Abstract
RecQ helicases have attracted considerable interest in recent years due to their role in the suppression of genome instability and human diseases. These atypical helicases exert their function by resolving a number of highly specific DNA structures. The crystal structure of a truncated catalytic core of the human RECQ1 helicase (RECQ149–616) shows a prominent β-hairpin, with an aromatic residue (Y564) at the tip, located in the C-terminal winged-helix domain. Here, we show that the β-hairpin is required for the DNA unwinding and Holliday junction (HJ) resolution activity of full-length RECQ1, confirming that it represents an important determinant for the distinct substrate specificity of the five human RecQ helicases. In addition, we found that the β-hairpin is required for dimer formation in RECQ149–616 and tetramer formation in full-length RECQ1. We confirmed the presence of stable RECQ149–616 dimers in solution and demonstrated that dimer formation favours DNA unwinding; even though RECQ1 monomers are still active. Tetramers are instead necessary for more specialized activities such as HJ resolution and strand annealing. Interestingly, two independent protein–protein contacts are required for tetramer formation, one involves the β-hairpin and the other the N-terminus of RECQ1, suggesting a non-hierarchical mechanism of tetramer assembly.
INTRODUCTION
Since the discovery of the first RecQ enzyme from Escherichia coli over 20 years ago (1), numerous other RecQ family helicase genes have been found in various organisms ranging from prokaryotes to eukaryotes. Unicellular organisms, such as bacteria and yeast have only one or two RecQ helicase genes per species, while higher eukaryotes generally express multiple RecQ enzymes (2–4). Five members of the RecQ family have been found in human cells: BLM, RECQ1 (alias RECQL or RECQL1), RECQ4 (RECQL4), RECQ5 (RECQL5) and WRN. Mutations in the genes encoding BLM, WRN and RECQ4 are linked to defined genetic disorders associated with genomic instability, cancer predisposition and features of premature aging (5–9). No heritable cancer predisposition disorder has been associated with mutations in the remaining two human RecQ helicase genes, RECQ1 and RECQ5 as yet. However, recent studies have linked a single-nucleotide polymorphism present in the RECQ1 gene to a reduced survival in pancreatic cancer patients (10). Recent reports showing that RECQ1 silencing has anti-cancer effect both in cellular and mouse xenograft models suggest that RECQ1 may have a role in cancer survival, and might be considered as a new suitable target for cancer therapy (11–13).
RecQ helicases unwind DNA with a 3′–5′ polarity and are capable of unwinding a variety of DNA structures in addition to standard B-form DNA duplexes, i.e. triple helices, 3- or 4-way junctions, and G-quadruplex DNA (14–17). Consistent with an ability to unwind various DNA structures, several cellular functions have been attributed to RecQ proteins, including roles in stabilization and repair of damaged DNA replication forks, telomere maintenance, homologous recombination and DNA damage checkpoint signaling (2,3,18). RECQ1 was the first RecQ helicase to be discovered in humans on the basis of its potent ATPase activity (19), but it is one of the less characterized in terms of enzymatic activity and function. Our previous studies demonstrated that RECQ1 has a substrate specificity that is distinct from that of BLM and WRN, suggesting that these helicases play non-overlapping functions in cells and that there must be key structural features that distinguish the catalytic domains of these enzymes (20). Of note, the substrate specificity of RECQ1 is also different from that of E. coli RecQ; for example, RECQ1 cannot resolve blunt-ended DNA duplexes or G-quadruplex DNA, both of which are efficiently unwound by the bacterial helicase.
Recently, a truncated form of RECQ1 lacking the first 48 residues and the last 33 amino acids at the C-terminus (RECQ149–616) was crystallized in presence of MgCl2 and ATPγS, and its structure was determined at a resolution of 2.0 Å (21). The truncated protein RECQ149–616 has an activity very similar to the full-length RECQ1 on a forked-duplex substrate, hence it can be seen as the catalytic core [similar to the crystallized fragment of the E. coli enzyme (22)]. However, it has no detectable strand annealing activity, consistent with our previous observation that higher order oligomers are responsible for strand annealing and that the N-terminus is required for higher order oligomer formation (20,23). Moreover, the N-terminally deleted mutant lacks Holliday junction (HJ) disruption activity, suggesting that the N-terminal domain, either directly or through the formation of higher order oligomers, is essential for the ability of RECQ1 to disrupt HJs (20).
A distinguishing feature of RecQ helicases is a RecQ-specific C-terminal domain (RecQ-Ct), which consists of a zinc-binding module and a winged-helix (WH) domain (24). The WH domains of E. coli RecQ, and the human RECQ1 and WRN helicases are structurally similar; however, the structures of E. coli RecQ and human RECQ1 differ in the orientation of the WH relative to the other helicase domains. In addition, human RECQ1 shows a prominent β-hairpin, with a tyrosine residue (Y564) at the tip, located in the wing of the WH domain. The corresponding β-hairpin in WRN is similarly topped by a phenylalanine at the same position; in contrast, the β-hairpin is much shorter and lacks an aromatic residue in the structure of the winged-helix (WH) domain of E. coli RecQ. Our previous mutagenesis studies indicated that this hairpin plays a crucial role in the DNA strand separation activity of RECQ149–616, as all mutants of the β-hairpin, including the single amino acid change of the aromatic residue at the tip (Y564A), are nearly devoid of DNA unwinding activity (21). These results suggest that the Tyr residue may act as a pin that abuts the end of the DNA duplex and hence promotes strand separation. A similar role has been previously ascribed to unrelated β-hairpin structures in other helicases of both the SF-1 and SF-2 superfamilies (25).
Here, we further investigated the contribution of the β-hairpin to the enzymatic activity of RECQ1 in the context of the full-length protein. Our mutational and structural data point to an additional role of the β-hairpin as an important interface for oligomer formation, and identify the distinct roles of different RECQ1 subunit interfaces in the formation of dimers and tetramers. Collectively, these studies provide new insight into the mechanism that regulates higher assembly state formation and controls the balance between the multiple enzymatic activities of RECQ1.
MATERIALS AND METHODS
Proteins
Full-length RECQ1 (RECQ1FL) and its mutants were purified from baculovirus/Sf9 cells as described previously (23,26). The truncated form of RECQ1 lacking the first 48 residues at the N-terminus and the last 33 amino acids at the C terminus (RECQ149-616, also termed RECQ1T1) and its mutants were instead produced in E. coli following a previously described procedure (21).
DNA substrates
All oligonucleotides were chemically synthesized and purified by reverse-phase high pressure liquid chromatography (RP-HPLC) (Sigma-Aldrich, Suffolk, UK). Each nucleotide was then resuspended in Tris–EDTA (TE) buffer (10 mM Tris–HCl, pH 7.5, 1 mM EDTA, pH 8.0). Oligonucleotide sequences used in helicase and annealing assays are reported in Table 1. For each substrate, a single oligonucleotide was 5′-end-labeled with [γ-32P] ATP using T4 polynucleotide kinase. The kinase reaction was performed in PNK buffer (70 mM Tris–HCl, pH 7.6, 10 mM MgCl2, 5 mM dithiothreitol) at 37°C for 45 min. For helicase assays, the [γ-32P]ATP-labeled oligonucleotides were then annealed to a 1.6-fold excess of the unlabeled complementary strands in annealing buffer (10 mM Tris–HCl, pH 7.5, 50 mM NaCl) by heating at 95°C for 8 min and then cooling slowly to room temperature. The purification of the forked duplex substrates was performed using Micro Bio-Spin columns (Bio-Rad), while the HJ substrates were purified using Sepharose-4B columns (Amersham Biosciences).
Table 1.
Sequences of oligonucleotides used for substrate preparation
| Oligonucleotide | ||
|---|---|---|
| Number | Name | Sequence 5′–3′ |
| 1 | Fork 20 (D) | GAACGAACACATCGGGTACGTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT |
| 2 | Fork 20 (U) | TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTCGTACCCGATGTGTTCGTTC |
| 8 | X12-1 | GACGCTGCCGAATTCTGGCTTGCTAGGACATCTTTGCCCACGTTGACCCG |
| 9 | X12-2 | CGGGTCAACGTGGGCAAAGATGTCCTAGCAATGTAATCGTCTATGACGTC |
| 10 | X12-3 | GACGTCATAGACGATTACATTGCTAGGACATGCTGTCTAGAGACTATCGC |
| 11 | X12-4 | GCGATAGTCTCTAGACAGCATGTCCTAGCAAGCCAGAATTCGGCAGCGTC |
Helicase assay
Helicase assays were performed in 20 μl of a reaction mixture containing helicase buffer (20 mM Tris–HCl, pH 7.5, 8 mM dithiothreitol, 5 mM MgCl2, 10 mM KCl, 10% glycerol, 80 μg/ml bovine serum albumin), 5 mM ATP and [γ-32P] ATP-labeled fork or synthetic HJ substrate (0.5 nM). The reactions were started by the addition of recombinant RECQ1 proteins (or its mutants) to a concentration indicated in the figures, and the mixture was incubated at 37°C for 20 min. The reactions were terminated by the addition of 20 µl of quench solution (0.4 M EDTA pH 8.0, 1% SDS, 10% glycerol). Reaction products were resolved using 10% native PAGE and the extent of DNA unwinding was quantified as described previously (26).
DNA binding assay
The DNA binding assays where performed by EMSA using the fork duplex and HJ substrates (Table 1). Briefly, the experiments were performed by incubating increasing concentrations of the purified protein (0–200 nM) with [γ32-P] labeled DNA (0.5 nM) in a 20 µl reaction mixture containing 20 mM Tris–HCl pH 7.8, 2 mM MgCl2, 50 mM NaCl, 1 mM DTT and 0.1 mg/ml BSA. After incubation for 30 min at room temperature, 2.5 µl of 30% glycerol were added and the reaction products were separated on a 6% non-denaturing polyacrylamide gel run at 4°C in TBE buffer. Labeled DNA fragments were detected by autoradiography (Cyclon, GE Healthcare). The apparent equilibrium dissociation constant (Kd) was determined from the mean of at least three independent experiments.
Size exclusion chromatography experiments
Size exclusion chromatography experiments were performed on a ÄKTA FPLC system (Amersham Biosciences) using a 10/30 Superdex 200 HR gel filtration column (GE Healthcare), as described previously (23). Briefly, the column was equilibrated at a flow rate of 0.5 ml/min with 20 mM Tris (pH 7.5), 150 mM KCl and 1 mM DTT. Approximately 150–200 μg of recombinant protein was loaded on the column and detected using an UV detector at 280 nm.
Analytical ultracentrifugation
Sedimentation velocity experiments were carried out at 8°C on a Beckman XL-I analytical ultracentrifuge equipped with a Ti-50 rotor. Protein samples were studied at a concentration of 0.5 mg/ml (or a mixture of two proteins, each at 0.5 mg/ml) in 10 mM HEPES pH 7.4, 150 mM NaCl, 0.5 mM TCEP, 2% (v/v) glycerol. After equilibration at 8°C for 1 h, 3000 r.p.m., the speed was increased to 50 000 r.p.m. for RECQ1T1 or 40 000 r.p.m. for RECQ1FL, and radial absorbance scans were taken every 2 min at 280 nm. Information from scans 5–50 was used for analysis. Data were analyzed using SEDFIT (27) to calculate c(s) distributions. The software package SEDNTERP (http://www.jphilo.mailway.com) was used in order to normalize the obtained sedimentation coefficient values to the corresponding values in water at 20°C, 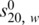 .
.
Analytical ultracentrifugation (AUC) sedimentation equilibrium experiments were performed at 8°C, with protein concentrations of 0.5, 0.3 and 0.2 mg/ml, dialysed against 10 mM HEPES pH 7.4, 150 mM NaCl, with addition of 1.0 mM TCEP. The samples were centrifuged at 7000 r.p.m. for 22 h and scanned; a further scan after 2 h was performed to confirm that equilibrium had been attained. The speed was then increased to 9000 r.p.m., and the chambers were scanned after 18 and 20 h. Finally, the speed was increased to 26 000, to achieve meniscus depletion, providing a baseline for the analysis.
Dynamic light scattering
The dynamic light scattering (DLS) measurements were carried out at 20°C at a scattering angle of 90° (Model 802 DLS, Viscotek, correlator resolution = 256 channels). A 12–μl cuvette was filled with 1 mg/ml of each sample in a buffer comprising 10 mM HEPES, pH 7.5, 150 mM NaCl, 2% (v/v) glycerol, after filtration (200 nm, NanoSep, Pall). Of each sample, at least 30 autocorrelations (duration: 2 s) were collected. A minimum of 10 similar autocorrelations representing the average were used to fit for the distribution of particles according to their hydrodynamic radius, N(Rh), from Laplace inversion (OmniSIZE, Viscotek), applying a buffer viscosity of 0.010825 poise, as calculated using SEDNTERP.
GST pull-down assay
We produced [35S]-labeled RECQ1 for in vitro binding assays by the TNT Reticulocyte Lysate System (Promega), using the corresponding pIRES FLAG-Hemagglutinin (FH)-RECQ1 vector as template. Glutathione S-transferase (GST) and GST-fused wild-type and truncated RECQ1 proteins were expressed in bacteria and purified according to a previously described procedure (28). Briefly, bacterial cultures harboring GST-fused proteins were induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) over night at room temperature. The pellets were resuspended in lysis buffer (50 mM Tris–HCl pH 8.0, 250 mM NaCl, 5% glycerol, 5 mM EDTA, 2 mM DTT and 1 mM PMSF plus protease inhibitor cocktail tablet) and sonicated. The cleared lysates were then loaded onto glutathione agarose beads (Sigma) and incubated for 1 h at 4°C. Following incubation and extensive washing with lysis buffer, proteins immobilized on beads were resuspended at 50% (v/v) in the same buffer and kept frozen until use. GST pull-down assays were performed as described (28,29). Briefly, immobilized GST-fused RECQ1 fragments were pretreated with DNase I and RNase A for 30 min at 25°C to remove bacterial nucleic acids. The beads were then washed twice with high salt solution (1 M NaCl) and equilibrated in binding buffer TNEN [20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1.0 mM EDTA pH 8.0, 0.5 % NP-40, 1 mM DTT 1, mM PMSF] supplemented with 0.1 mg/ml of ethidium bromide. The labeled in vitro translated RECQ1 protein was then added to 1–5 µg of proteins immobilized on beads in the same buffer in a final volume of 50 µl. The reaction mixture was incubated for 2 h at 4°C. The beads were subsequently washed two times in the ethidium bromide supplemented TNEN buffer, and three times with TNEN buffer. Finally, the beads were resuspended, heated in Laemmli buffer and separated by SDS–PAGE. Dried gels were visualized with a Phosphoimager. The two bands visible on the gels for the in vitro translated RECQ1 protein represent two protein products derived from two alternative translation initiation ATG codons. The first codon is located ‘upstream’ of FLAG-Hemagglutinin double tag, while the second corresponds to the ATG initiation codon of RECQ1 located ‘downstream’ of the double tag.
RESULTS
The β-hairpin of RECQ1FL is required for fork duplex unwinding and HJ branch migration
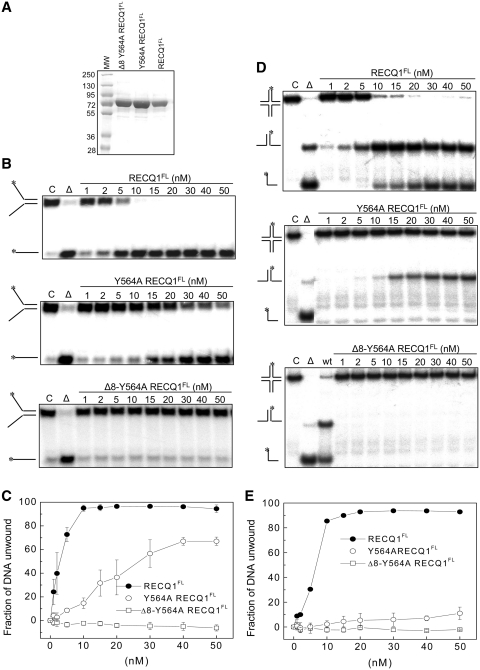
We have previously identified an essential role for the β-hairpin located in the WH domain in the forked-duplex DNA strand-separation activity of the truncated form of the RECQ1 protein (RECQ149-616). This truncated protein will be termed RECQ1T1 for simplicity (21). Here, we tested the impact of similar β-hairpin mutations on the ability of the full-length RECQ1 protein (RECQ1FL) to resolve forked and HJ substrates. In particular, we engineered a point mutant where Tyr564 was replaced with Ala (Y564A-RECQ1FL) and a deletion mutant lacking eight residues on both strands of the β-hairpin in addition to the Y564A substitution (Δ8-Y564A-RECQ1FL) (Table 2). Both proteins were expressed in SF9 cells and purified to near homogeneity using a previously established procedure (Figure 1A) (23,26). Using the mutant protein, we confirmed that the single amino acid substitution of the aromatic residue at the tip of the hairpin (Y564A) significantly affected the ability of RECQ1 to resolve a forked duplex substrate of 20 bp with ssDNA tails of 30 nt, as >95% of the substrate was unwound within 20 min using 10 nM RECQ1FL, while <40% is resolved using the same concentration of the Y564A-RECQ1FL mutant (Figure 1B and C). Interestingly, a more pronounced effect of the same mutation was observed using the HJ substrate since only a very limited amount of the splayed arm product (<15%) was observed using the Y564-RECQ1FL mutant even in the presence of 50 nM enzyme (Figure 1D and E). Deletion of the eight residues on both strands of the hairpin coupled to the replacement of Y564 with alanine had an even more dramatic effect since no unwinding of either DNA substrate was detected even at the highest protein concentrations. Collectively, these results confirm that the Tyr residue at the tip of the hairpin is a crucial structural determinant for the unwinding activity of RECQ1 also in the context of the full-length protein.
Table 2.
Hairpin mutants of full-length and truncated human RECQ1
| Mutants | Hairpin sequence |
|---|---|
| Full-length RECQ1 (RECQ1FL) | |
| RECQ1FL | 554YLKEDYSFTAYATISYLKIG573 |
| Y564A-RECQ1FL | 554YLKEDYSFTAAATISYLKIG573 |
| Δ8-Y564A-RECQ1FL | 554Y AA G573 |
| Truncated RECQ149–616 (RECQ1T1) | |
| RECQ1T1 | 554YLKEDYSFTAYATISYLKIG573 |
| Y564A-RECQ1T1 | 554YLKEDYSFTAAATISYLKIG573 |
| Δ2-Y564A-RECQ1T1 | 554YLKEDYS AA ISYLKIG573 |
| Δ3-Y564A-RECQ1T1 | 554YLKEDY AA SYLKIG573 |
| Δ8-Y564A-RECQ1T1 | 554Y AA G573 |
| Δ(F561-T562-T566)-RECQ1T1 | 554YLKEDYS AYA ISYLKIG573 |
| Y559A-S560A-F561A-RECQ1T1 | 554YLKEDAAATAYATISYLKIG573 |
| W227A-F231A-RECQ1T1 | (mutations not in hairpin) |
Figure 1.
Analysis of the unwinding activity of hairpin loop mutants of full-length RECQ1. (A) SDS–PAGE analysis of the purified RECQFL, Y564A-RECQ1FL and Δ8-Y564A-RECQ1FL proteins. (B) Unwinding assays using various concentrations of RECQ1FL, Y564A-RECQ1FL and Δ8-Y564A-RECQ1FL (0–50 nM) and a forked-duplex substrate of 20 bp with ssDNA tails of 30 nt (0.5 nM). All of the reactions were stopped after 20 min. (C) Plot of the unwinding activity as a function of protein concentration using the forked-duplex substrate. Concentration dependence experiments were performed using the mutants indicated in the figure (0–50 nM). The data points represent the mean of three independent experiments with the standard deviation indicated by error bars. (D) Unwinding assays using various concentrations of RECQ1FL, Y564A-RECQ1FL and Δ8-Y564A-RECQ1FL (0–50 nM) and a HJ substrate with a 12-bp homologous core (0.5 nM). All of the reactions were stopped after 20 min. (E) Plot of the unwinding activity as a function of protein concentration using the HJ substrate. Concentration dependence experiments were performed by using the mutants indicated in the figure (0–50 nM). The data points represent the mean of three independent experiments with the standard deviation indicated by error bars.
The β-hairpin is involved in RECQ1T1 dimer formation
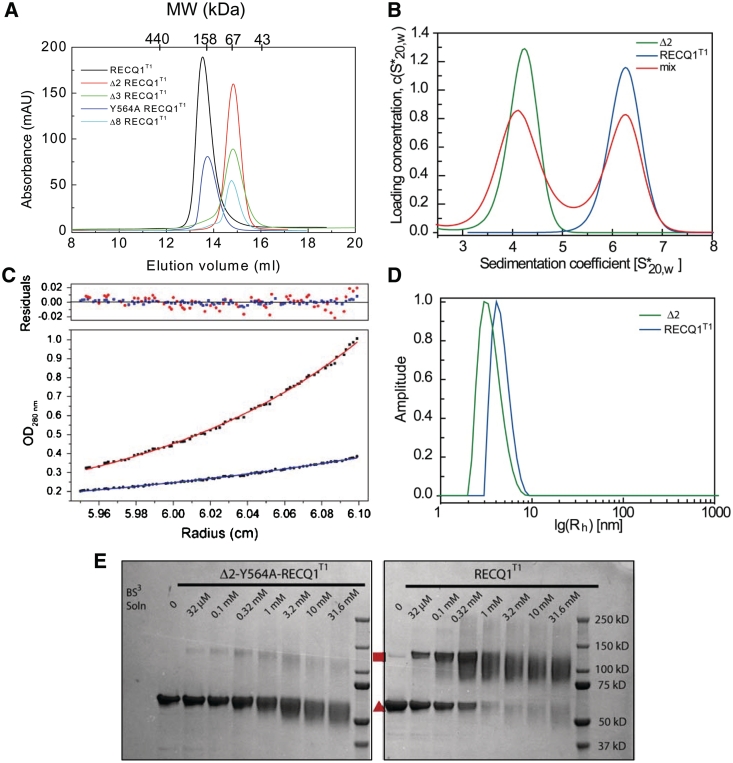
When purifying the β-hairpin mutants of RECQ1T1 we noticed that all mutants in which the hairpin was shortened by two or more amino acids on each strand eluted on gel filtration columns later than RECQ1T1 (Figure 2A). RECQT1 eluted from the gel filtration column with an apparent molecular mass of ∼150 kDa, while the mutants carrying deletions in the β-hairpin eluted with an apparent molecular mass of ∼67 kDa. Importantly, the Y564A mutation alone did not cause the same shift. This shift in chromatographic behavior could be due to changes in the conformation or in the oligomeric state of RECQ1T1. To resolve this, both the RECQ1T1 protein and the mutant denoted Δ2 (Δ2-Y564A-RECQ1T1), were analyzed by AUC, DLS and chemical cross-linking (Figure 2). The values of the sedimentation coefficients measured by analytical ultracentrifugation for Δ2-Y564A-RECQ1T1 and RECQ1T1 were 4.0 S and 6.0 S, respectively (Figure 2B). In agreement with the gel filtration profiles, each protein sedimented predominantly as a single peak. These sedimentation coefficients and the estimated diffusion coefficients correspond to molecular masses of 63 kDa and 117 kDa for Δ2-Y564A-RECQ1T1 and RECQ1T1, respectively (the molecular mass of a single chain or RECQ1T1 is 67.2 kDa). Equilibrium ultracentrifugation was used to calculate masses of 72 kDa and 132 kDa for Δ2-Y564A-RECQ1T1 and RECQ1T1, confirming that the two proteins behave as a monomer and a dimer, respectively (Figure 2C). DLS provided separate confirmation that both Δ2-Y564A-RECQ1T1 and RECQ1T1 proteins were monodisperse, with calculated masses of 67 kDa and 133 kDa, respectively, corresponding to a monomer and dimer (Figure 2D). This conclusion was further supported by chemical cross-linking, that showed a cross-linked species of double molecular weight for RECQ1T1, but not for Δ2-Y564A-RECQ1T1 (Figure 2E). Collectively, these experiments indicate that the RECQ1T1 protein is present predominantly as a dimer in solution, whereas the mutants deleted in the β-hairpin are mainly monomers. Furthermore, the similarity of the gel filtration profiles of RECQ1T1 obtained in the presence of ATPγS, and/or ssDNA indicated that the oligomeric state of RECQ1T1 was not affected by the interaction with a slowly hydrolyzable analog of ATP and/or ssDNA (21).
Figure 2.
RECQ1T1 is a dimer in solution. (A) Size exclusion chromatography profiles of RECQ1T1 (black), Y564A-RECQ1T1 (blue), Δ2-Y564A-RECQ1T1 (red), Δ3-Y564A-RECQ1T1 (green), Δ8-Y564A-RECQ1T1 (cyan). (B) Sedimentation velocity in AUC. Blue: RECQ1T1, green: Δ2-Y564A-RECQ1T1 mutant, red: 1:1 mixture of both proteins. (C) Absorbance distribution for RECQ1T1 in sedimentation equilibrium experiments shows dimerization. The ideal fit curves of both proteins at 9000 r.p.m. The red curve is an ideal fit of the data for RECQT1, giving a molecular weight of 132 kDa; the blue curve is an ideal fit for the Δ2-Y564A- RECQ1T1 mutant, giving a MW of 72 kDa. The residuals of both datasets are shown in the upper panel, showing absence of either aggregation or non-ideality. (D) Size distributions investigated with dynamic light scattering. The monodisperse distribution of RECQ1T1 (blue trace) shows a hydrodynamic radius equivalent to a mass of 133 kDa (calculated monomolecular weight: 67.3 kDa). The hydrodynamic radius observed for the Δ2-Y564A-RECQ1T1 mutant (green trace) was equivalent to a mass of 67 kDa (calculated monomolecular weight: 66.8 kDa). (E) Protein cross-linking. RECQ1T1 (right) and the Δ2-Y564A-RECQ1T1 mutant (left) were incubated with the crosslinker BS3 at the indicated concentrations for 30 min. The reactions were quenched and the proteins separated by denaturing SDS–PAGE. Monomers (red triangle) and cross-linked dimers (red square) are indicated.
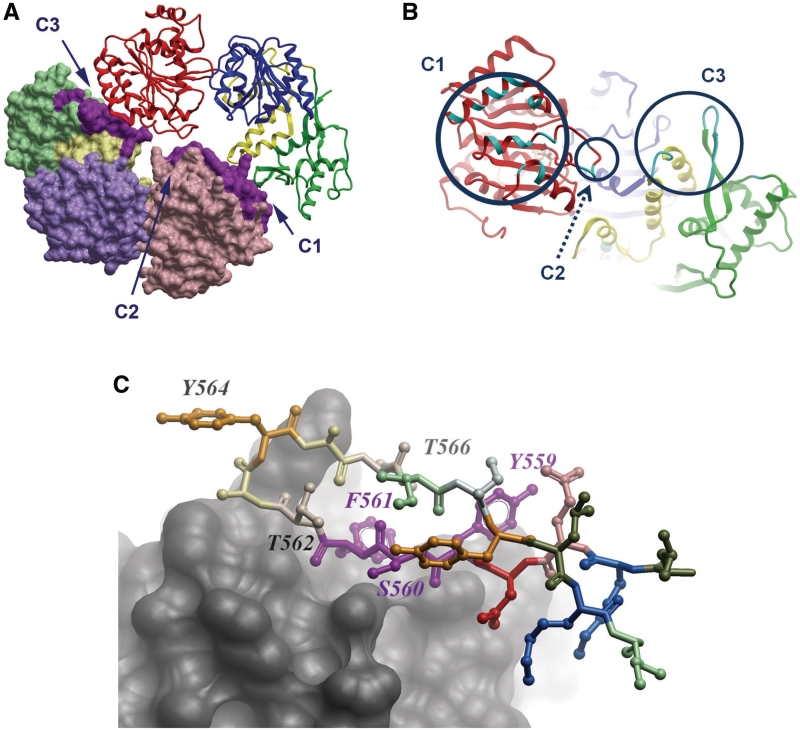
Our analysis of the crystal structure of RECQ1T1 (21) for potential oligomeric assemblies using the PISA server (30) revealed a significant interface with a neighboring molecule in the crystal lattice (Figure 3A). This dimeric interaction results in the burial of 1100 Å2 of accessible surface area from each monomer, which represents 4.7% of the available surface area. In contrast, all other crystal contacts typically involve <400 Å2 of buried surface. The same dimeric interactions occur in four different crystal forms of RECQ1T1 (PDB: 2V1X, 2WWY, and data not shown), in which other crystal contacts are not conserved. The dimer in the crystal structure is arranged in a head to tail organization, in which the main protein contacts are distributed in three patches, which we term C1, C2 and C3 (a list of interacting residues listed in Table 3; Figure 3A and B). C1 includes a patch of residues that are clustered on one face of the RecA-like domain D1; these interact with C3 region of the dimeric partner, which include residues in the C-terminal Zn and WH domains. The C2 region of each subunit, located at the tip of an aromatic-rich loop in domain D1 (31), lies at the centre of the interface and interacts with the same region in the other monomer.
Figure 3.
Organization of a RECQ1T1 dimer in the crystal structure. (A) Overall view, monomer A shown as backbone, monomer B shown in surface representation. The contact areas are indicated in purple. (B) The putative dimer interface, on monomer A, from the direction of monomer B. The circles indicate the contact areas C1, C2 and C3; the locations of amino acids involved in the interface are shown in cyan. (C) Detailed view of the β-hairpin (ball and stick representation) interacting with the opposing subunit (shown in skin representation). Some of the residues mutated in this study are marked. Specifically, Y559, S560 and F561 are involved in subunit interaction, whereas Y564 and subsequent residues are not directly involved, but are available for potential interaction with DNA.
Table 3.
Residues involved in dimer interactions in the crystal structure
| Mol1 | Mol2 |
|---|---|
| C1 | |
| I194 | I194 |
| A195 | A195 |
| K198 | K198 |
| M199 | M199 |
| M201 | M201 |
| S202 | S202 |
| E205 H | E205 H |
| – | E209 HS |
| C2 | |
| Q226 | Q226 |
| W227 | W227 |
| G228 | |
| F231 | F231 |
| K236 | K236 |
| A237 | A237 |
| G239 | – |
| I240 | I240 |
| R243 H | R243 H |
| Q244 H | Q244 H |
| F245 | |
| C3 | |
| E264 | |
| K267 H | |
| I268 H | I268 |
| C270 H | |
| V431 H | V431 H |
| M432 | M432 |
| D433 | D433 |
| N434 | N434 H |
| K509 | |
| D558 | |
| Y559 H | Y559 H |
| S560 H | S560 H |
| F561 H | F561 H |
| T562 | T562 |
| A563 | A563 |
| Y564 | Y564 |
| A565 | A565 |
| T566 |
The analysis is based on the PISA server (30). The residues that have been mutated or deleted are highlighted in bold. H following the residue number denotes residues involved in interchain hydrogen bonds; S indicates involvement in a salt bridge. The two chains are not symmetry related, and there are slight differences in the pattern of buried residues between the two monomers.
Interestingly, part of the dimer interface seen in the crystal involves exactly the same residues that are deleted in the hairpin mutants (denoted earlier as C3). Figure 3C shows the interface of the β-hairpin with the adjacent RECQ1 monomer; the most important residues involved in the contact are S560, F561 and T562; the tyrosine at the tip of the hairpin, Y564, plays a lesser role in the subunit interaction. Correspondingly, we found that the Y564A mutation did not disrupt the dimer, but further deletion of F561 caused the dimer to dissociate in solution. This suggests that the dimer interface seen in the crystal structure, or at least the region containing the β-hairpin, is indeed responsible for the dimeric association in solution.
RECQ1T1 dimers are not required for DNA unwinding
The fact that RECQ1T1 forms dimers in solution and in the crystal structure prompted us to investigate whether dimer formation is required for DNA unwinding. Thus, we engineered new β-hairpin mutants in which some of the key residues involved in the dimer formation were deleted or substituted with alanines, while maintaining the tyrosine residue (Y564) previously shown to be essential for DNA unwinding. In particular, we managed to successfully express and purify a deletion mutant Δ(F561-T562-T566)-RECQ1T1 lacking F561 and T562—both of which are involved in the dimer contact—and T566 (the latter was deleted to maintain the equal length of both β-strands of the hairpin). We also expressed the Y559A-S560A-F561A-RECQ1T1 mutant, in which three key residues responsible for the contact of the C3 region of one molecule with the C1 region of the other were substituted with Ala (Table 2).
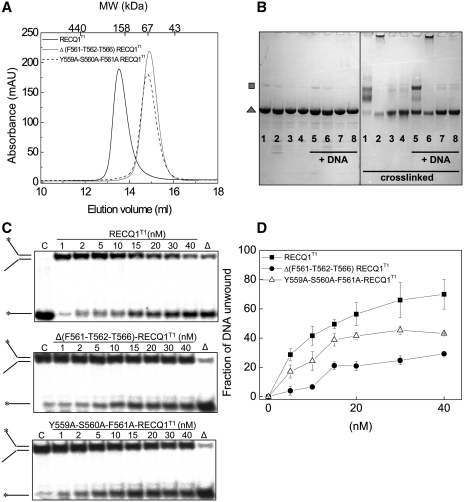
Size exclusion chromatography and chemical cross-linking experiments confirmed that the Δ(F561-T562-T566)-RECQ1T1 and Y559A-S560A-F561A-RECQ1T1 mutants were monomeric in solution (Figure 4A and B). Moreover, the chemical cross-linking experiments repeated in the presence of ssDNA indicated that these two mutants remained monomeric also in the presence of nucleic acids. The following analysis of the unwinding activity showed that both mutants retained the ability to unwind a forked DNA duplex in a concentration dependent fashion, although they were less active than the dimeric wild-type RECQ1T1 (Figure 4C and D). In addition, the Y559A-S560A-F561A-RECQ1T1 mutant was more active than Δ(F561-T562-T566)-RECQ1T1 possibly because the shortening of the β-hairpin might have affected the ability of Y564 to act as a strand separation pin or the ability of the mutant to interact with DNA. In particular, ∼60% of the substrate was unwound by RECQ1T1 using 20 nM enzyme, while ∼45% unwinding was observed using the same concentration of Y559A-S560A-F561A-RECQ1T1 mutant, and this percentage decreased to ∼20% using Δ(F561-T562-T566)-RECQ1T1. To test if the reduced unwinding activity of the monomers versus dimers was associated with impairment in DNA binding, we measured the apparent DNA binding affinity of wild-type RECQ1T1 and its β-hairpin mutants for the fork-duplex substrate by EMSA. All the β-hairpin mutations, with the notable exception of the Y564A mutation that does not disrupt the dimer interface, affected the DNA binding affinity of the variant proteins (Table 4). Of note, our previous CD spectroscopy and limited proteolysis experiments confirmed that mutations in the β-hairpin do not affect the overall structure of the WH domain (21), thus ruling out the possibility that the reduced DNA binding and unwinding activity is connected to a more extensive destabilization of the WH domain. Collectively, these results indicate that mutant RECQ1T1 monomers are still proficient in DNA unwinding, as long as the Y564 is conserved, and that dimers are more active than monomers in DNA unwinding probably because dimer formation provides an additional DNA binding interface that stabilizes the interaction with the substrate.
Figure 4.
RECQ1T1 monomers retain unwinding activity. (A) Size exclusion chromatography profiles of RECQ1T1, Δ(F561-T562-T566)-RECQ1T1 and Y559A-S560A-F561A-RECQ1T1. (B) Cross-linking experiments. Protein samples (10 µM) were incubated with (5–8) or without (1–4) a 42-nt DNA oligonucleotide (50 µM) for 10 min at room temperature. Samples were then treated with the cross-linker BS3 (60 µM, right panel) or control buffer (left panel) for 45 min at room temperature, followed by addition of 100 mM Tris–HCl, pH 7.5, 20 mM TCEP and SDS–PAGE sample buffer. 1, 5: RECQ1T1; 2, 6: Δ2-Y564A-RECQ1T1; 3, 7: Δ(F561-T562-T566)-RECQ1T1; 4, 8: Y559A S560A F561A-RECQ1T1. Monomers (grey triangle) and cross-linked dimers (grey square) are indicated. (C) Unwinding assays using various concentrations of RECQ1T1, Δ(F561-T562-T566)-RECQ1T1, and Y559A-S560A-F561A-RECQ1T1 (0–40 nM) and a forked-duplex substrate of 20 bp with ssDNA tails of 30 nt (0.5 nM). All of the reactions were stopped after 20 min. (D) Plot of the unwinding activity as a function of protein concentration. Concentration dependence experiments were performed using the mutants indicated in the figure (0–40 nM). The data points represent the mean of three independent experiments with the standard deviation indicated by error bars.
Table 4.
Summary of the fork–duplex unwinding, HJ branch migration activity and oligomeric properties of all the mutants of RECQ1T1 and RECQ1FL tested in this work
| Phenotype | RECQ1T1 |
RECQ1FL |
||||||
|---|---|---|---|---|---|---|---|---|
| Fork unwinding | Fork binding | HJ unwinding | HJ binding | Oligomeric state | Fork unwinding | HJ unwinding | Oligomeric state | |
| Hairpin mutation | ||||||||
| WT | Yes | 8 ± 2 | No | No | Dimer | Yes | Yes | Dimer + tetramer |
| Y564A | No | 11 ± 2 | No | No | Dimer | Partial (50%) | No | Dimer + tetramer |
| Y564A shortened (Δ2, Δ3, Δ8) | No | >70 | No | No | Monomer | No | No | Dimer* |
| Δ(561–562–566) | Partial (20%) | >70 | (ND) | No | Monomer | (ND) | (ND) | (ND) |
| AAA(559–560–561) | Partial (40%) | 25 ± 5 | (ND) | No | Monomer | (ND) | (ND) | (ND) |
| Aromatic-loop mutation | ||||||||
| AA(227–231) | No | 10 ± 2 | (ND) | No | Dimer | (ND) | (ND) | (ND) |
The apparent Kd values (nM) for the binding of RECQ1T1 to the fork–duplex substrate are also shown. The asterisk denotes a different kind of dimer held together by the N-terminus of RECQ1.
The aromatic-rich loop of domain D1 is not required for dimer formation, but is essential for DNA unwinding
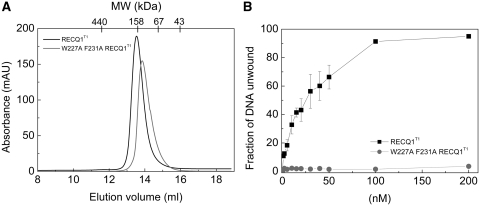
Our analysis of the crystal structure for additional regions that mediate dimer formation highlighted an aromatic-rich loop in domain D1 which lies at the centre of the interface and interacts with the same region in the other monomer (denoted earlier as C2) (Figure 3B). To test the impact of the C2 region in dimer formation, we expressed an additional mutant W227A-F231A were two aromatic residues involved in protein:protein interactions were substituted with Ala (Table 2). However, our size exclusion chromatography experiments showed that the elution profile did not change significantly relative to wild-type RECQ1T1 suggesting that the disruption of the C2 contacts is not sufficient to impair dimer formation (Figure 5A).
Figure 5.
Analysis of the oligomeric properties and unwinding activity of the aromatic-rich loop mutant W227A-F231A-RECQ1T1. (A) Size exclusion chromatography profiles of RECQ1T1(black) and W227A-F231A-RECQ1T1(gray). (B) Plot of the unwinding activity as a function of RECQ1T1 and W227A-F231A-RECQT1 concentration (0–200 nM) using a 20 bp fork-duplex substrate with 30-nt ssDNA tails at a final concentration of 0.5 nM. Reactions were stopped after 20 min. The data points represent the mean of three independent experiments with the standard deviation indicated by error bars.
Interestingly, this highly conserved aromatic-rich loop was originally identified by Keck and co-workers (31) as essential for DNA-coupled ATP hydrolysis by bacterial RecQ. Consistently, we found that the W227A-F231A mutant was unable to unwind a forked-duplex substrate even at the highest protein concentration used in the experiment (Figure 5B), even though it retained the same DNA binding affinity of wild-type RECQ1T1 (Table 4), suggesting that this loop plays an important role in coupling DNA binding to ATP-dependent DNA unwinding also in the case of RECQ1.
The β-hairpin is involved in RECQ1FL higher order oligomer formation
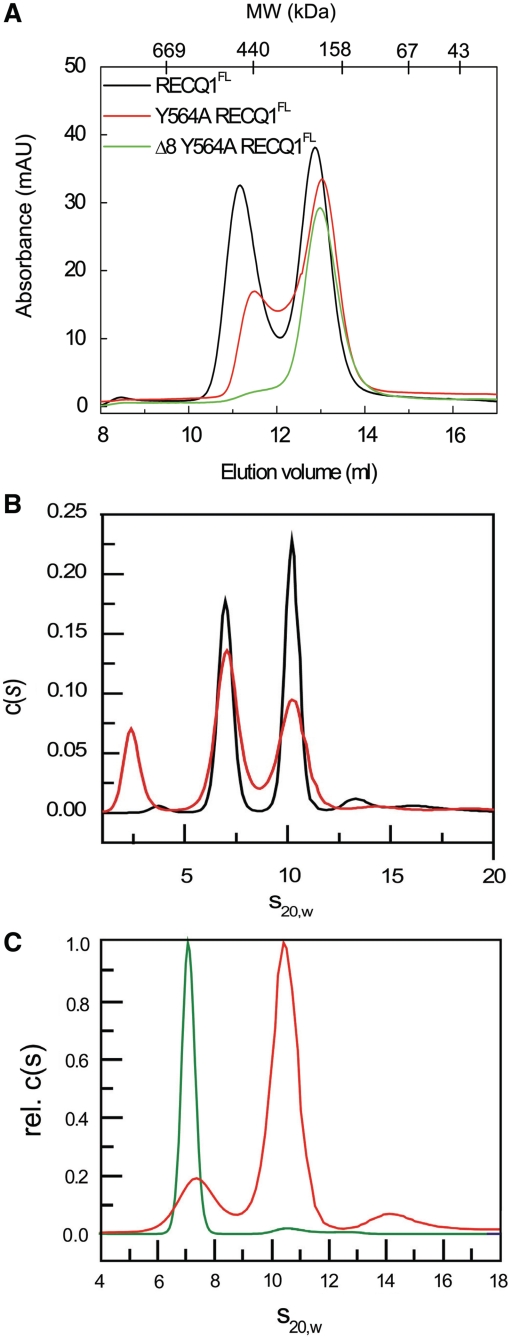
From the comparison of the elution profiles of RECQ1T1 and RECQ1FL, and from previous results, we infer that the smallest form of the full-length RECQ1 is also a dimer (21,26). This conclusion was also supported by velocity AUC experiments (Figure 6) confirming that RECQFL existed as a mix of two oligomeric forms: a smaller form of 140–160 kDa representing RECQ1 dimers, and a 250–280 kDa oligomeric form, which is probably a tetramer, rather than a pentamer or an hexamer as was previously suggested (23). Interestingly, our size exclusion chromatography experiments showed that the β-hairpin Δ8-Y564A-RECQ1FL mutant, that has lost the dimer interface seen in the RECQT1 structure, can still form dimers, but not tetramers (Figure 6A). In line with this observation, our AUC experiments showed that the β-hairpin Δ8-Y564A-RECQ1FL mutant exists in a single form of a dimer, while the Y564A-RECQ1FL is a mixture of dimers and tetramers. The fact that the Δ8-Y564A-RECQ1FL mutant can still form dimers can be explained considering that a protein interaction surface involving the N-terminal residues of RECQ1—which was previously shown to mediate tetramer formation (20,21)—is still functional in the Δ8 mutant that lacks the basic C3 dimer interface formed by the β-hairpin residues.
Figure 6.
The β-hairpin loop is required for tetramer formation in RECQ1FL. (A) Size exclusion chromatography profiles of the purified RECQFL, Y564A-RECQ1FL and Δ8-Y564A-RECQ1FL proteins. (B) Analytical ultracentrifugation of full-length RECQ1. RECQ1FL (subunit MW = 75 620) was analyzed in absence (black curve) or presence (red curve) of 50 µM ATPγS. Two peaks at 7S and 10S correspond to MW of 144 and 260, respectively; a low MW peak seen in the ATPγS experiment may be due to the nucleotide. (C) Analytical ultracentrifugation of Y564A-RECQ1FL (red curve) and Δ8-Y564A-RECQ1FL (green curve). The peaks at 7S and 10S correspond to dimers and tetramers; the Y564A-RECQ1FL protein migrates as a mixture of dimers, tetramers and a small amount of higher oligomer or aggregate; the hairpin deletion mutant Δ8-Y564A-RECQ1FLis almost entirely dimeric [note that the x-axis in 6A is in order of decreasing size, while the x-axes in (B) and (C) represent increasing protein size].
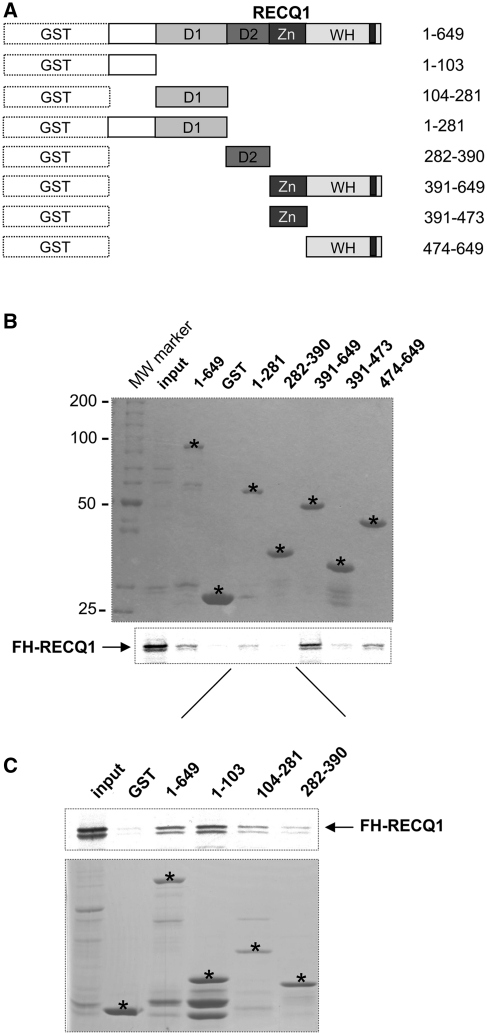
To map the domains involved in oligomer formation, we tested the ability of the in vitro translated RECQ1 protein to interact with a series of GST-tagged RECQ1 fragments mapping the different domains of the protein (Figure 7A). Our GST pull down experiments confirmed that RECQ1FL interacted with the fragment 1–281 containing the N-terminal tail plus domain D1 and with the fragment 391–649 containing the Zn-binding domain and the WH domain where the β-hairpin is located, but not with the fragment 282–390 containing the recA-like domain D2 or with the fragment 391–473 containing the Zn-binding domain alone (Figure 7B). The fragment 474–649 encompassing the WH domain, but lacking the Zn-binding domain, was also able to interact with RECQ1FL, although more weakly compared to fragment 391–649, suggesting that the deletion of residues 391–473 might affect the stability or the conformation of the WH domain. To explore the role of the N-terminus in more detail, we expressed two additional GST-tagged RECQ1 fragments: fragment 1–103 containing the N-terminal tail of RECQ1 and fragment 104–281 encompassing the D1 domain (Figure 7C). The results confirmed that RECQ1FL predominantly interacted with the first 103 aa containing the N-terminal tail previously shown to be involved in oligomer formation (20,21). Collectively, these results confirm that two different regions of RECQ1, the N-terminus and the C-terminal WH domain containing the β-hairpin module, are involved in higher-order oligomer assembly.
Figure 7.
In vitro RECCQ1 GST pull-down assay. (A) N-terminally GST tagged RECQ1 fragments (aa 1–103, 104–281, 1–281, 282–390, 391–649, 391–473, 474–649) used for the in vitro GST pull down assay. (B) Top: coomassie stained gel of the GST tagged RECQ1 fragments used in the assay. The stars indicate the location of the fragments in the gels. Bottom: autoradiography of the in vitro GST pull-down assay using the 35S-labeled full-length RECQ1 protein. (C) Top: autoradiography of the in vitro GST pull-down assay using additional GST tagged RECQ1 fragments covering the first 281 aa at the N-terminus of the protein and the 35S-labeled full-length RECQ1 protein. Bottom: coomassie stained gel of the additional GST tagged RECQ1 fragments used in the assay. The stars indicate the location of the fragments in the gels.
DISCUSSION
DNA and RNA helicases and translocases from superfamilies 1 and 2 (SF-1 and SF-2) share a core ATPase domain, which consists of tandem RecA-like folds, and is characterized by conserved sequence motifs (32). In addition to this conserved fold, most helicases include additional loops or domains that are more divergent, and may be conserved only within restricted subfamilies. These non-conserved regions may be important in DNA/RNA unwinding as well as in other enzymatic or regulatory functions. RecQ-family helicases share a characteristic C-terminal domain (termed RecQ-Ct), which includes a zinc-binding module and a WH domain. We have previously suggested, based on structural alignments with other DNA helicases, that the β-hairpin wing of the WH domain is poised to serve as a pin that aids DNA strand separation at a ssDNA–dsDNA junction (21). This prediction was supported by the observation that point mutations or deletions of the hairpin of a truncated form of the RECQ1 protein severely impaired DNA unwinding activity, with no effect on the ssDNA-dependent ATPase activity of the mutant proteins. Here, using β-hairpin mutants of the full-length RECQ1 helicase we show that this prominent β-hairpin is not only essential for fork duplexes unwinding, but it also required the HJ disruption activity of full-length RECQ1, which is one of the most characteristic activities of RecQ proteins (Table 4). In addition, we show that this β-hairpin is a key structural element that controls not only the enzymatic activity of RECQ1, but also the balance between the multiple oligomeric states of the protein. In particular, it mediates dimer formation in the truncated RECQ1 protein lacking 48 residues at the N-terminus and 33 residues at the C-terminus, and tetramer formation in the full-length protein.
Interestingly, a recently published structure of the WH domain of WRN in complex with a 14 base pair DNA duplex shows that the β-hairpin wedges apart the terminal base pair of the DNA duplex, resulting in loss of base–base stacking, thus supporting the notion that the aromatic residue at the tip of the hairpin acts as a pin that abuts the end of the DNA duplex and hence promotes strand separation (33). A recently solved structure of a complex of the human RECQ1 with DNA provides further support to this model (PDB ID 2WWY; Pike et al., manuscript in preparation). Similar β-hairpin motifs were previously found to be essential for the unwinding activity of other helicase members of both the SF-1 and SF-2 families. The structures of two SF-1 proteins, UvrD and PcrA, complexed with partial DNA duplexes with 7-nt 3′-tails, show that the bound DNA duplex unwinds in front of a ‘pin’ located in domain 2A (34,35). Similarly, the crystal structures of the SF-2 proteins, Hel308 from Archaeoglobus fulgidus and helicase NS3 of the hepatitis C virus, show a β-hairpin structure between motifs V and VI of the helicase domain (25,36). In particular, the structure of Hel308 bound to a 15-bp DNA duplex with a 10-nt 3′-tail shows that the β-hairpin is positioned exactly at the opening of the unwound tailed duplex (25). Thus, RecQ helicases might share an unwinding mechanism similar to that of these SF-1 and SF-2 helicases where the aromatic residue at the tip of the β-hairpin module sits at junction between ss- and dsDNA, and acts as a ‘pin’ that promotes strand separation. However, this common structural motif is derived from different regions in the primary structure of SF-1, Hel308 and RECQ1 helicases, and may have evolved independently.
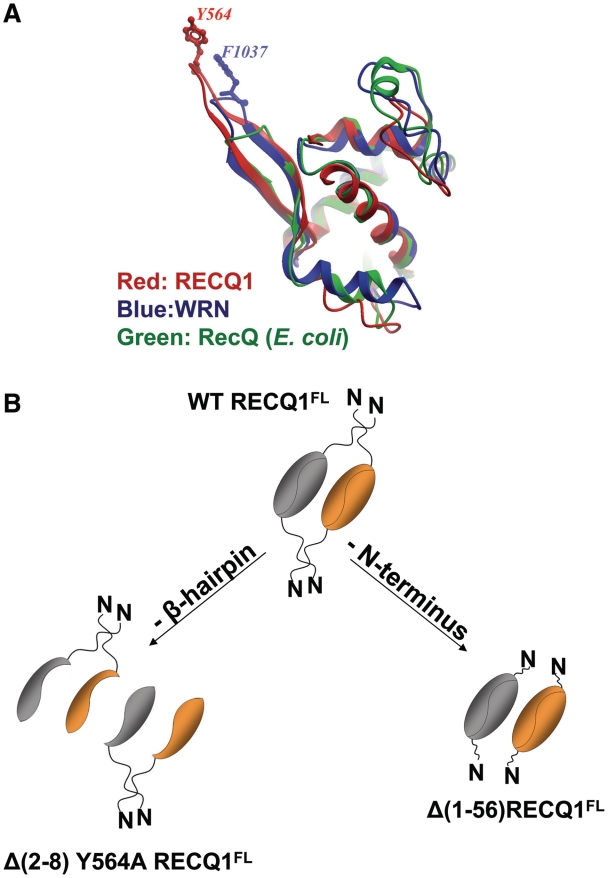
Intriguingly, the β-hairpin is not highly conserved between different RecQ-family proteins (21). In particular, the superimposition of the WH-domain structures of E. coli RecQ, RECQ1 and WRN reveals that the β-hairpin is significantly shorter in E. coli RecQ and WRN (Figure 8A). Moreover, the aromatic residue at the tip of the β-hairpin of RECQ1 and WRN is substituted with a His in E. coli RecQ, suggesting that the β-hairpin module might play different roles in the three helicases. Indeed, we previously showed that a His to Ala substitution at the tip of the hairpin in the bacterial RecQ, as well as deletion of the hairpin, do not affect the enzymatic activity of the bacterial protein (21). The putative β-hairpin in BLM helicase also lacks an aromatic residue. This structural variance indicates that RecQ proteins may utilize different mechanisms for substrate recognition and unwinding.
Figure 8.
(A) Superimposition of WH domains of the RecQ-Ct regions of human RECQ1 (red; 2V1X), E. coli RecQ (green; 1OYW) and WRN (blue; 3AAF). The β-hairpins are seen in the upper-left corner. Aromatic residues at the tip of the hairpin (Y564 in RECQ1 and F1037 in WRN) are shown; there is no aromatic residue at the tip of the shorter hairpin of the E. coli enzyme. (B) Model for RECQFL tetramer assembly. The RECQ1FL tetramer is represented at the top. Two independent protein–protein contacts are required for tetramer formation, one mediated by the β-hairpin and the other by the N-terminus of RECQ1, suggesting a non-hierarchical mechanism of tetramer assembly. Deletion of the β-hairpin leads to a dimeric structure were the two subunits are held together by the N-terminus (left), while deletion of the N-terminus leads to dimers held together by the β-hairpin mediated contacts (right).
This apparently straightforward interpretation of the role of the β-hairpin is complicated by our observations on the oligomeric state of RECQ1. The purified full-length RECQ1 protein (denoted here RECQ1FL) occurs as a mixture of a lower and higher oligomeric forms. We here determine, using analytical ultracentrifugation and gel filtration, that these correspond to dimers and tetramers of RECQ1. We have previously observed that deletion of a short amino-terminal segment (aa 1–48) leads to loss of the tetrameric form. A truncated protein (RECQ1T1, lacking the first 48 aa and the last 33 aa at the C-terminus), which purifies uniquely as a dimer, was used in crystallization experiments. The crystal structure identifies a potential dimerization interface between two adjacent RECQ1 molecules. The two molecules in the dimer are arranged in a head to tail orientation. The buried interaction surface may be divided into three patches: an amino-terminal region termed C1 interacts with a C-terminal patch (C3) in the opposite subunit. The second patch (termed C2) involves an aromatic-rich loop in domain D1 which interacts with the same region in the other monomer. The β-hairpin of the WH domain (specifically, residues S560, F561, and T562 in one β-strand) forms a crucial part of the C3 patch. The tyrosine at the tip of the hairpin, Y564, is not however involved in protein:protein interactions, which is consistent with its proposed role in DNA strand separation.
Detailed mutational analysis confirmed the important role of the β-hairpin in dimer formation. Deletions that shorten the hairpin, or substitute the interacting residues with alanines in RECQ1T1 cause the mutant proteins to purify exclusively as monomers (Figures 2 and 4A). The second region involved in dimer interaction (C2) includes an aromatic-rich loop in domain D1 that interacts with the same region in the other monomer. In contrast with the β-hairpin mutations, the substitution to Ala of two key residues of this loop, W227 and F231, is not sufficient to abrogate dimer formation. This could be due to the fact that the mutations of residues in the C2 region only disrupt one of the three patches involved in dimer formation, while mutations in the β-hairpin module simultaneously perturb two of the contacts involved in the ‘head-to-tail’ dimer interface. However, additional mutations in C2 region would be required to confirm this hypothesis. Of note, the aromatic-rich of region C2 was previously identified by Keck and co-workers as essential for DNA-coupled ATP hydrolysis (31). Consistently, we found that the W227A-F231A mutant of RECQ1 is unable to unwind DNA, although it retains the ability to bind DNA. Interestingly, this aromatic loop includes the highly conserved sequence CxSQWGHDFR which is characteristic of RecQ helicases—but is not present in other helicases of the SF-1 and SF-2 family—suggesting that it may present a unique aspect of the catalytic mechanism of RecQ helicases (37).
While RECQ1 is able to form dimers, the bacterial RecQ is a monomer in solution and does not exhibit a clear dimer interface in the crystal. Indeed, the different orientation of the WH domain, and the poor conservation of many of the residues involved in the dimer contact, seems to preclude a similar dimeric association. At this point, we can only speculate on the functional significance of the dimeric structure of RECQ1. To assess the functional significance of the dimerization of RECQ1T1, one needs to examine the activity of mutants that are unable to form dimers. The mutants affecting the dimeric state are alterations of the β-hairpin, which is now seen to have two functions (disruption of the DNA duplex and protein:protein interaction). To disentangle these two effects, we tested the DNA unwinding activity, the DNA binding properties and oligomeric state of several mutants, which are summarized in Table 4. The single substitution of the aromatic Y564 to Ala abolishes DNA unwinding, but does not affect DNA binding and dimer formation. Conversely, mutants in which the hairpin is shortened (Δ561–562–566) or substituted (AAA:559–560–561) without changing Y564, fail to form stable dimers, but are at least partially active in DNA unwinding. Interestingly, this reduced DNA unwinding activity is associated to a weaker DNA binding affinity of these mutants for the fork DNA duplexes. Thus, we conclude that dimerization is not essential for helicase activity, although it may regulate the efficiency of DNA unwinding by providing an additional interface that stabilizes the RECQ1–DNA complex. The relative activity of monomers and higher multimers of non-hexameric helicases has been the subject of some debate (38). Previous studies suggested that some helicases, such as the Escherichia coli Rep and UvrD helicases, need to oligomerize in order to achieve full DNA unwinding activity (39,40). The mode of cooperation of multiple helicases acting on a single substrate is not clear. It is likely that only one helicase molecule contacts the strand separation point, while the additional molecules tug on the 3′-ssDNA tail, increasing the force or processivity or preventing slippage. The stable dimeric structure of RECQ1 may be important in this context. The helicase domains of other RecQ-family proteins exist as monomers (22,41–43). Compared with these helicases, RECQ1 lacks the C-terminal HRDC domain, which is thought to play a role in DNA binding in other RecQ-family helicases. Thus, one could speculate that the dimerization of RECQ1 provides an additional DNA binding interface that is normally provided by the HRDC domain. On the other hand, some of the proposed functions of the HRDC domain of other RecQ helicases, such as HJ recognition (44–46), might not be simply ‘rescued’ by dimerization since none of truncated RECQ1T1 protein variants binds or unwinds HJ, independently on whether they form dimers or not (Table 4) (20).
The full-length RECQ1 exists in two oligomeric forms: a higher order oligomeric form which we here identify as a tetramer, and a smaller form which is a dimer. However, the fact that a deletion mutant lacking eight residues on both strands of the β-hairpin in addition to the Y564A substitution (Δ8-Y564A-RECQ1FL) in the full-length context still forms dimers suggests that the tetramers observed in the full-length contexts are not simply a ‘dimer of dimers’. Rather, tetramers are formed by two independent protein–protein contacts, one mediated by the C1–C3 contact, as we see in the crystal structure of the truncated protein, and another mediated by the N-terminus of RECQ1, as suggested from our previous size exclusion chromatography analysis of truncated versions of the protein lacking the first 48 or 56 residues (20,21). These results were also confirmed by GST pull-down assays showing that oligomer assembly is indeed mediated by the N-terminus and a C-terminal domain containing the β-hairpin (Figure 7). Thus, we can elaborate a model for tetramer formation that is not hierarchical: a protein interaction surface represented by the N-terminus of RECQ1, which normally mediates tetramer formation, is still functional in the (Δ8-Y564A-RECQ1FL) mutant and might hold the two subunits together even though they lack their basic dimer interface (Figure 8).
In summary, this study provides novel information on the structural determinants and the oligomeric forms of RECQ1 associated with DNA unwinding as well as on the mechanism that regulates higher assembly state formation. We show that the β-hairpin module is not only crucial for the DNA strand separation activity of RECQ1, but it is also essential to mediate dimer formation in the context of the truncated protein and tetramer formation in the context of full-length RECQ1. Interestingly, dimer formation promotes DNA unwinding, while tetramer formation seems to be required for more specialized activities. However, many questions remain. First, does the β-hairpin module work as a ‘pin’ to separate the complementary strands and as a ‘module’ to promote oligomer formation, in all RecQ helicases? Our mutagenesis studies suggest that this is not the case. When we compared the helicase activity of the β-hairpin mutants of RECQ1 and E. coli RecQ, we clearly saw that mutations in the β-hairpin of the bacterial protein did not have an impact on the helicase activity of E. coli RecQ (21). Moreover, E. coli RecQ is monomeric, suggesting that RecQ helicases might adopt different mechanisms and oligomeric structures to promote DNA unwinding (22,42). Second, what is the functional significance of the higher order oligomeric structures observed for several RecQ helicases? Our previous studies with the N-terminally deleted RECQ1 mutants indicated that these mutants were unable to resolve HJ structures or to promote the annealing of complementary ssDNA duplexes, suggesting that higher order oligomers are required for more ‘specialized’ activities. This notion is also supported by the previous observation made for BLM and WRN that connected their strand annealing activity to the ability to form higher order protein–DNA complexes (47,48). However, further work will be required to test the role of the β-hairpin module in other RecQ helicases and define the exact stoichiometry and functions of these higher order RecQ helicase complexes. These future studies will contribute to explain the distinct functions of the various RecQ helicase homologs present in eukaryotes and might shed light on how the lack of one or more of these ‘specialized’ RecQ helicase activities are linked to increased genomic instability and tumorigenesis.
FUNDING
Associazione Italiana per la Ricerca sul Cancro (AIRC; AIRC5088 to A.V.); Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from the Canadian Institutes for Health Research; the Canadian Foundation for Innovation; Genome Canada through the Ontario Genomics Institute; GlaxoSmithKline; Karolinska Institutet; the Knut and Alice Wallenberg Foundation; the Ontario Innovation Trust; the Ontario Ministry for Research and Innovation; Merck & Co., Inc.; the Novartis Research Foundation; the Swedish Agency for Innovation Systems; the Swedish Foundation for Strategic Research; the Wellcome Trust. Funding for open access charge: Associazione Italiana per la Ricerca sul Cancro (AIRC; AIRC5088).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The contribution of Drs Silvia Costantini, Venkateswarlu Popuri and Gianluca Triolo in providing the necessary guidelines for protein production and biochemical assays is gratefully acknowledged.
REFERENCES
- 1.Nakayama H, Nakayama K, Nakayama R, Irino N, Nakayama Y, Hanawalt PC. Isolation and genetic characterization of a thymineless death-resistant mutant of Escherichia coli K12: identification of a new mutation (recQ1) that blocks the RecF recombination pathway. Mol. Gen. Genet. 1984;195:474–480. doi: 10.1007/BF00341449. [DOI] [PubMed] [Google Scholar]
- 2.Bohr VA. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem Sci. 2008;33:609–620. doi: 10.1016/j.tibs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nat. Rev. Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 4.Hartung F, Puchta H. The RecQ gene family in plants. J. Plant Physiol. 2006;163:287–296. doi: 10.1016/j.jplph.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 5.Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 6.Kitao S, Lindor NM, Shiratori M, Furuichi Y, Shimamoto A. Rothmund-thomson syndrome responsible gene, RECQL4: genomic structure and products. Genomics. 1999;61:268–276. doi: 10.1006/geno.1999.5959. [DOI] [PubMed] [Google Scholar]
- 7.Siitonen HA, Kopra O, Kaariainen H, Haravuori H, Winter RM, Saamanen AM, Peltonen L, Kestila M. Molecular defect of RAPADILINO syndrome expands the phenotype spectrum of RECQL diseases. Hum. Mol. Genet. 2003;12:2837–2844. doi: 10.1093/hmg/ddg306. [DOI] [PubMed] [Google Scholar]
- 8.Van Maldergem L, Siitonen HA, Jalkh N, Chouery E, De Roy M, Delague V, Muenke M, Jabs EW, Cai J, Wang LL, et al. Revisiting the craniosynostosis-radial ray hypoplasia association: Baller-Gerold syndrome caused by mutations in the RECQL4 gene. J. Med. Genet. 2006;43:148–152. doi: 10.1136/jmg.2005.031781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, et al. Positional cloning of the Werner’s syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 10.Li D, Frazier M, Evans DB, Hess KR, Crane CH, Jiao L, Abbruzzese JL. Single nucleotide polymorphisms of RecQ1, RAD54L, and ATM genes are associated with reduced survival of pancreatic cancer. J. Clin. Oncol. 2006;24:1720–1728. doi: 10.1200/JCO.2005.04.4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Futami K, Kumagai E, Makino H, Goto H, Takagi M, Shimamoto A, Furuichi Y. Induction of mitotic cell death in cancer cells by small interference RNA suppressing the expression of RecQL1 helicase. Cancer Sci. 2008;99:71–80. doi: 10.1111/j.1349-7006.2007.00647.x. [DOI] [PubMed] [Google Scholar]
- 12.Futami K, Kumagai E, Makino H, Sato A, Takagi M, Shimamoto A, Furuichi Y. Anticancer activity of RecQL1 helicase siRNA in mouse xenograft models. Cancer Sci. 2008;99:1227–1236. doi: 10.1111/j.1349-7006.2008.00794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Futami K, Ogasawara S, Goto H, Yano H, Furuichi Y. RecQL1 DNA repair helicase: a potential tumor marker and therapeutic target against hepatocellular carcinoma. Int. J. Mol. Med. 25:537–545. doi: 10.3892/ijmm_00000375. [DOI] [PubMed] [Google Scholar]
- 14.Bachrati CZ, Hickson ID. RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem. J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bachrati CZ, Hickson ID. RecQ helicases: guardian angels of the DNA replication fork. Chromosoma. 2008;117:219–233. doi: 10.1007/s00412-007-0142-4. [DOI] [PubMed] [Google Scholar]
- 16.Bennett RJ, Keck JL. Structure and function of RecQ DNA helicases. Crit. Rev. Biochem. Mol. Biol. 2004;39:79–97. doi: 10.1080/10409230490460756. [DOI] [PubMed] [Google Scholar]
- 17.Opresko PL, Cheng WH, Bohr VA. Junction of RecQ helicase biochemistry and human disease. J. Biol. Chem. 2004;279:18099–18102. doi: 10.1074/jbc.R300034200. [DOI] [PubMed] [Google Scholar]
- 18.Ouyang KJ, Woo LL, Ellis NA. Homologous recombination and maintenance of genome integrity: cancer and aging through the prism of human RecQ helicases. Mech. Ageing Dev. 2008;129:425–440. doi: 10.1016/j.mad.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 19.Seki M, Yanagisawa J, Kohda T, Sonoyama T, Ui M, Enomoto T. Purification of two DNA-dependent adenosinetriphosphatases having DNA helicase activity from HeLa cells and comparison of the properties of the two enzymes. J. Biochem. 1994;115:523–531. doi: 10.1093/oxfordjournals.jbchem.a124369. [DOI] [PubMed] [Google Scholar]
- 20.Popuri V, Bachrati CZ, Muzzolini L, Mosedale G, Costantini S, Giacomini E, Hickson ID, Vindigni A. The Human RecQ helicases, BLM and RECQ1, display distinct DNA substrate specificities. J. Biol. Chem. 2008;283:17766–17776. doi: 10.1074/jbc.M709749200. [DOI] [PubMed] [Google Scholar]
- 21.Pike AC, Shrestha B, Popuri V, Burgess-Brown N, Muzzolini L, Costantini S, Vindigni A, Gileadi O. Structure of the human RECQ1 helicase reveals a putative strand-separation pin. Proc. Natl Acad. Sci. USA. 2009;106:1039–1044. doi: 10.1073/pnas.0806908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernstein DA, Zittel MC, Keck JL. High-resolution structure of the E.coli RecQ helicase catalytic core. EMBO J. 2003;22:4910–4921. doi: 10.1093/emboj/cdg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muzzolini L, Beuron F, Patwardhan A, Popuri V, Cui S, Niccolini B, Rappas M, Freemont PS, Vindigni A. Different quaternary structures of human RECQ1 are associated with its dual enzymatic activity. PLoS Biol. 2007;5:e20. doi: 10.1371/journal.pbio.0050020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vindigni A, Hickson ID. RecQ helicases: multiple structures for multiple functions? HFSP J. 2009;3:153–164. doi: 10.2976/1.3079540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buttner K, Nehring S, Hopfner KP. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat. Struct. Mol. Biol. 2007;14:647–652. doi: 10.1038/nsmb1246. [DOI] [PubMed] [Google Scholar]
- 26.Cui S, Arosio D, Doherty KM, Brosh RM, Jr, Falaschi A, Vindigni A. Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A. Nucleic Acids Res. 2004;32:2158–2170. doi: 10.1093/nar/gkh540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown PH, Schuck P. Macromolecular size-and-shape distributions by sedimentation velocity analytical ultracentrifugation. Biophys. J. 2006;90:4651–4661. doi: 10.1529/biophysj.106.081372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paolinelli R, Mendoza-Maldonado R, Cereseto A, Giacca M. Acetylation by GCN5 regulates CDC6 phosphorylation in the S phase of the cell cycle. Nat. Struct. Mol. Biol. 2009;16:412–420. doi: 10.1038/nsmb.1583. [DOI] [PubMed] [Google Scholar]
- 29.Marcello A, Massimi P, Banks L, Giacca M. Adeno-associated virus type 2 rep protein inhibits human papillomavirus type 16 E2 recruitment of the transcriptional coactivator p300. J. Virol. 2000;74:9090–9098. doi: 10.1128/jvi.74.19.9090-9098.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 31.Zittel MC, Keck JL. Coupling DNA-binding and ATP hydrolysis in Escherichia coli RecQ: role of a highly conserved aromatic-rich sequence. Nucleic Acids Res. 2005;33:6982–6991. doi: 10.1093/nar/gki999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 33.Kitano K, Kim SY, Hakoshima T. Structural basis for DNA strand separation by the unconventional winged-helix domain of RecQ helicase WRN. Structure. 18:177–187. doi: 10.1016/j.str.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 34.Lee JY, Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 36.Kim JL, Morgenstern KA, Griffith JP, Dwyer MD, Thomson JA, Murcko MA, Lin C, Caron PR. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 37.Vindigni A, Marino F, Gileadi O. Probing the structural basis of RecQ helicase function. Biophys. Chem. 2010;149:67–77. doi: 10.1016/j.bpc.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 38.Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat. Rev. Mol. Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 39.Ha T, Rasnik I, Cheng W, Babcock HP, Gauss GH, Lohman TM, Chu S. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 2002;419:638–641. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- 40.Maluf NK, Fischer CJ, Lohman TM. A dimer of Escherichia coli UvrD is the active form of the helicase in vitro. J. Mol. Biol. 2003;325:913–935. doi: 10.1016/s0022-2836(02)01277-9. [DOI] [PubMed] [Google Scholar]
- 41.Janscak P, Garcia PL, Hamburger F, Makuta Y, Shiraishi K, Imai Y, Ikeda H, Bickle TA. Characterization and mutational analysis of the RecQ core of the bloom syndrome protein. J. Mol. Biol. 2003;330:29–42. doi: 10.1016/s0022-2836(03)00534-5. [DOI] [PubMed] [Google Scholar]
- 42.Xu HQ, Deprez E, Zhang AH, Tauc P, Ladjimi MM, Brochon JC, Auclair C, Xi XG. The Escherichia coli RecQ helicase functions as a monomer. J. Biol. Chem. 2003;278:34925–34933. doi: 10.1074/jbc.M303581200. [DOI] [PubMed] [Google Scholar]
- 43.Zhang XD, Dou SX, Xie P, Hu JS, Wang PY, Xi XG. Escherichia coli RecQ is a rapid, efficient, and monomeric helicase. J. Biol. Chem. 2006;281:12655–12663. doi: 10.1074/jbc.M513089200. [DOI] [PubMed] [Google Scholar]
- 44.Kim YM, Choi BS. Structure and function of the regulatory HRDC domain from human Bloom syndrome protein. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq586. doi:10.1093/nar/gkq586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.von Kobbe C, Thoma NH, Czyzewski BK, Pavletich NP, Bohr VA. Werner syndrome protein contains three structure-specific DNA binding domains. J. Biol. Chem. 2003;278:52997–53006. doi: 10.1074/jbc.M308338200. [DOI] [PubMed] [Google Scholar]
- 46.Wu L, Chan KL, Ralf C, Bernstein DA, Garcia PL, Bohr VA, Vindigni A, Janscak P, Keck JL, Hickson ID. The HRDC domain of BLM is required for the dissolution of double Holliday junctions. EMBO J. 2005;24:2679–2687. doi: 10.1038/sj.emboj.7600740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheok CF, Wu L, Garcia PL, Janscak P, Hickson ID. The Bloom’s syndrome helicase promotes the annealing of complementary single-stranded DNA. Nucleic Acids Res. 2005;33:3932–3941. doi: 10.1093/nar/gki712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muftuoglu M, Kulikowicz T, Beck G, Lee JW, Piotrowski J, Bohr VA. Intrinsic ssDNA annealing activity in the C-terminal region of WRN. Biochemistry. 2008;47:10247–10254. doi: 10.1021/bi800807n. [DOI] [PMC free article] [PubMed] [Google Scholar]