Abstract
Lineage fate decisions underpin much of development as well as tissue homeostasis in the adult. A mechanistic paradigm for such decisions is the erythroid versus myeloid fate decision controlled by cross-antagonism between gata1 and pu.1 transcription factors. In this study, we have systematically tested this paradigm in blood-producing populations in zebrafish embryos, including the haematopoietic stem cells (HSCs), and found that it takes a different form in each population. In particular, gata1 activity varies from autostimulation to autorepression. In addition, we have added a third member to this regulatory kernel, tif1γ (transcription intermediate factor-1γ). We show that tif1γ modulates the erythroid versus myeloid fate outcomes from HSCs by differentially controlling the levels of gata1 and pu.1. By contrast, tif1γ positively regulates both gata1 and pu.1 in primitive erythroid and prodefinitive erythromyeloid progenitors. We therefore conclude that the gata1/pu.1 paradigm for lineage decisions takes different forms in different cellular contexts and is modulated by tif1γ.
Keywords: gata1, haematopoiesis, lineage fate decisions, pu.1, tif1γ
Introduction
The cross-antagonism between gata1 and pu.1 in haematopoietic progenitors, together with their positive autoregulation (autostimulation), has become a paradigm for lineage switching generally (Graf and Enver, 2009). Overexpression experiments have shown that gata1 and pu.1 suppress each other's activity by direct protein interaction (Rekhtman et al, 1999; Zhang et al, 1999; Nerlov et al, 2000). Consequently, forced expression of gata1 is sufficient to reprogramme myeloid into erythroid cells (Kulessa et al, 1995; Yamaguchi et al, 1998) and conversely overexpression of pu.1 reprogrammes erythroid cells into the myeloid lineage (Nerlov and Graf, 1998). Moreover, both transcription factors can positively regulate their own expression (Trainor et al, 1996; McDevitt et al, 1997; Yu et al, 2002). Collectively, these observations have led to a model whereby the positive autoregulation and cross-antagonistic activities of gata1 and pu.1 are critical to determine lineage commitment in haematopoietic progenitors (Cantor and Orkin, 2002; Graf, 2002; Chickarmane et al, 2009; Graf and Enver, 2009). This model, however, does not account for all interactions between gata1 and pu.1: in mast cells, pu.1 positively regulates gata1 expression (Takemoto et al, 2010). Furthermore, the testing of this model in primary cells in vivo has been limited to the early unipotent populations of either myeloid or erythroid progenitors (Galloway et al, 2005; Rhodes et al, 2005).
In the developing vertebrate embryo, haematopoiesis occurs in discrete waves, termed either primitive or definitive. In the mouse, the primitive wave takes place in yolk sac blood islands and gives rise mainly to transient populations of erythrocytes (Dzierzak and Speck, 2008 and references therein). In zebrafish, the primitive wave takes place in two distinct embryonic locations: the posterior lateral mesoderm (PLM), which later becomes the intermediate cell mass (ICM) where erythrocytes, and possibly thrombocytes, arise (Detrich et al, 1995; Warga et al, 2009), and the anterior lateral mesoderm (ALM), which gives rise to macrophages (Herbomel et al, 1999; Figure 1A–C). The definitive wave gives rise to haematopoietic stem cells (HSCs) that are capable of generating all the mature blood lineages throughout adult life. Definitive HSCs arise in the ventral wall of the dorsal aorta (DA) in a region known as the aorta–gonad–mesonephros in mammals (Medvinsky and Dzierzak 1996; de Bruijn et al, 2000, 2002). Lineage tracing experiments in zebrafish have shown that cells in the ventral wall of the DA give rise to rag1+ cells in the thymus, indicating that the DA itself is the source of HSCs in the aorta–gonad–mesonephros (Jin et al, 2007; Kissa et al, 2008). Very recently, elegant live imaging experiments captured the endothelial cells in the ventral wall of the DA transdifferentiating into cells that eventually give rise to T-cell derivatives in the embryonic thymus, and lymphoid, myeloid and erythroid cells in the adult (Bertrand et al, 2010; Kissa and Herbomel, 2010). A fourth population of haematopoietic cells, termed erythromyeloid progenitors (EMPs), has also been described in zebrafish (Bertrand et al, 2007). These progenitors arise at around 30 h post-fertilisation (hpf) in the posterior blood island (PBI) derived from the most posterior PLM (Figure 1A–C), and have the potential to give rise to erythroid and myeloid cells in vitro (Bertrand et al, 2007). This transient cell population, which may be equivalent to the second wave of haematopoiesis from the yolk sac in mammals, is thought to disappear by 48 hpf (Bertrand et al, 2007, 2008). In mammals, the main anatomical sites colonised by HSCs shift as development proceeds, with the fetal liver (FL) colonised first and the bone marrow just before birth (Mikkola and Orkin, 2006; Dzierzak and Speck, 2008). A similar shift in the sites of haematopoietic development occurs in zebrafish, with HSCs from the DA migrating to the caudal haematopoietic tissue (CHT), where they give rise to erythroid and myeloid progeny, and from there to the thymus, for T-cell development, and the kidney, the haematopoietic equivalent of the mammalian bone marrow (Murayama et al, 2006; Jin et al, 2007, 2009). Thus, the CHT has a role analogous to the FL in mammals (Murayama et al, 2006).
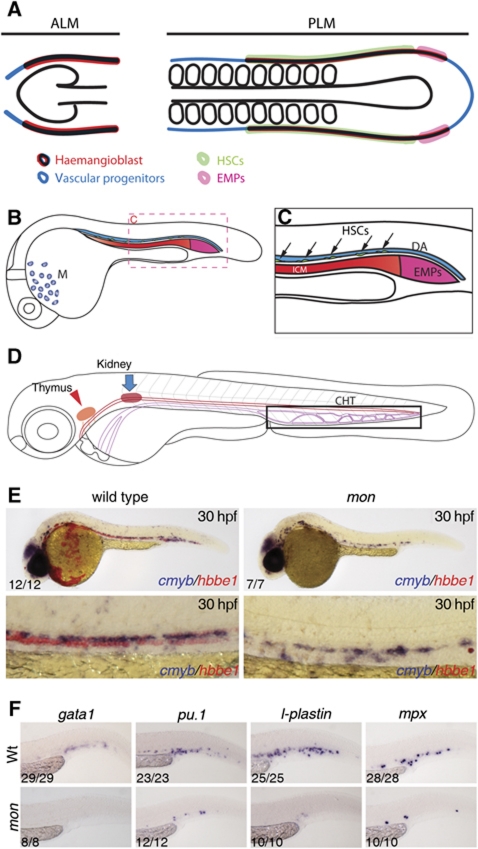
Figure 1.
Schematic representation of the different haematopoietic populations analysed in this study. Here, we depict the relative positions of haematopoietic and vascular progenitors in the lateral plate mesoderm populations, as well as the regions that are thought to give rise to haematopoietic stem cells (HSCs) and to erythromyeloid progenitors (EMPs). (A) Fate map of the anterior lateral plate mesoderm (ALM) and posterior lateral plate mesoderm (PLM) at 10 somites. (B) Fate map of ALM and PLM populations by 24 hpf. Haematopoietic progenitors from the ALM give rise to macrophages (M) in the head region, whereas the PLM gives rise predominantly to primitive erythrocytes in the intermediate cell mass (ICM). (C) Magnification of the trunk and tail region shown in (B). Definitive HSCs arise from the ventral wall of the dorsal aorta (DA, black arrows). EMPs arise from the most posterior PLM (posterior blood island, or PBI) from 24 to 30 hpf (Bertrand et al, 2007). (D) Schematic representation of the caudal haematopoietic tissue (CHT), the intermediate site of haematopoiesis, the thymus (red arrowhead) and the kidney (blue arrow). HSCs migrate from the DA to the CHT and, from 3 dpf onwards, they migrate to the thymus to give rise to T-lymphoid cells and to the kidney, the site of adult haematopoiesis. tif1γ loss-of-function suppresses primitive erythrocytes and EMPs, but does not affect HSC emergence. (E) Double in situ hybridisation for cmyb (blue) and hbbe1 (red) in wild-type (left panels) and moonshine (mon) embryos at 30 hpf. (F) At 36 hpf, gata1, pu.1, mpx and l-plastin are expressed in erythromyeloid progenitors (EMPs) in the PBI. mon embryos do not express gata1 and pu.1, mpx and l-plastin expression is absent or severely reduced. See also Supplementary Figure S1.
Transcription intermediate factor-1γ (tif1γ, ectodermin, TRIM33) is a RING domain E3 ubiquitin ligase containing an N-terminal RBCC (RING-finger-B box coil-coil) domain, a C-terminal PHD domain and a bromodomain, characteristic of chromatin-interacting proteins (Venturini et al, 1999). tif1γ localises to the nucleus (Ransom et al, 2004), binds to tif1α but not to DNA or tif1β (Peng et al, 2002), and can inhibit transcription when fused to a GAL4 DNA binding domain (Venturini et al, 1999). A truncating mutation in tif1γ in the bloodless mutant moonshine (montg234) leads to primitive erythroid progenitors being initially specified but failing to maintain adequate levels of haematopoietic transcription factors and undergoing apoptosis (Ransom et al, 2004). In contrast, the development of T cells in the thymus appeared to be unaffected in these mutants, suggesting that HSC emergence in the DA might also be unperturbed (Ransom et al, 2004).
With a view to studying the definitive lineage without the confounding presence of circulating primitive cells, we confirmed that HSCs emerge normally from the DA and then analysed the first signs of their differentiation in the CHT of montg234 mutants. We found that tif1γ favours the erythroid over the myeloid fate and in particular gata1 expression over pu.1 expression. We went on to establish the epistatic relationships between these three regulators in this cell population. We found that pu.1 antagonises gata1 activity and positively regulates its own expression, as predicted from the cell line studies, but it has no effect on tif1γ expression. gata1 antagonises pu.1 expression as predicted but, contrary to the cell line studies, gata1 negatively regulates itself and it also negatively regulates tif1γ. Having established this departure from the paradigm in this population of blood precursors, we analysed the status of this regulatory triad in the other blood populations of the early embryo. We found that it takes different forms in the different populations in ways which can account for the different output from these populations. In particular, we note that wherever myelopoiesis occurs, pu.1 antagonises gata1 along with stimulating its own expression. Likewise, wherever erythropoiesis occurs, gata1 antagonises pu.1 expression. However, the autoregulatory status of gata1 in the populations with erythroid output varied from stimulatory through no autoregulation to repressive. We therefore conclude that the regulatory interactions between gata1 and pu.1, modulated by tif1γ, are context dependent and in no case fully conform to the canonical view of the gata1/pu.1 cross-antagonism in any of the blood populations analysed.
Results
Tif1γ is required for primitive erythroid and EMP development, but not for primitive myeloid or HSC emergence
Loss of tif1γ function in mon mutant zebrafish embryos leads to complete loss of primitive erythrocytes, but seeding of the thymus by lymphoid progenitors is unaffected at 5 dpf (Ransom et al, 2004; Supplementary Figure S1A–C). In this study, we show that seeding of the thymus is unaffected from the outset (3 dpf, Supplementary Figure S1C) and that this indeed reflects apparently normal HSC emergence in the DA (Figure 1E and Supplementary Figure S1D–G′). The mutant status of these embryos was confirmed by the complete loss of hbbe1+ (primitive erythroid) cells.
We also analysed the role of tif1γ in the other two blood populations in the zebrafish embryo, namely, the primitive myeloid population in the ALM and the prodefinitive EMP population in the PBI (Figure 1A–C). To determine if EMPs are formed in mon mutants, we monitored expression of the markers originally used to identify them, namely, gata1, pu.1, l-plastin and mpx at 36 hpf (Bertrand et al, 2007). We found that expression of these genes was either lost or severely reduced (Figure 1F), suggesting that tif1γ is required for the emergence of EMPs. Morpholino knockdown of runx1 showed no effect on any of these EMP markers (Supplementary Figure S2A–N′), confirming that these progenitors are not derived from runx1-dependent HSCs. In contrast to the EMP population, generation of primitive myeloid cells was not affected by loss of tif1γ function (Supplementary Figure S1H, I and data not shown), indicating that tif1γ does not have a role in the specification or differentiation of the primitive myeloid population. Thus, tif1γ is not required for primitive myeloid development or HSC emergence, whereas it is critical for EMP and primitive erythroid development.
Tif1γ is required for erythroid differentiation of HSCs
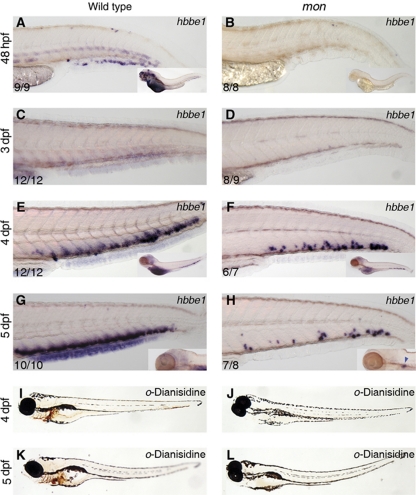
Circulating primitive erythrocytes express the haemoglobin gene hbbe1 at 48 hpf, but expression ceases by 3 dpf (Brownlie et al, 2003; Figure 2A and C). Erythroid derivatives of HSCs can first be detected by in situ hybridisation in the CHT at 3.5–4 dpf (Jin et al, 2007, 2009), reflected in strong re-expression of hbbe1 from 4 dpf onwards (Figure 2E and G). CHT expression of hbbe1 in mon mutants, however, was seen in only very few cells at this stage (Figure 2F and H), which were incapable of giving rise to terminally differentiated, circulating erythrocytes, as detected by o-dianisidine staining (Figure 2I–L). Similar results were observed up to 10 dpf (Supplementary Figure S1J). The absence of hbbe1 expression in runx1 morphants, which fail to develop HSCs (Gering and Patient, 2005), confirms that these erythroid cells are derived from HSCs (Supplementary Figure S2O, P). Thus, HSCs are incapable of generating mature erythrocytes in the absence of tif1γ, as seen for the primitive erythroid lineage. Mutant larvae died between 10 and 14 dpf, presumably due to lack of efficient oxygenation. Expression of tif1γ was found in the CHT from 48 hpf onwards (Supplementary Figure S3) and was lost in runx1 morphants (Supplementary Figure S2U, V), confirming that it is expressed in HSC derivatives when these are differentiating into erythroid cells. Taken together, these data indicate that tif1γ is required for the generation of erythrocytes from HSCs.
Figure 2.
Time course of hbbe1 expression from 2 to 5 dpf in wild-type (A, C, E, G) and mon (B, D, F, H) larvae. Only a few erythroid cells arise de novo in the CHT of mon mutants, but fail to terminally differentiate. At 48 hpf, hbbe1 is expressed in primitive erythrocytes in wild-type embryos (A), but not in mon (B). At 3 dpf, neither wild-type (C) nor mon (D) showed hbbe1 expression in the CHT. (E) At 4 dpf, robust expression of hbbe1 is found in wild type larvae. (F) mon larvae also show hbbe1+ cells, but far fewer. (G) Wild-type hbbe1 expression at 5 dpf. (H) Only a few cells in the mon mutant CHT are hbbe1+. o-Dianisidine staining for terminally differentiated erythrocytes at 4 and 5 dpf in wild-type (I, K) and mon larvae (J, L) confirmed the lack of circulating erythrocytes in mon. See also Supplementary Figure S1.
Loss of erythroid cells is accompanied by a dramatic increase in definitive myeloid cell numbers
The primitive erythroid population in mon mutants eventually undergoes apoptosis (Ransom et al, 2004; see also Supplementary Figure S4A, A′). To investigate whether this also occurs in the definitive erythroid population in the CHT, we performed fluorescent immunostaining with an antibody against activated caspase-3, a hallmark of apoptosis (Kratz et al, 2006 and references therein; Supplementary Figure S4A–D′). The increased apoptosis of the primitive erythroid cells in the ICM region of mon mutants at 24 hpf acted as a positive control (Supplementary Figure S4A, A′, arrow). However, we found few, if any, extra apoptotic cells in the CHT of mon mutants compared with wild-type embryos (Supplementary Figure S4B–D′). Apoptosis was also monitored by TUNEL in the CHT of 3 and 4 dpf zebrafish embryos, but the modest increase observed (1–2 cells) could not account for the dramatic loss of erythroid cells (Supplementary Figure S4E). We therefore determined if, instead, proliferation of haematopoietic progenitors had been decreased, by performing in situ hybridisation for proliferating cell nuclear antigen (PCNA, Koudijs et al, 2005) from 3 to 5 dpf (Supplementary Figure S4F–K). We found no evidence of decreased proliferation in the CHT of mon larvae from 3 dpf onwards compared with wild-type embryos. Similar results were obtained upon immunostaining for phosphorylated histone H3 (Supplementary Figure S4L–R). We therefore conclude that neither increased apoptosis nor decreased proliferation can account for the loss of erythroid cells in the CHT of mon mutants.
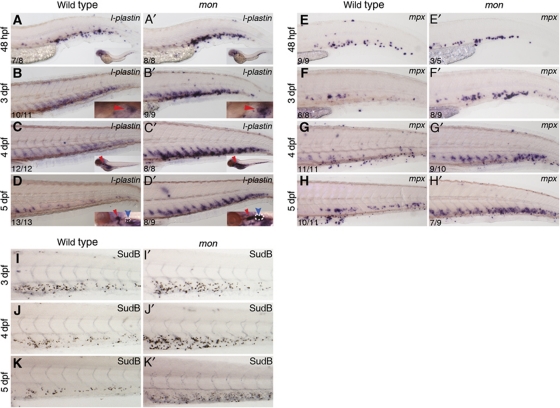
HSCs are thought to give rise to myeloid as well as erythroid cells in the CHT (Jin et al, 2009), and this is confirmed by the abolition of myeloid gene expression there in runx1 morphants (Supplementary Figure S2S, T). In view of this, and since cell numbers did not decrease in tif1γ morphants, we analysed the expression of the pan-leukocyte marker l-plastin (Moss et al, 2009) and the myeloid marker mpx (granulocytes; Herbomel et al, 1999), between 2 and 5 dpf (Figure 3A–H′). At 2 dpf, l-plastin+ and mpx+ cells were found in mon mutant CHT at levels similar to those in wild-type embryos (Figure 3A, A′ and E, E′). However, from 3 dpf onwards, expression of both markers was increased in the CHT of mon larvae (Figure 3B, B′ and F, F′), and this increase became more prominent by 4 and 5 dpf (Figure 3C–D′, G–H′). To determine whether the extra myeloid cells could terminally differentiate, we stained wild-type and mon larvae from 3 to 5 dpf with Sudan Black, which stains the granules in granulocytes (Figure 3I–K′). We found an increased number of Sudan Black+ cells in the mutant CHTs when compared with wild type from 3 dpf onwards, with the highest increase observed at 4 dpf (compare Figure 3I and J with I′ and J′). Furthermore, the increase in l-plastin and mpx expression was not limited to the CHT niche, but was also seen in the kidney (Figure 3B–D′, see blue arrowheads in insets) and in differentiating myeloid cells in the trunk (Supplementary Figure S5A–E′), which are HSC-derived 3 (Jin et al, 2007, 2009). We therefore conclude that all definitive myeloid differentiation is increased in the absence of tif1γ.
Figure 3.
tif1γ loss of function promotes expansion of (functional) myeloid cells in the CHT. (A–D) l-plastin is expressed in the wild-type CHT at 48 hpf (A), 3 dpf (B), 4 dpf (C) and 5 dpf (D). l-plastin is also present in the thymus from 3 dpf (B, C, see inset) and in the kidney at 5 dpf (D, blue arrow, see inset). (A′–D′) Expression of l-plastin is increased in the mon CHT at all stages analysed, with an expression peak by 4 dpf. In 5 dpf mon larvae, kidney l-plastin expression was also increased (D′, blue arrow, see inset). (E–H) mpx is expressed in the wild-type CHT from 48 hpf to 5 dpf. (E′–H′) From 3 dpf onwards (F′), mon mutants show increased mpx expression. Sudan Black staining for granulocytes was performed in wild type (I–K) and mon CHTs from 3 to 5 dpf (I′–K′). In insets, red arrowheads identify the thymus and blue arrowheads identify the kidney. White broken lines delimit the boundaries of l-plastin expression in the kidney. The number of embryos analysed are shown in each panel. See also Supplementary Figure S5.
Tif1γ differentially regulates the expression of gata1 and pu.1 in the CHT
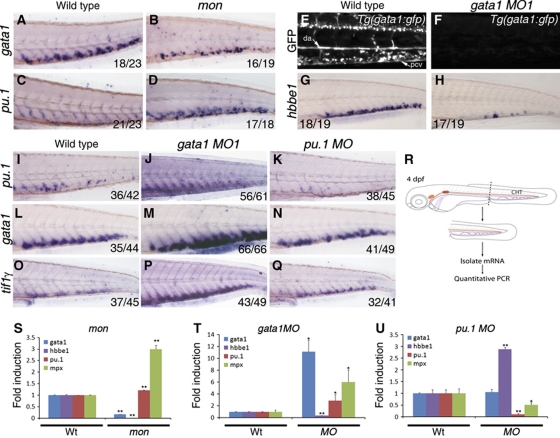
As tif1γ regulates the erythroid versus myeloid output from HSCs in the CHT, a lineage choice thought to be under the control of gata1 and pu.1, we analysed the epistatic relationships between these three genes (Figure 4). mon mutants contain few gata1+ cells in the CHT at 4 dpf (Figure 4A and B), whereas the number of pu.1+ cells is increased (Figure 4C and D), compared with their wild-type siblings. To quantify these changes in expression in the CHT, we performed quantitative RT–PCR (qPCR) on mRNA isolated from dissected tails (Figure 4R). In agreement with the in situ hybridisation, gata1 and hbbe1 expression were severely downregulated, whereas pu.1 and mpx were upregulated in mon mutants (Figure 4S).
Figure 4.
Tif1γ differentially regulates expression of gata1 and pu.1 in the CHT. Epistatic analysis of tif1γ, gata1 and pu.1 function in the CHT at 4 dpf. mon mutants were used to analyse tif1γ loss of function; gata1 MO1 (30 ng/nl) and pu.1 MO (30 ng/nl) were used to analyse gata1 and pu.1 loss of function, respectively. (A) At 4 dpf, gata1 is expressed in the CHT of wild-type embryos and (B) severely reduced in mon larvae. (C) pu.1 is expressed at low levels in the wild-type CHT, whereas (D) it is massively increased in the CHT of mon mutants. (E) Expression of the GFP transgene in Tg(gata1:gfp) larvae. Abundant GFP-expressing cells are found in circulation and in the CHT region between the dorsal aorta (DA) and the posterior cardinal vein (PCV). (F) gata1 MO1 injection induced a complete loss of GFP expression. (G) Expression of hbbe1 in wild-type larvae. (H) hbbe1 expression was significantly downregulated in gata1 morphants. (I) Expression of pu.1 in wild-type larvae. (J) gata1 morphants show increased pu.1 expression in the CHT, whereas (K) pu.1 morphants show little pu.1 expression. (L) Expression of gata1 in wild-type embryos. (M) gata1 morphants show increased gata1 expression, whereas (N) pu.1 morphants show no obvious change in gata1 expression in the CHT. (O) Expression of tif1γ in wild-type larvae. (P) tif1γ expression is upregulated in gata1 morphants. (Q) pu.1 MO knockdown does not appear to affect tif1γ expression in the CHT. All CHTs are shown as lateral views, anterior to the left. (R) Schematic representation of tail microdissection (containing the CHT) for mRNA isolation and quantitative PCR analysis. (S) Analysis of gata1, hbbe1, pu.1 and mpx expression in wild-type or mon mutants at 4 dpf. (T) Analysis of gata1, hbbe1, pu.1 and mpx expression in wild-type or gata1 morphants and (U) pu.1 morphants at 4 dpf. (A–Q) The number of embryos analysed are shown in each panel. (S–U) Expression of each gene in dissected wild-type tails was set to 1 for comparison. All data are shown as average±s.d. values; **P<0.0001; *P<0.007. See also Supplementary Figure S5.
To determine if gata1 and pu.1 are indeed controlling the erythroid/myeloid switch in HSC derivatives in the CHT, and to establish the relationship of tif1γ with them, we knocked down gata1 and pu.1 by morpholino injection and monitored the expression of pu.1, gata1 and tif1γ at 4 dpf (Figure 4E–Q). Suppression of gata1 activity in gata1 morpholino oligonucleotide (MO)-injected embryos was confirmed by loss of GFP fluorescence in Tg:gata1-gfp transgenic embryos (Figure 4E and F) and loss of hbbe1 expression (Figure 4G, H). Suppression of pu.1 activity was confirmed by the upregulation of gata1 expression anteriorly in pu.1 morphants at 24 hpf (Galloway et al, 2005) (Figure 6I, green arrowhead). In the CHT at 4 dpf, expression of pu.1 was increased in gata1 morphants and lost in pu.1 morphants (Figure 4I–K), as predicted by the cross-antagonism and autostimulation model. However, unexpectedly, gata1 expression was upregulated in gata1 morphants and unaffected in pu.1 morphants (Figure 4L–N), suggesting that gata1 rather than pu.1 negatively regulates gata1 expression in the CHT (Figure 7B). Quantitative PCR analysis of dissected tails confirmed the decrease in hbbe1 and the increase in pu.1, mpx and gata1 expression seen by in situ hybridisation in gata1 morphants (Figure 4T). Similarly, the severe decrease in both pu.1 and mpx seen by in situ hybridisation in pu.1 morphants was confirmed, as was the lack of effect on gata1 expression (Figure 4U). However, hbbe1 expression was clearly upregulated in pu.1 morphants (Figure 4U), suggesting that even though pu.1 does not inhibit expression of gata1 in the CHT, it inhibits gata1's stimulation of hbbe1 expression. As gata1 is autoinhibitory in the CHT and loss of pu.1 has no effect on gata1 expression there, these data indicate that pu.1 only inhibits transactivation by gata1 and not repression. Consistent with this, gata1 negatively regulates tif1γ as well as itself in the CHT, and tif1γ expression is also unaffected by the loss of pu.1 (Figure 4O–Q). Thus, we see a significant departure from the gata1/pu.1 paradigm in the CHT (compare Figure 7A and B), and also find that tif1γ maintains gata1 but suppresses pu.1 expression there, thereby favouring an erythroid over a myeloid lineage output.
Tif1γ functions cell-autonomously in HSC derivatives residing in the CHT
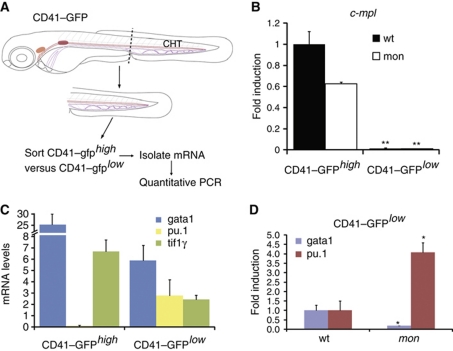
In zebrafish, high level expression of a CD41–gfp transgene is characteristic of thrombocytes, whereas HSCs are found within the CD41–gfplow population (Kissa et al, 2008). To confirm that tif1γ is acting cell-autonomously in HSC derivatives residing in the CHT, we isolated CD41–gfphigh and CD41–gfplow cells from 4 dpf dissected tails by flow cytometry and assessed tif1γ expression by qPCR (Figure 5). Expression of c-mpl, a marker enriched in thrombocytes (Lin et al, 2005; Bertrand et al, 2008), was assessed to verify the purity of the two cell populations. In contrast to CD41–gfphigh, CD41–gfplow cells expressed very little c-mpl (Figure 5B). This enrichment was also found in the CD41–gfphighmon cell population (Figure 5B), although c-mpl expression was lower than that in wild-type CD41–gfphigh. It has been suggested that thrombocytes are derived from bipotential progenitors residing in the ICM region at 24 hpf (Warga et al, 2009), which are absent in mon. However, we can still detect CD41–gfphigh cells in mon, suggesting that thrombocytes may also be HSC derived.
Figure 5.
Tif1γ cell-autonomously regulates gata1 and pu.1 in CD41–gfplow HSCs present in the CHT. (A) Schematic representation of the strategy for HSC isolation from the CHT. Tails of 4 dpf CD41–gfp or CD41–gfp;mon embryos were dissected and dissociated to obtain a single-cell suspension. This was followed by flow cytometry to sort the CD41–gfphigh (thrombocyte) and CD41–gfplow (HSC) cell populations. (B) Quantitative RT–PCR analysis of c-mpl expression in CD41–gfphigh and CD41–gfplow cell populations in wild-type or mon mutants. c-mpl was highly enriched in the CD41–gfphigh population, confirming the accuracy of the cell sorting. Expression of c-mpl in the CD41–gfphigh population was set to 1 for comparison. (C) Expression analysis of gata1, pu.1 and tif1γ in CD41–gfphigh and CD41–gfplow wild-type cells in the CHT. pu.1 mRNA was absent, whereas gata1 and tif1γ were expressed in CD41–gfphigh, all three genes were expressed at lower levels in CD41–gfplow (HSCs) cells. Absolute quantification is shown. (D) Expression analysis of gata1 and pu.1 in wild-type and mon CD41–gfplow cells in the CHT at 4 dpf. Note that gata1 expression is already downregulated, whereas pu.1 is upregulated, in this cell population at 3 dpf (see Supplementary Figure S5). Expression of each gene in wild-type CD41–gfplow cells was set to 1 for comparison. All data are shown as average±s.d. values; **P<0.0005; *P<0.007.
Importantly though, tif1γ as well as gata1 and pu.1 were found to be expressed, albeit at low levels, in CD41-gfplow cells, whereas pu.1 was absent and tif1γ and gata1 were more highly expressed in the CD41–gfphigh population (Figure 5C). Thus, tif1γ, gata1 and pu.1 are coexpressed in HSCs or their early derivatives in the CHT, a prerequisite for their interaction being cell-autonomous and tif1γ regulating gata1 and pu.1 directly in these cells. Consistent with such a relationship, we found that gata1 was severely downregulated in CD41–gfplow cells from dissected tails of mon larvae at 4 dpf, whereas pu.1 was upregulated (Figure 5D). Since erythroid cells can be detected in the CHT at 3.5 dpf (Jin et al, 2007, 2009), we confirmed that the effect was already apparent in CD41–gfplowmon cells at 3 dpf (Supplementary Figure S5F–H). Thus, tif1γ functions cell-autonomously in HSC derivatives to maintain gata1 and suppress pu.1 expression, thereby regulating the erythroid and myeloid output from HSCs.
Epistatic relationships between tif1γ, gata1 and pu.1 in the ALM, PLM/ICM and EMP haematopoietic populations
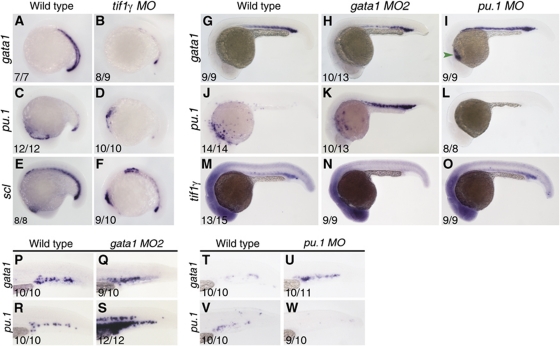
In view of the observed modifications to the classical gata1/pu.1 cross-antagonism paradigm in the CHT (compare Figure 7A and B), we also determined the epistatic relationships between tif1γ, gata1 and pu.1 in the earlier blood populations in developing embryos. Loss of tif1γ function by morpholino injection induced a marked decrease in the expression of both gata1 and pu.1 in the PLM/ICM at the 16 somite stage (Figure 6A–D). Scl expression was severely reduced in these morphants, but still present at low levels in the PLM/ICM (Figure 6E and F), indicating that the observed phenotype was due to loss of gene expression rather than cell death. gata1 knockdown induced gain of pu.1 and loss of tif1γ expression in the ICM by 24 hpf (Figure 6 J, K, M and N). However at 12 somites, neither gata1 nor tif1γ knockdown has any effect on haematopoietic gene expression in the PLM (Galloway et al, 2005 and results not shown). Thus, the activities of both tif1γ and gata1 in the PLM/ICM network only kick in after the initial induction of haematopoietic gene expression. Furthermore, although knocking down pu.1 induced ectopic expression of gata1 in the ALM (Figure 6I, green arrowhead) and loss of its own expression (Figure 6L), it had no effect on the expression of tif1γ in the PLM/ICM (Figure 6M and O). We therefore conclude that, in contrast to the situation in the HSC derivatives in the CHT, tif1γ maintains both gata1 and pu.1 expression in the PLM/ICM (summarised in Figure 7C). Furthermore, although gata1 negatively regulates tif1γ expression in the CHT, it has a positive effect in the PLM/ICM; this difference is also seen for gata1 autoregulation, which is negative in the CHT and positive in the PLM/ICM. Contrary to the predictions from the gata1/pu.1 paradigm, but in line with the CHT data, pu.1 does not regulate gata1 expression in the PLM/ICM (Figure 7C). These differences likely relate to the differential myeloid output from the two populations; the PLM/ICM mainly gives rise to erythrocytes, whereas HSCs generate both erythroid and myeloid cells in the CHT.
Figure 6.
Genetic regulatory interactions between tif1γ, gata1 and pu.1 in the PLM/ICM and EMP populations. Morpholinos were injected at the following concentrations: tifγ MO—1 ng/nl, gata1 MO2—5 ng/nl and pu.1 MO—20 ng/nl. (A) Expression of gata1 in wild-type and (B) tif1γ MO-injected embryos at 16 somites. (C) Expression of pu.1 in wild-type and (D) tif1γ MO-injected embryos at 16 somites. (E) Expression of scl in wild-type and (F) tif1γ MO-injected embryos at 16 somites. (G) gata1 expression in wild type, (H) gata1 morphants and (I) pu.1 morphants at 24 hpf. (J) pu.1 expression in wild type, (K) gata1 morphants and (L) pu.1 morphants at 24 hpf. (M) tif1γ expression in wild type, (N) gata1 morphants and (O) pu.1 morphants at 24 hpf. (P–W) Analysis of gata1 and pu.1 expression in the EMP population at 30 hpf in gata1- and pu.1 morphants. Expression of gata1 is similar in wild-type (P) and gata1 morphants (Q). (R) Expression of pu.1 in wild-type EMPs. (S) pu.1 expression is higher in gata1 morphants. Expression of gata1 in wild-type (T) and pu.1 morphants (U). (V) Expression of pu.1 in wild-type EMPs. (W) pu.1 expression is lost in pu.1 morphants. All embryos shown in lateral views, anterior to the left; numbers of embryos analysed are shown in each panel. See also Supplementary Figure S6.
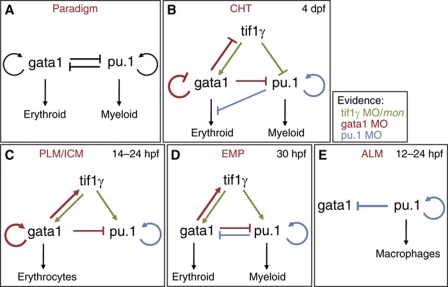
Figure 7.
Schematic representation of the genetic regulatory interactions between tif1γ, pu.1 and gata1 in the different haematopoietic populations analysed in this study. (A) Schematic representation of the classical gata1/pu.1 cross-antagonistic paradigm. (B) In the CHT, tifγ is required for gata1 expression, whereas it negatively regulates pu.1. Consequently, tifγ favours the erythroid and restricts the myeloid lineage in this haematopoietic niche. (C) tifγ is required for the expression of pu.1 and gata1 in the PLM/ICM. Here, gata1 represses pu.1 but pu.1 does not repress gata1 expression. (D) In the EMP compartment, like in the PLM/ICM, tifγ is required for the expression of pu.1 and gata1, while gata1 and pu.1 antagonise each other's expression. However, we found no evidence of gata1-positive autoregulation. (E) Genetic interactions between gata1 and pu.1 in the ALM. Note that pu.1 showed positive autoregulation in all the haematopoietic populations analysed. Regulatory interactions are shown in different colours to highlight which loss-of-function analysis originated the evidence: green (tif1γ MO knockdown/mon), red (gata1 MO knockdown) or blue (pu.1 MO knockdown).
EMPs found in the PBI are temporally located between the PLM/ICM and CHT populations during development (Figure 1A–C). Similar to the HSCs, EMPs have both erythroid and myeloid potential (Bertrand et al, 2007), but similar to the PLM/ICM population are only transient and are only detectable up to 48 hpf (Bertrand et al, 2007). In the EMPs, knocking down gata1, while upregulating pu.1 expression (Figure 6R and S), had little effect on gata1 expression (Figure 6P and Q). Thus, in these cells, in contrast to both the PLM/ICM and CHT populations, gata1 does not autoregulate at all, either positively or negatively (Figure 7D). pu.1 knockdown induced an increase in gata1 (Figure 6T and U) and loss of pu.1 expression in the EMP compartment (Figure 6V and W), as predicted by the cross-antagonism model. We conclude that tif1γ maintains gata1 and pu.1 expression in both the PLM/ICM and the EMP populations, whereas it differentially affects their expression in the CHT (Figure 7B–D). gata1 also maintains tif1γ expression in both the PLM/ICM and EMP compartments, whereas it is inhibitory in the CHT. Finally, whereas pu.1 is autostimulatory in all four compartments, gata1 ranges from autostimulatory through no autoregulation to autorepressive.
Discussion
We have demonstrated that tif1γ is a critical regulator of cell fate during haematopoietic ontogeny. We have shown that it is required in HSCs and their derivatives in the FL equivalent niche in zebrafish, the CHT, favouring erythroid over myeloid commitment. We show that it does this by inhibiting pu.1 expression and stabilising gata1 expression (Figure 7B). This situation contrasts with that in the earlier blood populations where tif1γ is required to stabilise both gata1 and pu.1 expression (Figure 7C and D). Thus, tif1γ is clearly a critical player in the development of blood cells. In elucidating the regulatory relationships between tif1γ and the gata1 and pu.1 master regulators of erythroid versus myeloid lineage choice in the haematopoietic system, it became clear that the relationships between gata1 and pu.1 themselves were more variable than expected. By perturbing all three regulators and monitoring the impact on their expression in each case, we have been able to assemble networks describing the genetic logic relating to the differing output from each of the four blood populations found in the developing zebrafish embryo (Figure 7). Surprisingly, none of the networks fully conforms to the cross-antagonism, autostimulation paradigm originally established using cell lines in vitro (Graf and Enver, 2009, Figure 7A).
The novel function of tif1γ in favouring erythroid over myeloid cell fate commitment, seen in the HSC population differentiating in the CHT, was not seen in the earlier primitive or transient definitive haematopoietic lineages produced by the ALM, the PLM/ICM or the EMP compartments (Figure 7). In the primitive myeloid compartment in the ALM, tif1γ seems to have no role, whereas in the predominantly primitive erythroid compartment in the PLM/ICM and in the bipotential EMP population, tif1γ maintains both gata1 and pu.1 expression. Thus, its input into the expression of pu.1 can be either positive or negative in a context-dependent manner, while its input into gata1 is positive independent of the context. The description of tif1γ in transcription assays as a repressor (Venturini et al, 1999) fits its activity towards pu.1 in the CHT; however, its activity in the other contexts and its activity towards gata1 would require an intermediate target repressor. Alternatively, very recent data assign a positive role for tif1γ in overcoming a block to transcriptional elongation of gata1 and scl transcripts in the PLM/ICM erythroid population by recruiting Cdk9 to the stalled transcriptional complex (Bai et al, 2010), suggesting that tif1γ may be acting directly on gata1 or pu.1 transcription when acting positively.
We have shown that gata1 negatively regulates expression of its own gene and tif1γ in the CHT, whereas that regulation is positive in the PLM/ICM compartment (Figure 7B and C). Clearly, these different activities of gata1 are likely to be mediated at least in part by different cofactors in the two cell populations. Bai et al (2010) have recently shown that tif1γ physically associates with the scl complex, which contains gata1 (Meier et al, 2006). Thus, we speculate that gata1 and tif1γ are part of a transcriptional complex required in HSCs to repress pu.1 expression and/or activity. Nevertheless, however it is being achieved, the regulatory logic reflects the need to restrict the erythroid fate in the CHT population to ensure a myeloid output, whereas in the PLM/ICM population no such restriction is required, with differentiation into erythroid cells predominating. This change of activity of gata1 accompanies a change of activity of tif1γ with respect to pu.1, which also goes from negative in the CHT to positive in the PLM/ICM. Thus, the negative activity of gata1 towards tif1γ in the multipotent cells in the CHT may be required to restrict the repression of pu.1 expression by tif1γ and protect a myeloid output.
gata1 also positively regulates tif1γ in EMPs, which retain myeloid potential (Figure 7D). However, in these cells gata1 does not autoregulate and, furthermore, pu.1 negatively regulates gata1. The negative regulation of gata1 expression and/or activity by pu.1 is also seen in the ALM and the CHT (Figure 7B and E). In other words, pu.1 antagonism of gata1 is always seen when there is a myeloid output. Conversely, gata1 antagonism of pu.1 expression is always seen when there is an erythroid output. Interestingly, pu.1 autostimulation is seen in all the populations regardless of significant myeloid output, suggesting that the antagonism is more important than the autostimulation in determining cell fate. Autostimulation may be more important when cells need to be maintained in a bipotential state (Chickarmane et al, 2009).
Overall, the regulatory relationships between gata1, pu.1 and tif1γ have been adjusted in each of the blood populations in the developing embryo, to tilt the output towards erythroid or myeloid, or to balance the two. We conclude that the genetic interactions between gata1 and pu.1 are dynamic and highly dependent on haematopoietic cell type, haematopoietic niche and the presence or activity of the critical modulating partner tif1γ.
Materials and methods
Fish maintenance and morpholino injections
Fish were bred and maintained as described (Westerfield, 2007). Wild-type or Tg(gata1:gfp) (Long et al, 1997) embryos were obtained by natural mating. Moonshine (montg234; Ransom et al, 1996) were crossed with Tg(CD41:gfp) (Lin et al, 2005) fish to give rise to Tg(CD41:gfp); montg234 heterozygous carriers. montg234 or Tg(CD41:gfp); montg234 heterozygous carriers were incrossed to obtain homozygous mutants (herein referred to as mon or CD41-gfp;mon mutants, respectively). Typically, the phenotypically normal embryos generated from these crosses were used as wild-type controls. A MO was used to target runx1 (Gering and Patient, 2005) and suppress runx1-dependent definitive haematopoiesis in mon mutants. To query the genetic relationship between tif1γ, gata1 and pu.1, we used morpholinos targeting gata1–gata1 MO1, gata1 MO2 (Galloway et al, 2005), pu.1 (Rhodes et al, 2005) and the 5′-UTR of tif1γ (5′-GCTCTCCGTACAATCTTGGCCTTTG-3′). To analyse gata1 and pu.1 function in the CHT, we have injected 30 ng/nl of both gata1 and pu.1 morpholinos. This MO concentration allowed us to circumvent the loss of MO efficiency at later stages and efficiently repress their target genes hbbe1 and mpx, respectively. The amount of morpholino used to target gata1 and pu.1 are specified in each figure. Injection of 1 ng/nl of the tif1γ MO phenocopied the mon mutant phenotype at 24 hpf (Supplementary Figure S6). We typically inject 1 nl MO per 1-cell stage embryo.
Whole mount in situ hybridisation
Whole mount in situ hybridisation was carried out as described (Jowett and Yan, 1996). A PCR fragment spanning ∼1 kb was amplified from 24 hpf embryo cDNA (tif1γ-F, TGCTCAACCTCGACTCAATG; tif1γ-R, CCGCAGTGGTCTGACTGTTA) and used as template to generate a probe for tifγ. DIG-labelled antisense RNA probes were transcribed from linearised templates using T3, T7 or Sp6 RNA polymerases (Roche, Burgess Hill, UK). After hybridisation, embryos were bleached in 5% formamide, 0.5% SSC, 10% H2O2 for 10–30 min, washed in PBST (PBS, 0.1% Tween-20) and transferred to 80% glycerol for imaging.
Immunochemistry, o-dianisidine and Sudan Black staining
To analyse proliferation, embryos collected at the desired stages were fixed in 4% PFA, dehydrated and then rehydrated and incubated with proteinase K (10 mg/ml in PBST) according to their stage. After a brief fixation (20 min, 4% PFA), embryos were washed with PBST for 5 × 5 min at room temperature, incubated in block solution (1% BSA in PBST) for 1 h at room temperature and then incubated with rabbit anti-phosphorylated histone H3 (ser10; Upstate, 1:400) at 4°C overnight. After removing the primary antibody, the embryos were washed with PBST and incubated with Alexa Fluor 488-labelled goat anti-rabbit IgG antibody (Molecular Probes, 1:400). Apoptotic cells were detected with antiactivated caspase3 antibody (Sigma, 1:250) essentially as described (Kratz et al, 2006). Twenty-four hpf mon embryos were used as an internal positive control for apoptosis (Supplementary Figure S4). Terminally differentiated erythrocytes were stained with o-dianisidine, and embryos were mounted for imaging as described (Detrich et al, 1995). Sudan Black is a classic lipid dye that stains the granules in granulocytes; staining was performed as described (Le Guyader et al, 2008). Following staining, the larvae were mounted in 80% glycerol for imaging.
Tail dissection, cell dissociation and sorting, RNA isolation and qPCR
To quantitate the effects of gata1, pu.1 or tif1γ loss of function in the CHT, tails from anaesthetised 3 or 4 dpf larvae were microdissected with a straight stab knife. Tails from anaesthetised 4 dpf CD41–gfp or CD41–gfp;mon larvae were collected in ice-cold PBSS (0.9 × PBS/5% heat-inactivated serum) and dissociated as described (Bertrand et al, 2007). Viable cells were sorted on a MoFlo cell sorter based on their fluorescence profile and forward scatter, then collected in 0.9 × PBS/serum. RNA was isolated with the RNEasy Micro kit (Qiagen) following the manufacturer's instructions. Total RNA (200–250 ng) was reverse transcribed into cDNA using a Superscript III RT–PCR enzyme (Invitrogen, Philadelphia, PA). Primers used to assess expression of c-mpl, ef1a, mpx, pu.1 and gata1 transcripts have been described elsewhere (Bertrand et al, 2007 and references therein); primers for tif1γ and hbbe1 were as follows: tif1γ F2—GACACGGAGACGAAAGAAGC and tif1γ R2—CATTAGGGGTCCCGTCTGTA; hbbe1—AACTGTGCTCAAGGGTCTGG and hbbe1—TACGTGGAGCTTCTCGGAGT. For quantification, gene expression was always normalised to that of ef1a.
Image acquisition and processing
Hybridised embryos were photographed with a Nikon DXM 1200 digital camera and Nikon ACT-1 software (version 2.12) mounted on a Nikon SMZ 1500 zoom stereomicroscope (Nikon, Melville, NY). Embryos were viewed at × 3 to × 11.25 magnification, depending on stage. Alternatively, fluorescence images were captured on a Zeiss Lumar V.12 Stereomicroscope with an AxioCam MRm (Zeiss) and AxioVision software. Images were processed with Adobe Photoshop CS4; schemes were assembled in Adobe Illustrator CS4 (Adobe Systems, San Jose, CA).
Supplementary Material
Acknowledgments
We thank Aldo Ciau-Uitz and Carla Galinha for discussions and critical reading of the paper. We are very grateful to the staff of the Biomedical Services Unit for aquatic support. This work was supported by the EuTRACC Consortium of the European Union Framework 7 Programme and by the MRC.
Author contributions: RM designed and performed experiments, analysed the data and wrote the paper; CP performed FACS analysis and RP designed experiments, analysed the data and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bai X, Kim J, Yang Z, Jurynec MJ, Akie TE, Lee J, LeBlanc J, Sessa A, Jiang H, DiBiase A, Zhou Y, Grunwald DJ, Lin S, Cantor AB, Orkin SH, Zon LI (2010) TIF1gamma controls erythroid cell fate by regulating transcription elongation. Cell 142: 133–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand JY, Kim AD, Teng S, Traver D (2008) CD41+ cmyb+ precursors colonize the zebrafish pronephros by a novel migration route to initiate adult hematopoiesis. Development 135: 1853–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand JY, Kim AD, Violette EP, Stachura DL, Cisson JL, Traver D (2007) Definitive hematopoiesis initiates through a committed erythromyeloid progenitor in the zebrafish embryo. Development 134: 4147–4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand JY, Chi NC, Santoso B, Teng S, Stainier DY, Traver D (2010) Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 464: 108–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlie A, Hersey C, Oates AC, Paw BH, Falick AM, Witkowska HE, Flint J, Higgs D, Jessen J, Bahary N, Zhu H, Lin S, Zon L (2003) Characterization of embryonic globin genes of the zebrafish. Dev Biol 255: 48–61 [DOI] [PubMed] [Google Scholar]
- Cantor AB, Orkin SH (2002) Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene 21: 3368–3376 [DOI] [PubMed] [Google Scholar]
- Chickarmane V, Enver T, Peterson C (2009) Computational modeling of the hematopoietic erythroid-myeloid switch reveals insights into cooperativity, priming, and irreversibility. PLoS Comput Biol 5: e1000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn MF, Ma X, Robin C, Ottersbach K, Sanchez MJ, Dzierzak E (2002) Hematopoietic stem cells localize to the endothelial cell layer in the midgestation mouse aorta. Immunity 16: 673–683 [DOI] [PubMed] [Google Scholar]
- de Bruijn MF, Speck NA, Peeters MC, Dzierzak E (2000) Definitive hematopoietic stem cells first develop within the major arterial regions of the mouse embryo. EMBO J 19: 2465–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detrich HW III, Kieran MW, Chan FY, Barone LM, Yee K, Rundstadler JA, Pratt S, Ransom D, Zon LI (1995) Intraembryonic hematopoietic cell migration during vertebrate development. Proc Natl Acad Sci USA 92: 10713–10717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzierzak E, Speck NA (2008) Of lineage and legacy: the development of mammalian hematopoietic stem cells. Nat Immunol 9: 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway JL, Wingert RA, Thisse C, Thisse B, Zon LI (2005) Loss of gata1 but not gata2 converts erythropoiesis to myelopoiesis in zebrafish embryos. Dev Cell 8: 109–116 [DOI] [PubMed] [Google Scholar]
- Gering M, Patient R (2005) Hedgehog signaling is required for adult blood stem cell formation in zebrafish embryos. Dev Cell 8: 389–400 [DOI] [PubMed] [Google Scholar]
- Graf T (2002) Differentiation plasticity of hematopoietic cells. Blood 99: 3089–3101 [DOI] [PubMed] [Google Scholar]
- Graf T, Enver T (2009) Forcing cells to change lineages. Nature 462: 587–594 [DOI] [PubMed] [Google Scholar]
- Herbomel P, Thisse B, Thisse C (1999) Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development 126: 3735–3745 [DOI] [PubMed] [Google Scholar]
- Jin H, Sood R, Xu J, Zhen F, English MA, Liu PP, Wen Z (2009) Definitive hematopoietic stem/progenitor cells manifest distinct differentiation output in the zebrafish VDA and PBI. Development 136: 647–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Xu J, Wen Z (2007) Migratory path of definitive hematopoietic stem/progenitor cells during zebrafish development. Blood 109: 5208–5214 [DOI] [PubMed] [Google Scholar]
- Jowett T, Yan YL (1996) Double fluorescent in situ hybridization to zebrafish embryos. Trends Genet 12: 387–389 [DOI] [PubMed] [Google Scholar]
- Kissa K, Murayama E, Zapata A, Cortés A, Perret E, Machu C, Herbomel P (2008) Live imaging of emerging hematopoietic stem cells and early thymus colonization. Blood 111: 1147–1156 [DOI] [PubMed] [Google Scholar]
- Kissa K, Herbomel P (2010) Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 464: 112–115 [DOI] [PubMed] [Google Scholar]
- Koudijs MJ, den Broeder MJ, Keijser A, Wienholds E, Houwing S, van Rooijen EM, Geisler R, van Eeden FJ (2005) The zebrafish mutants dre, uki, and lep encode negative regulators of the hedgehog signaling pathway. PLoS Genet 1: e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratz E, Eimon PM, Mukhyala K, Stern H, Zha J, Strasser A, Hart R, Ashkenazi A (2006) Functional characterization of the Bcl-2 gene family in the zebrafish. Cell Death Differ 13: 1631–1640 [DOI] [PubMed] [Google Scholar]
- Kulessa H, Frampton J, Graf T (1995) GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes Dev 9: 1250–1262 [DOI] [PubMed] [Google Scholar]
- Le Guyader D, Redd MJ, Colucci-Guyon E, Murayama E, Kissa K, Briolat V, Mordelet E, Zapata A, Shinomiya H, Herbomel P (2008) Origins and unconventional behavior of neutrophils in developing zebrafish. Blood 111: 132–141 [DOI] [PubMed] [Google Scholar]
- Lin HF, Traver D, Zhu H, Dooley K, Paw BH, Zon LI, Handin RI (2005) Analysis of thrombocyte development in CD41-GFP transgenic zebrafish. Blood 106: 3803–3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q, Meng A, Wang H, Jessen JR, Farrell MJ, Lin S (1997) GATA-1 expression pattern can be recapitulated in living transgenic zebrafish using GFP reporter gene. Development 124: 4105–4111 [DOI] [PubMed] [Google Scholar]
- McDevitt MA, Fujiwara Y, Shivdasani RA, Orkin SH (1997) An upstream, DNase I hypersensitive region of the hematopoietic-expressed transcription factor GATA-1 gene confers developmental specificity in transgenic mice. Proc Natl Acad Sci USA 94: 7976–7981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvinsky A, Dzierzak E (1996) Definitive hematopoiesis is autonomously initiated by the AGM region. Cell 86: 897–906 [DOI] [PubMed] [Google Scholar]
- Meier N, Krpic S, Rodriguez P, Strouboulis J, Monti M, Krijgsveld J, Gering M, Patient R, Hostert A, Grosveld F (2006) Novel binding partners of Ldb1 are required for haematopoietic development. Development 133: 4913–4923 [DOI] [PubMed] [Google Scholar]
- Mikkola HK, Orkin SH (2006) The journey of developing hematopoietic stem cells. Development 133: 3733–3744 [DOI] [PubMed] [Google Scholar]
- Moss LD, Monette MM, Jaso-Friedmann L, Leary JH III, Dougan ST, Krunkosky T, Evans DL (2009) Identification of phagocytic cells, NK-like cytotoxic cell activity and the production of cellular exudates in the coelomic cavity of adult zebrafish. Dev Comp Immunol 33: 1077–1087 [DOI] [PubMed] [Google Scholar]
- Murayama E, Kissa K, Zapata A, Mordelet E, Briolat V, Lin HF, Handin RI, Herbomel P (2006) Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity 25: 963–975 [DOI] [PubMed] [Google Scholar]
- Nerlov C, Graf T (1998) PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev 12: 2403–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerlov C, Querfurth E, Kulessa H, Graf T (2000) GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1-dependent transcription. Blood 95: 2543–2551 [PubMed] [Google Scholar]
- Peng H, Feldman I, Rauscher FJ III (2002) Hetero-oligomerization among the TIF family of RBCC/TRIM domain-containing nuclear cofactors: a potential mechanism for regulating the switch between coactivation and corepression. J Mol Biol 320: 629–644 [DOI] [PubMed] [Google Scholar]
- Ransom DG, Haffter P, Odenthal J, Brownlie A, Vogelsang E, Kelsh RN, Brand M, van Eeden FJ, Furutani-Seiki M, Granato M, Hammerschmidt M, Heisenberg CP, Jiang YJ, Kane DA, Mullins MC, Nüsslein-Volhard C (1996) Characterization of zebrafish mutants with defects in embryonic hematopoiesis. Development 123: 311–319 [DOI] [PubMed] [Google Scholar]
- Ransom DG, Bahary N, Niss K, Traver D, Burns C, Trede NS, Paffett-Lugassy N, Saganic WJ, Lim CA, Hersey C, Zhou Y, Barut BA, Lin S, Kingsley PD, Palis J, Orkin SH, Zon LI (2004) The zebrafish moonshine gene encodes transcriptional intermediary factor 1gamma, an essential regulator of hematopoiesis. PLoS Biol 2: E237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekhtman N, Radparvar F, Evans T, Skoultchi AI (1999) Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev 13: 1398–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes J, Hagen A, Hsu K, Deng M, Liu TX, Look AT, Kanki JP (2005) Interplay of pu.1 and gata1 determines myelo-erythroid progenitor cell fate in zebrafish. Dev Cell 8: 97–108 [DOI] [PubMed] [Google Scholar]
- Takemoto CM, Brandal S, Jegga AG, Lee YN, Shahlaee A, Ying Y, Dekoter R, McDevitt MA (2010) PU.1 positively regulates GATA-1 expression in mast cells. J Immunol 184: 4349–4361 [DOI] [PubMed] [Google Scholar]
- Trainor CD, Omichinski JG, Vandergon TL, Gronenborn AM, Clore GM, Felsenfeld G (1996) A palindromic regulatory site within vertebrate GATA-1 promoters requires both zinc fingers of the GATA-1 DNA-binding domain for high-affinity interaction. Mol Cell Biol 16: 2238–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturini L, You J, Stadler M, Galien R, Lallemand V, Koken MH, Mattei MG, Ganser A, Chambon P, Losson R, de Thé H (1999) TIF1gamma, a novel member of the transcriptional intermediary factor 1 family. Oncogene 18: 1209–1217 [DOI] [PubMed] [Google Scholar]
- Warga RM, Kane DA, Ho RK (2009) Fate mapping embryonic blood in zebrafish: multi- and unipotential lineages are segregated at gastrulation. Dev Cell 16: 744–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M (2007) THE Zebrafish Book, 5th edn. A Guide for the Laboratory Use of Zebrafish (Danio rerio). Eugene: University of Oregon Press [Google Scholar]
- Yamaguchi Y, Zon LI, Ackerman SJ, Yamamoto M, Suda T (1998) Forced GATA-1 expression in the murine myeloid cell line M1: induction of c-Mpl expression and megakaryocytic/erythroid differentiation. Blood 91: 450–457 [PubMed] [Google Scholar]
- Yu C, Cantor AB, Yang H, Browne C, Wells RA, Fujiwara Y, Orkin SH (2002) Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo. J Exp Med 195: 1387–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Behre G, Pan J, Iwama A, Wara-Aswapati N, Radomska HS, Auron PE, Tenen DG, Sun Z (1999) Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc Natl Acad Sci USA 96: 8705–8710 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.