Abstract
The capsaicin receptor TRPV1, a member of the transient receptor potential family of non-selective cation channels is a polymodal nociceptor. Noxious thermal stimuli, protons, and the alkaloid irritant capsaicin open the channel. The mechanisms of heat and capsaicin activation have been linked to voltage-dependent gating in TRPV1. However, until now it was unclear whether proton activation or potentiation or both are linked to a similar voltage-dependent mechanism and which molecular determinants underlie the proton gating. Using the whole-cell patch-clamp technique, we show that protons activate and potentiate TRPV1 by shifting the voltage dependence of the activation curves towards more physiological membrane potentials. We further identified a key residue within the pore region of TRPV1, F660, to be critical for voltage-dependent proton activation and potentiation. We conclude that proton activation and potentiation of TRPV1 are both voltage dependent and that amino acid 660 is essential for proton-mediated gating of TRPV1.
Keywords: gating, proton, TRP, TRPV1, voltage dependence
Introduction
The superfamily of transient receptor potential (TRP) channels has a major role in sensory transduction including thermosensation, taste sensation, hearing, vision, touch, osmosensation, and pain sensation (Clapham, 2003; Dhaka et al, 2006; Ramsey et al, 2006; Damann et al, 2008). The mammalian TRP channel family currently comprises 28 members which can further be classified into seven subfamilies based on sequence homologies (Montell et al, 2002; Clapham et al, 2003): TRPCs (canonical), TRPVs (vanilloid), TRPMs (melastatin), TRPAs (ankyrin), TRPPs (polycystin), and TRPMLs (mucolipin). TRP channels are cation channels: some are highly selective for divalent cations such as Ca2+, Mg2+, Fe2+, or Zn2+, others are non-selective for mono- and divalent cations, and still others are only permeable to monovalent cations. TRP channels can form homo- or heterotetramers, where each subunit consists of six transmembrane spanning domains (TMD1–TMD6), a pore domain between TMD5 and TMD6, and both C- and N-termini located intracellular. Besides these general characteristics, TRP channels display a plethora of different structural and functional features (Clapham, 2003; Ramsey et al, 2006; Damann et al, 2008). A subgroup of TRP channels, namely TRPV1–TRPV4, TRPM8, and TRPA1, are heat- or cold-sensitive (Clapham, 2003; Dhaka et al, 2006; Ramsey et al, 2006; Damann et al, 2008). Recent evidence suggests that physical stimuli such as temperature, but also the binding of various ligands shifts the voltage dependence of channel activation, for example, of TRPV1, TRPV3, TRPM5, or TRPM8, towards physiologically relevant potentials (Nilius et al, 2003; Voets et al, 2004, 2005, talavera et al, 2005). Attempts to identify molecular gating determinants of capsaicin or heat activation revealed that mutations in the pore region of TRPV1 lead to a selective loss of capsaicin or heat sensitivity (Kuzhikandathil et al, 2001; Grandl et al, 2010). Likewise, specific point mutations have been identified in recent mutagenesis experiments which selectively abolish heat activation of the TRPV1-related TRP channel TRPV3 (Grandl et al, 2008, 2010). In the TRPV1 study, the triple mutant N628K/N652T/Y653T showed the most pronounced effect on temperature activation. For the same mutant it could also be demonstrated that the temperature–voltage coupling was changed while allosteric coupling of capsaicin voltage was normal (at 20°C; Grandl et al, 2010). Another triple mutant with mutations in TMD6, TRPV1 (NML676FAP), had been reported previously to disrupt the ability of capsaicin and resiniferatoxin to activate TRPV1 while retaining the ability to respond to protons (Kuzhikandathil et al, 2001), suggesting that distinct amino acids close to or in TMD6 control gating in response to several modes of TRPV1 activation. However, until now it was unclear whether TRPV1 proton activation or potentiation or both would show voltage dependence similar to heat and capsaicin activation and what were the underlying molecular determinants.
TRPV1-mediated proton sensing in tissues is physiologically relevant under normal conditions and in disease states (Holzer, 2007; Sugiura et al, 2007). Tissue damage such as that associated with infection or inflammation produces a variety of chemical mediators that activate or sensitize nociceptor terminals to elicit pain and promote tenderness at the site of injury. TRPV1 channels are abundant on dorsal-root and nodose afferents innervating different organs, for example, stomach and colon (Holzer, 2007), and serve there as acidity sensors. By sensing protons with two different extracellular amino-acid residues (E600 and E648), pH has a dual effect on the TRPV1 channel: at low pH (<6), protons open the channel; at high pH (6–7), protons lower the threshold for TRPV1 activation, for example, by capsaicin (Jordt et al, 2000).
Based on the findings presented here, we propose that proton activation and potentiation of TRPV1 are both voltage dependent and that amino acid F660 in TMD6 is the key integrator of both the processes.
Results
Voltage dependence of proton activation of TRPV1
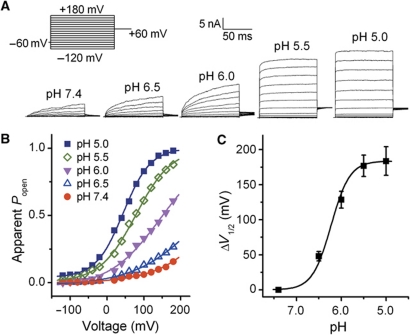
Temperature sensing is tightly linked to voltage-dependent gating in TRPV1 (Voets et al, 2004). Agonists of TRPV1 and TRPM8 such as capsaicin and menthol have been demonstrated to function as gating modifiers, shifting activation curves towards physiological membrane potentials, thus mimicking and potentiating the thermal responses (Voets et al, 2004). We, therefore, tested the hypothesis that TRPV1 is activated by protons in a similar way. In whole-cell patch-clamp experiments with HEK293 cells transiently expressing human TRPV1 currents were elicited using a voltage step protocol ranging from −120 to +180 mV at different extracellular pH values (Figure 1A). At pH 7.4, there was very little channel activity even at potentials above +100 mV, and the midpoint voltage of activation (V1/2) was 234±17 mV. Increasing proton concentration induced a gradual leftward shift of the activation curve resulting in channel activity at more negative voltages. V1/2 was shifted by 184±20 mV when the extracellular pH was reduced from 7.4 to 5.0 (Figure 1B and C, n=7). These data suggest that, similar to capsaicin and heat, protons activate TRPV1 by causing a large shift of the voltage dependence of activation.
Figure 1.
Voltage shift of activation curve of TRPV1 by protons. (A) Representative whole-cell current traces elicited by a voltage step protocol in response to protons as indicated. (B) Normalized steady-state maximal conductance curves at different pH values obtained from current traces shown in A. Lines represent Boltzmann fit to the data. (C) ΔV1/2 as a function of pH (n=7). Solid line represents Hill fit to the data. Error bars represent s.e.m.
TRPV1 mutant F660S ablates proton activation but not capsaicin or heat activation
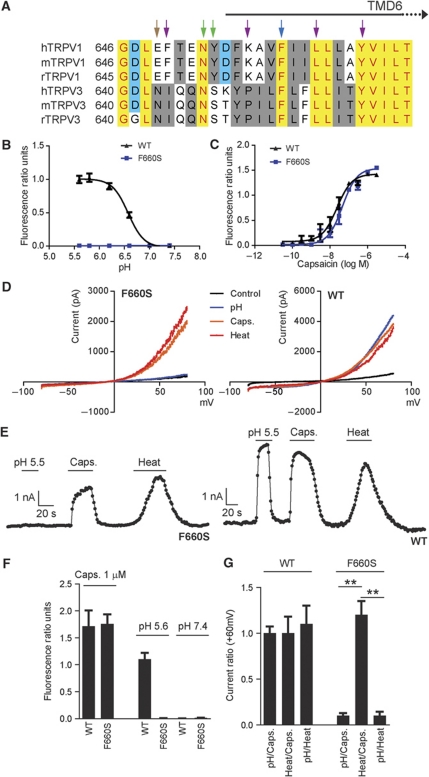
The pore regions of TRPV1 and TRPV1-related TRPV3 are packed with amino-acid residues involved in different modes of activation. Grandl et al (2008, 2010) have recently identified TRPV1 and TRPV3 mutants displaying selectively impaired heat activation profiles while maintaining their ability to be activated by protons and capsaicin (TRPV1) or 2-APB and camphor (TRPV3), respectively. Specifically, N652 and Y653 in TRPV1 and the equivalent amino acids in TRPV3, N647, and S648, have been shown to have a role in heat activation (green arrows, Figure 2A). In addition, N643 has been reported (Grandl et al, 2008) to affect heat activation of TRPV3 (purple arrows). The equivalent position in TRPV1, E648 (brown arrow, Figure 2A), was found to be critical for proton activation but not for proton potentiation (Jordt et al, 2000). Other amino acids shown to be required for heat activation of TRPV3 are indicated by purple arrows (I644, P451, L657, and Y661, Figure 2A, Grandl et al, 2008). In addition, amino acid N628 in the pore loop of TRPV1 was found to be involved in heat activation (Grandl et al, 2010), while mutations at position 633 affect proton activation of TRPV1 (Ryu et al, 2007).
Figure 2.
TRPV1 mutant F660S ablates proton activation but not capsaicin or heat activation. (A)An amino-acid sequence alignment of part of the pore region, including the upper third of TMD6 of human (h), mouse (m), and rat (r) TRPV1 and TRPV3. Residues reported to be specifically required for temperature activation of TRPV3 are indicated by purple arrows (I644, P651, L657, and Y661). Amino acids found to be required for temperature activation of both rTRPV1 and mTRPV3 are indicated by green arrows. The blue arrow indicates amino acid involved in heat activation of mTRPV3 and proton activation and potentiation of hTRPV1. The brown arrow indicates amino acid involved in heat activation of mTRPV3 and proton activation of hTRPV1. (B, C) Concentration response curves obtained from Fluo-4-based calcium flux experiments using HEK293 cells transiently expressing hTRPV1 wild-type or TRPV1 (F660S) mutant. Representative pH dose–response measurement (B) using MES-buffered solutions adjusted to different pH values. MES-buffered solutions were added to cells loaded with Fluo-4 calcium indicator dye in standard buffer solution without HEPES (adjusted to pH 7.4) resulting in final pH values of 5.6, 5.8, 6.2, 6.6, 7.0, and 7.4. Representative capsaicin dose–response measurement (C) performed in standard buffer solution containing 20 mM HEPES, adjusted to pH 7.4. Average EC50 values (mean±s.d.) of 25±12 nM were measured for F660S versus 12±6 nM for wild-type TRPV1 (n=10, each). (D) Steady-state current–voltage relationships obtained from whole-cell patch-clamp experiments with HEK293 cells transiently expressing hTRPV1 wild-type or F660S mutant. Cells were stimulated subsequently with protons (pH 5.5), capsaicin (1 μM), or heat (45°C). (E) Representative whole-cell current traces (at +60 mV) obtained from HEK293 cells transiently expressing hTRPV1 or F660S mutant after subsequent stimulation with protons (pH 5.5), capsaicin (100 nM), or heat (45°C). (F) Bar diagram summarizing data obtained from experiments as described in B and C. (G) Bar diagram showing ratio values calculated based on data as described in D and E. Statistical analysis was performed using one-way ANOVA followed by Tukey's post test; **P<0.001.
In an attempt to identify amino acids which may be selectively involved in pH-mediated channel gating and pH–voltage coupling of TRPV1, we therefore, decided to generate multiple TRPV1 isoforms with mutations in the pore loop and TMD6. Mutations at position F660 in TMD6 (the equivalent in TRPV3, F654, is involved in heat activation, Grandl et al, 2008) were found to selectively affect pH activation of TRPV1 (blue arrow, Figure 2A). Calcium flux experiments using HEK293 cells transiently expressing TRPV1 mutants or wild-type TRPV1 revealed a complete lack of activation of mutant F660S by protons (Figure 2B and F). In contrast, F660S maintained responsiveness to capsaicin with average EC50 values (mean±s.d.) of 25±12 versus 12±6 nM for wild-type TRPV1 at pH 7.4 final (n=10, each, HEPES-buffered; Figure 2C and F). Likewise, in whole-cell patch-clamp experiments cells expressing the F660S mutant were not activated by protons (pH 5.5). In contrast, activation by capsaicin (1 μM, pH 7.4) and heat (45°C, pH 7.4) was similar to wild-type TRPV1 (Figure 2D and E). To minimize variability in the transient expression system, the following current ratios were calculated (+60 mV): pH/capsaicin 0.1±0.03 at pH 5.5 for F660S versus 1.0±0.1 for wild-type TRPV1. Similarly, pH/heat current ratios were 0.1±0.02 for F660S versus 1.0±0.2 (+60 mV) for wild-type TRPV1. In contrast, heat/capsaicin current ratios were 1.2±0.2 for F660S versus 1.0±0.1 (+60 mV) for wild-type TRPV1 (Figure 2G).
Aromaticity at position 660 is critical for proton activation
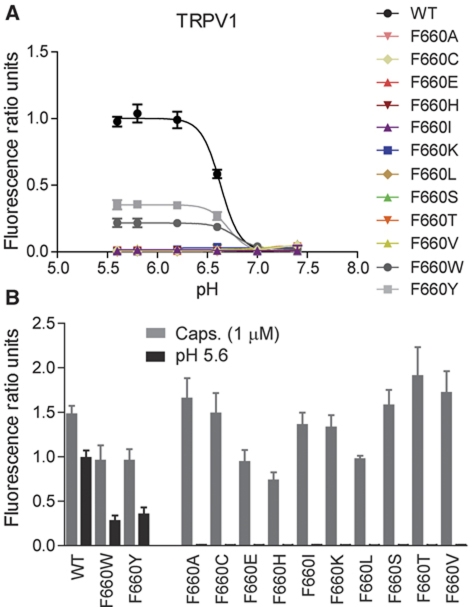
In subsequent experiments, we replaced amino acid F660 with a variety of different amino acids to determine the side-chain contribution to the proton activation of TRPV1. Proton activation was ablated by all amino-acid replacements with the exception of F660Y and F660W, the two alternative non-basic aromatic amino acids besides phenylalanine. Replacing phenylalanine with histidine, which contains a basic aromatic ring (imidazole) or non-aromatic amino acids caused complete loss of proton activation (Figure 3A and B). However, F660Y showed reduced sensitivity to proton activation compared with wild-type TRPV1 (Figure 3A and B). Although less pronounced, maximum effect values at 1 μM capsaicin were also reduced compared with wild type (Figure 3A and B), while capsaicin EC50 values at pH 7.4 were comparable: 14±6 nM for F660Y versus 17±9 nM (P>0.05, n=5) for wild-type TRPV1 (measured in MES-buffered solution). F660W showed a reduction in sensitivity to proton activation as well as capsaicin activation similar to F660Y (Figure 3A and B). These data suggest that a non-basic aromatic amino acid at position 660 is essential for proton activation.
Figure 3.
An aromatic amino acid at position 660 is required for proton activation. (A) Calcium flux experiments as described in Figure 2B, showing pH concentration response curves obtained from HEK293 cells transiently transfected with either hTRPV1 wild type or one of the following mutants: F660A, F660C, F660E, F660H, F660I, F660K, F660L, F660S, F660T, F660V, F660W, and F660Y. Only wild type, F660Y, and F660W responded to proton stimulation. (B) Bar diagram summarizing results as shown in A, obtained from at least five independent experiments, each.
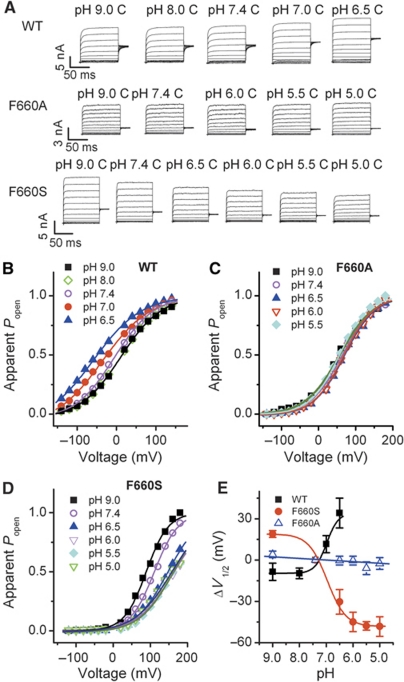
Mutations at position 660 affect the voltage dependence of proton activation
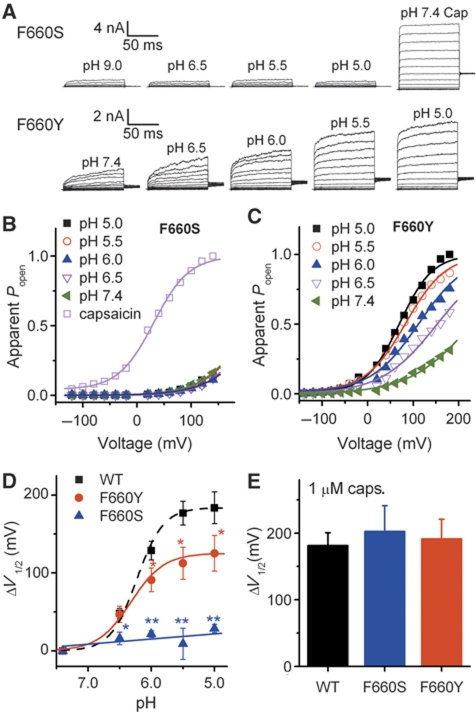
As shown in Figure 2B–I, mutation of phenylalanine at position 660 to serine completely ablates the response of TRPV1 to protons, while preserving the responsiveness to capsaicin and heat. Currents elicited with the voltage step protocol as applied in Figure 1 showed that 1 μM capsaicin induced a 203±38 mV shift of V1/2 voltage activation in F660S, which is very similar to that of wild-type TRPV1 (P>0.05, n=4–5). A shift of voltage dependence was not observed when F660S was activated in a pH range up to 5.0. Replacement of phenylalanine with tyrosine, another aromatic amino acid, showed a voltage-dependent proton activation similar to wild-type TRPV1 (Figure 4A–C). However, the V1/2 shift in Y660 was only 125±23 mV when the extracellular pH was reduced from 7.4 to 5.0, which is significantly smaller than the one found for wild-type TRPV1 (Figure 4D; P<0.01, n=5–6). These data indicate that position 660 of human TRPV1 is critical for the voltage dependence of proton activation.
Figure 4.
Mutations at position 660 affect the voltage dependence of proton activation. (A) Representative whole-cell current traces for TRPV1 (F660S) and TRPV1 (F660Y) mutants elicited by a voltage step protocol in response to protons as indicated. (B, C) Normalized steady-state maximal conductance curves at different pH values obtained from current traces shown in A. Lines represent Boltzmann fit to the data. (D) ΔV1/2 as a function of pH for wild type, F660S, and F660Y (n=5–6, *P<0.05, **P<0.01). Lines represent Hill fit to the data. Error bars represent s.e.m. (E) Comparison of ΔV1/2 for wild type, F660S, and F660Y in response to 1 μM capsaicin (n=4–5, P>0.05). Error bars represent s.e.m.
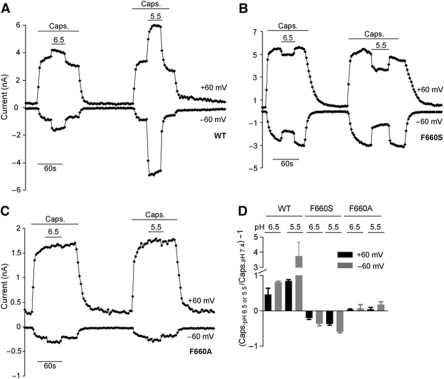
Position F660 is critical for the gating of proton-mediated potentiation of TRPV1
To assess the effect of mutations at position 660 also on proton-mediated potentiation, whole-cell patch-clamp experiments were performed using buffers adjusted to different pH values (pH 5.5, 6.5, and 7.4 final) in combination with 100 nM capsaicin. These experiments revealed an inhibitory effect of protons on TRPV1 (F660S) activation by capsaicin. Instead of potentiating capsaicin responses at low extracellular pH (pH 5.5 or 6.5) as seen with wild-type TRPV1 (Figure 5A and B), capsaicin responses were inhibited when switching from neutral pH to acidic pH. Average current ratio values of the responses (mean±s.e.m.) shown in Figure 5E were calculated based on the following equation: (capsaicin response at pH 6.8 or 5.5/capsaicin response at pH 7.4)−1. For TRPV1 (F660S), the calculated ratio values were −0.2±0.04 (+60 mV) and −0.36±0.06 (−60 mV) for pH 6.5, and −0.37±0.04 (+60 mV) and −0.59±0.03 (−60 mV) for pH 5.5 (Figure 5E). For wild-type TRPV1, the values were +0.46±0.18 (+60 mV) and +0.81±0.03 (−60 mV) for pH 6.5, and +0.84±0.04 (+60 mV) and +3.7±0.9 (−60 mV) for pH 5.5 (Figure 5E). While in TRPV1 (F660S) mutant expressing cells the potentiation effect on the capsaicin mode of activation was reversed compared with wild-type TRPV1, TRPV1 (F660A) did neither significantly inhibit nor significantly potentiate capsaicin responses in presence of protons (Figure 5C). Average ratio values were 0.03±0.02 (+60 mV) and 0.07±0.1 (−60 mV) for pH 6.5, and 0.04±0.05 (+60 mV) and 0.18±0.08 (−60 mV) for pH 5.5 (Figure 5E). Calcium flux experiments using capsaicin in combination with MES-buffered solutions adjusted to different pH values (pH 6.8 and 7.4 final) showed that capsaicin concentration response curves were shifted in the opposite direction at low extracellular pH in TRPV1 (F660S) expressing cells compared with wild type (Supplementary Figure S1A and B), further corroborating the effects on TRPV1 wild-type and mutant capsaicin activation under acidic conditions observed in patch-clamp experiments. The following average capsaicin EC50 values were calculated for F660S: 125±15 nM at pH 6.8 versus 35±8 nM at pH 7.4 (MES-buffered). Capsaicin concentration response curves in TRPV1 (F660A) expressing cells on the other hand did not significantly change with pH (Supplementary Figure S1C), likewise confirming patch-clamp experiments shown previously. The following average capsaicin EC50 values were calculated for F660A: 39±13 nM at pH 6.8 versus 41±12 nM at pH 7.4.
Figure 5.
Proton potentiation is affected in TRPV1 (F660) mutants. (A–C) Effect of acidic pH on capsaicin stimulation in HEK293 cells expressing either hTRPV1 wild-type (A), TRPV1 (F660S), or TRPV1(F660A) mutant (B, C). Shown are representative current traces (at +60 mV and −60 mV), each. Substitution of F660 to alanine caused a selective loss of activation and potentiation by protons (pH 5.5) but did not cause inhibition of capsaicin responses as shown for F660S. Substitution of F660 to tyrosine did not result in a complete loss of activation by protons, but caused an inhibition of capsaicin responses similar to F660S. (D) Summary of data obtained from experiments as shown in (A–C). All values are shown as mean±s.e.m.
Voltage dependence of proton-mediated potentiation of TRPV1
To investigate the mechanism of proton-mediated potentiation in TRPV1 wild-type and mutant channels, we applied the same voltage step protocol as in Figure 1. As in Figure 5, 100 nM capsaicin were used for activation and voltage dependence was tested at the indicated range of pH values (Figure 6). In wild-type TRPV1, there was a small but clear gradual shift of the voltage dependence when extracellular pH was decreased from 9.0 to 6.5 (Figure 6A and B). Owing to significant channel activation beyond pH 6.5 (Figure 1) in wild type, higher proton concentrations were not applied. In TRPV1 (F660A) mutant expressing cells voltage dependence of proton-mediated potentiation was completely lost (Figure 6A and C). In contrast, in F660S expressing cells, voltage dependence was reversed compared with wild-type TRPV1 (Figure 6D). The V1/2 voltage shift data of wild type and mutants are summarized in Figure 6E. These data demonstrate the correlation between different potentiation phenotypes (Figure 5) and the differing voltage dependence shifts found in wild-type TRPV1 (leftward shift), F660A mutant (no shift) and F660S mutant (rightward shift).
Figure 6.
Proton-mediated sensitization of TRPV1 capsaicin activation is voltage dependent. (A) Representative whole-cell current traces of wild-type and mutant TRPV1 (F660A and F660S) elicited by 100 nM capsaicin at different pH values as indicated. C means the indicated pH solution contained 100 nM capsaicin. (B–D) Normalized steady-state maximal conductance curves at different pH values obtained from current traces shown in A. Lines represent Boltzmann fit to the data. (E) ΔV1/2 as a function of pH for wild type, F660A and F660S currents elicited by 100 nM capsaicin (n=5–6). Lines represent Hill fit to wild type and F660S and linear fit to F660A data. Error bars represent s.e.m.
Discussion
The first key finding of our study is that proton sensing of human TRPV1 is associated with a shift in its voltage dependence. In a recent study, Voets et al (2004) found that temperature sensing is tightly linked to voltage-dependent gating in TRPV1. They also showed that capsaicin functions as a gating modifier, shifting activation curves, thus mimicking and potentiating the thermal responses. However, until now it was unclear whether proton activation or potentiation or both would show a similar voltage dependence. Here, we show that protons activate and potentiate TRPV1 by shifting the voltage dependence of the activation curves towards more physiological membrane potentials. Channel activity is determined by the product of the number of channels in the cell membrane (N), the unitary conductance (i), and the open probability (po). Extracellular protons have been shown to act primarily by increasing the probability of TRPV1 channel opening (Tominaga et al, 1998; Baumann and Martenson, 2000) rather than by increasing unitary conductance. In addition, Baumann and Martenson (2000) found that acidic solutions increase the po of TRPV1 channels. Our data demonstrate that the steady state apparent open probability of the channel is tightly linked to voltage-dependent gating when protons activate and potentiate TRPV1. The shift of the voltage dependence of potentiation in wild type, F660A and F660S at different pH values can be explained by either a proton-mediated change of channel gating or a proton-mediated change of capsaicin binding affinity or both (Colquhoun, 1998). The location of phenylalanine 660 (TMD6) is, however, quite distant from the reported capsaicin-binding site (between TMD3 and TMD4; Jordt and Julius, 2002). Also, voltage dependence of activation is already shifted to the left at pH 6.5 in wild-type TRPV1 but not in the F660S mutant. Finally, it was shown that low pH (5.5–7.0) does not increase the binding affinity of resiniferatoxin (Szallasi and Blumberg, 1993) which shares the same binding site on TRPV1 with capsaicin (Chou et al, 2004; Gavva et al, 2004). It is, therefore, more likely that mutations at this position affect the gating of the channel rather than the binding affinity of capsaicin. In summary, these data suggest that protons facilitate the voltage-dependent gating of TRPV1 resulting in the potentiation of TRPV1 capsaicin activation.
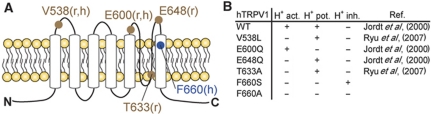
The second key finding of the present study is the identification of a distinct residue in TMD6 that is required for proton activation and potentiation of TRPV1. Mutating F660 in TMD6 of human TRPV1 to serine causes a loss of proton activation and potentiation while capsaicin and heat responsiveness are maintained. F660 is different from other residues, which have been reported to be involved in proton activation or potentiation as summarized in Figure 7. TRPV1 pH mutants identified previously were either mutants lacking the ability to potentiate capsaicin and/or heat effects or mutants with impaired proton activation. Jordt et al (2000) described a glutamic residue (E600) on the extracellular side of TMD5 as crucial for proton potentiation of capsaicin and heat responses. They also described another acidic residue, E648, located in the linker between selectivity filter and TMD6 to mediate direct response to protons and concluded that proton-evoked channel activation and proton-mediated potentiation were distinct processes. Ryu et al (2007) identified two sites, one involving the pore helix (T633) and the other one the extracellular loop between TMD3 and TMD4 (V538) as critical for proton activation. Finally, Wang et al (2010) reported that amino acid E536 in the extracellular loop between TMD3 and TMD4 is important for further stimulation of fully liganded TRPV1. However, none of these mutants showed a simultaneous role in activation and potentiation by protons as observed with F660 mutants, suggesting that amino acid 660 is the key integrator of proton activation and potentiation. The mutagenesis study at position 660 revealed that whenever phenylalanine was replaced by non-aromatic amino acids or histidine (aromatic but weakly basic) the resulting mutants could no longer be activated by protons but maintained their responsiveness to capsaicin. Based on these findings, we conclude that a non-aromatic amino acid or histidine at position 660 can be tolerated for the channel to be functional in the capsaicin activation mode; a non-basic aromatic side chain, however, appears to be required to maintain activation by protons.
Figure 7.
Summary of known and novel pH mutants of TRPV1. (A) Schematic topology showing the putative positions of TRPV1 point mutations involved in proton activation or potentiation (brown) or activation and potentiation (blue). Letter(s) in parentheses indicate whether the respective position is in human (h) and/or rat (r) TRPV1. (B) Published and novel TRPV1 point mutants with effect on proton activation and/or potentiation of TRPV1.
To clearly distinguish between ligand binding and gating (Colquhoun, 1998) is difficult as mutations affecting proton sensing or gating will both result in a shift of the voltage-dependent activation curve of TRPV1 channels. There is, however, a clear difference between our mutants and the mutants described previously (Jordt et al, 2000). Jordt et al showed a clear titration phenotype (exemplified by E600) which suggested proton sensing is affected. On the contrary, the loss of activation by protons when replacing F660 with charged amino acids and the absence of a titration phenotype suggest that phenylalanine is critical for the transduction of proton-mediated gating rather than voltage or proton sensing. In addition, F660S cannot be activated by protons but apparently can still sense pH because TRPV1 (F660S) capsaicin activation is inhibited by protons reflected by the shift to the right of the voltage-dependent activation curve. Interestingly, a similar role of phenylalanine in gating transduction in Shaker potassium channels has been reported very recently (Tao et al, 2010). In this elegant work, Tao et al showed that the side chain of Phe233 in transmembrane 3 is located in the centre of voltage sensors, and that only two substitutions at this position, Trp or Tyr, produce currents near wild-type levels. By replacing Phe233 with cyclohexylalanine, they further demonstrated that a rigid cyclic side chain is critical to catalyse the transfer of positive charges across the membrane. Because the cyclohexane ring is not well suited for direct interaction with gating charges, these results suggest that the native phenylalanine residue serves a structural role as part of the seal to assure smooth transmembrane domain 4 movement through the gating pore without ionic leakage (Catterall, 2010). Based on the similarity of the role of phenylalanine in Shaker potassium channels and in TRPV1, it may be interesting to focus on the regions surrounding the 660 side chain to indentify amino acids involved in voltage sensing in TRPV1.
It is intriguing to speculate, given the selective modulation of different modes of TRPV1 activation, that selective antagonism of the respective TRPV1 activation modes may generally be possible. Although our data underline the close proximity of structural domains involved in heat and pH activation of TRPV1, they also indicate that proton activation can be separated from heat activation on a molecular level. This is of mechanistic as well as therapeutic interest considering the observation of hyperthermia side-effects of polymodal TRPV1 antagonists in vivo and a potential correlation with block of proton activation as reported recently (Garami et al, 2010). If suggested correlation between block of proton activation and in vivo development of hyperthermia is translatable across species, it should be of advantage to design TRPV1 antagonists, which block heat and capsaicin modes of activation while being pH neutral. Alternatively, if heat block is the major cause of hyperthermia, compounds blocking proton and capsaicin but not heat activation could be advantageous. The presented data support the possibility of generating antagonists, which are more selective for capsaicin, heat or pH activation.
In summary, we conclude that proton activation and potentiation of TRPV1 are both voltage dependent and that amino acid 660 is the key residue regulating proton-mediated gating of human TRPV1.
Materials and methods
DNA constructs, cell culture, and transfection
Human wild-type TRPV1 (NM_080704) was subcloned into pcDNA6 V5 His B expression vector (Invitrogen Life Technologies, Breda, The Netherlands). All mutants were generated by in vitro mutagenesis (Quick-Change XL Mutagenesis Kit, Stratagene, La Jolla, CA, USA in combination with appropriate mutagenic primers) and verified by sequencing both strands entirely. HEK293 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% (v/v) fetal bovine serum. Cells were transiently transfected with Lipofectamine 2000 according to the manufacturer’s protocol (Invitrogen Life Technologies) and used for patch-clamp experiments 24–48 h after transfection. Alternatively, HEK293 Freestyle cells and 293fectin Transfection Reagent (Invitrogen Life Technologies) were used for transient transfection of wild-type and mutant TRPV1 isoforms.
Electrophysiology and calcium flux measurements
Patch-clamp experiments were performed in whole-cell configuration at room temperature (20–22°C) using an Axopatch 200B or Multiclamp 700A patch-clamp amplifier controlled by Pclamp 10 software (Molecular Devices). Patch pipettes had resistances between 2 and 4 MΩ. Basic extracellular solution contained (mM) 150 NaCl, 5 KCl, 1 CaCl2, 2 MgCl2, 20 HEPES, and 10 glucose; pH was adjusted to 7.4 with NaOH. For solutions with pH values <6.5, HEPES was replaced with MES. The intracellular (pipette) solution contained (mM) 150 CsF, 1 MgCl, 1 MgATP, 10 HEPES, and 10 BAPTA; pH was adjusted to 7.3 with CsOH. The osmolarity of all solutions was maintained at 300–315 mOsm/l. All chemicals were purchased from Sigma. Currents were sampled at 20 kHz and filtered at 5 kHz. The series resistance between 60 and 90% was compensated to reduce voltage errors. Two voltage protocols were used to elicit TRPV1 currents. A linear voltage ramp (−80 to +80 mV in 200 ms) was applied every 4 s from a holding potential of −60 mV. Current–voltage (I–V) relationships were obtained from the currents measured during the voltage ramp. A voltage step protocol was applied to study the voltage dependence of TRPV1 activation. The voltage protocol consisted of steps from −120 to +180 mV (with 20 mV increments) from a holding potential of −60 mV, followed by a step to +60 mV (Figure 1A). Whole-cell patch-clamp data were analysed using Clampfit 10 (Molecular Devices) and Origin 8.1 (Originlab). Results are presented as mean±s.e.m. Tests for statistical significance were performed using Student’s t-test or non-parametric ANOVA as noted. Apparent Popen was calculated from steady-state current–voltage relationships as G/Gmax, where Gmax represents the maximal steady-state conductance, which was obtained at strongly depolarized potentials (+180 mV) in response to pH 5.0 or 1 μM capsaicin (Voets et al, 2004). Heat responses of TRPV1 were recorded by heating perfusion solutions with a SC-20 dual in-line heater/cooler (Warner Instruments). Popen–voltage relationship data were fitted with the Boltzmann function and pH–response relationships were fitted with the Hill equation.
For calcium flux experiments transiently transfected cells were plated after 48 h into poly-D-lysine coated 384-well clear-bottom assay plates (Greiner) at a cell density of 20 000/well. Four to five hours after plating, cells were loaded with Fluo-4 calcium dye (FLIPR calcium 4 assay kit, Molecular Devices) for 1 h at room temperature in the dark. For analysis, a Functional Drug Screening System (FDSS6000, Hamamatsu) was used. Fluo-4 calcium dye solution was prepared according to manufacturer’s protocol using 1 × HBSS containing 1 mM CaCl2 and 1 mM MgCl2 but no HEPES or probenecid (pH 7.4). For pH measurements, MES (2-(N-morpholino)ethanesulfonic acid) buffered solutions were prepared in 1 × HBSS buffer: 4 ml of 1 M MES solution was diluted in 46 ml of HBSS buffer w/o HEPES and w/o probenecid (not adjusted to pH 7.4), giving a final concentration of 20 mM. Solutions were adjusted to respective pH values using 1 N NaOH as appropriate. In all calcium flux experiments fluorescence ratio units were calculated as follows: (Max/Min)−Min whereby Min means before and Max after agonist addition. Curve fitting and statistical analysis were performed using GraphPad Prism 5 (GraphPad Software Inc., San Diego, CA, USA).
Supplementary Material
Acknowledgments
We thank Dr Peter Cox, Dr Sarah Nickolls, and Dr Mike Trevethick (Pfizer Global R&D) for discussing data presented in this paper, Dr Peter Cox for kindly reading the manuscript, and Jade Strover and Dr James Turner (Pfizer Global R&D) for technical support.
Author contributions: EA, LC, and CG designed the study, and collected and analysed data. EA, LC, and CG wrote the manuscript. MP collected and analysed data. ES and SP edited the manuscript. All of the authors discussed the results and commented on the manuscript.
Footnotes
The authors declare that they have no conflict of interest. All authors are Pfizer employees.
References
- Baumann TK, Martenson ME (2000) Extracellular protons both increase the activity and reduce the conductance of capsaicin-gated channels. J Neurosci 20: RC80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall WA (2010) Ion channel voltage sensors: structure, function, and pathophysiology. Neuron 67: 915–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou MZ, Mtui T, Gao YD, Kohler M, Middleton RE (2004) Resiniferatoxin binds to the capsaicin receptor (TRPV1) near the extracellular side of the S4 transmembrane domain. Biochemistry 43: 2501–2511 [DOI] [PubMed] [Google Scholar]
- Clapham DE (2003) TRP channels as cellular sensors. Nature 426: 517–524 [DOI] [PubMed] [Google Scholar]
- Clapham DE, Montell C, Schultz G, Julius D (2003) International Union of Pharmacology. XLIII. Compendium of voltage-gated ion channels: transient receptor potential channels. Pharmacol Rev 55: 591–596 [DOI] [PubMed] [Google Scholar]
- Colquhoun D (1998) Binding, gating, affinity and efficacy: the interpretation of structure–activity relationships for agonists and of the effects of mutating receptors. Br J Pharmacol 125: 924–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damann N, Voets T, Nilius B (2008) TRPs in our senses. Curr Biol 18: R880–R889 [DOI] [PubMed] [Google Scholar]
- Dhaka A, Viswanath V, Patapoutian A (2006) TRP ion channels and temperature sensing. Annu Rev Neurosci 29: 135–161 [DOI] [PubMed] [Google Scholar]
- Garami A, Shimansky YP, Pakai E, Oliveira DL, Gavva NR, Romanovsky AA (2010) Contributions of different modes of TRPV1 activation to TRPV1 antagonist-induced hyperthermia. J Neurosci 30: 1435–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavva NR, Klionsky L, Qu Y, Shi L, Tamir R, Edenson S, Zhang TJ, Viswanadhan VN, Toth A, Pearce LV, Vanderah TW, Porreca F, Blumberg PM, Lile J, Sun Y, Wild K, Louis JC, Treanor JJ (2004) Molecular determinants of vanilloid sensitivity in TRPV1. J Biol Chem 279: 20283–20295 [DOI] [PubMed] [Google Scholar]
- Grandl J, Hu H, Bandell M, Bursulaya B, Schmidt M, Petrus M, Patapoutian A (2008) Pore region of TRPV3 ion channel is specifically required for heat activation. Nat Neurosci 11: 1007–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandl J, Kim SE, Uzzell V, Bursulaya B, Petrus M, Bandell M, Patapoutian A (2010) Temperature-induced opening of TRPV1 ion channel is stabilized by the pore domain. Nat Neurosci 13: 708–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzer P (2007) Role of visceral afferent neurons in mucosal inflammation and defense. Curr Opin Pharmacol 7: 563–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordt SE, Julius D (2002) Molecular basis for species-specific sensitivity to ‘hot’ chili peppers. Cell 108: 421–430 [DOI] [PubMed] [Google Scholar]
- Jordt SE, Tominaga M, Julius D (2000) Acid potentiation of the capsaicin receptor determined by a key extracellular site. Proc Natl Acad Sci USA 97: 8134–8139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzhikandathil EV, Wang H, Szabo T, Morozova N, Blumberg PM, Oxford GS (2001) Functional analysis of capsaicin receptor (vanilloid receptor subtype 1) multimerization and agonist responsiveness using a dominant negative mutation. J Neurosci 21: 8697–8706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C, Birnbaumer L, Flockerzi V, Bindels RJ, Bruford EA, Caterina MJ, Clapham DE, Harteneck C, Heller S, Julius D, Kojima I, Mori Y, Penner R, Prawitt D, Scharenberg AM, Schultz G, Shimizu N, Zhu MX (2002) A unified nomenclature for the superfamily of TRP cation channels. Mol Cell 9: 229–231 [DOI] [PubMed] [Google Scholar]
- Nilius B, Mahieu F, Prenen J, Janssens A, Owsianik G, Vennekens R, Voets T (2003) Voltage dependence of the Ca2+-activated cation channel TRPM4. J Biol Chem 278: 30813–30820 [DOI] [PubMed] [Google Scholar]
- Ramsey IS, Delling M, Clapham DE (2006) An introduction to TRP channels. Ann Rev Physiol 68: 619–647 [DOI] [PubMed] [Google Scholar]
- Ryu S, Liu B, Yao J, Fu Q, Qin F (2007) Uncoupling proton activation of vanilloid receptor TRPV1. J Neurosci 27: 12797–12807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura T, Bielefeldt K, Gebhart GF (2007) Mouse colon sensory neurons detect extracellular acidosis via TRPV1. Am J Physiol Cell Physiol 292: C1768–C1774 [DOI] [PubMed] [Google Scholar]
- Szallasi A, Blumberg PM (1993) [3H]resiniferatoxin binding by the vanilloid receptor: species-related differences, effects of temperature and sulfhydryl reagents. Naunyn Schmiedebergs Arch Pharmacol 347: 84–91 [DOI] [PubMed] [Google Scholar]
- Talavera K, Yasumatsu K, Voets T, Droogmans G, Shigemura N, Ninomiya Y, Margolskee RF, Nilius B (2005) Heat activation of TRPM5 underlies thermal sensitivity of sweet taste. Nature 438: 1022–1025 [DOI] [PubMed] [Google Scholar]
- Tao X, Lee A, Limapichat W, Dougherty DA, MacKinnon R (2010) A gating charge transfer center in voltage sensors. Science 328: 67–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga M, Caterina MJ, Malmberg AB, Rosen TA, Gilbert H, Skinner K, Raumann BE, Basbaum AI, Julius D (1998) The cloned capsaicin receptor integrates multiple pain-producing stimuli. Neuron 21: 531–543 [DOI] [PubMed] [Google Scholar]
- Voets T, Droogmans G, Wissenbach U, Janssens A, Flockerzi V, Nilius B (2004) The principle of temperature-dependent gating in cold- and heat-sensitive TRP channels. Nature 430: 748–754 [DOI] [PubMed] [Google Scholar]
- Voets T, Talavera K, Owsianik G, Nilius B (2005) Sensing with TRP channels. Nat Chem Biol 1: 85–92 [DOI] [PubMed] [Google Scholar]
- Wang S, Poon K, Oswald RE, Chuang HH (2010) Distinct modulations of human capsaicin receptor by protons and magnesium through different domains. J Biol Chem 285: 11547–11556 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.