Abstract
DNA non-homologous end joining (NHEJ) and homologous recombination (HR) function to repair DNA double-strand breaks (DSBs) in G2 phase with HR preferentially repairing heterochromatin-associated DSBs (HC-DSBs). Here, we examine the regulation of repair pathway usage at two-ended DSBs in G2. We identify the speed of DSB repair as a major component influencing repair pathway usage showing that DNA damage and chromatin complexity are factors influencing DSB repair rate and pathway choice. Loss of NHEJ proteins also slows DSB repair allowing increased resection. However, expression of an autophosphorylation-defective DNA-PKcs mutant, which binds DSBs but precludes the completion of NHEJ, dramatically reduces DSB end resection at all DSBs. In contrast, loss of HR does not impair repair by NHEJ although CtIP-dependent end resection precludes NHEJ usage. We propose that NHEJ initially attempts to repair DSBs and, if rapid rejoining does not ensue, then resection occurs promoting repair by HR. Finally, we identify novel roles for ATM in regulating DSB end resection; an indirect role in promoting KAP-1-dependent chromatin relaxation and a direct role in phosphorylating and activating CtIP.
Keywords: ATM, DSB repair, homologous recombination, non-homologous end joining, pathway choice
Introduction
DNA double-strand breaks (DSBs) are significant biological lesions that cause cell death if unrepaired but promote translocations and genomic instability if misrepaired. Regulating how DSBs are processed to limit genomic instability is critical for cancer avoidance. DSBs can arise from exogenous DNA-damaging agents and via endogenously generated reactive oxygen species, or following metabolic processes such as replication, V(D)J recombination and meiosis. These processes can generate DSBs directly, for example following exposure to ionising radiation (IR) or indirectly for example following replication fork collapse. DSBs can be structurally distinct depending on their origin; for example, one-ended DSBs arise at replication forks, IR generates two-ended DSBs and the DSBs generated during V(D)J recombination are four ended and held in a synaptonemal complex.
DSBs are repaired by two major pathways; DNA non-homologous end joining (NHEJ) or homologous recombination (HR). NHEJ repairs DSBs in all cell-cycle phases and represents the major pathway in G1, while HR functions in S/G2. HR also has a significant role at the replication fork in promoting replication fork recovery in a manner that does not necessitate DSB formation (Helleday et al, 2007). NHEJ involves the binding of the heterodimeric Ku protein to double-stranded DNA ends, recruitment of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) to generate the DNA-PK holoenzyme and DNA-PK kinase activation (Lieber, 2010). The assembled DNA-PK complex on the DNA end then helps to recruit a complex involving DNA ligase IV, XRCC4 and the less tightly associated XLF/Cernunnos protein, which effects the rejoining step (Lieber, 2010). HR is a more complex process involving 5′–3′ end resection to generate a 3′ single-stranded (ss)DNA overhang (Wyman and Kanaar, 2006). The ssDNA is rapidly bound by RPA, which is subsequently displaced by Rad51 in a process that involves BRCA2. Invasion of a homologous sequence to generate a Holliday junction and heteroduplex DNA then follows. Subsequent steps involve branch migration, fill-in of the ssDNA regions and Holliday junction resolution.
In addition to these core DSB repair processes, recent studies have shown that ∼15–20% of IR-induced DSBs require ATM, the nuclease Artemis, the MRN complex, γH2AX, MDC1, RNF8, RNF168 and 53BP1 for repair (Riballo et al, 2004; Noon et al, 2010). The DSBs whose repair requires these additional proteins are those located at or in close proximity to heterochromatin (HC) and the process requires ATM-dependent phosphorylation of Kruppel-associated box-associated protein-1 (KAP-1), an HC-building protein (Goodarzi et al, 2008). Since HC-DSBs appear to be repaired with slow kinetics in control cells, the findings suggest that HC is a barrier to DSB repair that ATM relieves via KAP-1 phosphorylation. Recent studies have shown that in G2 phase, the slowly repaired DSBs are ATM-Artemis-Rad51/BRCA2 dependent, showing that the process represents HR (Beucher et al, 2009).
A current critical question is what regulates the choice of DSB repair pathway usage. Studies have shown that the level of several critical HR proteins increases from S to G2 phase (e.g. Rad51, Rad52 and BRCA1) and that steps of HR are activated by CDKs (Shrivastav et al, 2008). Most significantly, CDK phosphorylation upregulates CtIP-dependent resection (Huertas et al, 2008). In yeast and chicken DT40 cells, the cell-cycle-dependent activation of CDK rigorously promotes the switch from NHEJ to HR during S/G2 phase (Aylon et al, 2004; Sonoda et al, 2006). However, in mammalian cells, recent studies have shown that, even though HR has the capacity to function in G2 phase, NHEJ represents the major DSB repair pathway in G2 as in G1 with ∼75–85% of IR-induced DSBs undergoing repair by NHEJ (Beucher et al, 2009). Thus, even though HR is activated and available in G2 phase, additional factors must influence which pathway is utilised.
In this study, we examine factors that regulate the choice between usage of HR and NHEJ. Since it is likely that the pathway chosen will differ depending upon whether the DSBs are one, two or four ended (i.e. their origin), we have focused our study on pathway usage at IR-induced two-ended DSBs in G2 phase. Moreover, chromatin superstructure may be distinct in S phase. Guided by the finding that in G2 phase, NHEJ rejoins the majority of DSBs, while HR predominantly rejoins the slowly repaired HC-DSBs, we examined whether the speed of DSB repair influences pathway choice. We show that both DNA damage complexity and chromatin complexity influence the magnitude of DSB end resection. Our findings suggest a model whereby NHEJ makes the first attempt to repair DSBs but allows access to the resection machinery when rejoining does not rapidly ensue.
Results
DNA end complexity determines the speed of DSB repair and the level of DSB end resection in G2 phase
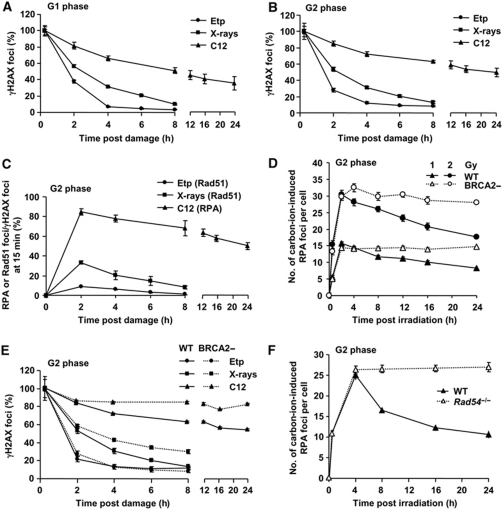
Our previous findings showed that HR preferentially repairs HC-DSBs in G2 phase (Beucher et al, 2009). Since HC influences the speed of DSB repair, we examined the impact of DSB repair kinetics on repair pathway usage in G2 phase (Goodarzi et al, 2008). Previous findings have suggested that the complexity of DNA damage is another parameter that influences DSB repair kinetics (Ritter and Durante, 2010). We compared the kinetics of DSB repair and the magnitude of end resection in G2 phase following exposure to high linear energy transfer (LET) carbon ions, X-rays and etoposide (Etp), a topoisomerase II inhibitor. Carbon ions induce highly complex DSBs compared with X-rays (Hada and Georgakilas, 2008; Tobias et al, 2010), while we have previously observed that Etp-induced DSBs are repaired with fast kinetics (Riballo et al, 2004). DSB repair was examined by γH2AX foci enumeration in unsynchronised cycling cells since cell-cycle synchronisation methods tested led to genomic damage. G2 phase cells were identified using CENP-F. Aphidicolin (APH) was added to prevent irradiated S phase cells progressing into G2 phase during analysis. S phase cells, identified by marked pan-nuclear γH2AX staining, were excluded from analysis. G1 phase cells represented those remaining (see Supplementary Figure S1 for details and Beucher et al (2009) for additional control experiments). DSBs induced by carbon ions were repaired with substantially slower kinetics compared with X-ray-induced DSBs in G1 consistent with the notion that damage complexity influences DSB repair kinetics (Figure 1A). We also observed slower repair of carbon-ion-induced DSBs in G2 compared with X-ray-induced DSBs (Figure 1B). In contrast, Etp-induced DSBs are rapidly repaired in G1 and G2 cells (Figure 1A and B). Thus, the magnitude of damage complexity correlates with DSB repair kinetics for these three agents in G1 and G2 phase. We next examined HR-processing steps following exposure to these agents by monitoring the percentage of induced DSBs that undergo resection or load Rad51 in G2 phase (Figure 1C). We assess resection or Rad51 loading by monitoring RPA or Rad51 foci numbers from 2 h after treatment and divide this by number of induced DSBs assessed from the γH2AX foci numbers at 15 min after treatment. We have previously observed that by 2 h post-IR, RPA/Rad51 foci predominantly (>90%) overlap with γH2AX and 53BP1 foci (see also Supplementary Figure S2) (Beucher et al, 2009). Surprisingly, RPA foci form at nearly all carbon-ion-induced DSBs in contrast to X-ray-induced DSBs, when only 20–30% undergo resection consistent with previous findings (Beucher et al, 2009). In contrast, only a low percentage of Etp-induced DSBs undergo resection. To exclude the possibility that end resection might be underestimated by ongoing HR repair, we enumerated RPA foci in BRCA2-defective cells, which form RPA foci but fail to load Rad51. Consistent with our data after X-rays (Beucher et al, 2009), while BRCA2-defective cells show sustained RPA foci after carbon ions, the number of foci present at 2 h, which we consider represents the maximum level, was similar in control and BRCA2-defective cells (Figure 1D; see Supplementary Figure S3A for representative images). Thus, the data for control cells in Figure 1C is a good representation of the maximum level of resection. To consolidate that RPA foci formation represent DSB repair by HR, we enumerated the rate of γH2AX foci loss in BRCA2-defective cells. Approximately 90% of γH2AX persist in BRCA2 cells 24 h after carbon-ion irradiation (Figure 1E). Although the high percentage of DSBs remaining unrepaired in control cells by 24 h precludes a full assessment of the contribution of HR, the finding nonetheless demonstrates a significant role for HR. DSB repair kinetics in BRCA2-deficient cells after X-rays is similar to our previous findings, suggesting that HR represents the slow DSB repair component repairing 20–30% of induced DSBs. In stark contrast, DSB repair kinetics in control and BRCA2-defective cells were similar following exposure to Etp. We further confirmed this result in mouse embryonic fibroblasts (MEFs), which show slightly faster repair kinetics than human cells. Rad54-deficient MEFs exhibit a substantial DSB repair defect and substantial persistent RPA foci (Figure 1F; DSB repair kinetics by γH2AX foci is shown in Supplementary Figure S3B). Taken together, these results provide evidence that the speed of DSB repair correlates with the level of DSB end resection.
Figure 1.
DSB end complexity determines the speed of DSB repair and the level of DSB end resection in G2 phase. (A, B) Positive correlation between DNA damage complexity and repair kinetics. The kinetics of DSB repair following exposure to 2 Gy carbon ions, 2 Gy X-rays and 20 (A) or 5 (B) μM Etp for 15 min, was monitored by enumerating γH2AX foci in HSF1 primary cells in G1 (A) or G2 (B) cells. The percent of γH2AX foci is normalised to γH2AX foci numbers at 15 min after damage (representing the number of DSBs induced). The dose of Etp chosen produced a similar number of DSBs to that induced by 2 Gy X-rays. Irradiated G1 and G2 cells were identified as CENP-F negative and positive, respectively. S phase cells identified by pan-nuclear γH2AX staining were excluded from analysis (Supplementary Figure S1). (C) The magnitude of resection correlates with the speed of DSB repair. RPA or Rad51 foci numbers were enumerated following exposure to 2 Gy carbon ions, 2 Gy X-rays and 5 μM Etp in HSF1 primary cells. The number of RPA or Rad51 foci at the times indicated was normalised to γH2AX foci numbers at 15 min after damage. RPA and Rad51 foci numbers were similar (data not shown). The actual numbers of γH2AX and RPA–Rad51 foci scored per cell in G2 phase are given in Supplementary Figure S3C and D. We have previously undertaken kinetic analyses of RPA foci formation and shown maximal numbers 2 h post-IR (Beucher et al, 2009). Here, we observe a similar finding for Rad51 foci formation (Supplementary Figure S2C). Further discussion of this analysis is given in Supplementary Figure S2 legend. (D) Total DSB end resection events in HSF1 (WT) and HSC62 (BRCA2) cells. Following exposure to 1 or 2 Gy carbon ions, the number of RPA foci was scored in G2 cells. (E) The contribution of BRCA2-dependent HR to G2 DSB repair following exposure to 2 Gy carbon ions, 2 Gy X-rays and 5 μM Etp. Approximately 90% of DSBs remained in HSC62 (BRCA2) cells at 24 h after carbon ions compared with ∼30% of X-ray-induced DSBs. There is no detectable DSB repair defect in BRCA2-deficient cells following exposure to Etp. (F) More than ∼90% of RPA foci persist in Rad54−/− MEFs up to 24 h after carbon-ion irradiation. The number of RPA foci was enumerated after 2 Gy carbon ions. G2 cells were identified as speckled p-histoneH3 Ser10 staining. Error bars represent the s.e.m. of three experiments.
Slowly repaired Etp-induced DSBs that localise to HC undergo resection
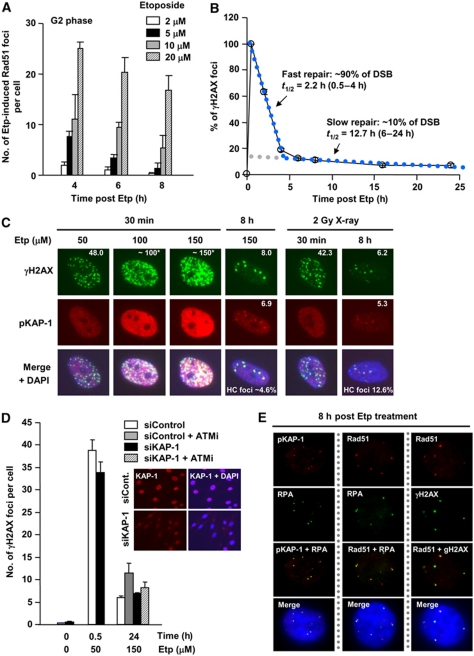
We reproducibly observed a significant number of Etp-induced Rad51 foci (∼10% of γH2AX foci at 15 min) in G2 cells (Figure 1C). To consolidate the notion that Etp induces a small percentage of Rad51 foci formation, we exposed cells to increasing Etp concentrations and enumerated Rad51 foci at varying times after exposure (Figure 2A). These results confirmed that Rad51 foci form in a dose-dependent manner after Etp treatment at a low percentage of DSBs. Since topoisomerase II introduces a uniform type of DSB end with a 4-bp 5′-overhang (Spitzner et al, 1995), we speculated that there must be a unique aspect that promotes DSB end resection at a subset of these DSBs. Given our finding that after X- or γ-rays, the slowly repaired HC-DSBs (∼15–25% of induced DSB) are preferentially repaired by HR in G2 phase, we examined whether the subset of Etp-induced DSBs that undergo resection might localise to HC regions. We used a high concentration of Etp to examine the kinetics of DSB repair in G1 cells and, importantly, observed biphasic kinetics (Figure 2B). (N.B. Etp treatment causes >10-fold more γH2AX foci in G2 than in G1, necessitating the use of higher concentrations to investigate G1 cells; G0/G1 cells were used to allow analysis up to 24 h after treatment). The fraction of slowly repaired DSBs was substantially lower than that induced by X-rays (∼90% of DSBs were repaired with the fast kinetics (t1/2=2.2 h); ∼10% were repaired more slowly (t1/2=12.7 h)) (Figure 2B; Supplementary Table S1). Moreover, there was a correlation between the fraction of slowly repaired DSBs and those harbouring RPA/Rad51 foci consistent with our proposed causal relationship (Figures 1C and 2B). To examine whether these might indeed represent HC-DSBs, we examined their co-localisation with pS824-KAP-1, a heterochromatic marker (Noon et al, 2010). Approximately 8–24 h following IR exposure, pS824-KAP-1 foci form uniquely at HC-DSBs in an ATM-dependent manner (Noon et al, 2010). We also confirmed that pKAP-1 foci do not form at all persistent DSBs by examining their formation in DSB repair defective, XLF cells (Supplementary Figure S4A). To facilitate quantification, we treated cells with a high concentration of Etp (150 μM) and observed ∼8 γH2AX foci and 6.9 pKAP-1 foci with strong co-localisation at 8 h, when the fast DSB repair process is completed (Figure 2C). The high number of DSBs present at 30 min after 150 μM Etp precluded an accurate estimation of DSB induction. However, exposure to 50 μM Etp gave ∼48 DSBs at 30 min, from which we estimated an induction level of 150 DSBs following 150 μM Etp assuming a dose-linear induction rate. Thus, a concentration of Etp that induced 150 DSBs yielded ∼7 DSBs, associating with pKAP-1 at 8 h (HC-DSBs); in contrast, 2 Gy IR induces ∼50 DSBs and ∼5 pKAP-1 foci at 8 h (Supplementary Table S1). Thus, the slow DSB repair component after Etp treatment is approximately three-fold lower compared with X-rays, correlating with the lower frequency of resection after Etp (Figures 1C and 2C). We further consolidated the notion that HC-DSBs form at a lower frequency after Etp compared with γ-rays using NIH3T3 cells, which have readily visualised dense DAPI regions, which represent HC regions (Supplementary Figure S4B). To substantiate that Etp generates HC-DSBs, we examined the requirement for ATM in DSB repair after Etp with or without KAP-1 siRNA, a treatment that has previously been shown to relieve the requirement for ATM for DSB repair (Goodarzi et al, 2008). We observed a small fraction of unrepaired DSBs following ATMi addition at 24 h after high-dose Etp treatment; further, the requirement for ATM could be relieved by KAP-1 siRNA (Figure 2D). Finally, we observed that RPA, Rad51 and γH2AX foci co-localise with pKAP-1 foci in G2 cells at 8 h after Etp treatment (Figure 2E). Collectively, these findings strongly suggest that Etp-induced DSBs that undergo end resection localise to HC regions as observed for IR-induced DSBs. Taken together, we suggest that two factors can influence the speed of DSB repair, DNA damage complexity and chromatin complexity, and that slowly repaired DSBs as a consequence of either damage or chromatin complexity preferentially undergo DSB end resection.
Figure 2.
Slowly repaired etoposide-induced DSBs localise to heterochromatin and undergo resection. (A) Rad51 foci arise following exposure to Etp for 30 min in G2 cells in a dose-dependent manner. Cells were exposed to increasing doses of Etp and Rad51 foci enumerated in HSF1 primary cells. The number of Rad51 foci in untreated cells (<2) was subtracted from results shown. (B) DSB repair kinetics in 48BR primary G0/G1 cells following exposure to 50 μM Etp for 30 min. Approximately 90% of DSBs are repaired with fast kinetics from 0.5 to 4 h (half life: 2.2 h); ∼10% of DSBs are repaired with slower kinetics from 6 to 24 h (half life: 12.7 h). Direct comparison of repair kinetics between Etp and IR is shown in Supplementary Table S1. (C) A low fraction of HC-associated DSBs arise following Etp treatment. 48BR primary G0/G1 cells were treated with 50, 100 and 150 μM Etp and foci enumerated at 30 min and 8 h. Total DSBs were detected with γH2AX foci, whereas HC-associated DSBs were detected with pKAP-1 foci. (N.B. after DSB induction at 30 min, pan-nuclear KAP-1 phosphorylation obscures the presence of pKAP-1 foci; thus foci were only scored at 8 h (Noon et al, 2010)). DSB induction was estimated from γH2AX present at 30 min assuming a dose-linear induction rate. The HC-associated DSBs at 8 h after Etp treatment was approximately three-fold lower than following X-ray exposure, 4.6 and 12.6%, respectively. (D) ATM-dependent DSB repair after Etp treatment is alleviated by KAP-1 siRNA. 48BR primary cells were exposed to 50 and 150 μM Etp for 30 min with or without ATMi. ATMi was readded after medium refreshing. γH2AX foci enumerated at 30 min and 24 h in G1 phase. Right panel shows KAP-1 knockdown efficiency. (E) RPA, Rad51 and γH2AX foci in Etp-treated G2 cells co-localise with the HC-DSB marker, pKAP-1. 48BR primary cells were exposed to 10 μM Etp and fixed 8 h after treatment. In these experiments (A–E), Etp was added for 30 min in medium at 37°C. Cells were washed with PBS three times before adding drug-free medium and cells were then incubated until the indicated time points.
DNA-PK competes with DSB end resection at all DSB ends
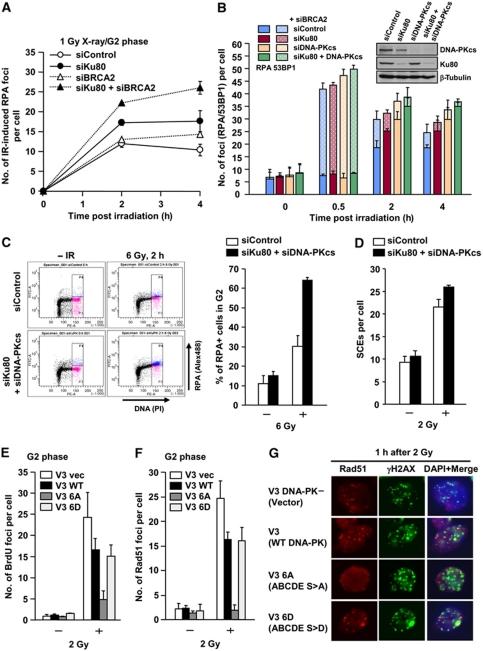
We next examined whether in the absence of NHEJ proteins, DSB ends destined for repair by NHEJ can undergo resection promoting a switch to HR. Previous studies involving I-Sce1-induced DSBs have suggested this but we aimed here to specifically examine pathway choice in the context of chromatin superstructure at two-ended DSBs in G2 phase cells (Pierce et al, 2001; Cui et al, 2005). To monitor all DSB end-resection events, we examined RPA foci formation following BRCA2 siRNA (Figure 3A). Consistent with the data in BRCA2-defective cells (Figure 1D), we observed a similar number of IR-induced RPA foci at 2 h with or without BRCA2 siRNA. BRCA2 siRNA cells, however, show sustained RPA foci (Figure 3A; Supplementary Figure S5A). Following Ku80 siRNA, we observed increased RPA foci at 2 and 4 h, demonstrating that more DSBs undergo resection (Figure 3A; Supplementary Figure S5A). To monitor the percentage of resected DSBs, we quantified the number of RPA and 53BP1 foci following Ku80 and/or DNA-PKcs siRNA plus BRCA2 siRNA (Figure 3B). In Ku and/or DNA-PKcs-defective cell lines, some DSB repair occurs by 2 h post-IR, which could reflect rejoining by back-up NHEJ or incomplete siRNA. However, of significance here, cells subjected to DNA-PKcs siRNA alone or in combination with Ku80 siRNA undergo enhanced end resection (Figure 3B). It is difficult to achieve efficient siRNA of Ku or DNA-PKcs, since both have abundant transcripts. Thus, the apparent additive impact of siRNA Ku and DNA-PKcs likely simply reflects the consequence of incomplete knockdown. Notwithstanding this, combined Ku80 and DNA-PKcs siRNA allowed >60–70% of induced DSBs to undergo resection, demonstrating that HR can occur at euchromatic (EC)-DSBs. Further, the results clearly show that loss of DNA-PKcs (which does not impact upon Ku80 protein stability or DNA-binding capacity) results in substantially increased DSB end resection (Drouet et al, 2005). We consolidated this finding using a FACS-based approach involving the detection of chromatin-bound RPA in G2 cells using α-RPA following detergent extraction. Consistent with our foci analysis, we observed increased RPA retention in G2 phase cells following Ku80+DNA-PKcs siRNA after IR (Figure 3C). Additionally, we observed increased IR-induced SCEs arising from G2 phase cells in Ku80+DNA-PKcs double-knockdown cells (Figure 3D). Thus, loss of either Ku or DNA-PKcs can enhance resection, demonstrating that the DNA-PK holoenzyme (Ku+DNA-PKcs) functions as a complex to ensure the appropriate regulation of resection at DNA ends.
Figure 3.
DNA-PK competes with DSB end resection. (A) Ku80 siRNA significantly increases RPA foci numbers post-IR. A549 cells were exposed to 1 Gy X-rays and RPA foci were enumerated as indicated either with or without BRCA2 siRNA. G2 cells were identified with CENP-F. Knockdown efficiency and typical images are shown in Supplementary Figure S5A. (B) Ku80 siRNA, DNA-PKcs siRNA or combined siRNA causes increased DSB end resection in A549 G2 cells. RPA and 53BP1 foci were enumerated after 1 Gy X-rays. Solid bars represent the number of RPA foci, and solid+hashed bars represent the total number of 53BP1 foci. The knockdown efficiencies are shown in the right panel. (C) Loss of Ku80 and DNA-PKcs increased IR-induced RPA retention in G2. RPA retention in G2 cells was analysed using α-RPA antibody after detergent extraction. Chromatin-associated RPA was detected as an Alex488 signal following FACS. APH alone does not induce detectable RPA signal by FACS (data not shown), although some level of RPA signal was detected by IF (Supplementary Figure S2). Percent of RPA positive was shown in the right panel. Error bars represent two independent experiments. (D) Increased IR-induced SCEs in Ku80 and DNA-PKcs double-knockdown G2 cells. IR-induced SCEs in G2 phase were analysed following 2 Gy X-rays. (E) A significant reduction of IR-induced BrdU foci formation in CHO cells expressing DNA-PKcs ABCDE S>A. In all, 20 μM BrdU was added 24 h before IR. Cells were extracted with 0.2% Triton for 1 min at 2 h after 2 Gy and stained with α-BrdU antibody without denaturation. (F, G) CHO cells expressing DNA-PKcs ABCDE S>A fail to form Rad51 foci at 1 h after 2 Gy X-rays in contrast to control cells and cells expressing the phosphorylation mimic, DNA-PKcs ABCDE S>D. G2 cells were identified by DAPI intensity using ImageJ. Similar results were obtained examining RPA foci (data not shown). DNA-PK expression levels in the V3 strains are shown in Supplementary Figure S5B. Further characterisation of the strains has been described previously (Chan and Lees-Miller, 1996; Cui et al, 2005; Meek et al, 2007).
The finding that there is increased DSB end resection in the presence of Ku but absence of DNA-PKcs implies that Ku can be removed from or vacates the DSB end to allow resection when NHEJ does not progress. To gain insight into this, we examined HR processing in a DNA-PKcs-defective cell line, V3, and derivatives expressing wild type (WT) DNA-PKcs, and DNA-PKcs mutants with alanine or aspartic acid substitutions at all six phosphorylation sites within the ABCDE autophosphorylation cluster (2609–2647) on DNA-PKcs (DNA-PKcs ABCDE S>A or S>D, respectively). It has been reported that the DNA-PKcs ABCDE S>A mutant binds DSBs but is released more slowly than WT DNA-PKcs. Cells expressing ABCDE S>A are substantially more radiosensitive than cells lacking DNA-PKcs, suggesting a dominant negative impact (Chan and Lees-Miller, 1996; Cui et al, 2005). First, V3 cells expressing WT DNA-PKcs form less ssDNA, as detected using α-BrdU antibody, and have fewer Rad51 foci compared with V3 cells expressing empty vector (Figure 3E and F) consistent with the findings in Figure 3B. Strikingly, cells expressing DNA-PKcs ABCDE S>A fail to form ssDNA, as detected using α-BrdU antibody (Figure 3E). Additionally, both Rad51 and RPA foci formation after IR were significantly reduced in cells expressing DNA-PKcs ABCDE S>A in contrast to cells expressing the phosphorylation mimic, DNA-PKcs ABCDE S>D (Figure 3F and G and data not shown). This result provides strong evidence that DNA-PK initially binds to HC-DSBs as well as EC-DSBs, suggesting that the Ku/DNA-PKcs complex is the dominant pathway that either binds all DSBs initially or ‘wins’ the competition because it is highly expressed. This strongly suggests that NHEJ makes an initial attempt to repair DSBs but that resection occurs if NHEJ cannot rapidly ensue.
CtIP-dependent DSB end resection determines the usage of HR
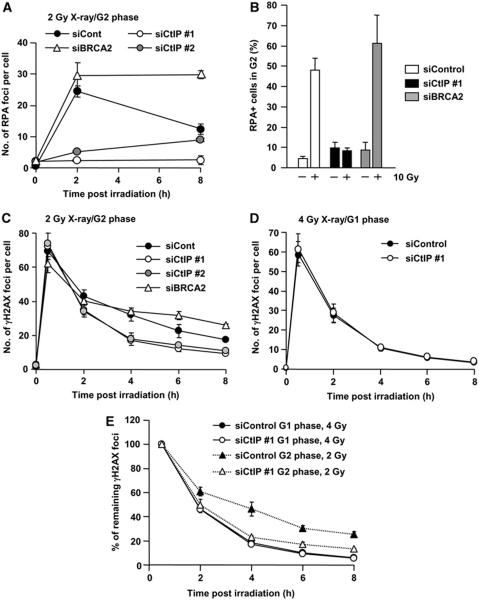
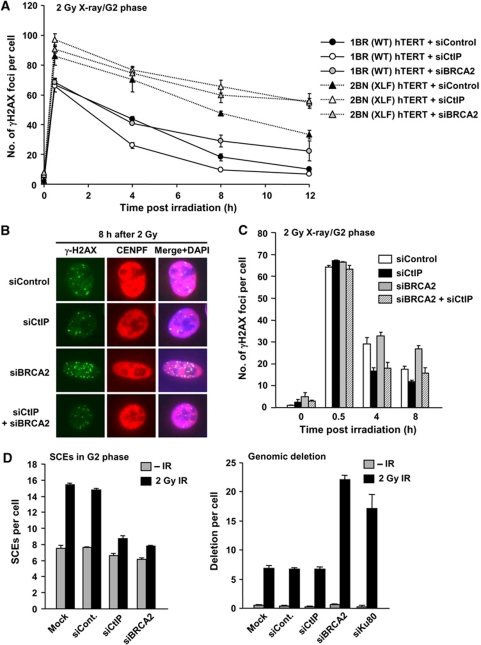
The above section provides evidence that in the absence of DNA-PK, DSBs normally destined for repair by NHEJ undergo resection and processing for HR. Here, we address the converse question; can DSBs normally repaired by HR undergo repair by NHEJ. We inhibit HR either by CtIP siRNA, which is required to initiate DSB end resection or by BRCA2 siRNA, which is dispensable for resection but required for Rad51 loading (Yuan et al, 1999; Yang et al, 2002). Consistent with previous reports, CtIP siRNA abolished IR-induced RPA/Rad51 foci formation and reduced Chk1 phosphorylation, which requires ATR activation at resected DSBs, but did not affect ATM signalling monitored by p1981-ATM, pChk2 and pKAP-1 (Figure 4; Supplementary Figure S6). The significant reduction of RPA foci numbers in CtIP-depleted cells was consolidated using FACS analysis to monitor RPA chromatin binding in G2 phase cells (Figure 4B). We next examined whether CtIP siRNA affects the rate of DSB repair by enumerating γH2AX foci in G2 cells post-IR. Surprisingly, we observed a normal rate of DSB repair between 0.5 and 2 h but faster kinetics between 4 and 8 h compared with control cells (Figure 4C). In contrast, BRCA2 siRNA showed the anticipated DSB repair defect in G2 at 6–8 h (Beucher et al, 2009) (Figure 4C). HR is a slower DSB repair pathway compared with NHEJ (Mao et al, 2008). We therefore speculated that following CtIP siRNA in G2, DSB repair switches from HR to NHEJ. To examine this, we compared DSB repair kinetics in G1 and G2 cells with or without CtIP siRNA using 4 and 2 Gy for G1 and G2 cells, respectively, to achieve similar DSB induction levels (Figure 4C–E). Strikingly, following CtIP siRNA, DSB repair in G2 showed similar kinetics compared to that observed in G1 phase cells. CtIP siRNA did not affect the rate of repair in G1 phase (Figure 4D). To substantiate that DSB repair occurs by NHEJ after CtIP siRNA in G2 cells, we examined CtIP siRNA or BRCA2 siRNA in XLF-defective (2BN) hTERT cells and observed an additive repair defect (Figure 5A). Thus, even though CtIP siRNA alone results in efficient repair with faster kinetics in G2, this is abolished when NHEJ fails to function. The finding that faster DSB repair kinetics in G2 are observed following CtIP siRNA but not following BRCA2 siRNA suggested that resection may restrict the ability to exploit NHEJ. We, therefore, examined CtIP siRNA+BRCA2 siRNA and observed that CtIP siRNA alleviated the BRCA2-dependent repair defect in G2 (Figure 5B and C). This interesting result suggests that resection, at least following BRCA2 siRNA, precludes the possibility to utilise NHEJ for DSB repair. Finally, we verified that following CtIP siRNA, DSBs are repaired by NHEJ using SCE formation as a monitor of HR, exploiting a method we developed previously to examine specifically SCEs arising following irradiation of G2 cells (Beucher et al, 2009). Both CtIP and BRCA2 siRNA abolished IR-induced SCE formation (Figure 5D, left panel). Examination of chromosome deletions, which are detectable in metaphase preparations, revealed a marked increase in deletion events following Ku80 or BRCA2 siRNA, but not following CtIP siRNA, consolidating the notion that DSB repair after CtIP siRNA occurs by NHEJ (Figure 5D, right panel).
Figure 4.
CtIP siRNA-treated G2 cells show faster DSB repair kinetics compared with control cells but similar to G1 phase DSB repair. (A) CtIP siRNA treatment abolishes DSB end resection, whereas RPA foci levels are sustained following BRCA2 siRNA. A549 cells were treated with control, BRCA2 or CtIP siRNA. RPA foci were enumerated after 2 Gy X-rays. Similar results were obtained using two distinct CtIP siRNA oligonucleotides. (B) Cells subjected to CtIP siRNA show impaired IR-induced RPA retention. RPA retention was analysed as described in Figure 3C. Consistent with RPA foci data, CtIP siRNA cells show loss of RPA signal. (C) CtIP siRNA cells show faster repair kinetics at 4–8 h compared with control cells, whereas BRCA2 siRNA cells show a DSB repair defect. Following exposure to 2 Gy X-rays, DSB repair kinetics in A549 G2 cells was measured by enumerating γH2AX foci. G2 cells were identified using CENP-F. (D) The kinetics of γH2AX disappearance is unaffected by CtIP siRNA after 4 Gy X-rays. A549 G1 cells were identified as CENP-F negative. (E) Direct comparison of DSB repair kinetics in A549 G1 and G2 cells following control and CtIP siRNA. CtIP siRNA in G2 results in similar repair kinetics to G1 phase, suggesting that DSB repair in G2 following CtIP siRNA occurs by NHEJ. Note that even though all DSBs are repaired by NHEJ, fast and slow kinetics are observed since HC-DSBs are repaired slowly even in G1. In all panels, error bars represent the mean and s.d. from three independent experiments except panel (B), which is from two experiments.
Figure 5.
CtIP siRNA allows repair to be switched from HR to NHEJ. (A) CtIP siRNA and BRCA2 siRNA causes a similar DSB repair defect in XLF-defective cells following IR. DSB repair was monitored by γH2AX foci enumeration after 2 Gy X-rays using the treatments and/or cells indicated. As in Figure 4, CtIP siRNA shows faster repair, whereas BRCA2 siRNA confers a repair defect in WT G2 cells. In contrast, CtIP or BRCA2 siRNA in XLF cells showed a similar DSB repair defect, demonstrating that DSB repair following CtIP siRNA occurs by NHEJ. (B, C) CtIP siRNA alleviates the BRCA2-dependent repair defect in A549 G2 cells after 2 Gy X-rays. The slightly higher γH2AX foci numbers following combined BRCA2 and CtIP siRNA than following CtIP siRNA alone at 8 h is probably a result of imperfect siRNA efficiency. (D) CtIP or BRCA2 siRNA inhibits IR-induced SCEs. Ku80 or BRCA2 siRNA but not CtIP siRNA causes increased deletion events. IR-induced SCEs in G2 phase were analysed following 2 Gy X-rays (left panel). Genomic deletions were examined in metaphase preparations following 2 Gy X-rays (right panel).
Collectively, these results suggest that whereas loss of NHEJ allows DSB repair to ensue by HR, CtIP-dependent DSB end resection precludes a switch to NHEJ. In contrast, a failure to effect CtIP-dependent resection allows DSB repair to ensue by NHEJ.
Roles for ATM in regulating HR
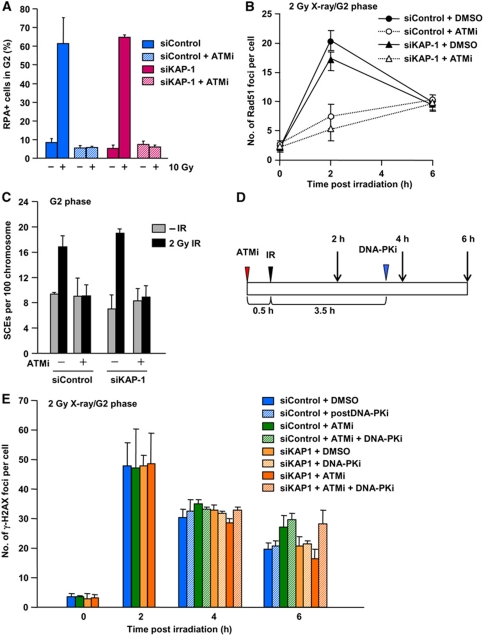
We next examined ATM’s role in regulating HR in G2 phase. Since the slow DSB repair component represents the repair of HC-DSBs and since this process represents HR in G2 phase, this implies that one role of ATM that promotes HR is the phosphorylation of KAP-1, which is required to allow HC-DSB repair to ensue (Goodarzi et al, 2008; Beucher et al, 2009). Consistent with this, we found that KAP-1 siRNA alleviates the ATM-dependent repair defect in G1 and G2 cells (Beucher et al, 2009). However, KAP-1 siRNA in the presence of ATMi did not rescue RPA retention, Rad51 foci formation or IR-induced SCEs, suggesting that repair likely occurs by NHEJ as observed following CtIP siRNA (Figure 6A–C). To examine whether NHEJ is utilised following KAP-1 siRNA+ATMi treatment, we added the DNA-PK inhibitor, DNA-PKi, at 3.5 h after 1 Gy X-rays, a time when most NHEJ but not HR is completed (Figure 6D and E). As expected, this DNA-PKi treatment did not affect DSB repair in control cells since most NHEJ is completed by 3.5 h; further it did not cause any additive impact following ATMi treatment. In contrast, cells subjected to KAP-1 siRNA+ATMi+DNA-PKi showed the same DSB repair defect as observed in ATMi-treated cells although KAP-1 siRNA+ATMi allowed normal repair kinetics (Figure 6E). This strongly suggests that DNA repair following KAP-1 siRNA+ATMi occurs by NHEJ. These results raise the possibility that in addition to phosphorylating KAP-1 to allow HC remodelling, ATM has another role in promoting the end resection step of HR.
Figure 6.
KAP-1 siRNA does not rescue DSB end resection nor IR-induced SCEs in the presence of ATMi, but DSB repair occurs by NHEJ. (A) ATMi treatment abolishes RPA retention after IR. Additionally, KAP-1 siRNA does not restore RPA retention in the presence of ATMi. Error bars represent the s.d. from two independent experiments. (B) ATMi treatment significantly reduced Rad51 foci numbers after X-ray exposure irrespective of KAP-1 status. Similar results were obtained for RPA foci (data not shown). (C) ATMi treatment abolishes IR-induced SCEs in G2 irrespective of KAP-1 status. IR-induced SCEs in G2 phase were analysed following KAP-1 siRNA with/without ATMi after 2 Gy X-rays. KAP-1 siRNA does not restore Rad51 or SCE formation in the presence of ATMi. (D) Diagram showing the procedure used to examine the role of NHEJ in DSB repair. A549 cells were treated with a DNA-PK inhibitor (DNA-PKi) at 3.5 h post-IR, when most NHEJ-dependent DSB repair is completed after 2 Gy X-rays. (E) IR-induced DSBs are repaired by NHEJ following KAP-1 siRNA in the presence of ATMi. A549 cells were exposed to 2 Gy X-rays. ATMi-treated cells showed a repair defect. Addition of DNA-PKi at 3.5 h post-IR does not affect DSB repair in a control background. KAP-1 siRNA alleviates ATM-dependent DSB repair in G2 (i.e. enhanced repair is observed following KAP-1 siRNA+ATMi) (despite the lack of Rad51 foci formation and SCEs). DNA-PKi addition at 3.5 h prevents the enhanced repair observed following KAP-1 siRNA+ATMi between 4 and 6 h, indicating that the DSBs are repaired by NHEJ.
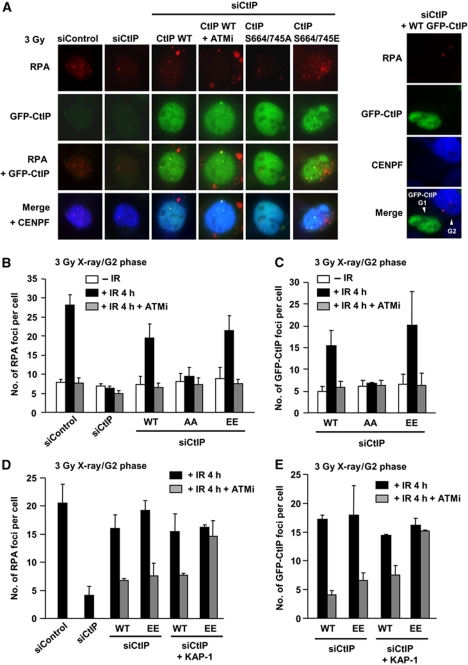
The ability to exploit NHEJ in place of HR was also observed following CtIP siRNA in control cells raising the possibility that ATM is required for CtIP function at DSBs in G2. CtIP is a CDK substrate and CDK-dependent CtIP phosphorylation is required for DSB end resection in S/G2 (Huertas et al, 2008). CtIP has previously been reported to be phosphorylated at S664/S745 by ATM (Li et al, 2000). To examine whether this phosphorylation might also be required for end resection, we overexpressed siRNA-resistant GFP-CtIP in CtIP siRNA knockdown cells. At 4 h post-IR, RPA and GFP-CtIP foci were observed in GFP-CtIP expressing G2 but not G1 cells (Figure 7A). Treatment with ATMi prevented GFP-CtIP as well as RPA foci formation (Figure 7A–C). We next examined the impact of expressing CtIP with alanine mutations at the two identified ATM phosphorylation sites. Interestingly, GFP-CtIP S664/745A expressing cells failed to form both GFP-CtIP and RPA foci post-IR independently of ATMi treatment. In marked contrast, expression of a phosphomimic form of CtIP, GFP-CtIP S664/745E, resulted in normal GFP-CtIP and RPA foci formation but failed to form GFP-CtIP and RPA foci following ATMi treatment. This could arise if ATM has two roles in resection, HC relaxation and CtIP phosphorylation. To examine this possibility, we carried out KAP-1 siRNA in CtIP S664/745E expressing cells (Figure 7D and E). Significantly, KAP-1 siRNA restored RPA and GFP-CtIP foci formation in ATMi-treated cells expressing GFP-CtIP S664/745E but not in ATMi-treated cells expressing WT CtIP. We conclude that ATM has dual functions in DSB end resection. First, ATM facilitates HC remodelling via KAP-1 phosphorylation, which is required to allow CtIP resection and recruitment/retention at HC-DSBs. Second, ATM has a more direct role in resection activating CtIP via phosphorylation at S664/745.
Figure 7.
ATM-dependent CtIP phosphorylation is required for DSB end resection in G2 cells. (A) Typical images of RPA and GFP-CtIP foci at 4 h in U2OS G2 cells after 3 Gy X-rays. In contrast, GFP-CtIP expressed in G1 cells does not form either RPA or GFP-CtIP foci (right panel). CtIP mutant proteins were expressed following CtIP siRNA. (B, C) CtIP siRNA abolished IR-induced RPA foci formation. GFP-CtIP was overexpressed in U2OS cells. GFP-CtIP S664/745A expressing cells fail to form RPA and GFP-CtIP foci after 3 Gy X-rays, whereas wild type and S664/745E GFP-CtIP overexpression rescued RPA/GFP-CtIP foci formation. (D, E) KAP-1 siRNA alleviates loss of GFP-CtIP and RPA foci formation in S664/745E mutant in the presence of ATMi following exposure to 3 Gy X-rays. GFP-CtIP was overexpressed in U2OS cells. KAP-1 siRNA does not affect GFP-CtIP or RPA foci formation in CtIP WT expressing cells, whereas it relieves DSB end resection in S664/745E expressing cells.
Discussion
DSB lesion and chromatin complexity influence pathway choice
Here, we examine factors that influence the choice between NHEJ and HR in G2 phase. We recently provided evidence that chromatin complexity influences repair pathway choice since X-ray-induced DSBs located at regions of HC predominantly undergo repair by HR (Beucher et al, 2009). Here, we show that DNA lesion complexity is a second factor influencing that decision since highly complex DSBs induced by high LET radiation have a greater propensity to undergo resection and Rad51 loading, whereas Etp-induced DSBs are predominantly repaired by NHEJ. Consistent with this notion, very few carbon-ion-induced DSBs undergo repair in BRCA2-defective cells, whereas only a minor repair defect is observed following Etp treatment. Furthermore, although most Etp-induced DSBs are repaired rapidly, a small fraction located within HC is repaired slowly and undergoes resection. It is currently unclear whether the lower fraction of slowly repaired DSBs induced by Etp compared with X-rays is a consequence of a lower induction level of HC-DSBs following Etp treatment, since topoII primarily targets active genes. Importantly, in all cases, the lesions that undergo resection and repair by HR are those repaired with slow kinetics in G1 phase. Collectively, our findings provide strong evidence that the speed of DSB repair influences pathway choice in G2 phase.
Relationship between NHEJ and HR factors and pathway choice
Our findings argue that NHEJ is the preferred pathway when DSB repair proceeds rapidly raising the possibility that NHEJ makes the first attempt to repair DSBs in G2 phase. We, therefore, examined how loss or stalling of NHEJ influences the ability to undergo HR and conversely, how loss or stalling of HR influences NHEJ usage. Although these studies have to some degree been previously undertaken following I-Sce1 cleavage, here we examine pathway choice in a setting that encompasses the range of chromatin complexity. siRNA of Ku or DNA-PKcs promotes elevated DSB end resection compared to that observed in control cells, consistent with previous models that NHEJ competes with HR albeit differently in mammalian versus chicken cells (Fukushima et al, 2001; Pierce et al, 2001). Since complete loss of Ku or DNA-PKcs is rarely achieved by siRNA (due to high mRNA expression), we have not attempted to closely dissect the rate of resection. Further, we and others observed that a back-up NHEJ pathway, designated B-NHEJ, can occur in the absence of Ku but not in the absence of DNA-PKcs (Wang et al, 2006) (data not shown) potentially providing an additional competing pathway. Notwithstanding these limitations, we observed that ∼60–70% of induced DSBs undergo resection following DNA-PKcs siRNA, which was similar to or even greater than that observed following Ku80 siRNA. This demonstrates that resection can occur at EC-DSBs. This notion is further supported by our finding that nearly all carbon-ion-induced DSBs, encompassing EC-DSBs and HC-DSBs, are resected. Further, the results show that although Ku completely blocks B-NHEJ, it does not block resection. Indeed, Ku binding but a failure to complete NHEJ efficiently due to loss of DNA-PKcs enhances resection. Importantly, expression of a DNA-PKcs mutant that fails to undergo autophosphorylation at the ABCDE cluster (DNA-PKcs ABCDE 6A) dramatically impairs resection at 1 h post-IR. Although some resection may occur at later times post-IR (data not shown), the significance here is that impairing DNA-PK dissociation from the DSB end dramatically impairs the initiating step of HR. These findings are consistent with a previous report that DNA-PKcs ABCDE S>A can inhibit HR resulting in increased radiosensitivity and sensitivity to MMC (Cui et al, 2005). Our findings here show that inhibition of HR can occur at HC-DSBs arguing that NHEJ represents the dominant initial pathway with the Ku/DNA-PKcs complex arriving first at all lesions either because it is abundantly expressed or has avid DSB end-binding capacity.
Examination of how a failure to undergo the initiating or a downstream step in HR impacts upon NHEJ was also revealing. Although CtIP siRNA dramatically impaired resection, DSB rejoining occurs efficiently and indeed more rapidly than in control cells. To verify that this represents NHEJ, we show that CtIP siRNA confers an additive repair defect following XLF siRNA, an NHEJ protein. Consistent with this, we observed that in G2 phase, the slow component of DSB repair occurs with subtly slower kinetics compared with G1 phase cells, which is most apparent when the dose is adjusted to yield similar DSB levels in G1 and G2 phase cells (since G2 phase cells have a 4N DNA content). Finally, we show that when HR cannot progress beyond the resection step due to loss of BRCA2, then NHEJ cannot rejoin the DSB. This suggests that excessive resection due to loss of BRCA2 precludes the ability to utilise NHEJ. Resection is not a simple process and requires the concerted actions of several nucleases. Thus, although it is currently unclear which precise resection step confers a commitment to HR and precludes NHEJ usage, our results strongly demonstrate that loss of CtIP, an early step in the resection process, is compatible with NHEJ activity.
Collectively, our findings show that two factors influence the speed of DSB repair, DNA lesion and chromatin complexity. These factors consequently influence whether HR or NHEJ is used for DSB repair. Additionally, in G2 phase, the pathway utilised influences the repair kinetics. Thus, in a WT cell, the slowly repaired DSBs undergo repair by HR, conferring slightly slower kinetics to the ‘slow’ repair component compared with G1 phase. This aspect of the speed of DSB repair, however, is a consequence of, rather than influencing, pathway choice.
NHEJ is the pathway of first choice
Based on these findings, we propose that DNA-PK binds rapidly to all DSBs, either because Ku has strong DSB-binding capacity or because it is highly abundant, and NHEJ makes a first attempt at repair. If the subsequent steps of NHEJ can progress, then NHEJ rapidly and efficiently repairs the DSB. However, if rapid rejoining cannot ensue either due to DNA lesion or chromatin complexity, then DSB end resection and HR occurs in G2 phase (Supplementary Figure S7). If resection cannot ensue, then NHEJ makes a further attempt to repair the DSB and, indeed, can function efficiently. However, resection or possibly a later stage in the progression to HR commits to HR and precludes the possibility of returning to NHEJ usage. We present indirect and direct evidence to support this. We show that the magnitude of NHEJ or HR usage depends upon the DSB-inducing agent; nearly all Etp-induced DSBs are repaired by NHEJ; conversely nearly all carbon-ion-induced DSBs undergo resection. This argues against a simple competition model, which might be expected to be independent of the type of DSB. Informatively, at HC-DSBs, although DNA-PK can initially bind (since DNA-PKcs ABCDE 6A mutant blocks resection), NHEJ does not rapidly ensue and resection occurs. However, if resection cannot take place due to loss of CtIP, then NHEJ can proceed and, indeed repair occurs more rapidly than by HR. It is noteworthy that in G1 phase, the HC-DSBs are repaired with slower kinetics than the EC-DSBs, demonstrating that the HC superstructure provides a barrier that slows the rate of repair (even though both are repaired by a process requiring NHEJ proteins). This supports the notion that it is the restriction to the progress of NHEJ posed by HC structure that promotes resection in G2 phase. Another informative finding is that elevated resection occurs following loss of DNA-PKcs (but presence of Ku). Thus, resection can occur in the presence of Ku. FRAP studies have suggested that Ku binding is dynamic at DSB ends and it is possible that Ku/DNA-PKcs is dynamically bound and released at the DNA end allowing a ‘competition’ with resection (Mari et al, 2006; Uematsu et al, 2007). Most importantly, as direct evidence that NHEJ proteins make the first attempt at DSB repair, we show that a mutant form of DNA-PKcs can prevent efficient resection at HC-DSBs providing direct evidence that DNA-PK binds to these ends. Thus, collectively, our data strongly suggest that NHEJ is the first choice pathway.
Roles for ATM in regulating HR
Finally, we examine the role of ATM in the regulation of pathway usage. We have previously shown that ATM phosphorylates KAP-1 at HC-DSBs to promote their relaxation (Goodarzi et al, 2008). Most significantly, in G1 and G2, KAP-1 siRNA allows DSB repair to ensue in the absence of ATM activity—that is it overcomes the requirement for ATM for DSB repair. Here, we show that although DSB repair occurs in ATMi+KAP-1 siRNA-treated G2 cells, DSB end resection is severely impaired. This strongly suggests that ATM has an additional role in HR in G2 phase. We show that this role represents the phosphorylation of CtIP. Thus, we observe reduced resection in CtIP S664/745A expressing cells; normal resection in cells expressing the phosphomimic form, GFP-CtIP S664/745E, but a failure to undergo resection under any of these situations in the presence of ATMi. Although KAP-1 siRNA did not relieve the resection defect caused by ATMi treatment in CtIP WT cells, it did promote resection following expression of the GFP-CtIP S664/745E. Thus, we conclude that HC superstructure poses a barrier to resection but that ATM-dependent CtIP phosphorylation is also required. The precise role of CtIP phosphorylation is unclear but it could regulate interaction with BRCA1 or the MRN complex (Li et al, 2000; Sartori et al, 2007) since CtIP interacts with both in G2 phase (Yu and Chen, 2004). In conclusion, we demonstrate that ATM has at least two roles in promoting resection, and hence HR, in G2 phase: (1) it relieves a barrier posed by the HC superstructure by phosphorylating KAP-1 and (2) it activates CtIP via phosphorylation of the S664/745 sites. The former represents a chromatin modifying role rather than the direct regulation of an HR step. The latter, however, represents a direct role for ATM in HR in G2 phase.
Materials and methods
Cell culture, X-irradiation and drug treatment
48BR and HSF1, human primary control fibroblasts, HSC62 and 2BN, defective in BRCA2 and XLF, respectively, were cultured in high-glucose Dulbecco's modification of Eagles medium with 15% fetal calf serum (FCS). DNA-PK CHO cell lines were provided by Dr S Lees-Miller. A549, U2OS and CHO cells were cultured in minimal essential medium (MEM) with 10% FCS. In all, 120 kV X-rays were delivered at 12 mA with aluminium filter (dose rate 0.5 Gy/min). Since X-rays cause an increase in dose for cells grown on glass cover slips relative to plastic surfaces, this was taken into account in the dose assessment (Kegel et al, 2007). Cells were treated with Etp (Sigma-Aldrich, Poole, UK) for 15 (experiments in Figure 1) or 30 min (all other experiments) in MEM at 37°C, then washed with PBS three times before adding fresh medium. The ATM inhibitor, KU55933, and the DNA-PK inhibitor, NU7441, were gifts from KuDOS Pharmaceuticals (Cambridge, UK). In all, 10 μM KU55933 was added 30 min before irradiation. In all, 10 μM NU7441 was added as stated. Etp was added to medium for 15 or 30 min at 37°C in Figures 1 and 2, respectively. Cells were then washed with PBS three times before adding drug-free medium and incubating as required.
Carbon-ion irradiation
Irradiation was done at the UNILAC facility at the GSI Helmholtzzentrum Schwerionenforschung (GSI, Darmstadt, Germany) with low-energy carbon ions (9.8 MeV/nucleon; LET 170 keV/μm on target). Cells grown on glass coverslips were mounted in 3 cm petri dishes with sterile vaseline and placed in a multi-sample rack. Irradiation was performed at an angle of ∼5° to generate linear stripes of γH2AX and RPA foci after immunodetection along the trajectory of the carbon ions.
siRNA knockdown and CtIP overexpression
siRNA transfection of 48BR, A549 and U2OS cells was undertaken using Hiperfect (Qiagen, Hilden, Germany). siRNA oligonucleotides for scrambled control, Ku80, DNA-PKcs, BRCA2, CtIP #1 were from the Dharmacon SMARTpool siRNA. The CtIP siRNA oligonucleotide used in the overexpression experiment is 5′-AAGCTAAAACAGGAACGAATC-3′ (designated CtIP #2). The KAP-1 siRNA oligonucleotide is 5′-CAGTGCTGCACTAGCTGTGAGGATA-3′. siRNA was carried out in suspended cells after trypsinisation. After 24 h, cells were retransfected with siRNA in suspended cells after trypsinisation. Cells were incubated for 48 h after the second transfection before analysis. Transient CtIP overexpression was carried out in U2OS cells by NanoJuice™ transfection kit (Novagen) at 48 h after the second siRNA transfection. DNA-transfection reagent mixture was added for 3 h, then removed and the MEM refreshed. After 24 h, cells were irradiated and processed for RPA and CtIP foci.
RPA retention assay using fluorescence-activated cell sorting
Cells were irradiated 48 h post-siRNA transfection. APH was added immediately after IR to block S to G2 progression. APH alone does not induce detectable RPA signal by FACS (data now shown). Irradiated cells were trypsinised and washed three times, and suspended cells were permeabilised using 0.2% Triton in PBS for 2 min on ice, followed by fixation with 3% PFA–2% sucrose. After incubation with primary α-RPA mouse antibody and Alex488-conjugated α-mouse secondary antibody for 45 min each at 37°C, cells were washed and resuspended in propidium iodide with RNase A (Sigma-Aldrich). Cells were analysed using a fluorescence-activated cell sorter (FACSCanto, BD Biosciences) with FACS Diva software. G2 cells were identified on the basis of their DNA content assessed by PI staining.
Antibodies for immunofluorescence and immunoblotting
Antibodies used were γH2AX (Upstate Biotechnology, Buckingham, UK), CENP-F (Abcam, Cambridge, UK), RPA (Calbiochem), Rad51 (Santa Cruz, Santa Cruz, CA), Ku80 (polyclonal rabbit antibody), BRCA2 (Calbiochem), β-tubulin (Abcam) and BrdU (BD Biosciences). α-pKAP-1 S824 was described previously (Noon et al, 2010). γH2AX foci and cell-cycle phase analysis was carried out as described previously (Beucher et al, 2009). Foci scoring was carried out blindly with >800 foci/sample being scored. Unless stated otherwise, all foci analysis represents the mean and s.d. of three experiments.
Analysis of sister chromatid exchanges and genomic deletions
SCEs analysis was carried out as previously described (Beucher et al, 2009). Briefly, cells were grown for 48 h in BrdU before IR exposure. Colcemid and 1 mM caffeine were added at 6 or 8 h until 9 or 12 h post-IR to collect cells in mitosis. Genomic deletions (chromatid breaks and gaps) and SCEs were scored in at least 100 metaphases from three independent experiments.
Supplementary Material
Acknowledgments
The ML laboratory is supported by the Deutsche Forschungsgemeinschaft (Lo 677/4-3) and the Bundesministerium für Bildung und Forschung (02S8335, 02S8355 and 03NUK001C). The PAJ laboratory is supported by the Medical Research Council, the Association for International Cancer Research, Department of Health and the Wellcome Research Trust.
Author contributions: AS, PJ and ML designed the experiments and wrote the paper. SC and VG performed repair analysis after carbon ions. OV made the CtIP overexpression vector. JB and AI participated in DSB repair analysis. AK performed SCE assay. KM established DNA-PKcs V3 cell lines. GTS provided the heavy ion irradiation source. AS oversaw and participated in all experiments except the carbon-ion analysis
Footnotes
The authors declare that they have no conflict of interest.
References
- Aylon Y, Liefshitz B, Kupiec M (2004) The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J 23: 4868–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beucher A, Birraux J, Tchouandong L, Barton O, Shibata A, Conrad S, Goodarzi AA, Krempler A, Jeggo PA, Lobrich M (2009) ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J 28: 3413–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DW, Lees-Miller SP (1996) The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. J Biol Chem 271: 8936–8941 [DOI] [PubMed] [Google Scholar]
- Cui X, Yu Y, Gupta S, Cho YM, Lees-Miller SP, Meek K (2005) Autophosphorylation of DNA-dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Mol Cell Biol 25: 10842–10852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouet J, Delteil C, Lefrancois J, Concannon P, Salles B, Calsou P (2005) DNA-dependent protein kinase and XRCC4-DNA ligase IV mobilization in the cell in response to DNA double strand breaks. J Biol Chem 280: 7060–7069 [DOI] [PubMed] [Google Scholar]
- Fukushima T, Takata M, Morrison C, Araki R, Fujimori A, Abe M, Tatsumi K, Jasin M, Dhar PK, Sonoda E, Chiba T, Takeda S (2001) Genetic analysis of the DNA-dependent protein kinase reveals an inhibitory role of Ku in late S-G2 phase DNA double-strand break repair. J Biol Chem 276: 44413–44418 [DOI] [PubMed] [Google Scholar]
- Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA (2008) ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell 31: 167–177 [DOI] [PubMed] [Google Scholar]
- Hada M, Georgakilas AG (2008) Formation of clustered DNA damage after high-LET irradiation: a review. J Radiat Res (Tokyo) 49: 203–210 [DOI] [PubMed] [Google Scholar]
- Helleday T, Lo J, van Gent DC, Engelward BP (2007) DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair (Amst) 6: 923–935 [DOI] [PubMed] [Google Scholar]
- Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP (2008) CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455: 689–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kegel P, Riballo E, Kuhne M, Jeggo PA, Lobrich M (2007) X-irradiation of cells on glass slides has a dose doubling impact. DNA Repair (Amst) 6: 1692–1697 [DOI] [PubMed] [Google Scholar]
- Li S, Ting NS, Zheng L, Chen PL, Ziv Y, Shiloh Y, Lee EY, Lee WH (2000) Functional link of BRCA1 and ataxia telangiectasia gene product in DNA damage response. Nature 406: 210–215 [DOI] [PubMed] [Google Scholar]
- Lieber MR (2010) The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 79: 181–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Bozzella M, Seluanov A, Gorbunova V (2008) Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair (Amst) 7: 1765–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari PO, Florea BI, Persengiev SP, Verkaik NS, Bruggenwirth HT, Modesti M, Giglia-Mari G, Bezstarosti K, Demmers JA, Luider TM, Houtsmuller AB, van Gent DC (2006) Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc Natl Acad Sci USA 103: 18597–18602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek K, Douglas P, Cui X, Ding Q, Lees-Miller SP (2007) Trans autophosphorylation at DNA-dependent protein kinase's two major autophosphorylation site clusters facilitates end processing but not end joining. Mol Cell Biol 27: 3881–3890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noon AT, Shibata A, Rief N, Lobrich M, Stewart GS, Jeggo PA, Goodarzi AA (2010) 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol 12: 177–184 [DOI] [PubMed] [Google Scholar]
- Pierce AJ, Hu P, Han M, Ellis N, Jasin M (2001) Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev 15: 3237–3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riballo E, Kuhne M, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, Parker AR, Jackson SP, Gennery A, Jeggo PA, Lobrich M (2004) A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol Cell 16: 715–724 [DOI] [PubMed] [Google Scholar]
- Ritter S, Durante M (2010) Heavy-ion induced chromosomal aberrations: a review. Mutat Res 701: 38–46 [DOI] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP (2007) Human CtIP promotes DNA end resection. Nature 450: 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastav M, De Haro LP, Nickoloff JA (2008) Regulation of DNA double-strand break repair pathway choice. Cell Res 18: 134–147 [DOI] [PubMed] [Google Scholar]
- Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S (2006) Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair (Amst) 5: 1021–1029 [DOI] [PubMed] [Google Scholar]
- Spitzner JR, Chung IK, Gootz TD, McGuirk PR, Muller MT (1995) Analysis of eukaryotic topoisomerase II cleavage sites in the presence of the quinolone CP-115,953 reveals drug-dependent and -independent recognition elements. Mol Pharmacol 48: 238–249 [PubMed] [Google Scholar]
- Tobias F, Durante M, Taucher-Scholz G, Jakob B (2010) Spatiotemporal analysis of DNA repair using charged particle radiation. Mutat Res 704: 54–60 [DOI] [PubMed] [Google Scholar]
- Uematsu N, Weterings E, Yano K, Morotomi-Yano K, Jakob B, Taucher-Scholz G, Mari PO, van Gent DC, Chen BP, Chen DJ (2007) Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J Cell Biol 177: 219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G (2006) PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res 34: 6170–6182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman C, Kanaar R (2006) DNA double-strand break repair: all's well that ends well. Annu Rev Genet 40: 363–383 [DOI] [PubMed] [Google Scholar]
- Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, Zheng N, Chen PL, Lee WH, Pavletich NP (2002) BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science 297: 1837–1848 [DOI] [PubMed] [Google Scholar]
- Yu X, Chen J (2004) DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol Cell Biol 24: 9478–9486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan SS, Lee SY, Chen G, Song M, Tomlinson GE, Lee EY (1999) BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res 59: 3547–3551 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.