Abstract
Ribosome biogenesis is a tightly controlled pathway that requires an intricate spatial and temporal interplay of protein networks. Most structural rRNA components are generated in the nucleolus and assembled into pre-ribosomal particles, which are transferred for further maturation to the nucleoplasm and cytoplasm. In metazoa, few regulatory components for these processes have been characterized. Previous work revealed a critical role for the SUMO-specific protease SENP3 in the nucleolar steps of ribosome biogenesis. We biochemically purified a SENP3-associated complex comprising PELP1, TEX10 and WDR18, and demonstrate that this complex is involved in maturation and nucleolar release of the large ribosomal subunit. We identified PELP1 and the PELP1-associated factor LAS1L as SENP3-sensitive targets of SUMO, and provide evidence that balanced SUMO conjugation/deconjugation determines the nucleolar partitioning of this complex. This defines the PELP1–TEX10–WDR18 complex as a regulator of ribosome biogenesis and suggests that its SUMO-controlled distribution coordinates the rate of ribosome formation. These findings contribute to the basic understanding of mammalian ribosome biogenesis and shed new light on the role of SUMO in this process.
Keywords: nucleolus, ribosome biogenesis, SUMO
Introduction
Ribosomes are specialized molecular machines designated for the synthesis of cellular proteins. Eukaryotic ribosomes are composed of a small 40S and a large 60S subunit, each of which contains distinct ribosomal proteins and ribosomal RNA (rRNA). The structural rRNA component of the 40S subunit is the 18S rRNA, while the 60S subunit contains the 28S (25S in yeast), 5.8S and 5S rRNA species. Ribosomal assembly and maturation proceed via a highly regulated cellular pathway that involves a large set of non-ribosomal proteins as well as non-coding RNA species (Kressler et al, 2010). Distinct stages of this biogenesis pathway are spatially separated in specific subcellular regions (Henras et al, 2008). The process is initiated in the nucleolus, where RNA polymerase I generates a 47S (35S in yeast) rRNA precursor, which contains the sequences for the 18S, 5.8S and 28S rRNA. These regions are flanked by the external transcribed spacers (5′-ETS and 3′-ETS) and separated by the internal transcribed spacers (ITS1 and ITS2). Cleavage and processing of these spacer regions mostly take place in the nucleolus and generate rRNA intermediates that assemble in the nucleolar pre-40S and pre-60S particles, respectively. The pre-40S particle is rapidly exported to the cytoplasm, whereas the pre-60S particle undergoes a complex maturation pathway in the nucleolus, nucleoplasm and cytoplasm, which is accompanied by the exchange of accessory factors and a stepwise remodelling of the particles.
Genetic experiments in the yeast Saccharomyces cerevisiae led to the identification of many critical components of the ribosome biogenesis pathway. Moreover, biochemical approaches enabled the isolation of specific ribosomal pre-40S and pre-60S subcomplexes, which seem to function as intermediates along the pathway (Tschochner and Hurt, 2003). In mammalian cells, however, only a limited number of components have been characterized. Additionally, in both higher and lower eukaryotes it is largely unclear what drives the spatial distribution and the exchange of protein components along the maturation pathway and what controls the assembly and disassembly of the subcomplexes.
Genetic studies in yeast also revealed an involvement of the ubiquitin-like SUMO system in the formation and nuclear export of pre-ribosomal particles (Panse et al, 2006). Recent work by our group and others has delineated a crucial role of the SUMO system in the nucleolar steps of mammalian ribosome biogenesis (Haindl et al, 2008; Yun et al, 2008). SUMO functions as a post-translational modifier that is covalently attached to lysine residues of target proteins (Geiss-Friedlander and Melchior, 2007). Human cells express three SUMO forms (SUMO1–3, with SUMO2/3 being almost identical), which are conjugated to target proteins in a pathway that requires the E1 activating enzyme Aos1/Uba2, the E2 conjugating enzyme Ubc9 and in many cases involves E3 SUMO ligases (Wang and Dasso, 2009). The attachment of SUMO to target proteins typically modulates the dynamics and specificity of protein–protein interactions by either repelling or attracting a binding partner, which is mediated through non-covalent binding of the SUMO conjugate to a specialized SUMO-interaction motif (SIM) in a binding partner. Importantly, SUMO modification is a reversible process, in which the demodification of a given SUMO conjugate is catalysed by SUMO-specific proteases of the SENP family. In humans six SENP members, SENP1–3 and SENP5–7, have been identified so far (Hay, 2007; Mukhopadhyay and Dasso, 2007; Yeh, 2009). SENP3 and SENP5 exhibit preferential activities towards SUMO2/3 conjugates and are specifically concentrated in the nucleolus implying that they control nucleolar functions (Nishida et al, 2000; Di Bacco et al, 2006; Gong and Yeh, 2006). In line with this assumption, we and others have shown that SENP3 is required for rRNA processing, in particular, for the conversion of the 32S rRNA intermediate to the mature 28S rRNA (Haindl et al, 2008; Yun et al, 2008). This function is connected to nucleophosmin (NPM1), a key factor of 28S rRNA maturation in mammals (Grisendi et al, 2006). NPM1 is a major binding partner of SENP3 that governs its stability and nucleolar localization (Yun et al, 2008). Moreover, SENP3 mediates desumoylation of NPM1, which appears to be critical for NPM1 function in rRNA processing (Haindl et al, 2008; Nishida and Yamada, 2008).
Given that eukaryotic ribosome synthesis requires the coordinated action of a series of cellular components, we anticipated that it involves SENP3-mediated desumoylation of additional factors in this pathway. Here, we describe the characterization of a novel SENP3-associated complex comprising PELP1, TEX10, WDR18 and two associated proteins, MDN1 and LAS1L. We provide compelling evidence that this complex is implicated in large ribosomal subunit maturation. We further show that PELP1 and LAS1L are dynamically modified by SUMO in a SENP3-regulated process, and suggest that SENP3-mediated desumoylation is required for nucleolar compartmentalization of this complex.
Results
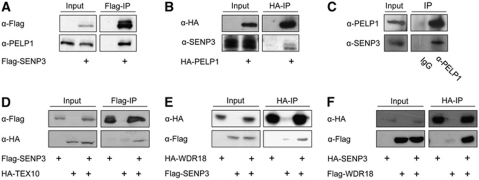
SENP3 is associated with a complex composed of PELP1, TEX10 and WDR18
To identify interaction partners and potential substrates of SENP3, we expressed Flag-tagged SENP3 in HEK293T cells and captured it together with associated proteins on a Flag-affinity column. Using mass spectrometry one major co-purifying protein was identified as NPM1 and others as PELP1, TEX10, WDR18 and LAS1L, respectively (Supplementary Figure 1A). While no function has been annotated for TEX10 and WDR18, PELP1 was mainly described as a transcriptional co-regulator of the oestrogen receptor (Vadlamudi and Kumar, 2007). Interestingly, LAS1L was very recently described as a factor involved in ribosome biogenesis (Castle et al, 2010). Moreover, Psi-Blast searches revealed a sequence similarity and conserved domains between PELP1, TEX10, WDR18 and the yeast proteins Rix1, Ipi1, Ipi3 (Supplementary Figures 1B and 2A–C), which are essential in yeast and form a complex that is required for ribosome biogenesis (Krogan et al, 2004; Nissan et al, 2004). In humans, no functional orthologue of this complex has been described so far. In order to establish that PELP1, TEX10 and WDR18 do indeed constitute a complex in mammalian cells and to confirm their association with SENP3 we performed directed co-immunoprecipitation experiments. We could validate the interaction of ectopically expressed Flag-SENP3 with endogenous PELP1 and reciprocally show binding of HA-PELP1 to endogenous SENP3 (Figure 1A and B). Furthermore, the association of both proteins at their endogenous levels of expression could be demonstrated (Figure 1C). In further agreement with the results from mass spectrometry Flag-SENP3 interacted with HA-TEX10 or HA-WDR18, and Flag-WDR18 bound HA-SENP3 (Figure 1D–F). Additionally, WDR18 and PELP1 were co-immunoprecipitated when expressed ectopically or at their endogenous levels of expression, irrespective of RNase treatment of cellular extracts (Figure 2A–C; Supplementary Figure 3A). Finally, we also detected binding of Flag-tagged TEX10 to HA-PELP1 and observed an interaction of HA-TEX10 with endogenous WDR18 (Figure 2D and E). From this network of mutual interactions, we conclude that PELP1, TEX10 and WDR18 are part of a common complex, which is physically linked to SENP3 (Figure 2F). The interdependency of components in this complex is further supported by the observation that siRNA-mediated depletion of WDR18 also led to a reduction in the amount of PELP1 and, reciprocally, loss of PELP1 results in lower amounts of WDR18 (Supplementary Figure 3B; see also Figure 4B, three bottom panels). The yeast Rix1–Ipi1–Ipi3 complex is transiently associated with the giant AAA ATPase Rea1 (Galani et al, 2004; Nissan et al, 2004; Ulbrich et al, 2009). Interestingly, mass-spectrometric analysis identified the human Rea1 orthologue MDN1 as part of the PELP1-associated proteome (Supplementary Figure 4A). Consistent with this finding, we observed an interaction of ectopically expressed Flag-PELP1 or endogenous PELP1 with endogenous MDN1 further supporting the idea that the mammalian PELP1–TEX10–WDR18 complex is related to the Rix1–Ipi1–Ipi3 complex (Supplementary Figure 4B and C). As mentioned above, the recently described rRNA processing factor LAS1L was also found to physically interact with SENP3 in the initial mass spectrometry-based proteomic analysis (Supplementary Figure 1A). Moreover, LAS1L was also identified as a binding partner of PELP1 in proteomic experiments and by directed co-immunoprecipitation (Supplementary Figure 4D and data not shown). This further points to an involvement of PELP1 and its associated components in the pathway of ribosome biogenesis.
Figure 1.
SENP3 is associated with PELP1, TEX10 and WDR18. (A, B) Flag-SENP3 or HA-PELP1 was expressed in HeLa cells as indicated. Proteins were captured on Flag- or HA-beads and immunoblotted using antibodies directed against endogenous PELP1 or SENP3 as indicated. (C) Endogenous PELP1 was immunoprecipitated from HeLa cells with a rabbit polyclonal antibody and immunocomplexes were probed for the presence of SENP3 by western blotting with an anti-SENP3 antibody. (D–F) Flag- or HA-tagged proteins were expressed in HeLa cells, captured on Flag- or HA-beads and subjected to immunoblotting as indicated. In all experiments, the inputs represent 2.5% of the total cell lysate.
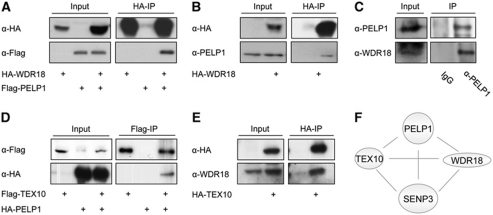
Figure 2.
WDR18 and TEX10 co-immunoprecipitate with PELP1. (A, B) Flag-PELP1 and HA-WDR18 were expressed in HeLa cells as indicated. HA-WDR18 was captured on HA-beads and the bound material was probed by immunoblotting for the presence of ectopically expressed Flag-tagged PELP1 or endogenous PELP1 as indicated. (C) Endogenous PELP1 was immunoprecipitated from HeLa cells with a rabbit polyclonal antibody and immunocomplexes were probed for the presence of endogenous WDR18 by western blotting with an anti-WDR18 antibody. (D) Flag-TEX10 was expressed with HA-PELP1 in HeLa cells, captured on Flag-beads and proteins were immunoblotted as indicated. (E) HA-TEX10 was expressed, captured on HA-beads and associated proteins were probed for the presence of endogenous WDR18 using immunoblotting. The inputs represent 2.5% of the total cell lysate. (F) Scheme of mutual interactions between PELP1, TEX10, WDR18 and SENP3.
Components of the complex localize to the nucleolus and are involved in ribosome biogenesis
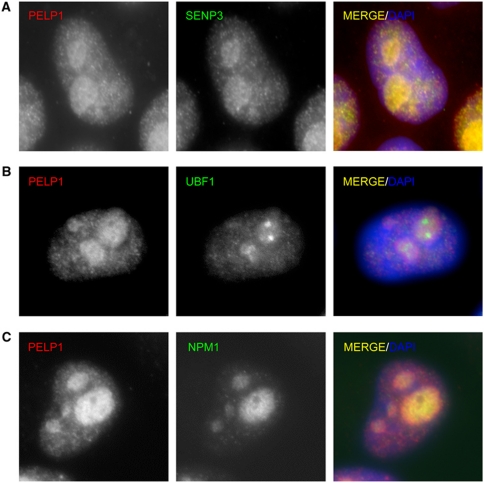
Because SENP3 is mainly compartmentalized in the nucleolus, we investigated the subcellular localization of PELP1, TEX10 or WDR18. Biochemical fractionation of cell extracts revealed that all three proteins are mainly found in the nucleoplasm with a subfraction concentrated in the nucleolus (Supplementary Figure 5). Immunofluorescence staining of HeLa cells with anti-PELP1 antibodies confirms that PELP1 indeed accumulates in the nucleolus, where it exhibits an overlapping localization with SENP3 (Figure 3A). The nucleolus can be subdivided into distinct functional subregions (Boisvert et al, 2007). The fibrillar centre (FC) is one such subregion and is the site of rDNA transcription. Co-staining of PELP1 with UBF1, which serves as a marker for the FC, shows that PELP1 is largely excluded from this region, indicating that it is most likely not directly involved in the transcription of rDNA (Figure 3B). NPM1 is a marker protein for the granular component (GC) subregion, where processing of the 32S rRNA precursor to the 28S rRNA and the assembly of pre-60S ribosome subunits take place. We observed an overlapping localization of PELP1 and NPM1 and additionally revealed an interaction of both proteins in co-immunoprecipitation experiments thus pointing to a functional role of PELP1 in the GC subregion (Figure 3C; Supplementary Figure 6A and B). Similarly to what observed for PELP1, the PELP1-associated components LAS1L and MDN1 exhibit strong nucleolar enrichment (Supplementary Figure 7A and B). TEX10 was also found in the nucleolus, but is more evenly distributed between nucleolus and nucleoplasm (Supplementary Figure 7C).
Figure 3.
PELP1 localizes to the granular region of the nucleolus. (A–C) Immunofluorescence staining of endogenous PELP1, SENP3, UBF1 and NPM1 in HeLa cells. Nuclei were visualized using DAPI staining.
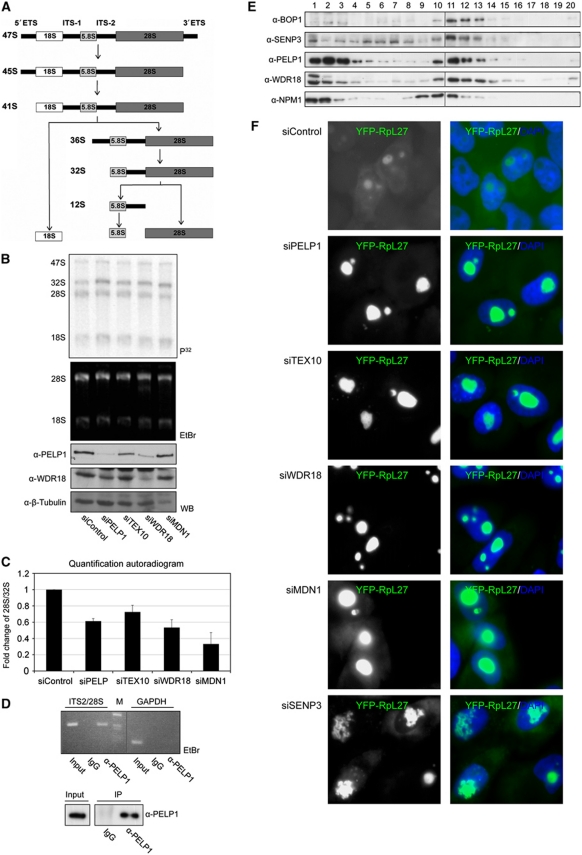
To investigate a potential involvement of PELP1 and its associated factors TEX10, WDR18 or MDN1 in rRNA processing, the proteins were individually depleted from HeLa cells by siRNA. rRNA processing—schematically depicted in Figure 4A—was studied by pulse-chase labelling of nascent rRNA with 32P-orthophosphate. Depletion of the respective proteins was verified by immunoblotting or quantitative RT–PCR (Figure 4B, three bottom panels; Supplementary Figure 8). To study pre-rRNA processing, RNA was prepared and separated by denaturing agarose gel electrophoresis. Metabolically labelled rRNA species were visualized by autoradiography (Figure 4B, upper panel). In cells transfected with a control siRNA, the mature 28S rRNA species is typically more abundant than its 32S rRNA precursor. By contrast, knock-down of PELP1, WDR18, TEX10 or MDN1 causes an accumulation of the 32S rRNA relative to the 28S form. Quantification of a series of experiments shows that the depletion of PELP1, TEX10, WDR18 or MDN1 reduces the ratio of the mature 28S rRNA to the 32S precursor to 60, 75, 50 or 30%, respectively, when compared with control cells (Figure 4C). This suggests that the PELP1–TEX10–WDR18 complex and its associated factor MDN1 are indeed involved in the conversion of the 32S rRNA intermediate to the mature 28S rRNA. To further substantiate this finding we wondered whether the complex contains rRNA species. To this aim, we isolated total RNA from anti-PELP1 immunoprecipitates (Figure 4D, bottom), reverse transcribed it to cDNA and performed a PCR with primers covering the region from the 3′-end of the ITS2 to the 5′-end of the 28S rRNA encoding region. A specific amplification product indicative for an ITS2-containing rRNA species was seen in anti-PELP1 precipitates, but not in control immunoprecipitates (Figure 4D, top). Importantly also, mRNA encoding for the housekeeping protein glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was absent from the anti-PELP1 immunoprecipitate excluding non-specific RNA association.
Figure 4.
PELP1, TEX10, WDR18 and MDN1 are involved in ribosome biogenesis. (A) Diagram summarizing the main steps of rRNA processing. (B) HeLa cells were transfected with siRNA duplexes targeting PELP1, TEX10, WDR18, MDN1 or a control siRNA as indicated. Downregulation of the respective proteins was analysed by western blotting as indicated (WB, three lower panels). At 72 h after transfection, cells were pulse labelled with 32P-orthophosphate for 1 h and chased for 2.5 h. RNA was separated on a denaturing agarose gel and detected by ethidium bromide (EtBr) staining and autoradiography after drying of the gel. (C) The signal intensities of the 28S and 32S rRNA forms were quantified by phosphoimager analysis and the ratio of 28S:32S rRNA was calculated. Values represent the average of four (two for siMDN1) independent experiments. Error bars indicate s.d. (D) PELP1 is associated with ITS2-containing RNA species. Endogenous PELP1 was immunoprecipitated from HeLa cells. In total, 10% of the immunoprecipitated material was analysed by western blotting (lower panel). The remaining material was used for RNA isolation and cDNA synthesis. The cDNA was used as template for PCR amplification using primers within the ITS2/28S region or GAPDH as a control. Input corresponds to 10% of the cell lysate. Vertical line in the upper panel indicates removal of irrelevant adjacent lanes. (E) Pre-ribosomal particles were separated by sucrose gradient centrifugation of nuclear extracts. Proteins were recovered from the fractions by TCA precipitation and further analysed by SDS–PAGE and western blotting using antibodies as indicated. Vertical lines solely indicate loading onto two separate gels. (F) Nucleolar export of pre-60S particles is strongly compromised in cells depleted of PELP1, TEX10, WDR18, MDN1 or SENP3. HeLa cells expressing YFP-RpL27 from a tetracycline-inducible promoter were transfected with the indicated siRNAs. After 24 h, expression of YFP-RpL27 was induced and 72 h later the localization of RpL27 was monitored. Pictures were taken using identical exposure times.
To monitor whether the PELP1–TEX10–WDR18 complex and SENP3 are associated with pre-ribosomes we isolated pre-ribosomal particles from cell nuclei by sucrose gradient centrifugation (Figure 4E; Supplementary Figure 9). Fractions of sucrose gradients were collected and the presence of BOP1, a component of the PES1–BOP1–WDR12 (PeBoW) rRNA processing complex (Lapik et al, 2004; Holzel et al, 2005) served as a marker for fractions containing pre-60S complexes (Figure 4E, fractions 10–13, note that in fractions 1–4 unbound proteins are found). This analysis revealed that endogenous PELP1 and WDR18 perfectly co-fractionate with BOP1 demonstrating that both proteins are associated with pre-60S particles. A significant fraction of SENP3 co-sediments with PELP1, WDR18 and BOP1, but SENP3 is also found in earlier fractions corresponding to pre-40S particles (Figure 4E, fractions 5–8; Supplementary Figure 9). Notably also, NPM1 is present in fractions overlapping with PELP1, WDR18 and BOP1, but was also reproducibly found in particles with lower density (Figure 4E, fractions 8 and 9).
To further validate the functional involvement of PELP1, TEX10, WDR18 and MDN1 in large ribosomal subunit maturation, we examined the localization of the YFP-tagged large ribosomal subunit protein L27 (YFP-RpL27) in HeLa cells depleted of the respective proteins (Figure 4F). In control siRNA-treated cells, YFP-RpL27 was detectable in the cytoplasm and nucleoli, which is in accordance with its presence in nucleolar pre-60S particles as well as mature 60S ribosomes (Andersen et al, 2005). Strikingly, however, in cells depleted of PELP1, TEX10, WDR18 or MDN1 YFP-RpL27 exclusively accumulated within nucleoli. Moreover, the nucleoli in these cells are dramatically enlarged, a phenotype associated with the depletion of other processing components (Rosby et al, 2009). Depletion of SENP3 induced a similar phenotype and led to the nucleolar accumulation of the YFP-RpL27 reporter and a complete loss of cytoplasmic or nucleoplasmic staining further confirming the crucial role of SENP3-mediated desumoylation for proper ribosome maturation. Notably, the nucleolar shape is also changed in a subset of SENP3 depleted cells.
Overall, these observations indicate that the PELP1–TEX10–WDR18 complex is required for the maturation and nucleolar release of the large ribosomal subunit.
PELP1 is modified by SUMO in a SENP3-controlled process
PELP1 has previously been identified as a non-covalent binding partner of SUMO2 (Rosendorff et al, 2006). Consistent with this finding we could define two hydrophobic SIM regions in PELP1, which are required for binding to SUMO2 in GST pull-down experiments (Supplementary Figure 10A). To investigate whether PELP1 can also be covalently modified by SUMO we initially used a reconstituted in vitro SUMO modification system. 35S-labelled PELP1 was generated by in vitro transcription/translation and incubated with the E1 activating enzyme, Ubc9, and either SUMO1 or SUMO2. In a control reaction, which lacked SUMO, PELP1 migrates at the predicted size of 180 kDa (Supplementary Figure 10B, lane 1). Addition of either SUMO1 or SUMO2, however, resulted in the formation of a SUMO conjugate with an apparent molecular weight of 220 kDa (Supplementary Figure 10B, lanes 2 and 5). Notably, addition of wild-type SENP3, but not a catalytically inactive version, led to the reduction of the PELP1–SUMO1 conjugates and an almost complete loss of the PELP1–SUMO2 forms suggesting that SENP3 acts on sumoylated PELP1 (Supplementary Figure 10B, lanes 3, 4 and 5, 7). The higher activity towards SUMO2 conjugates is in agreement with the preference of SENP3 towards SUMO2/3 forms (Haindl et al, 2008).
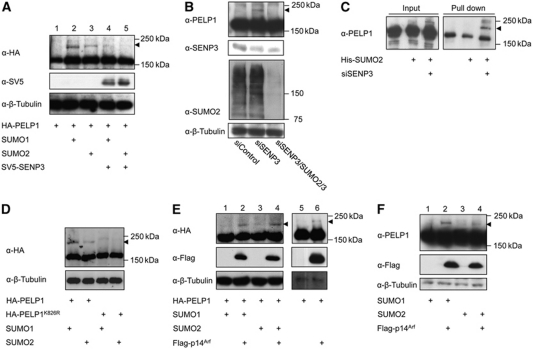
To examine whether SUMO paralogues can also modify PELP1 in vivo, HA-tagged PELP1 was expressed alone or together with SUMO1 or SUMO2. In all samples, anti-HA immunoblotting reveals a major band at 180 kDa corresponding to unmodified PELP1. Importantly, however, upon expression of SUMO1 or SUMO2 a modified form at 220 kDa corresponding to a potential SUMO–PELP1 conjugate is detected (Figure 5A, lanes 2 and 3). In support of this interpretation co-expression of SENP3 reverses the modification (Figure 5A, lanes 4 and 5). To see whether endogenous PELP1 can be modified by SUMO and whether this process is indeed controlled by endogenous SENP3, cells were depleted of SENP3 and PELP1 was detected by anti-PELP1 immunoblotting. Depletion of SENP3 led to the appearance of a 220-kDa anti-PELP1 reactive band, which became almost undetectable upon co-depletion of SUMO2/3, suggesting that it corresponds to a SUMO–PELP1 conjugate and mainly represents a SUMO2/3 modified form of PELP1 (Figure 5B). This was further validated by Ni-NTA pull-down experiments, in which His-tagged SUMO2–PELP1 conjugates could be specifically enriched on Ni-NTA beads from cells that are depleted of SENP3 and express His-SUMO2 (Figure 5C). The exchange of lysine 826 of PELP1 to arginine abrogated the modification both in vitro an in vivo, indicating that this residue serves as the major site for SUMO attachment (Figure 5D; Supplementary Figure 10C).
Figure 5.
PELP1 is modified by SUMO in a SENP3-controlled process. (A) HA-PELP1 was expressed alone or in combination with SUMO1 or SUMO2 in the presence or absence of SV5-tagged SENP3 in HeLa cells. Expression of the respective proteins was verified by western blotting. Detection of β-tubulin served as loading control. (B) SUMO modification of endogenous PELP1 was monitored by anti-PELP1 immunoblotting in HeLa cells transfected with siRNAs as indicated. Depletion of the respective proteins was verified by western blotting. (C) HeLa cells were transfected with siRNAs and plasmids as indicated. His-SUMO conjugates were recovered on Ni-NTA beads and subjected to western blotting using anti-PELP1 antibodies. (D) HA-PELP1 or HA-PELP1K826R was expressed in the presence of SUMO1 or SUMO2 in HeLa cells. Expression of the respective proteins was analysed by western blotting. Detection of β-tubulin served as loading control. (E, F) Flag-p14ARF and SUMO were expressed in HeLa cells and SUMO modification of ectopically expressed HA-PELP1 or endogenous PELP1 was monitored by immunoblotting with anti-HA or anti-PELP1 antibodies as indicated. Arrowheads indicate SUMO–PELP1 conjugates.
To understand how sumoylation of PELP1 is regulated we studied the potential role of p14ARF because it has been reported to counter SENP3 activity (Kuo et al, 2008). Co-expression of HA-PELP1 with p14ARF alone (Figure 5E, lane 6) or together with either SUMO1 or SUMO2 (Figure 5E, lanes 2 and 4) induced sumoylation of PELP1. Similarly, p14ARF enhances SUMO conjugation to endogenous PELP1 (Figure 5F, lanes 2 and 4) further supporting the idea that PELP1 is a physiological substrate of p14ARF-mediated sumoylation.
Nucleolar partitioning of PELP1 is controlled by the SUMO system
The data described above identify PELP1 as a nucleolar substrate of SUMO and provide evidence that the modification state of PELP1 is dynamically regulated by SENP3. Considering that SUMO is frequently involved in the control of subcellular compartmentalization we investigated whether a change in the cellular sumoylation status affects the nucleolar/nucleoplasmic distribution of PELP1 (Heun, 2007). We therefore initially ectopically expressed SUMO2 and monitored the localization of endogenous PELP1 by immunofluorescence (Figure 6A). SUMO2 shows the described localization in the nucleoplasm, where it is occasionally found in nuclear speckles, but is excluded from the nucleolus (Ayaydin and Dasso, 2004). Importantly, in all cells ectopically expressing SUMO2 PELP1 was also largely excluded from the nucleolus (Figure 6A, green arrowheads), whereas in adjacent untransfected cells it shows the typical distribution in the nucleoplasm and the nucleolus (Figure 6A, white arrowheads). This indicates that enhanced sumoylation prevents nucleolar accumulation of PELP1.
Figure 6.
Nucleolar partitioning of PELP1 is controlled by the SUMO system. (A) Flag-SUMO2 was expressed in HeLa cells by transient transfection. Flag-SUMO2-expressing cells were visualized by indirect immunofluorescence. The localization of endogenous PELP1 was monitored in Flag-SUMO2-positive cells (green arrowheads) or untransfected cells (white arrowheads) using anti-PELP1 antibody. (B–D) HeLa cells were transfected with siRNAs as indicated. After fixation and permeabilization, the localization of PELP1, LAS1L, TEX10 or PES1 was monitored by indirect immunofluorescence using the corresponding antibodies. (E) HeLa cells were depleted of endogenous PELP1 and reconstituted with siRNA resistant constructs expressing either wild-type HA-tagged PELP1 or a Flag-tagged PELP1–SUMO2 fusion protein. The localization of the respective proteins was monitored by indirect immunofluorescence using either anti-HA- or anti-Flag-antibodies. (F) The localization of LAS1L or SENP3 was analysed in cells expressing Flag-PELP1–SUMO2. Indirect immunofluorescence was done using anti-LAS1L or anti-SENP3 antibodies. Nuclei were counterstained with DAPI.
In order to expand upon the above finding we monitored how depletion of SENP3, which is paralleled by an increase in PELP1 sumoylation, affects the localization of PELP1. Again, in cells treated with the control siRNA PELP1 exhibits a nucleoplasmic and nucleolar localization pattern (Figure 6B). Strikingly, however, the absence of SENP3 causes an almost complete release of the nucleolar fraction of PELP1 to the nucleoplasm, indicating that desumoylation by SENP3 is required for its nucleolar partitioning (Figure 6B). Noteworthy, depletion of SENP3 did not affect the nucleolar accumulation of the 18S regulator WDR50/hUtp18, assuring that loss of SENP3 does not generally affect nucleolar integrity (Supplementary Figure 11A). Next, we monitored the localization of the PELP1-associated components LAS1L, TEX10 and MDN1, as well as the unrelated PeBoW component PES1, upon depletion of SENP3. Importantly, TEX10, LAS1L and MDN1 were largely excluded from the nucleolus upon loss of SENP3 (Figure 6C, rows 1–4; Supplementary Figure 11B), while the nucleolar accumulation of PES1, which does not appear to be an integral component of the PELP1–TEX10–WDR18 complex, was unaffected (Figure 6C, rows 5 and 6; Supplementary Figure 12).
To further strengthen the idea that the nucleolar compartmentalization of PELP1 is controlled through SUMO2/3 modification, PELP1 localization was studied after siRNA-mediated knock-down of SUMO2/3. Remarkably, in the absence of SUMO2/3, but not SUMO1, PELP1 strongly accumulates in the nucleolus, indicating that SUMO2/3 is indeed required for the nucleolar exclusion of PELP1 (Figure 6D).
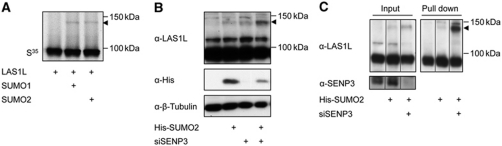
Based on these findings, we wished to determine whether SUMO modification of PELP1 directly mediates its subnuclear partitioning. To mimic constitutive modification, we linearly fused SUMO2 to the C-terminus of PELP1 and expressed it in cells depleted of the endogenous protein (Figure 6E). Importantly, in contrast to PELP1WT, which accumulated in the nucleolus, the PELP1–SUMO2 fusion protein exhibited a nucleoplasmic distribution and was largely excluded from the nucleolus. Moreover, PELP1-associated factors, such as LAS1L and SENP3, were also released from the nucleolus in the presence of PELP1–SUMO2 (Figure 6F). The SIM-deficient PELP1–SUMO2 fusion exhibits a similar localization, suggesting that the covalent attachment of SUMO to PELP1 is sufficient to determine its subnuclear distribution (Supplementary Figure 13A). Notably, however, PELP1 is not the only critical target of SENP3 in nucleolar partitioning of this complex because the non-sumoylatable PELP1K826R as well as the SUMO-binding deficient mutant (PELP1IV790/1AA,VI880/1AA) are excluded from the nucleolus upon depletion of SENP3 (Supplementary Figure 13B). Although we cannot totally exclude residual sumoylation of PELP1K826R in the absence of SENP3, we favoured the hypothesis that the nucleolar exclusion of PELP1K826R may result from the modification of an additional SENP3-sensitive SUMO2/3 substrate that is associated with PELP1. In support of this idea in vitro and in vivo experiments demonstrated that LAS1L is modified by SUMO (Figure 7A–C). Moreover, the modification of endogenous LAS1L by endogenous SUMO2 or ectopically expressed SUMO2 is drastically induced upon depletion of SENP3 (Figure 7B, compare lanes 1–3 and 2–4, Figure 7C). We therefore propose a model, in which SENP3 controls nucleolar partitioning by desumoylating multiple components of this ribosome biogenesis complex.
Figure 7.
LAS1L is modified by SUMO in a SENP3-controlled manner. (A) LAS1L was generated by in vitro transcription/translation and incubated with recombinant E1 and E2 enzymes and SUMO1 or SUMO2, respectively, in the presence of ATP. In the control reaction (lane 1) SUMO was not added. (B) HeLa cells were transfected with siRNAs and plasmids as indicated. Expression of the respective proteins was verified by western blotting. Detection of β-tubulin served as loading control. (C) HeLa cells were transfected with siRNAs and plasmids as indicated. His-SUMO2 conjugates were captured on magnetic Ni-NTA beads and subjected to western blotting using anti-LAS1L antibodies. Vertical lines indicate removal of irrelevant neighbouring lanes from the initial gel (see Supplementary Figure 14).
Discussion
Proper coordination of ribosome biogenesis requires an intricate spatial and temporal interplay of protein networks. Here, we define the PELP1–TEX10–WDR18 complex as a novel regulator in the biogenesis pathway of the 60S ribosomal subunit in mammalian cells and provide evidence that the SUMO system controls the spatial distribution of this complex and the associated factors MDN1 and LAS1L.
We focused on a function of PELP1–TEX10–WDR18 in ribosome biogenesis, because sequence comparison unravelled a relationship between PELP1, TEX10, WDR18 and the yeast Rix1, Ipi1, Ipi3 proteins, which comprise a three-component protein subcomplex of the pre-60S ribosomal particle (Galani et al, 2004; Krogan et al, 2004; Nissan et al, 2004). Although the overall homology between the respective human and yeast proteins is limited, they clearly share conserved regions and domains. Human PELP1 is about 20% identical to Rix1 in a region between amino acids 150 and 500. Additionally, both proteins possess a characteristic highly acidic C-terminal region. TEX10 and Ipi1 are 30% identical within a conserved 200 residue N-terminal region, which includes a domain—annotated as Ipi1N domain—that defines a eukaryotic protein family. Notably, however, TEX10 has an additional 600 amino-acid extension, which is missing in Ipi1. WDR18 and Ipi3 belong to the WD40 protein family and are the most conserved subunits of the complex sharing an overall identity of about 25% along the entire sequence.
The yeast Rix1/Ipi core complex and its associated factor Rea1 have been connected to large subunit maturation. Krogan et al (2004) revealed a requirement of the complex for processing of the primary rRNA transcript within the ITS2 region, which separates the 25S rRNA (28S in mammals) encoding sequence from the 5.8S region within the yeast 27S precursor (32S rRNA in mammals). Distinct cleavage reactions in ITS2 that mainly take place in the GC region of the nucleolus generate the mature 25S rRNA species and a 7S pre-rRNA intermediate (12S in mammals), which is further trimmed to the 5.8S form by the exosome in the nucleoplasm. Individual depletion of the Rix1–Ipi1–Ipi3 proteins resulted in a reduction of the mature 25S and 5.8S rRNA species and a concomitant accumulation of the corresponding 27S and 7S precursors, suggesting that the Rix/Ipi complex affects both nucleolar and nucleoplasmic stages of ITS2 cleavage (Wu et al, 2002; Peng et al, 2003; Krogan et al, 2004). Wu et al (2002) reported similar phenotypes upon depletion of Ipi3. Using slightly different experimental approaches, Hurt and co-workers have placed the activity of the complex primarily at the extranucleolar stage of the assembly pathway and show that it is specifically required for the nucleoplasmic 7S pre-rRNA processing step and the nuclear export of late pre-60S particles from the nucleoplasm to the cytoplasm (Galani et al, 2004; Ulbrich et al, 2009). Notably, however, more recent work showed that at least Rea1 functions at multiple stages of the biogenesis pathway and also drives exit of early pre-60S particles from the nucleolus (Bassler et al, 2010).
Our results demonstrate that the mammalian PELP1–TEX10–WDR18 complex as well as the Rea1 orthologue MDN1 are involved in nucleolar steps of the 60S maturation pathway. Depletion of either component of the complex affects the processing of the 32S rRNA and concomitantly prevents the nucleolar export of the pre-60S ribosomal subunit. In line with a nucleolar function, a subfraction of PELP1–TEX10–WDR18 was detected in the nucleolus corroborating proteomic studies of the nucleolus (Andersen et al, 2005; Ahmad et al, 2009; Boisvert et al, 2010) (see also http://www.lamondlab.com/NOPdb/). The distinct localization of PELP1 to the GC region and its association with ITS2-containing 28S rRNA precursor forms and pre-60S particles are also in perfect agreement with a function in 32S processing. Additional evidence for an involvement of PELP1 in this process comes from the observation that it binds to the recently described 28S rRNA maturation factor LAS1L (Castle et al, 2010). Collectively, these data argue for an evolutionary conserved function of the PELP1–TEX10–WDR18 complex and the Rix1–Ipi1–Ipi3 complex in ITS2 processing and large subunit maturation, with PELP1–TEX10–WDR18 having a pronounced role in the nucleolar steps of this process.
Notably, however, PELP1, TEX10 and WDR18 as well as MDN1 and LAS1L are also found in the nucleoplasm, indicating that they are dynamically distributed between both compartments. Importantly, SENP3, which is physically linked to PELP1–TEX10–WDR18, is required for the nucleolar partitioning of this complex and the associated proteins LAS1L and MDN1. This suggests that the balanced SUMO conjugation/deconjugation governs the spatial distribution of these ribosome biogenesis factors. Consistent with this idea constitutive modification of PELP1 by SUMO2, which was mimicked by linearly fusing SUMO2 to PELP1 leads to nucleolar exclusion of PELP1 and LAS1L. Endogenous PELP1 undergoes a robust sumoylation in the absence of SENP3 and thus represents one important SENP3-controlled target in the PELP1–TEX10–TEX18 complex. One other SENP3-sensitive target is the PELP1-interacting protein LAS1L. We propose that enhanced sumoylation of LAS1L accounts for the nucleolar exclusion of the non-sumoylatable variant of PELP1 in SENP3-depleted cells. We thus favour a model, in which SENP3 controls nucleolar partitioning by desumoylating multiple components of this complex. Importantly, we show that the constitutive modification of one component of the complex, that is PELP1, affects the subnuclear partitioning of other members. This scenario also explains why removal of the SUMO site on a single component is not sufficient to interfere with SENP3-controlled subnuclear distribution.
Quantitative proteomic studies revealed that loss of SENP3 does not affect the interaction of PELP1 with either WDR18 or LAS1L, indicating that enhanced sumoylation does not trigger disassembly of the complex (Finkbeiner and Muller, unpublished results). We rather hypothesize that the modification promotes binding to a nucleoplasmic protein or prevents the interaction with a nucleolar binding partner or rRNA. Considering that a SUMO-dependent nucleolar release upon DNA damage has been previously described for mammalian topoisomerase I and yeast Rad52 (Mo et al, 2002; Torres-Rosell et al, 2007), our data expand the concept of SUMO-mediated nucleolar exclusion as a general mechanism to control nucleolar protein dynamics.
The SUMO-regulated dynamic distribution of PELP1 and its binding partners likely provides an important mechanism to coordinate the rate of ribosome formation with the physiological state of the cell. We propose that enhanced sumoylation shifts the activity of these proteins from the nucleolus to the nucleoplasm thereby compromising 60S maturation and limiting ribosome biogenesis. We speculate that the balanced SUMO modification assures the timely association of PELP1 and its binding partners with 60S pre-ribosomal particles. We hypothesize that sumoylation acts as a signal to release the PELP1–TEX10–WDR18 complex from these structures. Loss of SENP3 would therefore prevent binding to the pre-60S particles or induce a premature release from these structures. The regulated, dynamic sumoylation would thus provide a mechanism to control the remodelling of pre-ribosomal particles in order to assure the proper timely and spatial regulation of the maturation process.
Intriguingly, the p14ARF tumour suppressor—a known inhibitor of ribosome biogenesis—acts as an inducer of PELP1 sumoylation and thus shifts the balance of sumoylation/desumoylation towards the modified state. Several lines of evidence indicate that the ARF-induced inhibition of ribosome biogenesis contributes to its p53-independent anti-proliferative and tumour suppressive functions and recent data suggest that this is mechanistically linked to ARFs ability to induce sumoylation of nucleolar proteins by antagonizing SENP3 (Haindl et al, 2008). Our results further substantiate this view and suggest that PELP1 is a functionally relevant target of the ARF/SENP3 circuit. The inhibitory action of ARF on ribosome biogenesis might thus be connected to an improper spatial control of the PELP1–TEX10–WDR18 complex.
While p14ARF acts as a negative regulator of ribosome biogenesis, the proto-oncogene c-myc promotes ribosome biogenesis by inducing the expression of nucleolar proteins involved in rRNA transcription or processing (van Riggelen et al, 2010). This assures an increase in ribosome supply when cell growth and proliferation are enhanced, but also contributes to the ability of c-myc to initiate tumourigenesis. Notably, PELP1 and WDR18 have been identified as target genes of c-myc, suggesting that the PELP1–TEX10–WDR18 complex is also under the control of c-myc (Schlosser et al, 2003; Zeller et al, 2006). Several reports show that the expression of PELP1 is deregulated in a variety of human tumours and provide evidence that PELP1 acts itself as a potential proto-oncogene (Rajhans et al, 2007; Vadlamudi and Kumar, 2007; Dimple et al, 2008). The growth-promoting effects of PELP1 have been connected to its role in nuclear hormone receptor-mediated genomic and non-genomic signalling pathways (Vadlamudi and Kumar, 2007). Based on our new findings, it is now tempting to speculate that the pathological role of PELP1 in tumourigenesis might also be related to a deregulation of ribosome production, which is a hallmark of many tumours (Ruggero and Pandolfi, 2003).
Materials and methods
Cell culture and transfection
HeLa cells were grown under standard conditions. OCI-AML3 cells were grown in RPMI media containing 20% FBS. Plasmid transfections were carried out with FuGeneHD (Roche) using 5 μl transfection reagent and 2 μg DNA per 105 HeLa cells in a 35-mm diameter well. For siRNA-mediated knock-down 105 HeLa cells per 35 mm diameter well were transfected with the respective siRNAs (120 pmol) using Oligofectamine (Invitrogen) according to the manufacturer's instructions. For the in vivo labelling assays, HeLa cells were transfected with siRNA using the Amaxa Cell Line Nucleofector Kit R (Lonza). In all experiments, 106 cells were transfected with 900 pmol of the corresponding siRNA and one fifth of the cells were seeded per 35 mm diameter well.
The following siRNA sequences (sense) were used:
Control: CGUACGCGGAAUACUUCGAdTdT
PELP1: GGAAUGAAGGCUUGUAUGAdTdT
TEX10: AGAUGCUAAUGUACGAUUAdTdT
WDR18: AUCGGGACCUGUUCGACUUdTdT
SENP3: CUGGCCCUGUCUCAGCCAUdTdT
MDN1: GGAAUGCCGAAGCCAUUAAdTdT
SUMO2/3: GUCAAUGAGGCAGAUCAGAdTdT
Cloning and mutagenesis
cDNAs encoding for PELP1, WDR18 and LAS1L was amplified by PCR from EST clones provided by ImaGenes (PELP1: IRATp970H0593D, WDR18: IRAUp969B034D, LAS1L: IRAUp969CO677D). A cDNA for TEX10 was generated by PCR amplification from a human testis cDNA library (Marathon-Ready, Invitrogen). For transient expression of Flag- or HA-epitope-tagged proteins in mammalian cells the respective cDNA sequences were inserted into the pCI vector (Invitrogen). For bacterial expression of GST-fusion proteins cDNA sequences were inserted into pGEX-4T-1 (GE Healthcare). All other plasmids are described previously (Ledl et al, 2005; Haindl et al, 2008; Klein et al, 2009; Stehmeier and Muller, 2009). Site-directed mutagenesis was carried out using the QuickChange Mutagenesis Kit (Stratagene) according to the manufacturer's instructions.
Expression of recombinant proteins and GST pull-down experiments
GST-fusion proteins were expressed in Escherichia coli BL21 after induction of cells with IPTG for 3 h. Bacteria were harvested, resuspended in lysis buffer (PBS, 1% Triton X-100, 1 mM DTT and 100 μM PMSF) and homogenized with a cell disruptor (Emulsi-Flex C3). The lysate was incubated for 3 h with Glutathione Sepharose 4B (GE Healthcare). The beads were washed three times with lysis buffer and stored at −20°C. GST pull-down experiments were done as described (Schmidt and Muller, 2002).
Generation of YFP-RpL27-expressing cells
HeLa cells were transfected with a tetracycline-inducible episomal pRTS1 vector encoding eYFP-RpL27 and cells were selected with 400 μg/ml Hygromycin B for 14 days to generate a homogenous cell population (Bornkamm et al, 2005). Expression of eYFP-RpL27 was induced by the addition of 1 μg/ml Doxycycline and the localization of RpL27 was monitored by immunofluorescence.
Immunoprecipitation, Ni-NTA pull-down and western blotting
For the immunoprecipitation of ectopically expressed or endogenous proteins 9 × 106 HeLa cells were used. For the immunoprecipitation performed in OCI-AML3 cells 3 × 107 cells were used. Cells were lysed in 50 mM HEPES pH 7.4, 150 mM NaCl, 2 mM EDTA, 0.5% NP-40 containing Complete Protease Inhibitor (Roche). Cleared cell lysates were incubated overnight with anti-Flag beads (Sigma-Aldrich), anti-HA agarose beads (Roche) or specific antibodies directed against the respective antigens. Bead-coupled immunocomplexes were directly collected by centrifugation and washed three times with lysis buffer. Soluble antibody complexes were captured on Protein A/G beads (Roche) and processed identically. Bound proteins were eluted by boiling the beads with SDS–PAGE loading buffer. Purification of Flag-SENP3- or Flag-PELP1-associated proteins and subsequent mass spectrometry as well as Ni-NTA pull-down experiments were carried out as described (Haindl et al, 2008). The initial purification of Flag-SENP3-associated proteins was done in Nocodazole-treated cells. Western blotting was done using ECL detection reagents (GE Healthcare).
For immunoprecipitations done in the presence of RNase A 100 μg/ml of the enzyme (Roche) was added to the lysis buffer. To control for complete digestion, RNA was isolated by TRIZOL reagent (Invitrogen) and subjected to random hexamer-primed reverse transcription using Transcriptor First Strand cDNA Synthesis Kit (Roche). The cDNA was used as a template for PCR amplification of the ITS2/28S region or GAPDH as a control (primer sequences see below).
Immunofluorescence
For immunofluorescence, HeLa cells were fixed in 3.4% paraformaldehyde, permeabilized with 0.5% Triton X-100 and processed using standard protocols. Images were acquired with an AX10 microscope (Zeiss). Antibodies are listed below. As secondary antibodies Alexa Fluor 555 goat anti-rabbit (Invitrogen), Alexa Fluor 488 goat anti-rat (Invitrogen), Alexa Fluor 488 goat anti-rabbit (Invitrogen), Cy3 goat anti-mouse (Jackson ImmunoResearch) and FITC donkey anti-mouse (Jackson ImmunoResearch) were used.
32P in vivo labelling and RNA analysis
Metabolic labelling of rRNA and analysis of RNA was carried out as described previously (Haindl et al, 2008).
RNA immunoprecipitation
For the isolation of PELP1-associated RNA species 8 × 107 HeLa cells were used for immunoprecipitation. Cells were lysed in lysis buffer (25 mM Tris–HCl pH 7.2, 100 mM KCl, 2.5 mM EDTA, 10% glycerol, 1.4% NP-40, 0.2% sodium deoxycholate, 2 mM DTT, 20 U/ml RNase inhibitor (Roche), Complete Protease Inhibitor (Roche)) and immunoprecipitation was performed as described above using a polyclonal anti-PELP1 antibody. In total, 10% of the immunoprecipitated material was kept for immunoblotting to control for efficient IP, while the remainder was used for RNA extraction using TRIZOL reagent (Invitrogen) according to the manufacturer's instructions. RNA was digested with DNase (Turbo DNA-free, Ambion) and subjected to random hexamer-primed reverse transcription using Transcriptor First Strand cDNA Synthesis Kit (Roche). For each sample a control reaction without reverse transcriptase was included. The cDNA was used as a template for PCR amplification of the ITS2/28S region or GAPDH as a control. Twenty-five cycles were performed using the following primers:
ITS2/28S forward: 5′-GCTCTCTCTCCCGTCGCCTCTC, reverse: 5′-CCTGTTCACTCGCCGTTACTGAGG
GAPDH forward: 5′-TGACAACTTTGGTATCGTGGAAG, reverse: 5′-CAGTAGAGGCAGGGATGATGTT
Nucleolar isolation
Cells were fractionated according to the following protocol http://www.lamondlab.com/f7nucleolarprotocol.htm
Sucrose gradients
The analysis of pre-ribosomal ribonucleoprotein complexes was carried out as described according to published procedures with minor modifications (Pestov et al, 2008). Fractions of 600 μl were manually collected and A254 of each fraction was measured photometrically (Nanodrop, Peqlab). After TCA precipitation of proteins pellets were air dried and dissolved with 25 μl urea sample buffer. In total, 7 μl of each fraction was used for SDS–PAGE.
In vitro sumoylation and desumoylation
35S-labelled PELP1 and LAS1L were generated by in vitro transcription/translation using the TNT Quick Coupled T7 Kit (Promega, Madison, WI, USA) and sumoylation was carried out as described previously (Schmidt and Muller, 2002). For demodification SENP3, generated by in vitro transcription/translation, was added to the in vitro modification reactions and samples were incubated for an additional 90 min at 30°C. Proteins were separated on SDS gels and detected by autoradiography.
Antibodies used for western blotting and immunofluorescence
The following antibodies were used for western blotting, immunoprecipitation and immunofluorescence: anti-HA (clone 16B12, Covance), anti-Flag (clone M2, Sigma-Aldrich), anti-Flag (#F7425, Sigma-Aldrich), anti-SV5 (clone R960-25, Invitrogen), anti-β-Tubulin (clone E7, Developmental Studies Hybridoma Bank), anti-Vinculin (clone hVIN-1, Sigma-Aldrich), anti-PELP1 (#A300-180A, Bethyl Laboratories), anti-WDR18 (#15165-1-AP, ProteinTech Group), anti-TEX10 (#17372-1-AP, ProteinTech Group), anti-SENP3 (clone G3, Santa Cruz), anti-U2AF65 (#ab37483, Abcam), anti-Fibrillarin (#ab5821, Abcam), anti-UBF1 (clone F9, Santa Cruz), anti-NPM/B23 (clone NA24, Santa Cruz), anti-NPM (#32-5200, Invitrogen), anti-GFP (clone B2, Santa Cruz), anti-SUMO2/3 (clone 1E7, MBL), anti-MDN1 (#HP A02 9666, Sigma-Aldrich), anti-LAS1L (#AV34629, Sigma-Aldrich), anti-RGS-(His)6 (#34610, Qiagen), anti-PES1 (rat monoclonal antibody, gift from Dirk Eick, Helmholtz Zentrum, Munich), anti-BOP1 (rat monoclonal antibody, gift from Dirk Eick, Helmholtz Zentrum) and anti-WDR50 (rat monoclonal antibody, gift from Dirk Eick, Helmholtz Zentrum). Polyclonal anti-SENP3 antibody was raised in rabbits by immunization as described (Haindl et al, 2008). An anti-TEX10 antibody was raised in rabbits by immunization with the peptide QLKEDGTLPTNNRKL (Eurogentech SA, Belgium). The crude serum was affinity-purified against the antigenic peptide.
Supplementary Material
Acknowledgments
We thank Dirk Eick for reagents and Uschi Kagerer for technical assistance, John Weir for comments on the manuscript and Stefan Jentsch for discussions and continuous support. This work was supported by the ‘Deutsche Forschungsgemeinschaft’ SPP1365 and SFB684. Author contributions: MH performed the initial biochemical interaction screen (Supplementary Figure S1A). EF and SM designed the experiments and EF performed all experiments. EF and SM wrote the manuscript. MH contributed to editing the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ahmad Y, Boisvert FM, Gregor P, Cobley A, Lamond AI (2009) NOPdb: Nucleolar Proteome Database—2008 update. Nucleic Acids Res 37: D181–D184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M (2005) Nucleolar proteome dynamics. Nature 433: 77–83 [DOI] [PubMed] [Google Scholar]
- Ayaydin F, Dasso M (2004) Distinct in vivo dynamics of vertebrate SUMO paralogues. Mol Biol Cell 15: 5208–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler J, Kallas M, Pertschy B, Ulbrich C, Thoms M, Hurt E (2010) The AAA-ATPase Rea1 drives removal of biogenesis factors during multiple stages of 60S ribosome assembly. Mol Cell 38: 712–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, Lam YW, Lamont D, Lamond AI (2010) A quantitative proteomics analysis of subcellular proteome localization and changes induced by DNA damage. Mol Cell Proteomics 9: 457–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI (2007) The multifunctional nucleolus. Nat Rev Mol Cell Biol 8: 574–585 [DOI] [PubMed] [Google Scholar]
- Bornkamm GW, Berens C, Kuklik-Roos C, Bechet JM, Laux G, Bachl J, Korndoerfer M, Schlee M, Holzel M, Malamoussi A, Chapman RD, Nimmerjahn F, Mautner J, Hillen W, Bujard H, Feuillard J (2005) Stringent doxycycline-dependent control of gene activities using an episomal one-vector system. Nucleic Acids Res 33: e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle CD, Cassimere EK, Lee J, Denicourt C (2010) Las1L is a nucleolar protein required for cell proliferation and ribosome biogenesis. Mol Cell Biol 30: 4404–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bacco A, Ouyang J, Lee HY, Catic A, Ploegh H, Gill G (2006) The SUMO-specific protease SENP5 is required for cell division. Mol Cell Biol 26: 4489–4498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimple C, Nair SS, Rajhans R, Pitcheswara PR, Liu J, Balasenthil S, Le XF, Burow ME, Auersperg N, Tekmal RR, Broaddus RR, Vadlamudi RK (2008) Role of PELP1/MNAR signaling in ovarian tumorigenesis. Cancer Res 68: 4902–4909 [DOI] [PubMed] [Google Scholar]
- Galani K, Nissan TA, Petfalski E, Tollervey D, Hurt E (2004) Rea1, a dynein-related nuclear AAA-ATPase, is involved in late rRNA processing and nuclear export of 60 S subunits. J Biol Chem 279: 55411–55418 [DOI] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F (2007) Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 8: 947–956 [DOI] [PubMed] [Google Scholar]
- Gong L, Yeh ET (2006) Characterization of a family of nucleolar SUMO-specific proteases with preference for SUMO-2 or SUMO-3. J Biol Chem 281: 15869–15877 [DOI] [PubMed] [Google Scholar]
- Grisendi S, Mecucci C, Falini B, Pandolfi PP (2006) Nucleophosmin and cancer. Nat Rev Cancer 6: 493–505 [DOI] [PubMed] [Google Scholar]
- Haindl M, Harasim T, Eick D, Muller S (2008) The nucleolar SUMO-specific protease SENP3 reverses SUMO modification of nucleophosmin and is required for rRNA processing. EMBO Rep 9: 273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay RT (2007) SUMO-specific proteases: a twist in the tail. Trends Cell Biol 17: 370–376 [DOI] [PubMed] [Google Scholar]
- Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y (2008) The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci 65: 2334–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heun P (2007) SUMOrganization of the nucleus. Curr Opin Cell Biol 19: 350–355 [DOI] [PubMed] [Google Scholar]
- Holzel M, Rohrmoser M, Schlee M, Grimm T, Harasim T, Malamoussi A, Gruber-Eber A, Kremmer E, Hiddemann W, Bornkamm GW, Eick D (2005) Mammalian WDR12 is a novel member of the Pes1-Bop1 complex and is required for ribosome biogenesis and cell proliferation. J Cell Biol 170: 367–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein UR, Haindl M, Nigg EA, Muller S (2009) RanBP2 and SENP3 function in a mitotic SUMO2/3 conjugation-deconjugation cycle on Borealin. Mol Biol Cell 20: 410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, Hurt E, Bassler J (2010) Driving ribosome assembly. Biochim Biophys Acta 1803: 673–683 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Peng WT, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP, Beattie BK, Lalev A, Zhang W, Davierwala AP, Mnaimneh S, Starostine A, Tikuisis AP, Grigull J, Datta N, Bray JE, Hughes TR, Emili A, Greenblatt JF (2004) High-definition macromolecular composition of yeast RNA-processing complexes. Mol Cell 13: 225–239 [DOI] [PubMed] [Google Scholar]
- Kuo ML, den Besten W, Thomas MC, Sherr CJ (2008) Arf-induced turnover of the nucleolar nucleophosmin-associated SUMO-2/3 protease Senp3. Cell Cycle 7: 3378–3387 [DOI] [PubMed] [Google Scholar]
- Lapik YR, Fernandes CJ, Lau LF, Pestov DG (2004) Physical and functional interaction between Pes1 and Bop1 in mammalian ribosome biogenesis. Mol Cell 15: 17–29 [DOI] [PubMed] [Google Scholar]
- Ledl A, Schmidt D, Muller S (2005) Viral oncoproteins E1A and E7 and cellular LxCxE proteins repress SUMO modification of the retinoblastoma tumor suppressor. Oncogene 24: 3810–3818 [DOI] [PubMed] [Google Scholar]
- Mo YY, Yu Y, Shen Z, Beck WT (2002) Nucleolar delocalization of human topoisomerase I in response to topotecan correlates with sumoylation of the protein. J Biol Chem 277: 2958–2964 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Dasso M (2007) Modification in reverse: the SUMO proteases. Trends Biochem Sci 32: 286–295 [DOI] [PubMed] [Google Scholar]
- Nishida T, Tanaka H, Yasuda H (2000) A novel mammalian Smt3-specific isopeptidase 1 (SMT3IP1) localized in the nucleolus at interphase. Eur J Biochem 267: 6423–6427 [DOI] [PubMed] [Google Scholar]
- Nishida T, Yamada Y (2008) SMT3IP1, a nucleolar SUMO-specific protease, deconjugates SUMO-2 from nucleolar and cytoplasmic nucleophosmin. Biochem Biophys Res Commun 374: 382–387 [DOI] [PubMed] [Google Scholar]
- Nissan TA, Galani K, Maco B, Tollervey D, Aebi U, Hurt E (2004) A pre-ribosome with a tadpole-like structure functions in ATP-dependent maturation of 60S subunits. Mol Cell 15: 295–301 [DOI] [PubMed] [Google Scholar]
- Panse VG, Kressler D, Pauli A, Petfalski E, Gnadig M, Tollervey D, Hurt E (2006) Formation and nuclear export of preribosomes are functionally linked to the small-ubiquitin-related modifier pathway. Traffic 7: 1311–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng WT, Robinson MD, Mnaimneh S, Krogan NJ, Cagney G, Morris Q, Davierwala AP, Grigull J, Yang X, Zhang W, Mitsakakis N, Ryan OW, Datta N, Jojic V, Pal C, Canadien V, Richards D, Beattie B, Wu LF, Altschuler SJ, Roweis S, Frey BJ, Emili A, Greenblatt JF, Hughes TR (2003) A panoramic view of yeast noncoding RNA processing. Cell 113: 919–933 [DOI] [PubMed] [Google Scholar]
- Pestov DG, Lapik YR, Lau LF (2008) Assays for ribosomal RNA processing and ribosome assembly. Curr Protoc Cell Biol, Chapter 22: Unit 22.11 [DOI] [PubMed] [Google Scholar]
- Rajhans R, Nair S, Holden AH, Kumar R, Tekmal RR, Vadlamudi RK (2007) Oncogenic potential of the nuclear receptor coregulator proline-, glutamic acid-, leucine-rich protein 1/modulator of the nongenomic actions of the estrogen receptor. Cancer Res 67: 5505–5512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosby R, Cui Z, Rogers E, deLivron MA, Robinson VL, DiMario PJ (2009) Knockdown of the Drosophila GTPase nucleostemin 1 impairs large ribosomal subunit biogenesis, cell growth, and midgut precursor cell maintenance. Mol Biol Cell 20: 4424–4434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosendorff A, Sakakibara S, Lu S, Kieff E, Xuan Y, DiBacco A, Shi Y, Gill G (2006) NXP-2 association with SUMO-2 depends on lysines required for transcriptional repression. Proc Natl Acad Sci USA 103: 5308–5313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D, Pandolfi PP (2003) Does the ribosome translate cancer? Nat Rev Cancer 3: 179–192 [DOI] [PubMed] [Google Scholar]
- Schlosser I, Holzel M, Murnseer M, Burtscher H, Weidle UH, Eick D (2003) A role for c-Myc in the regulation of ribosomal RNA processing. Nucleic Acids Res 31: 6148–6156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Muller S (2002) Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc Natl Acad Sci USA 99: 2872–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehmeier P, Muller S (2009) Regulation of p53 family members by the ubiquitin-like SUMO system. DNA Repair (Amst) 8: 491–498 [DOI] [PubMed] [Google Scholar]
- Torres-Rosell J, Sunjevaric I, De Piccoli G, Sacher M, Eckert-Boulet N, Reid R, Jentsch S, Rothstein R, Aragon L, Lisby M (2007) The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat Cell Biol 9: 923–931 [DOI] [PubMed] [Google Scholar]
- Tschochner H, Hurt E (2003) Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol 13: 255–263 [DOI] [PubMed] [Google Scholar]
- Ulbrich C, Diepholz M, Bassler J, Kressler D, Pertschy B, Galani K, Bottcher B, Hurt E (2009) Mechanochemical removal of ribosome biogenesis factors from nascent 60S ribosomal subunits. Cell 138: 911–922 [DOI] [PubMed] [Google Scholar]
- Vadlamudi RK, Kumar R (2007) Functional and biological properties of the nuclear receptor coregulator PELP1/MNAR. Nucl Recept Signal 5: e004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Riggelen J, Yetil A, Felsher DW (2010) MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer 10: 301–309 [DOI] [PubMed] [Google Scholar]
- Wang Y, Dasso M (2009) SUMOylation and deSUMOylation at a glance. J Cell Sci 122: 4249–4252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LF, Hughes TR, Davierwala AP, Robinson MD, Stoughton R, Altschuler SJ (2002) Large-scale prediction of Saccharomyces cerevisiae gene function using overlapping transcriptional clusters. Nat Genet 31: 255–265 [DOI] [PubMed] [Google Scholar]
- Yeh ET (2009) SUMOylation and De-SUMOylation: wrestling with life′s processes. J Biol Chem 284: 8223–8227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun C, Wang Y, Mukhopadhyay D, Backlund P, Kolli N, Yergey A, Wilkinson KD, Dasso M (2008) Nucleolar protein B23/nucleophosmin regulates the vertebrate SUMO pathway through SENP3 and SENP5 proteases. J Cell Biol 183: 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT, Ooi HS, Orlov YL, Shahab A, Yong HC, Fu Y, Weng Z, Kuznetsov VA, Sung WK, Ruan Y, Dang CV, Wei CL (2006) Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci USA 103: 17834–17839 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.