Abstract
A number of external and internal insults disrupt nucleolar structure, and the resulting nucleolar stress stabilizes and activates p53. We show here that nucleolar disruption induces acetylation and accumulation of p53 without phosphorylation. We identified three nucleolar proteins, MYBBP1A, RPL5, and RPL11, involved in p53 acetylation and accumulation. MYBBP1A was tethered to the nucleolus through nucleolar RNA. When rRNA transcription was suppressed by nucleolar stress, MYBBP1A translocated to the nucleoplasm and facilitated p53–p300 interaction to enhance p53 acetylation. We also found that RPL5 and RPL11 were required for rRNA export from the nucleolus. Depletion of RPL5 or RPL11 blocked rRNA export and counteracted reduction of nucleolar RNA levels caused by inhibition of rRNA transcription. As a result, RPL5 or RPL11 depletion inhibited MYBBP1A translocation and p53 activation. Our observations indicated that a dynamic equilibrium between RNA generation and export regulated nucleolar RNA content. Perturbation of this balance by nucleolar stress altered the nucleolar RNA content and modulated p53 activity.
Keywords: MYBBP1A, nucleolar disruption, p53, RPL11, RPL5
Introduction
The tumour suppressor protein p53 responds to diverse stresses and regulates many target genes, the products of which induce cell-cycle arrest, apoptosis, senescence, and DNA repair (Levine, 1997; Prives and Hall, 1999). In unstressed cells, p53 is maintained at low levels by its inhibitor HDM2, an ubiquitin ligase that ubiquitinates p53, thereby targeting the protein for proteasome-mediated degradation through a feedback mechanism (Michael and Oren, 2003; Brooks and Gu, 2006). In response to various stresses, such as DNA damage or oncogene expression, p53 is stabilized and activated (Prives and Hall, 1999).
DNA damage is widely believed to activate p53 as a transcription factor through post-translational modifications, such as phosphorylation, ubiquitination, and acetylation (Appella and Anderson, 2001; Brooks and Gu, 2003; Bode and Dong, 2004; Olsson et al, 2007; Vousden and Lane, 2007; Carter and Vousden, 2009; Kruse and Gu, 2009). Phosphorylation of p53 in response to DNA damage is mediated by several kinases (Bode and Dong, 2004; Olsson et al, 2007) and produces a number of downstream effects by changing the interaction between p53 and HDM2 or p300 (Shieh et al, 1997; Lambert et al, 1998; Chehab et al, 1999; Dumaz and Meek, 1999; Unger et al, 1999). However, an accumulation of evidence has shown that the requirement for the aforementioned phosphorylation is probably not universal for p53 stabilization or activation (Ashcroft et al, 1999, 2000; Blattner et al, 1999; Prives and Hall, 1999; Wu et al, 2002; Thompson et al, 2004).
The p53 protein is acetylated at multiple lysine residues (Lys120, Lys164, Lys305, Lys320, Lys370, Lys372, Lys373, Lys381, Lys382, Lys386) by the p300/CBP complex, the p300/CBP-associated factor (PCAF), or the Tip60 (Gu and Roeder, 1997; Sakaguchi et al, 1998; Tang et al, 2008). Acetylation of p53 is sufficient to abrogate its ubiquitination by HDM2. There is a direct competition between acetylation and ubiquitination of the same C-terminal lysine residues in p53 (Li et al, 2002). In addition, the acetylation of p53 enhances sequence-specific DNA binding of p53 (Gu and Roeder, 1997; Sakaguchi et al, 1998; Luo et al, 2004) and recruitment of a coactivator complex to promoter regions for activation of p53-targeted gene expression (Barlev et al, 2001). Moreover, recent studies suggest that HDM2 represses transcription not only by mediating ubiquitination of p53, but also by forming a suppressor complex with p53 on the promoters of specific p53-responsive genes (Minsky and Oren, 2004; Arva et al, 2005; Ohkubo et al, 2006). Acetylation blocks the interaction of p53 with HDM2 on DNA, which activates p53 regardless of its phosphorylation status. The p53-8KR mutant, in which the eight known acetylated sites are substituted with arginine, is unable to activate p21 and induce cell-cycle arrest and apoptosis (Tang et al, 2008). From these observations, p53 acetylation is believed to be indispensable for p53 activation. However, in a mouse model, the C-terminal lysine residues of p53 are not required for either stability or transactivation (Krummel et al, 2005). Therefore, the contributions of acetylation at the C-terminus to p53 stability and transactivation may differ between mice and humans.
The p53 protein induces cell-cycle arrest or apoptosis under a wide variety of cellular stresses. However, it is not known how a single protein can integrate such a diversity of signals. Recently, a number of external and internal insults were shown to induce nucleolar stress by disrupting nucleolar structure (Rubbi and Milner, 2003; Mayer and Grummt, 2005). Previous reports demonstrated that nucleolar disruption, induced by either anti-upstream binding factor (UBF) antibody microinjection or depletion of the transcription initiation factor-IA (TIF-IA), caused p53 stabilization (Rubbi and Milner, 2003; Yuan et al, 2005). Thus, the existence of a stress sensor that monitors nucleolar structure and function and regulates p53 levels was proposed. Recent studies of the cellular response to nucleolar stress demonstrated that several nucleolar proteins, including nucleophosmin (NPM; also called B23), nucleolin (NCL; also called C23), nucleostemin (NS), ARF, and ribosomal proteins, such as RPL5, RPL11, RPL23, and RPS7, can bind to HDM2 and inhibit its activity towards p53 (Marechal et al, 1994; Kamijo et al, 1998; Pomerantz et al, 1998; Lohrum et al, 2003; Zhang et al, 2003; Dai and Lu, 2004; Dai et al, 2004, 2008; Jin et al, 2004; Kurki et al, 2004; Saxena et al, 2006; Chen et al, 2007; Zhu et al, 2009).
Here, we show that nucleolar disruption induces acetylation and accumulation of p53 without phosphorylation. We screened nucleolar proteins involved in acetylation and stabilization of p53 and identified Myb-binding protein 1a (MYBBP1A). Our results demonstrated that nucleolar disruption led to translocation of MYBBP1A from the nucleolus to the nucleoplasm. MYBBP1A then bound to p53 and facilitated binding between p53 and p300 to enhance p53-mediated transcription. We also found that knockdown of RPL5 and RPL11 significantly abrogated the accumulation and acetylation of p53 protein by inhibiting the translocation of MYBBP1A induced by nucleolar disruption.
Results
Nucleolar protein MYBBP1A is necessary for phosphorylation-independent p53 acetylation
Phosphorylation of p53 at Ser15 enhances its acetylation by increasing the interaction with p300/CBP acetyltransferase (Lambert et al, 1998; Dumaz and Meek, 1999). Recent reports, however, indicated that phosphorylation-independent p53 acetylation occurred during low-dose actinomycin D (ActD) treatment, which specifically inhibited RNA polymerase I- but not RNA polymerase II-driven transcription (Ashcroft et al, 2000; Ito et al, 2001; Tang et al, 2008). In agreement with the reports, we observed that low-dose ActD treatment increased the p53 acetylation levels at Lys382 without inducing phosphorylation at Ser15 (Supplementary Figure S1A) in the MCF-7 cell line, which expresses wild-type p53. These results suggest that there may be pathways that induce acetylation of p53 in the absence of its phosphorylation at Ser15.
Recently, it was shown that activation of p53 was induced by nucleolar disruption, which also occurred during low-dose ActD treatment (Supplementary Figure S1B) (Rubbi and Milner, 2003; Yuan et al, 2005). To test the relationship between Ser15 phosphorylation-independent p53 acetylation and nucleolar disruption, we induced nucleolar disruption by TIF-IA depletion. In agreement with previous reports, after TIF-IA siRNA treatment, nucleolar structure disappeared and translocation of the nucleolar marker protein NPM from the nucleolus to the nucleoplasm was observed in the MCF-7 and LNCaP cell lines, both of which express wild-type p53 (Supplementary Figures S2A and B, S3A and B). Knockdown of TIF-IA induced the accumulation of p53 and elevated the acetylation levels at the lysine residues that were known to be acetylated by p300/CBP without phosphorylation at Ser15 (Figure 1A; Supplementary Figures S2C and D and S3C). These results suggest that nucleolar disruption induces phosphorylation-independent acetylation of p53.
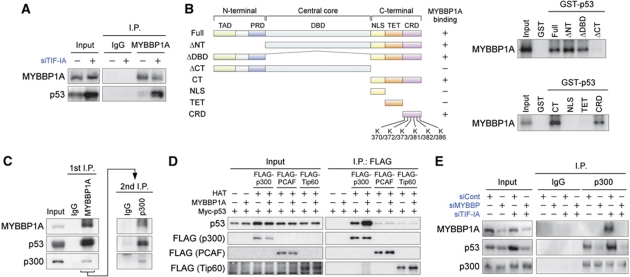
Figure 1.
MYBBP1A is necessary for p53 acetylation induced by nucleolar disruption. (A) Knockdown of TIF-IA induces acetylation at multiple lysine residues in p53. MCF-7 cells were treated with siCont or siTIF-IA for 48 h, and the cell lysates were analysed by immunoblot using the indicated antibodies. (B) Nucleolar disruption releases the nucleolar protein MYBBP1A into the nucleoplasm. MCF-7 cells were treated with siCont or siTIF-IA for 48 h, and the localization of MYBBP1A and UBF was visualized by immunofluorescence using anti-MYBBP1A and anti-UBF antibodies. (C) Knockdown of MYBBP1A antagonizes the acetylation of p53 proteins caused by nucleolar disruption. MCF-7 cells were treated with the siCont or MYBBP1A siRNA (siMYBBP) with or without siTIF-IA for 48 h, and cell lysates were analysed by immunoblot using the indicated antibodies. (D) Knockdown of MYBBP1A decreases the acetylation levels at multiple lysine residues in the p53 protein. (Left) MCF-7 cells were treated with the siCont or two independent siMYBBPs (siMYBBP and siMYBBP-2) with or without siTIF-IA for 48 h, and cell lysates were analysed by immunoblot using site-specific acetylated p53 antibodies. (Right) Relative quantification of acetylation levels of the p53 protein. The intensities of the acetylated p53 proteins were corrected using the p53 protein level. The intensity of the siCont-treated cells was normalized to 1.0. (E) Knockdown of MYBBP1A antagonizes the acetylation of p53 induced by low-dose ActD treatment. MCF-7 cells were treated with siCont or siMYBBP for 48 h, followed by 5 nM ActD treatment for indicated times. The cell lysates were analysed by immunoblot using the indicated antibodies. (F) Recovery of the acetylation levels of p53 protein by introducing siRNA-resistant MYBBP1A along with siRNA for MYBBP1A. p53-deficient H1299 cells were treated with the siCont or siMYBBP with or without siTIF-IA for 24 h before transfection with Myc-p53 and siRNA-resistant MYBBP1A (MYBBP1A-simut). Twenty-four hours after transfection, cell lysates were analysed by immunoblot with the indicated antibodies.
The acetylation levels of p53 have been shown to increase significantly in response to stress and correlate well with p53 activation and stabilization (Luo et al, 2000, 2001; Ito et al, 2001, 2002; Vaziri et al, 2001; Knights et al, 2006; Li et al, 2007; Kim et al, 2008; Zhao et al, 2008). An accumulated body of evidence supports the conclusion that acetylation stabilizes p53 and is indispensable for p53 activation (Li et al, 2002; Tang et al, 2008). Therefore, we searched for nucleolar proteins that could regulate p53 acetylation. Using a quantitative proteomics approach, Matthias Mann and coworkers characterized the flux of 489 endogenous nucleolar proteins, and found that nearly 300 proteins translocated from the nucleolus to the nucleoplasm following nucleolar disruption (Andersen et al, 2005). Among these 300 proteins, we excluded 200 proteins based on their known function and selected 107 candidates for investigation. We generated a library of siRNAs against the mRNAs for these proteins (Supplementary Table S1) and examined the effects of each siRNA on both the acetylation status and quantity of p53 proteins in cells that had been treated with TIF-IA siRNA to increase p53 acetylation. As shown in Supplementary Figure S4, treatment of MCF-7 cells with siRNA against MYBBP1A, RPL5, or RPL11 reduced the protein levels and acetylation status of p53.
RPL5 and RPL11 have been shown to be involved in the stabilization of p53 by binding to HDM2 and blocking its function (Lohrum et al, 2003; Zhang et al, 2003; Dai and Lu, 2004). Thus, we first examined the function of MYBBP1A. MYBBP1A is a 160-kDa protein present mainly in the nucleolus (Tavner et al, 1998). The role of MYBBP1A in the nucleolus was previously unknown. Immunostaining of cells with anti-MYBBP1A antibodies confirmed the nucleolar localization of MYBBP1A (Figure 1B, upper panel). TIF-IA depletion released MYBBP1A into the nucleoplasm (Figure 1B, lower panel). To confirm that knockdown of MYBBP1A antagonized the acetylation of p53 proteins caused by nucleolar disruption, we evaluated the effects of MYBBP1A knockdown on p53 acetylation status when nucleolar disruption was triggered by siRNA for TIF-IA. As shown in Figure 1C, acetylation at Lys382 in p53, induced by nucleolar disruption, was significantly abrogated by MYBBP1A knockdown. MYBBP1A knockdown also decreased the acetylation levels at other lysine residues (Lys305 and Lys373) of p53 in MCF-7 and LNCaP cells (Figure 1D; Supplementary Figure S3D) and partially counteracted the accumulation of p53 proteins induced by nucleolar disruption. The same result was obtained when a different siRNA for MYBBP1A (siMYBBP-2) was used (Figure 1D). Furthermore, MYBBP1A knockdown also reduced the acetylation levels of p53 during low-dose ActD treatment (Figure 1E).
To confirm that MYBBP1A was involved in the acetylation of p53 caused by nucleolar disruption, we performed a rescue experiment. An expression plasmid containing p53 was transfected into p53-deficient H1299 cells (Figure 1F). Western blot analysis measured the basal expression and acetylation levels of p53 in p53-transfected H1299 cells without stress. TIF-IA depletion induced p53 acetylation that was reduced by treatment with siRNA for MYBBP1A (Figure 1F). The reduction of p53 acetylation by MYBBP1A siRNA was recovered by coexpression of mutated MYBBP1A (MYBBP1A-simut), which was not downregulated by the siRNA for MYBBP1A (Figure 1F), demonstrating that MYBBP1A was necessary for the efficient acetylation of p53 protein under nucleolar disruption.
We next evaluated the contributions of MYBBP1A to p53 acetylation induced by DNA-damaging stress, resulting from exposure to adriamycin (ADR), UV light, or high-dose ActD. These stresses are known to induce nucleolar disruption (Rubbi and Milner, 2003), translocation of MYBBP1A into the nucleoplasm (Yamauchi et al, 2008), and p53 modifications, including phosphorylation and acetylation (Shieh et al, 1997; Tibbetts et al, 1999; Ito et al, 2001; Saito et al, 2003). Depletion of MYBBP1A did not alter phosphorylation but did reduce the acetylation levels of p53 (Supplementary Figure S5). The reduction of p53 acetylation by MYBBP1A depletion was more significant in cells treated with TIF-IA siRNA than in those treated with DNA-damaging agents. Thus, our results indicated that while MYBBP1A was also involved in the DNA damage-induced acetylation of p53, its contribution to p53 acetylation was higher in cells subjected to TIF-IA depletion than in cells subjected to DNA damage.
MYBBP1A increases the binding of p300 to p53
To investigate the molecular basis for the functions of MYBBP1A, the interaction between MYBBP1A and p53 was first investigated. Coimmunoprecipitation experiments revealed binding of endogenous MYBBP1A to p53 in MCF-7 cells under nucleolar stress conditions (Figure 2A). GST pull-down experiments showed that MYBBP1A directly bound to the C-terminal 34 amino-acid region of p53, which contains six lysine residues that are acetylated by p300/CBP (Figure 2B).
Figure 2.
MYBBP1A directly binds to p53, increasing the binding of p300 to p53. (A) Endogenous MYBBP1A associates with p53. MCF-7 cells were treated with siTIF-IA for 48 h. The cell lysates were immunoprecipitated with normal rabbit IgG or anti-MYBBP1A antibodies and analysed by immunoblot using antibodies against MYBBP1A and p53. (B) (Left) Domain structure of the full-length p53 and various deletion mutants. TAD, transactivation domain; PRD, proline-rich domain; DBD, DNA-binding domain; NLS, nuclear localization signal; TET, tetramerization domain; CRD, C-terminal regulatory domain; K, lysine residues that can be acetylated in the CRD region. (Right) MYBBP1A directly binds to the CRD region in p53. 35S-labelled MYBBP1A was incubated with the GST-fused full-length or truncated p53 proteins. After extensive washing, bound proteins were analysed by SDS–PAGE and autoradiography. (C) MYBBP1A forms a ternary complex with p53 and p300. MCF-7 cells were treated with siTIF-IA for 48 h. The cell lysates were sequentially immunoprecipitated with anti-MYBBP1A and anti-p300 antibodies, and immunoprecipitates were detected by immunoblot using antibodies against MYBBP1A, p53, and p300. (D) MYBBP1A increases the binding of p300, but not PCAF and Tip60, to p53. H1299 cells were transfected with expression vectors for FLAG-p300, FLAG-PCAF, FLAG-Tip60, MYBBP1A, and Myc-p53 as indicated. Twenty-four hours after transfection, the cells were treated with ADR (0.5 μg/ml) for 12 h to induce nucleolar disruption. FLAG-p300, FLAG-PCAF, and FLAG-Tip60 were immunoprecipitated using anti-FLAG antibodies, and the immunoprecipitates were eluted using FLAG peptides, then analysed by immunoblot using antibodies against FLAG and p53. HAT, histone acetyltransferases. (E) Knockdown of MYBBP1A decreases the binding of p300 to p53. MCF-7 cells were treated with siCont or siMYBBP with or without siTIF-IA for 48 h. The cell lysates were prepared, immunoprecipitated with normal rabbit IgG or anti-p300 antibodies, and analysed by immunoblot using antibodies against MYBBP1A, p53, and p300.
Because MYBBP1A is required for effective acetylation at p300/CBP-mediated acetylation sites in p53, the interactions among p53, MYBBP1A, and p300 were investigated next (Figure 2C). Nuclear extracts derived from MCF-7 cells treated with TIF-IA siRNA were prepared for this purpose. Proteins in the extracts were sequentially immunoprecipitated with anti-MYBBP1A and anti-p300 antibodies. Immunoblot analysis demonstrated the presence of p53 in the final immunoprecipitant, indicating that p53 formed a ternary complex with MYBBP1A and p300 in the TIF-IA siRNA-treated cells (Figure 2C).
We next tested the effects of MYBBP1A on the interaction between p53 and p300. p53 and p300 expression plasmids were introduced into H1299 cells with or without the MYBBP1A expression plasmid. Coimmunoprecipitation experiments showed that the association of p53 with p300 was significantly enhanced by MYBBP1A expression in the presence of ADR (Figure 2D). Conversely, knockdown of MYBBP1A in MCF-7 cells abrogated the nucleolar disruption-induced binding between p53 and p300 (Figure 2E). In contrast, the binding of p53 to PCAF and Tip60, both of which are known to be histone acetyltransferases for p53, was not enhanced by MYBBP1A (Figure 2D). These results indicated that MYBBP1A selectively stabilized the binding between p53 and p300.
Knockdown of MYBBP1A decreases the transactivating capabilities of p53
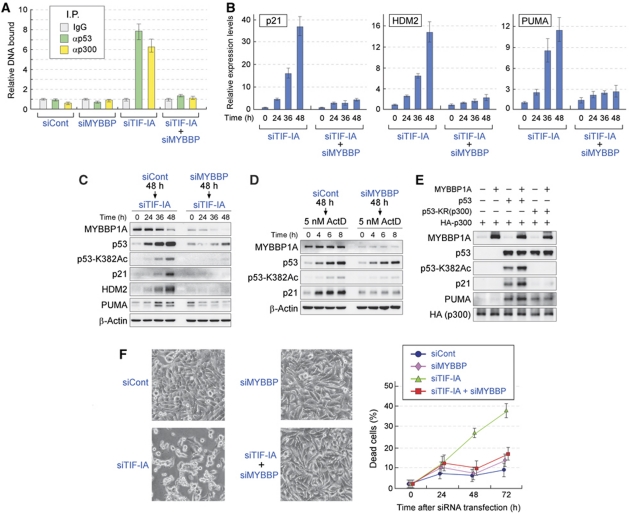
Our results indicated that MYBBP1A enhanced the binding between p300 and p53 and increased p53 acetylation levels. p300 also functions as a coactivator for p53 by acetylating histones and enhancing the transcription of p53-target genes. Thus, we investigated the recruitment of p53 and p300 to the promoter region of the p21 gene by a chromatin immunoprecipitation (ChIP) assay. As shown in Figure 3A and Supplementary Figure S6A, treatment of cells with TIF-IA siRNA, ADR, or ActD enhanced the recruitment of p53 and p300 to the p21 promoter. The recruitment of p53 and p300 was abrogated by knockdown of MYBBP1A (Figure 3A; Supplementary Figure S6A).
Figure 3.
Knockdown of MYBBP1A decreases the transactivating capacity of p53 and protects cells from nucleolar stress-induced apoptosis. (A) Knockdown of MYBBP1A reduces the recruitment of p53 and p300 to the p21 promoter in siTIF-IA-treated cells. MCF-7 cells were treated with siCont or siMYBBP for 48 h before treatment with siTIF-IA for 48 h. A ChIP assay was performed using normal rabbit IgG, anti-p53, and anti-p300 antibodies. The p53-binding region of the p21 promoter was amplified and analysed by qPCR. Values are given as the mean±s.d. for triplicate experiments. (B) Knockdown of MYBBP1A reduces the elevation of p53-target gene expression induced by siTIF-IA treatment. MCF-7 cells were treated with siCont or siMYBBP for 48 h before treatment with siTIF-IA for the indicated times. The total RNAs were prepared and expression of the indicated genes was analysed by RT–qPCR. Values are given as the mean±s.d. for triplicate experiments. (C) Knockdown of MYBBP1A reduces the elevation of p53-target gene products induced by siTIF-IA treatment. MCF-7 cells were treated with siCont or siMYBBP for 48 h before treatment with siTIF-IA for the indicated times. The cell lysates were prepared and analysed by immunoblot using the indicated antibodies. (D) Knockdown of MYBBP1A reduces the elevation of p21 protein levels induced by low-dose ActD treatment. MCF-7 cells were treated with siCont or siMYBBP for 48 h before treatment with 5 nM ActD for the indicated times. The cell lysates were prepared and analysed by immunoblot using the indicated antibodies. (E) The activation of p53 by MYBBP1A was mediated by acetylation of p53. H1299 cells were transfected with combination of the expression vectors for MYBBP1A, p53, p53-KR(300), and/or HA-p300, as indicated. Twenty-four hours after transfection, the cell lysates were prepared and analysed by immunoblot using the indicated antibodies. (F) Knockdown of MYBBP1A decreases the level of apoptosis induced by TIF-IA depletion. (Left) The phase-contrast images of MCF-7 cells were treated with the indicated siRNAs and cultured for 72 h. Representative images are shown. (Right) Percentage of dead cells. MCF-7 cells were transfected with the indicated siRNAs for the indicated times, and the percentage of dead cells was measured by trypan blue exclusion assay. Values are given as the mean±s.d. for triplicate experiments.
The effects of MYBBP1A on the induction levels of p53-target gene products were examined. Consistent with the results obtained from ChIP analysis, depletion of TIF-IA increased the mRNA and protein levels of p21, HDM2, and PUMA in MCF-7 and LNCaP cells (Figure 3B and C; Supplementary Figure S3E). The elevation of these mRNA and protein levels was impaired in cells in which MYBBP1A was knocked down by siRNA (Figure 3B and C; Supplementary Figure S3E). Low-dose ActD treatment also increased p21 protein levels (Figure 3D), which were also reduced by MYBBP1A siRNA treatment (Figure 3D). Similar results were observed for ADR and high-dose ActD treatments (Supplementary Figure S6B).
To confirm that the activation of p53 by MYBBP1A was mediated by p53 acetylation, we constructed a p53 acetylation-deficient mutant, p53-KR(p300). This mutant contained eight amino-acid substitutions (K164R, K305R, K370R, K372R, K373R, K381R, K382R, and K386R) at the lysine residues (Supplementary Figure S7A), which were not acetylated by p300. Introduction of these substitutions did not reduce the interaction between MYBBP1A and p53 (Supplementary Figure S7B). To test the effects of MYBBP1A expression on p53 acetylation and p53-target gene expression, an expression plasmid containing wild-type p53 or p53-KR(p300) was introduced into the H1299 cells (Figure 3E). Exogenously expressed wild-type p53 was acetylated, and protein levels of p21 and PUMA were found to increase (Figure 3E). Expression of MYBBP1A with wild-type p53 enhanced p53 acetylation and further increased the protein levels of p21 and PUMA (Figure 3E). In contrast, expression of MYBBP1A with p53-KR(p300) neither changed the acetylation status of p53-KR(p300) nor potentiated the elevation of p21 and PUMA (Figure 3E). These results support our model that MYBBP1A enhanced transcriptional activity of p53 by increasing p53 acetylation by p300.
We next investigated the effects of MYBBP1A knockdown on nucleolar disruption-induced apoptosis. In agreement with previous results, treatment of MCF-7 or LNCaP cells with siRNA for TIF-IA increased the number of dead cells (Figure 3F; Supplementary Figure S3F) and cell death caused by TIF-IA knockdown was dependent on the presence of p53 (Supplementary Figure S8). Cell death induced by TIF-IA siRNA was significantly suppressed by knockdown of MYBBP1A (Figure 3F; Supplementary Figure S3F). The contribution of MYBBP1A to apoptosis induced by ADR or ActD treatment was evaluated. These stresses increased the number of dead cells (Supplementary Figure S6C). Depletion of MYBBP1A decreased the number of dead cells (Supplementary Figure S6C), indicating that MYBBP1A was also involved in cell death induced by ADR and ActD treatment. Consistent with the results shown in Figure 1 and Supplementary Figure S5, MYBBP1A depletion had a greater affect on p53-mediated responses in cells treated with TIF-IA siRNA than in those treated with DNA-damaging agents.
MYBBP1A is anchored in the nucleolus through binding to nucleolar RNA
To further investigate the function of MYBBP1A, a protein complex containing MYBBP1A was purified. We generated MCF-7 cells that stably expressed FLAG-tagged MYBBP1A. Nucleolar extract fractions were then prepared from these cells, and the protein complexes, including FLAG-MYBBP1A, were precipitated using anti-FLAG antibody-conjugated M2-agarose beads. The MYBBP1A-interacting proteins were identified by mass spectrometry as NOL1, DHX15, p68, YB-1, NPM, and EBP2 (Figure 4A).
Figure 4.
Nucleolar localization of MYBBP1A depends on nucleolar RNA. (A) Purification and identification of MYBBP1A-associated proteins. Nucleolar extracts prepared from MCF-7 cells (Control) or those stably expressing FLAG-tagged MYBBP1A (FLAG-MYBBP1A) were incubated with anti-FLAG antibody-conjugated agarose beads, and the bound proteins were eluted by FLAG peptides. Using mass spectrometry, MYBBP1A-interacting proteins were identified. NPM and EBP2 were identified in the same protein band. *Proteins that showed nonspecific binding. (B) MYBBP1A was translocated from the nucleolus to the nucleoplasm by RNase treatment. (Upper) Permeabilized MCF-7 cells were incubated in the absence (−) or presence (+) of RNase. They were subsequently fixed and stained with the indicated antibodies or DAPI. (Lower) The percentage of cells that showed translocation of UBF, MYBBP1A, or NPM from the nucleolus. Values are given as the mean±s.d. for triplicate experiments.
Some of these proteins are known to be involved in rRNA processing and are capable of RNA binding (Gustafson et al, 1998; Gonda et al, 2006; Grisendi et al, 2006; Fukuda et al, 2007; Hirano et al, 2009), whereas MYBBP1A does not contain obvious candidate RNA-binding domains. These results, together with the observation that suppression of rRNA gene transcription induced translocation of MYBBP1A from the nucleolus, suggested that MYBBP1A may indirectly bind to RNA as part of an RNA-binding complex and may be anchored to the nucleolus through RNA. To test this possibility, MCF-7 cells were permiabilized and incubated with RNase. As shown in Figure 4B, RNase treatment did not affect the localization of UBF, which directly bound to the rDNA promoter region; however, the treatment caused translocation of MYBBP1A and NPM from the nucleolus to the nucleoplasm (Figure 4B). These results indicated that RNA acted as a scaffold for MYBBP1A and NPM. Taken together, our results suggested that reduction of rRNA production through inhibition of rRNA gene transcription decreased the MYBBP1A scaffold in the nucleolus and caused its translocation from the nucleolus to the nucleoplasm.
RPL5 and RPL11 are necessary for MYBBP1A transport from the nucleolus to the nucleoplasm
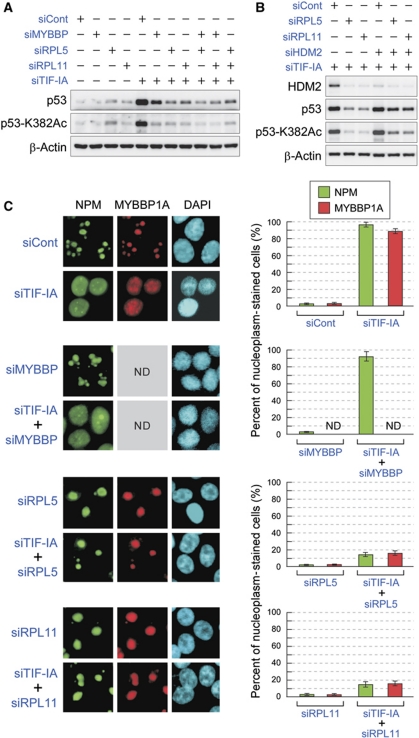
Using an siRNA library against nucleolar proteins, we found that three gene products, MYBBP1A, RPL5, and RPL11, were involved in p53 accumulation and acetylation (Supplementary Figure S4). Depletion of MYBBP1A, RPL5, or RPL11 reduced both the protein levels and the acetylation status of p53, which had been elevated by TIF-IA knockdown (Figure 5A). Reductions in the protein levels and acetylation status of p53 were more effective by RPL5 or RPL11 knockdown than by MYBBP1A knockdown (Figure 5A). To investigate the relationships among MYBBP1A, RPL5, and RPL11, MYBBP1A was knocked down together with either RPL5 or RPL11. Treatment of cells with siRNAs for MYBBP1A and RPL5 or for MYBBP1A and RPL11 did not result in additive effects on the protein or acetylation levels of p53, which had been reduced by RPL5 or RPL11 siRNA (Figure 5A). These results suggested that RPL5 and RPL11 are positioned upstream of MYBBP1A in the pathway that regulates p53 accumulation and acetylation.
Figure 5.
RPL5 and RPL11 are required for MYBBP1A translocation from the nucleolus. (A) The protein and acetylation levels of p53 were decreased by siRNA for MYBBP1A, RPL5, or RPL11. MCF-7 cells were treated with a combination of the indicated siRNAs for 48 h, and the cell lysates were analysed by immunoblot with the indicated antibodies. (B) Knockdown of HDM2 partially recovered the reductions in p53 accumulation and acetylation caused by RPL5 or RPL11 knockdown. MCF-7 cells were treated with a combination of the indicated siRNAs for 48 h, and the cell lysates were analysed by immunoblot with the indicated antibodies. (C) MYBBP1A translocation induced by TIF-IA knockdown was hampered by the knockdown of RPL5 or RPL11. (Left) MCF-7 cells were treated with the indicated siRNAs for 48 h, and the cells were stained with the indicated antibodies or DAPI. (Right) The percentage of cells that showed translocation of NPM or MYBBP1A from the nucleolus after treatment with the indicated siRNAs. Values are given as the mean±s.d. for triplicate experiments. ND, not determined.
It is well known that RPL5 and RPL11 stabilize p53 by binding to HDM2 and blocking its function. Knockdown of HDM2 partially recovered the reductions in p53 protein and acetylation levels caused by RPL5 or RPL11 knockdown (Figure 5B, compare lanes 2 and 3, respectively, with lanes 5 and 6). These results confirmed the involvement of HDM2 in p53 regulation by RPLs. However, the partial HDM2-mediated recovery of reductions in p53 accumulation and acetylation induced by RPL5 or RPL11 knockdown suggested the presence of an HDM2-independent pathway for p53 activation. Therefore, we examined the effects of RPL5 or RPL11 siRNAs on MYBBP1A localization. Immunostaining showed that TIF-IA depletion caused translocation of both MYBBP1A and NPM from the nucleolus to the nucleoplasm in MCF-7 and LNCaP cells (Figure 5C; Supplementary Figure S9). Surprisingly, while MYBBP1A depletion did not affect the localization of NPM, knockdown of RPL5 or RPL11 significantly inhibited MYBBP1A and NPM translocation from the nucleolus to the nucleoplasm, which had been induced by TIF-IA knockdown (Figure 5C; Supplementary Figure S9). Depletion of RPL5 or RPL11 also abrogated the ADR and ActD-induced translocation of MYBBP1A into the nucleoplasm (Supplementary Figure S10). In contrast, knockdown of RPL23 or RPL26 could not inhibit the translocation of MYBBP1A and NPM induced by nucleolar disruption (Supplementary Figure S11).
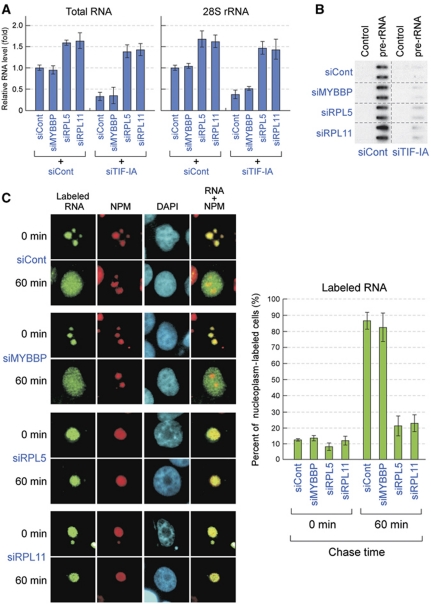
These observations, together with the results showing that MYBBP1A and NPM were anchored to the nucleolus through RNA, suggested that the nucleolar RNA content could be retained by RPL5 or RPL11 depletion, even when rRNA transcription was abrogated by TIF-IA depletion. Thus, we purified the nucleolus from cells treated with a combination of the siRNAs indicated in Figure 6A and determined the nucleolar RNA content. As shown in Figure 6A, TIF-IA depletion decreased the content of both the total and 28S rRNA in the nucleolus. MYBBP1A knockdown did not affect the reduction of nucleolar RNA content induced by TIF-IA depletion (Figure 6A). In agreement with our hypothesis, knockdown of RPL5 or RPL11 increased the RNA content in the nucleolus and maintained high RNA levels, even when the cells were treated with TIF-IA siRNA (Figure 6A). We also found that the nucleoli in RPL5 or RPL11 siRNA-treated cells appeared to be larger than those in the control siRNA-treated cells. This result may reflect an increase in nucleolar RNA content in RPL5 or RPL11-depleted cells (Figure 6A). The same result was obtained when cells were treated with ADR (Supplementary Figure S12).
Figure 6.
Knockdown of RPL5 or RPL11 retains nucleolar RNA by preventing its export. (A) Knockdown of RPL5 or RPL11 retained RNA content in the nucleolus. The total RNA was isolated from the isolated nucleoli of MCF-7 cells transfected with the indicated siRNAs for 48 h. The total RNA (left) and 28S rRNA (right) levels were quantified by spectrophotometry and RT–qPCR, respectively. The level of the siCont-treated cells was normalized to 1.0. (B) Downregulation of MYBBP1A, RPL5, or RPL11 did not affect the transcription levels of pre-rRNA that had been reduced by TIF-IA knockdown. Nuclear run-on assays were performed to measure the transcription levels of pre-rRNA in MCF-7 cells transfected with the indicated siRNAs for 48 h. Transcription from the rRNA gene was measured by hybridization of in vitro-synthesized 32P-labelled run-on transcripts to immobilized plasmids containing no insert (Control) or cDNA corresponding to 28S rRNA (pre-rRNA). The assays were performed in duplicate. (C) Knockdown of RPL5 or RPL11 prevents nucleolar RNA export from the nucleolus. (Left) MCF-7 cells were transfected with the indicated siRNAs for 48 h. The RNA polymerase II was inhibited by α-amanitin and the newly synthesized rRNA in the cells was pulse labelled with BrUTP. These cells were chased in the BrUTP-free medium for 0 or 60 min, fixed, and stained with anti-BrdU (green), anti-NPM (red) antibodies, or DAPI (blue). (Right) The percentage of cells that showed BrUTP staining in the nucleoplasm at 0 or 60 min after chase. Values are given as the mean±s.d. for triplicate experiments.
Because knockdown of RPL5 or RPL11 abrogates the reduction of nucleolar RNA content induced by TIF-IA depletion, we first hypothesized that knockdown of RPL5 or RPL11 counteracted the suppression of rRNA transcription caused by TIF-IA depletion. To test this, we performed nuclear run-on assays (Figure 6B). The results showed that the treatment of cells with TIF-IA siRNA significantly suppressed the transcription of rRNA genes, and this suppression was not restored by knockdown of RPL5 or RPL11 (Figure 6B). These results indicated that retention of nucleolar RNA content by RPL5 or RPL11 depletion was not due to the recovery of rRNA transcription.
Recently, it was reported that rpL5 and rpL11, orthologs of mammalian RPL5 and RPL11, were required for export of pre-ribosomes with rRNA in Saccharomyces cerevisiae (Zhang et al, 2007). Therefore, we next examined the effects of RPL5 or RPL11 knockdown on rRNA transport. We labelled newly synthesized rRNA in MCF-7 cells with BrUTP in the presence of α-amanitin, which specifically inhibits RNA polymerase II. These cells were then cultured in a BrUTP-free chase medium. The localization of labelled RNA was determined by immunostaining using anti-BrdU antibodies at the time points indicated in Figure 6C. At the beginning of the chase (0 min), the labelled RNA was located in the nucleolus in each siRNA-treated cell. After 60 min incubation, the labelled RNA was exported into the nucleoplasm in the control or MYBBP1A siRNA-treated cells; however, the labelled RNA was retained in the nucleolus in RPL5 or RPL11-depleted cells (Figure 6C). These results indicated that RPL5 and RPL11 were necessary for RNA transport from the nucleolus in mammalian cells. Knockdown of RPL5 or RPL11 inhibited rRNA transport from the nucleolus and counteracted the reduction of RNA levels in the nucleolus that had been induced by inhibition of rRNA transcription (Figure 6C). As a result, RPL5 or RPL11 knockdown inhibited MYBBP1A translocation.
Our observations indicated that the nucleolar RNA content was maintained by a dynamic equilibrium between RNA generation and export. The loss of this balance due to stress altered the nucleolar RNA content and modulated p53 activity.
Discussion
DNA damage has been shown to induce phosphorylation at multiple serine residues, including Ser15, in p53 (Siliciano et al, 1997; Ashcroft et al, 1999, 2000). Phosphorylation of p53 was shown to enhance its ability to interact with p300/CBP, thereby enhancing p53 acetylation (Lambert et al, 1998; Dumaz and Meek, 1999). As loss of acetylation completely abolishes p53-dependent growth arrest and apoptosis, acetylation of p53 is an indispensable event for p53 activation (Tang et al, 2008).
Grummt's group showed that nucleolar disruption was sufficient for induction of p53-dependent apoptosis without DNA damage (Yuan et al, 2005). The nucleolar disruption did not enhance p53 phosphorylation, but instead induced its acetylation. Our observations indicated that disruption of the nucleolus led to translocation of the nucleolar protein MYBBP1A into the nucleoplasm. MYBBP1A then bound to the C-terminus of p53 and stabilized binding between p53 and p300. As a result, MYBBP1A enhanced acetylation of p53 and increased its transcriptional activity. Several nucleolar proteins, including RPL5, RPL11, RPL23, NPM, NCL, NS, and ARF, are known to be involved in the stabilization of p53 proteins. Ribosomal proteins RPL5, RPL11, and RPL23, which are usually assembled into the 60S ribosome subunit and exported to the rough endoplasmic reticulum for protein synthesis, have been identified as proteins that directly bind to HDM2 and inhibit HDM2-mediated p53 ubiquitination (Marechal et al, 1994; Lohrum et al, 2003; Zhang et al, 2003; Dai and Lu, 2004; Dai et al, 2004; Jin et al, 2004). Likewise, the nucleolar proteins NPM, NCL, NS, and ARF also directly bind to HDM2 and inhibit p53 ubiquitination (Kamijo et al, 1998; Pomerantz et al, 1998; Kurki et al, 2004; Saxena et al, 2006; Dai et al, 2008). Thus, the mode of action of MYBBP1A was found to be different from those of RPLs, NPM, NCL, NS, or ARF. In contrast to these nucleolar proteins, MYBBP1A directly binds to the C-terminus of p53 and enhances its acetylation. In a mouse model, the C-terminal lysine residues of p53 are not required for either stability or transactivation (Krummel et al, 2005). On the other hand, many biochemical and human cell line studies suggest that p53 acetylation at the C-terminus stabilizes p53 and activates its transcriptional activity. MYBBP1A is also regulated differently in mice and humans. Mouse Mybbp1a was processed to a short form by ActD treatment (Yamauchi et al, 2008), although human MYBBP1A was not (data not shown). Therefore, the mechanisms of p53 activation by MYBBP1A may differ between species.
Our results indicated that MYBBP1A binds to the C-terminus of p53. An accumulation of evidence supports the suggestion that the core domain and C terminus may be the docking sites for several p53 coactivators, such as ASPPs, hCAS/CSE1L, HZF, MUC1, and YB-1, which are all critically involved in the induction of different targets by p53 (Takenaka et al, 1995; Samuels-Lev et al, 2001; Homer et al, 2005; Wei et al, 2005; Trigiante and Lu, 2006; Das et al, 2007; Tanaka et al, 2007; Kruse and Gu, 2009; Tian et al, 2009; Vousden and Prives, 2009). Considering that MYBBP1A complex contains YB-1, MYBBP1A may also be involved in the selective recruitment of these coactivators to the promoter regions of p53-target genes. Some studies suggest that MYBBP1A has a role in RNA polymerase II-dependent transcription regulation. It has been shown to bind to the transcription factors c-Myb, c-Jun, aryl hydrocarbon receptor, PPARγ coactivator 1α (PGC-1α), the RelA/p65 subunit of NF-κB, Prep1, and mCRY1 (Favier and Gonda, 1994; Tavner et al, 1998; Jones et al, 2002; Fan et al, 2004; Diaz et al, 2007; Owen et al, 2007; Hara et al, 2009), although the influence of MYBBP1A on transcription was variable. Considering these reports together with our results, MYBBP1A may provide a physical signalling link between RNA polymerase II-dependent transcription and nucleolar function.
It is thought that RNA polymerase I-driven transcription is responsible for the maintenance of the steady-state nucleolar structure (Schwarzacher and Wachtler, 1993; Scheer and Hock, 1999). Indeed, transcriptional inhibition of RNA polymerase I by TIF-IA depletion or DNA-damaging agents led to dissociation of several nucleolar proteins, including a nucleolar marker NPM and MYBBP1A from the nucleolus (Figure 5C; Supplementary Figure S10) (Rubbi and Milner, 2003; Mayer et al, 2005; Yuan et al, 2005; Yamauchi et al, 2008). In our experiment, we found that MYBBP1A associated with several rRNA processing proteins that possessed RNA-binding abilities. RNase treatment of the cells caused dissociation of MYBBP1A as well as NPM from the nucleolus. Knockdown of RPL5 or RPL11 retained the RNA content in the nucleolus and inhibited the translocation of MYBBP1A and NPM from the nucleolus to the nucleoplasm, even when rRNA transcription was stopped by TIF-IA depletion. These observations imply that the presence of nucleolar RNA, not the RNA polymerase I-driven transcription, was responsible for maintenance of the nucleolar structure.
A previous report showed that in Xenopus laevis, the recruitment of rRNA processing proteins to the nucleolus was dependent on the presence of pre-rRNAs but not on RNA polymerase I-dependent transcription (Verheggen et al, 2000) In human cell lines, it was demonstrated that pre-rRNA was predominantly synthesized near the G2/M transition. The synthesized pre-rRNA persisted throughout mitosis and participated in the assembly of the daughter cell nucleolus by recruiting rRNA processing complexes. This recruitment was independent of RNA polymerase I-driven transcription (Dousset et al, 2000). Gonda et al (2003, 2006) showed that nucleolar disruption was induced by the specific intrinsic proteins FRGY2a and FRGY2b, both of which contained RNP1-like and RNP2-like RNA-binding motifs, under conditions in which RNA polymerase I transcription in X. laevis was ongoing. These findings were consistent with our model that nucleolar RNA content, rather than RNA polymerase I-dependent transcription, was important for maintaining nucleolar structure.
The siRNA library screen showed that depletion of RPL5 or RPL11 significantly abrogated the accumulation and acetylation of p53. Vousden’s and other groups have demonstrated that RPL5 and RPL11 bind to the central acidic regions of HDM2, and inhibit the HDM2 E3 ligase activity towards p53 protein (Lohrum et al, 2003; Zhang et al, 2003; Dai and Lu, 2004; Lindstrom et al, 2007; Horn and Vousden, 2008). Depletion of either RPL5 or RPL11 significantly impeded the stabilization of p53 in response to ribosomal stress, and enforced expression of RPL5 or RPL11 resulted in p53-dependent cell-cycle arrest (Lohrum et al, 2003; Zhang et al, 2003; Bhat et al, 2004; Dai and Lu, 2004). In agreement with these reports, we also found that depletion of RPL5 or RPL11 was more effective at inhibiting p53 accumulation than was depletion of MYBBP1A. In addition to these functions, we found that RPL5 and RPL11 were necessary for RNA export from the nucleolus to the nucleoplasm. Our results demonstrated that knockdown of RPL5 or RPL11 downregulated rRNA transport and retained RNA in the nucleolus after suppression of rRNA transcription. As a result, RPL5 or RPL11 knockdown inhibited MYBBP1A translocation. RPL5 and RPL11 are reportedly components of the 5S rRNA–protein complex (Spierer and Zimmermann, 1978; Steitz et al, 1988; Nariai et al, 2005; Zhang et al, 2007; Horn and Vousden, 2008). Incorporation of this complex into pre-ribosomes is an important step in the assembly of 60S ribosomal subunits (Van Ryk et al, 1992; Dechampesme et al, 1999). RPL5 and RPL11 are essential for the incorporation step of the 5S rRNA complex into pre-ribosomes (Zhang et al, 2007). It is predicted from our results that incorporation of 5S rRNA into pre-ribosomes may be an essential step for rRNA transport from the nucleolus.
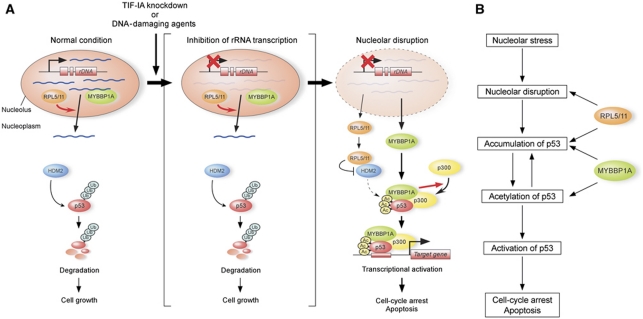
Our observations indicated that nucleolar RNA content is maintained by its generation and export. The loss of balance between RNA generation and export due to stress alters nucleolar RNA content and modulates p53 activity. When the nucleolar RNA content was reduced, MYBBP1A and RPLs were released from the nucleolus, an event that subsequently enhanced p53 activity (Figure 7).
Figure 7.
Proposed model for the role of RNA content on p53 acetylation by MYBBP1A following nucleolar stress. (A) Model for the role of RNA content in the nucleolus on p53 regulation. See the text. (B) Role of RPL5, RPL11, and MYBBP1A on p53 regulation. See the text.
Materials and methods
Cell culture and treatments
MCF-7 human breast cancer cells, LNCaP human prostate cancer cells, H1299 p53-deficient human lung cancer cells were maintained in DMEM (Sigma). All media were supplemented with 10% fetal bovine serum and penicillin–streptomycin mixed solution (Nacalai tesque). Cells were maintained at 37°C in an atmosphere containing 5% CO2 and 100% humidity. DNA damage was induced by ADR (0.5 μg/ml), UV irradiation (25 J/m2), or ActD (40 nM).
siRNA and plasmid DNA transfection
For transfection of siRNAs, cells at 30–50% confluency were transfected with 20 nM of siRNA using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s protocol. Protein and RNA were extracted at 48 h after transfection of siRNA. For transfection of plasmid DNA, cells at 70–80% confluency were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Protein was extracted at 24 h after transfection of plasmid DNA.
Immunoprecipitation
To monitor the interaction of MYBBP1A, p300, PCAF, Tip60, and p53 in vivo, H1299 cells were transfected with expression vectors encoding the respective proteins (pcDNA3-HA-MYBBP1A, pcDNA3-FLAG-p300, pcDNA3-FLAG-PCAF, pcDNA3-FLAG-Tip60, and pcDNA3-Myc-p53). Twenty-four hours after transfection, cells were lysed in TNE buffer (20 mM Tris–HCl at pH 7.5, 150 mM NaCl, 2 mM EDTA, and 1% NP-40) at 4°C for 30 min. The cleared lysates were incubated for 2 h with 10 μl of anti-FLAG M2-agarose beads. After washing four times with the same buffer, FLAG-tagged proteins were eluted with 0.25 mg/ml of FLAG peptide in TNE buffer at 4°C for 30 min, separated on 7.5% SDS–polyacrylamide gels, and analysed by immunoblotting with indicated antibodies. For coimmunoprecipitation of the endogenous proteins, MCF-7 cells were lysed in TNE buffer at 4°C for 30 min. The cleared lysate was incubated for 4 h with 2 μg of antibodies against the indicated proteins, 10 μl of protein G sepharose was added, and the sample was rotated for 1 h at 4°C. After washing four times with the same buffer, immunoprecipitates were separated on 7.5% SDS–polyacrylamide gels, and analysed by immunoblotting with the indicated antibodies. For detection of a ternary complex consisting of MYBBP1A, p53, and p300, MCF-7 cells were treated with TIF-IA siRNA for 48 h. The cell lysates were first immunoprecipitated with anti-MYBBP1A antibody and eluted from the beads using the peptide with raised anti-MYBBP1A antibody. The eluted sample was sequentially immunoprecipitated using anti-p300 antibody, and immunoprecipitates were detected by immunoblotting using antibodies against MYBBP1A, p53, and p300.
Chromatin immunoprecipitation
ChIP assays were performed as described (Saavalainen et al, 2005; Sinkkonen et al, 2005; Saramaki et al, 2006) with a minor modification. Anti-p53 (FL393) or anti-p300 (N-15) antibody (Santa Cruz Biotechnology) was used for immunoprecipitation. Real-time PCR was performed using the Thermal Cycler Dice™ TP800 (Takara) and SYBER Premix Ex Taq (Takara). The primers for real-time PCR were as follows: 5′-GTGGCTCTGATTGGCTTTCTG-3′ and 5′-CTGAAAACAGGCAGCCCAAG-3′ for p21-p53RE.
RNase treatment
Cells grown on chamber slides were permeabilized with 0.1% Triton X-100 in PBS, washed with PBS, and treated for 10 min with RNase A (1 mg/ml). After washing, cells were fixed 3.7% formaldehyde in PBS for 10 min and immunofluorescence analysis was performed.
Nucleoli purification and quantitative determination of RNA content
Nucleoli were isolated from 1.2 × 107 MCF-7 cells in high purity by density gradient fractionation as previously described (Andersen et al, 2005). Total RNA was prepared from the isolated nucleoli and quantified by spectrophotometry. 28S rRNA levels were determined by RT–qPCR and normalized to DNA content, which was determined by qPCR for p21 promoter region (p21-p53RE) using the nuclear fraction.
Pulse-chase analysis
Pulse-chase analysis was performed as previously described (Thiry et al, 2008). Briefly, cells were grown on chamber slides and pretreated with 20 μg/ml α-amanitin for 2 h 30 min. The BrUTP–FuGENE HD complexes were prepared by mixing 5 mM BrUTP (Sigma) and FuGENE HD (Roche) and incubated for 15 min. Then, the complexes were added to the cells for 15 min at 37°C. After this pulse, the complexes were removed and the cells were either directly fixed or incubated in the BrUTP-free medium containing 10 μg/ml α-amanitin for 60 min at 37°C. The labelled RNA was analysed by immunofluorescence using anti-BrdU antibody.
Supplementary Material
Acknowledgments
This work was supported by grants from Precursory Research for Embryonic Science and Technology (PRESTO), Japan Science and Technology Agency (JST), and from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Author contribution: TK performed most of the experiments; AM performed ChIP and some IP experiments; NK performed some immunostain experiments; YO and EF performed some experiments; HM, ME, ST, and KK discussed the data; JY planned the project and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M (2005) Nucleolar proteome dynamics. Nature 433: 77–83 [DOI] [PubMed] [Google Scholar]
- Appella E, Anderson CW (2001) Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem 268: 2764–2772 [DOI] [PubMed] [Google Scholar]
- Arva NC, Gopen TR, Talbott KE, Campbell LE, Chicas A, White DE, Bond GL, Levine AJ, Bargonetti J (2005) A chromatin-associated and transcriptionally inactive p53-Mdm2 complex occurs in mdm2 SNP309 homozygous cells. J Biol Chem 280: 26776–26787 [DOI] [PubMed] [Google Scholar]
- Ashcroft M, Kubbutat MH, Vousden KH (1999) Regulation of p53 function and stability by phosphorylation. Mol Cell Biol 19: 1751–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashcroft M, Taya Y, Vousden KH (2000) Stress signals utilize multiple pathways to stabilize p53. Mol Cell Biol 20: 3224–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD, Berger SL (2001) Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol Cell 8: 1243–1254 [DOI] [PubMed] [Google Scholar]
- Bhat KP, Itahana K, Jin A, Zhang Y (2004) Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J 23: 2402–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner C, Tobiasch E, Litfen M, Rahmsdorf HJ, Herrlich P (1999) DNA damage induced p53 stabilization: no indication for an involvement of p53 phosphorylation. Oncogene 18: 1723–1732 [DOI] [PubMed] [Google Scholar]
- Bode AM, Dong Z (2004) Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer 4: 793–805 [DOI] [PubMed] [Google Scholar]
- Brooks CL, Gu W (2003) Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr Opin Cell Biol 15: 164–171 [DOI] [PubMed] [Google Scholar]
- Brooks CL, Gu W (2006) p53 ubiquitination: Mdm2 and beyond. Mol Cell 21: 307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter S, Vousden KH (2009) Modifications of p53: competing for the lysines. Curr Opin Genet Dev 19: 18–24 [DOI] [PubMed] [Google Scholar]
- Chehab NH, Malikzay A, Stavridi ES, Halazonetis TD (1999) Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage. Proc Natl Acad Sci USA 96: 13777–13782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Zhang Z, Li M, Wang W, Li Y, Rayburn ER, Hill DL, Wang H, Zhang R (2007) Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene 26: 5029–5037 [DOI] [PubMed] [Google Scholar]
- Dai MS, Lu H (2004) Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem 279: 44475–44482 [DOI] [PubMed] [Google Scholar]
- Dai MS, Sun XX, Lu H (2008) Aberrant expression of nucleostemin activates p53 and induces cell cycle arrest via inhibition of MDM2. Mol Cell Biol 28: 4365–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H (2004) Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol 24: 7654–7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Raj L, Zhao B, Kimura Y, Bernstein A, Aaronson SA, Lee SW (2007) Hzf determines cell survival upon genotoxic stress by modulating p53 transactivation. Cell 130: 624–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechampesme AM, Koroleva O, Leger-Silvestre I, Gas N, Camier S (1999) Assembly of 5S ribosomal RNA is required at a specific step of the pre-rRNA processing pathway. J Cell Biol 145: 1369–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz VM, Mori S, Longobardi E, Menendez G, Ferrai C, Keough RA, Bachi A, Blasi F (2007) p160 Myb-binding protein interacts with Prep1 and inhibits its transcriptional activity. Mol Cell Biol 27: 7981–7990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dousset T, Wang C, Verheggen C, Chen D, Hernandez-Verdun D, Huang S (2000) Initiation of nucleolar assembly is independent of RNA polymerase I transcription. Mol Biol Cell 11: 2705–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumaz N, Meek DW (1999) Serine15 phosphorylation stimulates p53 transactivation but does not directly influence interaction with HDM2. EMBO J 18: 7002–7010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M, Rhee J, St-Pierre J, Handschin C, Puigserver P, Lin J, Jaeger S, Erdjument-Bromage H, Tempst P, Spiegelman BM (2004) Suppression of mitochondrial respiration through recruitment of p160 myb binding protein to PGC-1alpha: modulation by p38 MAPK. Genes Dev 18: 278–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favier D, Gonda TJ (1994) Detection of proteins that bind to the leucine zipper motif of c-Myb. Oncogene 9: 305–311 [PubMed] [Google Scholar]
- Fukuda T, Yamagata K, Fujiyama S, Matsumoto T, Koshida I, Yoshimura K, Mihara M, Naitou M, Endoh H, Nakamura T, Akimoto C, Yamamoto Y, Katagiri T, Foulds C, Takezawa S, Kitagawa H, Takeyama K, O’Malley BW, Kato S (2007) DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol 9: 604–611 [DOI] [PubMed] [Google Scholar]
- Gonda K, Fowler J, Katoku-Kikyo N, Haroldson J, Wudel J, Kikyo N (2003) Reversible disassembly of somatic nucleoli by the germ cell proteins FRGY2a and FRGY2b. Nat Cell Biol 5: 205–210 [DOI] [PubMed] [Google Scholar]
- Gonda K, Wudel J, Nelson D, Katoku-Kikyo N, Reed P, Tamada H, Kikyo N (2006) Requirement of the protein B23 for nucleolar disassembly induced by the FRGY2a family proteins. J Biol Chem 281: 8153–8160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisendi S, Mecucci C, Falini B, Pandolfi PP (2006) Nucleophosmin and cancer. Nat Rev Cancer 6: 493–505 [DOI] [PubMed] [Google Scholar]
- Gu W, Roeder RG (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90: 595–606 [DOI] [PubMed] [Google Scholar]
- Gustafson WC, Taylor CW, Valdez BC, Henning D, Phippard A, Ren Y, Busch H, Durban E (1998) Nucleolar protein p120 contains an arginine-rich domain that binds to ribosomal RNA. Biochem J 331 (Pt 2): 387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara Y, Onishi Y, Oishi K, Miyazaki K, Fukamizu A, Ishida N (2009) Molecular characterization of Mybbp1a as a co-repressor on the Period2 promoter. Nucleic Acids Res 37: 1115–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano Y, Ishii K, Kumeta M, Furukawa K, Takeyasu K, Horigome T (2009) Proteomic and targeted analytical identification of BXDC1 and EBNA1BP2 as dynamic scaffold proteins in the nucleolus. Genes Cells 14: 155–166 [DOI] [PubMed] [Google Scholar]
- Homer C, Knight DA, Hananeia L, Sheard P, Risk J, Lasham A, Royds JA, Braithwaite AW (2005) Y-box factor YB1 controls p53 apoptotic function. Oncogene 24: 8314–8325 [DOI] [PubMed] [Google Scholar]
- Horn HF, Vousden KH (2008) Cooperation between the ribosomal proteins L5 and L11 in the p53 pathway. Oncogene 27: 5774–5784 [DOI] [PubMed] [Google Scholar]
- Ito A, Kawaguchi Y, Lai CH, Kovacs JJ, Higashimoto Y, Appella E, Yao TP (2002) MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J 21: 6236–6245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito A, Lai CH, Zhao X, Saito S, Hamilton MH, Appella E, Yao TP (2001) p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J 20: 1331–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin A, Itahana K, O’Keefe K, Zhang Y (2004) Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol Cell Biol 24: 7669–7680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LC, Okino ST, Gonda TJ, Whitlock JP Jr (2002) Myb-binding protein 1a augments AhR-dependent gene expression. J Biol Chem 277: 22515–22519 [DOI] [PubMed] [Google Scholar]
- Kamijo T, Weber JD, Zambetti G, Zindy F, Roussel MF, Sherr CJ (1998) Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci USA 95: 8292–8297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JE, Chen J, Lou Z (2008) DBC1 is a negative regulator of SIRT1. Nature 451: 583–586 [DOI] [PubMed] [Google Scholar]
- Knights CD, Catania J, Di Giovanni S, Muratoglu S, Perez R, Swartzbeck A, Quong AA, Zhang X, Beerman T, Pestell RG, Avantaggiati ML (2006) Distinct p53 acetylation cassettes differentially influence gene-expression patterns and cell fate. J Cell Biol 173: 533–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krummel KA, Lee CJ, Toledo F, Wahl GM (2005) The C-terminal lysines fine-tune P53 stress responses in a mouse model but are not required for stability control or transactivation. Proc Natl Acad Sci USA 102: 10188–10193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse JP, Gu W (2009) Modes of p53 regulation. Cell 137: 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurki S, Peltonen K, Latonen L, Kiviharju TM, Ojala PM, Meek D, Laiho M (2004) Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell 5: 465–475 [DOI] [PubMed] [Google Scholar]
- Lambert PF, Kashanchi F, Radonovich MF, Shiekhattar R, Brady JN (1998) Phosphorylation of p53 serine 15 increases interaction with CBP. J Biol Chem 273: 33048–33053 [DOI] [PubMed] [Google Scholar]
- Levine AJ (1997) p53, the cellular gatekeeper for growth and division. Cell 88: 323–331 [DOI] [PubMed] [Google Scholar]
- Li AG, Piluso LG, Cai X, Gadd BJ, Ladurner AG, Liu X (2007) An acetylation switch in p53 mediates holo-TFIID recruitment. Mol Cell 28: 408–421 [DOI] [PubMed] [Google Scholar]
- Li M, Luo J, Brooks CL, Gu W (2002) Acetylation of p53 inhibits its ubiquitination by Mdm2. J Biol Chem 277: 50607–50611 [DOI] [PubMed] [Google Scholar]
- Lindstrom MS, Jin A, Deisenroth C, White Wolf G, Zhang Y (2007) Cancer-associated mutations in the MDM2 zinc finger domain disrupt ribosomal protein interaction and attenuate MDM2-induced p53 degradation. Mol Cell Biol 27: 1056–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH (2003) Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 3: 577–587 [DOI] [PubMed] [Google Scholar]
- Luo J, Li M, Tang Y, Laszkowska M, Roeder RG, Gu W (2004) Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc Natl Acad Sci USA 101: 2259–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W (2001) Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 107: 137–148 [DOI] [PubMed] [Google Scholar]
- Luo J, Su F, Chen D, Shiloh A, Gu W (2000) Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature 408: 377–381 [DOI] [PubMed] [Google Scholar]
- Marechal V, Elenbaas B, Piette J, Nicolas JC, Levine AJ (1994) The ribosomal L5 protein is associated with mdm-2 and mdm-2-p53 complexes. Mol Cell Biol 14: 7414–7420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Bierhoff H, Grummt I (2005) The nucleolus as a stress sensor: JNK2 inactivates the transcription factor TIF-IA and down-regulates rRNA synthesis. Genes Dev 19: 933–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Grummt I (2005) Cellular stress and nucleolar function. Cell Cycle 4: 1036–1038 [DOI] [PubMed] [Google Scholar]
- Michael D, Oren M (2003) The p53-Mdm2 module and the ubiquitin system. Semin Cancer Biol 13: 49–58 [DOI] [PubMed] [Google Scholar]
- Minsky N, Oren M (2004) The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol Cell 16: 631–639 [DOI] [PubMed] [Google Scholar]
- Nariai M, Tanaka T, Okada T, Shirai C, Horigome C, Mizuta K (2005) Synergistic defect in 60S ribosomal subunit assembly caused by a mutation of Rrs1p, a ribosomal protein L11-binding protein, and 3′-extension of 5S rRNA in Saccharomyces cerevisiae. Nucleic Acids Res 33: 4553–4562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkubo S, Tanaka T, Taya Y, Kitazato K, Prives C (2006) Excess HDM2 impacts cell cycle and apoptosis and has a selective effect on p53-dependent transcription. J Biol Chem 281: 16943–16950 [DOI] [PubMed] [Google Scholar]
- Olsson A, Manzl C, Strasser A, Villunger A (2007) How important are post-translational modifications in p53 for selectivity in target-gene transcription and tumour suppression? Cell Death Differ 14: 1561–1575 [DOI] [PubMed] [Google Scholar]
- Owen HR, Elser M, Cheung E, Gersbach M, Kraus WL, Hottiger MO (2007) MYBBP1a is a novel repressor of NF-kappaB. J Mol Biol 366: 725–736 [DOI] [PubMed] [Google Scholar]
- Pomerantz J, Schreiber-Agus N, Liegeois NJ, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee HW, Cordon-Cardo C, DePinho RA (1998) The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell 92: 713–723 [DOI] [PubMed] [Google Scholar]
- Prives C, Hall PA (1999) The p53 pathway. J Pathol 187: 112–126 [DOI] [PubMed] [Google Scholar]
- Rubbi CP, Milner J (2003) Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J 22: 6068–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavalainen K, Pasonen-Seppanen S, Dunlop TW, Tammi R, Tammi MI, Carlberg C (2005) The human hyaluronan synthase 2 gene is a primary retinoic acid and epidermal growth factor responding gene. J Biol Chem 280: 14636–14644 [DOI] [PubMed] [Google Scholar]
- Saito S, Yamaguchi H, Higashimoto Y, Chao C, Xu Y, Fornace AJ Jr, Appella E, Anderson CW (2003) Phosphorylation site interdependence of human p53 post-translational modifications in response to stress. J Biol Chem 278: 37536–37544 [DOI] [PubMed] [Google Scholar]
- Sakaguchi K, Herrera JE, Saito S, Miki T, Bustin M, Vassilev A, Anderson CW, Appella E (1998) DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev 12: 2831–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels-Lev Y, O’Connor DJ, Bergamaschi D, Trigiante G, Hsieh JK, Zhong S, Campargue I, Naumovski L, Crook T, Lu X (2001) ASPP proteins specifically stimulate the apoptotic function of p53. Mol Cell 8: 781–794 [DOI] [PubMed] [Google Scholar]
- Saramaki A, Banwell CM, Campbell MJ, Carlberg C (2006) Regulation of the human p21(waf1/cip1) gene promoter via multiple binding sites for p53 and the vitamin D3 receptor. Nucleic Acids Res 34: 543–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena A, Rorie CJ, Dimitrova D, Daniely Y, Borowiec JA (2006) Nucleolin inhibits Hdm2 by multiple pathways leading to p53 stabilization. Oncogene 25: 7274–7288 [DOI] [PubMed] [Google Scholar]
- Scheer U, Hock R (1999) Structure and function of the nucleolus. Curr Opin Cell Biol 11: 385–390 [DOI] [PubMed] [Google Scholar]
- Schwarzacher HG, Wachtler F (1993) The nucleolus. Anat Embryol (Berl) 188: 515–536 [DOI] [PubMed] [Google Scholar]
- Shieh SY, Ikeda M, Taya Y, Prives C (1997) DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 91: 325–334 [DOI] [PubMed] [Google Scholar]
- Siliciano JD, Canman CE, Taya Y, Sakaguchi K, Appella E, Kastan MB (1997) DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev 11: 3471–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkkonen L, Malinen M, Saavalainen K, Vaisanen S, Carlberg C (2005) Regulation of the human cyclin C gene via multiple vitamin D3-responsive regions in its promoter. Nucleic Acids Res 33: 2440–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spierer P, Zimmermann RA (1978) Stoichiometry, cooperativity, and stability of interactions between 5S RNA and proteins L5, L18, and L25 from the 50S ribosomal subunit of Escherichia coli. Biochemistry 17: 2474–2479 [DOI] [PubMed] [Google Scholar]
- Steitz JA, Berg C, Hendrick JP, La Branche-Chabot H, Metspalu A, Rinke J, Yario T (1988) A 5S rRNA/L5 complex is a precursor to ribosome assembly in mammalian cells. J Cell Biol 106: 545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka I, Morin F, Seizinger BR, Kley N (1995) Regulation of the sequence-specific DNA binding function of p53 by protein kinase C and protein phosphatases. J Biol Chem 270: 5405–5411 [DOI] [PubMed] [Google Scholar]
- Tanaka T, Ohkubo S, Tatsuno I, Prives C (2007) hCAS/CSE1L associates with chromatin and regulates expression of select p53 target genes. Cell 130: 638–650 [DOI] [PubMed] [Google Scholar]
- Tang Y, Zhao W, Chen Y, Zhao Y, Gu W (2008) Acetylation is indispensable for p53 activation. Cell 133: 612–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavner FJ, Simpson R, Tashiro S, Favier D, Jenkins NA, Gilbert DJ, Copeland NG, Macmillan EM, Lutwyche J, Keough RA, Ishii S, Gonda TJ (1998) Molecular cloning reveals that the p160 Myb-binding protein is a novel, predominantly nucleolar protein which may play a role in transactivation by Myb. Mol Cell Biol 18: 989–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiry M, Lamaye F, Thelen N, Chatron-Colliet A, Lalun N, Bobichon H, Ploton D (2008) A protocol for studying the kinetics of RNA within cultured cells: application to ribosomal RNA. Nat Protoc 3: 1997–2004 [DOI] [PubMed] [Google Scholar]
- Thompson T, Tovar C, Yang H, Carvajal D, Vu BT, Xu Q, Wahl GM, Heimbrook DC, Vassilev LT (2004) Phosphorylation of p53 on key serines is dispensable for transcriptional activation and apoptosis. J Biol Chem 279: 53015–53022 [DOI] [PubMed] [Google Scholar]
- Tian C, Xing G, Xie P, Lu K, Nie J, Wang J, Li L, Gao M, Zhang L, He F (2009) KRAB-type zinc-finger protein Apak specifically regulates p53-dependent apoptosis. Nat Cell Biol 11: 580–591 [DOI] [PubMed] [Google Scholar]
- Tibbetts RS, Brumbaugh KM, Williams JM, Sarkaria JN, Cliby WA, Shieh SY, Taya Y, Prives C, Abraham RT (1999) A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev 13: 152–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trigiante G, Lu X (2006) ASPP and cancer. Nat Rev Cancer 6: 217–226 [DOI] [PubMed] [Google Scholar]
- Unger T, Juven-Gershon T, Moallem E, Berger M, Vogt Sionov R, Lozano G, Oren M, Haupt Y (1999) Critical role for Ser20 of human p53 in the negative regulation of p53 by Mdm2. EMBO J 18: 1805–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ryk DI, Lee Y, Nazar RN (1992) Unbalanced ribosome assembly in Saccharomyces cerevisiae expressing mutant 5 S rRNAs. J Biol Chem 267: 16177–16181 [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA (2001) hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107: 149–159 [DOI] [PubMed] [Google Scholar]
- Verheggen C, Almouzni G, Hernandez-Verdun D (2000) The ribosomal RNA processing machinery is recruited to the nucleolar domain before RNA polymerase I during Xenopus laevis development. J Cell Biol 149: 293–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Lane DP (2007) p53 in health and disease. Nat Rev Mol Cell Biol 8: 275–283 [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C (2009) Blinded by the light: the growing complexity of p53. Cell 137: 413–431 [DOI] [PubMed] [Google Scholar]
- Wei X, Xu H, Kufe D (2005) Human MUC1 oncoprotein regulates p53-responsive gene transcription in the genotoxic stress response. Cancer Cell 7: 167–178 [DOI] [PubMed] [Google Scholar]
- Wu Z, Earle J, Saito S, Anderson CW, Appella E, Xu Y (2002) Mutation of mouse p53 Ser23 and the response to DNA damage. Mol Cell Biol 22: 2441–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi T, Keough RA, Gonda TJ, Ishii S (2008) Ribosomal stress induces processing of Mybbp1a and its translocation from the nucleolus to the nucleoplasm. Genes Cells 13: 27–39 [DOI] [PubMed] [Google Scholar]
- Yuan X, Zhou Y, Casanova E, Chai M, Kiss E, Grone HJ, Schutz G, Grummt I (2005) Genetic inactivation of the transcription factor TIF-IA leads to nucleolar disruption, cell cycle arrest, and p53-mediated apoptosis. Mol Cell 19: 77–87 [DOI] [PubMed] [Google Scholar]
- Zhang J, Harnpicharnchai P, Jakovljevic J, Tang L, Guo Y, Oeffinger M, Rout MP, Hiley SL, Hughes T, Woolford JL Jr (2007) Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev 21: 2580–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y (2003) Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol 23: 8902–8912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W (2008) Negative regulation of the deacetylase SIRT1 by DBC1. Nature 451: 587–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Poyurovsky MV, Li Y, Biderman L, Stahl J, Jacq X, Prives C (2009) Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol Cell 35: 316–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.