Abstract
Plants and many other eukaryotes can make use of two major pathways to cope with mutagenic effects of light, photoreactivation and nucleotide excision repair (NER). While photoreactivation allows direct repair by photolyase enzymes using light energy, NER requires a stepwise mechanism with several protein complexes acting at the levels of lesion detection, DNA incision and resynthesis. Here we investigated the involvement in NER of DE-ETIOLATED 1 (DET1), an evolutionarily conserved factor that associates with components of the ubiquitylation machinery in plants and mammals and acts as a negative repressor of light-driven photomorphogenic development in Arabidopsis. Evidence is provided that plant DET1 acts with CULLIN4-based ubiquitin E3 ligase, and that appropriate dosage of DET1 protein is necessary for efficient removal of UV photoproducts through the NER pathway. Moreover, DET1 is required for CULLIN4-dependent targeted degradation of the UV-lesion recognition factor DDB2. Finally, DET1 protein is degraded concomitantly with DDB2 upon UV irradiation in a CUL4-dependent mechanism. Altogether, these data suggest that DET1 and DDB2 cooperate during the excision repair process.
Keywords: DNA repair, NER, plants, ubiquitin, UV stress
Introduction
Most organisms are exposed to the damaging effects of sunlight and need to repair UV-induced DNA lesions to maintain genome integrity. Among the repertoire of DNA repair pathways, plants and many other multicellular organisms can make use of two major mechanisms to remove UV-induced DNA photoproducts, photoreactivation and nucleotide excision repair (NER). Photoreactivation is a fast, efficient and error-free mechanism that involves specific cyclobutane pyrimidinone dimer (CPD)- and 6,4-photolyase enzymes to remove CPDs and pyrimidine (6,4) pyrimidinone dimers (6,4PPs), respectively, using the light energy from a photon in the UV-A/blue light range (Britt, 1999). This light-dependent repair mechanism allows direct conversion of pyrimidine dimers to monomers without a DNA excision step (reviewed in Sancar et al, 2004). The pathway is not found in placental mammals such as humans, and is supplemented in all eukaryotes by NER, a light-independent mechanism that allows removal of a wide spectrum of helix-distorting base lesions, including UV-induced CPDs and 6,4PPs (Svejstrup, 2002; Gillet and Scharer, 2006).
NER entails a multistep process that involves ∼30 proteins for damage recognition, helix opening, dual incision of the 25–30 nt damaged strand, gap-filling DNA synthesis and ligation of the newly synthesized DNA (Wood et al, 2000). The damage detection step is differentially achieved depending on the location of the lesion and thereby leads to two sub-pathways. When located within transcribed regions UV-DNA lesions stall RNA polymerase II. This serves as a damage recognition signal by the Cockayne Syndrome type A or B (CSA or CSB) factors, thereby initiating transcription-coupled repair (TCR; Svejstrup, 2002; Fousteri and Mullenders, 2008). In non-transcribed regions, damage detection relies on the damaged DNA binding protein 2 (DDB2 protein; Wittschieben et al, 2005) and on the XPC–HR23B–CEN2 complex (Volker et al, 2001), which act as sensors to detect DNA conformational changes and initiate the global genome repair pathway (GGR; Hanawalt et al, 2003). Genetic deficiencies in the recognition factors CSA and DDB2 lead to hereditary diseases such as Cockayne syndrome (CS) and xeroderma pigmentosum (XP), which are marked by cutaneous hypersensitivity to sunlight exposure and high susceptibility to UV-induced skin cancer (reviewed in Shuck et al, 2008).
Although acting in the two different TCR and GGR sub-pathways, CSA and DDB2 have been found to assemble within nearly identical complexes containing DDB1, CULLIN4 (CUL4) and regulator of cullin 1 (ROC1 or RBX1) to form typical cullin-RING ubiquitin E3 ligases (CRL) whose activity is regulated by the COP9 signalosome (CSN) (Groisman et al, 2003; Bernhardt et al, 2006; Chen et al, 2006). The current accepted model establishes that CUL4–DDB1 either directly docks substrates to the ubiquitylation machinery or acts indirectly by recruiting a third protein harbouring WD40-repeats with conserved WDxR motifs (such as DDB2), which is responsible for substrate specificity (Angers et al, 2006; He et al, 2006; Higa et al, 2006). This structure positions CUL4 as a modular protein that serves as a scaffold to assemble multiple CRL complexes (reviewed in Petroski and Deshaies, 2005), with several targets during NER.
In humans, the DDB2 protein has strong affinity for bulky DNA lesions such as 6,4PPs and CPDs (Scrima et al, 2008). Upon UV irradiation, DDB2 dissociates from the CSN, detects and directly binds the DNA lesion where it recruits XPC by protein–protein interaction. Together with CUL4 neddylation, this contributes to activate the E3 ligase activity of a CUL4–DDB1 CRL that poly-ubiquitylates several targets such as XPC, various histones, as well as DDB2 and CUL4 themselves, triggering different protein fates. While poly-ubiquitylation induces DDB2 proteolytic degradation and triggers histone release from nucleosomes, it also facilitates the DNA-binding activity of XPC. Because DDB2 exhibits a high affinity for DNA photoproducts, its proteolysis potentiates its displacement by XPC and other downstream GG-NER factors (Chen et al, 2001; Nag et al, 2001; Rapic-Otrin et al, 2002; Sugasawa et al, 2005; El-Mahdy et al, 2006; Kapetanaki et al, 2006; Wang et al, 2006; Guerrero-Santoro et al, 2008). In this process, DDB2 is therefore the target of a CUL4–DDB1-containing ubiquitin ligase for proteasome-mediated degradation.
Although these mechanisms have been poorly studied in plants, it was recently shown that the roles of CUL4–DDB1DDB2 and CUL4–DDB1CSA E3 ligases in NER are conserved in Arabidopsis thaliana and interconnect to the highly efficient light-dependent photoreactivation system as well as with the checkpoint kinase ataxia telangiectasia and Rad3-related (ATR) factor (Molinier et al, 2008; Biedermann and Hellmann, 2010; Zhang et al, 2010). These observations are of interest in light of other studies that have revealed the role of CUL4–DDB1 E3 ligases in plant development and in particular in the control of photomorphogenesis (reviewed in Jackson and Xiong, 2009). More specifically, intensive efforts in Arabidopsis have revealed the existence of several CUL4–DDB1-containing CRLs that contribute to repress light-responsive genes in darkness by associating with negative regulators such as COnstitutive-Photomorphogenic 1 (COP1) and suppressor of phytochrome A (SPA) (Chen et al, 2010), as well as with the CDD complex in both darkness and under light conditions (Bernhardt et al, 2006; Chen et al, 2006). The Arabidopsis CDD complex comprises DDB1A, De-Etiolated 1 (DET1) and COP10, a plant-specific ubiquitin-conjugating E2 variant (Schroeder et al, 2002; Yanagawa et al, 2004; Lau and Deng, 2009). This ∼350 kDa complex can associate with the CSN and COP1 complexes, and the three factors act together to control specific steps of plant development (Yanagawa et al, 2004). The role of COP1 and DET1 in the inhibition of photomorphogenesis in darkness has been proposed to mainly rely on ubiquitin-mediated proteolytic degradation of target factors such as the bZIP factor LONG HYPOCOTYL5 (HY5) (Chory, 1992; Osterlund et al, 2000). After its discovery through genetic screens for plants affected in etiolated development (Chory et al, 1989; Pepper et al, 1994), DET1 was shown to be conserved in humans and also to associate with COP1 and a CUL4–DDB1 E3 ligase to target c-jun for degradation (Wertz et al, 2004; Pick et al, 2007).
Recent analyses of mutants in Arabidopsis COP1, DET1 and CSN subunits revealed that these photomorphogenic mutants display elevated levels of single- and/or double-strand DNA breaks, as evidenced in situ using a TUNEL assay (Dohmann et al, 2008). By contrast, we have recently observed that the det1-1 mutant exhibits hyposensitivity to UV-C irradiation that mainly relies on two cooperative effects directly linked to its photomorphogenic phenotype: (i) UV-induced DNA damage is reduced as a consequence of the overaccumulation of UV-absorbing compounds acting as ‘sunscreens’ and (ii) photoreactivation is enhanced due to the strong overexpression of the two photolyase genes (Castells et al, 2010). To better assess this apparent discrepancy, we investigated the potential impact of DET1 on DNA damage responses in light-independent repair mechanisms. We present evidence that appropriate dosage of the DET1 protein is necessary for efficient removal of UV-induced DNA lesions through the GGR pathway, and that DET1 is required for CUL4–DDB1-mediated proteolytic degradation of DDB2. We further show that DET1 is degraded upon UV irradiation in a CUL4-dependent manner, leading us to propose that DET1 and DDB2 cooperate during the DNA excision repair process.
RESULTS
DET1 protein dosage influences UV-C sensitivity
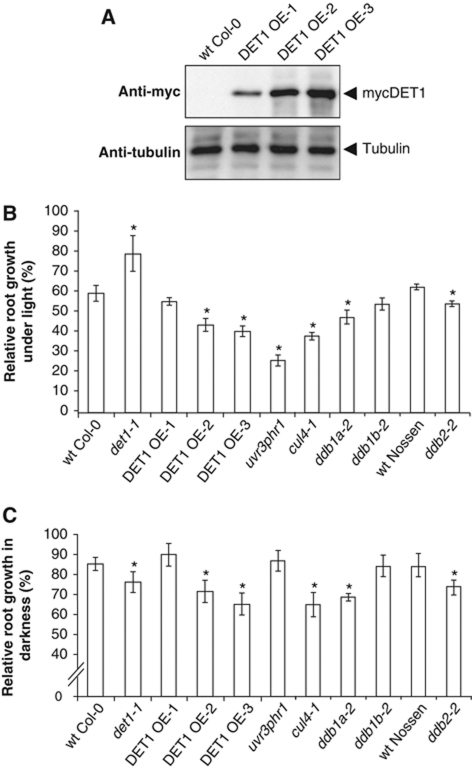
Arabidopsis plants bearing null mutations in the DET1 gene are lethal at early stages of embryo or seedling development (Misera et al, 1994) and therefore cannot be tested for UV sensitivity. Nevertheless, most individuals bearing the hypomorphic det1-1 allele can survive, and display a constitutive photomorphogenic phenotype (Pepper et al, 1994). We produced Arabidopsis transgenic lines overexpressing myc-tagged DET1 protein (DET1 OE-1, OE-2 and OE-3 lines) and used them together with the det1-1 mutant to better determine how DET1 protein dosage affects UV-C sensitivity. As estimated using an antibody for the MYC-epitope tag (Figure 1A) and a rabbit antiserum raised against the full-length Arabidopsis DET1 protein (Supplementary Figure S1), the abundance of the mycDET1 protein ranges from endogenous DET1 levels in the OE-1 line to about 10-fold in DET1 OE-3. The mycDET1 fusion protein is functional since it can efficiently rescue the pale and dwarf phenotype of det1-1 mutant seedlings and partially complement its skotomorphogenic phenotype by restoring normal hypocotyl elongation in darkness (Supplementary Figure S2).
Figure 1.
DET1 dosage influences UV-C sensitivity upon recovery in both light and dark conditions. (A) mycDET1 overexpression in DET1 OE transgenic lines. Equal amounts of whole protein extracts from epitope-tagged mycDET1 lines OE-1, OE-2 and OE-3 were loaded on a 10% SDS–PAGE and analysed by immunoblot using an anti-myc antibody. The same blot was analysed with an anti-tubulin antibody as loading control. (B, C) Root growth inhibition upon UV-C exposure. Four-day-old seedlings with the indicated genotypes were exposed to UV-C (600 J/m2) and immediately returned to normal light conditions (B) or to complete darkness (C). Relative root growth was determined 24 h after irradiation by comparison with the respective non-irradiated control of the same genotype (100% root growth). Error bars represent standard deviations from three replicate experiments (n>20). Asterisks indicate t-test significant differences at P⩽0.05 relative to wild-type control at same dose.
To test sensitivity of these genotypes to UV, 3-day-old seedlings were first exposed to a single dose of UV-C irradiation and kept 24 h under normal light conditions for recovery before determining relative primary root growth inhibition. A mutant deficient for the two photolyase genes UVR3 and PHRI as well as the cul4-1 mutant were used as controls for photoreactivation and GGR defects, respectively. By contrast to the effect of DET1 knock-down, the two strong DET1 overexpressing lines were significantly more UV sensitive than wild-type seedlings (Figure 1B). Although they do not reach the high level of sensitivity of the uvr3phrI photolyase double mutant, in agreement with the major role played by photoreactivation in the removal of UV-DNA photoproducts under light conditions, the sensitivity of DET1 OE-2 and OE-3 lines is comparable to the effect of the cul4-1 mutation. The ddb1a-2 and ddb2-2 mutants are slightly less affected, as previously observed (Molinier et al, 2008), and no significant defect is observed in a ddb1b-2 mutant, a knockout for the second orthologue of DDB1 in Arabidopsis. These data indicate that DET1 dosage influences UV-C sensitivity following recovery under light conditions. Because this effect might be explained by repression of photolyase gene expression as a consequence of DET1 overexpression, we examined mRNA levels of UVR3 and PHRI by RT–qPCR in DET1 OE seedlings. In contrast to their overexpression in det1-1, no significant difference with wild-type seedlings was observed in the three DET1 OE lines (Supplementary Figure S3A). The alternative possibility that UV-absorbing compounds such as flavonoids are decreased in DET1 OE lines was also tested and shown not to be the case (Supplementary Figure S3B). We conclude from these data that disturbing DET1 cellular content affects DNA repair at a level other than sunscreen effect or photoreactivation.
To avoid the confounding effect of photoreactivation and to better assess the possible implication of NER, we tested UV-C sensitivity following recovery in darkness. Under these conditions, the cul4-1, ddb1a-2 and ddb2-2 mutants defective in GGR are significantly more sensitive than wild type, while the uvr3phrI photolyase mutant is not (Figure 1C). In both light and dark conditions, cul4-1, ddb1a-2 but not ddb1b-2 mutants are sensitive (Figure 1B and C), suggesting that DDB1A but not DDB1B is involved in NER. These two proteins share a high sequence similarity (Schroeder et al, 2002) and both can interact with DET1 (Supplementary Figure S4), indicating that the Arabidopsis CDD complex may contain one molecule either of DDB1A or DDB1B in addition to COP10. Like in light conditions, the strong DET1 OE lines exhibited significant UV-C sensitivity in darkness, at levels that correlate with DET1 overexpression levels (Figure 1C). Importantly, these experiments revealed that, following recovery in darkness, det1-1 mutant plants are more sensitive than the wild type, even though these plants display an enhanced level of UV-protecting compounds and are therefore significantly less damaged than wild type (Castells et al, 2010). Altogether, these data suggest that appropriate DET1 dosage may be necessary for an efficient light-independent DNA repair mechanism.
DET1 is required for efficient DNA photoproduct removal through a light-independent pathway
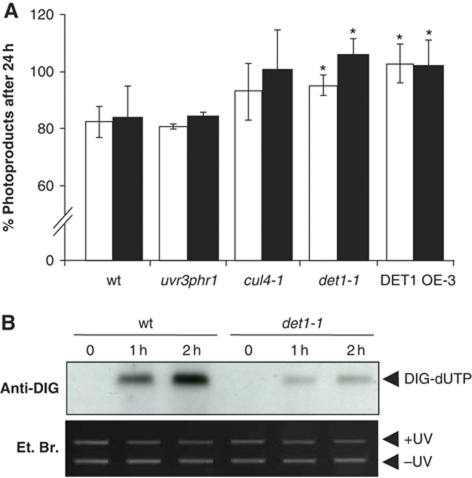
In order to determine whether UV-C hypersensitivity of the det1-1 mutant and of DET1 OE lines is related to a defect in DNA repair, we measured the removal efficiency of UV-induced DNA photoproducts in the different plant lines. The amount of photoproducts was quantified by immunodot-blot of genomic DNA using anti-6,4PP and anti-CPD antibodies. For each genotype, the remaining amount of CPDs and 6,4PPs was determined after 24 h in darkness and compared with the initial damage immediately after UV irradiation (Castells et al, 2010). This approach avoids biases due to reduced DNA damage in det1-1 mutant seedlings. As expected, upon recovery in darkness similar amounts of both CPDs and 6,4PPs remained in wild type and in the photoreactivation mutant uv3phrI (Figure 2A). Also, most of the DNA photoproducts persisted 24 h after irradiation in the cul4-1 mutant that is impaired in GGR (92 and 96% of CPDs and 6,4PPs remaining, respectively). Removal of photoproducts was also significantly impaired in the det1-1 mutant and was abolished in the strong DET1 OE line (∼100% remaining). Altogether, these data confirm that DET1 dosage can affect the efficient removal of UV-induced DNA photoproducts through a light-independent DNA repair pathway.
Figure 2.
DET1 is required for synthesis-dependent repair of UV-induced DNA lesions. (A) Modulation of DET1 level affects the removal of 6,4PP and CPD photoproducts by light-independent DNA repair. Fourteen-day-old wild-type (Col-0), uvr3phrI, cul4-1, det1-1 and DET1 OE-3 seedlings were irradiated with UV-C (1000 J/m2) and harvested immediately after irradiation or allowed to repair for 24 h in darkness. Serial dilutions of genomic DNA were subjected to immunodot-blot analysis using anti-CPD (white bars) or anti-6,4PP (black bars) antibodies, respectively, and normalized relative to 5-methylcytosine content. For each genotype, the percent of CPDs and 6,4PPs remaining after 24 h was calculated relative to the initial level immediately after UV irradiation. Error bars represent standard deviations from three independent experiments. Asterisks indicate t-test significant differences at P⩽0.05 relative to wild-type controls at the same dose. (B) Arabidopsis DET1-defective plants are impaired in synthesis-dependent repair of UV-induced DNA lesions in vitro. Cell extracts from wild-type and det1-1 mutant plants were incubated for 0, 1 or 2 h with UV-C damaged (+UV) and undamaged control plasmid (−UV) in the presence of DIG-dUTP to monitor synthesis-dependent DNA repair efficiency.
DET1-defective plants are impaired in synthesis-dependent repair of UV-induced DNA lesions
In order to further characterize the role of DET1 in light-independent DNA repair, we tested the capacity of the det1-1 mutant to perform synthesis-dependent repair of UV-induced DNA lesions using an in vitro DNA repair assay. This assay evaluates the efficiency of DIG-dUTP incorporation by plant extracts in a UV-C damaged plasmid, reflecting the ability to perform efficient NER (Li et al, 2002). Figure 2B shows that nuclear extracts of det1-1 plants were less efficient than wild type in incorporating DIG-dUTP, following 1 or 2 h of incubation. This observation indicates that UV-C hypersensitivity of det1-1 mutant plants correlates with a defect in the excision repair process, as previously shown for cul4-1 and ddb2-2 mutants (Molinier et al, 2008). As DET1 knock-down and overexpressing plants both exhibit impaired removal of UV-DNA photoproducts and enhanced UV-C sensitivity, the DET1 OE-3 line was further tested using the same assay. This line also displayed a defect in synthesis-dependent DNA repair (Supplementary Figure S5), further indicating that DET1 dosage is critical for efficient NER. These observations suggest that DET1 may have a direct function in the GGR repair process together with its known partners CUL4 and DDB1A.
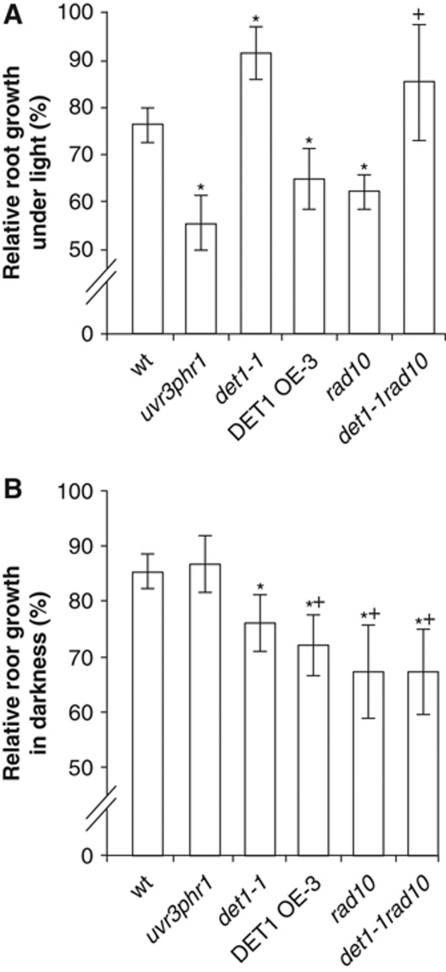
Epistatic interaction of det1-1 and rad10 mutations
To further understand the positioning of DET1 in light-independent DNA repair pathways, we introgressed the det1-1 allele in a mutant for RAD10 (Molinier et al, 2008), which encodes an endonuclease that excises bulky DNA lesions as part of the NER process. UV-C sensitivity of the det1-1rad10 double mutant was determined by root growth assay upon recovery in light or darkness. As expected, under light conditions the det1-1 single mutant exhibited a clear UV-C hyposensitivity with a dominant effect over the secondary mutation observed in the det1-1rad10 double mutant (Figure 3A). This is consistent with increased UV-protecting compounds and exacerbated photoreactivation in det1-1 (Castells et al, 2010). Conversely, upon recovery in darkness the det1-1rad10 double mutant was significantly more sensitive than wild type, with a reduction of relative root growth that is similar to the det1-1 and rad10 single mutants and to the DET1 overexpressing line (Figure 3B). These data indicate that the det1-1 mutation is epistatic to rad10, and are consistent with DET1 being involved in the same UV-DNA repair pathway as the RAD10 endonuclease. We further tested whether the det1-1 mutant was affected in other repair pathways by assessing its sensitivity to cisplatin, an agent that produces DNA inter-crosslinks. By contrast to ddb2-2, we observed that det1-1 plants do not exhibit hypersensitivity to this genotoxic agent (Supplementary Figure S6), suggesting that DET1 may act more specifically in UV-DNA damage repair. Surprisingly, the DET1 OE-3 overexpressing line was found to be sensitive, which we can explain only through possible indirect effects. Finally, like the ddb2-2 mutant, we observed that det1-1 mutation does not increase sensitivity to hydrogen peroxide, in agreement with the fact that H2O2-induced DNA lesions are not predominantly repaired by the NER pathway (data not shown).
Figure 3.
Genetic interactions between det1-1 and rad10 and their effect on UV-C sensitivity. (A, B) Root growth inhibition upon UV-C exposure. Four-day-old seedlings with the indicated genotypes were exposed to UV-C (600 J/m2) and immediately returned to normal light conditions (A) or to complete darkness (B) for 24 h. Relative root growth was determined 24 h after irradiation by comparison with the respective non-irradiated control of the same genotype (100% root growth). Error bars represent standard deviations from three replicate experiments (n>20). Asterisks indicate t-test significant differences at P⩽0.05 relative to wild-type controls at the same dose, while plus signs (+) indicate no significant difference with respect to the det1-1 mutant.
CUL4-dependent DET1 protein degradation upon UV-C irradiation
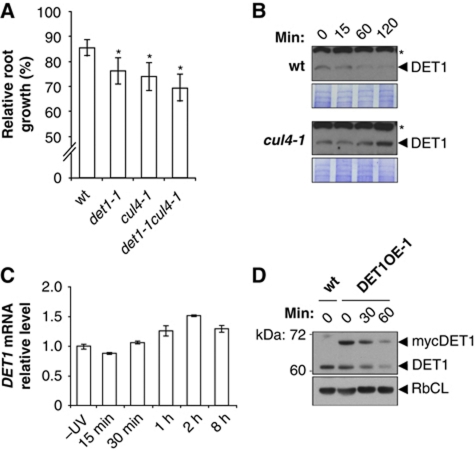
In order to determine whether DET1 acts together with CUL4 in the GGR pathway, we first tested the genetic interaction between DET1 and CUL4 genes in UV sensitivity. Following recovery in darkness, det1-1cul4-1 double mutants displayed a UV-induced root growth reduction similar to det1-1 and cul4-1 single mutants (Figure 4A), consistent with an epistatic interaction between the two alleles and implying that DET1 and CUL4 act in the same DNA repair pathway.
Figure 4.
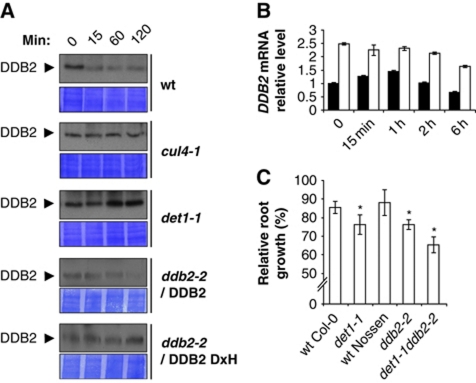
CUL4 interconnects with DET1 and triggers DET1 protein degradation upon UV-C exposure. (A) CUL4 and DET1 mutations are epistatic for UV-C sensitivity. Seedlings with the indicated genotypes were exposed to UV-C (600 J/m2) and immediately placed in complete darkness. Relative root growth was determined 24 h after irradiation by comparison with respective non-irradiated controls of the same genotype (100% root growth). Error bars represent standard deviations from three replicate experiments (n>20). Asterisks indicate t-test significant differences at P⩽0.05 relative to wild-type controls at the same dose. (B) Immunoblot analysis of endogenous DET1 content before (0) or 15, 60 and 120 min after UV-C exposure (3000 J/m2) in wild-type and cul4-1 seedlings. Coomassie blue staining (lower panels) is shown as loading controls. Asterisks indicate cross-reacting bands. (C) Analysis of DET1 expression upon UV-C exposure. Quantitative RT–PCR analysis was used to monitor DET1 mRNA levels in 10-day-old wild-type seedlings (Col-0) harvested at the indicated times after UV-C exposure (900 J/m2). Error bars indicate standard deviations from two biological replicates. (D) Immunoblot analysis of mycDET1 protein content before (0) or 30 and 60 min after UV-C exposure (3000 J/m2). Equal amounts of whole protein extracts were loaded on a 10% SDS–PAGE and analysed by immunoblot using anti-DET1 antibody. The same blot was probed with anti-RbCL antibody as a loading control.
Considering that CUL4–DDB1DDB2 ubiquitylates and triggers the proteolytic degradation of several proteins in the NER process, including CUL4 and DDB2 themselves, we questioned whether DET1 stability was affected following UV irradiation. The DET1 antibody was therefore used to analyse endogenous DET1 protein levels upon UV-C exposure. To this end, seedlings were immediately kept in darkness after UV-C irradiation and total proteins were extracted after 15, 60 or 120 min for immunoblot analysis. Interestingly, in wild-type plants DET1 steady-state levels rapidly decreased after UV-C treatment and the protein was barely detectable after 2 h (Figure 4B). Because this reduction in DET1 content was fast, it was unlikely to result from reduced expression of the DET1 gene upon UV exposure. This was confirmed by RT–qPCR analysis of DET1 mRNA levels, which revealed that DET1 gene expression was instead slightly induced upon UV-C exposure (Figure 4C), a mechanism that may allow reconstituting DET1 protein content.
To further confirm decay of the DET1 protein upon UV-C exposure, we analysed protein levels of the constitutively expressed mycDET1 fusion protein in the DET1 OE-1 transgenic line, which is not affected in UV-C sensitivity (Figure 1). Both endogenous and mycDET1 protein levels rapidly decreased upon UV-C exposure (Figure 4D), confirming the post-transcriptional nature of this decay. We further observed that endogenous DET1 protein was visibly less degraded in the strong mycDET1 overexpressor line OE-3, in agreement with the reduced DNA repair functionality in these plants (Supplementary Figure S7A). Size-exclusion chromatography analysis of the DET1 OE-3 line additionally revealed that a significant fraction of the mycDET1 protein is found within a high-molecular weight (HMW) complex that may correspond to aggregated forms (Supplementary Figure S7B).
DET1 protein content was subsequently examined in the cul4-1 mutant in order to determine if, similarly to DDB2, its UV-induced degradation depends on a CUL4 E3 ligase. As shown in Figure 4B, degradation of DET1 upon UV-C treatment was abolished in the cul4-1 mutant, and the DET1 protein even accumulated after 2 h. Altogether, these data suggest that DET1 is degraded upon UV exposure in a CUL4 CRL-mediated pathway, therefore positioning DET1 in close vicinity to the repair process.
Arabidopsis DDB2 degradation is impaired in the det1-1 mutant
Considering that DDB2 and DET1 have been shown to interact with CUL4–DDB1 complexes with ubiquitylation activities in both plant and human cells and are both necessary for GGR, we investigated through genetic and biochemical approaches whether these two factors cooperate in this process. In humans, the DDB2 protein has strong affinity for UV photoproducts and is released from the DNA lesion through CUL4–DDB1-mediated degradation (Matsuda et al, 2005; El-Mahdy et al, 2006). Patients with XPE DDB2 alleles suffer from deficient NER that correlates with impaired binding to DDB1 substrate adaptor and consequent impaired DDB2 proteolysis (Nichols et al, 2000). This clearly indicates that DDB2 protein turnover is a crucial step in GGR in mammals. We therefore tested if this was conserved in plants and whether DET1 could be involved in this step of NER.
First, to confirm the relevance of UV-induced degradation of DDB2 in plant systems, we investigated the effect of mimicking human XPE mutation on DDB2 function in planta. Plant vectors allowing stable expression of native DDB2 protein or harbouring a point mutation within the WDxR motif at the homologous position of the XPE allele (Molinier et al, 2008) were used to transform the ddb2-2 mutant and complementation assays for UV sensitivity were performed. We found that plants expressing native DDB2 had a restored UV-induced root growth inhibition (Supplementary Figure S8), which was also associated with normal DDB2 protein degradation upon UV-C exposure (Figure 5A). In contrast, DDB2 with mutated WDxR motif expressed at a similar level as native DDB2 tends to overaccumulate during the repair process (Figure 5A) and could not complement the ddb2-2 mutant for UV sensitivity (Supplementary Figure S8). As expected, DDB2 protein level remained stable in cul4-1 mutants (Figure 5A). The observation that mutation of either CUL4 or the WDxR motif of DDB2 both stabilize steady-state levels of DDB2 suggests that the mechanism of UV-induced DDB2 degradation mediated by CUL4–DDB1 is conserved in plants.
Figure 5.
DDB2 degradation following UV exposure is dependent on DET1. (A) Immunoblot analysis of endogenous DDB2 content upon UV-C exposure (3000 J/m2) in seedlings from wild-type, cul4-1, det1-1 and ddb2-2 mutant expressing native DDB2 protein or DDB2 protein with mutated WDxR motif (WDxH). Coomassie blue staining is shown as loading control (lower panels). (B) Analysis of DDB2 gene expression upon UV-C exposure. Quantitative RT–PCR analysis was used to monitor DDB2 mRNA levels in 10-day-old wild-type Col-0 (black bars) and det1-1 (white bars) seedlings harvested at the indicated times after UV-C exposure (600 J/m2). Error bars indicate standard deviations from two biological replicates. (C) DDB2 and DET1 mutations show additive effects for UV-C sensitivity. Seedlings with the indicated genotypes were exposed to 600 J/m2 of UV-C and immediately placed in complete darkness. Relative root growth was determined 24 h after irradiation by comparison with respective non-irradiated controls of the same genotype (100% root growth). Error bars represent standard deviations from three replicate experiments (n>20). Asterisks indicate t-test significant differences at P⩽0.05 relative to wild-type controls at the same dose.
Then to test the possible role of DET1, this experiment was performed in parallel using the det1-1 mutant. This revealed that DDB2 protein degradation upon UV-C exposure was abolished in this mutant (Figure 5A). To exclude the possibility that overall DDB2 steady-state levels could result from a concomitant increase in DDB2 gene expression in the det1-1 mutant, we determined DDB2 mRNA levels in the same samples. As recently reported (Biedermann and Hellmann, 2010), we observed that DDB2 mRNA levels transiently increased upon UV irradiation. Interestingly, DDB2 mRNA levels overaccumulated by a two-fold ratio in det1-1 mutants before UV irradiation and remained stable afterwards (Figure 5B). Altogether, this indicates that increased stability of DDB2 protein upon UV irradiation in det1-1 does not result from transcriptional responses. Both DDB2 and DET1 are therefore degraded upon UV-C exposure, and DDB2 proteolytic degradation depends on both CUL4 and DET1. These data further suggest that the role of DDB2 degradation in NER is conserved in plants, and that its impairment in cul4-1 and det1-1 mutants may therefore contribute to defective DNA repair.
To further support these findings, a double mutant for DET1 and DDB2 genes was generated and phenotypically characterized for UV-induced root growth inhibition in darkness. The det1-1ddb2-2 double mutant plants exhibit an enhanced UV-C sensitivity compared with the respective single mutants, while null segregants from the cross were not affected (Figure 5C; Supplementary Figure S9). This additive effect suggests that, like CUL4, DET1 may have a wider role in DNA repair than DDB2.
Dynamics of the DET1 complex upon UV-C irradiation
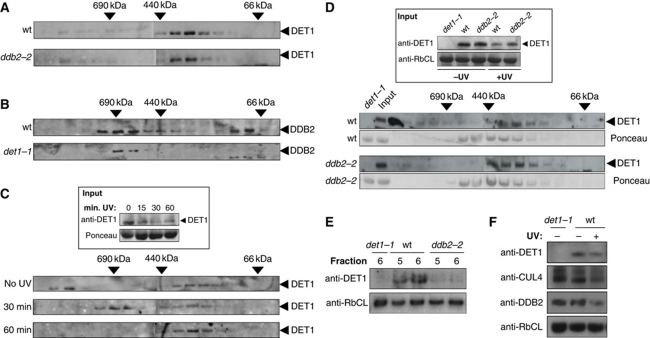
To determine whether the CDD complex remains stable upon the decay of DET1 protein, we examined DET1 complex size after UV irradiation. Size-exclusion chromatography was first performed on protein extracts from non-irradiated seedlings. DET1 fractionated predominantly in an ∼350-kDa complex (Figure 6A), which corresponds to the previously described size of the CDD complex in which DET1 is stably assembled with DDB1A and COP10 (Schroeder et al, 2002; Yanagawa et al, 2004). The same fractions were analysed using a DDB2 antibody, revealing that DDB2 predominantly fractionates as a larger complex of ∼600–800 kDa (Figure 6B). Most of the DET1 and DDB2 proteins therefore do not elute in the same fractions. Furthermore, while both DET1 protein level and complex formation are destabilized in a mutant for the COP10 subunit of the CDD complex (Chen et al, 2010), we observed that DET1 and DDB2 protein levels and complex sizes are unaffected by the absence of the other respective protein in the ddb2-2 and det1-1 mutants (Figure 6A, B and F). Together with the lack of direct interaction observed by BiFC (Supplementary Figure S4), these data are consistent with a model in which DET1 and DDB2 stably associate within distinct complexes under normal growth conditions.
Figure 6.
A high-molecular weight complex containing DET1 coelutes transiently with DDB2 following UV-C exposure. (A, B) DET1 and DDB2 fractionate in different protein complexes under normal light conditions. (A) Size-exclusion chromatography analysis of DET1 complex in protein extracts from wild-type (Nossen) and ddb2-2 mutant seedlings. Fractions were analysed by immunoblot using DET1 and DDB2 antibodies. Arrows indicate elution of molecular-weight standards in the same conditions. (B) Size-exclusion chromatography analysis of DDB2 complex in protein extracts from wild-type (Col-0) and det1-1 mutant. (C) A DET1 high-molecular weight complex is transiently detected upon UV-C irradiation. Experiment was performed as in (A) except that seedlings were exposed to UV-C (900 J/m2) and harvested after 30 or 60 min for protein extraction and size-exclusion chromatography. The inset shows DET1 protein content in input samples before chromatography separation. (D) Size-exclusion chromatography of protein extracts from wild-type Nossen and ddb2-2 mutant plants irradiated at 900 J/m2 UV-C and harvested 30 min after UV exposure. The inset shows DET1 input content in wild-type, det1-1 and ddb2-2 seedlings harvested before (−UV) or 30 min after UV-C exposure (+UV). Ponceau staining is used as control for equal fractionation of the two samples. Arrows indicate elution of molecular-weight standards in the same conditions. (E) Side-by-side analysis of fractions 5 and 6 containing DET1 UV-induced HMW complex. Fractions 5 and 6 (50 μl) from size-exclusion separation of wild-type Nossen, ddb2-2 and det1-1 extracts were analysed by immunoblot using a DET1 antibody. The same blot was probed with anti-RbCL antibody as a loading control. (F) Protein content of DET1, DDB2 and CUL4 in det1-1 mutant and wild type before and 30 min after UV-C exposure. Whole cell protein extracts (40 μg) were separated on 10% SDS–PAGE and analysed with the indicated antibodies.
Considering that DET1 and DDB2 proteins may transiently interact with additional factors during the precocious step of the excision repair process, we determined their respective sizes upon UV-C exposure. Seedlings were UV-C irradiated and soluble proteins were extracted at different time points. Upon UV irradiation, both DET1 and DDB2 protein levels decreased but the different complexes could nevertheless be detected. While DDB2 complex size was not visibly affected by UV irradiation (Supplementary Figure S10), an additional 800 kDa DET1-containing complex was detected 30 min after UV-C treatment (Figure 6C, fractions 6–8) and was not observed at later time points. This HMW DET1 complex therefore coelutes with DDB2-containing fractions, suggesting that both proteins may transiently interact.
To determine whether the HMW DET1 complex requires DDB2 to assemble, the experiment was repeated using the ddb2-2 mutant and its respective wild type (Nossen accession). Again, global DET1 protein levels significantly decreased 30 min after UV exposure in both wild-type and ddb2-2 input protein extracts, albeit to a slightly lesser extent in the ddb2 mutant (Figure 6D, upper panel). While the 800-kDa DET1 complex was detected in wild-type extracts, it was not visible in ddb2-2 (Figure 6D). This was confirmed through side-by-side migration of fractions 5 and 6 from wild-type, ddb2-2 and det1-1 mutants (Figure 6E). Altogether, these data indicate that DDB2 is required for the formation of a transient UV-dependent DET1 HMW complex in a timeframe that coincides with CUL4- and DET1-dependent degradation of DDB2.
Discussion
In this study, we tested the effect of modulating DET1 protein level on the capacity of plants to recover from UV-C irradiation through light-dependent and light-independent DNA repair pathways. DET1 overexpressing and knock-down plants both exhibited an enhanced sensitivity to UV-C following recovery in darkness, as shown in vivo by increased root growth inhibition and decreased removal of CPD and 6,4PP photoproducts. This defect has been further characterized and shown to be genetically linked to a pathway involving CUL4 and the RAD10 endonuclease, two factors central to GGR. Moreover, an in vitro assay confirmed that the det1-1 mutant is affected in synthesis-dependent DNA repair. It can be concluded from these data that the det1-1 mutant is affected in the GGR pathway, a deficiency that is masked under light conditions by enhanced photoreactivation, the predominant pathway for UV-DNA photoproduct removal in the light in plants (Sancar et al, 2004). These two properties reveal a new interface between plant photomorphogenesis and DNA repair acting through DET1.
In mammals, Schizosaccharomyces pombe and plants, CUL4 proteins serve as platforms to assemble multiple specific cullin-ring ligases (CRLs). In Arabidopsis, these CRLs are involved in various developmental processes such as the repression of photomorphogenesis when involving DET1, SPAs or COP1 (Yanagawa et al, 2004; Chen et al, 2006, 2010), but also in DNA repair mechanisms when involving DDB2, XPC or CSA (Liang et al, 2006; Molinier et al, 2008; Biedermann and Hellmann, 2010; Zhang et al, 2010). Post-translational modifications by covalent binding of ubiquitin chains typically result in the functional regulation of the substrate or in its proteasomal degradation (Ciechanover et al, 2000; Pickart, 2001; Smalle and Vierstra, 2004). In the NER process, CUL4–DDB1 has been shown to operate on multiple targets by triggering different protein fates in mammals, that is, eviction of DDB2 from the damaged region by proteolytic degradation followed by promotion of XPC affinity for the lesion (reviewed in Huang and D’Andrea, 2006). Many central factors of the recognition and excision steps have been shown to be conserved in Arabidopsis (Molinier et al, 2004, 2008; Liang et al, 2006; Biedermann and Hellmann, 2010; Zhang et al, 2010), suggesting that NER is conserved between mammals and plants. Our data indicate that UV-induced DDB2 degradation is affected in both cul4-1 and det1-1 mutants. Considering that DET1 associates with CUL4–DDB1 complexes and with ubiquitin ligase activities in plants and mammals (Osterlund et al, 2000; Wertz et al, 2004; Yanagawa et al, 2004; Chen et al, 2006), DET1 may cooperate with CUL4–DDB1 to induce DDB2 proteolytic degradation during NER. Consistent with this hypothesis, we observed that DET1 transiently associates with a HMW complex upon UV irradiation, which co-fractionates with DDB2-containing fractions and is sensitive to DDB2 depletion.
Analysis of endogenous DET1 content further shows that it is degraded upon UV irradiation by a CUL4-dependent mechanism, concomitant with DDB2 decay. At first glance, it could be envisaged that DDB2 degradation may disrupt a common complex with DET1, subsequently affecting the stability of DET1. Our data rule out this possibility because DET1 and DDB2 are present in different protein complexes before UV irradiation, and each complex is resistant to the absence of the other respective protein. Genetic analysis further indicates that DET1 and CUL4 operate in the same pathway for light-independent DNA repair and that each of them is necessary for DDB2 protein degradation during GGR. Altogether, these data support a model in which CUL4–DDB1 and DET1 are required for DDB2 degradation, a crucial step for subsequent NER processes (Figure 7). In the absence of DET1, unmodified DDB2 might remain immobilized on damaged DNA thereby impairing subsequent NER steps. This defective mechanism is also likely to be reproduced when mutating DDB2 at a WDxR motif in our complementation experiments. Whether CUL4 and DET1 assemble as a proper CUL4–DDB1DET1 ubiquitin ligase targeting DDB2 for degradation remains an interesting aspect to investigate, although it is likely to be challenging because biochemical and functional cooperation between CUL4, DDB1 and DET1 for ubiquitylation activity already occurs in the absence of UV stress (Wertz et al, 2004; Yanagawa et al, 2004; Chen et al, 2006).
Figure 7.
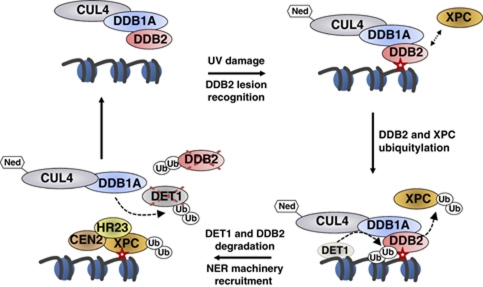
A working model for DET1 cooperation with CUL4–DDB1DDB2 in DNA repair. UV-DNA photoproduct (red star) is detected and bound by DDB2, which focalizes neddylated (Ned) CUL4–DDB1 ubiquitin ligase on the damaged region. In mammals, XPC is recruited and poly-ubiquitylated by CUL4–DDB1DDB2, thereby enhancing its binding to DNA. DDB2 protein is subsequently ubiquitylated and degraded by a CUL4–DDB1- and DET1-dependent mechanism. This involves the formation of a transient large DET1 complex presumably containing CUL4 in addition to DDB1, which is a stable partner of DET1 through direct protein–protein interaction. DET1 would then also be targeted by a CUL4–DDB1 ubiquitin ligase for degradation. Proteolysis of DDB2 and DET1 may allow their eviction from chromatin to facilitate recruiting the NER machinery, which is initiated by binding of the heterotrimeric XPC–HR23–CEN2 (plant RAD4-RAD23-CEN2) factor onto the lesion. DET1 has the capacity to bind histone H2B, represented here on an adjacent nucleosome for simplicity, but whether it acts as a soluble or histone-bound factor for DDB2 degradation remains to be determined.
An intriguing observation is the analogous effect of DET1 overexpression and DET1 knock-down on UV-C sensitivity, observed both in vivo and in vitro for synthesis-dependent DNA repair. Similar effects have recently been observed with the Arabidopsis CSA homologue (Biedermann and Hellmann, 2010; Zhang et al, 2010), and also in the context of photomorphogenesis, because both gain and loss of DDB1A function aggravate the photomorphogenic phenotype of the det1-1 mutant (Zhang and Schroeder, 2010). This is also the case for the E3 ligase component ROC1, whose overexpression yielded similar phenotypes as those observed in loss of function lines (Gray et al, 2002; Lechner et al, 2002; Schwechheimer et al, 2002). A possible cross-talk between DET1, DDB1A and DDB2 gene expression has recently been observed (Al Khateeb and Schroeder, 2009; Zhang and Schroeder, 2010), as for the slight overexpression of DDB2 mRNA in the det1-1 mutant. It has been proposed that excessive abundance of one CUL4–DDB1 adaptor or substrate specifier might affect the access of other proteins to DDB1-binding sites and therefore compromise the assembly of other CUL4-based CRLs that share the major subunits. This model, however, would predict a general and severe CUL4 loss of function effect resulting from the overexpression of any one DDB1-binding protein such as DET1, DDB2 or CSA, and should theoretically lead to lethal phenotypes such as in CUL4 knockout mutants. Here, we favour a model in which formation of specific CUL4 complexes may require correct dosage of each component to allow proper E3 ligase activity. In the particular case of DNA repair, the dosage of DDB2 and DET1 protein levels may be critical to allow their efficient eviction from the damaged chromatin through proteolytic degradation, thereby facilitating the recruitment of the NER machinery (Figure 7). Such a mechanism would explain why strong DET1 overexpressor lines affect UV-dependent DET1 degradation and DNA repair, while weak overexpressors do not. In a broader view, the control of DDB2 protein content is crucial for proper cell growth in humans, as shown by overexpression of DDB2 into oestrogen receptor (ER)-negative cells leading to growth stimulation and colony formation (Kattan et al, 2008). This recent study further revealed that DDB2 content is higher in ER-positive compared with ER-negative tumour samples of patients exhibiting breast carcinoma, suggesting that DDB2 could have oncogenic effects (Kattan et al, 2008). The role of DET1 in the control of c-Jun, CDT1 and DDB2 protein contents in humans and plants, respectively (Wertz et al, 2004; Pick et al, 2007; this study), strongly suggests that this conserved factor may be a regulator of cell growth and of the cellular response to UV irradiation, thereby contributing to genome stability.
Much effort has been devoted to deciphering DET1 function in the context of plant development, and in particular during photomorphogenesis (Chory et al, 1989; Chory and Peto, 1990; Pepper et al, 1994; Schroeder et al, 2002; Ma et al, 2003; Chen et al, 2004). Ubiquitin-mediated protein degradation is known to have a central role in this developmental transition, notably by regulating the abundance of transcription factors (Chen et al, 2004; Lorrain et al, 2006; Jiao et al, 2007). While COP1 and DET1 both contribute to inhibit photomorphogenesis in darkness, COP1 is slowly excluded from the nucleus upon illumination while DET1 is constitutively nuclear (Serino and Deng, 2003; Chen et al, 2004). This property renders DET1 more susceptible to cope with the harmful effects of sudden light exposure on etiolated seedlings and positions DET1 as a multifaceted factor in the switch from dark- to light-dependent development in plants. Further elucidation of the multiple roles of DET1 may now require the integration of chromatin dynamics, as plant DET1 has been shown to interact with histone H2B (Benvenuto et al, 2002). This capacity may indeed be relevant, given that poly-ubiquitylation of various histones has been shown recently to involve CUL4–DDB1 and to have a role in NER in mammalian cells (Kapetanaki et al, 2006). The study by Wang et al (2006) furthermore revealed that CUL4A knock-down cells had reduced UV-induced histone H3 and H4 ubiquitylation, impaired recruitment of XPC to damaged foci, and defective DNA repair. Histone poly-ubiquitylation may loosen association with DNA and trigger histone eviction to enable access of the NER machinery. Because the precise mechanism by which CUL4A–DDB1 CRLs can influence histone ubiquitylation is poorly understood, it remains possible that other CUL4–DDB1-associated proteins such as DET1 may target one or several CUL4-based CRLs to chromatin. The possible loading of DET1 onto chromatin at damaged foci and the precise role of chromatin-bound DET1 remain fascinating questions that will need to be addressed in future studies.
Materials and methods
Plant materials
Arabidopsis mutants atr-2, rad10, ddb1a-2, as well as phrI and uvr3 mutants, have been described in Molinier et al (2008), det1-1 in Chory et al (1989), cul4-1 in Bernhardt et al (2006) and ddb1b-2 in Bernhardt et al (2010). All mutants are in the Columbia-0 (Col-0) background except the ddb2-2 mutant in Nossen (Molinier et al, 2008). Arabidopsis seeds were surface sterilized and plated on Murashige and Skoog medium containing 0.9% agar and stratified for 3 days at 4°C before transfer to growth chambers with a 16 h/8 h light/dark photoperiod (23°C/19°C) with 100 μmol m2 per second of light. All plant samples were harvested and extracted at mid photoperiod (12–16 h).
Generation of transgenic plants
Construction of the pK35S::mycDET1 binary vector has been described in Dubin et al (2008), while the generation of DDB2 R343H mutation was described in Molinier et al (2008). The binary plasmids were mobilized using Agrobacterium tumefaciens to transform wild-type Col-0, Nossen and det1-1 or ddb2-2 A. thaliana plants by floral dipping (Clough and Bent, 1998). Transformants with a single insertion were selected based on kanamycin antibiotic resistance. Expression of the mycDET1 fusion protein was confirmed by immunoblot using a mouse anti-myc antibody (05-724, Millipore) and anti-DET1 antibody.
Phenotype and UV-C sensitivity analyses
Photomorphogenic phenotype analyses and root growth assays were performed as in Castells et al (2010) with the exception that for root length measurements upon UV-C exposure seedlings were either kept for 24 h under white light or in complete darkness. At least 20 plants were used per replicate and the experiments were triplicated.
Determination of 6,4PP and CPD removal in vivo by immunodot-blot
Two-week-old seedlings were irradiated with UV-C (1000 J/m2, λ=254 nm) using a UVC 500 Crosslinker (Amersham). Samples were harvested just after irradiation or kept in darkness and harvested 24 h later. Genomic DNA was extracted and analysed by immunodot-blot as described in Castells et al (2010). The amounts of CPDs or 6,4PPs remaining after 24 h were determined for each specific genotype by comparing with the initial level immediately after UV-C irradiation assuming that repair reaction rate is independent of substrate concentration.
Quantitative RT–PCR
Quantitative RT–PCR (qRT–PCR) analysis was performed on total RNA extracted using RNeasy plant minikit (Qiagen) from 10-day-old seedlings exposed to 900 J/mol UV with a UVC 500 Crosslinker (Amersham). Reverse transcription was performed on 1 μg of total RNA using random hexamers and a cDNA reverse transcription kit (Applied Biosystems). Quantitative PCR was performed using a LightCycler 480 and LightCycler 480 SYBR green I Master mix (Roche). To exclude any contamination of the samples by genomic DNA, PCR was also performed using primers flanking one intron of ACTIN2 and the size of the amplicons was checked on agarose gels. Data were normalized relative to At4g29130 and At5g13440 genes with invariant expression over dozens of publicly available transcriptome analyses. Sequences of the primers are detailed in Supplementary Table 1.
In vitro synthesis DNA repair assay
In vitro repair assay was performed on plant nuclear extracts according to Li et al (2002). Linearized pGEX plasmid was UV-C damaged (450 J/m2) using a Stratalinker (Stratagene) and used as repair substrate. Twenty micrograms of protein extracts were used per time point and mixed with 200 ng of damaged plasmid and 300 ng of non-damaged pBSK linearized plasmid as internal control. The reaction was stopped after 1 or 2 h by flash freezing in liquid nitrogen. Plasmids were gel purified and separated by electrophoresis on a 0.8% agarose gel. DNA was transferred onto a nylon membrane (Roche) by capillary transfer and detection was performed using the DIG Nucleic Acid Detection kit (Roche).
DET1 antibody production
The full-length DET1 coding sequence was amplified by RT–PCR from wild-type Arabidopsis Col-0 RNAs and cloned into the pET-15 Escherichia coli expression vector in frame with a 6 × HIS amino-terminal tag using BamHI and XhoI restriction sites. Upon induction with 1 mM IPTG for 30 min at 37°C, bacterial cells were broken using a French press and inclusion bodies were purified according to Barneche et al (2006). DET1 recombinant protein was further purified from low level contaminants by SDS–PAGE followed by electroelution of the excised Coomassie-stained bands. In total, 100 μg of the purified protein was combined with Freund's adjuvant to form a stable emulsion that was injected subcutaneously in rabbit. This procedure was repeated after 3 weeks and 1 week after injection blood was collected from the central ear artery and the antibody's titre was checked by immunoblot. Two additional injections with 20 μg of recombinant protein were performed at week 7 and 10; in the last one the protein was mixed with 0.5 ml of PolyA-PolyU (2 mg/ml, polyadenylic-polyuridylic acid, Sigma) and injected intravenously. At week 11, blood was collected and the serum was separated and clarified by centrifugation. The serum was further affinity purified on DET1 recombinant protein using a SulfoLink Immobilization Kit for Peptides using the manufacturer's instruction (Thermo Scientific Pierce).
Gel filtration
Fifteen-day-old light-grown seedlings were ground in liquid nitrogen and resuspended in 1 ml of extraction buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.1% NP-40, 10% glycerol and protease inhibitors). The extract was cleared by centrifugation for 5 min at 5000 r.p.m., 5 min at 10 000 r.p.m. and for 1 h at 14 000 r.p.m. Soluble fraction was subsequently concentrated for 2 h at 14 000 r.p.m. using YM-10 centricons (Amicon). Five-hundred micrograms of cleared extract was then injected in a pre-calibrated Superdex 200 (Amersham) gel filtration column with the same extraction buffer at 0.4 ml/min using an AKTA FPLC system. Twenty fractions of 0.5 ml were collected, and 50 μl of each was analysed by immunoblotting with the indicated antibodies.
Protein immunoblotting
Analysis of DET1 and DDB2 content after UV-C irradiation was done according to Molinier et al (2008). Two-week-old seedlings were UV-C irradiated (at 1500 or 3000 J/m2) using a UV-Crosslinker (GE Healthcare, Amersham Biosciences). Total protein extract was prepared using a denaturing buffer (Büche et al, 2000) and boiled for 5 min. Twenty micrograms of total protein were separated by 10% SDS–PAGE and transferred onto Immobilon-P membranes (Millipore) and blotted with anti-DDB2 and anti-DET1 antibodies.
Supplementary Material
Acknowledgments
We are very grateful to David Stroebel for technical contribution at the IBENS Proteomic platform, to Diana Molino and Daniele Silvestro (Stazione Zoologica, Naples, Italy) for help with DNA repair assays, and to Roberto Bassi (Verona University) for expertise with DET1 serum production, as well as to Josep Casacuberta for critical reading of the manuscript. This work was supported by grants ANR-07-BLAN-0216 from the French Agence Nationale pour la Recherche (ANR) and EU-SOL project FOOD-CT-2006-016214 from the European Union to CB, by a fellowship LT00299/2005 from the Human Frontier Science Program (HFSP) to F.B., and by a post-doctoral fellowship from the Fondation Pierre-Gilles de Gennes pour la Recherche to E.C.
Author contributions: EC and JM performed the UV sensitivity and DNA repair assays; EC, JM, AZ, GZ and FB performed the biochemical experiments; FB, GB and JM generated mutants and transgenic lines; SC generated the DET1 antibody, Clara B performed the gene expression analyses; JM, EC, GB, PG, FB and Chris B designed the experiments; EC and FB wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Al Khateeb WM, Schroeder DF (2009) Overexpression of Arabidopsis damaged DNA binding protein 1A (DDB1A) enhances UV tolerance. Plant Mol Biol 70: 371–383 [DOI] [PubMed] [Google Scholar]
- Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N (2006) Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 443: 590–593 [DOI] [PubMed] [Google Scholar]
- Barneche F, Winter V, Crevecoeur M, Rochaix JD (2006) ATAB2 is a novel factor in the signalling pathway of light-controlled synthesis of photosystem proteins. EMBO J 25: 5907–5918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benvenuto G, Formiggini F, Laflamme P, Malakhov M, Bowler C (2002) The photomorphogenesis regulator DET1 binds the amino-terminal tail of histone H2B in a nucleosome context. Curr Biol 12: 1529–1534 [DOI] [PubMed] [Google Scholar]
- Bernhardt A, Lechner E, Hano P, Schade V, Dieterle M, Anders M, Dubin MJ, Benvenuto G, Bowler C, Genschik P, Hellmann H (2006) CUL4 associates with DDB1 and DET1 and its downregulation affects diverse aspects of development in Arabidopsis thaliana. Plant J 47: 591–603 [DOI] [PubMed] [Google Scholar]
- Bernhardt A, Mooney S, Hellmann H (2010) Arabidopsis DDB1a and DDB1b are critical for embryo development. Planta 232: 555–566 [DOI] [PubMed] [Google Scholar]
- Biedermann S, Hellmann H (2010) The DDB1a interacting proteins ATCSA-1 and DDB2 are critical factors for UV-B tolerance and genomic integrity in Arabidopsis thaliana. Plant J 62: 404–415 [DOI] [PubMed] [Google Scholar]
- Britt AB (1999) Molecular genetics of DNA repair in higher plants. Trends Plant Sci 4: 20–25 [DOI] [PubMed] [Google Scholar]
- Büche C, Poppe C, Schäfer E, Kretsch T (2000) eid1: a new Arabidopsis mutant hypersensitive in phytochrome A-dependent high-irradiance responses. Plant Cell 12: 547–558 [PMC free article] [PubMed] [Google Scholar]
- Castells E, Molinier J, Drevensek S, Genschik P, Barneche F, Bowler C (2010) det1-1-induced UV-C hyposensitivity through UVR3 and PHR1 photolyase gene over-expression. Plant J 63: 392–404 [DOI] [PubMed] [Google Scholar]
- Chen H, Huang X, Gusmaroli G, Terzaghi W, Lau OS, Yanagawa Y, Zhang Y, Li J, Lee JH, Zhu D, Deng XW (2010) Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell 22: 108–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Shen Y, Tang X, Yu L, Wang J, Guo L, Zhang Y, Zhang H, Feng S, Strickland E, Zheng N, Deng XW (2006) Arabidopsis CULLIN4 forms an E3 ubiquitin ligase with RBX1 and the CDD complex in mediating light control of development. Plant Cell 18: 1991–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C (2004) Light signal transduction in higher plants. Annu Rev Genet 38: 87–117 [DOI] [PubMed] [Google Scholar]
- Chen X, Zhang Y, Douglas L, Zhou P (2001) UV-damaged DNA-binding proteins are targets of CUL-4A-mediated ubiquitination and degradation. J Biol Chem 276: 48175–48182 [DOI] [PubMed] [Google Scholar]
- Chory J (1992) A genetic model for light-regulated seedling development in Arabidopsis. Development 115: 337–354 [Google Scholar]
- Chory J, Peto C, Feinbaum R, Pratt L, Ausubel F (1989) Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell 58: 991–999 [DOI] [PubMed] [Google Scholar]
- Chory J, Peto CA (1990) Mutations in the DET1 gene affect cell-type-specific expression of light-regulated genes and chloroplast development in Arabidopsis. Proc Natl Acad Sci USA 87: 8776–8780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A, Orian A, Schwartz AL (2000) Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays 22: 442–451 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dohmann EM, Levesque MP, De Veylder L, Reichardt I, Jürgens G, Schmid M, Schwechheimer C (2008) The Arabidopsis COP9 signalosome is essential for G2 phase progression and genomic stability. Development 135: 2013–2022 [DOI] [PubMed] [Google Scholar]
- Dubin MJ, Bowler C, Benvenuto G (2008) A modified Gateway cloning strategy for overexpressing tagged proteins in plants. Plant Methods 4: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Mahdy MA, Zhu Q, Wang QE, Wani G, Praetorius-Ibba M, Wani AA (2006) Cullin 4A-mediated proteolysis of DDB2 protein at DNA damage sites regulates in vivo lesion recognition by XPC. J Biol Chem 281: 13404–13411 [DOI] [PubMed] [Google Scholar]
- Fousteri M, Mullenders LH (2008) Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res 18: 73–84 [DOI] [PubMed] [Google Scholar]
- Gillet LC, Scharer OD (2006) Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem Rev 106: 253–276 [DOI] [PubMed] [Google Scholar]
- Gray WM, Hellmann H, Dharmasiri S, Estelle M (2002) Role of the Arabidopsis RING-H2 protein RBX1 in RUB modification and SCF function. Plant Cell 14: 2137–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, Kisselev AF, Tanaka K, Nakatani Y (2003) The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113: 357–367 [DOI] [PubMed] [Google Scholar]
- Guerrero-Santoro J, Kapetanaki MG, Hsieh CL, Gorbachinsky I, Levine AS, Rapic-Otrin V (2008) The cullin 4B-based UV-damaged DNA-binding protein ligase binds to UV-damaged chromatin and ubiquitinates histone H2A. Cancer Res 68: 5014–5022 [DOI] [PubMed] [Google Scholar]
- Hanawalt PC, Ford JM, Lloyd DR (2003) Functional characterization of global genomic DNA repair and its implications for cancer. Mutat Res 544: 107–114 [DOI] [PubMed] [Google Scholar]
- He YJ, McCall CM, Hu J, Zeng Y, Xiong Y (2006) DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev 20: 2949–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa LA, Wu M, Ye T, Kobayashi R, Sun H, Zhang H (2006) CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat Cell Biol 8: 1277–1283 [DOI] [PubMed] [Google Scholar]
- Huang TT, D’Andrea AD (2006) Regulation of DNA repair by ubiquitylation. Nat Rev Mol Cell Biol 7: 323–334 [DOI] [PubMed] [Google Scholar]
- Jackson S, Xiong Y (2009) CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem Sci 34: 562–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Kapetanaki MG, Guerrero-Santoro J, Bisi DC, Hsieh CL, Rapic-Otrin V, Levine AS (2006) The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc Natl Acad Sci USA 103: 2588–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattan Z, Marchal S, Brunner E, Ramacci C, Leroux A, Merlin JL, Domenjoud L, Dauca M, Becuwe P (2008) Damaged DNA binding protein 2 plays a role in breast cancer cell growth. PLoS One 3: e2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OS, Deng XW (2009) Effect of Arabidopsis COP10 ubiquitin E2 enhancement activity across E2 families and functional conservation among its canonical homologues. Biochem J 418: 683–690 [DOI] [PubMed] [Google Scholar]
- Lechner E, Xie D, Grava S, Pigaglio E, Planchais S, Murray JA, Parmentier Y, Mutterer J, Dubreucq B, Shen WH, Genschik P (2002) The AtRbx1 protein is part of plant SCF complexes, and its down-regulation causes severe growth and developmental defects. J Biol Chem 277: 50069–50080 [DOI] [PubMed] [Google Scholar]
- Li A, Schuermann D, Gallego F, Kovalchuk I, Tinland B (2002) Repair of damaged DNA by Arabidopsis cell extract. Plant Cell 14: 263–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Flury S, Kalck V, Hohn B, Molinier J (2006) CENTRIN2 interacts with the Arabidopsis homolog of the human XPC protein (AtRAD4) and contributes to efficient synthesis-dependent repair of bulky DNA lesions. Plant Mol Biol 61: 345–356 [DOI] [PubMed] [Google Scholar]
- Lorrain S, Genoud T, Fankhauser C (2006) Let there be light in the nucleus! Curr Opin Plant Biol 9: 509–514 [DOI] [PubMed] [Google Scholar]
- Ma L, Zhao H, Deng XW (2003) Analysis of the mutational effects of the COP/DET/FUS loci on genome expression profiles reveals their overlapping yet not identical roles in regulating Arabidopsis seedling development. Development 130: 969–981 [DOI] [PubMed] [Google Scholar]
- Matsuda N, Azuma K, Saijo M, Iemura S, Hioki Y, Natsume T, Chiba T, Tanaka K (2005) DDB2, the xeroderma pigmentosum group E gene product, is directly ubiquitylated by Cullin 4A-based ubiquitin ligase complex. DNA Repair 4: 537–545 [DOI] [PubMed] [Google Scholar]
- Misera S, Muller AJ, Weiland-Heidecker U, Jurgens G (1994) The FUSCA genes of Arabidopsis: negative regulators of light responses. Mol Gen Genet 244: 242–252 [DOI] [PubMed] [Google Scholar]
- Molinier J, Lechner E, Dumbliauskas E, Genschik P (2008) Regulation and role of Arabidopsis CUL4-DDB1A-DDB2 in maintaining genome integrity upon UV stress. PLoS Genet 4: e1000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinier J, Ramos C, Fritsch O, Hohn B (2004) CENTRIN2 modulates homologous recombination and nucleotide excision repair in Arabidopsis. Plant Cell 16: 1633–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A, Bondar T, Shiv S, Raychaudhuri P (2001) The xeroderma pigmentosum group E gene product DDB2 is a specific target of cullin 4A in mammalian cells. Mol Cell Biol 21: 6738–6747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols AF, Itoh T, Graham JA, Liu W, Yamaizumi M, Linn S (2000) Human damage-specific DNA-binding protein p48. Characterization of XPE mutations and regulation following UV irradiation. J Biol Chem 275: 21422–21428 [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Pepper A, Delaney T, Washburn T, Poole D, Chory J (1994) DET1, a negative regulator of light-mediated development and gene expression in arabidopsis, encodes a novel nuclear-localized protein. Cell 78: 109–116 [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ (2005) Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6: 9–20 [DOI] [PubMed] [Google Scholar]
- Pick E, Lau OS, Tsuge T, Menon S, Tong Y, Dohmae N, Plafker SM, Deng XW, Wei N (2007) Mammalian DET1 regulates Cul4A activity and forms stable complexes with E2 ubiquitin-conjugating enzymes. Mol Cell Biol 27: 4708–4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70: 503–533 [DOI] [PubMed] [Google Scholar]
- Rapic-Otrin V, McLenigan MP, Bisi DC, Gonzalez M, Levine AS (2002) Sequential binding of UV DNA damage binding factor and degradation of the p48 subunit as early events after UV irradiation. Nucleic Acids Res 30: 2588–2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S (2004) Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem 73: 39–85 [DOI] [PubMed] [Google Scholar]
- Schroeder DF, Gahrtz M, Maxwell BB, Cook RK, Kan JM, Alonso JM, Ecker JR, Chory J (2002) De-etiolated 1 and damaged DNA binding protein 1 interact to regulate Arabidopsis photomorphogenesis. Curr Biol 12: 1462–1472 [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Serino G, Deng XW (2002) Multiple ubiquitin ligase-mediated processes require COP9 signalosome and AXR1 function. Plant Cell 14: 2553–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrima A, Konickova R, Czyzewski BK, Kawasaki Y, Jeffrey PD, Groisman R, Nakatani Y, Iwai S, Pavletich NP, Thoma NH (2008) Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell 135: 1213–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serino G, Deng XW (2003) The COP9 signalosome: regulating plant development through the control of proteolysis. Annu Rev Plant Biol 54: 165–1682 [DOI] [PubMed] [Google Scholar]
- Shuck SC, Short EA, Turchi JJ (2008) Eukaryotic nucleotide excision repair: from understanding mechanisms to influencing biology. Cell Res 18: 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Sugasawa K, Okuda Y, Saijo M, Nishi R, Matsuda N, Chu G, Mori T, Iwai S, Tanaka K, Hanaoka F (2005) UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell 121: 387–400 [DOI] [PubMed] [Google Scholar]
- Svejstrup JQ (2002) Mechanisms of transcription-coupled DNA repair. Nat Rev Mol Cell Biol 3: 21–29 [DOI] [PubMed] [Google Scholar]
- Volker M, Mone MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA, Mullenders LH (2001) Sequential assembly of the nucleotide excision repair factors in vivo. Mol Cell 8: 213–224 [DOI] [PubMed] [Google Scholar]
- Wang H, Zhai L, Xu J, Joo HY, Jackson S, Erdjument-Bromage H, Tempst P, Xiong Y, Zhang Y (2006) Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol Cell 22: 383–394 [DOI] [PubMed] [Google Scholar]
- Wertz IE, O’Rourke KM, Zhang Z, Dornan D, Arnott D, Deshaies RJ, Dixit VM (2004) Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 303: 1371–1374 [DOI] [PubMed] [Google Scholar]
- Wittschieben BO, Iwai S, Wood RD (2005) DDB1-DDB2 (xeroderma pigmentosum group E) protein complex recognizes a cyclobutane pyrimidine dimer, mismatches, apurinic/apyrimidinic sites, and compound lesions in DNA. J Biol Chem 280: 39982–39989 [DOI] [PubMed] [Google Scholar]
- Wood RD, Araujo SJ, Ariza RR, Batty DP, Biggerstaff M, Evans E, Gaillard PH, Gunz D, Koberle B, Kuraoka I, Moggs JG, Sandall JK, Shivji MK (2000) DNA damage recognition and nucleotide excision repair in mammalian cells. Cold Spring Harb Symp Quant Biol 65: 173–182 [DOI] [PubMed] [Google Scholar]
- Yanagawa Y, Sullivan JA, Komatsu S, Gusmaroli G, Suzuki G, Yin J, Ishibashi T, Saijo Y, Rubio V, Kimura S, Wang J, Deng XW (2004) Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes Dev 18: 2172–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Schroeder DF (2010) Effect of overexpression of Arabidopsis damaged DNA-binding protein 1A on de-etiolated 1. Planta 231: 337–348 [DOI] [PubMed] [Google Scholar]
- Zhang C, Guo H, Zhang J, Guo G, Schumaker KS, Guo Y (2010) Arabidopsis cockayne syndrome a-like proteins 1A and 1B form a complex with CULLIN4 and damage DNA binding protein 1A and regulate the response to UV irradiation. Plant Cell 22: 2353–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.