Abstract
Meiosis-specific mRNAs are transcribed in vegetative fission yeast, and these meiotic mRNAs are selectively removed from mitotic cells to suppress meiosis. This RNA elimination system requires degradation signal sequences called determinant of selective removal (DSR), an RNA-binding protein Mmi1, polyadenylation factors, and the nuclear exosome. However, the detailed mechanism by which meiotic mRNAs are selectively degraded in mitosis but not meiosis is not understood fully. Here we report that Red1, a novel protein, is essential for elimination of meiotic mRNAs from mitotic cells. A red1 deletion results in the accumulation of a large number of meiotic mRNAs in mitotic cells. Red1 interacts with Mmi1, Pla1, the canonical poly(A) polymerase, and Rrp6, a subunit of the nuclear exosome, and promotes the destabilization of DSR-containing mRNAs. Moreover, Red1 forms nuclear bodies in mitotic cells, and these foci are disassembled during meiosis. These results demonstrate that Red1 is involved in DSR-directed RNA decay to prevent ectopic expression of meiotic mRNAs in vegetative cells.
Keywords: cleavage body, fission yeast, meiosis, mRNA degradation
Introduction
Various kinds of compartments, called cellular bodies, are present in the nucleus and cytoplasm of eukaryotic cells (Spector, 2001; Anderson and Kedersha, 2009; Bond and Fox, 2009; Buchan and Parker, 2009; Zhao et al, 2009). It is believed that nuclear bodies, composed of proteins and RNAs, have important roles in many nuclear functions. For example, cleavage bodies contain factors involved in 3′-end processing of pre-mRNAs, and the processing machinery cleaves the 3′-end of pre-mRNAs and adds poly(A) tails to maturing mRNAs (de Jong et al, 1996). In addition, snRNAs with trimethylguanosine (m3G snRNAs) are transported to Cajal bodies where they are subjected to additional modification, and these modified m3G snRNAs associate with snRNP-specific proteins, resulting in the assembly of mature snRNPs (Hebert, 2010). Thus, it seems that a variety of nuclear proteins involved in specific tasks are assembled in discrete nuclear compartments to perform their function efficiently. However, the mechanisms by which nuclear bodies are formed and the exact functions of these structures are not understood fully (Matera et al, 2009).
Meiosis is the process of cell division that produces gametes from germ cells. Meiosis involves (1) the reduction of chromosome number by half when a single round of DNA replication is followed by two consecutive rounds of chromosome segregation and (2) the diversification of genetic information by meiotic recombination (Nebreda and Ferby, 2000; Yanowitz, 2010). Therefore, meiosis is thought to enable offsprings to adapt to various environments and to be a driving force in the evolution of life (Coop and Przeworski, 2007). The fission yeast, Schizosaccharomyces pombe, is one of the best-studied organisms with regard to the regulation of meiosis (Yamamoto, 1996). In fission yeast, nitrogen deprivation is an essential signal that switches the mitotic cell division to meiotic sexual differentiation. Nitrogen starvation ultimately induces two key events: the activation of the transcription factor Ste11, which enhances the transcription of various meiosis-specific genes including mei2+, and the inactivation of kinases that inhibit Mei2/Ste11 functions (Yamamoto, 1996). Although meiosis in S. pombe was long believed to be regulated only at the transcriptional and post-translational levels, at least some of meiosis-specific genes, including mei4+ and crs1+ that are transcribed even in vegetative cells, are negatively controlled at the post-transcriptional level to prevent the ectopic expression of meiotic mRNAs (Harigaya et al, 2006; McPheeters et al, 2009). Mmi1, an RNA-binding protein with a YTH domain, forms nuclear dots and promotes the selective removal of meiotic mRNAs containing determinant of selective removal (DSR) sequences by the nuclear exosome in mitotic cells (Harigaya et al, 2006). During meiosis, activated Mei2 traps and inactivates Mmi1 to allow meiotic mRNAs to be translated (Harigaya et al, 2006). Recent studies have also shown that polyadenylation by the canonical poly(A) polymerase (PAP; Pla1 in fission yeast) and a nuclear poly(A)-binding protein (PABP), Pab2, is required for selective elimination of meiotic mRNAs (St-Andre et al, 2010; Yamanaka et al, 2010). These findings suggest that the DSR-Mmi1 system represents a distinct pathway of poly(A)-dependent mRNA degradation and that with this decay system the inappropriate expression of meiosis-specific genes is strictly prevented in mitotic cells. However, the detailed mechanism by which the polyadenylation-dependent DSR-Mmi1 system specifically activates RNA degradation by the exosome is not yet established, and it is likely that additional, unidentified factors function in this system.
Using a localization-based approach, we have identified several factors that localize to nuclear or cytoplasmic foci in fission yeast. Here we report on one such protein, Red1, which localizes to nuclear foci in vegetatively growing cells. Cells lacking red1+ are defective in mating and sporulation. Expression profiling of red1Δ indicates that numerous meiotic mRNAs accumulate in mitotic red1Δ cells. These results are reminiscent of Mmi1, and Red1 co-localizes and associates with Mmi1; moreover, Red1 is required for DSR-mediated mRNA elimination. In addition, Red1 foci coincide with the foci of the canonical PAP, Pla1; a 3′-cleavage factor, Pcf11; Rrp6, a subunit of nuclear exosome. Unlike Mmi1 foci, Red1 foci disassemble in early meiosis independently of Mei2/Mei3/Mei4/meiRNA, and this disassembly of Red1 apparently coincides with the period when meiotic genes are expressed. These results led us to propose that Red1 is a novel factor that is essential for the selective elimination of meiotic mRNAs with DSR sequences.
Results
Identification of Red1, a novel protein that forms nuclear dots
We sought novel factors that form nuclear or cytoplasmic bodies in fission yeast by concentrating on candidate proteins selected from the localization database reported previously (Matsuyama et al, 2006). We constructed a fission yeast library that expressed these candidate proteins with carboxy-terminal GFP (Ogawa et al, 2004) or tdTomato (Shaner et al, 2004) fusions from their native promoters. Microscopic analyses demonstrated that many proteins localized as nuclear dots (TS, data not shown). One such protein, encoded by a previously uncharacterized gene SPAC1006.03c, localized to the nucleus mainly in 1–4 foci (Figure 1A). We named this gene red1+ (RNA elimination defective 1). Theoretical translation of the red1+ gene resulted in a 712 amino-acid protein with coiled-coil domains and a putative zinc-finger domain (Supplementary Figure S1A). The putative zinc-finger domain had the consensus sequence CX7−8CX4C3H, which closely resembles the typical CCCH zinc-finger domain (CX8CX5C3H) often found in DNA/RNA-binding proteins (Brown, 2005; Hall, 2005). A BLAST search with the predicted Red1 sequence revealed that the Red1 zinc finger-like domain is found in proteins of metazoans, including human, mouse, rat, and chicken. One such protein is human CCDC131, also called proline/serine-rich coiled-coil 2; however, further analyses are required to determine whether human CCDC131 is a homologue of Red1 as the similarity between these proteins is limited to the zinc-finger motif (Supplementary Figure S1B). Based on a previous report, Red1 interacts with the fission yeast Polo-like kinase (Plo1) in a two-hybrid system, and this interaction depends on the integrity of three polo boxes in Plo1 (Reynolds and Ohkura, 2003). However, the authors of that report did not examine the biological significance of the Red1–Plo1 interaction. From these data, we speculated that Red1 is a conserved protein in eukaryotes and that the zinc-finger motif of Red1 is critical for Red1 function.
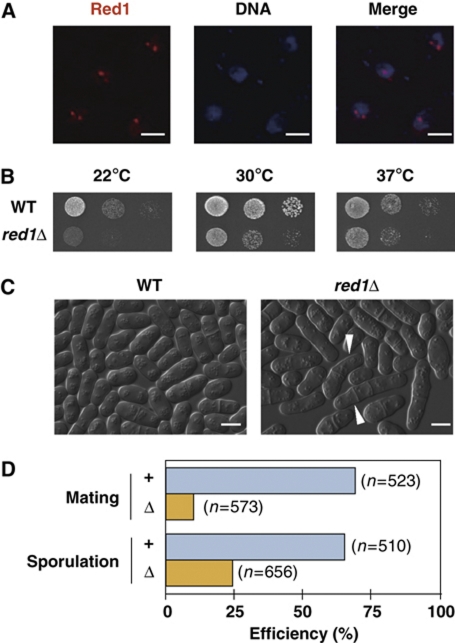
Figure 1.
Red1 localizes as nuclear dots and is required for normal growth and meiotic processes. (A) Red1 forms nuclear bodies in vegetatively growing cells. Yeast cells expressing Red1 tagged with tdTomato were fixed and stained with DAPI. Representative deconvolved images are shown. Bars, 2 μm. (B) red1Δ cells display growth retardation. Serial dilutions of wild-type and red1Δ cells were spotted onto YEA plates and then incubated at various temperatures for 3–5 days before taking pictures. (C) Some red1Δ cells display abnormal cell morphology. Differential interference contrast (DIC) images of wild type (WT) and red1Δ show that red1Δ cells are elongated relative to wild-type cells. Arrow heads indicate cells with abnormalities, including multi-septa and altered cell polarity. Bars, 5 μm. (D) Mating and sporulation efficiencies are reduced in red1Δ cultures. The efficiencies of mating and sporulation in wild-type and red1Δ cultures were measured as described in the Materials and methods.
Loss of Red1 function leads to growth retardation and mating/sporulation deficiencies
To investigate the role(s) of Red1, we generated a red1Δ strain and examined the phenotypes of this strain. We found that red1Δ cells grew slowly compared with parental wild-type cells, particularly at low temperature (Figure 1B). Elongated cells or cells with more than one septum occurred in red1Δ culture (Figure 1C), thus suggesting that Red1 is required for normal cell growth. In addition, iodine staining of red1Δ cells after the induction of meiosis was decreased (data not shown), implying that Red1 is involved in meiotic processes, including mating, sporulation, or mating-type switching. To discriminate these possibilities, we calculated the mating and sporulation efficiencies of red1Δ cells, and found that both mating and sporulation efficiencies were significantly lower in red1Δ cultures than in red1+ cultures (Figure 1D). These results strongly suggest that Red1 has an important role in mating and sporulation.
Red1 is required for meiotic mRNA suppression
To further explore the role of Red1, we carried out the expression profiling of red1Δ cells to identify genes affected by red1Δ. Using microarray techniques, we identified 123 mRNAs that were increased more than two-fold in red1Δ cells (Figure 2A; Supplementary Table SII). Interestingly, the vast majority (88%) of the RNAs that were upregulated in mitotic red1Δ cells were previously reported as meiotic mRNAs that were increased during meiosis (Mata et al, 2002; Chikashige et al, 2006). The most upregulated transcript in red1Δ cells was SPBPB2B2.03c, a pseudogene homologous to a membrane transporter, and most of the upregulated mRNAs listed in Figure 2B were classified as early meiotic mRNAs (Mata et al, 2002) (Figure 2B). A previous study showed that meiotic mRNAs accumulate in vegetative mmi1-deficient cells (Harigaya et al, 2006). We therefore compared Red1- and Mmi1-regulated genes and found that 11 of the 12 Mmi1-regulated genes, such as crs1+, mcp5+, rec8+, and ssm4+ (Harigaya et al, 2006), were upregulated in red1Δ cells (Figure 2B). These results suggest that Red1 mainly downregulates meiotic mRNAs and that Red1 coordinates with Mmi1 to remove meiotic mRNAs in vegetative cells.
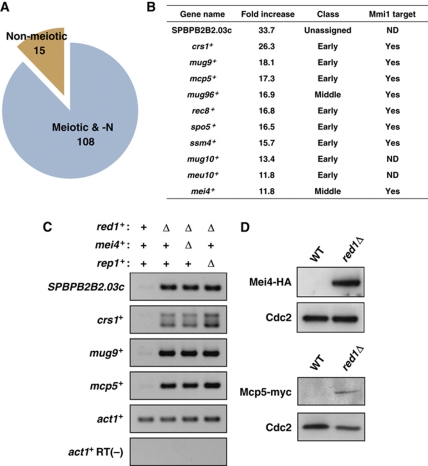
Figure 2.
Meiosis-specific mRNAs are downregulated by Red1 in mitotic cells. (A) A large fraction of the mRNAs accumulating in growing red1Δ are meiotic mRNAs. Expression analyses using a microarray technique demonstrate that 88% of increased (more than two-fold) transcripts have previously been reported as genes upregulated in response to nitrogen starvation/pheromone and during meiosis. (B) The 11 mRNAs exhibiting the greatest increase in red1Δ cells. These genes were classified according to the timing of gene expression in meiosis (Mata et al, 2002). The fold increases shown in the list are the averages of two independent experiments. The Mmi1 target genes have been identified previously (Harigaya et al, 2006). (C) Meiotic mRNAs accumulated in red1Δ cells independently of both Mei4 and Rep1 transcription factors. Total RNA samples from wild-type, red1Δ, red1Δmei4Δ, and red1Δrep1Δ cells were subjected to RT–PCR analyses. RT–PCR of SPBPB2B2.03c, crs1+, mug9+, and mcp5+ indicates that the increased levels of these meiotic transcripts also accumulate in red1Δmei4Δ and red1Δrep1Δ cells. (D) Mei4 and Mcp5 proteins are produced in mitotically growing red1Δ cells. Protein extracts of mitotic wild-type and red1Δ cells expressing Mei4-HA and Mcp5-myc were resolved on a polyacrylamide gel and then analysed by western blotting using anti-HA (top panels) and anti-myc (bottom panels) antibodies. Cdc2 was monitored as a loading control.
To confirm the expression array results, we carried out RT–PCR to assess the steady-state levels of four mRNAs: SPBPB2B2.03c, crs1+, mug9+, and mcp5+. RT–PCR data demonstrated that these meiotic RNAs were indeed increased in red1Δ cells (Figure 2C). As the microarray analyses revealed that red1 deficiency led to the upregulation of mei4+ and rep1+, both of which activate the transcription of many meiotic genes, the accumulation of meiotic mRNAs observed in mitotic red1Δ cells was possibly due to misexpression of these transcription factors. To test this, we combined red1Δ with mei4Δ or rep1Δ and examined the steady-state levels of meiotic mRNAs. RT–PCR results clearly indicated that neither mei4Δ nor rep1Δ suppressed the increases in meiotic mRNAs caused by red1Δ (Figure 2C), suggesting that the upregulated meiotic mRNAs in red1Δ cells were direct targets of Red1 and that Mei4 and Rep1 did not mediate the effects of red1Δ.
Because the levels of meiotic mRNAs were stable in mitotic red1Δ cells, it is possible that these meiotic mRNAs were translated into proteins. To test whether meiosis-specific proteins were present in the mitotic red1Δ cells, we introduced mei4+-HA and mcp5+-myc individually into the red1Δ cells and performed western blot assays to detect the fusion proteins. Mei4-HA and, to a lesser extent, Mcp5-myc proteins were detected in mitotic cells lacking Red1 (Figure 2D). These results suggest that at least some meiotic proteins are expressed in mitotic red1Δ cells, and this could explain the growth defects observed in vegetative red1Δ cultures because meiotic proteins, including Crs1 and Rem1, cause growth arrest in mitotically dividing cells (Averbeck et al, 2005; Malapeira et al, 2005).
Retrotransposons are repressed by Red1 during mitosis
Previous studies have demonstrated that transposable elements, including wtf3 and wtf11 (also known as meu8+ and meu24+, respectively), are upregulated in meiotic cells (Watanabe et al, 2001; Mata et al, 2002). Similarly, our expression analyses demonstrated that transcripts derived from wtf elements increased significantly in red1Δ cells. We carried out RT–PCR to confirm our microarray results, and observed that the levels of wtf, Tf2 long terminal repeat (LTR), and Tf2 open reading frame (ORF) RNAs were increased in red1Δ relative to red1+ cells (Supplementary Figure S2A). The S. pombe genome contains retrotransposons and their remnants, including Tf2, wtf, and solo LTRs (Bowen et al, 2003). A previous study showed that transposase-related centromeric protein CENP-B homologues, Abp1, Cbh1, and Cbh2, bind to these elements and recruit two histone deacetylases, Clr3 and Clr6, to silence them at the level of transcription in S. pombe (Cam et al, 2008). Intriguingly, Tf2 elements are clustered into discrete nuclear foci, termed ‘Tf bodies’, and this clustering depends on CENP-B proteins (Cam et al, 2008). We compared the localization patterns of Red1 and Abp1 and found that Red1 did not co-localize with Abp1 (Supplementary Figure S2B). These data suggest that Red1 directly or indirectly contributes to the suppression of retrotransposons.
Red1 is required for the expression of various genes
The expression analyses identified a relatively small number of RNAs, representing 30 genes, with decreased expression in red1Δ cells (Supplementary Figure S3A; Supplementary Table SIII). Approximately 30% of the genes identified, including mfm2+, ste4+, and sep11+, are involved in conjugation and meiosis (Kjaerulff et al, 1994; Barr et al, 1996; Grallert et al, 1999) or are upregulated during meiosis (Mata et al, 2002; Chikashige et al, 2006), and the downregulation of these genes could explain the defective mating and sporulation seen in red1Δ cells. SPBPB10D8.03, a pseudogene, was the most downregulated transcript in red1Δ cells (Supplementary Figure S3B). We performed RT–PCR to confirm part of our expression array results (Supplementary Figure S3C). We found that many of these downregulated genes are clustered; specifically, 20 of these mRNAs are located in the regions close to telomeres of chromosomes 1 and 2 (Supplementary Figure S3D). These results suggest that Red1 directly or indirectly maintains the expression of several genes, in particular, subtelomeric genes.
Red1 co-localizes and interacts with Mmi1 in mitotic cells
The findings that Red1 and Mmi1 form nuclear foci in vegetatively growing cells (Harigaya et al, 2006) (Figure 1A) and that both proteins are essential for meiotic RNA elimination (Figure 2) prompted us to examine whether Red1 co-localizes with Mmi1. Microscopic analysis indicated that Red1 does co-localize with Mmi1 in mitotic cells (Figure 3A). Furthermore, reciprocal immunoprecipitation experiments demonstrated that Red1 co-immunoprecipitates with Mmi1 and vice versa; however, only small fractions of Red1 and Mmi1 were found to interact with each other (Figure 3B). It is possible that the Red1 interaction with Mmi1 is mediated by RNA molecules. To address this, we examined the sensitivity of Red1–Mmi1 interaction to RNase treatment. We found that Red1 does in fact co-immunoprecipitate with Mmi1 even after RNase treatment (Supplementary Figure S4A), suggesting that RNA molecules do not mediate this interaction.
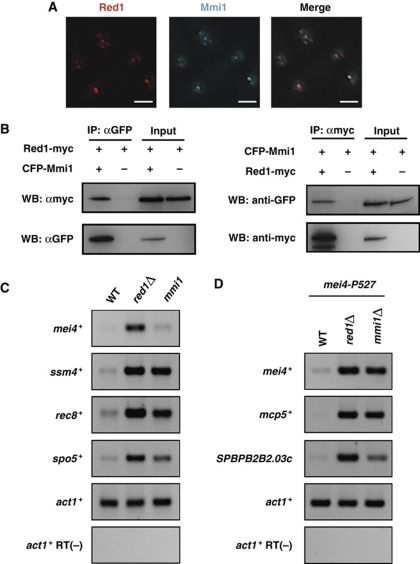
Figure 3.
Red1 co-localizes and interacts with Mmi1 in vegetative cells. (A) Red1 foci coincide with Mmi1 dots. Deconvolved images of a strain expressing both Red1-tdTomato and CFP-Mmi1 are shown. Bars, 2 μm. (B) Red1 co-immunoprecipitates with Mmi1. Total cell lysates from strains expressing CFP-Mmi1, Red1-myc, and Red1-myc/CFP-Mmi1 were subjected to immunoprecipitation, followed by western blotting. (C) Red1 and Mmi1 have common target genes. RT–PCR of four DSR-containing genes (mei4+, ssm4+, rec8+, and spo5+) indicates that either a red1 deficiency or a mmi1 mutation (mmi1-619) resulted in increased levels of these meiotic mRNAs. (D) Meiotic mRNAs accumulate in conditional mmi1Δ cells. Total RNA samples from mei4-P527, red1Δmei4-P527, and mmi1Δmei4-P527 cells were subjected to RT–PCR analyses. RT–PCR of mei4+, mcp5+, and SPBPB2B2.03c indicates that red1Δ or mmi1Δ in a mei4-P527 background results in increased levels of these meiotic transcripts. Note that mmi1+ was deleted in the mei4-P527 mutant strain because mei4 deficiency suppresses the severe growth defect caused by mmi1Δ.
We also confirmed that four DSR-containing meiotic mRNAs, mei4+, ssm4+, rec8+, and spo5+, accumulated in red1Δ cells (Figure 3C). We observed a slight increase of mei4+ mRNA in the mmi1-619 mutant. This modest response probably resulted from the weak penetrance of the mmi1-619 mutation because the phenotypes of mmi1-619 are milder than those of other mutations such as mmi1-ts3 and mmi1-ts6 (Harigaya et al, 2006). Consistent with this idea, we observed the clear upregulation of mei4+ mRNA in a conditional mmi1Δ strain (Figure 3D). These data suggest that Red1 and Mmi1 work in the same pathway or overlapping pathways to remove meiotic mRNAs from vegetatively growing cells.
Red1 co-localizes with pre-mRNA 3′-processing factors and facilitates polyadenylation of meiotic mRNAs
A recent study has shown that Mmi1 interacts and co-localizes with polyadenylation factors and the nuclear exosome and that the polyadenylation of DSR-containing mRNAs is a prerequisite for selective mRNA degradation by the exosome (Yamanaka et al, 2010). Therefore, we tested whether Red1 foci coincided or overlapped with pre-mRNA 3′-processing factors and the exosome. As we anticipated, Red1 well co-localized with Pla1, the canonical PAP; Rrp6, a subunit of the nuclear exosome; Pab2, a nuclear PABP; and Pcf11 (SPAC4G9.04c), a component of 3′-end processing machinery, which is homologous to budding yeast PCF11 (Figure 4A), but we also noticed that there were some Red1 foci that did not co-localize with 3′-processing factors. Immunoprecipitation experiments demonstrated that Red1 co-immunoprecipitated with Pla1 and Rrp6, but not with Pab2 (Figure 4B; Supplementary Figure S5). In addition, Red1 association with Pla1 and Rrp6 was refractory to RNase treatment (Supplementary Figure S4B), suggesting that Red1 physically binds to Pla1 and Rrp6.
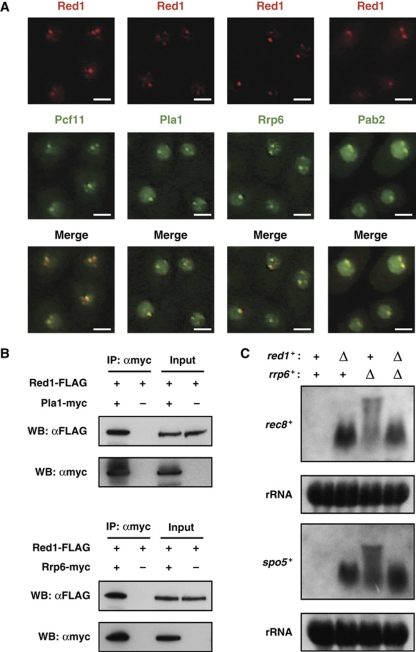
Figure 4.
Red1 localizes to cleavage bodies in mitotically dividing cells and cooperates with polyadenylation factors to hyperadenylate meiotic mRNAs. (A) Red1 dots co-localize with cleavage factor, Pcf11; the canonical poly(A) polymerase, Pla1; a nuclear exosome subunit, Rrp6; and a nuclear poly(A)-binding protein, Pab2. Microscopic images of strains expressing both Red1-tdTomato and either Pcf11-, Pla1-, Rrp6-, or Pab2-GFP are shown. Bars, 2 μm. (B) Red1 co-immunoprecipitates with Pla1 and Rrp6. Total cell extracts prepared from strains expressing both Red1-FLAG and Rrp6/Pla1-myc were subjected to immunoprecipitation, followed by western blotting. (C) Meiotic mRNAs in red1Δ do not have highly elongated poly(A) tails. Northern blotting was performed to analyse rec8+ and spo5+ mRNAs in wild type (WT), red1Δ, rrp6Δ, and red1Δrrp6Δ. The same blots were stained with methylene blue to visualize ribosomal RNAs (rRNA), which serve as a loading control.
The rec8+ and spo5+ mRNAs have highly elongated poly(A) tails in a rrp6 mutant, but not in mmi1 or pla1 mutants (Yamanaka et al, 2010). Therefore, we performed northern blot analyses to determine which step, intensive adenylation or degradation, was affected in red1Δ cells. As shown in Figure 4C, rec8+ and spo5+ mRNAs in red1Δ were evident as distinct bands; in contrast, these mRNAs in rrp6Δ were evident in smeary and slowly migrating bands as described previously (Yamanaka et al, 2010). We also observed distinct bands of rec8+ and spo5+ mRNAs in red1Δrrp6Δ cells. These results suggest that Red1 and Mmi1 facilitate the polyadenylation of DSR-containing RNAs by Pla1 and/or degradation by the exosome.
Red1 is involved in DSR-directed mRNA degradation
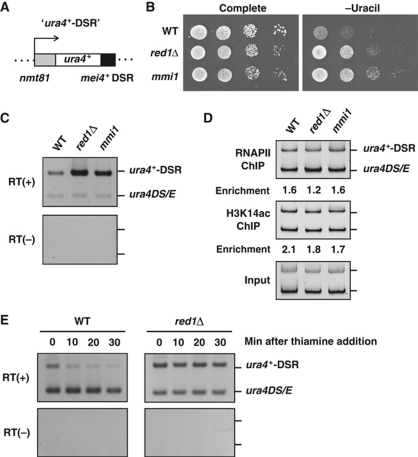
Although we demonstrated that Red1 negatively regulates meiotic mRNAs (Figures 2 and 3), it was still unclear whether Red1 was required for DSR-mediated selective RNA elimination. To address this issue, we utilized the ura4+-DSR construct, which is composed of the thiamine-repressive nmt81 promoter and the ura4+ gene fused with the DSR region derived from mei4+ (Figure 5A). We generated wild-type, red1Δ, and mmi1-619 mutant strains carrying the ura4+-DSR construct; we spotted serial dilutions of these strains onto complete or uracil-deficient (−Ura) plates. Wild-type cells did not grow on −Ura plates because of selective elimination of ura4+-DSR mRNA (Figure 5B). However, red1Δ as well as mmi1-619 mutants with the ura4+-DSR construct grew on the −Ura plates, indicating that the ura4+-DSR transcript accumulated in these strains (Figure 5B), consistent with the idea that DSR-containing mRNAs were translated into proteins in the absence of Red1 function (Figure 2D). The accumulation of ura4+-DSR mRNA in red1Δ and mmi1-619 was also confirmed by RT–PCR (Figure 5C). To determine whether red1Δ resulted in a higher rate of ura4+-DSR transcription, we performed chromatin immunoprecipitation (ChIP) with ura4+-DSR to assess RNA polymerase II (Pol II) occupancy and histone H3K14 acetylation (H3K14ac), which are both associated with active transcription (Sugiyama et al, 2007). ChIP results showed that there was no significant difference in the levels of Pol II and H3K14ac at the ura4+-DSR locus among the wild-type, red1Δ, and mmi1-619 strains (Figure 5D), suggesting that red1Δ did not enhance the transcriptional activity of ura4+-DSR. We therefore hypothesized that red1Δ increased the stability of DSR-containing meiotic mRNAs, and compared the stability of the ura4+-DSR transcript between wild type and red1Δ. We found that in wild-type cells, the level of ura4+-DSR transcript decreased rapidly, within 10 min of shutting down the nmt81 promoter with thiamine, whereas in red1Δ cells the ura4+-DSR mRNA levels persisted even 30 min after shutting down transcription of ura4+-DSR (Figure 5E). Therefore, we conclude that Red1 promotes meiotic mRNA degradation by destabilizing DSR-containing RNAs.
Figure 5.
Red1 promotes the destabilization of DSR-containing mRNA. (A) Schematic representation of the ura4+-DSR construct. The DSR region derived from mei4+ was fused to the ura4+ gene and this ura4+-DSR mRNA was driven by the nmt81 promoter, which was repressed in the presence of thiamine. This construct was integrated into the lys1+ locus. (B) red1Δ led to the ‘ura4+-on’ state in cells carrying the ura4+-DSR construct. Serial dilutions of wild-type (WT), red1Δ, and mmi1-619 (mmi1) cells containing the ura4+-DSR construct were spotted onto a complete or uracil-lacking plate and then incubated for 3–4 days at 30°C. (C) The ura4+-DSR transcript accumulates in red1Δ and mmi1-619 cells. Wild-type, red1Δ, and mmi1-619 cells carrying both the ura4+-DSR construct and a ura4+ minigene (ura4DS/E) were grown in the absence of thiamine and then subjected to RT–PCR analysis using a ura4+ primer set that amplifies both ura4+-DSR and ura4DS/E. The ura4DS/E was used as the internal control. (D) red1Δ does not increase RNA polymerase II (Pol II) occupancy or the levels of histone H3 Lys14 acetylation (H3K14ac). Wild-type, red1Δ, and mmi1-619 cells expressing both ura4+-DSR and ura4DS/E were subjected to chromatin immunoprecipitation using anti-Pol II and anti-H3K14ac antibodies. The precipitated DNAs were analysed by PCR using the same primer described in (C). (E) The ura4+-DSR became more stable in red1Δ compared with wild-type cells. Thiamine was added to the culture of wild-type or red1Δ cells grown without thiamine, and then the levels of ura4+-DSR and ura4DS/E transcripts were examined by RT–PCR.
Red1 downregulates meiotic mRNAs independently of Mei2/meiRNA
Taken together, our data suggest that Red1 is most likely functioning in concert with Mmi1 and other RNA processing factors to remove meiotic mRNAs. However, it is possible that Red1, like Pat1 kinase and 14-3-3 protein Rad24 (Kitamura et al, 2001; Sato et al, 2002), inhibits Mei2 activity in mitosis, allowing Mmi1 to eliminate meiotic RNAs (Supplementary Figure S6A). To rule out this possibility, we combined red1Δ with mei2Δ or sme2Δ and examined the steady-state levels of mei4+ and rec8+ mRNAs in red1Δmei2Δ and red1Δsme2Δ cells. RT–PCR data clearly indicated that mei4+ and rec8+ transcripts accumulated in red1Δ, red1Δmei2Δ, and red1Δsme2Δ cells, but not in mei2Δ or sme2Δ cells (Supplementary Figure S6B). These observations indicate that Red1 promotes meiotic mRNA elimination rather than inactivating Mei2/meiRNA function.
Red1 zinc-finger domain is essential for mRNA elimination but not for Red1 localization
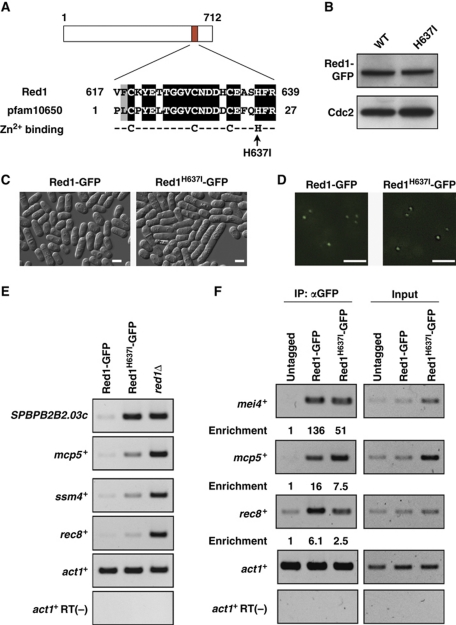
As Red1 contains a putative CCCH-type zinc-finger domain conserved in many species, we hypothesized that this zinc-finger domain is critical for Red1 function. To test this hypothesis, we introduced a point mutation (His637 to Ile) to the zinc-finger domain of Red1 (Figure 6A). The same strategy (H to I substitution) has previously been utilized to abrogate Zfs1 function (Kanoh et al, 1995). Western blotting demonstrated that the protein level of Red1H637I-GFP was comparable to that of Red1-GFP (Figure 6B), suggesting that the mutation did not significantly affect the stability of the mutant fusion protein. With microscopic analyses, we observed that the elongated cells were more frequently found in the Red1H637I-GFP cell cultures than in the Red1-GFP cell cultures (Figure 6C), suggesting that the H637I substitution mimics, at least to some extent, red1Δ in terms of cell morphology. We next tested whether the mutation altered Red1 localization and found that Red1H637I-GFP and Red1-GFP localized as nuclear dots (Figure 6D), suggesting that the integrity of the zinc-finger domain was dispensable for the formation of Red1 dots in the nucleus. We then compared the expression levels of meiotic mRNAs between Red1-GFP and Red1H637I-GFP cells. RT–PCR experiments demonstrated that meiotic mRNAs, SPBPB2B2.03c, mcp5+, ssm4+, and rec8+, increased in the Red1H637I-GFP cells, but the increased levels of mcp5+ and ssm4+ were not as high as in red1Δ cells (Figure 6E), indicating that the H637I mutation affected but not completely abolished Red1 activity. Given that the canonical CCCH zinc-finger domain is found in many RNA/DNA-binding proteins, we assumed that the Red1 zinc-finger might be involved in binding target mRNAs. We therefore carried out an RNA-IP assay to investigate whether Red1 associated with meiotic mRNAs. In this assay, Red1-GFP or Red1H637I-GFP was immunoprecipitated, and the mRNAs co-precipitated with Red1 or Red1H637I were analysed by RT–PCR. As shown in Figure 6F, meiotic mRNAs were enriched in the Red1-GFP pull-down fraction compared with the Red1H637I-GFP pull-down.
Figure 6.
Red1 zinc-finger motif is required for meiotic RNA elimination. (A) Red1 possesses a putative zinc-finger domain. A BLAST search reveals homology between Red1 and a conserved domain termed pfam10650, a CCCH-type zinc-finger that has the CX8CX4C3H motif as a consensus sequence. The arrow indicates the conserved histidine residue, which is substituted for isoleucine. (B) The histidine637 to isoleucine mutation (H637I) does not affect the protein level of Red1. Protein extracts from Red1-GFP and Red1H637I-GFP strains were examined by western blotting using anti-GFP or anti-Cdc2 antibody. Cdc2 was probed as a loading control. (C) DIC images of vegetative cells expressing Red1-GFP and Red1H637I-GFP strains. Bars, 5 μm. (D) Fluorescent microscopy indicates that both Red1-GFP and Red1H637I-GFP form nuclear dots. Bars, 2 μm. (E) The H637I mutation leads to the accumulation of meiotic RNAs. Total RNAs isolated from Red1-GFP, Red1H637I-GFP, and red1Δ strains were subjected to RT–PCR using primers specific for four meiotic mRNAs, SPBPB2B2.03c, mcp5+, ssm4+, and rec8+. (F) RNA-immunoprecipitation of Red1-GFP and Red1H637I-GFP. Cell lysates from untagged, Red1-GFP, and Red1H637I-GFP strains were subjected to immunoprecipitation using anti-GFP, and precipitated RNAs were analysed by RT–PCR. The values of relative enrichment are shown.
Taken together, these results suggest that the Red1 CCCH zinc-finger domain, which might have RNA-binding activity, is important for the destabilization of meiotic mRNAs but is dispensable for Red1 localization.
The formation of Red1 foci is dynamically regulated during meiosis
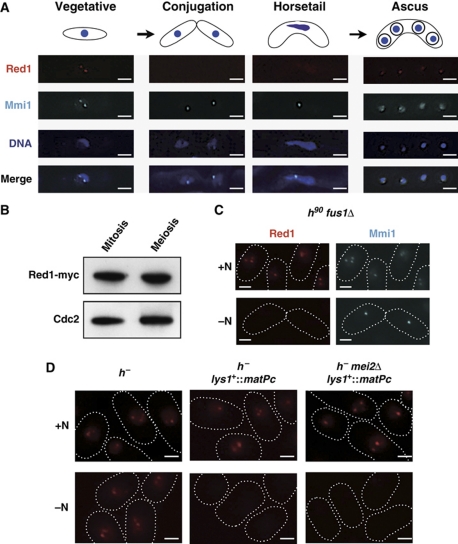
As described above, Red1 and Mmi1 cooperate to eliminate meiotic mRNAs in vegetative cells. Once cells enter meiosis, these factors should be inactivated to allow meiotic mRNAs to be translated, otherwise Red1/Mmi1 could interfere with the expression of meiotic genes. It has been proposed that from the conjugation stage to meiosis I, the Mei2-meiRNA ribonucleoprotein physically captures and inactivates Mmi1 (Harigaya et al, 2006). We hypothesized that Red1 co-localizes with Mmi1 and Red1 function is under the control of Mei2-meiRNA in meiosis. To explore this hypothesis, we examined the Red1 localization pattern during meiosis. Unexpectedly, Red1 foci disappeared before the completion of conjugation and re-appeared in spores (Figure 7A). These Red1 localization patterns were clearly different from those of Mmi1. In addition, the disappearance of Red1 foci occurred even in mei2Δ, mei3Δ, mei4Δ, and sme2Δ cells (Supplementary Figure S7). This suggests that the localization patterns of Red1 and Mmi1 are regulated independently during meiosis. We next performed western blotting to determine whether Red1 is dispersed or was degraded during meiosis. Western blotting revealed that Red1 protein is in fact present, even in the middle of meiosis when Red1 foci are not evident (Figure 7B). These data indicate that Red1 foci disassemble at an early stage of meiosis and then reassemble after meiotic chromosome segregation is completed, and that Red1, unlike Mmi1, is not under the control of Mei2/meiRNA during meiosis.
Figure 7.
Red1 foci are dynamic during meiosis. (A) Red1 signals disappeared at early in meiosis and then reappear in spores. Representative deconvolved images of a strain expressing both Red1-tdTomato and CFP-Mmi1 during meiosis are shown. DNA was stained with DAPI. Bars, 2 μm. (B) The protein level of Red1 does not change during meiosis. Cell lysates were prepared from a myc-tagged Red1-expressing strain in a vegetatively growing state or arrested at meiotic metaphase I and then analysed by western blotting. Cdc2 levels were also examined as a loading control. (C) Cell conjugation was not required for the loss of Red1 foci. fus1Δ cells expressing both Red1-tdTomato and CFP-Mmi1 were cultured with (+N) or without (−N) a nitrogen source at 26°C. Representative images are shown. The shapes of cells are indicated with white dotted lines. Bars, 2 μm. (D) Pheromone signaling but not nitrogen starvation triggers the disassembly of Red1 foci. h− strains carrying Red1-tdTomato, Red1-tdTomato/matPc, or Red1-tdTomato/matPc/mei2Δ were grown in the presence or absence of nitrogen for 12 h at 26°C. The white dotted lines indicate cell shape. Bars, 2 μm.
The disassembly of Red1 foci might be initiated by nitrogen starvation, pheromone signaling, or cell conjugation. To discriminate among these possibilities, we first determined whether cell fusion would lead to Red1 dispersion. As shown in Figure 7C, nitrogen starvation caused the dispersion of Red1 dots in fus1Δ cells, which are able to carry out cell contact and agglutination but not conjugation (Petersen et al, 1995). We next utilized two strains: h− and h− carrying matPc (h− lys1+::matPc). With lys1+::matPc, nitrogen starvation is reported to activate pheromone signaling even in h− strains (Yamamoto and Hiraoka, 2003). When we eliminated the nitrogen source, Red1 signals were evident in h− cells, but not in h− lys1+::matPc cells (Figure 7D). In addition, the loss of Red1 foci caused by nitrogen deprivation in h− lys1+::matPc was not suppressed by mei2Δ (Figure 7D), supporting the notion that Red1 localization is not under the control of Mei2 during meiosis. We also monitored Red1 foci and Red1 protein levels during the activation of pheromone signaling by microscopic analyses and western blotting, respectively. As shown in Supplementary Figure S8, we did not observe the significant changes in Red1 protein levels when Red1 foci disappeared, suggesting that Red1 had dispersed in response to the pheromone. These results clearly demonstrate that activation of pheromone signaling, rather than nitrogen starvation or conjugation, elicits the disassembly of Red1 dots.
Discussion
In this report, we have shown that Red1, which forms nuclear bodies, is essential for meiotic mRNA degradation during mitosis. We also demonstrated that Red1 foci disassemble in response to activation of pheromone signaling. Therefore, we have identified Red1 as another key factor in the process of meiotic mRNA elimination in mitotically dividing fission yeast.
We have demonstrated that Red1 is essential for DSR-mediated mRNA decay and that Red1 co-localizes and interacts with Mmi1 in vegetatively growing cells. These results indicate that Red1 is a novel factor working with Mmi1 to suppress meiotic mRNAs. We also found that Red1 foci disappeared during meiosis in mei2Δ, mei3Δ, mei4Δ, or sme2Δ cells, and that this is quite different to the localization dynamics of Mmi1, which co-localizes with the Mei2 dot from the conjugation stage to meiosis I (Harigaya et al, 2006). This difference suggests that Red1 and Mmi1 are independently regulated in meiosis. Moreover, the period when Red1 dots are lost, which largely extends from the activation of pheromone signaling to the completion of chromosome segregation, may coincide with meiotic gene expression. This implies that the localization of Red1 with cleavage bodies is crucial for its mRNA elimination activity. Furthermore, red1+ is dispensable for cell viability but mmi1+ is essential, and we did not observe Red1 interaction with Pab2, which associates with Mmi1 (Yamanaka et al, 2010). These differences may represent the complexity of the selective removal of meiotic mRNAs.
Several questions remain unanswered. First, is the localization of Red1 to cleavage bodies essential for its function? Second, why does Red1 delocalize during meiosis, and is this delocalization necessary for Red1 function? Third, why do Red1 and Mmi1 show distinct localization patterns in meiosis? To address these questions, it would be useful to isolate several red1 mutations, for example, a mutant that co-localizes with Mmi1 in meiosis, a mutant that does not localize to cleavage bodies in mitosis, and another that does not bind to Mmi1. By isolating various red1 mutants, the detailed functions of this protein will be revealed.
We have found that Red1 foci disperse in response to the activation of pheromone signaling and that this disassembly does not depend on Mei2, Mei3, Mei4, or meiRNA. Pheromone signaling eventually activates the Byr2/Byr1/Spk1 MAP kinase cascade (Yamamoto, 1996), and a domain search revealed that Red1 contains several MAP kinase docking motifs ([R/K]XXXXØXØ, Ø: hydrophobic residue) and MAPK phosphorylation sites ([S/T]P) (Tanoue and Nishida, 2003). Therefore, we suggest that Red1 localization is under the control of a MAP kinase cascade and that Red1 itself and/or, if any, Red1-binding protein(s) are phosphorylated, which results in the loss of Red1 foci in meiotic cells.
Our analyses demonstrated that Red1 associates with two essential factors in DSR-mediated mRNA decay: Pla1, a canonical PAP that polyadenylates both meiotic and non-meiotic mRNAs; and Rrp6, which is responsible for the degradation of DSR-containing mRNAs with poly(A) tails. It has been also shown that the meiosis-specific processing of crs1+ mRNA is strictly prevented during mitosis by Mmi1 and that Mmi1 physically interacts with Rna15, a component of the cleavage stimulation subcomplex CF IA (McPheeters et al, 2009; Yamanaka et al, 2010). A previous study demonstrated that in budding yeast, Rrp6 physically interacts with the canonical PAP Pap1 and is proposed to degrade aberrantly processed pre-mRNAs (Burkard and Butler, 2000). This degradation system, called nuclear RNA surveillance, is activated by defects in pre-mRNA processing steps, including 5′-capping, splicing, 3′-end formation, and messenger ribonucleoprotein (mRNP) assembly (Schmid and Jensen, 2008). Based on these observations, we speculate that Mmi1 and Red1 may affect one or more steps in pre-mRNA processing, in particular, splicing and/or export-competent mRNP formation of meiotic mRNAs carrying DSR sequences, resulting in the activation of nuclear exosome-dependent RNA elimination. The nuclear surveillance system not only degrades aberrantly processed mRNAs but also retains transcripts at the sites of transcription, resulting in the formation of intranuclear foci in the presence of the functional nuclear exosome (Hilleren and Parker, 2001; Hilleren et al, 2001; Jensen et al, 2001). Therefore, it would be intriguing to test whether Mmi1/Red1 dots coincide with the transcriptional sites of meiotic genes. We are currently investigating the detailed mechanism of this elimination system.
As described above, it is most likely that DSR-mediated RNA elimination represents a distinct class of polyadenylation-dependent RNA degradation systems. The findings that mRNA degradation as signaled by DSR requires the canonical PAP, a nuclear PABP, and hyperadenylation are reminiscent of ‘host-shutoff’, the global mRNA degradation caused by lytic Kaposi's sarcoma-associated herpesvirus (KSHV) infection (Glaunsinger and Ganem, 2004b). Host-shutoff, mediated by the viral factor SOX, decreases ∼95% of host mRNAs (Glaunsinger and Ganem, 2004a; Chandriani and Ganem, 2007). The following results have been reported: SOX directly or indirectly increases poly(A) length of mRNAs; PAPII, one of the canonical PAPs, and PABPN1, a nuclear PABP, are responsible for SOX-mediated hyperadenylation and mRNA decay; mRNAs lacking poly(A) tails are refractory to mRNA elimination by SOX (Lee and Glaunsinger, 2009). These features are strikingly similar to DSR-directed mRNA decay (St-Andre et al, 2010; Yamanaka et al, 2010), and we anticipate that polyadenylation-dependent RNA decay systems are evolutionally conserved in eukaryotes and that KSHV takes over the conserved mRNA degradation system using SOX to downregulate host mRNAs. It would be of considerable interest to test whether mammalian homologues of Red1 and Mmi1 are involved in SOX-directed mRNA degradation.
Meiotic mRNAs are subjected to selective elimination in vegetatively dividing fission yeast, and the inactivation of this elimination system results in misexpression of meiosis-specific mRNAs in mitotic cells. A BLAST search indicated that the Red1 zinc-finger domain is conserved from fission yeast to human; therefore, it is possible that Red1 homologues prevent the expression of meiosis/germline-specific genes in other organisms. Indeed, a similar phenomenon, termed soma-to-germline transformation, has been reported in Caenorhabditis elegans. In the worm, germline-specific genes are transcriptionally repressed by the Mi-2 complex, composed of MEP-1, LET-418, and HDA-1, and by factors involved in the Rb pathway including LIN-36, HPL-1, and DPL-1 (Unhavaithaya et al, 2002; Wang et al, 2005). A recent report has also demonstrated that insulin-like signaling mutants such as daf-2 and age-1 express germline genes in somatic cells (Curran et al, 2009). Although the misexpression of germline genes in mutant worms that are defective in Mi-2 function, Rb pathway, or insulin-like signaling is the result of transcriptional derepression, we speculate that post-transcriptional regulation also contributes to the repression of germ-specific genes. Moreover, we expect that the selective removal of certain kinds of mRNA maintains the undifferentiated state of stem cells in mammals.
Materials and methods
Yeast strains and growth media
Supplementary Table SI provides a complete list of the S. pombe strains used in this study. Standard conditions and procedures were used for growth, sporulation, and tetrad analysis (Moreno et al, 1991). Mating and sporulation efficiencies were measured as described previously (Kanoh et al, 1995). Strains carrying the CFP-Mmi1, ura4+-DSR, or mmi1-619 mutation were provided by M Yamamoto (University of Tokyo), and Mei4-HA and Mcp5-myc strains were obtained from the National Bioresource Project (NBRP/YGRC) of Japan. Other strains expressing tdTomato (Shaner et al, 2004), GFP (Ogawa et al, 2004), FLAG, or myc epitope-tagged proteins and deletion strains were constructed using PCR-based methods (Bahler et al, 1998; Kawashima et al, 2007). To generate the red1H637I mutant, a wild-type strain was transformed with a DNA fragment consisting of mutated red1 fragment fused to a GFP tag and the kanMX6 cassette. The mutation was confirmed by genomic DNA sequencing.
RNA analyses
Total RNA samples were prepared from yeast cells growing exponentially in YEA (YE supplemented with 75 mg/l adenine) using mirVana™ miRNA isolation kit (Applied Biosystems). DNA contamination in the RNA samples was removed using TURBO DNA-free™ (Applied Biosystems). The DNase-treated total RNA samples were then subjected to microarray analyses using the S. pombe expression 4x72K array (090408 Spom exp X4, Roche-NimbleGen), northern blotting using the Alkphos Direct™ Labelling and Detection System with CDP-Star™ (GE Healthcare), or RT–PCR using the PrimeScript® II 1st strand cDNA synthesis kit (TaKaRa Bio Inc.) and TaKaRa ExTaq® (TaKaRa Bio Inc.). The microarray analyses were independently preformed twice according to the manufacturer's protocol. Briefly, total RNAs were converted to double-stranded cDNAs and then labeled with Cy3. The S. pombe expression 4x72K array was hybridized with Cy3-labeled cDNAs, washed, and scanned. Scanned images were imported and the array data were extracted using the NimbleScan software (Roche-NimbleGen). The extracted data of wild-type and red1Δ cells were compared, and the transcripts showing more than a two-fold increase or a 0.5-fold decrease in both experiments were selected as ‘increased’ or ‘decreased’ RNAs, respectively. The total number of increased and decreased RNAs were 123 and 30, respectively. The microarray data are provided in Supplementary data and can be accessed at NCBI GEO under the accession number GSE23083. The lists of increased and decreased genes are shown in Supplementary Tables SII and SIII.
Immunoprecipitation and western blotting
Exponentially growing fission yeast strains expressing tagged proteins were harvested, resuspended in 1 × IP buffer (50 mM HEPES (pH 7.5), 150 mM KCl, 1 mM EDTA, 20% glycerol, 0.1% NP-40, 1 mM PMSF) supplemented with protease and phosphatase inhibitor cocktails (EDTA-free, Nacalai tesque), and disrupted with acid-washed glass beads (Sigma). Cell lysates were incubated with anti-c-myc (A-14, Santa Cruz) or anti-GFP (RQ2, MBL) antibody. Immunoprotein complexes were recovered by incubation with protein A/G-Sepharose beads (GE Healthcare) and then washed three times with 1 × IP buffer. Immunoprecipitation experiments with RNase treatment were performed as described previously (Lemieux and Bachand, 2009). Briefly, immunoprotein complexes captured with protein A/G-Sepharose beads were washed once and incubated for 30 min at room temperature with the RNase Cocktail™ Enzyme Mix (Applied Biosystems), which contains RNase A and RNase T1. After the RNase treatment, the beads were washed three times. The precipitated proteins were analysed by western blotting using anti-c-myc (9E10, Covance), anti-GFP (Roche), or anti-FLAG (M2, Sigma) antibody.
For western blotting of Mei4-HA, Mcp5-myc, Red1-myc, or Red1-GFP, protein extracts from exponentially growing yeast cultures were prepared using 20% trichloroacetic acid and acid-washed glass beads. Precipitated proteins were washed with ethanol and then resuspended in 1 M Tris–HCl (pH 8.8) and SDS–PAGE sample buffer. Anti-GFP (Roche), anti-c-myc (9E10, Covance), anti-HA (HA124, Nacalai tesque), or anti-Cdc2 (Y100.4, Santa Cruz) antibody was used for probing the epitope-tagged proteins or Cdc2.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was carried out as previously described (Sugiyama et al, 2005) with some modifications. Briefly, yeast cells were fixed with 1% formaldehyde for 30 min at room temperature. Chromatin fractions from yeast cells were immunoprecipitated with anti-Pol II (8WG16, Covance) or anti-histone H3 Lys14 acetylated (EP964Y, Abcam) antibody. DNA samples recovered from the immunoprecipitated fractions were analysed by PCR using TaKaRa ExTaq (TaKaRa Bio Inc.).
RNA-immunoprecipitation
Exponentially growing fission yeast strains expressing tagged proteins or the parental untagged strain were harvested, resuspended in 1 × RNA-IP buffer (50 mM HEPES (pH 7.5), 140 mM NaCl, 10% glycerol, 1 mM EDTA, 0.1% Triton X-100, 0.1% NP-40, 1 mM PMSF, 2 mM vanadyl ribonucleoside complex, and 400 U/ml RNasin Plus RNase inhibitor) supplemented with protease and phosphatase inhibitor cocktails (EDTA-free, Nacalai tesque), and disrupted with acid-washed glass beads (Sigma). Cell lysates were incubated with anti-GFP magnetic beads (RQ2, MBL). Immunoprotein complexes were washed three times with 1 × RNA-IP buffer, and then precipitated RNAs were recovered using Sepazol RNA I Super G (Nacalai tesque). The pull-down RNAs were subjected to DNase treatment and RT–PCR as described in RNA analyses.
Microscopy
An AxioImager M1 microscope (Carl Zeiss MicroImaging) was used for fluorescence microscopy and differential interference contrast (DIC) imaging. The raw images were processed using the AxioVision software (Carl Zeiss MicroImaging).
Supplementary Material
Acknowledgments
We thank S Grewal for the strains and for his support. We also thank M Yamamoto and A Yamashita for the strains and protocols, R Tsien and Y Watanabe for the tdTomato-tagging vector, H Ogawa for the GFP-expression vector, T Tani for the valuable discussions, S Fujishiro for technical assistance, and the National Bioresource Project (NBRP/YGRC) for the strains. This work was supported by a Grant-in-Aid for Young Scientists (A) of the Ministry of Education, Culture, Sports, Science and Technology, a Grant for Basic Science Research Projects of the Sumitomo Foundation and special coordination funds of JST (to TS). TS is a JST PRESTO researcher.
Author contributions: TS designed all of the experiments; TS and RS performed the experiments; TS and RS wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Anderson P, Kedersha N (2009) RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol 10: 430–436 [DOI] [PubMed] [Google Scholar]
- Averbeck N, Sunder S, Sample N, Wise JA, Leatherwood J (2005) Negative control contributes to an extensive program of meiotic splicing in fission yeast. Mol Cell 18: 491–498 [DOI] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A III, Steever AB, Wach A, Philippsen P, Pringle JR (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951 [DOI] [PubMed] [Google Scholar]
- Barr MM, Tu H, Van Aelst L, Wigler M (1996) Identification of Ste4 as a potential regulator of Byr2 in the sexual response pathway of Schizosaccharomyces pombe. Mol Cell Biol 16: 5597–5603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond CS, Fox AH (2009) Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol 186: 637–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen NJ, Jordan IK, Epstein JA, Wood V, Levin HL (2003) Retrotransposons and their recognition of pol II promoters: a comprehensive survey of the transposable elements from the complete genome sequence of Schizosaccharomyces pombe. Genome Res 13: 1984–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RS (2005) Zinc finger proteins: getting a grip on RNA. Curr Opin Struct Biol 15: 94–98 [DOI] [PubMed] [Google Scholar]
- Buchan JR, Parker R (2009) Eukaryotic stress granules: the ins and outs of translation. Mol Cell 36: 932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard KT, Butler JS (2000) A nuclear 3′-5′ exonuclease involved in mRNA degradation interacts with Poly(A) polymerase and the hnRNA protein Npl3p. Mol Cell Biol 20: 604–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam HP, Noma K, Ebina H, Levin HL, Grewal SI (2008) Host genome surveillance for retrotransposons by transposon-derived proteins. Nature 451: 431–436 [DOI] [PubMed] [Google Scholar]
- Chandriani S, Ganem D (2007) Host transcript accumulation during lytic KSHV infection reveals several classes of host responses. PLoS One 2: e811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y, Tsutsumi C, Yamane M, Okamasa K, Haraguchi T, Hiraoka Y (2006) Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell 125: 59–69 [DOI] [PubMed] [Google Scholar]
- Coop G, Przeworski M (2007) An evolutionary view of human recombination. Nat Rev Genet 8: 23–34 [DOI] [PubMed] [Google Scholar]
- Curran SP, Wu X, Riedel CG, Ruvkun G (2009) A soma-to-germline transformation in long-lived Caenorhabditis elegans mutants. Nature 459: 1079–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong L, Grande MA, Mattern KA, Schul W, van Driel R (1996) Nuclear domains involved in RNA synthesis, RNA processing, and replication. Crit Rev Eukaryot Gene Expr 6: 215–246 [DOI] [PubMed] [Google Scholar]
- Glaunsinger B, Ganem D (2004a) Highly selective escape from KSHV-mediated host mRNA shutoff and its implications for viral pathogenesis. J Exp Med 200: 391–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaunsinger B, Ganem D (2004b) Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Mol Cell 13: 713–723 [DOI] [PubMed] [Google Scholar]
- Grallert A, Grallert B, Zilahi E, Szilagyi Z, Sipiczki M (1999) Eleven novel sep genes of Schizosaccharomyces pombe required for efficient cell separation and sexual differentiation. Yeast 15: 669–686 [DOI] [PubMed] [Google Scholar]
- Hall TM (2005) Multiple modes of RNA recognition by zinc finger proteins. Curr Opin Struct Biol 15: 367–373 [DOI] [PubMed] [Google Scholar]
- Harigaya Y, Tanaka H, Yamanaka S, Tanaka K, Watanabe Y, Tsutsumi C, Chikashige Y, Hiraoka Y, Yamashita A, Yamamoto M (2006) Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature 442: 45–50 [DOI] [PubMed] [Google Scholar]
- Hebert MD (2010) Phosphorylation and the Cajal body: modification in search of function. Arch Biochem Biophys 496: 69–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH (2001) Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413: 538–542 [DOI] [PubMed] [Google Scholar]
- Hilleren P, Parker R (2001) Defects in the mRNA export factors Rat7p, Gle1p, Mex67p, and Rat8p cause hyperadenylation during 3′-end formation of nascent transcripts. RNA 7: 753–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen TH, Patricio K, McCarthy T, Rosbash M (2001) A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol Cell 7: 887–898 [DOI] [PubMed] [Google Scholar]
- Kanoh J, Sugimoto A, Yamamoto M (1995) Schizosaccharomyces pombe zfs1+ encoding a zinc-finger protein functions in the mating pheromone recognition pathway. Mol Biol Cell 6: 1185–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima SA, Tsukahara T, Langegger M, Hauf S, Kitajima TS, Watanabe Y (2007) Shugoshin enables tension-generating attachment of kinetochores by loading Aurora to centromeres. Genes Dev 21: 420–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura K, Katayama S, Dhut S, Sato M, Watanabe Y, Yamamoto M, Toda T (2001) Phosphorylation of Mei2 and Ste11 by Pat1 kinase inhibits sexual differentiation via ubiquitin proteolysis and 14-3-3 protein in fission yeast. Dev Cell 1: 389–399 [DOI] [PubMed] [Google Scholar]
- Kjaerulff S, Davey J, Nielsen O (1994) Analysis of the structural genes encoding M-factor in the fission yeast Schizosaccharomyces pombe: identification of a third gene, mfm3. Mol Cell Biol 14: 3895–3905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Glaunsinger BA (2009) Aberrant herpesvirus-induced polyadenylation correlates with cellular messenger RNA destruction. PLoS Biol 7: e1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux C, Bachand F (2009) Cotranscriptional recruitment of the nuclear poly(A)-binding protein Pab2 to nascent transcripts and association with translating mRNPs. Nucleic Acids Res 37: 3418–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malapeira J, Moldon A, Hidalgo E, Smith GR, Nurse P, Ayte J (2005) A meiosis-specific cyclin regulated by splicing is required for proper progression through meiosis. Mol Cell Biol 25: 6330–6337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Lyne R, Burns G, Bahler J (2002) The transcriptional program of meiosis and sporulation in fission yeast. Nat Genet 32: 143–147 [DOI] [PubMed] [Google Scholar]
- Matera AG, Izaguire-Sierra M, Praveen K, Rajendra TK (2009) Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev Cell 17: 639–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, Kobayashi Y, Hashimoto A, Hamamoto M, Hiraoka Y, Horinouchi S, Yoshida M (2006) ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol 24: 841–847 [DOI] [PubMed] [Google Scholar]
- McPheeters DS, Cremona N, Sunder S, Chen HM, Averbeck N, Leatherwood J, Wise JA (2009) A complex gene regulatory mechanism that operates at the nexus of multiple RNA processing decisions. Nat Struct Mol Biol 16: 255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194: 795–823 [DOI] [PubMed] [Google Scholar]
- Nebreda AR, Ferby I (2000) Regulation of the meiotic cell cycle in oocytes. Curr Opin Cell Biol 12: 666–675 [DOI] [PubMed] [Google Scholar]
- Ogawa H, Yu RT, Haraguchi T, Hiraoka Y, Nakatani Y, Morohashi K, Umesono K (2004) Nuclear structure-associated TIF2 recruits glucocorticoid receptor and its target DNA. Biochem Biophys Res Commun 320: 218–225 [DOI] [PubMed] [Google Scholar]
- Petersen J, Weilguny D, Egel R, Nielsen O (1995) Characterization of fus1 of Schizosaccharomyces pombe: a developmentally controlled function needed for conjugation. Mol Cell Biol 15: 3697–3707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N, Ohkura H (2003) Polo boxes form a single functional domain that mediates interactions with multiple proteins in fission yeast polo kinase. J Cell Sci 116(Part 7): 1377–1387 [DOI] [PubMed] [Google Scholar]
- Sato M, Watanabe Y, Akiyoshi Y, Yamamoto M (2002) 14-3-3 protein interferes with the binding of RNA to the phosphorylated form of fission yeast meiotic regulator Mei2p. Curr Biol 12: 141–145 [DOI] [PubMed] [Google Scholar]
- Schmid M, Jensen TH (2008) Quality control of mRNP in the nucleus. Chromosoma 117: 419–429 [DOI] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY (2004) Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22: 1567–1572 [DOI] [PubMed] [Google Scholar]
- Spector DL (2001) Nuclear domains. J Cell Sci 114(Part 16): 2891–2893 [DOI] [PubMed] [Google Scholar]
- St-Andre O, Lemieux C, Perreault A, Lackner DH, Bahler J, Bachand F (2010) Negative regulation of meiotic gene expression by the nuclear poly(a)-binding protein in fission yeast. J Biol Chem 285: 27859–27868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Cam H, Verdel A, Moazed D, Grewal SI (2005) RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc Natl Acad Sci USA 102: 152–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, Grewal SI (2007) SHREC, an effector complex for heterochromatic transcriptional silencing. Cell 128: 491–504 [DOI] [PubMed] [Google Scholar]
- Tanoue T, Nishida E (2003) Molecular recognitions in the MAP kinase cascades. Cell Signal 15: 455–462 [DOI] [PubMed] [Google Scholar]
- Unhavaithaya Y, Shin TH, Miliaras N, Lee J, Oyama T, Mello CC (2002) MEP-1 and a homolog of the NURD complex component Mi-2 act together to maintain germline-soma distinctions in C. elegans. Cell 111: 991–1002 [DOI] [PubMed] [Google Scholar]
- Wang D, Kennedy S, Conte D Jr, Kim JK, Gabel HW, Kamath RS, Mello CC, Ruvkun G (2005) Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature 436: 593–597 [DOI] [PubMed] [Google Scholar]
- Watanabe T, Miyashita K, Saito TT, Yoneki T, Kakihara Y, Nabeshima K, Kishi YA, Shimoda C, Nojima H (2001) Comprehensive isolation of meiosis-specific genes identifies novel proteins and unusual non-coding transcripts in Schizosaccharomyces pombe. Nucleic Acids Res 29: 2327–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Hiraoka Y (2003) Monopolar spindle attachment of sister chromatids is ensured by two distinct mechanisms at the first meiotic division in fission yeast. EMBO J 22: 2284–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M (1996) Regulation of meiosis in fission yeast. Cell Struct Funct 21: 431–436 [DOI] [PubMed] [Google Scholar]
- Yamanaka S, Yamashita A, Harigaya Y, Iwata R, Yamamoto M (2010) Importance of polyadenylation in the selective elimination of meiotic mRNAs in growing S. pombe cells. EMBO J 29: 2173–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanowitz J (2010) Meiosis: making a break for it. Curr Opin Cell Biol 22: 744–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Bodnar MS, Spector DL (2009) Nuclear neighborhoods and gene expression. Curr Opin Genet Dev 19: 172–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.