% wild-type root hairs were sytox green positive and showed cell shrinkage, whereas only 30–40% of root hairs in the two mutant backgrounds underwent PCD (Figure 2B and C). This reduced PCD phenotype is consistent with the proposition that KOD regulates PCD.
KOD transient expression in tobacco leaf induces cell death
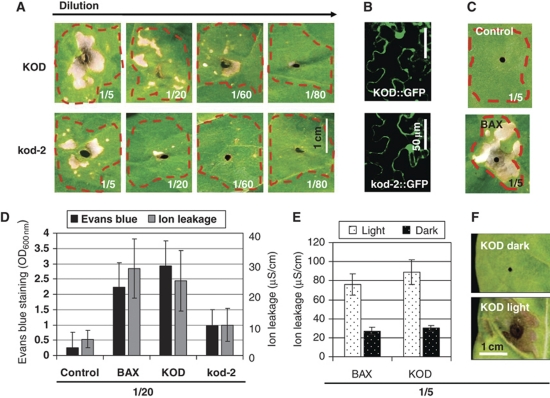
To test KOD function in a PCD assay, we transfected tobacco leaves with KOD∷GFP using Agrobacterium infiltration (Figure 3A). BAX∷YFP was used as a positive control because of its ability to induce plant PCD in several experimental systems including Agrobacterium-mediated infiltration. BAX, a pro-apoptotic member of the mammalian Bcl-2 gene family, despite having no known plant homologue can induce PCD in tobacco leaves (Lacomme and Santa Cruz, 1999). Both KOD∷GFP and BAX∷YFP were expressed under the control of the 35S promoter and induced a lesion of collapsed, bleached tissue in the infiltrated area, within 3–4 days post-infiltration (dpi) (Figure 3A–C). The GFP control showed no cell death (Figure 3C). The relative PCD-inducing capabilities of KOD∷GFP and kod-2∷GFP were then directly compared using a dilution series of Agrobacterium culture (Figure 3A). For the three most concentrated dilutions (1/5–1/60) there was visually, significantly more cell death in the KOD∷GFP infiltrations compared with kod-2∷GFP. This difference in cell death was quantified using Evans blue and ion leakage on the 1/20 dilutions, revealing that the level of cell death for kod-2 was half of that for KOD (Figure 3D).
Figure 3.
KOD expression in N. tabacum leaves induces death. (A) Agrobacterium-mediated transfection of tobacco leaves using 35S∷KOD∷GFP and 35S∷kod2∷GFP. OD600 for each sample was adjusted to 1 and further diluted to create the series, that is 1/5=OD 0.2. Phenotypes were seen 3 dpi. (B) Fluorescent confocal microscopy confirming GFP expression in the infiltrated samples, scale bar=50 μm. (C) 35S∷GFP control and 35S∷BAX∷YFP infiltrations. (D) Quantification of cell death for 1/20 dilutions, using leaf discs for both ion leakage and Evans blue in triplicate ±2 × s.e. (E) Tobacco leaves were infiltrated with the OD 0.2 (1/5) dilution of KOD∷GFP, BAX∷YFP and GFP. Leaves either kept in 8 h light/16 h dark or wrapped in tin foil to exclude light. Ion leakage was taken at 4 dpi; measurements are minus the average reading for the GFP control. Values are taken in triplicate, ±2 × s.e. (F) Photos (4 dpi) of KOD infiltrations.
A number of PCD pathways in plants have been demonstrated to be light dependent in photosynthetic tissues, for example cryptochrome1-mediated singlet-oxygen-induced PCD (Danon et al, 2006) and BAX-induced PCD (Yoshinaga et al, 2005). Using tobacco leaves infiltrated with KOD∷GFP, the level of cell death for leaves kept in the dark for 3 dpi was found to be considerably less than for those kept in light conditions (8 h light/16 h dark) (Figure 3F). This difference between light and dark incubated infiltrations was confirmed using ion leakage measurements (Figure 3E).
KOD expression induces cell death and caspase3 activity in Arabidopsis
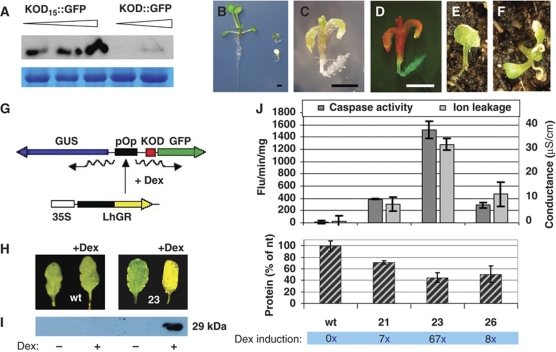
We generated Arabidopsis plants expressing KOD∷GFP or KOD15∷GFP truncated of aa 16–25, under the 35S promoter. A population of 75 T1 transformants were analysed for each construct. In the KOD population, 28% of the T1 plants had a severely affected development compared with 0% in the KOD15 population. A third of these affected plants (10% of the total) showed missing cotyledons, no true leaves at the shoot apex or necrotic regions at the margins of cotyledons and leaves (Figure 4B–F). GFP expression was qualitatively assessed under a dissecting scope. KOD15∷GFP seedlings showed low, medium, or high fluorescence. In KOD∷GFP, affected seedlings correlated with KOD∷GFP expression (Figure 4C and D), whereas the remaining unaffected plants all showed undetectable or very low KOD∷GFP expression. This was confirmed using western blot analysis and a GFP antibody: overall GFP expression was much lower in KOD∷GFP plants than in KOD15∷GFP plants, possibly indicative of counter-selection (Figure 4A). Thus, we concluded that KOD∷GFP expression correlated with severe developmental defects, including cell death, during early seedling development. The first 15 aa of KOD were unable to cause those defects.
Figure 4.
KOD overexpression in Arabidopsis correlates with PCD. (A) Western blot analysis using anti-GFP antibody on 35S∷KOD∷GFP and 35S∷KOD15∷GFP Arabidopsis lines. T1 plants were grouped according to their GFP signal intensity. Equal loading was controlled with Coomassie blue staining. (B–F) F1 35S∷KOD∷GFP plants. (B) Unaffected seedling (left) compared with most affected kanamycinR seedlings (top right) and to kanamycinS seedlings (bottom right). Magnified view in bright field of an affected kanamycinR seedling (C) and under blue light excitation (D), scale bars=1 mm. (G) Cartoon of the pOp-LhGR/dex 2-component inducible system for conditional expression. (H) Leaves after 3 days of induction in continuous light. Individual leaves were kept in 1.5 ml Eppendorf tubes with their petioles submerged in 30 μM dex. (I) Western using anti-GFP on protein extracted from wt leaves (±dex) and high expresser line 23 (±dex), after 3 days with 30 μM dex. (J) (Top panel) Quantification of cell death using ion leakage and caspase3-like activity in three inducible KOD lines after 3 days of induction. (Middle panel) Quantification of total protein loss measured with the Bradford assay (7 days of induction), values from triplicate with ±2 × s.e. (Bottom panel) Fold dex induction of expression using a GUS fluorometric assay.
Given the results obtained with constitutive expression, we generated several Arabidopsis lines using the pOpOn2/LhGR system (Craft et al, 2005) to drive dexamethasone-inducible expression of KOD∷GFP (Figure 4G). Three lines were selected: two medium expressers (lines 21 and 26) and a high expresser (line 23) (Figure 4J). As a positive control, we obtained an Arabidopsis line containing a dex-inducible BAX that undergoes PCD (Kawai-Yamada et al, 2001; Yoshinaga et al, 2005). Detached leaves from each line, a wt control and the dex-BAX line were incubated in Eppendorfs with the petiole submerged in 30 μM dex. At 3 days of induction, western blot analysis confirmed KOD∷GFP expression for the expresser lines, only in the presence of dex (Figure 4I). After 3–4 days of continuous light, the leaves of the three KOD expressers and the dex-BAX line turned yellow and the tissue began to degrade (Figure 4H; Supplementary Figure S4). We concluded that ectopic expression of KOD∷GFP is sufficient to induce cell death in Arabidopsis leaves. This cell death correlated with a reduction in total protein content (Figure 4J) and with increased ion leakage (Figure 4J).
Caspase3-like activity is a strong hallmark for PCD. This activity has been implicated in a wide range of plant PCD pathways, including self-incompatibility and HR (Bonneau et al, 2008). To see if KOD∷GFP can induce this activity in dex-induced leaves, an enzymatic assay using the caspase3-substrate DEVD-Rh110 was carried out. The wt leaves with and without dex and KOD lines leaves without dex showed negligible DEVDase activity. The dex-induced KOD lines leaves (≠21, 26 and 23) all showed an increase in the levels of DEVDase activity (Figure 4J), suggesting that KOD-induced cell death is mediated by proteases with caspase3-like activity. Overall, ion leakage, caspase3-like activity and protein content measurements correlated with the relative level of dex induction in the three lines (Figure 4J).
KOD localizes in the cytosol
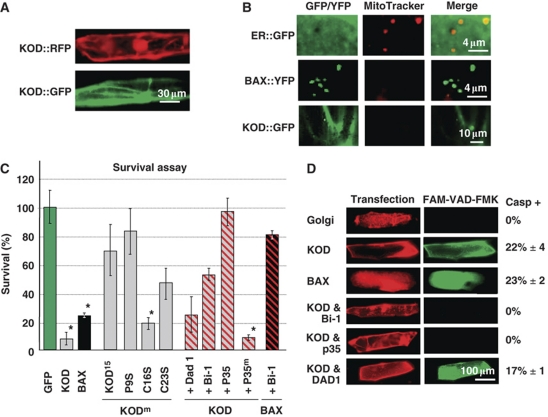
KOD c-terminal fusions to either GFP or RFP were bombarded into onion epidermal cells in order to visualize subcellular localization. At 15 h post-bombardment (pb), both KOD∷GFP and KOD∷RFP localized to the cytoplasm and nuclei of transfected cells (Figure 5A). There was no specific pattern that could have been suggestive of organelle localization or of a membrane localization in these onion cells, in tobacco leaf epidermal cells expressing transiently KOD∷GFP (Figure 3B) or in dex-inducible Arabidopsis lines expressing KOD∷GFP (data not shown). This is suggestive of a cytosolic localization.
Figure 5.
KOD-induced PCD pathway. (A) Subcellular localization of KOD∷GFP and KOD∷RFP in onion epidermal cells 15 h post-bombardment (pb). (B) Onion cells 24 h pb, incubated in 100 nM of MitoTracker Red for 5 min. Absence of MitoTracker signal indicates loss of ψmit. ER∷GFP, ER-targeted GFP. (C) Survival assay in onion cells quantified as the percentage of cells with no pH shift (fluorescent cells). KODm are P9S, C16S and C23S single missense mutations. AtBi-1, p35 and Dad1 are used as suppressors of PCD; p35m is the null mutant allele D87A of p35. All suppressor plasmids were co-bombarded in a two-fold molar excess over KOD or BAX. Values from triplicates ±2 × s.e. (D) In situ fluorescent caspase assay in transfected onion cells. Onion cells incubated with FAM-VAD-FMK inhibitor at 30 h pb. A small number of cells with caspase activity was observed in the negative control (golgi∷RFP) and was subtracted from all other samples. Values from triplicates with ±2 × s.e.
KOD expression results in mitochondrial depolarization
It has been proposed that mitochondrial dysfunction is an early step in the progression of PCD. For example, in heat-shocked tobacco cells undergoing PCD, a loss of mitochondrial membrane potential (ψmit) was shown 4 h after heat treatment and 44 h before DNA laddering (Vacca et al, 2004). A loss of ψmit has also been seen in other plant PCD pathways, for example in BAX and nitric oxide (NO)-induced PCD and during tracheary element formation (Saviani et al, 2002; Yu et al, 2002; Yoshinaga et al, 2005). With regards to the BAX and NO studies, this loss of ψmit was detected by a reduction of fluorescence emitted by the dye MitoTracker Red. This dye is a cationic lipophilic fluorochrome, which acts by accumulating in the negatively charged matrix of the mitochondria. The accumulation of this probe in the mitochondria is dependent upon the strength of the ψmit, the loss of which results in a proportional loss of MitoTracker fluorescence (Kerry and Mark, 1999). Transfected cells expressing KOD∷GFP were incubated with MitoTracker Red. At 24 h pb, mitochondria from both KOD∷GFP and BAX∷YFP expressing cells failed to fluoresce with MitoTracker Red, whereas the mitochondria of the GFP control and all untransfected cells clearly fluoresced red in the presence of the dye (Figure 5B).
The PCD-suppressor genes AtBi-1 and p35 downregulate KOD-induced PCD
To analyse the pathway activated by KOD, we tested three PCD-suppressor genes against KOD in transfected onion cells using biolistics. Here, the pH sensitivity of GFP fluorescence is used to score the cytosolic acidification that occurs during plant PCD. The loss of fluorescence can be reversed when cells are experimentally buffered back up to pH 7 (Young et al, 2010). BAX or KOD were fused to GFP and transfected to provide both proof of expression at 16 h and survival score at 48 h. The number of transfected cells in each experiment can be scored by counting GFP-positive cells at 16 h. Alternatively, a 35S∷GUS plasmid can be co-transfected and GUS histochemical staining carried out after scoring GFP at 48 h. BAX∷GFP at 48 h pb yielded 24% of fluorescent cells compared with 100% for the BAX∷GFP cells at 16 h or for the GFP cells at 48 h (Figure 5C). BAX-induced PCD could be prevented in 55% of the cells by co-bombarding a two-molar plasmid excess of the Arabidopsis orthologue of the PCD-suppressor BAX inhibitor 1 (AtBi-1), a suppression effect seen in other plant systems (Kawai-Yamada et al, 2001). Similarly at 48 h pb, KOD∷GFP expression yielded 7% of fluorescent cells compared with 100% for the GFP control or KOD∷GFP at 16 h, confirming that KOD is sufficient to induce cell death in this assay. Co-expression of AtBi-1 with KOD prevented 46% of the transfected cells from undergoing PCD (Figure 5C). Further, strengthening the notion that KOD-induced cell death is PCD, the pan caspase inhibitor p35 (Danon et al, 2004) prevented cell death in 93% of the transfected cells. Conversely, the null allele p35D87A (p35m) failed to prevent cell death in any cells, suggesting a specific interaction of p35 with protease(s) possessing caspase-like activity in onion cells. KOD was also tested alongside defender against apoptotic death 1 (AtDAD1), an inhibitor of UVC-induced PCD in Arabidopsis (Danon et al, 2004). This particular suppressor only had a small effect by increasing survival from 7 to 23% of cells, suggesting that DAD1 at least in this system is unable to prevent KOD-induced cell death.
To link KOD expression with caspase-like activity in situ, cells were incubated with the pan-caspase inhibitor FAM-VAD-FMK. This fluorescent probe irreversibly binds to the active site of proteases with caspase-like activities and allows caspase-like activity to be scored under the microscope. At 30 h pb, 23% of BAX+golgi∷RFP co-transfected cells had caspase activity compared with the negative control (golgi∷RFP) (Figure 5D). KOD∷RFP transfected cells exhibited 22% of cells with caspase activity, similar to BAX. Co-transfection of KOD∷RFP with the PCD suppressors AtBi-1 and p35 suppressed caspase activity while DAD1 did not (Figure 5D). Suppression of caspase-like activity by AtBi-1 suggested that this protein acts upstream of caspase-like activity in KOD-induced PCD. The number of cells with caspase activity in Figure 5D was lower than the number of cells undergoing intracellular acidification in Figure 5C, as caspase activity was measured at the earlier time point of 30 h pb compared with 48 h pb.
Proline-9 and cysteine-23 are required for KOD-induced PCD
The kod-2 (P9S) mutation was tested in the onion transient assay and it induced cell death in only 20% of transfected cells compared with the 93% obtained with wild-type KOD (Figure 5C). Also in this assay, the truncated KOD15∷GFP had a large loss of function with only 30% death, consistent with the constitutive expression study (Figure 4A–F). The two cysteines in the KOD sequence (C16 and C23) were mutated to serine as cysteine are often important for protein function. The C16 to serine (C16S) mutation (80% death) introduced a very small loss of function compared with wild type (93% death). By contrast, cysteine-23 appeared to be more important than cysteine-16 for KOD function, as the C23 mutation (C23S) had a greater effect than C16S with a clear loss of KOD activity (50% death).
Discussion
We present here evidence that the KOD locus is involved in the positive regulation of PCD. The KOD transcript contains a short ORF of 75 bp. The point mutations P9S and C23S both affect KOD function and are consistent with the peptide being the active molecule rather than the mRNA. There are several known plant examples of short signalling peptides, with a size ranging from 5 to 76 aa generated by the cleavage of a larger pro-peptide (Lindsey et al, 2002). Short signalling peptides encoded by short ORF are less frequently described in plants and it can be argued that it is because they have been overlooked as a consequence of their size. Nevertheless, there are already several examples of such small genetic units with demonstrated functions during development. POLARIS is a 36-aa peptide that appears to modulate the ethylene/auxin response (Chilley et al, 2006) and the DVL peptide family (∼50 aa) has a role in Arabidopsis development (Wen et al, 2004). There is no known short ORF peptide involved in PCD regulation. The closest example to KOD is the GRIM REAPER (GRI) protein in Arabidopsis. This precursor protein generates a 60–70 aa peptide by cleavage that is involved in the initiation of cell death induced by extracellular ROS (Wrzaczek et al, 2009). The peptide is active in the extracellular space.
The involvement of KOD with PCD is clear as the P9S loss of function in overexpression assays correlated well with both the phenotypes in the mutant kod-2 of reduced PCD in suspensor and reduced PCD in root hairs. It is not possible to establish whether PCD is delayed or suppressed in embryos of kod-2, the P9S mutant line, as the surviving suspensors are crushed between the embryo root tip and the seed teguments during the later phase of embryogenesis. Nevertheless, our results suggest that KOD may define a novel class of short peptides, regulators of plant PCD. The root hair phenotype after heat shock and the induction of KOD expression by biotic and abiotic stress, in particular in an HR situation, suggest a regulatory role for KOD in PCD outside embryogenesis. The fact that KOD is inducible by stresses associated with PCD strengthens the observation that KOD overexpression induces PCD.
We have established here that KOD is above caspase-like activity as the caspase inhibitor p35 was able to block the effect of KOD overexpression. Furthermore, KOD overexpression in transient assays and in stably transformed Arabidopsis lines resulted in the induction of VADase and DEVDase caspase-like activities, respectively. In addition, PCD suppression by AtBi-1 is indicative of an implication of the ER downstream of KOD. AtBi-1 has been shown to be involved with ER calcium efflux during PCD and to interact with the calcium-binding protein CALMODULIN (Ihara-Ohori et al, 2007). Cells co-expressing KOD and AtBi-1 showed reduced caspase-like activity, indicating that the observed suppression by AtBi-1 occurs upstream of the caspase-like activity detected. This may place calcium efflux from the ER before caspase-like activity in the KOD PCD cascade.
We show that both KOD∷RFP and KOD∷GFP are localized to the cytosol and nucleus. The nuclear localization of KOD is possibly simply a result of KOD∷GFP and KOD∷RFP being under the 40-kDa exclusion size of nuclear pores. There was no evidence of a mitochondrial localization for KOD∷GFP. KOD, however, acted upstream of mitochondrial membrane depolarization since a loss of ψmit was time wise the earliest detectable change observed so far in cells overexpressing KOD. This loss of ψmit was also described as an early event in BAX and NO-induced PCD in plants and has been shown to occur as a result of the formation of the permeability transition pore (Saviani et al, 2002; Yoshinaga et al, 2005).
Finally, we show that KOD-induced lesion formation in tobacco leaves is light dependent, an effect already observed for BAX-induced PCD in tobacco cells (Yoshinaga et al, 2005). The effect of light upon plant PCD in leaf tissue is still relatively uncharacterized; its requirement is however undeniable. Light has been shown to have a significant role in the initiation of the HR, with the absence of light resulting in a reduced response (Guo et al, 1993). In this instance, light enhances an oxidative burst via alteration in the photosynthetic electron transport (Allen et al, 1999). In support of this, our expression data suggest that KOD expression may be under the regulation of cellular H2O2. Another possibility is a requirement for blue light sensing as PCD induced by singlet oxygen in Arabidopsis protoplasts was found to be dependent upon a functional Cryptochrome1, a blue light/UVA-specific photoreceptor (Danon et al, 2006).
Further experiments will be required to define the mode of action of KOD and to identify the death signal that is mediated by KOD. KOD may interact genetically or physically with negative regulators of PCD as the GUS trap line 276S suggests expression as early as the globular stage, a stage at which there is no PCD of the suspensor. Nevertheless, our results already provide evidence that the KOD peptide is a novel component of the PCD machinery. KOD is inducible by biotic and abiotic stress and its overexpression is sufficient to induce PCD. Overexpression of KOD provides an experimental system with which PCD can be induced in plants in the absence of these stresses. This should allow dissociating the plant PCD process from peripheral stress responses thereby providing a simplified experimental system in Arabidopsis for the analysis of PCD. In summary, the identification of KOD provides a significant contribution to the understanding of the early stages of the PCD machinery in plants.
Materials and methods
Plant material
The trap collection was generated in the C24 ecotype background using the binary construct pDeltaGUS (Devic et al, 1995). Line 276S was backcrossed twice to C24. kod-1 GABI-Kat # K029908 was provided by CeBiTec (Germany) (Rosso et al, 2003); kod-2 accession CS87682 was provided by the Seattle Tilling Project (Till et al, 2003). The dex-inducible BAX line was provided by Maki Kawai-Yamada (Tokyo, Japan). Arabidopsis thaliana was grown on soil, 16 h light photoperiod, 21°C. For seedlings in vitro, seeds were surface sterilized and plated on agar containing Murashige and Skoog salts, Gamborg's vitamins supplemented with 15 g/l glucose and selection antibiotics if required.
Mapping the KOD locus
Genomic DNA and sequences were obtained using standard procedures. Southern analysis was carried out to select restriction enzymes producing fragments of a size compatible with inverse PCR. Ligation and inverse PCR amplification were carried out using a PstI digest and the GUSI and GUS3 primers (Supplementary Table S1). A 1750-bp fragment upstream of the T-DNA insertion was cloned into pCRII, sequenced and used to screen a Col-0 genomic library; two clones were isolated and sequenced.
Embryos, GUS staining and microscopy
Seeds were assayed for GUS activity using 1 mM of the substrate 5-bromo-4-chloro-3-indolyl-glucuronic acid (X-Gluc), vacuum infiltrated for 5 min and incubated 4–48 h at 37°C. Seeds were then incubated in ethanol:acetic acid (1:1) o/n, transferred to Hoyer's solution (7.5 g gum arabic, 100 g chloral hydrate and 5 ml glycerol in 30 ml water), and cleared for 16 h. For early embryo stages, siliques were split longitudinally with a needle along the septum and cleared as above (except using 50 g chloral hydrate). Embryos were visualized using a Zeiss Axioplan microscope equipped with DIC optics. For suspensor phenotyping, 100 embryos at the bent-cotyledon stage were scored for each line.
Root hair PCD
PCD assay developed in Paul McCabe's laboratory, Dublin (BV Hogg et al, in preparation). Seedlings were grown on MS agar plates for 3 days, submerged in 3 ml distilled water in a six-well plate and floated in a water bath at 55°C for 10 min. Morphology and SYTOX green (Molecular Probes) was scored at 6 h after heat shock. The SYTOX molecule is excluded from living cells and stains DNA in dead cells. The commercial stock was diluted to 670 nM and applied to cells. After 5-min incubation, cells were washed twice in 3 ml of sterile distilled water before fluorescent microscopy (FITC filter).
Structural analysis
NMR data were acquired at 2 mM in 65% CF3C2H2O2H, 35% water (v/v) according to Volpon et al (2004). W2 and W3 spin systems were used to start the sequence-specific assignment. The 22 converged models showed a complete agreement with the NMR constraints.
Plasmid construction and plant transformation
pPK100 was obtained by cloning EGFP from pEGFP1 (Clontech) NcoI/NotI in pRTL2-GUS kindly provided by Dr J Carrington. pKOD15∷GFP (first 15 aa) and pKOD∷GFP were obtained by sequentially digesting pPK100 with NcoI, Nuclease S1, and XhoI, then, respectively, ligating adaptors Spep5/Spep3 and pep5/pep3 (Supplementary Table S1). BAX∷YFP was provided by Dr A Gilmore (Manchester) and cloned as an Eco47–XhoI fragment into pDH51 (ID 455497) SmaI–XhoI. AtBAX inhibitor 1 (At5g47120) was obtained from EST ATTS1836, with the full-coding sequence obtained by adding the N-term sequence coding for MDAFSSF at the 5′ end. The ORF was excised out of pBluescript SK+ cloned into pDH51 using BamHI–SalI. Details about p35, p35m (D87A) and DAD1 vectors can be found in Danon et al (2004). KOD∷GFP and KOD15∷GFP XhoI/XbaI fragments were cloned into pART27 and transformed into Agrobacterium tumefaciens GV3101 for floral dip infiltration of Arabidopsis (Bechtold and Pelletier, 1998). Transformants were selected on kanamycin 50 mg/l and examined under a Zeiss fluorescence dissecting scope equipped with a GFP filter. All point mutations (P9S, C16S and C23S) were created using the QuikChange™ Site-Directed Mutagenesis Kit (Stratagene) in compliance with the manufacturer's manual. For the dex-inducible lines, KOD∷GFP was first subcloned from pART27 as a NotI/XhoI fragment into pENTR1A (Invitrogen). The Gateway Clonase reaction was carried out to transfer KOD∷GFP from pENTR1A into the pOpOn2.1 vector provided by Dr I Moore (Craft et al, 2005). This vector was then transformed into A. tumefaciens GV3101 for floral dip infiltration of Arabidopsis, and selected as described above.
Expression analysis
RNA was extracted using RNeasy Plant mini kit (Qiagen). PolyA+ mRNAs were obtained using the polyAtract kit (Promega). Reverse transcription was carried out using ProSTAR™ First-Strand RT–PCR kit (Stratagene) on 10 μg of DNAse RQI-treated total RNA or 0.1 μg polyA+ RNA. RT–PCR was carried out using primers, oSUPR and oGUSJ or oexTi15 or oTi05 (Supplementary Table S1) for 35 cycles. The gel was blotted on Nylon N+ membrane and hybridized with a PCR-radiolabelled probe corresponding to the oSUPR-oexTi15 fragment. Primers used for reference genes: ACTIN2 (act2.5, act2.3), ACTIN11 (act11F, act11R), XPO1a (oxpo4, oxpo7). 3′RACE was carried out using Race-3 and Race-anchor primers in conjunction with two nested primers in the KOD sequence: ToexF and KOD-ATGF (Supplementary Table S1). Q-PCR was carried out in triplicates using TaqmanR technology with primers KOD-267F, KOD-353R, KOD-293T; 18S-29F, 18S-102R, 18S-58T; Act2-123F, Act2-190R, Act2-148T (Supplementary Table S1). Genomic DNA was eliminated from RNA samples using DNAse RQI (Promega) and RT was carried out using MMLV (Promega) and primer Race-3 increased to 4 μM. Absolute transcript copy numbers were calculated using a genomic DNA standard curve.
Pathogen inoculations
P. syringae pv. tomato DC3000 wild-type (virulent) or expressing AvrRPM1 (avirulent) (both gift of Cyril Zipfel, The Sainsbury Laboratory) were grown overnight at 28°C in King's medium (OD 1.3), resuspended in 10 mM MgCl2 at OD 0.2 (108 c.f.u./ml) and infiltrated in leaves of 4-week-old plants. Leaf tissue was harvested at various times for analysis and spare infiltrated leaves examined for disease symptoms at 20 hpi.
H2O2 induction
H2O2 induction was obtained essentially as described in He et al (2008). Leaf discs were punched out of 4-week-old plants and floated on 3 ml sterile distilled water (SDW) or 30 mM H2O2.
Heat shock
Three-day-old seedlings were floated on 3 ml SDW in a six-well plate and the plate was floated in a waterbath at 55°C, 10 min without shaking. Seedlings were then kept in a light cabinet at 23°C until used to score root hair cell death under the microscope or harvested for RNA extraction. For heat shock of leaves, similar leaves were detached from 4-week-old plants and floated on 3 ml SDW and treated as above. Control samples were treated under the same conditions except for the heat shock.
Immunological detection
Leaf tissue (0.1 g) was homogenized in 200 μl of boiling PREXBU/Laemli buffer 4:1 (PREXBU: 100 mM MOPS pH 7.6, 100 mM NaCl, 5% (v/v) Glycerol, 1 mM EDTA, 14 mM β-MercaptoEthanol, 1 mM PMSF, 2 μg/ml Pepstatin A, 0.2 μg/ml Leupeptin, 1 μg/ml Aprotinin) using a MagNA Lyser at 6500 r.p.m. for 50 s. Extracts were cleared 10 min at 8000 g. In all, 10 μg were separated on a 12% SDS–PAGE, blotted on a nitrocellulose membrane and incubated with anti-GFP polyclonal antibody (Santa Cruz Biotech).
Transient expression
Tobacco leaf infiltrations were carried out on soil grown Nicotiana tabacum (Wisconsin 38) plants. Agrobacteria were infiltrated into tobacco leaves as described by Sparkes et al (2006). Onion epidermal cells were bombarded using a PDS-1000/He (Bio-Rad) and 10 μg total DNA loaded onto gold aliquots according to Hull et al (1996). The onion slices were incubated after bombardment in the dark for 15–50 h at 22°C, the epidermis peeled off and GFP-positive cells were observed with a Zeiss Axioplan fluorescence microscope equipped with an FITC filter. For dual RFP and GFP images, a Leica DM5500 fitted with a Photometrics cascade II 512B EMCCD camera (Photometrics UK) and a dual filter YFP/dsRED (part 51019; Chroma Technology Corp) was used. Pictures were captured using the SPOT Advanced suite and were edited with ImageJ. Immediately after counting fluorescent cells, epidermal pieces were incubated in GUS stain for 15 h at 37°C to score GUS-positive cells.
MitoTracker staining for mitochondrial depolarization
Onion epidermal pieces were incubated for 5 min with 100 nM of MitoTracker Red CMXRos (Invitrogen) prior to microscopic analysis.
Fluorescent caspase activity
Activity was detected in situ using the APO LOGIX Carboxyfluorescein Caspase Detection Kit, FAM-VAD-FMK. Onion cells were incubated for 10 min in 4 μl of the 30 × working dilution of FAM-VAD-FMK diluted to 300 μl with SDW. Cells were washed twice in 5 ml of SDW before microscopic analysis.
Fluorometric caspase assay
Individual leaves were ground in 250 μl of SDW before being pelleted (20 min, 8000 g/4°C). Resultant supernatants were transferred to fresh tubes and protein concentrations were measured using Bradford Reagent (Bio-Rad). Enzymatic assays were carried out using microtitre plates. For each well, 2 μg of protein was added to 100 μl final of assay buffer (200 mM NaCl, 50 mM NaAc pH 5.5, DTT 3 mM and 50 μM DEVD-Rhodamine110 (Bachem Ltd). Relative fluorescence units/min were measured using a Fluoroskan Ascent Fluorometre (Labsystems, DYNEX Technologies), with an excitation wavelength of 485 nm and emission of 530 nm. Data were analysed using the Fluoroskan Ascent software.
Ion leakage measurements
Measurements were carried out using a 24-well plate with each well containing 1 ml of SDW and one 1-cm leaf disc punched out from infiltrated area. After 1 h equilibration, the conductivity of 100 μl of water from each well was measured using a Horiba Twin Cond conductivity meter B-173 (HORIBA Ltd, Kyoto, Japan).
Evans blue readings
In all, 1-cm leaf discs were punched out from infiltrated area and were placed in 1.5 ml microcentrifuge tubes containing 1 ml of 0.025% Evans blue (Sigma). Samples were then vacuum infiltrated for 10 min to aid penetration of the stain. After rinsing, the discs were ground in 200 μl of 0.1% SDS, before being pelleted at 10 000 g for 5 min. The OD for 50 μl of the supernatant was then measured at 600 nm.
Supplementary Material
Acknowledgments
We thank C Berger for the discovery of line 276S, A Gilmore for BAX, A Day for the use of his particle gun, R Whitman for help with microscopes, D Puertolas for protoplast assays, S de Vries for hosting FV to screen seed cDNA libraries, SR Turner for critical reading of the manuscript, JA Hickman for helpful suggestions. This work was supported by the EU EPEN network, the CNRS, the French Ministry for Education and Research, and the University of Manchester. BY holds a BBSRC studentship.
Footnotes
The authors declare that they have no conflict of interest.
References
- Allen LJ, MacGregor KB, Koop RS, Bruce DH, Karner J, Bown AW (1999) The relationship between photosynthesis and a mastoparan-induced hypersensitive response in isolated mesophyll cells. Plant Physiol 119: 1233–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82: 259–266 [DOI] [PubMed] [Google Scholar]
- Bonneau L, Ge Y, Drury GE, Gallois P (2008) What happened to plant caspases? J Exp Bot 59: 491–499 [DOI] [PubMed] [Google Scholar]
- Bozhkov P, Filonova L, Suarez M (2005) Programmed cell death in plant embryogenesis. Curr Topics Dev Biol 67: 135–179 [DOI] [PubMed] [Google Scholar]
- Bozhkov PV, Filonova LH, Suarez MF, Helmersson A, Smertenko AP, Zhivotovsky B, von Arnold S (2003) VEIDase is a principal caspase-like activity involved in plant programmed cell death and essential for embryonic pattern formation. Cell Death Differ 11: 175–182 [DOI] [PubMed] [Google Scholar]
- Chilley PM, Casson SA, Tarkowski P, Hawkins N, Wang KLC, Hussey PJ, Beale M, Ecker JR, Sandberg GK, Lindsey K (2006) The POLARIS peptide of arabidopsis regulates auxin transport and root growth via effects on ethylene signaling. Plant Cell 18: 3058–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craft J, Samalova M, Baroux C, Townley H, Martinez A, Jepson I, Tsiantis M, Moore I (2005) New pOp/LhG4 vectors for stringent glucocorticoid-dependent transgene expression in Arabidopsis. Plant J 41: 899–918 [DOI] [PubMed] [Google Scholar]
- Danon A, Gallois P (1998) UV-C radiation induces apoptotic-like changes in Arabidopsis thaliana. FEBS Lett 437: 131–136 [DOI] [PubMed] [Google Scholar]
- Danon A, Rotari VI, Gordon A, Mailhac N, Gallois P (2004) Ultraviolet-C overexposure induces programmed cell death in arabidopsis, which is mediated by caspase-like activities and which can be suppressed by caspase inhibitors, p35 and defender against apoptotic death. J Biol Chem 279: 779–787 [DOI] [PubMed] [Google Scholar]
- Danon A, Sanchez Coll N, Apel K (2006) Cryptochrome-1-dependent execution of programmed cell death induced by singlet oxygen in Arabidopsis thaliana. Proc Natl Acad Sci USA 103: 17036–17041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devic M, Hecht V, Berger C, Delseny M, Gallois P (1995) An assessment of promoter trapping as a tool to study plant zygotic embryogenesis. CR Acad Sci Life Sci 318: 121–128 [Google Scholar]
- Grant M, Brown I, Adams S, Knight M, Ainslie A, Mansfield J (2000) The RPM1 plant disease resistance gene facilitates a rapid and sustained increase in cytosolic calcium that is necessary for the oxidative burst and hypersensitive cell death. Plant J 23: 441–450 [DOI] [PubMed] [Google Scholar]
- Groover A, DeWitt N, Heidel A, Jones A (1997) Programmed cell death of plant tracheary elements differentiating in vitro. Protoplasma 196: 197–211 [Google Scholar]
- Gunawardena AHLAN, Greenwood JS, Dengler NG (2004) Programmed cell death remodels lace plant leaf shape during development. Plant Cell 16: 60–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo A, Reimers PJ, Leach JE (1993) Effect of light on incompatible interactions between Xanthomonas oryzae pv oryzae and rice. Physiol Mol Plant Pathol 42: 413–425 [Google Scholar]
- He R, Drury GE, Rotari VI, Gordon A, Willer M, Farzaneh T, Woltering EJ, Gallois P (2008) Metacaspase-8 modulates programmed cell death induced by ultraviolet light and H2O2 in Arabidopsis. J Biol Chem 283: 774–783 [DOI] [PubMed] [Google Scholar]
- Heath MC (2000) Hypersensitive response-related death. Plant Mol Biol 44: 321–334 [DOI] [PubMed] [Google Scholar]
- Hoeberichts FA, Woltering EJ (2003) Multiple mediators of plant programmed cell death: interplay of conserved cell death mechanisms and plant-specific regulators. BioEssays 25: 47–57 [DOI] [PubMed] [Google Scholar]
- Hull G, Garrido JMG, Parcy F, Menossi M, Martinez-Izquierdo JA, Gallois P (1996) Use of the lacZ reporter gene as an internal control for GUS activity in microprojectile bombarded plant tissue. Plant Sci 120: 153–160 [Google Scholar]
- Ihara-Ohori Y, Nagano M, Muto S, Uchimiya H, Kawai-Yamada M (2007) Cell death suppressor arabidopsis Bax inhibitor-1 is associated with calmodulin binding and ion homeostasis. Plant Physiol 143: 650–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J, Fukuda H (2002) ZEN1 is a key enzyme in the degradation of nuclear DNA during programmed cell death of tracheary elements. Plant Cell 14: 3201–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai-Yamada M, Jin L, Yoshinaga K, Hirata A, Uchimiya H (2001) Mammalian Bax-induced plant cell death can be down-regulated by overexpression of Arabidopsis Bax Inhibitor-1 (AtBI-1). Proc Natl Acad Sci USA 98: 12295–12300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerry G, Mark W (1999) The use of chloromethyl-X-rosamine (MitoTracker Red) to measure loss of mitochondrial membrane potential in apoptotic cells is incompatible with cell fixation. Cytometry 36: 355–358 [DOI] [PubMed] [Google Scholar]
- Kim M, Kang Y (1999) Positional preference of proline in alpha-helices. Protein Sci 8: 1492–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacomme C, Santa Cruz S (1999) Bax-induced cell death in tobacco is similar to the hypersensitive response. Proc Natl Acad Sci USA 96: 7956–7961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey K, Casson S, Chilley P (2002) Peptides: new signalling molecules in plants. Trends Plant Sci 7: 78–83 [DOI] [PubMed] [Google Scholar]
- Lukowitz W, Roeder A, Parmenter D, Somerville C (2004) A MAPKK kinase gene regulates extra-embryonic cell fate in arabidopsis. Cell 116: 109–119 [DOI] [PubMed] [Google Scholar]
- McCabe PF, Leaver CJ (2000) Programmed cell death in cell cultures. Plant Mol Biol 44: 359–368 [DOI] [PubMed] [Google Scholar]
- Nakaune S, Yamada K, Kondo M, Kato T, Tabata S, Nishimura M, Hara-Nishimura I (2005) A vacuolar processing enzyme, delta-VPE, is involved in seed coat formation at the early stage of seed development. Plant Cell 17: 876–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara K, Kuriyama H, Fukuda H (2001) Direct evidence of active and rapid nuclear degradation triggered by vacuole rupture during programmed cell death in Zinnia. Plant Physiol 125: 615–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reape TJ, Molony EM, McCabe PF (2008) Programmed cell death in plants: distinguishing between different modes. J Exp Bot 59: 435–444 [DOI] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Saviani EE, Orsi CH, Oliveira JFP, Pinto-Maglio CAF, Salgado I (2002) Participation of the mitochondrial permeability transition pore in nitric oxide-induced plant cell death. FEBS Lett 510: 136–140 [DOI] [PubMed] [Google Scholar]
- Souèges R (1919) Les premières étapes de la division de l’oeuf et les différenciations du suspenseur chez le Capsella bursa-pastoris Moench. Ann des ScBot 10: 1–28 [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc 1: 2019–2025 [DOI] [PubMed] [Google Scholar]
- Suarez MF, Filonova LH, Smertenko A, Savenkov EI, Clapham DH, von Arnold S, Zhivotovsky B, Bozhkov PV (2004) Metacaspase-dependent programmed cell death is essential for plant embryogenesis. Curr Biol 14: R339–R340 [DOI] [PubMed] [Google Scholar]
- Till BJ, Reynolds SH, Greene EA, Codomo CA, Enns LC, Johnson JE, Burtner C, Odden AR, Young K, Taylor NE, Henikoff JG, Comai L, Henikoff S (2003) Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res 13: 524–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacca RA, de Pinto MC, Valenti D, Passarella S, Marra E, De Gara L (2004) Production of reactive oxygen species, alteration of cytosolic ascorbate peroxidase, and impairment of mitochondrial metabolism are early events in heat shock-induced programmed cell death in tobacco bright-yellow 2 cells. Plant Physiol 134: 1100–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpon L, Lamthanh H, Barbier J, Gilles N, Molgó J, Ménez A, Lancelin J (2004) NMR solution structures of delta-conotoxin EVIA from Conus ermineus that selectively acts on vertebrate neuronal Na+ channels. J Biol Chem 279: 21356–21366 [DOI] [PubMed] [Google Scholar]
- Wen J, Lease KA, Walker JC (2004) DVL, a novel class of small polypeptides: overexpression alters Arabidopsis development. Plant J 37: 668–677 [DOI] [PubMed] [Google Scholar]
- Wrzaczek M, Brosche M, Kollist H, Kangasjarvi J (2009) Arabidopsis GRI is involved in the regulation of cell death induced by extracellular ROS. Proc Natl Acad Sci USA 106: 5412–5417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga K, Arimura S-i, Hirata A, Niwa Y, Yun D-J, Tsutsumi N, Uchimiya H, Kawai-Yamada M (2005) Mammalian Bax initiates plant cell death through organelle destruction. Plant Cell Rep 24: 408–417 [DOI] [PubMed] [Google Scholar]
- Young B, Wightman R, Blanvillain R, Purcel S, Gallois P (2010) pH-sensitivity of YFP provides an intracellular indicator of programmed cell death. Plant Methods 6: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Perdue T, Heimer Y, Jones A (2002) Mitochondrial involvement in tracheary element programmed cell death. Cell Death Differ 9: 189–198 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.