Abstract
Chromatin domains are believed to spread via a polymerization-like mechanism in which modification of a given nucleosome recruits a modifying complex, which can then modify the next nucleosome in the polymer. In this study, we carry out genome-wide mapping of the Sir3 component of the Sir silencing complex in budding yeast during a time course of establishment of heterochromatin. Sir3 localization patterns do not support a straightforward model for nucleation and polymerization, instead showing strong but spatially delimited binding to silencers, and weaker and more variable Ume6-dependent binding to novel secondary recruitment sites at the seripauperin (PAU) genes. Genome-wide nucleosome mapping revealed that Sir binding to subtelomeric regions was associated with overpackaging of subtelomeric promoters. Sir3 also bound to a surprising number of euchromatic sites, largely at genes expressed at high levels, and was dynamically recruited to GAL genes upon galactose induction. Together, our results indicate that heterochromatin complex localization cannot simply be explained by nucleation and linear polymerization, and show that heterochromatin complexes associate with highly expressed euchromatic genes in many different organisms.
Keywords: chromatin, genomics, epigenetics
Introduction
Eukaryotic genomes are packaged into a nucleoprotein complex known as chromatin. Classically, chromatin has been divided into a relatively accessible active compartment termed euchromatin, and an inaccessible compartment termed heterochromatin. In budding yeast, heterochromatin formation requires the silent information regulator (SIR) complex, which is involved in silencing of the silent mating loci, subtelomeric genes and ribosomal RNA gene repeats (Rusche et al, 2003).
The now-classic model for Sir complex recruitment is a nucleation–polymerization model. Sir is first recruited to silencers—at the silent mating loci these are the E and I silencers (reviewed in Rusche et al, 2003), while at subtelomeres Sir recruitment occurs via Rap1 binding to the telomeric repeats and ORC and Abf1 binding to the XCS sequence elements (Pryde and Louis, 1997, 1999). After recruitment, Sir spreads via a cycle of histone modification and binding—Sir2 is an H4K16 deacetylase and Sir3 is a K16-sensitive nucleosome binding protein (Hecht et al, 1996; Rusche et al, 2002; Moazed et al, 2004; Norris and Boeke, 2010).
The details of Sir complex spreading are unknown, but a number of observations have lead to the idea that Sir spreading can be considered in some ways analogous to polymerization. First, silencing of subtelomeric reporter genes has been observed to decay with increasing distance from the telomere (Renauld et al, 1993; Pryde and Louis, 1999). Second, overexpression of Sir3 leads to increased Sir3 binding at sites relatively distal to the telomere (Hecht et al, 1996), suggesting that Sir protein levels are limiting for the extent of spreading. Third, upon reintroduction of Sir3 into sir3Δ yeast, Sir3 localization extends further into the chromosome at increasing times after Sir3 reintroduction (Katan-Khaykovich and Struhl, 2005). Finally, Sir3 oligomerizes in vitro, and the Sir complex exhibits dramatic structural alterations in the presence of the metabolic by-product of Sir2 action, O-acetyl ADP-ribose, suggesting that chromatin binding is likely to be cooperative in vivo (Liou et al, 2005; Martino et al, 2009). In contrast, in vivo studies of Sir-dependent silencing often reveal evidence for discontinuous silencing (Fourel et al, 1999; Pryde and Louis, 1999; Valenzuela et al, 2008; Zill et al, 2010), which calls linear polymerization models for Sir spreading into question.
Here, we investigated the kinetics of Sir complex spreading by inducing expression of an epitope-tagged Sir3 subunit in a sir3Δ mutant, and mapping Sir3 localization patterns genome-wide via deep sequencing. We find that Sir3 localization patterns do not exhibit expected polymerization behaviour. Instead, Sir3 binds to ∼5–6 nucleosomes around silencers rapidly, then slowly associates with novel secondary silencing sites such as the seripauperin (PAU) genes. We show that the PAU genes have Sir recruitment activity, and that Sir binding to the PAU genes is Ume6 dependent. We further find that the Sir complex binds numerous euchromatic sites, largely at highly expressed genes. Binding to highly expressed genes is dynamic, as Sir3 relocalizes to GAL gene bodies upon their induction. Together, these results suggest that the mechanism of heterochromatin spreading cannot be described as a simple linear polymerization reaction, and provide a window into novel heterochromatic loci.
Results
Bidirectional and ‘cooperative’ Sir3 spreading from the HMR-E silencer
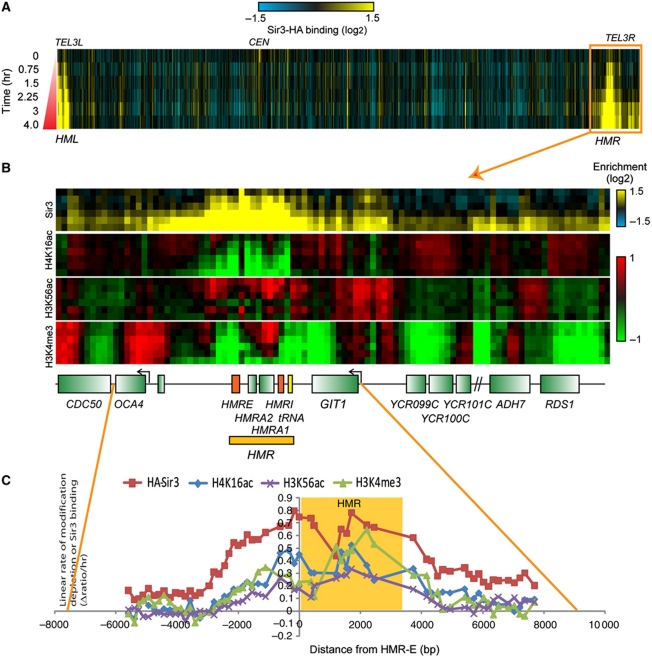
sir3Δ mutant yeast containing a galactose-inducible HA-tagged SIR3 construct (Cheng and Gartenberg, 2000) were grown to midlog in YP raffinose media. At varying intervals after induction of Sir3-HA with galactose, cells were crosslinked, and chromatin was digested to mononucleosomes and immunoprecipitated with anti-HA, anti-H4K16ac, anti-H3K4me3 and anti-H3K56ac antibodies. Chromatin immunoprecipitation (ChIP) material was amplified and hybridized to a 20-bp resolution tiled oligonucelotide array covering chromosome III and 223 additional promoters (Yuan et al, 2005) (Figure 1). As expected, Sir3-HA exhibited strong association with known heterochromatic loci, the silent mating loci and subtelomeric regions (Figure 1A). At HMR, Sir3 bound rapidly to an ∼10 nucleosome domain (Figure 1B and C) between the HMR-E silencer and the tRNA-Thr gene previously described as a heterochromatin boundary (Donze et al, 1999). H4K16 and H3K56 deacetylation as well as H3K4me3 demethylation appear to follow the dynamics of Sir3-HA binding, as observed previously (Katan-Khaykovich and Struhl, 2005; Xu et al, 2007), although H3K56 deacetylation was not as complete as were H4K16 deacetylation or H3K4 demethylation.
Figure 1.
Dynamics of Sir3 binding and histone modifications at HMR. (A) Time course of Sir3 binding to Chr3. Log2 (Sir3/IP input) values were obtained from anti-HA immunoprecipitation from THC70 cells after galactose induction of C-terminal HA-tagged Sir3, followed by microarray hybridization. Lines are results from IP experiments 0, 0.75, 1.5, 2.25, 3 and 4 h after galactose addition. Each column represents one nucleosome (Yuan et al, 2005). (B) Sir3 binding and histone modifications at HMR. Each panel represents a different ChIP-chip dataset, from top to bottom: Anti-HA (Sir3), anti-H4K16ac, anti-H3K56ac and anti-H3K4me3. Each line within each panel is a time point after galactose addition as in (A). (C) Rates of Sir3 binding and histone modification dynamics at HMR. Each data point is the slope of the linear fit to the log2 enrichment ratio time course for each nucleosome (see Supplementary Figure S1). The curves are centred at the HMR-E silencer.
Interestingly, we observed Sir3-HA binding at late time points at HMR-adjacent regions beyond the known heterochromatin boundaries. The secondary boundaries for this slower Sir3-HA spread appear to be located near the promoter regions of OCA4 on the centromeric and GIT1 on the telomeric side of HMR. By examining the kinetics of Sir3-HA binding across HMR and adjacent regions, we identify two modes for Sir3-HA spreading. The region spanning from 2 kb upstream to 4 kb downstream of HMR-E is characterized by a uniformly high binding rate (Supplementary Figure S1) most consistent with near-simultaneous binding of Sir3-HA across the region, suggesting either cooperative binding or very rapid polymerization out to boundary elements. Conversely, beyond this region of ‘cooperative’ Sir3-HA binding are ∼2 kb regions with Sir3-HA binding rates that decrease from one nucleosome to the next, which could be explained either by cell-to-cell variability in the level of Sir3-HA expression and resulting breadth of Sir3-HA-bound domains (Xu et al, 2006) or by slow polymerization. Rusché and colleague recently noted the rapidity of Sir3 binding at HMR, which they showed was in contrast to relatively slow Sir3 binding and spreading at TEL6R (Lynch and Rusche, 2009). As these qualitatively distinct Sir3 spreading behaviours are linked to the presence of different silencer sequences, we sought to globally characterize Sir3 spreading behaviour across the entire genome.
Dynamics of Sir3 binding at yeast telomeres
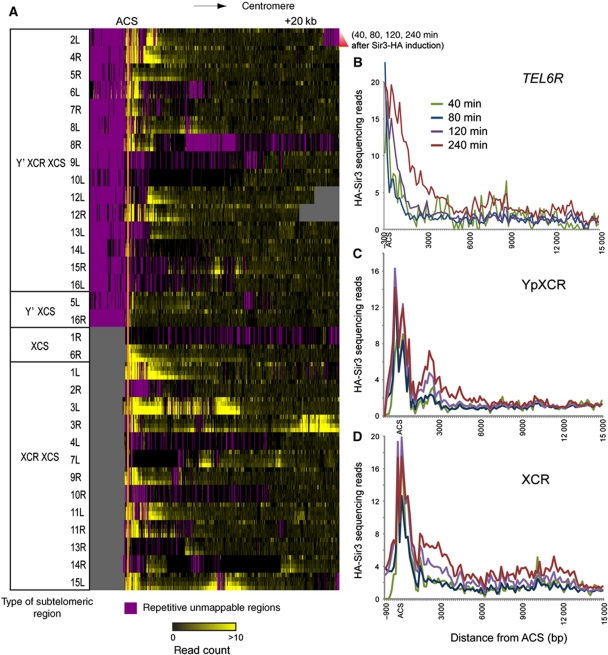
We therefore extended our results to whole-genome coverage by characterizing the distribution of Sir3 via deep sequencing (Albert et al, 2007; Shivaswamy et al, 2008) of Sir3-HA ChIP at varying times after Sir3-HA induction. Figure 2A shows Sir3-HA binding at all 32 yeast telomeres, with Sir3-HA binding shown in yellow and unmappable repeat regions shown in purple. Telomeres are categorized by three sequence elements internal to the telomeric TG repeats: Y’, XCR (X combinatorial repeats) and XCS (X-core sequence) (Louis, 1995), which are present in four combinations (Figure 2A). Y’ and XCR have been shown to possess insulator activity, and XCS functions as a proto-silencer—it promotes and stabilizes Sir-dependent gene silencing but it is not an autonomous silencer (Fourel et al, 1999; Pryde and Louis, 1999). All telomeres are aligned by the ARS consensus sequence (ACS) within the XCS.
Figure 2.
Dynamics of Sir3 binding to yeast telomeric regions. (A) Heat map of Sir3-binding dynamics measured by ChIP-seq of galactose induced anti-HA Sir3 IPs. Telomeres are grouped by type: Y’ XCR XCS, Y’ XCS, XCS and XCR XCS. Lines within each telomere panel are results from the galactose induction time course. From top to bottom: 40, 80, 120 and 240 min after galactose addition. Each column is a 30-bp region centred on the ACS element. The value of each data point is the normalized sequence read count number, which is proportional to the amount of Sir3-HA binding. (B) Sir3-HA-binding profiles at TEL6R. (C) Average Sir3-binding profiles for all Y’ XCR XCS telomeres. Data shown are a 150-bp window average. (D) Average Sir3-HA-binding profiles for all XCR XCS telomeres.
Several features are apparent in Figure 2A. First, this map is incomplete due to unmappable repetitive segments (purple regions in Figure 2A) such as Y’ elements, and due to sequence differences between our experimental strain and the reference genome at 9L, 10L, 7L, 14R, 1R and 13R (black no-read regions in Figure 2A—see Materials and methods and Supplementary Figure S2). Second, the silent mating loci are apparent in the 3L and 3R maps as prominent Sir3-bound regions ∼10 and ∼25 kb away from the XCS region, respectively. Interestingly, a previously undescribed region similar in Sir-binding profile to the silent mating loci is observed at 15L (see below, Figures 2A and 3). Finally, we note that even after 4 h of Sir3-HA expression, Sir3 does not generally spread further than 5–10 kb downstream of the ACS, with highest and fastest binding concentrated 1–2 kb around the ACS. Figure 2B shows Sir3-HA binding over time at the commonly studied telomere 6R.
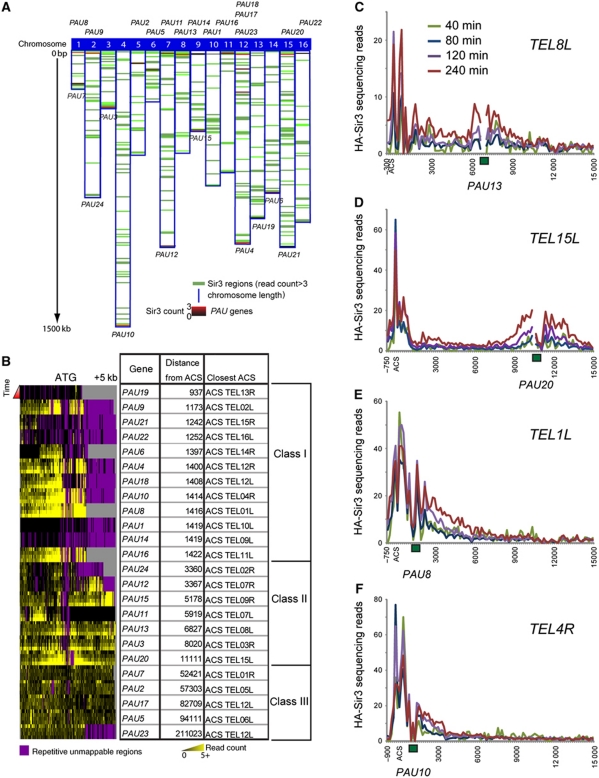
Figure 3.
Seripauperin genes as novel Sir recruitment sites. (A) Sir3-HA-enriched regions (with a read count number >3 at t=240 min after Sir3-HA induction) on all 16 yeast chromosomes are shown in green. PAU gene locations are shown in red/black, the intensity of the red bands is proportional to the Sir3 count number at a PAU gene. Chromosomes are aligned by their left telomeres along the top of the figure. Left and right telomere-proximal PAU gene names are listed on top and bottom of their respective chromosomes. (B) Heat map of Sir3-HA binding to PAU genes. Lines within each PAU gene panel are results from the galactose induction time course. From top to bottom: 40, 80, 120 and 240 min after galactose addition. PAU genes are ordered by distance from the ACS. Data shown are the same as in Figure 2A. PAU genes are aligned by their ATG and the +5 kb mark indicates distance from the ATG. (C–F) Sir3-HA-binding profiles at telomere 8L, 15L, 1L and 4R, as indicated. Locations of PAU genes are depicted as green rectangles.
Sir3 ChIP-seq maps and ChIP–qPCR experiments from a wild-type strain obtained using an antibody against Sir3 show a similar pattern of Sir3 binding to those obtained with the Sir3-HA overexpression system, with the most notable difference being a somewhat increased distance of Sir3-HA association from the telomeres in the overexpression system (Supplementary Figures S2 and S3). Interestingly, in wild-type cells some of the binding we observe at secondary recruitment sites seems to be stronger in galactose than in dextrose (see below).
What can be inferred about Sir3 spreading from these time courses? We averaged data for the two largest classes of subtelomeres, those with XCR sequences and those with both XCR and Y’ elements (Figure 2C and D, respectively). Interestingly, in both cases we observe two modes of Sir3-HA binding. As at HMR, Sir3-HA binds simultaneously to 1–2 kb around the ACS at XCR telomeres, and this binding is essentially saturated at the earliest time point after Sir3-HA induction. Beyond that region on the centromeric side there is a short but consistent dip in Sir3 binding, followed by decaying Sir3 binding with increasing distance into the chromosome. Unlike the ACS-proximal binding, Sir3 binding in this region increases gradually with time, as noted above for the Sir3 binding beyond the HMR boundaries.
Conversely, at telomeres that lack an XCR, Sir3-HA binding does not show any evidence for ‘cooperativity’, as Sir3-HA only binds ∼1–2 nucleosomes around the ACS at the Y’ XCS telomeres 5L and 16R. At the XCS telomere 6R, Sir3-HA binds initially close to the TG repeats at the end of the chromosome and then spreads slowly across the XCS up to 3 kb downstream from the ACS (Figure 2B). While it is tempting to assign the ability of Sir3-HA to bind cooperatively to the presence of the XCR, the sample size of XCR-less telomeres (only four, one of which—1R—has no available data) prevents us from drawing a more definite conclusion. Mutation analysis of XCR sites would shed more light on the role of XCR in Sir3-HA spreading.
Seripauperin genes as novel Sir3 recruitment sites
Beside XCS regions and the known Sir nucleation sites HML and HMR, new Sir3-binding sites are apparent at several telomeric regions. There is a strong novel binding/nucleation site at 15L about 10 kb from the ACS, and weaker additional sites at 3R, 2R, 8L, 9R, 15R, 7L, 11L and 14R (Figure 2A; Supplementary Figure S4). The observed Sir3 binding was not an artifact of overexpression of Sir3-HA, as we observed the same binding locations, albeit with weaker intensity, when mapping native Sir3 in wild-type yeast grown in dextrose or galactose (Supplementary Figures S2 and S5, see below). To determine whether these loci were associated with the Sir complex or solely with Sir3, we asked whether Sir2 is associated with a subset of these loci (at 8L, 7L and 15L) using a strain carrying HA-tagged Sir2. Consistent with binding of the Sir complex to these regions, we found that these loci associate with both Sir2-HA and Sir3 (Supplementary Figure S6C). Furthermore, Sir3 binding to these loci is Sir2 dependent, as anti-Sir3 ChIP–qPCR in a sir2Δ background gives only non-specific binding signals (Supplementary Figure S6B). These results strongly suggest that Sir3 proteins bound to these sites are part of the Sir complex as previously shown for the canonical subtelomeric heterochromatic sites and the silent mating type loci.
What is the nature of these secondary recruitment sites? Interestingly, we found that most of the secondary recruitment sites occur adjacent to members of the repetitive class of little-studied genes known as the seriPAUperin genes (Figure 3). For instance, the locus underlying the TEL15L secondary recruitment site corresponds to PAU20. Our data suggest that PAU genes have three classes of effects on Sir3 spreading based on their distance from the ACS (Figure 3B). One of these, which we termed class II, includes most of the secondary Sir recruitment sites (Figure 3B)—the strongest examples being the clearly separated sites 15L and 8L (Figure 3C and D). Interestingly, we note that many telomeres exhibiting extended spreading behaviours of the Sir complex have PAU genes located very close to the ACS (‘class I’), suggesting that the PAU gene at these telomeres acts as an accessory recruiting site that extends the ‘cooperative’-like Sir3 spreading region (Figure 3B, E and F). Conversely, telomeres that do not contain a PAU gene in close proximity to their ACS typically exhibit much shorter Sir domains or lack ‘cooperative’ Sir spreading around the ACS (telomeres: 5R, 6L, 7R, 8L, 8R, 14L, 3R, 9R, 11R, 15L, 6R, 5L, 16R and 3L; Figure 2A; Supplementary Figure S7). Indeed, PAU genes located >10 kb away from ACS sites (class III in Figure 3B) are not Sir3-binding sites (though they are nonetheless transcribed poorly), suggesting that proximity to silencers might be necessary for PAU genes to promote Sir3 association (see below). Furthermore, PAU genes are derepressed in sir3Δ mutant yeast, and derepression is higher for class I and class II PAU genes than for class III genes, indicating that the Sir complex is involved in seripauperin gene silencing (Supplementary Figure S8).
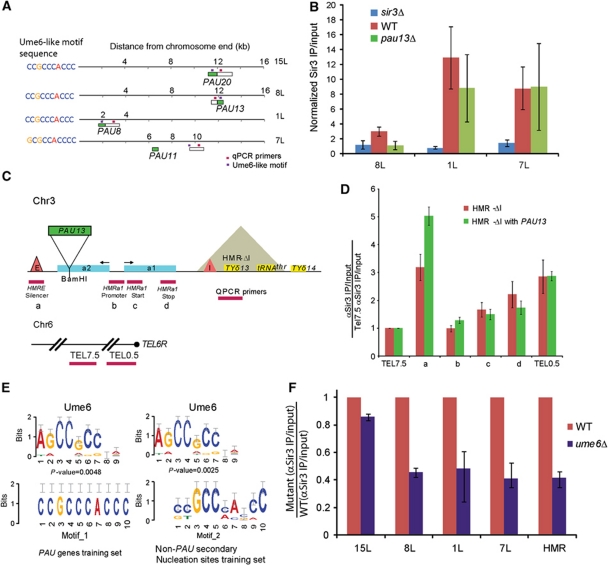
The association of Sir3 and Sir2 with PAU genes and flanking DNA suggested that these loci might directly recruit the Sir complex. We tested two predictions of this hypothesis. First, to determine whether the PAU genes were responsible for Sir3 binding in nearby genomic regions, we examined Sir3 binding adjacent to PAU13 (TEL 8L) in pau13Δ yeast. Consistent with some aspect of the PAU13 locus having a direct role in Sir3 recruitment, we found that deletion of PAU13 at telomere 8L eliminates Sir3 binding in the adjacent genome (Figure 4A and B). As a second test of the role of PAU genes in Sir recruitment, we introduced PAU13 into a sensitized HMR locus lacking the HMR-I silencer (Figure 4C). PAU13 incorporation at this locus increased Sir3 binding to the HMR-E silencer, again suggesting that the PAU genes act as accessory Sir3 recruitment sites (Figure 4D).
Figure 4.
Effect of pau13Δ and ume6Δ on Sir3 binding. (A) Diagram of PAU gene ORF locations (green rectangles) at telomeres 15L, 8L, 1L and 7L. Ume6 motif sequences (left) and their locations (lavender squares) with respect to PAU genes (green rectangles) at telomeres 15L, 8L, 1L and 7L are also shown. qPCR primer locations are shown as magenta squares. (B) αSir3 ChiP–qPCR for indicated genotypes. Results were normalized to signals from the SSL2 gene locus. Cells were grown in galactose O/N. Data are shown as mean±s.d. (C) Schematic of PAU13 insertion (green rectangle) at HMRa2 (blue rectangle) gene in the pRO869 plasmid (Valenzuela et al, 2009). Quantitative chromatin immunoprecipitation (qChIP) amplicons are shown as magenta rectangles below the diagram. The control amplicons are from TEL 6R, 7.5 and 0.5 kb away from the chromosome end for the negative and the positive control, respectively. (D) qChIP of Sir3 at HMR in GRY662 (hmrΔ;pau13Δ) with or without PAU13 inserted as in (C). ΔCt values were derived from the difference in real-time amplification of equal amounts of immunoprecipitated input DNA, and are normalized to negative control TEL7.5. Data are shown as mean±s.d. for four replicates (two biological replicates). TEL0.5 is used as a positive control. (E) MEME motif search results for a training set with all PAU genes (left) and a training set containing non-PAU gene secondary nucleation sites (right). Note the CCGCC sequence present in both motifs 1 and 2, as well as the Ume6 consensus binding sequence. (F) As in (B), except data are shown as a fraction of the WT-binding signal.
How do the PAU genes recruit Sir3? Intriguingly, we found that PAU gene regions with good Sir3 recruitment properties harbour Ume6-like DNA motifs (see Supplementary Figure S9; Figure 4E). Conversely, PAU genes with poor Sir3 recruitment properties all lack the CCGCCCACC sequences observed at strongly Sir-bound PAU genes. Furthermore, some of the observed secondary nucleation sites do not correspond to PAU genes (including those at 7L, 11L, 14R and 15R) (Figure 2A), but these sites also appear to have Ume6-like binding motifs (Figure 4E; Supplementary Figure S9B; see Discussion). As described above, deletion of PAU13 (at 8L) eliminates the Ume6-like binding motif, and eliminates Sir3 binding nearby (Figure 4B). To more specifically determine whether Ume6 is required for Sir3 recruitment to PAU genes, we carried out Sir3 ChIP–qPCR experiments in ume6Δ mutants, finding that Sir3 binding was reduced at PAU gene loci at 8L and 1L, as well as the 7L secondary nucleation site and HMR (Figure 4F; Supplementary Figure S10). Curiously, the deletion of UME6 had no effect on Sir3 binding to PAU20 (15L).
Sir3-dependent nucleosome positioning at subtelomeric genes
What is the nature of Sir3 ‘cooperative’ binding? As has been proposed earlier, the Sir complex probably promotes secondary structure chromatin folding (Shogren-Knaak et al, 2006; Johnson et al, 2009; Lynch and Rusche, 2009; Sperling and Grunstein, 2009). Chromatin folding into a 30-nm-like fibre or some other higher-order state could account for near-simultaneous association of Sir3 with a dozen nucleosomes. As nucleosomes in a higher-order structure might be expected to exhibit altered spacing, we therefore investigated the effects of Sir3 on nucleosome positioning at telomeres by whole-genome nucleosome mapping (Albert et al, 2007; Kaplan et al, 2008; Shivaswamy et al, 2008; Weiner et al, 2010) in WT and sir3Δ yeast.
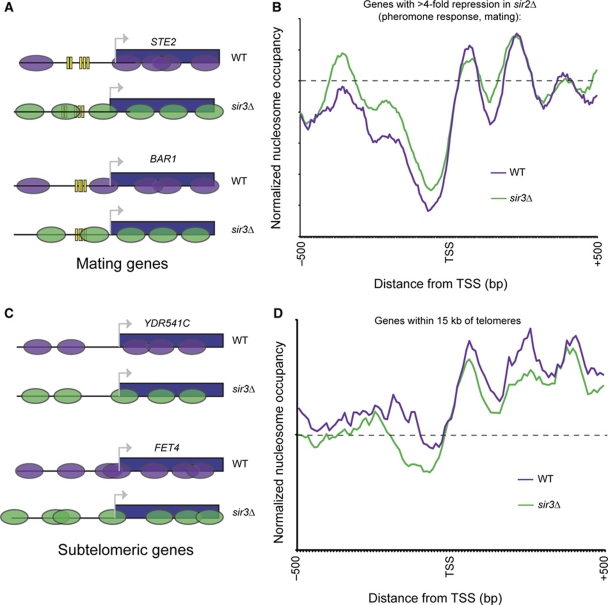
Overall, we found that nucleosome maps were quite consistent between WT and sir3Δ yeast—the two datasets exhibit a 0.8 correlation. Genes exhibiting the most dramatic chromatin differences between WT and sir3Δ were enriched for genes involved in pheromone signaling and other haploid-specific functions (Figure 5A and B; Supplementary Figure S11). This is consistent with many prior observations that loss of Sir-dependent silencing of the silent mating loci results in haploid yeast that exhibit ‘pseudo-diploid’ gene expression patterns (including downregulation of haploid-specific genes such as pheromone response genes) as a result of expression of the a1/α2 transcription factors. As shown in Figure 5A and B, loss of SIR3 results in significant increases in nucleosome occupancy over the promoters of pheromone response genes, likely as a secondary consequence of decreased TF binding to these promoters or decreased expression of these genes.
Figure 5.
Nucleosome mapping in sir3Δ cells. (A) Examples of mating-related gene with Sir-dependent chromatin structure. Schematic chromatin structure of the indicated genes for WT and sir3Δ yeast is shown. Ovals indicate nucleosomes and vertical bars indicate TF-binding sites. (B) Mating-related genes generally close in sir3Δ pseudo-diploid yeast. Average TSS-aligned chromatin structure of all genes whose mRNA abundance changes more than four-fold in sir2Δ. (C) Changes in subtelomeric promoter packaging in sir3Δ, as in (A). (D) Subtelomeric genes are underpackaged in sir3Δ. As in (B), for all genes within 15 kb of a telomere. A full-colour version of this figure is available at The EMBO Journal Online.
To identify more direct effects of the Sir complex on chromatin structure, we focused on the chromatin structure of subtelomeric genes shown to bind Sir3 above. Average nucleosome occupancy profiles for the promoters of all subtelomeric genes (within 15 kb from chromosome ends) show a wider nucleosome-free region upstream of the transcription start site (TSS) in the sir3Δ strain compared with WT (Figure 5C and D). In other words, the nucleosome immediately upstream of the TSS (−1 nucleosome) is missing on average in the sir3Δ strain, suggesting that the Sir complex has a role in stabilizing nucleosomes at these locations (although an indirect role via effects on RNA polymerase cannot be ruled out). Furthermore, overall nucleosome occupancy over subtelomeric regions appears to be decreased in sir3Δ, consistent with results observed upon Sir relocalization in ageing cells (Dang et al, 2009). The higher nucleosome occupancy and more uniform spacing in WT cells might be expected to stabilize secondary chromatin structure folding in subtelomeres, and to have a role in subtelomeric gene repression.
Sir3 binds to actively transcribed euchromatic genes
One unanticipated feature of our Sir3 maps was the presence of Sir3 at numerous euchromatic sites—some of these can be seen in Figure 1A. For instance, in addition to telomeric regions, we detected strong Sir3 binding for both Sir3-HA and wild-type Sir3 at the rDNA locus: 20–50 read counts/rDNA repeat (data not shown). This was unexpected given that while Sir2 is known to have a key role in silencing of a subset of rDNA repeats (Straight et al, 1999; Cockell et al, 2000), Sir3 is typically only thought to migrate to the nucleolus in aged cells (Kennedy et al, 1997; Gasser et al, 1998a), which should comprise a small fraction of our actively growing population of yeast.
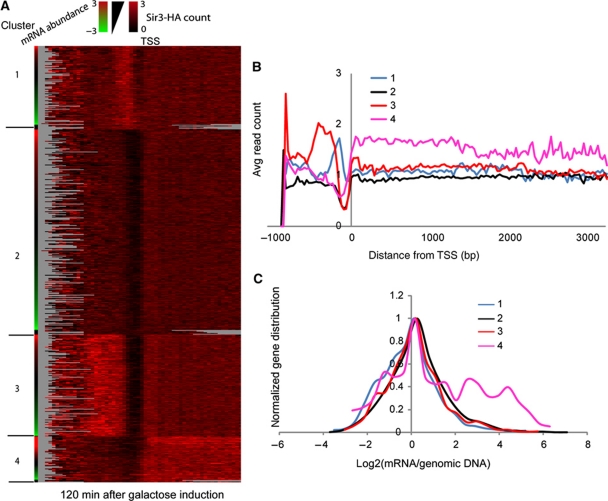
To identify patterns of Sir3 binding to euchromatic genes, we aligned all genes by TSS and carried out K-means clustering with K=4. Three major patterns emerge from this analysis—genes with background levels of Sir3-HA binding (cluster 2), genes with Sir3-HA bound at promoters (clusters 1 and 3) and genes with Sir3-HA relatively uniformly associated with the gene body (cluster 4) (Figure 6A and B). Genes with Sir3-HA bound to their promoters had few features in common, although they were slightly enriched for divergent gene orientations (not shown). We therefore focused on genes where Sir3-HA was bound throughout the coding region (cluster 4). Genes associated with Sir3-HA over coding regions were enriched for gene ontology categories such as ‘translation’ and ‘ribosome biogenesis’, categories of genes that are very highly expressed in actively growing yeast. Indeed, we found that Sir3-HA-bound genes were significantly enriched for genes expressed at high levels (Figure 6C), a seemingly paradoxical observation for a heterochromatin complex protein. These results do not reflect sequencing biases, as control deep sequencing datasets (Teytelman et al, 2009) do not show similar enrichment patterns (Supplementary Figure S12).
Figure 6.
Sir3 localizes to highly expressed genes. (A) K-means (K=4) clustering of Sir3-HA bound euchromatic genes 120 min after Sir3 induction by galactose. Genes within each cluster are ordered by mRNA abundance (high to low) shown in the first column of the heat map. Each line is a gene aligned by the TSS (Xu et al, 2009). (B) Average Sir3-binding profiles for each cluster in (A). (C) Normalized mRNA abundance distribution for each cluster in (A).
Previous studies have identified Sir proteins binding to ribosomal genes, in one case in wild-type yeast (Tsankov et al, 2006), in another case in yeast where the Sir complex was detached from its typical location at the nuclear periphery (Taddei et al, 2009). Given that the Rap1 transcription factor binds both to telomeres and to ribosomal protein promoters (Lieb et al, 2001), there is a natural link between these otherwise disparate types of loci.
Dynamic relocalization of Sir3 in response to active transcription
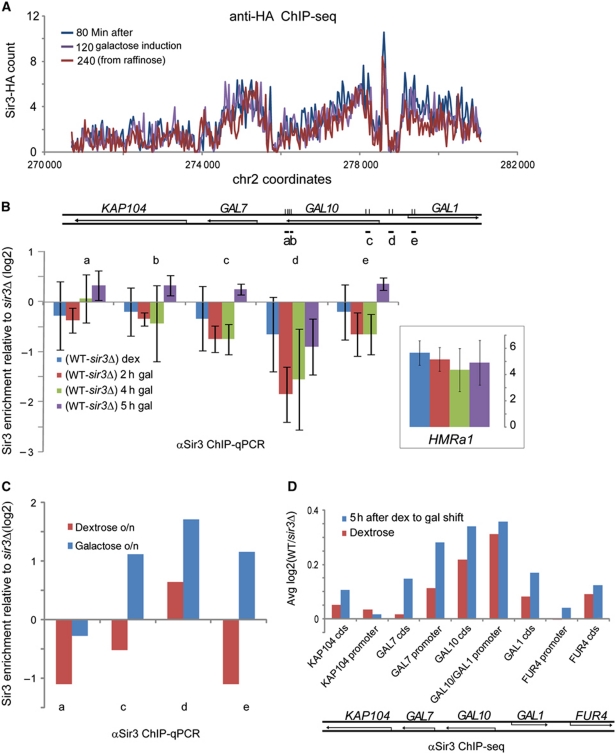
One major distinction between our culture conditions and prior studies showing Sir binding to active genes is that our yeast were grown in galactose to induce Sir3-HA expression. Interestingly, we found Sir3-HA binding throughout the active GAL7/10/1 locus (Figure 7A). Sir binding to the GAL genes had not been observed in prior studies of yeast grown in glucose (Lieb et al, 2001; Sperling and Grunstein, 2009), suggesting that Sir3 might dynamically relocalize to active genes in response to transcriptional activity.
Figure 7.
Sir3 binding to the GAL locus. (A) Sir3-HA-binding profiles at the GAL locus from the Sir3 induction ChIP-seq time course (Figure 2). (B) Sir3 binding to GAL10 and GAL1 measured by anti-Sir3 ChIP–qPCR of WT yeast (BY4741) and shown as the log2 of IP enrichment over input normalized to the SSL2 amplicon signal and corrected for sir3Δ negative control anti-Sir3 ChIP–qPCR results. The values shown are the averages of three biological replicates, and error bars represent s.d. HMRa1 provides a positive control (inset). Locations of the primer pairs (a, b, c and d) are shown in the diagram in (A). Dextrose to galactose shift was chosen for this experiment based on the observation that sir3Δ mutants exhibited delayed GAL induction kinetics under these conditions, although further analysis revealed that this effect was lost in hmlΔ background (Supplementary Figure S13). (C) As in (B), using overnight cultures of WT cells (BY4741) grown in dextrose (red) or galactose (blue). One biological replicate is shown. (D) Sir3 binding to the GAL locus in WT yeast cells in dextrose (red) and 5 h after a dextrose to galactose shift (blue). Bar graph shows average Sir3 binding to GAL gene bodies (cds) and promoters. Values are obtained from anti-Sir3 ChIP-seq of WT and sir3Δ cells.
To test this, and to determine whether the Sir3 localization we observed was a result of pGAL-mediated Sir3-HA overexpression, we carried out ChIP of native Sir3 in wild-type and sir3Δ cells using a Sir3 antibody. We confirmed that Sir3 binds to GAL gene bodies in cells grown in galactose, but not in cells grown in dextrose (Figure 7B and C). Furthermore, deep sequencing of Sir3 ChIP material from dextrose and galactose-grown wild-type yeast shows increased Sir3 binding in the GAL gene body when grown in galactose (Figure 7D). Note that experiments using Sir3-HA were carried out following a raffinose to galactose shift, whereas experiments with native Sir3 antibody were carried out following a dextrose to galactose shift, which results in much slower GAL gene induction.
The binding of heterochromatin complexes to highly expressed genes is seemingly paradoxical, raising the question of what role silencing complexes have at active genes. We therefore asked whether GAL gene induction dynamics or expression levels were affected by deletion of SIR genes. We found no detectable effects on levels of GAL genes in yeast grown in galactose, and were unable to identify any effects of SIR3 deletion on the dynamics of GAL1,10,7 induction when yeast were shifted from raffinose (not shown) or lactate/glycerol to galactose (Supplementary Figure S13). Interestingly, we did observe a reproducible acceleration of GAL10 and GAL1 induction in sir3Δ yeast upon dextrose to galactose shift. However, this effect was not reproduced in a sir3Δ hmlΔ strain (Supplementary Figure S13), indicating that this effect is likely secondary to the pseudo-diploid physiological state of sir3Δ mutant yeast.
Discussion
The Sir heterochromatin complex has been a paradigm for chromatin complex spreading and for gene silencing by chromatin regulators for decades. Here, we carried out dynamic mapping of Sir3 binding across the entire budding yeast genome during a time course of Sir3 expression. We did not observe slow spreading of Sir3 from silencers, instead finding rapid binding to ∼6–10 nucleosomes surrounding silencers with little lateral spread. We found a novel class of Sir-binding sites, the PAU genes, which appear to act as secondary silencing sites from which Sir3 exhibited slow spreading with time. Sir3 also associated with a surprising number of highly transcribed euchromatic loci. Interestingly, euchromatic binding by Sir3 was dynamic, with Sir3 redistributing to GAL genes upon galactose induction. Together, these results identify several novel aspects of Sir complex biology, and raise questions regarding the mechanism of Sir complex spreading from silencers in vivo.
Sir3 spreading dynamics
As noted above, the Sir complex is thought to polymerize along chromatin in a linear manner: the Sir complex nucleates at the silencer and then spreads in one nucleosome steps (Hoppe et al, 2002; Rusche et al, 2002). We find, however, that Sir3 has two modes of binding and spreading from a nucleation site: ‘cooperative’-like rapid spreading ∼1 kb downstream and upstream of a silencer, and slower linear spreading.
Cooperative-like and linear Sir3 binding have been reported recently at the HMR and telomere 6R, respectively (Lynch and Rusche, 2009). That study concluded that the type of silencer determines the mode of Sir3 spreading: cooperative versus linear. HMR-E, which is a strong silencer, promotes cooperative Sir complex binding while the telomere 6R XCS, a weaker proto-silencer, only initiates linear Sir3 spreading. The results from Figure 1, which displays the entire HMR locus at single nucleosome resolution, add a new twist, showing that cooperative and linear binding both occur at the HMR locus. HMR-E initiates bidirectional cooperative Sir3-HA binding over several kilobases, which then decays into linear Sir3-HA spreading outside the known HMR boundary elements. It is therefore possible that the type of silencer does not determine the mode of Sir3 binding, but rather the extent of cooperative binding. Strong silencers like HMR-E can promote cooperative binding over several kilobases, while cooperative binding from weak silencers like the TEL6R XCS would only extend over a few nucleosomes (300–500 bp) and rapidly decay into the linear binding mode, making cooperative binding hard to detect. Also, the weak HMR-I silencer on the telomeric side of HMR-E possibly assists HMR-E in extending the Sir cooperative binding over the entire HMR—a hypothesis supported by recent evidence of HMR-E and HMR-I physical interactions (Valenzuela et al, 2008).
Sir binding at telomeres also exhibits two types of spreading kinetics. Sir3 binding/polymerization around ACSs is rapid and ‘cooperative’-like. Beyond this region Sir spreading decays into the linear polymerization mode seen surrounding HMR. Interestingly, the presence of seripauperin genes extends the ‘cooperative’-like binding region (Figure 3B, E and F; Supplementary Figure S7) in much the same way seen for HMR-I at the HMR locus. The observation that a number of novel secondary Sir recruitment sites located further from the XCS region (class II in Figure 3B) also contain seripauperin genes further supports the argument for seripauperin genes as novel Sir nucleation sites (see below). Together with recent Sir4 ChIP-seq results (Zill et al, 2010), these results do not support a simple polymerization model for Sir complex spreading.
The PAU genes as novel silencers
The near-ubiquitous presence of seripauperin genes at yeast telomeres and the observed correlation between PAU gene presence and secondary Sir3 nucleation site formation (Figure 3), raises the question whether elements in PAU gene promoters or gene bodies can act as silencers or proto-silencers. Two lines of evidence argue that PAU genes can act as accessory Sir recruitment elements. First, deletion of the PAU13 sequence eliminates nearby Sir3 binding (Figure 4B). Second, introduction of the PAU13 sequence at a sensitized HMR locus enhances Sir3 binding to this locus (Figure 4D). Thus, PAU sequences are necessary for nearby Sir3 binding when in a subtelomeric context, and can function heterologously to enhance Sir3 binding at HMR. Importantly, PAU genes >10 kb away from XCS elements do not bind Sir3 (Figure 3), suggesting that PAU genes are not fully sufficient to recruit Sir3 and therefore act as accessory recruitment elements rather than fully autonomous silencers (although proving this would require swapping genomic locations for two PAU loci).
What effect does Sir complex binding have on the PAU genes? Seripauperins belong to a large gene family (24 members) encoding cell wall mannoproteins (Marguet et al, 1988) that are typically expressed during wine fermentation or anaerobic growth and can be stress induced by stimuli such as low temperature or chlorpromazine (Ai et al, 2002; Luo and van Vuuren, 2009). Presumably, Sir3 binding over these genes in aerobic conditions would ensure PAU gene silencing, which would be alleviated by Sir3 loss induced by changes in growth conditions. Interestingly, growth conditions that induce PAU genes also induce Sir3 hyperphosphorylation (Stone and Pillus, 1996; Ai et al, 2002), which either increases or decreases subtelomeric silencing depending on the locus under study. Such opposing effects of Sir3 phosphorylation on subtelomeric silencing could be explained if recruitment of Sir3 to PAU genes were phosphorylation sensitive, in contrast to XCS-dependent Sir3 nucleation. This way seripauperin gene regulation could be uncoupled from general subtelomeric silencing. Mapping phosphorylated Sir3 at subtelomeric loci and determining whether phosphorylation-defective Sir3 remains bound to PAU genes even after stress induction are needed to test this hypothesis.
How do PAU genes recruit the Sir complex? We found that PAU genes with strong Sir3 binding were associated with a UME6-related CCGCCCACC motif. Furthermore, MEME motif search (Bailey and Elkan, 1994) in XCSs and several secondary Sir nucleation sites that do not contain PAU genes (such as the one at 7L—Figure 4) produced two similar motifs, both related to the UME6-binding site (Supplementary Figure S9). Thus, we hypothesized that Ume6 might have a role in Sir3 recruitment to the PAU genes. Consistent with this hypothesis, UME6 deletion reduced Sir3 binding at several secondary nucleation sites, including the one at TEL7L that lacks a PAU gene, and at several PAU genes tested including PAU13 and PAU8 (but not PAU20, which interestingly was associated with an additional GC rich motif located downstream—Supplementary Figure S9). It is also worth noting that the ‘anomalous’ telomeres 5L, 16R, 1R and 6R that lack ‘cooperative’ Sir spreading also lack a Ume6-like binding motif in their XCS.
Ume6 is a master regulator of early meiotic genes (Williams et al, 2002), and it recruits Sin3 and Rpd3L, which repress transcription through histone H3 and H4 deacetylation (Kadosh and Struhl, 1997). Our results here show that Ume6 stimulates Sir3 binding to several secondary nucleation sites and HMR, but paradoxically, previous studies have shown that rpd3 mutants exhibit increased subtelomeric silencing in yeast and flies (De Rubertis et al, 1996). Rpd3 has also previously been implicated in heterochromatin boundary formation (Dhillon et al, 2009; Zhou et al, 2009; Ehrentraut et al, 2010). We do not currently know if Ume6's effect on Sir3 recruitment is Rpd3 dependent—if it is, then we speculate that competition between PAU genes and other heterochromatic loci for limiting Sir complex could reconcile the effects of ume6Δ and rpd3Δ on Sir complex localization, similar to what has been seen for dot1Δ and set1Δ mutants (van Leeuwen et al, 2002; Venkatasubrahmanyam et al, 2007).
The role described here for Ume6 in Sir association with subtelomeric loci is particularly intriguing in light of recent findings linking Ume6 to silent mating type loci silencing in Kluyveromyces lactis (Barsoum et al, 2010). While we did not find a Ume6-like binding motif at HMR, we cannot exclude that Ume6 binds to HMR, especially as we observe a reduction in Sir3 binding to HMR in ume6Δ cells. These results suggest the possibility that the K. lactis and Saccharomyces cerevisiae last common ancestor had a Ume6-dependent silencing system that, in S. cerevisiae, diverged to ORC/Abf1/Rap1/Sum1-based silencers at the mating type loci and ORC/Abf1/Ume6 silencers at telomeres.
Taken together, our results identify the PAU genes as novel secondary recruitment sites for the Sir complex, and identify a surprising role for Ume6 in Sir complex localization in S. cerevisiae.
Non-telomeric Sir3 localization
Finally, our genome-wide Sir3 maps reveal a large number of novel non-telomeric Sir3-binding sites. Sir3 was previously believed to relocate from subtelomeres to the nucleolus only in ageing cells (Kennedy et al, 1997; Sinclair et al, 1997; Dang et al, 2009; Kozak et al, 2010). Our data, however, show Sir3 binding to the rDNA locus in mixed populations, where the majority of cells are ‘young’: only ∼0.2% of cells have gone through more than eight divisions.
Most surprisingly, we found widespread binding of Sir3 to highly transcribed genes. Similar results have been observed for heterochromatin proteins in other systems—in flies and mice, HP1 is associated with actively transcribed genes (Vakoc et al, 2005; Johansson et al, 2007). Consistent with our data, Sir proteins have been localized to ribosomal genes in wild-type yeast (Tsankov et al, 2006) and in mutant yeast where heterochromatin association with the nuclear periphery has been disrupted (Taddei et al, 2009). Thus, our results are consistent with prior studies, but extend them in an important way: Sir binding to active genes is responsive to transcriptional patterns, as Sir3 binding to the coding regions of GAL genes is higher in galactose than in dextrose (Figure 7).
How is Sir3 recruited to actively transcribed genes? Prior studies have suggested a potential role for Rap1 or Abf1 in euchromatic Sir recruitment (Taddei et al, 2009). However, these factors have not been linked to the GAL genes, suggesting that additional mechanisms likely exist for Sir recruitment. Sir binding to nucleosomes is regulated by H4K16 acetylation—Sir3 and the intact Sir complex have both been shown to preferentially bind nucleosomes lacking H4K16 acetylation (Johnson et al, 2009; Martino et al, 2009). We have previously shown that H4K16ac is depleted over coding regions of highly transcribed genes (Liu et al, 2005), probably as a consequence of the rapid nucleosome replacement observed at such genes (Dion et al, 2007). We therefore favour a simple mechanism in which high transcription rates result in loss of H4K16ac from coding regions, creating a favourable binding site for Sir3. It is unlikely that this H4K16ac depletion is due to Sir2 activity at these sites, as Sir3 binding to highly expressed GAL genes was not dependent on Sir2 (Supplementary Figure S6D). Similar effects could explain the binding of Sir3 to many divergent promoter regions, as these regions are subject to somewhat higher histone turnover rates than are tandemly oriented promoters. An alternative, though not mutually exclusive, scenario is based on the observation that active genes localize to nuclear pores (Casolari et al, 2004, 2005). The Sir complex is associated with the nuclear periphery in between nuclear pores (Gasser et al, 1998b; Laroche et al, 2000), and it is possible that movement of active genes to the nuclear pores might result in ‘accidental’ Sir binding to these genes.
Does Sir3 binding have any effect on the transcription of active genes? We found no evidence of a role for the Sir complex in overall expression levels of the GAL genes in galactose. We did find that loss of Sir3 enabled faster GAL induction kinetics in shifts from dextrose to galactose, but this effect was lost in hmlΔ mutants where the loss of the Sir complex would not result in a pseudo-diploid state. Thus, we have not been able to uncover any reproducible effects of Sir3 on regulation of the GAL genes. Similar results have been reported for Sir-bound euchromatic sites in yeast (Sperling and Grunstein, 2009), and for HP1-bound euchromatic genes in flies (de Wit et al, 2007), raising the vexing question of whether the evolutionarily conserved heterochromatin complex binding to euchromatic genes has any functional role. In future studies, it will be interesting to see whether sir mutants have defects in transcriptional ‘memory’ of previous galactose exposure (Brickner et al, 2007; Kundu et al, 2007), or whether Sir binding to GAL genes affects some genomic process beyond transcription, such as replication fork progression, DNA damage repair or recombination.
Overall our dynamic genome-wide Sir3 maps have uncovered new aspects of Sir complex biology and opened new leads for the study of heterochromatin formation and spreading.
Materials and methods
Strain genotypes and construction
sir3Δ (MATa ura3Δ leu2Δ his3Δ met15Δ sir3Δ∷KanR) and ume6Δ (MATa ura3Δ leu2Δ his3Δ met15Δ ume6Δ∷KanR) were constructed by replacing the target gene with a KanR resistance gene by homologous recombination into wild-type BY4741 (MATa ura3Δ leu2Δ his3Δ met15Δ), followed by PCR confirmation.
THC70 (MATa HMLa HMRa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 matΔ∷TRP1 hmr∷rHMRa hmlΔ∷kanMX ura3-1∷GAL10P–SIR3HA∷URA3 bar1Δ∷hisG lys2Δ sir3Δ∷HIS3) was obtained from K Struhl (Cheng and Gartenberg, 2000; Katan-Khaykovich and Struhl, 2005).
PAU13Δ and sir2Δ are from the Saccharomyces Genome Deletion collection (BY4741 pau13Δ∷KanR) and (BY4741 sir2Δ∷KanR). Sir2-HA is the ROY1394 (MATa leu2 trp1 ura3-52 prb1-1122 pep4-3 prc1-407 gal2 3xHA-SIR2) strain from R Kamakaka.
The hmlΔ strains (W303 background) are LPY2709 (MATa ade2-1 his3-11,15, leu2-3,112 trp1-1 ura3-1 can1-100 bar1 sir3∷HIS3 hmlΔ∷TRP1 ADE2) and LPY4441 (MATa ade2-1 his3-11,15, leu2-3,112 trp1-1 ura3-1 can1-100 bar1 hmlΔ∷TRP1) (Stone et al, 2000) obtained from L. Pillus.
Yeast culture
For the Sir3-HA induction time course six flasks of THC70 cells were grown in YPraffinose at 30°C, shaking (220 r.p.m.), overnight. Formaldehyde (2% final concentration) was added to one of the flasks (time point 0) to fix the cells and galactose was added to the remaining five flasks (final concentration 0.5%). Cells in each flask were then fixed 45 min (or 40 min), 90 min, 135 min (or 120 min), 180 or 270 min (or 240 min) after galactose addition, respectively (values in parenthesis refer to the ChIP-seq experiment). Initial inoculation of the flasks was adjusted so that the cells OD at fixation was ∼0.5. After fixation, cells were incubated for 20 min at 30°C, followed by quenching with 125 mM glycine. The cells were then pelleted, washed with MilliQ water, and subjected to MNase digestion and immunoprecipitation (see below).
For nucleosome mapping in sir3Δ and WT cells or anti-Sir3 and anti-HA ChIP–qPCR of mutant cells, cells were grown in YPdextrose or YPgalactose at 30°C, 220 r.p.m., overnight to OD ∼0.5. Cells were then fixed and pelleted as above. For the dextrose to galactose shift experiment, sir3Δ and BY4741 were grown in YPdextrose at 30°C, 220 r.p.m., overnight to OD ∼0.3. Part of the culture was fixed and harvested as above and the rest was filtered (0.8 μm), the filters were washed with sterile MilliQ water and put in fresh YPgalactose media and grown at 30°C for 1.5, 2, 2.5, 3 and 4 h. Aliquots were taken at indicated times and cells were fixed and harvested as above. For RNA extraction, 50–100 ml of cells was pelleted without fixing and the pellets were flash frozen in liquid nitrogen.
Micrococcal nuclease digestion
Cell pellets (from 100 ml cells) were resuspended in 8.8 ml Buffer Z (1 M sorbitol, 50 mM Tris–Cl pH 7.4), with addition of 6.5 μl β-ME (14.3 M, final concentration 10 mM) and of 350 μl zymolyase solution (10 mg/ml in Buffer Z; Seikagaku America), and the cells were incubated at 30°C shaking at 220 r.p.m. After spinning at 3000 g, 10 min, 4°C, spheroplast pellets were resuspended in 600 μl NP-S buffer (0.5 mM spermidine, 1 mM β-ME, 0.075% NP-40, 50 mM NaCl, 10 mM Tris pH 7.4, 5 mM MgCl2, 1 mM CaCl2) per 100 ml cell culture equivalent. In total, 5–20 units (depending on yeast strain) of micrococcal nuclease (Worthington Biochemical) were added and spheroplasts were incubated at 37°C for 20 min. The digestion was halted by shifting the reactions to 4°C and by adding 0.5 M EDTA to a final concentration of 10 mM.
Chromatin immunoprecipitation
All steps were done at 4°C unless otherwise indicated. For each aliquot, Buffer L (50 mM Hepes-KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate) components were added from concentrated stocks (10–20 ×) for a total volume of 0.8 ml per aliquot. Each aliquot was rotated for 1 h with 100 μl 50% Sepharose Protein A Fast-Flow bead slurry (Sigma) previously equilibrated in Buffer L. The beads were pelleted at 3000 g for 30 s, and ∼100 μl of the supernatant was set aside for the input sample. With the remainder, antibodies were added to each aliquot (equivalent to 100 ml of cell culture) in the following volumes: 7–8 μl anti-HA (Abcam, polyclonal), or 8 μl anti-H4K16ac, anti-H3K56ac, or anti-H3K36me3 (all polyclonal Upstate (currently Millipore)), or 5 μl anti-H3K4me3 (monoclonal, Upstate (currently Millipore)) or 4 μl anti-Sir3 (provided by R Kamakaka). Immunoprecipitation, washing, protein degradation and DNA isolation were performed as previously described (Liu et al, 2003). For microarray experiments, data values are averages of 3–5 independent experiments, depending on the time point and IP set.
Linear amplification of DNA
The samples were amplified, with a starting amount of up to 75 ng for ChIP samples, using the DNA linear amplification method described previously (Liu et al, 2005).
Microarray hybridization of ChIP-ed material
In total, 2.5 μg of aRNA produced from the linear amplification was labeled via the amino-allyl method as described on http://www.microarrays.org. Labeled probes (a mixture of Cy5-labeled input and Cy3-labeled ChIP-ed material) were hybridized onto a yeast tiled oligonucleotide microarray (Yuan et al, 2005) at 55°C for 16 h and washed as described on http://www.microarrays.org. The arrays were scanned at 5 μ resolution with an Axon Laboratories GenePix 4000B scanner running GenePix 5.1. Image analysis and data normalization were performed as previously described (Liu et al, 2005).
Microarray hybridization of mRNA samples
mRNA was isolated from frozen THC70 cell pellets using the NEB magnetic mRNA isolation kit. Frozen cell pellets were resuspended in lysis buffer from the kit and subject to two rounds of flash freezing and boiling (2 min each) to break the cell walls. Subsequent steps were done according to the kit protocol.
Approximately 2 μg of mRNA was reverse transcribed using random hexamers and oligo-dT (0.15 μg/μl each) as primers in the presence of 6 μg/ml actinomycin D (SIGMA) at 42°C for 2 h. RNA was then degraded with NaOH at 65°C, and cDNA was purified using YM-30 centricon spin columns and dye-coupled with Cy5 or Cy3 NHS-esters as described previously (Liu et al, 2005). The Cy5 or Cy3-labeled cDNA was mixed with Cy3 or Cy5-labeled genomic DNA (Klenow-labeled as in Yuan et al, 2005), respectively. The labeled mixture was hybridized to home printed yeast expression arrays at 65°C for 16 h as described above. Images were scanned and analysed as described above.
Deep sequencing of ChIP material
ChIP DNA was treated with calf alkaline phosphatase (CIP NEB; in 1 × NEB buffer3, 0.25 U/μl CIP; 45 min at 37°C, reaction clean up with Qiagen MinElute spin columns).
Libraries for SOLEXA platform deep sequencing were prepared following manufacturer's instructions, with 18 cycles of PCR amplification. Gel-purified (200–300 bp in size) PCR products were ethanol precipitated, resuspended in TE buffer and sent for SOLEXA sequencing at the UMass Worcester core deep sequencing facility.
ChIP–qPCR
DNA from WT (BY4741) and sir3Δ, pau13Δ, ume6Δ, sir2Δ and Sir2-HA cells grown as indicated was chromatin immunoprecipitated as described above with the anti-Sir3 antibody from R Kamakaka. Approximately 1 ng DNA was used per qPCR reaction with IQ SYBR green Supermix (BioRad). Primer sequences are given in Supplementary Table S1. Observed Ct values were normalized to Ct from the SSL2 locus (which has no detectable Sir3 binding in the ChIP-seq data). HMRa1 was used as a positive control.
The experiment in Figure 4D was carried out as previously described in Dhillon et al (2009) and Valenzuela et al (2009). Chromatin was sheared by sonication into ∼300-bp fragments and immunoprecipitated using polyclonal anti-Sir3 antibodies (Dhillon et al, 2009). Primers are given in Supplementary Table S1.
RNA extraction
Frozen WT and sir3Δ cell pellets (equivalent of 100 ml culture) were resuspended in 0.75 ml of Trizol (Invitrogen) and subject to beading to break the cell walls. In total, 0.2 ml chloroform was then added to the cell suspension and spun at 12 000 g for 10 min at 4°C. An equal volume of isopropanol was added to the aqueous phase, incubated 10 min at RT and spun at 12 000 g for 10 min at 4°C. RNA pellets were washed with 80% ethanol, air dried and resuspended in 100 μl RNAse-free water. Total RNA was then treated with DNase I using the Qiagen RNAeasy mini kit.
Reverse transcription and microarray hybridization
cDNA was obtained by reverse transcription (Superscript II, Invitrogen) of total DNAse I-treated RNA (25 μg) with oligo-dT primers (0.5 μg/μl) in the presence of actinomycin D. RNA was degraded by NaOH treatment at 65°C and excess nucleotides were purified by buffer exchange in YM10 Amicon spin columns. Labeled cDNA was mixed with labeled genomic DNA as described above and hybridized to custom modified Agilent yeast gene expression arrays following the Agilent microarray hybridization protocol.
Reverse transcription and qPCR
Total DNase I-treated RNA was reverse transcribed and mixed with the relevant primers (Supplementary Table S1). The amount of cDNA used for each qPCR reaction with IQ SYBR green Supermix (BioRad) was equivalent to 10 ng of starting total RNA. The observed GAL loci Ct values were normalized to Ct from the SSL2 locus (which has no detectable Sir3 binding in our THC70 ChIP-seq experiment).
Deep sequencing data analysis
Sequences were aligned to S. cerevisiae genome by BLAT or in the ILLUMINA eland pipeline. We kept the uniquely aligned 100% match reads. We counted the number of reads for each base pair and the read counts were then normalized to 1 by dividing each base pair count with the genome-wide average base pair count number. Normalized read counts were extended by the cross correlation length between forward and reverse reads, forward and reverse reads were added up and then averaged in 30 bp windows. We use this 30 bp average normalized count number as a measure of Sir3 binding or nucleosome occupancy in the Sir3-HA induction ChIP-seq or the WT and sir3Δ nucleosome mapping experiments, respectively.
Sir3 enrichment loci from anti-Sir3 ChIP-seq dataset of WT yeast cells have been determined by dividing the WT averaged 30 bp read count regions by the equivalent regions from a sir3Δ ChIP-seq dataset and filtering out regions with a WT count <1 (those read count values were reset to 0).
The repetitive regions map was constructed by ‘blating’ all the possible 36 bp yeast genome sequences and determining all the unique 36 bp sequences. All the base coordinates that were not in those unique sequences were considered repetitive. In all, 30 bp regions that contained >10 ‘repetitive’ bases were considered repetitive.
Supplementary Material
Acknowledgments
We thank P Kaufman and A Hughes and other members of the Rando lab for comments and discussions. OJR is supported in part by a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund. This research was supported by NIGMS grant GM079205 and the HFSP (OJR), NIGMS grant GM078068 (RK), NIH Training Grant Award T32-GM008646 (GR), and the US-Israel Binational Foundation (NF and OJR).
Author contributions: OJR and MRL designed the experiments, and MRL carried out the experiments. GR and RK designed and carried out experiments for Figure 4D. MRL and OJR analysed the data, with initial help from AW and NF. MRL and OJR wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ai W, Bertram PG, Tsang CK, Chan TF, Zheng XF (2002) Regulation of subtelomeric silencing during stress response. Mol Cell 10: 1295–1305 [DOI] [PubMed] [Google Scholar]
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF (2007) Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446: 572–576 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Barsoum E, Sjostrand JO, Astrom SU (2010) Ume6 is required for the MATa/MAT{alpha}-cellular identity and transcriptional silencing in Kluyveromyces lactis. Genetics 184: 999–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH (2007) H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol 5: e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Drubin DA, Rando OJ, Silver PA (2005) Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes Dev 19: 1188–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA (2004) Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117: 427–439 [DOI] [PubMed] [Google Scholar]
- Cheng TH, Gartenberg MR (2000) Yeast heterochromatin is a dynamic structure that requires silencers continuously. Genes Dev 14: 452–463 [PMC free article] [PubMed] [Google Scholar]
- Cockell MM, Perrod S, Gasser SM (2000) Analysis of Sir2p domains required for rDNA and telomeric silencing in Saccharomyces cerevisiae. Genetics 154: 1069–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, Kaeberlein M, Kennedy BK, Berger SL (2009) Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature 459: 802–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubertis F, Kadosh D, Henchoz S, Pauli D, Reuter G, Struhl K, Spierer P (1996) The histone deacetylase RPD3 counteracts genomic silencing in Drosophila and yeast. Nature 384: 589–591 [DOI] [PubMed] [Google Scholar]
- de Wit E, Greil F, van Steensel B (2007) High-resolution mapping reveals links of HP1 with active and inactive chromatin components. PLoS Genet 3: e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon N, Raab J, Guzzo J, Szyjka SJ, Gangadharan S, Aparicio OM, Andrews B, Kamakaka RT (2009) DNA polymerase epsilon, acetylases and remodellers cooperate to form a specialized chromatin structure at a tRNA insulator. EMBO J 28: 2583–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ (2007) Dynamics of replication-independent histone turnover in budding yeast. Science 315: 1405–1408 [DOI] [PubMed] [Google Scholar]
- Donze D, Adams CR, Rine J, Kamakaka RT (1999) The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev 13: 698–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrentraut S, Weber JM, Dybowski JN, Hoffmann D, EhrenhoferMurray AE (2010) Rpd3-dependent boundary formation at telomeres by removal of Sir2 substrate. Proc Natl Acad Sci USA 107: 5522–5527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourel G, Revardel E, Koering CE, Gilson E (1999) Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J 18: 2522–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser SM, Gotta M, Renauld H, Laroche T, Cockell M (1998a) Nuclear organization and silencing: trafficking of Sir proteins. Novartis Found Symp 214: 114–126; discussion 126–132 [DOI] [PubMed] [Google Scholar]
- Gasser SM, Paro R, Stewart F, Aasland R (1998b) The genetics of epigenetics. Cell Mol Life Sci 54: 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A, Strahl-Bolsinger S, Grunstein M (1996) Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature 383: 92–96 [DOI] [PubMed] [Google Scholar]
- Hoppe GJ, Tanny JC, Rudner AD, Gerber SA, Danaie S, Gygi SP, Moazed D (2002) Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol Cell Biol 22: 4167–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson AM, Stenberg P, Pettersson F, Larsson J (2007) POF and HP1 bind expressed exons, suggesting a balancing mechanism for gene regulation. PLoS Genet 3: e209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A, Li G, Sikorski TW, Buratowski S, Woodcock CL, Moazed D (2009) Reconstitution of heterochromatin-dependent transcriptional gene silencing. Mol Cell 35: 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D, Struhl K (1997) Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89: 365–371 [DOI] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, Leproust EM, Hughes TR, Lieb JD, Widom J, Segal E (2008) The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katan-Khaykovich Y, Struhl K (2005) Heterochromatin formation involves changes in histone modifications over multiple cell generations. EMBO J 24: 2138–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BK, Gotta M, Sinclair DA, Mills K, McNabb DS, Murthy M, Pak SM, Laroche T, Gasser SM, Guarente L (1997) Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. Cell 89: 381–391 [DOI] [PubMed] [Google Scholar]
- Kozak ML, Chavez A, Dang W, Berger SL, Ashok A, Guo X, Johnson FB (2010) Inactivation of the Sas2 histone acetyltransferase delays senescence driven by telomere dysfunction. EMBO J 29: 158–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu S, Horn PJ, Peterson CL (2007) SWI/SNF is required for transcriptional memory at the yeast GAL gene cluster. Genes Dev 21: 997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroche T, Martin SG, Tsai-Pflugfelder M, Gasser SM (2000) The dynamics of yeast telomeres and silencing proteins through the cell cycle. J Struct Biol 129: 159–174 [DOI] [PubMed] [Google Scholar]
- Lieb JD, Liu X, Botstein D, Brown PO (2001) Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet 28: 327–334 [DOI] [PubMed] [Google Scholar]
- Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D (2005) Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell 121: 515–527 [DOI] [PubMed] [Google Scholar]
- Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ (2005) Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol 3: e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CL, Schreiber SL, Bernstein BE (2003) Development and validation of a T7 based linear amplification for genomic DNA. BMC Genomics 4: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis EJ (1995) The chromosome ends of Saccharomyces cerevisiae. Yeast 11: 1553–1573 [DOI] [PubMed] [Google Scholar]
- Luo Z, van Vuuren HJ (2009) Functional analyses of PAU genes in Saccharomyces cerevisiae. Microbiology 155: 4036–4049 [DOI] [PubMed] [Google Scholar]
- Lynch PJ, Rusche LN (2009) A silencer promotes the assembly of silenced chromatin independently of recruitment. Mol Cell Biol 29: 43–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marguet D, Guo XJ, Lauquin GJ (1988) Yeast gene SRP1 (serine-rich protein). Intragenic repeat structure and identification of a family of SRP1-related DNA sequences. J Mol Biol 202: 455–470 [DOI] [PubMed] [Google Scholar]
- Martino F, Kueng S, Robinson P, Tsai-Pflugfelder M, van Leeuwen F, Ziegler M, Cubizolles F, Cockell MM, Rhodes D, Gasser SM (2009) Reconstitution of yeast silent chromatin: multiple contact sites and O-AADPR binding load SIR complexes onto nucleosomes in vitro. Mol Cell 33: 323–334 [DOI] [PubMed] [Google Scholar]
- Moazed D, Rudner AD, Huang J, Hoppe GJ, Tanny JC (2004) A model for step-wise assembly of heterochromatin in yeast. Novartis Found Symp 259: 48–56; discussion 56–62, 163–169 [PubMed] [Google Scholar]
- Norris A, Boeke JD (2010) Silent information regulator 3: the Goldilocks of the silencing complex. Genes Dev 24: 115–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde FE, Louis EJ (1997) Saccharomyces cerevisiae telomeres. A review. Biochemistry (Mosc) 62: 1232–1241 [PubMed] [Google Scholar]
- Pryde FE, Louis EJ (1999) Limitations of silencing at native yeast telomeres. EMBO J 18: 2538–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renauld H, Aparicio OM, Zierath PD, Billington BL, Chhablani SK, Gottschling DE (1993) Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev 7: 1133–1145 [DOI] [PubMed] [Google Scholar]
- Rusche LN, Kirchmaier AL, Rine J (2002) Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol Biol Cell 13: 2207–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche LN, Kirchmaier AL, Rine J (2003) The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem 72: 481–516 [DOI] [PubMed] [Google Scholar]
- Shivaswamy S, Bhinge A, Zhao Y, Jones S, Hirst M, Iyer VR (2008) Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol 6: e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL (2006) Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311: 844–847 [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Mills K, Guarente L (1997) Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science 277: 1313–1316 [DOI] [PubMed] [Google Scholar]
- Sperling AS, Grunstein M (2009) Histone H3 N-terminus regulates higher order structure of yeast heterochromatin. Proc Natl Acad Sci USA 106: 13153–13159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone EM, Pillus L (1996) Activation of an MAP kinase cascade leads to Sir3p hyperphosphorylation and strengthens transcriptional silencing. J Cell Biol 135: 571–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone EM, Reifsnyder C, McVey M, Gazo B, Pillus L (2000) Two classes of sir3 mutants enhance the sir1 mutant mating defect and abolish telomeric silencing in Saccharomyces cerevisiae. Genetics 155: 509–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Shou W, Dowd GJ, Turck CW, Deshaies RJ, Johnson AD, Moazed D (1999) Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell 97: 245–256 [DOI] [PubMed] [Google Scholar]
- Taddei A, Van Houwe G, Nagai S, Erb I, van Nimwegen E, Gasser SM (2009) The functional importance of telomere clustering: global changes in gene expression result from SIR factor dispersion. Genome Res 19: 611–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teytelman L, Ozaydin B, Zill O, Lefrancois P, Snyder M, Rine J, Eisen MB (2009) Impact of chromatin structures on DNA processing for genomic analyses. PLoS One 4: e6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankov AM, Brown CR, Yu MC, Win MZ, Silver PA, Casolari JM (2006) Communication between levels of transcriptional control improves robustness and adaptivity. Mol Syst Biol 2: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakoc CR, Mandat SA, Olenchock BA, Blobel GA (2005) Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell 19: 381–391 [DOI] [PubMed] [Google Scholar]
- Valenzuela L, Dhillon N, Dubey RN, Gartenberg MR, Kamakaka RT (2008) Long-range communication between the silencers of HMR. Mol Cell Biol 28: 1924–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela L, Dhillon N, Kamakaka RT (2009) Transcription independent insulation at TFIIIC-dependent insulators. Genetics 183: 131–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F, Gafken PR, Gottschling DE (2002) Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109: 745–756 [DOI] [PubMed] [Google Scholar]
- Venkatasubrahmanyam S, Hwang WW, Meneghini MD, Tong AH, Madhani HD (2007) Genome-wide, as opposed to local, antisilencing is mediated redundantly by the euchromatic factors Set1 and H2A.Z. Proc Natl Acad Sci USA 104: 16609–16614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N (2010) High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res 20: 90–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RM, Primig M, Washburn BK, Winzeler EA, Bellis M, Sarrauste de Menthiere C, Davis RW, Esposito RE (2002) The Ume6 regulon coordinates metabolic and meiotic gene expression in yeast. Proc Natl Acad Sci USA 99: 13431–13436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu EY, Zawadzki KA, Broach JR (2006) Single-cell observations reveal intermediate transcriptional silencing states. Mol Cell 23: 219–229 [DOI] [PubMed] [Google Scholar]
- Xu F, Zhang Q, Zhang K, Xie W, Grunstein M (2007) Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Mol Cell 27: 890–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM (2009) Bidirectional promoters generate pervasive transcription in yeast. Nature 457: 1033–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ (2005) Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309: 626–630 [DOI] [PubMed] [Google Scholar]
- Zhou J, Zhou BO, Lenzmeier BA, Zhou JQ (2009) Histone deacetylase Rpd3 antagonizes Sir2-dependent silent chromatin propagation. Nucleic Acids Res 37: 3699–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zill OA, Scannell D, Teytelman L, Rine J (2010) Co-evolution of transcriptional silencing proteins and the DNA elements specifying their assembly. PLoS Biol 8: e1000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.