Abstract
Ionizing radiation (IR) triggers adaptive changes in gene expression. Here, we show that survival after IR strongly depends on the checkpoint kinase Chk2 acting upon its substrate HuR, an RNA-binding protein that stabilizes and/or modulates the translation of target mRNAs. Microarray analysis showed that in human HCT116 colorectal carcinoma cells (WT), IR-activated Chk2 triggered the dissociation of virtually all of HuR-bound mRNAs, since IR did not dissociate HuR target mRNAs in Chk2-null (CHK2−/−) HCT116 cells. Accordingly, several HuR-interacting mRNAs encoding apoptosis- and proliferation-related proteins (TJP1, Mdm2, TP53BP2, Bax, K-Ras) dissociated from HuR in WT cells, but remained bound and showed altered post-transcriptional regulation in CHK2−/− cells. Use of HuR mutants that were not phosphorylatable by Chk2 (HuR(3A)) and HuR mutants mimicking constitutive phosphorylation by Chk2 (HuR(3D)) revealed that dissociation of HuR target transcripts enhanced cell survival. We propose that the release of HuR-bound mRNAs via an IR-Chk2-HuR regulatory axis improves cell outcome following IR.
Keywords: Chk2, HuR phosphorylation, radiotherapy, ribonucleoprotein complex, RNA-binding protein
Introduction
The cell's DNA is exposed to damage arising endogenously from normal cellular processes such as DNA replication, and exogenously by exposure to genotoxic agents like chemotherapeutic factors and ultraviolet light (UV) and ionizing radiation (IR). Impairment of the DNA-damage response (DDR) causes genomic instability and predisposes cells to malignancy (Harper and Elledge, 2007; Jackson and Bartek, 2009). A key aspect of the DDR is altered transcription factor activity leading to changes in the subsets of transcribed mRNAs. In this regard, transcription factor activity is potently controlled by the checkpoint pathways, which are conserved signalling cascades activated during the DDR. A major component of DDR checkpoints is the checkpoint kinase 2 (Chk2), originally identified in yeast as a kinase essential for cell-cycle arrest following DNA replication blockage (Murakami and Okayama, 1995). The kinase ataxia-telangietasia mutated (ATM) phosphorylates Chk2 at T68, which becomes fully activated by homodimerization and trans-phosphorylation of the kinase domain (T383, T387). In turn, activated Chk2 phosphorylates protein substrates that implement many aspects of the cellular response to genotoxic damage (Matsuoka et al, 1998; Kastan and Lim, 2000; Melchionna et al, 2000). Through the phosphorylation of substrate proteins, including transcription factors BRCA1, E2F-1, and p53, cell-cycle regulatory proteins Cdc25A and Cdc25C, and the p53-regulatory protein Mdm2, Chk2 elicits its actions on DNA repair, apoptosis, senescence, and growth arrest (Bartek and Lukas, 2003; Ahn et al, 2004).
However, the DDR also affects gene expression potently through post-transcriptional processes. RNA-binding proteins (RBPs) and microRNAs (small non-coding RNAs) are strong regulators of gene expression following genotoxic damage (reviewed by Abdelmohsen et al, 2008; Gorospe and de Cabo, 2008; Pothof et al, 2009; Wagner-Ecker et al, 2010). The RBPs involved in the DDR associate with specific subsets of mRNAs and often regulate their post-transcriptional fate by eliciting turnover and translation regulatory functions (hence they are sometimes called TTR-RBPs (Abdelmohsen et al, 2008)). For example, the response to DNA-damaging agents like oxidants and UV involves TTR-RBPs that promote mRNA stability such as the Hu proteins (the ubiquitous HuR and the primarily neuronal HuB, HuC, and HuD), TTR-RBPs that enhance mRNA decay (including the AU-binding factor 1 (AUF1/hnRNP D) and tristetraprolin), and TTR-RBPs that suppress translation, including the T-cell-restricted intracellular antigen 1 (TIA-1) and the TIA-1-related protein TIAR (Stoecklin et al, 2004; López de Silanes et al, 2004; Lal et al, 2006; Hinman and Lou, 2008).
One of the best-studied TTR-RBPs, HuR regulates gene expression in cells responding to mitogenic, differentiation, immune, and stress-causing agents (Hinman and Lou, 2008). Following exposure to these stimuli, HuR influences the expression of target mRNAs via two main mechanisms. First, by translocation of the almost-exclusively nuclear HuR to the cytoplasm, where it stabilizes and/or modulates the translation rate of target mRNAs (Keene, 1999; Hinman and Lou, 2008). HuR nucleocytoplasmic movement is mediated by transport proteins like CRM1, transportins 1 and 2, and importin-α1 (Gallouzi and Steitz, 2001; Guttinger et al, 2004; Rebane et al, 2004; Wang et al, 2004), and is controlled via phosphorylation by protein kinase C (PKC), the mitogen-activated protein kinase p38, and cyclin-dependent kinase 1 (cdk1) (Doller et al, 2008a; Kim et al, 2008a, 2008b; Lafarga et al, 2009). Second, by increasing the association of HuR with target mRNAs, many of which bear a U-rich signature motif (López de Silanes et al, 2004); these interactions are also modulated via phosphorylation of HuR by kinases PKC, p38, and Chk2 (Abdelmohsen et al, 2007; Doller et al, 2007, 2008b; Lafarga et al, 2009). With dozens of HuR ligand mRNAs identified thus far, HuR has been implicated in key cellular processes (e.g. cell-cycle progression, apoptosis) and in processes affecting tissues and organs, including muscle development, epithelial integrity, immune function, and carcinogenesis (Hinman and Lou, 2008; Ghosh et al, 2009; Abdelmohsen and Gorospe, 2010).
Original screens of HuR activity revealed that many genotoxic agents promoted the translocation of HuR to the cytoplasm, but IR (γ irradiation) did not affect HuR abundance in the cytoplasm (Wang et al, 2000). Since HuR binding (not its subcellular distribution) was recently shown to be controlled by the prominent DDR kinase Chk2, we hypothesized that HuR binding to mRNA, instead of HuR cytoplasmic levels, might be influenced by IR. Moreover, since the influence of Chk2 phosphorylation of HuR at S88, S100, and T118 appears complex, with some phosphorylated residues promoting and others inhibiting HuR binding to mRNA, and with individual mRNA differences in these interactions (Abdelmohsen et al, 2007; Wilusz and Wilusz, 2007), we further anticipated a complex pattern of Chk2 influence on HuR–mRNA binding after IR. We examined these questions by comparing the response of human colorectal carcinoma HCT116 cells (WT) with that of isogenic HCT116 cells bearing somatic deletions of both CHK2 alleles (CHK2−/− cells) (Jallepalli et al, 2003). To our surprise, microarray analysis revealed that almost all of HuR-interacting mRNAs dissociated following IR in WT cells but remained associated in CHK2−/− cells. In turn, this pattern of HuR–mRNA interactions caused alterations in abundance of apoptotic and proliferative proteins. The enhanced IR toxicity encountered by CHK2−/− cells was linked to the inability to dissociate HuR–mRNA complexes, since a non-phosphorylatable HuR mutant retained mRNA binding after IR and conferred reduced survival to WT cells. By contrast, a phosphomimic HuR mutant displayed reduced mRNA interactions and improved the survival of both WT and CHK2−/− cells following IR. Our findings underscore a central role for HuR in regulating gene expression patterns and survival in response to IR, via a novel mechanism of global dissociation of bound mRNA targets.
Results
Differential association of HuR with target mRNAs in normal versus Chk2-null cells after IR
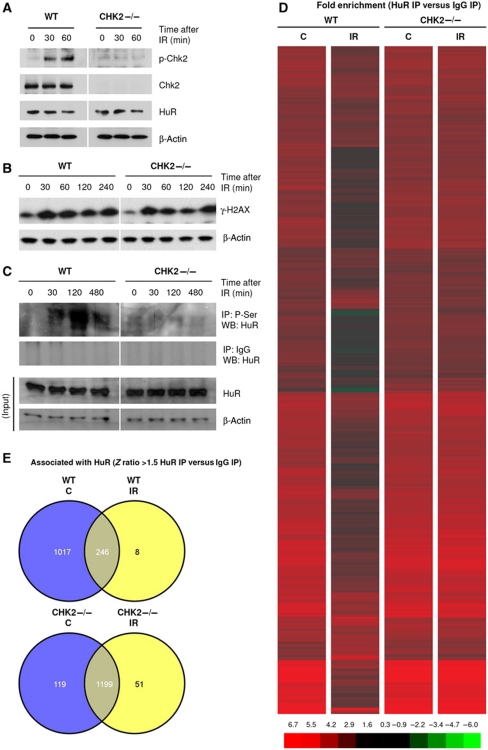
Western blot analysis indicated that IR triggered the phosphorylation of Chk2 at T68 (an activating modification) in HCT116 cells with wild-type Chk2 (WT) by 30 and 60 min after treatment with 10 Gy of IR; Chk2 and p-Chk2 were undetectable in Chk2-null HCT116 (CHK2−/−) cells (Figure 1A). The levels of HuR protein were comparable in both populations and were unaffected by IR treatment (Figure 1A). Increased levels of γ-H2AX, a marker of DNA double-strand (ds) breaks, indicated comparable dsDNA damage in both cell lines (Figure 1B). In keeping with earlier results (Abdelmohsen et al, 2007), HuR phosphorylation increased by IR in WT cells, but not in CHK2−/− cells (Figure 1C).
Figure 1.
Global dissociation of HuR–mRNA complexes after IR (10 Gy). (A, B) Wild-type (WT) and Chk2-null (CHK2−/−) human colorectal carcinoma HCT116 cells were irradiated with 10 Gy; at the times indicated, the levels of Chk2 phosphorylated at T68 ((p)CHK2), total Chk2, HuR (A), the levels of DNA-damage marker γ-H2AX (B), and loading control β-actin were assessed by western blot analysis. (C) After treatment of HCT116 cells as explained in (A), whole-cell lysates were subjected to HuR IP (or control IgG IP) followed by western blot (WB) analysis using anti-phospho-serine antibody; (Input) denotes western blot analysis of lysates before IP. (D) Heatmap image of the differential association of HuR with mRNAs in untreated (C) relative to IR-treated (IR) cells, either WT or CHK2−/− (see Supplementary Tables T1,T2,T3,T4 for complete array results). All of the transcripts showing enrichment in HuR IP in at least one of the groups are shown; differences in association were calculated by Z-ratio analysis. (E) Venn diagrams to summarize the mRNAs which associated with HuR significantly (Z-ratio differences ⩾±1.5, P-value<0.05) in HuR IP compared with IgG IP.
To study globally if IR changed the association of HuR with target mRNAs in a Chk2-dependent manner, ribonucleoprotein (RNP) immunoprecipitation (IP) using an anti-HuR antibody was followed by detection of HuR-associated mRNAs by microarray analysis. The association of mRNAs with HuR by 30 min after IR (10 Gy) was assessed by estimating the enrichment of mRNAs in HuR IP samples compared with those in control immunoglobulin G (IgG) IP samples. As shown in Figure 1D, IR triggered a broad reduction in the association of HuR with target mRNAs in WT cells, but not in CHK2−/− cells, where mRNAs remained bound (see Supplementary Tables T1,T2,T3,T4 for a complete list of transcripts and differences in abundance). As summarized using Venn diagrams, in WT cells many mRNAs associated with HuR in untreated WT cells (C), but most mRNAs dissociated from HuR after IR: of 1263 mRNAs bound in untreated cells (1017+246), only 246 mRNAs remained bound after IR and only 8 mRNAs were bound exclusively after IR (Figure 1E, top). By contrast, in CHK2−/− cells most HuR targets remained associated: some 1199 mRNAs stayed bound after IR out of a total of 1318 (Figure 1E, bottom). IR triggered the dissociation of 119 mRNAs from HuR in CHK2−/− cells and new association of HuR with 51 mRNAs, suggesting that additional, Chk2-independent pathways regulate HuR–mRNA interactions (Figure 1E, bottom). In untreated cells, the subsets of HuR-associated mRNAs in WT and CHK2−/− cells overlapped extensively (Supplementary Tables T1,T2,T3,T4).
These results suggested that a large subset of HuR-bound mRNAs dissociated via IR-triggered activation of Chk2, linked to the modification of HuR binding to target mRNAs following phosphorylation by Chk2. Interestingly, among the top mRNAs showing significant changes in association with HuR after IR in WT cells, we found several that encoded proteins with key functions in proliferation and apoptosis, which we set out to study in detail.
IR-triggered changes in association of HuR with target mRNAs
Among the transcripts that showed IR-triggered reduced association with HuR in WT cells compared with CHK2−/− cells (Figure 1D), we selected seven cancer-related targets for detailed analysis: TJP1 (also known as zona occludens-1), SOX4, SOX9, MDM2, KRAS, TP53BP2 (also known as apoptosis-stimulating protein of p53 (ASPP2)), and BAX mRNAs (Ghosh et al, 2009; Supplementary Table T5). The encoded proteins had important roles in tumour development by modulating processes such as cell proliferation and apoptosis. TJP1 has a key role in the structural organization of tight junctions at the plasma membrane, modulates cell growth and differentiation, and is considered a tumour suppressor (Tsukita et al, 1993; Fanning and Anderson, 2009). The transcription factor Sox4 can trigger apoptosis and may act as a tumour suppressor (Hur et al, 2004; Liu et al, 2006). Sox9, also a transcription factor, has a key role in sex determination and chondrogenesis during development (Foster et al, 1994; Kwok et al, 1995); its overexpression in melanoma cells induced cell-cycle arrest by increasing p21 transcription and inhibited tumourigenicity (Passeron et al, 2009). Although elevated levels of Mdm2 can inhibit apoptosis by reducing p53 abundance, Mdm2 can also promote apoptosis via p53-independent effects, which include Mdm2's action on the E2F-1, IGF-1R, and NF-κB pathways (reviewed by Bouska and Eischen, 2009; Manfredi, 2010). TP53BP2 binds the tumour suppressor p53 and induces apoptosis (Samuels-Lev et al, 2001). Bax (BCL2-associated protein X) is a pro-apoptotic member of the BCL2 family of regulators of programmed cell death (Cory and Adams, 2002). KRAS, a Kirsten Ras oncogene homologue and member of the mammalian Ras gene family of small GTPases, bears a single amino-acid substitution that renders it strongly transforming; K-Ras is implicated in various carcinomas (Malumbres and Barbacid, 2003; Siddiqui and Piperdi, 2010).
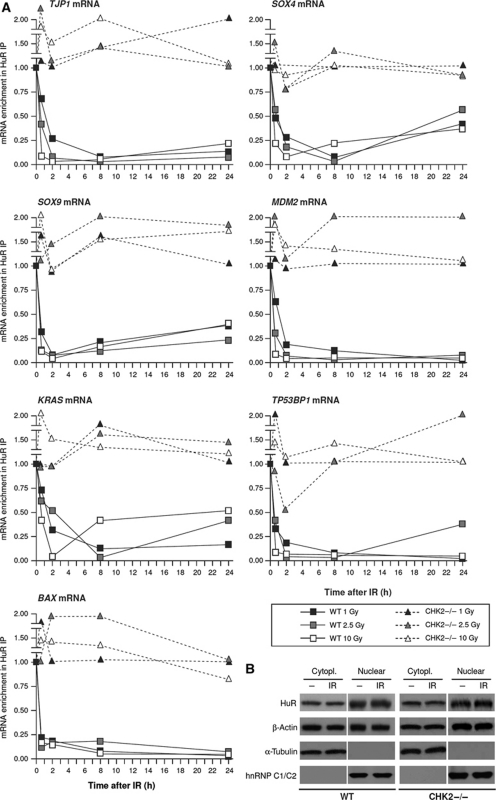
The association of these HuR target transcripts was first validated by RNP IP followed by reverse transcription (RT) and real-time, quantitative (q)PCR analysis of each individual target in WT and CHK2−/− cells. In agreement with the microarray results (Supplementary Table T5), HuR binding to TJP1, SOX4, SOX9, MDM2, KRAS, TP53BP2, and BAX mRNAs in WT cells decreased by 30 min and remained low for the following 24 h at the IR doses tested (1, 2.5, 10 Gy), while in CHK2−/− cells HuR remained bound to these mRNAs (in some instances increasing or decreasing up to ∼two-fold) for up to 24 h after IR treatment at the three doses (Figure 2A). Several stress agents, including oxidants and UV, triggered the translocation of HuR to the cytoplasm, linked to enhanced HuR binding to target mRNAs (Wang et al, 2000). Interestingly, however, exposure of HCT116 cells to 10 Gy IR did not affect the cytoplasmic abundance of HuR (Figure 2B), indicating that the levels of HuR–mRNA associations after IR were unrelated to changes in the subcellular concentration of HuR. The data further suggested that HuR phosphorylation by p38, PKC, and Cdk1, previously shown to influence its subcellular distribution (Doller et al, 2008a; Kim and Gorospe, 2008; Lafarga et al, 2009), likely had little or no influence on HuR during the IR response.
Figure 2.
Validation of HuR–mRNA interactions and subcellular localization after IR. (A) The microarray results (Figure 1; Supplementary Table T5) were verified by assessing the association of HuR with seven target transcripts (TJP1, SOX4, SOX9, MDM2, KRAS, TP53BP2, and BAX mRNAs) encoding proliferative/apoptotic proteins by 0.5, 2, 8, and 24 h after IR (1, 2.5, and 10 Gy). HuR–mRNA interactions were measured by RNP IP analysis followed by RT–qPCR amplification and expressed as enrichment of individual mRNAs in HuR IP relative to IgG IP; data were normalized to the levels of GAPDH mRNA, an abundant mRNA that is not a target of HuR and is present as a low-level contaminant in all IP samples. Shown are representative results out of four independent experiments performed. (B) The levels of HuR, loading control β-actin, cytoplasmic marker α-tubulin, and nuclear marker hnRNP C1/C2 were assessed by western blot analysis in nuclear and cytoplasmic fractions prepared from WT and CHK2−/− cells that were either left untreated (−) or irradiated (10 Gy) and collected 30 min later.
Minor influence of IR on steady-state levels of HuR target mRNAs
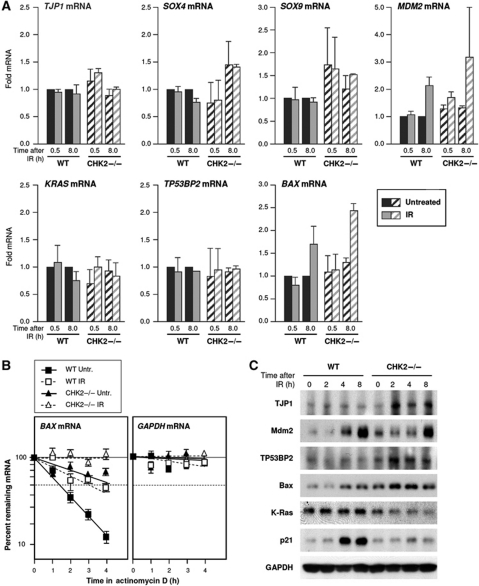
To investigate the consequences of the differential HuR–mRNA associations in WT relative to CHK2−/− cells, we first studied whether IR influenced total mRNA levels. Total RNA was prepared at 0.5 and 8 h after IR (10 Gy) and the levels of the seven mRNAs were tested by RT–qPCR analysis. IR did not significantly influence the steady-state abundance of most mRNAs in WT or CHK2−/− cells, although it did increase moderately the levels of BAX and MDM2 mRNAs in CHK2−/− cells (Figure 3A). The increase in BAX mRNA was due, at least in part, to increased stability of the BAX mRNA in CHK2−/− cells. Treatment with actinomycin D to block de novo gene transcription, followed by measurement of the rate of clearance of BAX mRNA in each cell population revealed that the half-life for BAX mRNA was the shortest in WT-untreated cells (1.3 h), it was longer in WT IR cells (3 h), and longer still in CHK2−/− untreated (4 h) and in CHK2−/− cells after IR (>4 h). These results indicate that the presence of Chk2 and IR treatment stabilized the BAX mRNA. MDM2 mRNA and the other mRNAs were relatively more stable and their half-lives could not be compared due to excessive toxicities observed beyond 4 h of IR and actinomycin D treatment (not shown).
Figure 3.
Levels of HuR target mRNAs in WT and CHK2−/− cells. (A) The levels of seven HuR target mRNAs in WT and CHK2−/− cells by 0.5 and 8 h after IR (10 Gy) were measured by RT–qPCR analysis. (B) The half-life of BAX mRNA was measured by treating cells with actinomycin D (2 μg/ml) and collecting total RNA at the times shown, whereupon the levels of BAX mRNA and GAPDH mRNA (a stable, housekeeping control mRNA) were measured by RT–qPCR analysis. mRNA half-life was calculated as the time needed to reduce transcript levels to one-half (50%, discontinuous line) of its initial abundance at time 0. The data in (A, B) represent the means and s.e.m. from three independent experiments. (C) Western blot analysis of the expression of TJP1, Mdm2, K-Ras, TP53BP2, Bax, positive control for IR in WT cells p21, and loading control GAPDH in WT and CHK2−/− cells at the times shown after IR (10 Gy). Data are representative of three independent experiments.
Next, the proteins encoded by these mRNAs were assessed by western blot analysis (except for Sox4 and Sox9, which gave poor protein western signals, not shown). Compared with WT cells, CHK2−/− cells showed constitutively higher levels of Mdm2 and Bax, and IR triggered more pronounced increases in TJP1 and TP53BP2 (Figure 3C). Only the levels of the proliferative protein K-Ras were reduced in CHK2−/− relative to WT cells. Analysis of the control protein p21, a cdk inhibitor whose levels increase by IR, showed elevated abundance in WT cells but not CHK2−/− cells (Figure 3C).
IR modulates the translation status of HuR target mRNAs
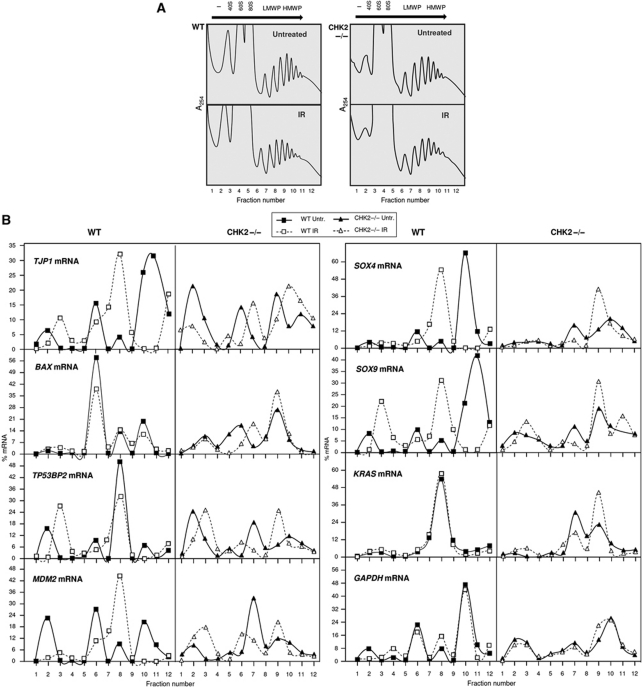
Since there were no dramatic changes in steady-state levels of HuR targets TJP1, MDM2, KRAS, TP53BP2, and BAX mRNAs, we examined if their translation status was affected by CHK2 status and/or IR. By 3 h following IR (10 Gy) or no treatment, lysates of WT and CHK2−/− cells were fractionated through sucrose gradients (Figure 4A). The relative abundance of each mRNA in fractions 1 and 2 (containing mRNAs not bound to ribosome components), fractions 3–5 (mRNAs bound to ribosome subunits or monosomes), fractions 6–8 (mRNAs forming low-molecular weight polysomes, LMWP), and fractions 9–12 (mRNAs forming high-molecular weight polysomes, HMWP) was calculated. This analysis revealed moderately different distribution patterns for each mRNA, but it was generally consistent with higher translation rates (rightward shifts in distribution) in CHK2−/− cells. TJP1 mRNA was abundant in HMWP (peaking at fraction 11) before IR, shifting leftward after IR (with peaks at fractions 3 and 8, consistent with lower translation rates) in WT cells; in CHK2−/− cells, TJP1 mRNA increased in polysomes, shifting polysome peaks at fractions 6 and 9 to 7 and 10 after IR. BAX mRNA showed peak abundance at fraction 6 in WT cells; in CHK2−/− cells, besides being more stable, BAX mRNA was more abundant in actively translating fractions and after IR shifted to larger polysomes (fractions 6–7) and had a larger polysome peak in fraction 9. In WT cells, the polysome peaks for TP53BP2 and MDM2 mRNAs (at fractions 8 and 10, respectively) were smaller after IR, while in CHK2−/− cells, both mRNAs were more abundant in HMWP and the peaks at fraction 7 shifted to fraction 9. SOX4 and SOX9 mRNAs in WT cells formed polysomes peaking in the HMWP fractions, which shifted to LMWP fractions after IR; in CHK2−/− cells, by contrast, IR triggered increases in the polysome sizes for both mRNAs. KRAS mRNA was present in LMWP region in WT cells, peaking at fraction 7 before and after IR treatment; in CHK2−/− cells, the peaks at fractions 7 and 9 shifted in relative size after IR. Thus, the reduced expression of K-Ras (Figure 3C) might not be due to marked decreases in KRAS mRNA polysomes, and other regulatory mechanisms (e.g. lower rates of KRAS mRNA translation elongation or lower stability of K-Ras protein) are likely responsible. The translation of a control transcript (GAPDH mRNA) showed little changes after IR in WT and in CHK2−/− cells (Figure 4B).
Figure 4.
Polysome analysis of HuR target mRNAs. (A) Lysates prepared from WT or CHK2−/− cells that were either left untreated or treated with IR and collected 4 h later were fractionated through sucrose gradients to generate polysome profiles by measuring the absorbance at 254 nm. (B) The relative distribution of HuR targets (TJP1, MDM2, BAX, SOX4, SOX9, KRAS, and TP53BP2 mRNAs), and housekeeping GAPDH mRNA on polysome gradients was studied by RT–qPCR analysis of the RNA present in each of 12 gradient fractions, and represented as percentage of total mRNA. Arrow indicates the direction of sedimentation; −, fractions with no ribosomal components; 40S and 60S, small and large ribosome subunits, respectively; 80S, monosomes; LMWP (fractions 6–8) and HMWP (fractions 9–12), low- and high-molecular weight polysomes, respectively. Data are representative of three independent experiments.
Elucidation of the specific influence of HuR on each mRNA, including the mapping of HuR-binding site(s) on each mRNA, HuR's relative contribution to mRNA stability and translational regulation, and reporter construct analyses, await further study. However, these results indicate that the persistent association of HuR with target mRNAs in CHK2−/− cells after IR correlates with altered (generally higher, except for K-Ras) expression of the encoded proteins in the CHK2−/− population.
Chk2 promotes cell survival following IR
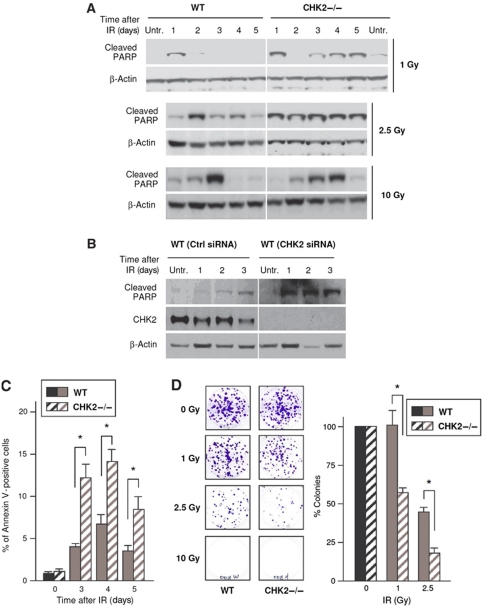
The impact of the differential HuR–mRNA associations in WT and CHK2−/− cells was first assessed by measuring the outcome of both cell types after IR. Cleavage of PARP (poly(ADP ribose) polymerase), a marker of apoptosis, was more extensive in CHK2−/− cells at virtually all days and doses of IR tested (Figure 5A). The specific influence of Chk2 on cell survival was further assessed by silencing Chk2 in WT cells; as shown, PARP cleavage was more pronounced in Chk2-silenced cells (Figure 5B). The percentages of cells that were positive for Annexin V (another apoptosis marker) were also significantly higher in CHK2−/− cells compared with WT cells following IR (2.5 Gy, Figure 5C). Furthermore, colony formation assay revealed that significantly more colonies formed from WT cells than from CHK2−/− cells following IR (Figure 5D). These four sets of experiments indicate that CHK2 promotes the survival of HCT116 cells in response to IR.
Figure 5.
Enhanced cytotoxicity of CHK2−/− cells following IR. (A) Western blot analysis of cleaved PARP (and loading control GAPDH) in whole-cell lysates prepared from cells that were treated with IR (1, 2.5, 10 Gy) and collected at the times shown. (B) WT cells were transfected with control (Ctrl) or CHK2-directed siRNAs; 48 h later, cells were irradiated with 10 Gy and harvested at the times shown for western blot analysis of cleaved PARP and CHK2. (C) The levels of the apoptotic marker Annexin V were measured in WT and CHK2−/− cells after 2.5 Gy of IR; the percentages of Annexin V-positive cells are shown. (D) Colony formation assay in WT and CHK2−/− cells that were either left untreated (0 Gy) or treated with IR (1, 2.5, or 10 Gy); 10 days later, the number of colonies developing from 500 cells/well were visualized (left) and counted (graph). (C, D) Values are the mean+s.e.m. from two independent experiments; *Significantly different compared with IR-treated WT cells (P<0.05 by two-tailed Student's t-test).
HuR dissociation from target mRNAs is lost in HuR mutants not phosphorylatable by Chk2
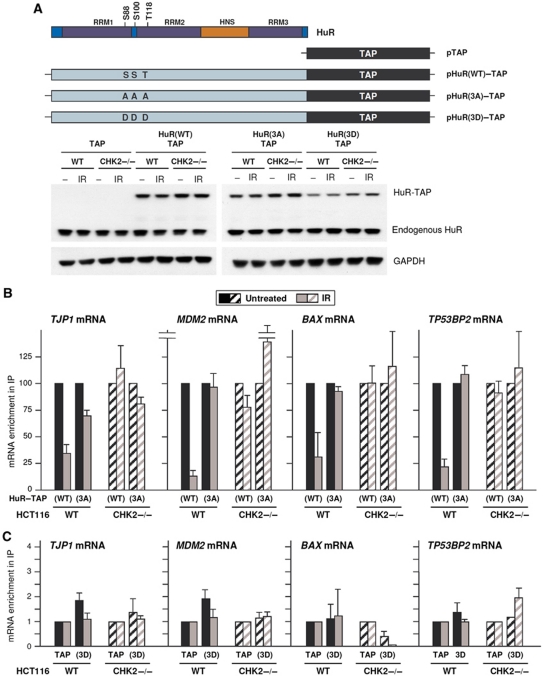
To further analyse the effects of HuR phosphorylation on its mRNA binding, we prepared HuR mutants with modified residues S88, S100, T118 (Figure 6A). The plasmid vector expressing a tagged mutant HuR that was not phosphorylatable by Chk2 (pHuR(S88A)(S100A)(T118A)–TAP) was termed pHuR(3A)–TAP; the plasmid expressing a mutant HuR with the three sites modified to aspartic acid to resemble constitutive phosphorylation by Chk2 (pHuR(S88D)(S100D)(T118D)–TAP) was termed pHuR(3D)–TAP. We examined the binding of the HuR–TAP fusion proteins to TJP1, MDM2, BAX, and TP53BP2 mRNAs. As shown in Figure 6B, in WT cells, IR caused dissociation of the mRNAs from HuR(WT) but not (or to a much lesser degree) from HuR(3A), suggesting that phosphorylation at S88, S100, and/or T118 was important for the release of these mRNAs. In agreement with this notion, both HuR(WT) and HuR(3A) retained binding to the mRNAs before and after IR in CHK2−/− cells (Figure 6B). According to the same notion, all four mRNAs associated with HuR(3D) at background levels, considered as the binding to the TAP tag alone (i.e. in cells transfected with the control vector pTAP) (Figure 6C). These results indicated that HuR phosphorylation by Chk2 was necessary for the release of these HuR target mRNAs in response to IR.
Figure 6.
Association of mutant HuR with target mRNAs. (A) Schematic of plasmids pTAP, pHuR(WT)–TAP, pHuR(3A)–TAP (carrying three non-phosphorylatable mutations, S88A, S100A, T118A), and pHuR(3D)–TAP (carrying three phosphomimic mutations, S88D, S100D, T118D). (B) The chimeric HuR(WT)–TAP proteins and HuR(3A)–TAP were expressed by transfection of the corresponding plasmids into WT and CHK2−/− cells. Thirty minutes after IR (10 Gy), binding of HuR–TAP proteins to the indicated HuR target mRNAs was tested by RNP IP and RT–qPCR, and represented as relative enrichment in control cells compared with IR-treated cells. (C) The enrichment of mRNAs in chimeric HuR(3D)–TAP relative to background binding (TAP) was calculated and compared as in (B). (B, C) The results are the mean+s.e.m. from three independent experiments.
Dissociation of HuR–mRNA complexes is required for cell survival in response to IR
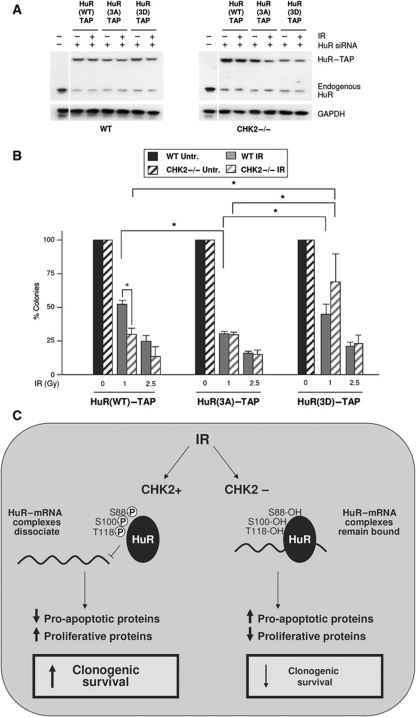
Finally, we investigated if HuR was an effector of the CHK2-dependent influence on cell survival after IR (Figure 5). To test this possibility, we silenced the endogenous HuR using small interfering (si)RNA directed to the 3′UTR of HuR and examined the effect of reintroducing HuR(WT), HuR(3A), or HuR(3D) vectors expressing only the HuR coding region and thus refractory to the HuR siRNA (Figure 7A). At 48 h after transfection of the respective plasmids, cells were subjected to IR. Colony formation assay revealed that expression of the non-phosphorylatable HuR (HuR(3A)), which retained binding to target mRNAs (Figure 6B), reduced significantly the number of surviving colonies (Figure 7B). By contrast, IR treatment of cells expressing the phosphomimic HuR mutant (HuR(3D)), which did not bind mRNAs (Figure 6B), significantly increased the number of colonies compared with those seen in the HuR(3A) transfection group (Figure 7B). Collectively, these results indicate that Chk2 phosphorylation of HuR after IR, causing the dissociation of HuR–mRNAs, helps to implement appropriate gene expression patterns that enhance the survival of cells with damaged DNA (Figure 7C).
Figure 7.
Enhanced cytotoxicity of CHK2−/− cells to IR. (A, B) Endogenous HuR was silenced using siRNA directed to the 3′UTR or HuR (Materials and methods), and different HuR–TAP mutants were overexpressed in WT and CHK2−/− cells that were then either left untreated or subjected to IR (1 or 2.5 Gy). Western blot analysis of endogenous HuR and overexpressed chimeric HuR–TAP proteins. Cells were first transfected with siRNA and 16 h later with plasmids (using Lipofectamine-2000); 48 h after plasmid transfection, whole-cell lysates were prepared for western blot analysis (A); quantification of colony formation assay in cells (500 per plate), counted on day 10. Values represent the mean+s.e.m. from two independent experiments. *Significantly different compared with IR-treated WT cells (P<0.05 by ANOVA and Bonferroni/Dunn test). (C) Model depicts proposed mechanism whereby HuR is cytoprotective in response to IR. In WT cells, Chk2 triggers the dissociation of HuR from target mRNAs encoding apoptotic and mitogenic proteins, such as TJP1, SOX4, SOX9, MDM2, KRAS, and TP53BP2 mRNAs; the ensuing changes in expressed proteins enhance clonogenic survival. In CHK2−/− cells, these mRNAs remain bound to HuR, leading to aberrantly elevated production of proteins such as TJP1, Mdm2, and TP53BP2, and reduced K-Ras; the dysregulated gene expression results in reduced clonogenic survival.
Discussion
We have investigated the influence of HuR phosphorylation by Chk2 within the cellular IR response. Using human colon cancer cells that differed in their CHK2 status, we discovered that IR triggered the dissociation of the majority of HuR target mRNAs in WT cells while HuR target mRNAs remained bound in CHK2−/− cells. The patterns of HuR–mRNA interactions determined subsequent protein expression patterns (Figure 3), primarily by affecting the translational profiles of the target mRNAs, and only minimally the mRNA steady-state abundance (Figures 3 and 4). These interactions significantly affected cell survival after IR, with WT cells showing reduced apoptotic markers and enhanced clonogenic survival compared with CHK2−/− cells (Figure 5). Importantly, HuR was a key factor in enhancing cell survival, since expression of mutant HuR that could not be phosphorylated (HuR(3A)) sensitized WT cells to IR toxicity, while expression of a phosphomimic of HuR (HuR(3D)) conferred enhanced resistance to CHK2−/− cells (Figure 7). Together, our results indicate that phosphorylation of HuR by CHK2 can engender cellular resistance to IR stress by dissociating HuR from subsets of target mRNAs.
Triple HuR(S88, S100, T118) mutants
Accumulating evidence indicates that phosphorylation of HuR by a number of kinases (PKCα, PKCδ, Cdk1, p38MAPK, and Chk2) can influence HuR subcellular localization and HuR–mRNA association in a variety of conditions (Abdelmohsen et al, 2007; Doller et al, 2007, 2008b; Kim et al, 2008b; Lafarga et al, 2009). Our earlier studies revealed that H2O2 activated Chk2, which in turn phosphorylated HuR; phosphorylation at each of the three HuR substrate residues (S88, S100, T118) had a distinct influence on the association of HuR with SIRT1 mRNA, with S88 having little effect, S100 reducing binding, and T118 promoting binding (Abdelmohsen et al, 2007; Wilusz and Wilusz, 2007). Here, we found that the IR-activated Chk2 similarly phosphorylated HuR, but we examined the influence of this phosphorylation process on new engineered HuR mutants bearing all three modified residues (triple Ser/Thr → Ala, triple Ser/Thr → Asp). The triple non-phosphorylatable mutant (HuR(3A)–TAP) could not release the bound mRNAs in response to IR (Figure 6B), an effect that did not recapitulate the results seen with individual non-phosphorylatable mutants (not shown). Conversely, HuR(3D)–TAP showed very little binding before and after IR (Figure 6C); it remains to be tested whether HuR(3D) retains the ability to bind any other target mRNAs. While replacement of endogenous HuR with HuR(3A) sensitized WT cells to IR toxicity, HuR(3D) instead conferred resistance to apoptosis in both WT and CHK2−/− cells. These results suggest that HuR phosphorylation by Chk2 directly affects gene expression programs and impacts upon cell survival. Structure–function analyses of the interaction of HuR and phospho-HuR with mRNAs will help to understand the molecular associations whereby HuR influences gene expression and cell survival.
HuR inhibits K-Ras expression
HuR has been shown to enhance the stability and translation of target mRNAs by numerous mechanisms, including competition with RBPs that promote mRNA decay (e.g. AUF1 (Lal et al, 2004)), with RBPs that inhibit translation (such as TIA-1 (Kawai et al, 2006)), or with microRNAs (Bhattacharyya et al, 2006). These or other mechanisms of action likely underlie the positive influence of HuR on the expression of the targets tested in CHK2−/− cells (Figures 2 and 3). In fewer instances, however, HuR has also been found to repress gene expression. HuR's negative influence on KRAS mRNA was reminiscent of HuR's repression of other target mRNAs, either via interference with 5′UTR sequences (p27, IGF-IR (Kullmann et al, 2002; Meng et al, 2008)) or via recruitment of microRNAs (Kim et al, 2009). Future studies will be needed to ascertain in detail how HuR represses K-Ras expression.
Role of Chk2 → HuR on clonogenicity, tumourigenesis, cancer therapy
While a broad spectrum of dissociated HuR targets may be necessary to engender resistance to IR, we examined several mRNAs encoding proteins with key roles in cell proliferation and apoptosis. In CHK2−/− cells exposed to IR, HuR binding to mRNAs encoding the apoptotic proteins TJP1, Mdm2, TP53BP2, and Bax was maintained or increased, as was the abundance of the corresponding proteins when compared with WT cells, which showed generally reduced mRNA binding and lower protein levels (Figures 2 and 3). Binding of HuR to KRAS mRNA declined in WT cells (although to a lesser extent than other mRNAs, Figure 2A) and increased in CHK2−/− cells, but expression of K-Ras, a protein that increases proliferation, declined only in CHK2−/− cells. The reduced abundance of K-Ras in CHK2−/− cells may contribute to the decreased colony forming ability of the CHK2−/− cell population. Together with the induction of the cdk inhibitor p21 in WT cells, our results support the notion that Chk2 activation leading to the dissociation of HuR–mRNA complexes causes growth arrest and enhances cell survival during the IR response.
The role of Chk2 in tumourigenesis has been controversial. A few germline mutations of CHK2 have been identified in familial cancers and some infrequent somatic mutations have been described (reviewed by Antoni et al, 2007). Chk2 can prevent tumourigenesis by promoting genomic stability, enhancing DNA repair or triggering apoptosis or senescence. Despite these roles, Chk2 does not appear to function as a bona fide tumour suppressor. For example, reduced Chk2 abundance (e.g. by CHK2 haploinsufficiency) is more strongly associated with tumourigenesis than is complete loss of CHK2 (Sodha et al, 2006), and some alleles protect against the development of certain tumours (Brennan et al, 2007). Consequently, CHK2 is now generally considered to function as a multi-organ tumour susceptibility gene (Antoni et al, 2007; Perona et al, 2008).
Regardless of Chk2's role in tumour development, there is heightened interest in exploiting CHK2 status in cancer therapy. In cells with mutated or reduced Chk2, inhibiting other DNA repair proteins may be advantageous in order to enhance the lethality of chemotherapies. In cells with functional Chk2, inhibiting Chk2 could elicit a number of responses, depending on the magnitude, duration, and type of damage. To gain in-depth knowledge of whether Chk2 will favour DNA repair, cellular senescence, cell-cycle arrest, or cell death, there has been much interest in understanding Chk2's downstream effectors (e.g. Cdc25, p53, PML, BRCA1, E2F-1, Mdm4, HuR). Our results that CHK2−/− HCT116 cells encounter more toxicity after IR contrast with earlier reports in different cell types and/or after different stress agents. For example, thymocytes and neurons of Chk2−/− mice were preferentially resistant to IR-induced apoptosis; in addition, these mice displayed no abnormalities in the gastrointestinal tract, although the phenotype was analysed just 24 h after IR (Takai et al, 2002), perhaps the effects are seen only after long-term analysis. They also differ from results that Chk2 promoted apoptosis of etoposide-treated breast cancer cells via phosphorylation of Chk2 substrate E2F-1 (Stevens et al, 2003) and IR-treated leukemia cells after Chk2 phosphorylated substrate PML (Yang et al, 2002). However, our findings agree with several other reports describing a protective function of tumour cells by Chk2 in response to IR. For example, Chk2 prevented apoptosis of several carcinoma lines and promoted tumourigenesis of HCT116 cells in a xenograft model (Ghosh et al, 2006). Similarly, Chk2 protected cervical, colorectal, and breast carcinoma cells from apoptosis, resulting from hypoxic stress (Gibson et al, 2005). Chk2 was also found to be protective against DNA damage arising from reoxygenation after hypoxia in colorectal carcinoma cells, an effect that was linked to the fact that cells with low or no Chk2 could not trigger necessary cell-cycle checkpoints (Freiberg et al, 2006). Finally, inhibition of Chk2 by UCN-01 sensitized colon cancer cells to IR-triggered apoptosis regardless of p53 status (Yu et al, 2002).
Perspective
The influence of Chk2 on cell outcome after DNA damage is elicited by its phosphorylation of downstream effector proteins. One such effector, HuR, is a pivotal regulator of cancer-related gene expression (Abdelmohsen and Gorospe, 2010). Accordingly, our present results link the heightened IR sensitivity of CHK2−/− cells to the impaired regulation of HuR function in these cells. Our findings further suggest that targeting HuR by preventing its phosphorylation may help to sensitize cancer cells to therapies like IR. For example, enhancing the activity of the (as-yet unknown) phosphatase that dephosphorylates residues S88, S100, and T118 may promote the toxicity seen with HuR(3A) and thus increase IR efficacy. Together with Chk2 substrates involved in DNA repair and cell-cycle progression, we propose that the influence of Chk2 on HuR critically modulates the role of Chk2 on cancer development and therapy.
Materials and methods
Cell culture, transfection, and treatment
Human colorectal cancer cells HCT116 (WT) and CHK2-null HCT116 cell line (CHK2−/−) were maintained in Dulbecco's modified essential medium, supplemented with 10% (v/v) fetal bovine serum and penicillin/streptomycin (Invitrogen, Carlsbad, CA). Cells were mock-irradiated or exposed to 1, 2.5, or 10 Gy of IR and lysed at the times indicated. CHK2 siRNA was obtained from Santa Cruz Biotechnology; siRNAs targeting the HuR CR (AAGAGGCAATTACCAGTTTCA), the HuR 3′UTR (AACGACTCAATTGTCCC GATA), and a control siRNA (AATTCTCCGAACGTGTCACGT) were obtained from Qiagen; siRNAs were used at 10 nM. Transfection of cells was performed using Lipofectamine-2000 (Invitrogen) when plasmids were used, or Lipofectamine RNAiMax (Invitrogen) when siRNAs were used. pHuR–TAP bearing three non-phosphorylatable mutations (HuR(3A); S88A, S100A, and T118A) and pHuR–TAP bearing three phospho-mimic mutations (HuR(3D); S88D, S100D, and T118D) were generated by site-directed mutagenesis.
RNP IP analysis
IP of RNP complexes was performed as previously described (Kuwano et al, 2010). Briefly, whole-cell lysates, prepared from either untreated or IR-treated cells, were divided into two equal parts and incubated (1 h, 4°C) with 80 μl of a 50% (v/v) suspension of protein A-Sepharose beads precoated with 20 μg each of mouse IgG1 (BD Pharmingen) or mouse anti-HuR (Santa Cruz Biotechnology). The beads were washed five times with NT2 buffer (50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1 mM MgCl2, and 0.05% Nonidet P-40 (NP-40)). For RNA analysis, the beads were incubated with 100 μl NT2 buffer containing 20 U of RNase-free DNase I (15 min, 30°C), washed twice with 1 ml NT2 buffer, and further incubated in 100 μl NT2 buffer containing 0.1% SDS and 0.5 mg/ml proteinase K (15 min, 55°C) to digest the proteins bound to the beads. RNA was extracted using phenol and chloroform and precipitated in the presence of glycoblue. For TAP IP of RNP complexes, lysates were prepared from cells that had been transfected with pTAP, pHuR(WT)–TAP, pHuR(3A)–TAP, or pHuR(3D)–TAP were subjected to IP using rabbit IgG-agarose (Sigma) as described above.
For analysis of individual mRNAs, the RNA isolated from IP materials was subjected to RT using random hexamers, and SSII reverse transcriptase (Invitrogen). Amplification and quantification of the PCR products were performed using the Applied Biosystems 7900 System (Applied Biosystems) and Power SYBR Green PCR Master Mix (Applied Biosystems). Background binding of the abundant GAPDH mRNA was used as a loading control. Each reaction was carried out in triplicate and three independent experiments were performed.
For microarray analysis, RNA from IP material was labelled using Illumina TotalPrep RNA Amplification Kit (Ambion, Austin, TX). Human HT-12 v1.0 gene expression BeadChips containing 48 000 RefSeq transcripts (Illumina, San Diego, CA) were used for microarray analysis. Raw microarray data were filtered by the detection P-value⩽0.02, normalized by Z-score transformation and tested for significant differences in signal intensity. The sample quality was analysed by scatter plot, principal component analysis, and gene sample Z-scores-based hierarchy clustering to exclude possible outliers. ANOVA test was used to eliminate the genes with larger variances within each comparison group. Genes were considered to be significantly changed after calculating Z-ratio, indicating fold difference (Z>1.5), false discovery rate (fdr), which controls for the expected proportion of false rejected hypothesis (fdr⩽0.3) and P<0.05.
Protein analysis
For western blot analysis, lysates were size-fractionated by SDS–PAGE and transferred onto PVDF membranes. Monoclonal antibodies recognizing HuR, Chk2, TP53BP2, K-Ras, GAPDH, or α-tubulin (a control cytoplasmic protein), as well as polyclonal antibodies recognizing hnRNP C1/C2 (a control nuclear protein), Bax, p21, and phospho-Chk2 (T68) were from Santa Cruz Biotechnology. A monoclonal antibody recognizing β-actin was from Abcam, a monoclonal antibody recognizing TJP1 was from Sigma, a monoclonal antibody recognizing γ-H2AX was from Upstate Biotechnologies, a monoclonal antibody recognizing Mdm2 was from Calbiochem, and a polyclonal antibody recognizing cleaved PARP was from Cell Signaling Technology. After secondary antibody incubations, signals were detected by enhanced chemiluminescence.
For co-IP analysis, protein A-Sepharose beads were precoated with IgG1 or anti-HuR antibodies and incubated with 0.5 μg protein (16 h, 4°C). After washes with NT2 buffer, samples were fractionated by SDS–PAGE and analysed by western blotting using an anti-phospho-Ser antibody (Assay Design).
Polyribosome fractionation
Cells at <80% confluence were incubated for 15 min in 0.1 mg/ml cycloheximide and then lifted in 1 ml PEB lysis buffer (0.3 M NaCl, 15 mM MgCl2, 15 mM Tris–HCl, pH 7.6, 1% Triton X-100, and 0.1 mg/ml cycloheximide) by scraping and lysed on ice for 10 min. Nuclei were pelleted at 10 000 g for 10 min, and the resulting supernatant was fractionated through a 10–50% linear sucrose gradient, as described previously (Kuwano et al, 2010). The eluted fractions were prepared with a fraction collector (Brandel), and their quality was monitored at 254 nm using a UV-6 detector (ISCO). RNA in each fraction was extracted with 8 M guanidine-HCl or Triazol.
Oligonucleotides used for real-time RT–qPCR analysis of HuR target mRNAs
Real-time, quantitative (q)PCR analysis of selected HuR targets was performed using the following gene-specific primer pairs: GGAGAGGTGTTCCGTGTTGT and GGCTAGCTGCTCAGCTCTGT for TJP1 mRNA, AAGCTTCAGCAACCAGCATT and CCCTCTCTCTCGCTCTCTCA for SOX4 mRNA, AGGCAAGCAAAGGAGATGAA and TGGTGTTCTGAGAGGCACAG for SOX9 mRNA, TCTGATAGTATTTCCCTTTCCTTTG and TGTTCACTTACACCAGCATCAA for MDM2 mRNA, TGGACGAATATGATCCAACAAT and TCCCTCATTGCACTGTACTCC for KRAS mRNA, ATCACCGGGCAGGTCTCT and GCTCTGAGCCAGTTTTACGC for TP53BP2 mRNA, ATGTTTTCTGACGGCAACTTC and ATCAGTTCCGGCACCTTG for BAX mRNA, and AGCCACATCGCTCAGACAC and GCCCAATACGACCAAATCC for GAPDH mRNA.
Colony formation assay and Annexin V stain
Either mock-irradiated (untreated) or IR-treated cells (500 cells/well) were plated in six-well plates and returned to the incubator; 10 days later, the colonies in each well were stained using crystal violet and counted. Annexin V staining was performed using Annexin V-FITC Apoptosis Detection Kit (BioVision, Mountain View, CA); Annexin V-positive cells were viewed under a fluorescence microscope using a filter for FITC, and the percentages of positive cells were calculated; >300 cells were counted for each determination.
Supplementary Material
Acknowledgments
This research was funded by the Intramural Research Program of the National Institute on Aging, National Institutes of Health. We thank A Lohani for his assistance with the irradiation experiments.
Author contributions: KM, KA, MMK, SS, RS, HHK, and MG designed the experiments; KM, KA, MMK, SS, EKL, KT, JLM, EL, YZ, and HHK performed the experiments; KM, KA, SS, KT, RS, EL, YZ, KGB, JYW, HHK, and MG analysed and/or interpreted the data; EKL, KT, XY, and KGB developed or provided key reagents; and KM, KA, and MG wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abdelmohsen K, Gorospe M (2010) Posttranscriptional regulation of cancer traits by HuR. WIRES RNA 1: 214–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelmohsen K, Kuwano Y, Kim HH, Gorospe M (2008) Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol Chem 389: 243–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelmohsen K, Pullmann R Jr, Lal A, Kim HH, Galban S, Yang X, Blethrow JD, Walker M, Shubert J, Gillespie DA, Furneaux H, Gorospe M (2007) Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol Cell 25: 543–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J, Urist M, Prives C (2004) The Chk2 protein kinase. DNA Repair 3: 1039–1047 [DOI] [PubMed] [Google Scholar]
- Antoni L, Sodha N, Collins I, Garrett MD (2007) CHK2 kinase: cancer susceptibility and cancer therapy—two sides of the same coin? Nat Rev Cancer 7: 925–936 [DOI] [PubMed] [Google Scholar]
- Bartek J, Lukas J (2003) Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell 3: 421–429 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W (2006) Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125: 1111–1124 [DOI] [PubMed] [Google Scholar]
- Bouska A, Eischen CM (2009) Murine double minute 2: p53-independent roads lead to genome instability or death. Trends Biochem Sci 34: 279–286 [DOI] [PubMed] [Google Scholar]
- Brennan P, McKay J, Moore L, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, Mates D, Bencko V, Foretova L, Janout V, Chow WH, Rothman N, Chabrier A, Gaborieau V, Odefrey F, Southey M, Hashibe M, Hall J, Boffetta P, Peto J, Peto R, Hung RJ (2007) Uncommon CHEK2 mis-sense variant and reduced risk of tobacco-related cancers: case control study. Hum Mol Genet 16: 1794–1801 [DOI] [PubMed] [Google Scholar]
- Cory S, Adams JM (2002) The Bcl2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer 2: 647–656 [DOI] [PubMed] [Google Scholar]
- Doller A, Akool el-S, Huwiler A, Müller R, Radeke HH, Pfeilschifter J, Eberhardt W (2008b) Posttranslational modification of the AU-rich element binding protein HuR by protein kinase Cdelta elicits angiotensin II-induced stabilization and nuclear export of cyclooxygenase 2 mRNA. Mol Cell Biol 28: 2608–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doller A, Huwiler A, Müller R, Radeke HH, Pfeilschifter J, Eberhardt W (2007) Protein kinase Cα-dependent phosphorylation of the mRNA-stabilizing factor HuR: implications for posttranscriptional regulation of cyclooxygenase-2. Mol Biol Cell 18: 2137–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doller A, Pfeilschifter J, Eberhardt W (2008a) Signalling pathways regulating nucleo-cytoplasmic shuttling of the mRNA-binding protein HuR. Cell Signal 20: 2165–2173 [DOI] [PubMed] [Google Scholar]
- Fanning AS, Anderson JM (2009) Zonula occludens-1 and -2 are cytosolic scaffolds that regulate the assembly of cellular junctions. Ann NY Acad Sci 1165: 113–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster JW, Dominguez-Steglich MA, Guioli S, Kwok C, Weller PA, Stev M, Weissenbach J, Mansour S, Young ID, Goodfellow PN, Brook JD, Schafer AJ (1994) Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature 372: 525–530 [DOI] [PubMed] [Google Scholar]
- Freiberg RA, Hammond EM, Dorie MJ, Welford SM, Giaccia AJ (2006) DNA damage during reoxygenation elicits a Chk2-dependent checkpoint response. Mol Cell Biol 26: 1598–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallouzi IE, Steitz JA (2001) Delineation of mRNA export pathways by the use of cell-permeable peptides. Science 294: 1895–1901 [DOI] [PubMed] [Google Scholar]
- Ghosh JC, Dohi T, Raskett CM, Kowalik TF, Altieri DC (2006) Activated checkpoint kinase 2 provides a survival signal for tumor cells. Cancer Res 66: 11576–11579 [DOI] [PubMed] [Google Scholar]
- Ghosh M, Aguila HL, Michaud J, Ai Y, Wu MT, Hemmes A, Ristimaki A, Guo C, Furneaux H, Hla T (2009) Essential role of the RNA-binding protein HuR in progenitor cell survival in mice. J Clin Invest 119: 3530–3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson SL, Bindra RS, Glazer PM (2005) Hypoxia-induced phosphorylation of Chk2 in an ataxia telangiectasia mutated-dependent manner. Cancer Res 65: 10734–10741 [DOI] [PubMed] [Google Scholar]
- Gorospe M, de Cabo R (2008) AsSIRTing the DNA damage response. Trends Cell Biol 18: 77–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttinger S, Muhlhausser P, Koller-Eichhorn R, Brennecke J, Kutay U (2004) Transportin2 functions as importin and mediates nuclear import of HuR. Proc Natl Acad Sci USA 101: 2918–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ (2007) The DNA damage response: ten years after. Mol Cell 28: 739–745 [DOI] [PubMed] [Google Scholar]
- Hinman MN, Lou H (2008) Diverse molecular functions of Hu proteins. Cell Mol Life Sci 65: 3168–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur EH, Hur W, Choi JY, Kim IK, Kim HY, Yoon SK, Rhim H (2004) Functional identification of the pro-apoptotic effector domain in human Sox4. Biochem Biophys Res Commun 325: 59–67 [DOI] [PubMed] [Google Scholar]
- Jackson SP, Bartek J (2009) The DNA-damage response in human biology and disease. Nature 461: 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallepalli PV, Lengauer C, Vogelstein B, Bunz F (2003) The Chk2 tumor suppressor is not required for p53 responses in human cancer cells. J Biol Chem 278: 20475–20479 [DOI] [PubMed] [Google Scholar]
- Kastan MB, Lim DS (2000) The many substrates and functions of ATM. Nat Rev Mol Cell Biol 1: 179–186 [DOI] [PubMed] [Google Scholar]
- Kawai T, Lal A, Yang X, Galban S, Mazan-Mamczarz K, Gorospe M (2006) Translational control of cytochrome c by RNA-binding proteins TIA-1 and HuR. Mol Cell Biol 26: 3295–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD (1999) Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc Natl Acad Sci USA 96: 5–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HH, Abdelmohsen K, Lal A, Pullmann R Jr, Yang X, Galban S, Srikantan S, Martindale JL, Blethrow J, Shokat KM, Gorospe M (2008b) Nuclear HuR accumulation through phosphorylation by Cdk1. Genes Dev 22: 1804–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HH, Gorospe M (2008) Phosphorylated HuR shuttles in cycles. Cell Cycle 7: 3124–3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M (2009) HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev 23: 1743–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HH, Yang X, Kuwano Y, Gorospe M (2008a) Modification at HuR(S242) alters HuR localization and proliferative influence. Cell Cycle 7: 3371–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullmann M, Göpfert U, Siewe B, Hengst L (2002) ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′UTR. Genes Dev 16: 3087–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwano Y, Pullmann R Jr, Marasa BS, Abdelmohsen K, Lee EK, Yang X, Martindale JL, Zhan M, Gorospe M (2010) NF90 selectively represses the translation of target mRNAs bearing an AU-rich signature motif. Nucleic Acids Res 38: 225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok C, Weller PA, Guioli S, Foster JW, Mansour S, Zuffardi O, Punnett HH, Dominguez-Steglich MA, Brook JD, Young ID, Goodfellow PN, Schafer AJ (1995) Mutations in SOX9, the gene responsible for Campomelic dysplasia and autosomal sex reversal. Am J Hum Genet 57: 1028–1036 [PMC free article] [PubMed] [Google Scholar]
- Lafarga V, Cuadrado A, Lopez de Silanes I, Bengoechea R, Fernandez-Capetillo O, Nebreda AR (2009) p38 Mitogen-activated protein kinase- and HuR-dependent stabilization of p21(Cip1) mRNA mediates the G(1)/S checkpoint. Mol Cell Biol 29: 4341–4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Abdelmohsen K, Pullmann R, Kawai T, Galban S, Yang X, Brewer G, Gorospe M (2006) Posttranscriptional derepression of GADD45alpha by genotoxic stress. Mol Cell 22: 117–128 [DOI] [PubMed] [Google Scholar]
- Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M (2004) Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J 23: 3092–3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Ramachandran S, Ali Seyed M, Scharer CD, Laycock N, Dalton WB, Williams H, Karanam S, Datta MW, Jaye DL, Moreno CS (2006) Sex-determining region Y box 4 is a transforming oncogene in human prostate cancer cells. Cancer Res 66: 4011–4019 [DOI] [PubMed] [Google Scholar]
- López de Silanes I, Zhan M, Lal A, Yang X, Gorospe M (2004) Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci USA 101: 2987–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M (2003) RAS oncogenes: the first 30 years. Nat Rev Cancer 3: 459–465 [DOI] [PubMed] [Google Scholar]
- Manfredi JJ (2010) The Mdm2-p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev 24: 1580–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Huang M, Elledge SJ (1998) Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science 282: 1893–1897 [DOI] [PubMed] [Google Scholar]
- Melchionna R, Chen XB, Blasina A, McGowan CH (2000) Threonine 68 is required for radiation-induced phosphorylation and activation of Cds1. Nat Cell Biol 2: 762–765 [DOI] [PubMed] [Google Scholar]
- Meng Z, Jackson NL, Choi H, King PH, Emanuel PD, Blume SW (2008) Alterations in RNA-binding activities of IRES-regulatory proteins as a mechanism for physiological variability and pathological dysregulation of IGF-IR translational control in human breast tumor cells. J Cell Physiol 217: 172–183 [DOI] [PubMed] [Google Scholar]
- Murakami H, Okayama H (1995) A kinase from fission yeast responsible for blocking mitosis in S phase. Nature 374: 817–819 [DOI] [PubMed] [Google Scholar]
- Passeron T, Valencia JC, Namiki T, Vieira WD, Passeron H, Miyamura Y, Hearing VJ (2009) Upregulation of SOX9 inhibits the growth of human and mouse melanomas and restores their sensitivity to retinoic acid. J Clin Invest 119: 954–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perona R, Moncho-Amor V, Machado-Pinilla R, Belda-Iniesta C, Sánchez Pérez I (2008) Role of CHK2 in cancer development. Clin Transl Oncol 10: 538–542 [DOI] [PubMed] [Google Scholar]
- Pothof J, Verkaik NS, van IJcken W, Wiemer EA, Ta VT, van der Horst GT, Jaspers NG, van Gent DC, Hoeijmakers JH, Persengiev SP (2009) MicroRNA-mediated gene silencing modulates the UV-induced DNA-damage response. EMBO J 28: 2090–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebane A, Aab A, Steitz JA (2004) Transportins 1 and 2 are redundant nuclear import factors for hnRNP A1 and HuR. RNA 10: 590–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels-Lev Y, O’Connor DJ, Bergamaschi D, Trigiante G, Hsieh JK, Zhong S, Campargue I, Naumovski L, Crook T, Lu X (2001) ASPP proteins specifically stimulate the apoptotic function of p53. Mol Cell 8: 781–794 [DOI] [PubMed] [Google Scholar]
- Siddiqui AD, Piperdi B (2010) KRAS mutation in colon cancer: a marker of resistance to EGFR-I therapy. Ann Surg Oncol 17: 1168–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodha N, Mantoni TS, Tavtigian SV, Eeles R, Garrett MD (2006) Rare germ line CHEK2 variants identified in breast cancer families encode proteins that show impaired activation. Cancer Res 66: 8966–8970 [DOI] [PubMed] [Google Scholar]
- Stevens C, Smith L, La Thangue NB (2003) Chk2 activates E2F-1 in response to DNA damage. Nat Cell Biol 5: 401–409 [DOI] [PubMed] [Google Scholar]
- Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby WF, Blackwell TK, Anderson P (2004) MK2-induced tristetraprolin: 14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J 23: 1313–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H, Naka K, Okada Y, Watanabe M, Harada N, Saito S, Anderson CW, Appella E, Nakanishi M, Suzuki H, Nagashima K, Sawa H, Ikeda K, Motoyama N (2002) Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. EMBO J 21: 5195–5205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita S, Itoh M, Nagafuchi A, Yonemura S, Tsukita S (1993) Submembranous junctional plaque proteins include potential tumor suppressor molecules. J Cell Biol 123: 1049–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner-Ecker M, Schwager C, Wirkner U, Abdollahi A, Huber PE (2010) MicroRNA expression after ionizing radiation in human endothelial cells. Radiat Oncol 5: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Furneaux H, Cheng H, Caldwell MC, Hutter D, Liu Y, Holbrook N, Gorospe M (2000) HuR regulates p21 mRNA stabilization by UV light. Mol Cell Biol 20: 760–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yang X, Kawai T, López de Silanes I, Mazan-Mamczarz K, Chen P, Chook YM, Quensel C, Köhler M, Gorospe M (2004) AMP-activated protein kinase-regulated phosphorylation and acetylation of importin alpha1: involvement in the nuclear import of RNA-binding protein HuR. J Biol Chem 279: 48376–48388 [DOI] [PubMed] [Google Scholar]
- Wilusz CJ, Wilusz J (2007) HuR-SIRT: the hairy world of posttranscriptional control. Mol Cell 25: 485–487 [DOI] [PubMed] [Google Scholar]
- Yang S, Kuo C, Bisi JE, Kim MK (2002) PML-dependent apoptosis after DNA damage is regulated by the checkpoint kinase hCds1/Chk2. Nat Cell Biol 4: 865–870 [DOI] [PubMed] [Google Scholar]
- Yu Q, La Rose J, Zhang H, Takemura H, Kohn KW, Pommier Y (2002) UCN-01 inhibits p53 up-regulation and abrogates gamma-radiation-induced G(2)-M checkpoint independently of p53 by targeting both of the checkpoint kinases, Chk2 and Chk1. Cancer Res 62: 5743–5748 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.