Abstract
Brassinosteroids (BRs) are steroid hormones that are essential for the development of plants. A tight control of BR homeostasis is vital for modulating their impact on growth responses. Although it is recognized that the rapid adaptation of de novo synthesis has a key role in adjusting required BR levels, our knowledge of the mechanisms governing feedback control is limited. In this study, we identify the transcription factor CESTA as a regulator of BR biosynthesis. ces-D was isolated in a screen of Arabidopsis mutants by BR over-accumulation phenotypes. Loss-of-function analysis and the use of a dominant repressor version revealed functional overlap among CESTA and its homologues and confirmed the role of CESTA in the positive control of BR-biosynthetic gene expression. We provide evidence that CESTA interacts with its homologue BEE1 and can directly bind to a G-box motif in the promoter of the BR biosynthesis gene CPD. Moreover, we show that CESTA subnuclear localization is BR regulated and discuss a model, in which CESTA interplays with BEE1 to control BR biosynthesis and other BR responses.
Keywords: brassinosteroids, homeostasis, signalling, steroid, transcription factor
Introduction
Plant hormones are organic substances that function as signalling molecules and act at low concentrations to regulate the growth and development of plants. One group of plant hormones is the brassinosteroids (BRs), which are sterol derivatives, structurally similar to mammalian sex steroid hormones and ecdysteroids of insects (Grove et al, 1979). BRs act as essential regulators of cell elongation, cell division and differentiation and participate in many aspects of development (Clouse, 2001; Bishop and Koncz, 2002; Haubrick and Assmann, 2006). Mutants impaired in synthesizing, perceiving or signalling of BRs consequently display dramatic growth defects, such as decreased cell elongation resulting in pleiotropic dwarf phenotypes (Clouse, 2001). Conversely, BR over-accumulation or hyper-responses enhance cell elongation (Wang et al, 2001; Mora-Garcia et al, 2004).
Biosynthesis of brassinolide (BL), the biologically most active BR, is well characterized. In a complex set of pathways, the product of general sterol synthesis campesterol is converted by several cytochrome P450 monooxygenases to other BRs and ultimately to BL (Fujioka and Yokota, 2003; Bishop, 2007). Research on BR signal transduction events, linking the hormones to their numerous biological effects, has made significant progress in the last few years. The current model of BR signalling suggests that BL is perceived by a cell surface receptor complex containing the receptor kinases BRI1 and BAK1, which initiates BR signalling by promoting an interaction of BSKI with BSU1, a serine/threonine phosphatase (Kim et al, 2009). BSU1 mediates dephosphorylation and thereby inactivation of the shaggy-like kinase BIN2, that acts to suppress transcription factors of the BES1/BZR1 family, which in turn regulate the expression of BR target genes (Kim et al, 2009). To date, BES1, BZR1 and their homologues are the only known substrates of BIN2 in planta (He et al, 2005; Yin et al, 2005; Rozhon et al, 2010).
BR signalling controls a wide range of target genes, with the magnitude of variation in gene expression being small, on average only two- to three-fold (Goda et al, 2002; Müssig et al, 2002). Among the factors most responsive to BRs is a set of basic helix-loop-helix (bHLH) transcription factors termed brassinosteroid enhanced expression (BEE), which were identified in Arabidopsis due to their rapid upregulation by BL. The BEEs are thought to act downstream of BRI1 in BR signalling; their function however has remained elusive (Friedrichsen et al, 2002). Another class of BR-regulated genes encodes BR-biosynthetic enzymes such as the cytochrome P450s DWF4/CYP90B1, CPD/CYP90A1 and ROT3/CYP90C1. The expression of these genes is tightly feedback controlled from the signalling pathway, with their transcription strongly repressed following BR application and significantly upregulated in response to the inhibition of BR biosynthesis (Mathur et al, 1998; Bancos et al, 2002; Shimada et al, 2003; Lisso et al, 2005; Tanaka et al, 2005). Thus, feedback regulation of BR biosynthesis is a BR response and seemingly essential for the control of BR action.
So far, three genes that, in addition to other roles, also participate in the control of BR biosynthesis have been characterized: BZR1 (Wang et al, 2002), which directly represses DWF4 and CPD transcription (He et al, 2005), BRX (BREVIS RADIX) that stimulates CPD expression in Arabidopsis roots (Mouchel et al, 2006) and Pra2, a Rab GTPase of pea, that regulates BR C2 hydroxylation by enhancing DDWF1 activity (Kang et al, 2001). Here, we present evidence for a function of the bHLH transcription factor CESTA (CES) in regulating BR-biosynthetic gene expression. CES is a nuclear protein that is preferentially expressed in vascular tissues. Analyses of CES gain and loss-of-function mutants, as well as the use of a dominant repressor version, revealed that CES acts as an activator of BR-biosynthetic gene expression and controls cell elongation. We show that CES can bind to G-box motifs present in the promoters of the BR biosynthesis gene CPD and another cytochrome P450, the CYP718, and that CES interacts with its close homologue BEE1 in vivo. Moreover, we present evidence indicating that CES nuclear localization is affected by BR signalling and that CES is a substrate of the BR-regulated GSK3 shaggy-like kinase BIN2. We discuss a model in which CES is regulated by BIN2 action, to allow for a control of BR biosynthesis and also of other BR responses.
Results
cesta-D, an activation-tagged mutant with phenotypes reminiscent of plants with increased BR accumulation or BR responses
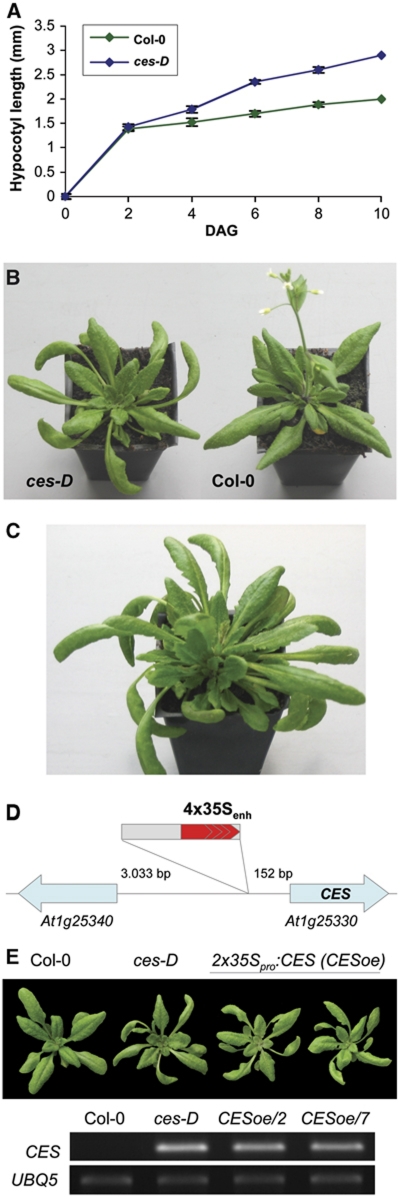
We isolated cesta-D (ces-D), a dominant mutant, in a collection of activation-tagged Arabidopsis thaliana T-DNA insertional mutants. The ces-D mutant's developmental phenotypes were already visible in light-grown seedlings, since hypocotyl growth was enhanced (Figure 1A), and became most pronounced after plants had formed first true leaves. The name for the mutant was chosen due to the adult morphology of its rosette leaves, which had elongated petioles, displayed a proximodistal lengthening, were serrated as well as outwardly curving and epinastic (Figure 1B), giving them a cesta-like appearance (the cesta, Spanish for basket, is used in the Basque ball game Pelota as a throwing and catching tool). Adult ces-D plants were furthermore characterized by prolonged vegetative development of axillary shoot meristems. Secondary rosettes were formed in the axils of rosette leaves in a basal–apical direction, which resulted in a markedly increased number of rosette leaves (Figure 1C) and inflorescences, upon conversion from the vegetative to the reproductive phase. In contrast to wild type, ces-D mutant plants continued to grow beyond 35 days after germination (DAG), with their flowering and senescence being delayed (Figure 1B and C).
Figure 1.
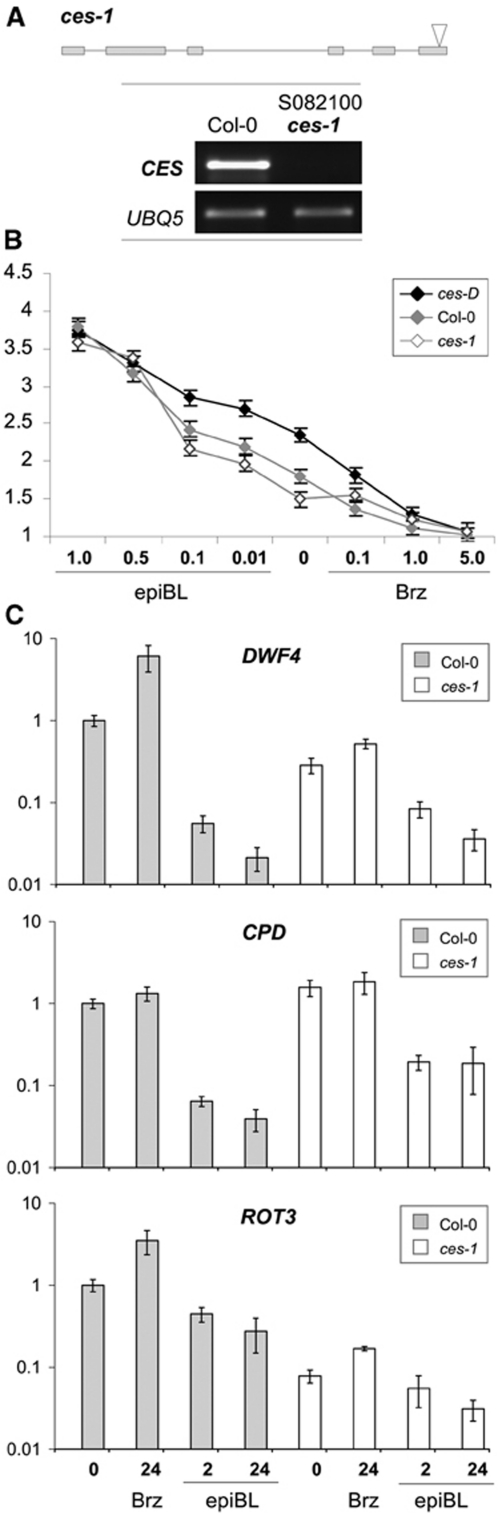
Phenotypic and molecular characterization of the ces-D mutant. (A) Hypocotyl length of light-grown wild-type Col-0 (green) and ces-D (blue) seedlings at different time points after germination. Data points are the average of three independent experiments. The standard error is shown. (B) Representative ces-D (left) and wild-type Col-0 (right) plants 30 DAG grown in long-day conditions. (C) Representative adult ces-D plant, grown in the same conditions as in (B), at 42 DAG. (D) Schematic representation of the ces-D mutation. (E) Recapitulation of the ces-D phenotypes. (Top) 4-week-old plants grown in the same conditions as in (B). (From left) Wild type, ces-D and two independent homozygous lines transformed with a 2x35Sp:CES construct. (Bottom) Semi-quantitative RT–PCR analysis of CES expression in 10-day-old seedlings of the plant lines shown. UBQ5 served as an internal control.
Many of the phenotypic features of light-grown ces-D plants, such as increased hypocotyl elongation, long petioles, outwardly curving leaf growth and increased leaf axillary meristem activity have previously been described to be characteristic for Arabidopsis plants that either over-accumulate BRs (Choe et al, 2001) or exhibit constitutive BR signalling responses (Wang et al, 2001; Yin et al, 2002; Mora-Garcia et al, 2004). Therefore, the set of phenotypes displayed by ces-D at different developmental stages indicates that BR homeostasis and/or BR responses are altered in the mutant.
CES is a homologue of BR responsive genes encoding bHLH proteins
The ces-D phenotypes were genetically linked to the BASTA resistance locus of a single T-DNA insertion and were dominant to wild type. To define the molecular nature of the mutant phenotypes, genomic DNA flanking both the right and the left border of the T-DNA was cloned by plasmid rescue and the border regions were sequenced. This revealed that the T-DNA was inserted on chromosome I in the 5′ UTR of a putative bHLH transcription factor (locus At1g25330), 152 bp upstream of the ATG, with the 35S enhancer element facing the start codon (Figure 1D) and that six basepairs at the insertion site (−159 CTTAAC −152) were deleted. Semi-quantitative RT–PCR analysis showed that the expression of At1g25330 was significantly increased in the ces-D mutant as compared with wild type (Figure 1E), whereas the expression of two neighbouring genes (At1g25320 and At1g25340) located on either side of the T-DNA was not altered (data not shown).
To determine if overexpression of At1g25330 caused the ces-D phenotypes, the cDNA of the gene was cloned into a binary vector under control of the constitutive CaMV 35S promoter and transformed into Arabidopsis wild-type Columbia-0 (Col-0) plants. Transgenic lines overexpressing At1g25330 to high levels recapitulated the characteristic ces-D phenotypes, elongated petioles and outwardly curving leaf growth in adult plants (Figure 1E), confirming that overexpression of CES resulted in the ces-D mutant phenotypes.
The CES gene consists of six exons, coding for a protein of 223 aa, that contains the bHLH signature domain (Toledo-Ortiz et al, 2003). bHLH transcription factors are represented by >160 members in the Arabidopsis genome (Bailey et al, 2003) and in a phylogenetic study CES, was named bHLH075 and assigned to bHLH subfamily 18 (Toledo-Ortiz et al, 2003). Its closest homologues are BEE1 and BEE3 (34 and 36% amino-acid identity, respectively), which belong to a group of BR early response genes and were previously identified as redundantly acting positive regulators of BR responses (Friedrichsen et al, 2002). Thus, CES encodes a close relative of bHLH proteins implicated in BR signalling.
CES is expressed in all organs and enriched in vascular tissues
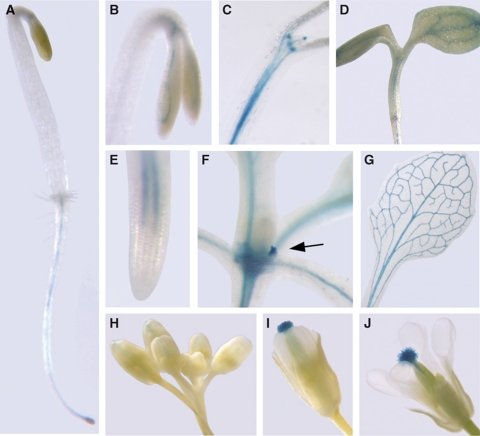
Expression patterns of CES were investigated using a transcriptional reporter in which 1.5 kb of the CES-promoter region was fused to the β-glucuronidase gene (CESpro:GUS). CES-promoter activity was detected histochemically in plants homozygous for CESpro:GUS at different developmental stages. Reporter expression was found to be present in all organs, especially in young tissues and vascular bundles, and was developmentally regulated (Figure 2). In young seedlings, GUS staining was detected in the vascular cylinder of roots, hypocotyls and cotyledons (Figure 2A–D). In dark-grown seedlings, CES expression increased later in development, becoming especially pronounced in the hook region (Figure 2C). In adult plants, the CESpro:GUS reporter was active in roots and hypocotyls and staining was also found in the vasculature of petioles and leaves as well as in leaf axillary meristems (Figure 2E–G). Floral organs showed strong CESpro:GUS expression specifically in the stigma (Figure 2H–J). Analysis of accessible transcriptome data (Zimmermann et al, 2004) confirmed our GUS reporter data.
Figure 2.
CES-GUS expression is present in all organs and is developmentally regulated. A homozygous line expressing a CES-promoter GUS fusion that showed a characteristic staining pattern was chosen for histochemical analysis of CESpro:GUS expression in different organs and developmental stages. (A, B) Dark-grown seedlings 2 DAG and (C) 10 DAG. (D) Light-grown seedling 3 DAG. (E) Root of a light-grown seedling 3 DAG. (F) Shoot of a 14-day-old plant; the arrow indicates a leaf axillary meristem. (G) Leaf of an adult plant. (H–J) Buds and flowers at stages 9–12 (as defined by Smyth et al (1990)).
In summary, CES expression was predominant in vascular tissues especially during early developmental stages. This expression pattern largely overlaps with those of key BR biosynthesis genes including CPD and ROT3 (Mathur et al, 1998; Kim et al, 2005).
CES is required for regulating BR-biosynthetic gene expression
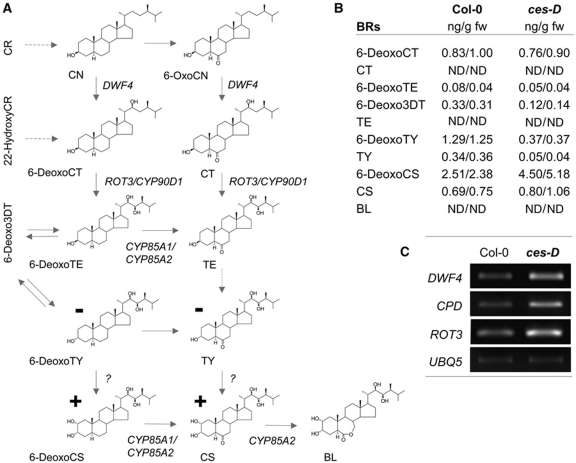
To test whether the ces-D-specific phenotypes correlate with altered BR levels in the mutant, we determined BR amounts by GC/MS. Light-grown ces-D plants contained decreased amounts of 3-dehydro-6-deoxoteasterone (6-Deoxo3DT), 6-deoxotyphasterol (6-DeoxoTY) and typhasterol (TY) (Figure 3A and B). In contrast, the BRs 6-deoxocastasterone (6-DeoxoCS) and castasterone (CS), which are formed late in biosynthesis, were significantly increased in ces-D. BL was below the detection limit in both ces-D and wild-type plants (Figure 3A and B).
Figure 3.
ces-D acts on BR biosynthesis. (A) Illustration of the BR-biosynthetic pathway (according to Bishop (2007)), indicating changes in the ces-D mutant as compared with wild-type plants (for values see (B)). − denotes decreased; + denotes increased. (B) Endogenous BR levels of adult ces-D plants as compared with those of wild type. Aerial parts of 4-week-old plants were analysed for free BR levels (ng/g fresh weight). For each line, two independent sets of samples were measured and are shown. n.d., not detected (below the detection limit). (C) Semi-quantitative RT–PCR analysis of the expression of DWF4, CPD and ROT3 in 10-day-old ces-D and wild-type seedlings. UBQ5 was used as an internal control.
Changes in BR concentrations in ces-D suggested that the expression of genes involved in BR biosynthesis might be altered. Therefore, we analysed DWF4, CPD and ROT3 transcript levels using semi-quantitative RT–PCR and quantitative real-time PCR analysis and found that the transcript levels of all three genes were elevated in ces-D seedlings as compared with those of wild type (Figures 3C and 5C).
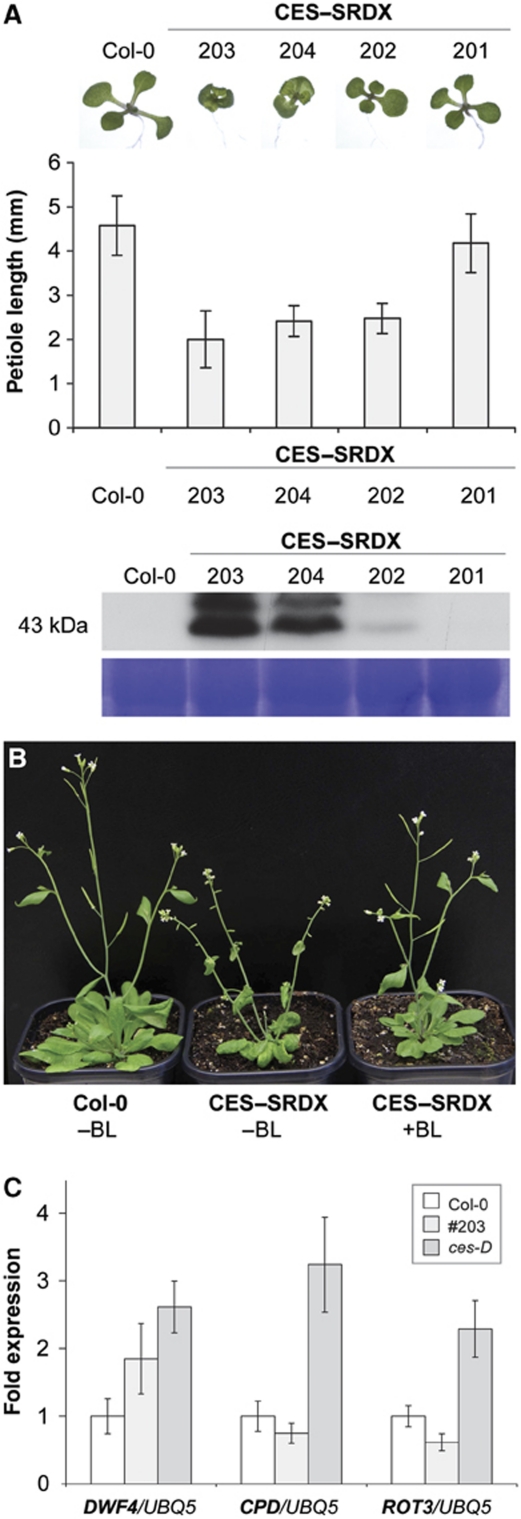
Figure 5.
Generation and characterization of 35Sp:c-Myc-CES-SRDX plants. (A) Phenotype of plants constitutively expressing a c-Myc–CES–SRDX fusion protein. (Top) Seedlings grown in long-day conditions 5 DAG. (From left to right) Wild type and four independent homozygous lines transformed with a 35Sp:c-Myc-CES-SRDX construct. (Middle) Petiole length of 12-day-old seedlings in mm, measured in three replicates. (Bottom) Western blot analysis of the plants shown using an anti-c-Myc antibody. (B) Adult phenotypes of 35Sp:c-Myc-CES-SRDX/203 plants. Four-week-old plants of wild type, of an untreated 35Sp:c-Myc-CES-SRDX/203 plant, and of a 35Sp:c-Myc-CES-SRDX/203 plant treated with 24-epiBL are shown. (C) Quantitative real-time PCR analysis of the expression of DWF4, CPD and ROT3 in 2-week-old 35Sp:c-Myc-CES-SRDX ces-D and wild-type seedlings. UBQ5 was used as an internal control.
To further determine if CES function is required for the regulation of BR biosynthesis, we identified and analysed a CES loss-of-function allele. Three T-DNA insertion lines were initially attained from the SALK database (Alonso et al, 2003). However, semi-quantitative RT–PCRs demonstrated that only the T-DNA insertion in line S082100 interfered completely with the formation of a CES full-length transcript, indicating that this mutant is a likely null allele (Figure 4A). The identified ces-1 allele was subjected to an analysis of its effects on hypocotyl elongation and the expression of DWF4, CPD and ROT3, particularly also in response to external application of the BR 24-epiBL or the BR biosynthesis inhibitor Brz2001 (Sekimata et al, 2001). Homozygous ces-1 seedlings showed significantly reduced hypocotyl length in the light, which could be rescued by exogenous application of 24-epiBL (Figure 4B). Adult ces-1 plants did not show any obvious BR-deficient phenotypes and measurements of BR levels in these plants did not reveal any statistically significant alterations (data not shown). However, the reduced hypocotyl elongation in ces-1 seedlings correlated with decreased transcript levels of DWF4 and ROT3 (Figure 4C), indicating that CES is required for maintaining BR levels balanced at an early stage of development. Moreover, and consistent with a requirement of CES as an activator of BR-biosynthetic gene expression, Brz2001-mediated induction of DWF4, CPD and ROT3 expression in ces-1 seedlings was reduced, when compared with wild type (Figure 4C).
Figure 4.
Identification and characterization of a ces-ko line. (A) (Top) Schematic illustration of the ces-1 mutant. Coding regions are indicated as boxes. The arrow shows the predicted location of the T-DNA insertion. (Bottom) Semi-quantitative RT–PCR analysis of CES expression in 10-day-old seedlings of the ces-1 and those of wild-type Col-0. UBQ5 served as an internal control. (B) Response of ces-1 and ces-D seedlings to externally applied 24-epiBL and Brz2001. Seeds of ces-D, ces-1 and wild-type plants were germinated on medium supplemented with different concentrations of 24-epiBL or Brz2001 and incubated in 50 μmol/m2/s of continuous white light at 21±1°C for 7 days. Data points represent the average of 20 measured hypocotyls. Error bars show the s.e. (C) Response of DWF4, CPD and ROT3 expression in ces-1 and wild-type seedlings to external application of 24-epiBL or Brz2001 (performed as in (B)), analysed by quantitative real-time PCR. CDKA1 was used as an internal control.
To summarize, phenotypes characteristic for BR over-accumulation correlated with altered BR levels and an enhanced expression of DWF4, CPD and ROT3 in ces-D plants suggestive of a role of CES as a positive regulator of BR biosynthesis. Consistently, a loss-of-function mutant of CES showed phenotypes indicative of BR deficiency as well as reduced expression of specific BR biosynthesis genes.
Expression of a dominant negative CES–SRDX fusion protein
The subtle developmental phenotypes observed in ces-1 plants suggested that CES loss-of-function might be complemented by functional homologues. Therefore, we chose a dominant repression approach, which has previously been used successfully to facilitate the analysis of functionally redundant transcription factors (Hiratsu et al, 2003; Mitsuda et al, 2007; Guo et al, 2009), to further analyse the effects of altered CES activity on BR responses. To this end, the EAR repression domain (Hiratsu et al, 2003) and a c-Myc epitope tag were fused to CES and expressed under the control of the CaMV 35S promoter in transgenic Arabidopsis plants. Several independent transgenic lines expressing the c-Myc–CES–SRDX fusion protein to high levels were selected. Interestingly, all of these plants showed characteristic BR-deficient phenotypes, which were already present in the seedling stage and were characterized by dwarf growth and reduced petiole elongation (Figure 5A); these phenotypes also correlated in severity with the amount of recombinant protein detected (Figure 5A). In adult plants, the phenotypes of 35Sp:c-Myc-CES-SRDX plants became even more pronounced (Figure 5B). To test if the phenotypes observed could be rescued by external application of BR, adult 35Sp:c-Myc-CES-SRDX/203 plants were sprayed twice a week with a 1 μM 24-epiBL solution. As shown in Figure 5B, this treatment reverted the phenotypes of the transgenic plants to wild type like growth morphologies, indicating that the phenotypes in 35Sp:c-Myc-CES-SRDX plants were caused by reduced levels of BRs.
To investigate if the morphological evidence for BR deficiency could be verified at the molecular level, the expression of DWF4, CPD and ROT3 was analysed using quantitative real-time PCRs in a line with high recombinant protein expression, namely 35Sp:c-Myc-CES-SRDX/203, and in ces-D plants as a control. As shown in Figure 5C, the expression of CPD and ROT3 was slightly reduced in 35Sp:c-Myc-CES-SRDX/203 plants, whereas DWF4 expression did not appear to be significantly altered.
In summary, dominant transcriptional repression of CES-dependent targets resulted in phenotypes opposing those of CES overexpression lines.
Transcriptome analysis of ces-D and 35Sp:c-Myc-CES-SRDX plants
To further reveal the molecular mechanisms underlying the ces-D and 35Sp:c-Myc-CES-SRDX/203 constitutive phenotypes, we performed expression-profiling experiments using the commercially available whole-genome Arabidopsis Affymetrix Gene Chip. Seedlings of wild type, ces-D or 35Sp:c-Myc-CES-SRDX/203 plants were grown for 10 days and were analysed in three independent biological experiments. The data obtained were then screened for the presence of genes with significantly changed expression (FDR Q-value of <0.10) in ces-D and 35Sp:c-Myc-CES-SRDX/203 as compared with wild-type plants.
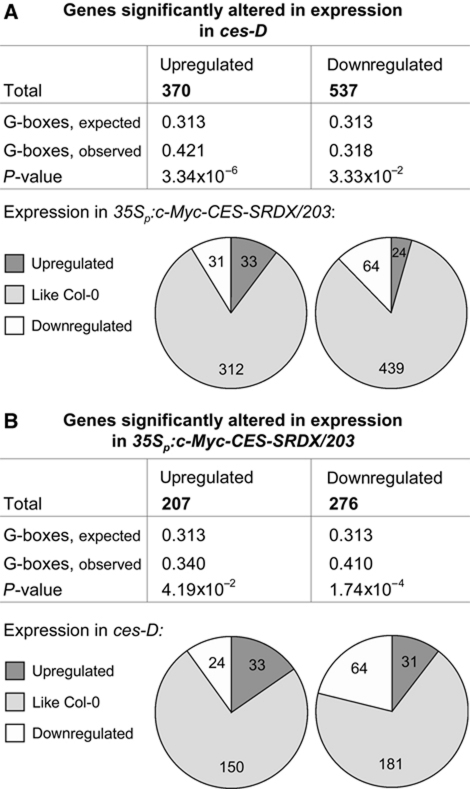
The results are presented in Supplementary Table S1 (raw data in Supplementary Table S2) and Figure 6 and show that in ces-D 370, genes were at least two-fold upregulated and 527 genes were at least two-fold downregulated in their expression. Very interestingly, when the presence of G-box motifs (5′-CACGTG-3′), known binding sites of bHLH proteins (Toledo-Ortiz et al, 2003), was determined in the 5′ UTRs of genes induced in ces-D, it was found that G-boxes were significantly enriched (expected 0.313, observed 0.421, P-value 3.34 × 10−6). On the contrary, G-boxes were hardly enriched in the 5′ UTRs of ces-D repressed genes (Figure 6A). Next, we determined if the expression of genes previously published to be BR responsive was altered in ces-D seedlings. A statistical analysis based on the data set of He et al (2005) revealed that of 370 BR-induced genes, 52 were also upregulated in ces-D. This overlap is statistically highly significant (P-value 1.74 × 10−61). Using additional data sets (Goda et al, 2002, 2004; Müssig et al, 2002), we could identify in total 57 of the 370 genes upregulated in ces-D as BR-induced genes (Supplementary Table S3). Moreover, a significant share (8 of 23 genes; P-value 5.3 × 10−4) of genes highly induced in ces-D (⩾5-fold) encodes proteins with known or predicted transcriptional activity (Supplementary Tables S1 and S4). When ces-D-induced genes were analysed for their expression levels in 35Sp:c-Myc-CES-SRDX/203 seedlings, it was revealed that ∼8.4% were repressed in transcription by CES–SRDX. Of 527 ces-D repressed genes, ∼4.5%, were increased in expression in 35Sp:c-Myc-CES-SRDX/203 seedlings (Figure 6A).
Figure 6.
Evaluation of ces-D and 35Sp:c-Myc-CES-SRDX/203 transcriptome analysis. The upstream sequences (3000 bp) were analysed for an enrichment of G-boxes. The default settings of the program motiffinder were used. The data to compile the pie charts were taken from Supplementary Table S1. (A) Illustration of the evaluation of ces-D transcriptome changes. (B) Illustration of the evaluation of 35Sp:c-Myc-CES-SRDX/203 transcriptome changes.
In 35Sp:c-Myc-CES-SRDX/203 seedlings, the transcript abundance of 207 genes was significantly increased by at least two-fold, while 276 genes showed a more than two-fold reduction in mRNA levels. Interestingly, G-box motifs were highly significantly enriched in 35Sp:c-Myc-CES-SRDX/203 repressed genes (expected 0.313, observed 0.410, P-value 1.74 × 10−6), whereas they were only slightly over-represented in genes induced in 35Sp:c-Myc-CES-SRDX expressing plants. Of 207 35Sp:c-Myc-CES-SRDX/203 induced genes, 11.6% were decreased in ces-D, and of 276 35Sp:c-Myc-CES-SRDX/203 repressed genes, 11.2% were increased in ces-D (Figure 6B).
Thus, in summary, constitutive induction of CES expression in ces-D plants results in a missexpression of ∼4.9% of the transcriptome, whereby ces-D acts to both activate and repress gene transcription. Consistent with CES being an activator of BR responses, a highly significant number of genes upregulated in ces-D are also BR-induced genes. Moreover, ces-D-induced genes are characterized by an enrichment of G-box motifs in their promoters. 35Sp:c-Myc-CES-SRDX expressing plants also show complex changes in whole-genome gene expression. The promoters of 35Sp:c-Myc-CES-SRDX/203 repressed genes are characterized by a highly significant enrichment of G-box motifs, suggesting that CES–SRDX acts to directly suppress transcription.
CES binds to G-box motifs in the promoters of CPD and CYP718 in planta
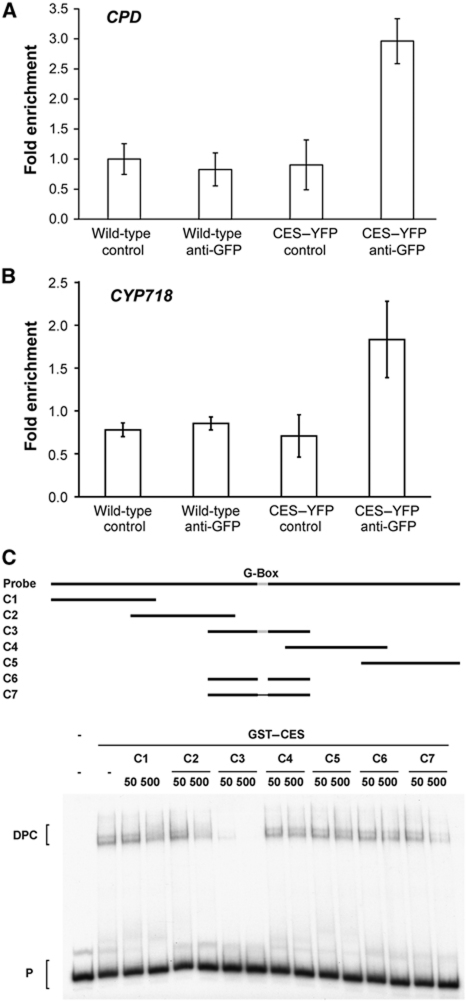
Since CES encodes a bHLH transcription factor and impacts on the regulation of gene expression, we were interested in analysing a promoter to which CES could bind. CPD was chosen as a putative target for this analysis, since it had proven to be significantly upregulated in ces-D (and downregulated in 35Sp:c-Myc-CES-SRDX/203) both by qPCRs and in microarray analyses, and contains G-box motifs, suspected CES-binding sites, in its promoter. Moreover, nine additional genes, that were significantly upregulated in their expression in ces-D and contain G-box motifs in their promoters, were selected: COR15a, COR15b, CYP718, CYP724A1, DIN11, DWF4, JR2/CORI3, KIN1 and PHE2. Arabidopsis plants stably expressing a 35Sp:CES-YFP construct were generated and chromatin immunoprecipitation (ChIP) experiments were carried out using an anti-GFP antibody to investigate CES binding to the G-box motifs in the sequences 5′ of the named genes. Very interestingly, of 10 genes analysed, CES bound specifically to fragments containing G-box motifs in the promoters of CPD and CYP718, a cytochrome P450 with a currently unknown function (Figure 7A and B). Of five G-box motifs present upstream of the CPD-coding sequence, CES bound specifically to one (data not shown).
Figure 7.
CES binds to G-box motifs. (A, B) ChIP experiments with wild-type and 35Sp:CES-YFP plants using an anti-GFP antibody. G-box containing fragments of the promoters of CPD and CYP718 were quantified by real-time PCR amplification from immunoprecipitated samples, with the primer pairs listed in Supplementary Table S4. The primer pair 5S-F/5S-R (Li et al, 2010) was used for standardization. The standard deviation of at least three measurements is shown. (C) EMSAs analysing CES binding to a fragment of the CPD promoter. A radioactively labelled probe representing the same part of the CPD promoter as in (A) was incubated with GST–CES in the absence or presence of cold competitor oligonucleotides. The competitors C1–C5 contain different regions (indicated by a solid line; upper panel) while the G-box (CACGTG; light grey) was deleted in C6 or mutated to AAAAAA in C7. The competitors were used in 50 and 500-fold molar excess to the probe. P, probe; DPC, DNA–protein complexes.
To unequivocally determine if CES can bind to G-box motifs, electrophoretic mobility shift assays (EMSAs) using the CPD-promoter fragment to which CES had bound in ChIPs as a probe (radioactively labelled), recombinant glutathione S-transferase (GST)–CES and cold competitor oligonucleotides were performed. The results confirmed that only the competitor containing the functional G-box (C3) could compete with the probe for CES binding. Other oligonucleotides, which did either not contain the G-box (C1, C2, C4 and C5) or harboured mutations in it (C6 and C7) could not out-compete the radioactively labelled fragment, showing specific interaction of CES with this motif (Figure 7C).
Thus, CES acts as a transcription factor that can bind to G-box motifs in the promoters of the cytochrome P450s CPD and CYP718 in planta.
CES localizes to the nucleus
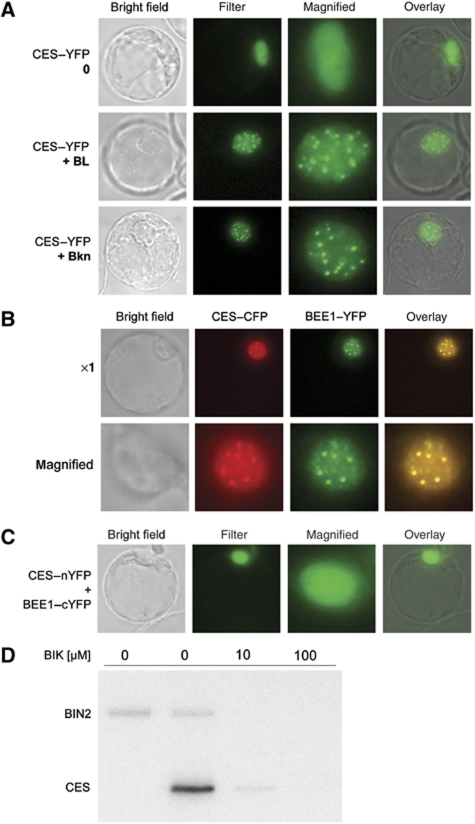
In an attempt to gain further insight into the role of CES, its subcellular localization was investigated. A yellow fluorescent protein (YFP) fusion to the C-terminus of full-length CES was expressed under the control of the CaMV 35S promoter and was analysed in Arabidopsis protoplasts. As shown in Figure 8A, CES–YFP expression was present diffusely in the nucleus. Interestingly, upon BR treatment, CES–YFP localization in protoplasts reorganized to display a speckled nuclear expression pattern (Figure 8A). To analyse if the BL-induced CES–YFP localization to subnuclear foci was dependant on BR signalling, protoplasts expressing the fusion protein were treated with Bikinin (Bkn), which constitutively activates BR signalling by inhibiting GSK3s that negatively regulate BR signal transduction (De Rybel et al, 2009). Similar to BR treatment, Bkn promoted a speckled CES–YFP localization pattern (Figure 8A). To verify the ability of BR treatment to alter CES–YFP subnuclear localization also in planta we analysed 35Sp:CES-YFP plants for BR-induced nuclear compartmentalization. Untreated 35Sp:CES-YFP plants showed a diffuse nuclear YFP localization (Supplementary Figure S1). As opposed to protoplasts, a 2 h BL treatment was not sufficient to induce CES–YFP nuclear compartmentalization in Arabidopsis. However, when plants were pretreated with a BR biosynthesis inhibitor for 24 h, a 2 h BL treatment induced relocalization of CES–YFP to subnuclear compartments, suggesting that CES nuclear localization is altered specifically in response to a rapid induction of BR biosynthesis/signalling (Supplementary Figure S1).
Figure 8.
CES is a nuclear protein that interacts with BEE1 and is phosphorylated by BIN2 in vitro. (A) CES–YFP reporter expression in Arabidopsis protoplasts treated with 24-epiBL (1 μM) or Bkn (30 μM) for 2 h as compared with an untreated control. (B) Colocalization of CES–CFP and BEE1–YFP. Images of a representative protoplast coexpressing 35Sp:CES-CFP and 35Sp:BEE1-YFP, treated for 2 h with 1 μM of 24-epiBL. (C) Bimolecular fluorescence complementation assay showing a representative protoplast cotransformed with CES–nYFP and BEE1–cYFP constructs. (D) In vitro kinase assays using 0.1 μg of GST–BIN2 and 1.0 μg of GST–CES. The reactions were treated for 2 h with increasing concentrations of Bkn. A reaction to which only BIN2 was added served as a negative control.
Previously, it was shown that the expression of BEE1 and BEE3, close homologues of CES, is BR inducible (Friedrichsen et al, 2002). To obtain information on the cellular localization of BEE1 and BEE3, which had not been investigated before, BEE1–YFP and BEE3–YFP fusion constructs under the control of the CaMV 35S promoter were generated. Analysis of 35Sp:BEE1–YFP and 35Sp:BEE3–YFP subcellular distribution in Arabidopsis protoplasts revealed that both fusion proteins showed diffuse nuclear localization in the absence of externally applied BR. However, in response to both BL and Bkn treatment, 35Sp:BEE1–YFP and 35Sp:BEE3–YFP also relocalized to distinct nuclear compartments (Supplementary Figure S2A and B). Moreover, coexpression of CES–CFP and BEE1–YFP fusions demonstrated that upon BL and Bkn treatment both reporters relocalized to the same nuclear compartments (Figure 8B). CES–CFP and BEE3–YFP similarly relocalized to the same subnuclear foci upon BL and Bkn treatment (Supplementary Figure S2C).
CES interacts with BEE1 in vivo
bHLH proteins typically act either as homodimers or as heterodimers in interaction with close homologues to regulate expression of their targets (Toledo-Ortiz et al, 2003). As the closest homologues of CES, BEE1 and BEE3 had previously been characterized as BL inducible, positive regulators of BR signalling (Friedrichsen et al, 2002), we tested for interactions between CES and the BEEs. Yeast two-hybrid assays showed that CES can homodimerize as well as interact with both BEE1 and BEE3 to induce β-gal activity in yeast (Supplementary Figure S2D). To test for the specificity of these interactions, we also analysed the ability of CES to interact with PIF3, a bHLH transcription factor involved in light signalling (Ni et al, 1998). In contrast to BEE1 and BEE3, PIF3 did not interact with CES in yeast.
To analyse if an interaction of CES with BEE1 or BEE3 may also occur in planta bimolecular fluorescence complementation assays (Walter et al, 2004) were carried out. For this purpose, CES fused to the N-terminal portion of YFP and both BEE1 and BEE3 fused to the C-terminal portion of YFP were coexpressed in protoplasts. As shown in Figure 8C, yellow fluorescence was seen diffusely in the nucleus when protoplasts were cotransformed with CES and BEE1. In contrast, protoplasts cotransformed with CES and BEE3 split-YFP constructs as well as in the controls did not exhibit detectable fluorescence under our experimental conditions (data not shown).
CES is phosphorylated by BIN2
Bkn-induced nuclear redistribution of CES–YFP in protoplasts provided evidence for an immanent role of CES in BR signalling, and that these potential regulatory events take place either at the level of BIN2 or downstream of it. To investigate if such potential predicted effects of BIN2 on CES may be direct, in vitro kinase assays were performed. Both BIN2 and CES were purified as GST fusion proteins from Escherichia coli and used in phosphorylation reactions in the presence of radioactively labelled ATP. As a specific inhibitor, Bkn was added in increasing concentrations to block BIN2 activity. These assays demonstrated that BIN2 was able to utilize CES as a substrate in vitro (Figure 8D).
Discussion
In plants, steroid signalling relies on a phosphorylation-dependent signal transduction cascade that is initiated upon hormone binding to a plasma membrane-localized receptor complex and results in the nuclear acquisition of transcription factors to regulate genomic responses (Nemhauser and Chory, 2004; Wang and He, 2004; Belkhadir and Chory, 2006). Genomic BR effects not only mediate BR-controlled growth and development, but are also essential for the adjustment of BR biosynthesis. To attain BR cellular homeostasis, BR signalling is utilized to create a feedback regulatory loop that allows adjusting the expression of genes that participate in BR biosynthesis (Bancos et al, 2002; Shimada et al, 2003; Lisso et al, 2005; Tanaka et al, 2005).
Here, we identify and characterize the bHLH transcription factor CES as a novel positive regulator of BR-specific growth responses and BR-biosynthetic gene expression. CES was identified by gain-of-function phenotypes of the ces-D mutant: drastic morphological changes indicative of BR over-accumulation/BR hyper-responses that correlated with elevated levels of DWF4, CPD and ROT3 mRNAs and increased amounts of late BR biosynthesis intermediates 6-DeoxoCS and CS. Interestingly, earlier intermediates, namely 6-DeoxoTY and TY levels, were decreased in ces-D, suggesting that CES regulates a gene(s) essential for BR C2 hydroxylation. Whereas it is known that in pea C2 hydroxylation of BRs is mediated by the cytochrome P450 DDWF1 (Kang et al, 2001), the enzyme catalysing this reaction in Arabidopsis is as yet unidentified. Notably, in this context, CYP718, a predicted cytochrome P450 strongly upregulated in ces-D, was here identified as a direct CES target. CYP718 is a close homologue of the cytochrome P450s CYP85A1 and CYP85A2, enzymes that catalyse C6 oxydation of BRs (Bishop, 2007). Thus, it is conceivable that CYP718 may be involved in BR biosynthesis and we are currently investigating this possibility.
The fact that in ces-D, in spite of increased levels of late pathway BRs, feedback control did not set in, but on the contrary DWF4, CPD and ROT3 were induced, suggested that CES also impacts on the regulation of these genes. An analysis of CES loss-of-function plants in which DWF4 and ROT3 mRNA levels were reduced provided further evidence that CES positively regulates BR-biosynthetic gene expression. Decreased DWF4 and ROT3 transcript levels and an attenuated reduction of DWF4, CPD and ROT3 mRNA levels in response to BR treatment also correlated with impaired hypocotyl elongation, supporting the idea that CES function is necessary for maintaining BR homeostasis.
The subtle constitutive phenotypes observed in ces-1 argue for the activity of CES homologues that can bypass a loss of CES in certain tissues, developmental stages and/or physiological processes. Indeed, functional redundancy is a hallmark of BR signalling (Thummel and Chory, 2001) and has also hindered the characterization of the BEE genes. Only when a bee1bee2bee3 triple knockout was generated, subtle phenotypes indicating impaired BR responses could be revealed, whereas single and double knockouts had no obvious morphological phenotypes (Friedrichsen et al, 2002). To circumvent the problem of refining CES function in the context of redundancy, we chose to use a chimeric CES–SRDX repressor version. EAR repression motif fusions have previously been used successfully to convert transcriptional activators into repressors, leading to a dominant downregulation of not only specific target genes, but also of targets of functional homologues (Hiratsu et al, 2003; Mitsuda et al, 2007; Guo et al, 2009). Consistent with ces-D and ces-1 phenotypes, when overexpressed in plants, CES–SRDX induced BR-deficient phenotypes, which correlated with the transcriptional downregulation of CPD and ROT3, providing further support to the notion that CES and its redundant factors affect BR biosynthesis.
In addition to its role in positively regulating BR-biosynthetic gene expression, there is evidence that CES also impacts on other BR responses. The results of a whole-genome expression analysis revealed, that a large number of BR-induced genes are constitutively upregulated in ces-D plants. There are different, not mutually exclusive explanations, how ces-D could alter BR responses. First, it is conceivable that increased BR levels in ces-D may induce BR signalling and thereby BR responses may be altered. Second, ces-D may stimulate indirect effects that alter BR responses and third, in addition to BR-biosynthetic genes, CES may also directly regulate other BR-responsive genes in ces-D. Different pieces of evidence suggest that secondary effects may account for the complex transcriptome changes observed in both ces-D and CES–SRDX expressing plants. On the one hand, although ces-D also has repressive effects on gene transcription, G-box motifs, which are CES-binding sites, are hardly significantly enriched in ces-D repressed genes. It seems thus likely that ces-D indirectly suppresses gene expression by activating transcriptional repressors. In agreement, transcription factors are significantly enriched among genes highly upregulated in ces-D and may account for the complex whole-genome expression changes observed in ces-D plants.
Altered transcriptional networks may also hold responsible for the fact that only a relatively small anti-overlap between genes significantly upregulated in ces-D and significantly downregulated in 35Sp:c-Myc-CES-SRDX/203 plants was observed. Secondary changes in gene expression in the constitutively expressing lines may mask CES and CES–SRDX primary effects. Moreover, it seems likely that CES and CES–SRDX do not exhibit comparable transcriptional activities in ces-D and 35Sp:c-Myc-CES-SRDX/203 plants. For example, CES expression in ces-D is approximately five times higher, than CES–SRDX expression in 35Sp:c-Myc-CES-SRDX/203 seedlings (Supplementary Table S1). These differences in expression are likely to impact on the extend, by which target genes are regulated and may also explain the fact that genes significantly upregulated by at least two-fold in ces-D regularly do not show a correspondingly strong downregulation in 35Sp:c-Myc-CES-SRDX/203 plants.
To add complexity, conditional interactions of CES with additional proteins could be decisive for the regulatory output mediated by this transcriptional regulator. bHLH transcription factors such as CES accomplish considerable diversity in recognition and regulation of target gene expression through dimerization, resulting in either homodimeric or heterodimeric regulatory complexes (Toledo-Ortiz et al, 2003). Consistently, CES could be demonstrated to interact with BEE1 in vivo. Heterodimerization has already been shown to be of significance in the control of BR-regulated genes. BES1 interacts with the bHLH transcription factor BIM1 to bind to the SAUR-AC promoter in vivo (Yin et al, 2005). Moreover, very recently, physical interactions of bHLH transcription factors and atypical, non-DNA binding bHLH proteins, have been found to mediate BR signalling (Wang et al, 2009; Zhang et al, 2009). CES and BEE1 are further examples of bHLH proteins, which may act as heterodimers to regulate BR responsive gene expression.
BEE1 has previously been shown to act as a positive regulator in BR signalling (Friedrichsen et al, 2002), having been identified as a factor that is strongly regulated by BL. In contrast, CES transcript abundance is not BL regulated. However, results from protoplast cultures and from stably transformed plants suggested that CES nuclear localization is altered by BRs, and more specifically also by application of Bkn, an inhibitor of GSK3 shaggy-like kinase function that explicitly activates BR signalling (De Rybel et al, 2009). Moreover, our finding that CES is a substrate of BIN2 in vitro supports the idea that CES action may be controlled by BIN2. Interestingly, CES does not contain a classical GSK3 consensus motif, tandemly repeated S/T/xxxS/T sequences (Cohen and Frame, 2001), which is present in the BES1/BZR1 family of transcription factors, the only in planta substrates of GSK3 shaggy-like kinases known to date (He et al, 2005; Yin et al, 2005; Rozhon et al, 2010).
At present, the functional significance of BIN2-mediated CES phosphorylation is not known. If CES acts in a feedback-regulated manner one could assume that in a BR-depleted state BIN2 would enhance CES activity to upregulate BR production and alter other BR responses. This would be in opposition to the roles of BIN2 in regulating BES1/BZR1 activity, which is a negative regulatory process. How phosphorylation inhibits BES1/BZR1 action has been a matter of debate. On the one hand, it has been suggested that BIN2-mediated phosphorylation leads to a decrease in BES1 and BZR1 protein stabilities and to altered subcellular expression patterns (Gampala et al, 2007; Ryu et al, 2007). However, other work provided evidence that, rather than regulating the nuclear translocation and accumulation of BES1, phosphorylation abolishes BES1 DNA-binding capacity and interferes with multimerization (Vert and Chory, 2006). Interestingly, BIN2 catalysed phosphorylation does not inhibit the DNA-binding abilities of CES in vitro (data not shown), supporting the idea that BIN2 may have distinct roles in the regulation of BES1/BZR1 versus CES activities. In mammals, it is already known that GSK3 shaggy-like kinases can act to suppress or enhance the activity of transcription factor targets (Cohen and Frame, 2001).
Collectively, our findings indicate that heterodimerization of CES with BEE1 might constitute a regulatory module essential for the positive regulation of BR-biosynthetic genes and other BR responses. The activity of this complex might be affected further by BR-dependent transcriptional control of BEE1 expression as well as by post-translational regulation via BIN2-mediated phosphorylation of CES and possibly BEE1. These regulatory switches could be essential for a fine-tuning of CES–BEE1 complex activity.
Materials and methods
Mutant screen and cloning of CES
ces-D was originally isolated in the eir1-1 mutant background (Luschnig et al, 1998) when screening a collection of T-DNA insertional mutants, generated with the T-DNA construct pSK115 (Weigel et al, 2000) corresponding to ∼14 000 independent transformants (Sieberer et al, 2003). When backcrossed into Col-0, ces-D was found to segregate in a 3:1 ratio. All further analyses were performed in the Col-0 background.
Southern blot and segregation analysis indicated that the ces-D phenotype was genetically linked to the BASTA resistance locus of a single T-DNA insertion. Genomic DNA flanking both the right and the left border of the T-DNA was cloned by plasmid rescue (Weigel et al, 2000) and the exact position of the T-DNA was determined by sequencing using the SOER2 and SOEL2 primers (all primers used are listed in the Supplementary Table S5).
Recapitulation of the ces-D phenotypes and generation of CES–SRDX expressing lines
For recapitulation of the ces-D mutant phenotypes, a vector was constructed that allowed constitutive overexpression of CES under control of the 35S promoter in plants and conferred resistance to the antibiotic gentamycin. The ORF of At1g25330 was PCR amplified from Col-0 cDNA (synthesized from flowers, developmental stages 10–12; as defined by Smyth et al, 1990) using gene-specific primers with integrated XhoI and BamHI restriction sites (CESp2RT-fw and CESp2RT-rv) and subcloned into the pGEM-T easy vector (Promega). After sequencing, the CES fragment was cloned into the plant expression vector p235a (Poppenberger et al, 2003) downstream of the 2x35S promoter. Twenty independent lines homozygous for 2x35Sp:CES were generated and analysed for CES expression using semi-quantitative RT–PCR.
The CES dominant repression construct (35Sp:c-Myc-CES-SRDX) was created by fusing the full-length CES cDNA in frame with the dominant EAR repression sequence (Hiratsu et al, 2003), which was ligated downstream of the CaMV 35S promoter into the binary plant expression vector pGWR8 (Rozhon et al, 2010).
Reporter construct generation and analysis
For the construction of a transcriptional CESpro:GUS reporter line, 1.5 kb of genomic sequence upstream of the CES start codon was amplified from genomic DNA using specific primers (CESfusions-fw-c and CEStranscGUS-rv-a) and cloned into pPZP-GUS.1 (Diener et al, 2000). The construct was introduced into a wild-type Col-0 background and 30 independent homozygous lines were analysed for their GUS activities to identify a line with representative staining patterns. The selected line was then analysed for its GUS activity.
For the generation of YFP and CFP reporter constructs, cDNAs of CES, BEE1 and BEE3 were amplified by PCR with the primers indicated in Supplementary Table S4 and cloned into pGWR8. cDNAs were subsequently tagged with YFP or CFP or with the N-terminal or C-terminal part of YFP for investigation of protein–protein interactions by bimolecular fluorescence (Walter et al, 2004). The sequenced constructs were used for transient transfection of A. thaliana protoplasts as described previously (Cardinale et al, 2002).
Stably transformed A. thaliana seedlings expressing 35Sp:CES-YFP treated with 1 μM 24-epiBL for 2 h were investigated with an Axioplan II fluorescence microscope (Zeiss, Oberkochen, Germany). If necessary, BL was depleted by application of 2.5 μM Brz for 24 h.
Transcript and transcriptome analysis
For semi-quantitative RT–PCR, DNaseI-treated total RNA isolated from plant tissue was used to synthesize cDNA using the RevertAid H minus first-strand cDNA synthesis kit (Fermentas, St Leon-Rot, Germany). PCR reactions were performed using gene-specific primers that amplified 250–400 bp large fragments located in the C-terminal parts of the genes investigated. UBQ5 was used as an internal template control (Poppenberger et al, 2005).
qPCR was performed with a StepONE Plus Real-Time PCR System (Applied Biosystems, Carlsbad, CA). Each reaction contained 10 μl 2 × Power PCR Master Mix (Applied Biosystems), 4 pmol of each primer and 5 μl cDNA (prepared as described and diluted 1:10) in a total volume of 20 μl. Cycling was performed as recommended by the manufacturer (initial denaturation: 94°C for 10 min; 40 cycles at 94°C for 15 s and 60°C for 1 min) and finally a melting curve was recorded. A dilution series of cloned cDNA was run under the same conditions and the results were used to plot a calibration curve, which served to calculate the relative transcript abundance in the samples. The relative expression levels were calculated from four replicates after normalization to UBQ5 (At3g62250) or CDKA1 (At3g48750).
For transcriptome analysis, 10-day-old seedling of wild type, ces-D and 35Sp:CES-SRDX-c-Myc, grown on ATS media in long-day conditions, were analysed in three independent biological replicates using the commercially available whole-genome Arabidopsis Affymetrix Gene Chip of NASC (Nottinhgam, UK). Genes with a very low-signal intensity were excluded from further analysis. A signal intensity of at least 5.0 on average in at least one set was set as a trash-hold. Genes were considered as upregulated if (i) the corresponding signal intensity was at least two-fold increased as compared with wild type and if (ii) the FDR Q-value was below 0.10. Similarly, genes were considered as downregulated in ces-D or 35Sp:CES-SRDX-c-Myc plants if (i) their signal intensity was at least half than that in wild type and (ii) the Q-value below 0.10. Enrichment of hexamer nucleotide motifs was analysed using the program motiffinder available from the TAIR homepage (http://www.arabidopsis.org/tools/bulk/motiffinder/index.jsp). Enrichment of gene ontology and overlaps of expression data sets were calculated with Excel and Gorilla (http://cbl-gorilla.cs.technion.ac.il/). The P-values of a two-tailed t-test were calculated with Excel and converted to the FDR Q-values using the qval spreadsheet (http://www.rowett.ac.uk/~gwh/qval.xls).
BR measurements
Plants were grown under long-day conditions (16 h of 120 μmol/(m2.s) white light/8 h dark; 21±1°C/17±1°C) for 30 days, before tissue of aerial plant parts was harvested. Quantification of BRs was performed as described previously (Noguchi et al, 1999; Fujioka et al, 2002).
ChIP and EMSAs
For ChIP, 10-day-old plants were treated with ice-cold 1% formaldehyde solution in PBS for 30 min. After rinsing three times with cold PBS, the plant material was ground to a fine powder in liquid nitrogen and nuclei isolated as described previously (Aufsatz, 2005). The nuclei were lysed and the ChIP was performed with an anti-GFP antibody (Roche Diagnostics, Indianapolis, IN) and a ChIP Assay Kit (Millipore Cooperation, Bedford, MA) as recommended by the manufacturer. Enrichment of specific fragments was investigated by semi-quantitative PCR and qPCR. The same conditions as for transcript analysis were used. A dilution series of genomic A. thaliana DNA was used to plot a calibration curve, which served to calculate the relative abundance of the fragment in the samples. According to Li et al (2010), the primer pair 5S-F/5S-R was used as internal control for qPCR to calculate the enrichment of the gene-specific fragment.
The 196-bp CPD-ChIP-9/CPD-ChIP-10 amplicon of the CPD promoter that had shown a clear enrichment in the ChIP assay was used for EMSA probe preparation: 1 ng of this amplicon was mixed with 5 μl 5 × PCR buffer, 0.2 μl 25 mM dNTPs, 1.5 μl 5 μM CPD-ChIP-9, 1.5 μl 5 μM CPD-ChIP-10, 80 μCi [α-32P]-dCTP, 2 U GoTaq DNA polymerase (Promega, Madison, WI) and water added to 25 μl. After an initial denaturization step at 94°C for 5 min, the reaction was cycled 35 times (94°C for 30 s, 45°C for 1 min and 72°C for 2 min) prior a final extension step at 72°C for 10 min. Binding of CES to DNA was tested in 10 μl reactions containing 0.5 μg GST–CES, 1 μl 10 × binding buffer (250 mM HEPES/KOH pH 8.0, 500 mM KCl, 20 mM MgSO4, and 1 mM DTT), 2 μl 50% glycerol, 0.2 fmol probe and, if desired, 10 or 100 fmol competitor DNA. After incubation at 0°C for 30 min, 3 μl loading buffer was added (50% glycerol, 0.05% bromophenol blue) and the samples loaded onto a 6% PAGE gel that was run at 0°C in 1 × TBE buffer at 10 V/cm for 3 h. Band was detected by autoradiography.
Supplementary Material
Acknowledgments
We thank Professor Dianna Bowles for her highly valued support of this work and many interesting and fruitful discussions. Professor Pierre Broun, Professor Heribert Hirt and Dr Fabian Vaistij are acknowledged for helpful suggestions. We also thank Dr Suguru Takatsuto (Joetsu University of Education, Japan) for supplying deuterated BRs, Dr Tadao Asami and Dr Shigeo Yoshida for providing Brz2001 and Dr Ference Nagy for the PIF3 Y2H constructs. Moreover, we thank the horticultural staff of the University of York and the MFPL for excellent plant care and Renata Milčevičová, Merete Tschokert and Andrij Belokurow for technical support. The Arabidopsis Biological Resource Center (ABRC) and SIGnAL are acknowledged for the T-DNA insertion line used. This work was supported by grants from the Hochschuljubiläumsstiftung of the City of Vienna, Austria, from the Austrian Science Fund FWF and by a Grant-in-Aid for Scientific Research (B) from the Ministry of Education, Culture, Sports, Science and Technology of Japan to SF (Grant No. 19380069). BP and TS received fellowships from the Austrian Academy of Sciences and the FWF. MK received a fellowship from the Higher Education Commission of Pakistan.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alonso JM, Stephanova AN, Leiss TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Aufsatz W (2005) Chromatin immunoprecipitation protocol to analyze histone modifications in Arabidopsis thaliana (PROT12) http://www.epigenome-noe.net/WWW/researchtools/protocol.php?protid=13
- Bailey PC, Martin C, Toledo-Ortiz G, Quail PH, Huq E, Heim MA, Jakoby M, Werber M, Weisshaar B (2003) Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell 15: 2497–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancos S, Nomura T, Sato T, Molnar G, Bishop GJ, Koncz C, Yokota T, Nagy F, Szekeres M (2002) Regulation of transcript levels of the Arabidopsis cytochrome p450 genes involved in brassinosteroid biosynthesis. Plant Physiol 130: 504–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir Y, Chory J (2006) Brassinosteroid signaling: a paradigm for steroid hormone signaling from the cell surface. Science 314: 1410–1411 [DOI] [PubMed] [Google Scholar]
- Bishop GJ (2007) Refining the plant steroid hormone biosynthesis pathway. Trends Plant Sci 12: 377–380 [DOI] [PubMed] [Google Scholar]
- Bishop GJ, Koncz C (2002) Brassinosteroids and plant steroid hormone signaling. Plant Cell 14(Suppl): S97–S110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale F, Meskiene I, Ouaked F, Hirt H (2002) Convergence and divergence of stress-induced mitogen-activated protein kinase signaling pathways at the level of two distinct mitogen-activated protein kinase kinases. Plant Cell 14: 703–711 [PMC free article] [PubMed] [Google Scholar]
- Choe S, Fujioka S, Noguchi T, Takatsuto S, Yoshida S, Feldmann KA (2001) Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J 26: 573–582 [DOI] [PubMed] [Google Scholar]
- Clouse S (2001) Brassinosteroids. In The Arabidopsis Book, Somerville C, Meyerowitz EM (eds) Rockville, MD: American Society of Plant Biologists [Google Scholar]
- Cohen P, Frame S (2001) The renaissance of GSK3. Nat Rev Mol Cell Biol 2: 769–776 [DOI] [PubMed] [Google Scholar]
- De Rybel B, Audenaert D, Vert G, Rozhon W, Mayerhofer J, Peelman F, Coutuer S, Denayer T, Jansen L, Nguyen L, Vanhoutte I, Beemster GT, Vleminckx K, Jonak C, Chory J, Inzé D, Russinova E, Beeckman T (2009) Chemical inhibition of a subset of Arabidopsis thaliana GSK3-like kinases activates brassinosteroid signaling. Chem Biol 16: 594–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener AC, Li H, Zhou W, Whoriskey WJ, Nes WD, Fink GR (2000) Sterol methyltransferase 1 controls the level of cholesterol in plants. Plant Cell 12: 853–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen DM, Nemhauser J, Muramitsu T, Maloof JN, Alonso J, Ecker JR, Furuya M, Chory J (2002) Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics 162: 1445–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S, Takatsuto S, Yoshida S (2002) An early C-22 oxidation branch in the brassinosteroid biosynthetic pathway. Plant Physiol 130: 930–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S, Yokota T (2003) Biosynthesis and metabolism of brassinosteroids. Annu Rev Plant Biol 54: 137–164 [DOI] [PubMed] [Google Scholar]
- Gampala SS, Kim TW, He JX, Tang W, Deng Z, Bai MY, Guan S, Lalonde S, Sun Y, Gendron JM, Chen H, Shibagaki N, Ferl RJ, Ehrhardt D, Chong K, Burlingame AL, Wang ZY (2007) An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev Cell 13: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S (2004) Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 134: 1555–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S (2002) Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 130: 1319–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove MD, Spencer GF, Rohwedder WK, Mandava NB, Worley JF, Warthen JD, Steffens GL, Flippen-Anderson JL, Cook JC (1979) A unique growth promoting steroid from Brassica napus pollen. Nature 281: 216–217 [Google Scholar]
- Guo Y, Qin G, Gu H, Qu LJ (2009) Dof5.6/HCA2, a Dof transcription factor gene, regulates interfascicular cambium formation and vascular tissue development in Arabidopsis. Plant Cell 21: 3518–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubrick LL, Assmann SM (2006) Brassinosteroids and plant function: some clues, more puzzles. Plant Cell Environ 29: 446–457 [DOI] [PubMed] [Google Scholar]
- He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY (2005) BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307: 1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Kang JG, Yun J, Kim DH, Chung KS, Fujioka S, Kim JI, Dae HW, Yoshida S, Takatsuto S, Song PS, Park CM (2001) Light and brassinosteroid signals are integrated via a dark-induced small G protein in etiolated seedling growth. Cell 105: 625–636 [DOI] [PubMed] [Google Scholar]
- Kim GT, Fujioka S, Kozuka T, Tax FE, Takatsuto S, Yoshida S, Tsukaya H (2005) CYP90C1 and CYP90D1 are involved in different steps in the brassinosteroid biosynthesis pathway in Arabidopsis thaliana. Plant J 41: 710–721 [DOI] [PubMed] [Google Scholar]
- Kim TW, Guan S, Sun Y, Deng Z, Tang W, Shang JX, Sun Y, Burlingame AL, Wang ZY (2009) Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Biol 11: 1254–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ye H, Guo H, Yin Y (2010) Arabidopsis IWS1 interacts with transcription factor BES1 and is involved in plant steroid hormne brassinosteroid regulated gene expression. Proc Nat Acad Sci USA 107: 3918–3923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisso J, Steinhauser D, Altmann T, Kopka J, Mussig C (2005) Identification of brassinosteroid-related genes by means of transcript co-response analyses. Nucleic Acids Res 33: 2685–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig C, Gaxiola RA, Grisafi P, Fink GR (1998) EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev 12: 2175–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Molnar G, Fujioka S, Takatsuto S, Sakurai A, Yokota T, Adam G, Voigt B, Nagy F, Maas C, Schell J, Koncz C, Szekeres M (1998) Transcription of the Arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J 14: 593–602 [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme-Takagi M (2007) NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19: 270–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Garcia S, Vert G, Yin Y, Cano-Delgado A, Cheong H, Chory J (2004) Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev 18: 448–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchel CF, Osmont KS, Hardtke CS (2006) BRX mediates feedback between brassinosteroid levels and auxin signaling in root growth. Nature 443: 458–461 [DOI] [PubMed] [Google Scholar]
- Müssig C, Fischer S, Altmann T (2002) Brassinosteroid-regulated gene expression. Plant Physiol 129: 1241–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Chory J (2004) BRing it on: new insights into the mechanism of brassinosteroid action. J Exp Bot 55: 265–270 [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667 [DOI] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE (1999) Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol 121: 743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppenberger B, Berthiller F, Lucyshyn D, Sieberer T, Schuhmacher R, Krska R, Kuchler K, Glossl J, Luschnig C, Adam G (2003) Detoxification of the Fusarium mycotoxin deoxynivalenol by a UDP-glucosyltransferase from Arabidopsis thaliana. J Biol Chem 278: 47905–47914 [DOI] [PubMed] [Google Scholar]
- Poppenberger B, Fujioka S, Sueno K, George GL, Vaistij FE, Seto H, Hiranuma S, Takastuto S, Adam G, Yoshida S, Bowles D (2005) The UGT73C5 of Arabidopsis thaliana glucosylates brassinosteroids. Proc Nat Acad Sci USA 102: 15253–15258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhon W, Mayerhofer J, Petuschnig E, Fujioka S, Jonak C (2010) Arabidopsis GSK3, functions in the brassinosteroid signalling pathway. Plant J 62: 215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Kim K, Cho H, Park J, Choe S, Hwang I (2007) Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 19: 2749–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimata K, Kimura T, Kaneko I, Nakano T, Yoneyama K, Takeuchi Y, Yoshida S, Asami T (2001) A specific brassinosteroid biosynthesis inhibitor, Brz2001: evaluation of its effects on Arabidopsis, cress, tobacco, and rice. Planta 213: 716–721 [DOI] [PubMed] [Google Scholar]
- Shimada Y, Goda H, Nakamura A, Takatsuto S, Fujioka S, Yoshida S (2003) Organ-specific expression of brassinosteroid-biosynthetic genes and distribution of endogenous brassinosteroids in Arabidopsis. Plant Physiol 131: 287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieberer T, Hauser MT, Seifert GJ, Luschnig C (2003) PROPORZ1, a putative Arabidopsis transcriptional adaptor protein, mediates auxin and cytokinin signals in the control of cell proliferation. Curr Biol 13: 837–842 [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Asami T, Yoshida S, Nakamura Y, Matsuo T, Okamoto S (2005) Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Physiol 138: 1117–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel CS, Chory J (2001) Steroid signaling in plants and insects—common themes, different pathways. Genes Dev 16: 3113–3129 [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Chory J (2006) Downstream nuclear events in brassinosteroid signalling. Nature 441: 96–100 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, Harter K, Kudla J (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang ZY, He JX (2004) Brassinosteroid signal transduction—choices of signals and receptors. Trends Plant Sci 9: 91–96 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T, Chory J (2002) Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2: 505–513 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J (2001) BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410: 380–383 [DOI] [PubMed] [Google Scholar]
- Wang H, Zhu Y, Fujioka S, Asami T, Li J, Li J (2009) Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix-loop-helix proteins. Plant Cell 21: 3781–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil EJ, Neff MM, Nguyen JT, Sato S, Wang ZY, Xia Y, Dixon RA, Harrison MJ, Lamb CJ, Yanofsky MF, Chory J (2000) Activation tagging in Arabidopsis. Plant Physiol 122: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J (2005) A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120: 249–259 [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191 [DOI] [PubMed] [Google Scholar]
- Zhang LY, Bai MY, Wu J, Zhu JY, Wang H, Zhang Z, Wang W, Sun Y, Zhao J, Sun X, Yang H, Xu Y, Kim SH, Fujioka S, Lin WH, Chong K, Lu T, Wang ZY (2009) Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and arabidopsis. Plant Cell 21: 3767–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.