Abstract
Bulk replicative DNA synthesis in eukaryotes is highly accurate and efficient, primarily because of two DNA polymerases (Pols): Pols δ and ε. The high fidelity of these enzymes is due to their intrinsic base selectivity and proofreading exonuclease activity which, when coupled with post-replication mismatch repair, helps to maintain human mutation rates at less than one mutation per genome duplication. Conditions that reduce polymerase fidelity result in increased mutagenesis and can lead to cancer in mice. Whereas yeast Pol ε has been well characterized, human Pol ε remains poorly understood. Here, we present the first report on the fidelity of human Pol ε. We find that human Pol ε carries out DNA synthesis with high fidelity, even in the absence of its 3′→5′ exonucleolytic proofreading and is significantly more accurate than yeast Pol ε. Though its spectrum of errors is similar to that of yeast Pol ε, there are several notable exceptions. These include a preference of the human enzyme for T→A over A→T transversions. As compared with other replicative DNA polymerases, human Pol ε is particularly accurate when copying homonucleotide runs of 4–5 bases. The base pair substitution specificity and high fidelity for frameshift errors observed for human Pol ε are distinct from the errors made by human Pol δ.
INTRODUCTION
Replication of the eukaryotic nuclear genome appears to be carried out primarily by three DNA polymerases, Pols α, δ and ε. Current evidence has led to a model for division of labor of these enzymes at the bidirectional replication fork (1). Pol α and its associated primase subunits have been implicated in the initiation of de novo synthesis on the leading and lagging strands (2). Genetic studies in yeast had shown that Pols δ and ε operate on opposite strands during replication (3,4). Subsequent work using combined genetic and biochemical analyses of polymerase mutator alleles showed that Pol ε is normally responsible for leading strand synthesis (5) while Pol δ functions primarily on the lagging strand (6). In addition, Pol ε is found at replication origins prior to replication (7) and in a replication complex containing Go-Ichi-Ni-San (GINS) (8), which is itself part of the Cdc45-MCM2-7-GINS (CMG) putative replicative helicase (9,10). A fourth DNA polymerase, Pol ζ, has also been shown to play an increasingly important, yet less well understood, role in replication (11).
An important and open question is whether a specific division of labor between DNA polymerases is a general rule or one of a number of replication fork assemblies for these enzymes. The stronger effect on mutation rate consistently seen in Pol δ exonuclease-deficient as compared with Pol ε exonuclease-deficient yeast (12,13) is in support of the latter idea. Furthermore, the ability of Pol δ to compensate for catalytic deletion of Pol ε, but not vice versa (14,15), helped lead to a model in which Pol ε begins leading strand replication but dissociates in response to a trigger that allows Pol δ to complete leading strand replication (16). This model is not inconsistent with the original interpretation that did not exclude a role for Pol δ on the leading strand (5). However, the validity of either model warrants further study. Another major unresolved issue is whether either replication fork model holds true for humans. Based on the observation that human Pol ε colocalizes with PCNA only late in S phase (17), it has been proposed that the Pol ε replication function might become temporally separated from Pol δ in response to a physiological event, such as heterochromatin replication or DNA damage during late replication.
In addition to its involvement in replication, Pol ε plays vital roles in a number of other essential cellular processes including repair of damaged DNA, control of cell cycle progression and maintenance of epigenetic inheritance. Pol ε was initially identified as a soluble factor able to correct a repair defect in human cells (18–20). Both Pols δ and ε are able to support nucleotide excision repair (NER) reconstitution (21–23) and mutants of both show reduced NER activity (24). Recently Pol ε was shown to function in concert with the CTF18-RFC clamp loader, accounting for half the repair synthesis in non-dividing human cells, with the remaining repair synthesis performed by Pols δ and κ working with the RFC1-RFC clamp loader (25). While Pol β serves as the primary base excision repair (BER) polymerase in mammalian cells (26), Pol ε, along with Pol δ, has been implicated in a backup BER pathway that synthesizes long patches of DNA (27). Early evidence in yeast suggested that Pol ε plays a role in proper maintenance of S phase progression (28,29). Pol ε also interacts with the yeast checkpoint mediator protein Mrc1 (30), which is phosphorylated by both Mec1 and Rad53 (31). Mrc1 plays an important role regulating normal replication fork progression (32). The same is true for the Xenopus Mrc1 homolog, xClaspin (33). Yeast Pol ε is also involved in gene silencing at both the HMR locus (34) and near telomeres (35). The structural integrity of the multisubunit holoenzyme may play an important part in these functions as one of the small subunits is implicated in gene silencing at the rDNA locus (36) and the other is also found as part of a chromatin remodeling complex (37–39).
Pol ε is comprised of four subunits in all eukaryotes studied so far. The human POLE1 gene encodes the large 261 kDa subunit (40,41). This p261 subunit contains the DNA polymerase and exonuclease active site motifs in the N-terminal half of the protein. This N-terminal half of the enzyme is stable and enzymatically active in a variety of species (42–44) and is created by caspase-3 and calpain cleavage during apoptosis (45). Surprisingly, the catalytic half of the enzyme was shown to be non-essential in yeast (15). The C-terminal half contains no known enzymatic activity, but rather contains interaction sites for the three smaller subunits (46,47). The 20 Å structure of the four subunit yeast holoenzyme determined by cryoelectron microscopy shows an asymmetric structure with the globular large catalytic subunit and a tail made up the smaller subunits in unknown orientation (48). This tail is able to wrap around a 40-bp double-strand DNA modeled onto the structure, suggesting a mechanism to describe the requirement of a 40-bp primer for Pol ε to catalyze processive DNA synthesis. This high processivity would likely be advantageous for continuous leading strand synthesis in eukaryotes. Initial comparison of the endogenous purified human enzymes showed Pol ε activity to be stimulated by the PCNA clamp, but still highly processive in its absence, whereas Pol δ is strictly PCNA-dependent (19,49). A direct comparison of yeast Pol ε with yeast Pol δ confirmed a weak Pol ε-PCNA interaction, but showed similar and remarkably low processivities for both replicative polymerases (50).
As a replicative DNA polymerase, Pol ε is responsible for a large amount of DNA synthesis during genome duplication. Left uncorrected, errors made during replication contribute to carcinogenesis. Proofreading inactivation of both mouse Pols δ and ε have recently been shown to lead to genome instability and carcinogenesis in mice, though differences in tumor spectra of the two enzymes are also observed (51). The mechanisms through which Pol ε replication errors contribute to this tumor spectrum remain unknown. Thus, it is of great interest to determine the replication fidelity of human Pol ε.
The replication fidelity of human Pol ε has not been studied because the exonuclease-deficient enzyme has been unavailable for biochemical analysis. To that end, we have expressed and purified the catalytic half of human Pol ε in both an exonuclease-proficient and exonuclease-deficient form. We first measured the DNA polymerization and exonuclease activities of both forms of human Pol ε. We then characterized the replication fidelity of the proofreading-proficient human Pol ε along with its intrinsic replication fidelity using the exonuclease-deficient form. We find that human Pol ε is highly faithful, due to both the exonuclease proofreading and a high intrinsic base selectivity. It is extremely accurate for deletions, especially in repetitive sequences and the pattern of DNA synthesis errors can be distinguished from those made by human Pol δ.
MATERIALS AND METHODS
Materials
Oligonucleotides were from Invitrogen (La Jolla, CA, USA) and were purified by PAGE. Radioactive nucleotides were from Perkin Elmer (Boston, MA, USA). The cloning vectors used were pENTR/TEV/D-TOPO from Invitrogen and pET60-DEST from EMD Chemicals (Gibbstown, NJ, USA). pRK603 contains an inducible TEV protease and was a kind gift of Thang Chiu, LSU Health Sciences Center.
Expression and purification of the catalytic fragment of human Pol ε
The DNA encoding residues 1–1189 of the catalytic subunit of human Pol ε was amplified by PCR and cloned into pENTR/TEV/D-TOPO by using baculovirus containing the open reading frame of p261 [described in (41)]. The human Pol ε construct was then cloned into pET60-DEST using LR Clonase (Invitrogen) to generate GST-hPolε-His6 construct. Human Pol ε was overexpressed in Escherichia coli using autoinduction medium (52) at 25°C until the culture was saturated. After the cells were harvested by centrifugation and the periplasmic fraction removed as described (53), they were resuspended in lysis buffer (100 mM sodium phosphate buffer, pH 8.0, 500 mM NaCl, 1 mM DTT) supplemented with protease inhibitors (Roche) and lysed in a French Press. Insoluble material was removed by centrifugation at 110 000g for 20 min and the resulting supernatant was adjusted to pH 8.0 and 5% glycerol and applied to a HisTrap column (GE Healthcare). The column was washed with binding buffer (lysis buffer supplemented with 20 mM imidazole) and eluted with a linear gradient of imidazole to 300 mM final concentration. Peak fractions were pooled and bound to a GSTrap column, washed with lysis buffer and eluted with elution buffer (50 mM Tris, pH 8.0, 50 mM glutathione, 500 mM NaCl, 1 mM DTT, 5% glycerol). Fractions were monitored by OD280 and peak fractions were pooled and dialyzed overnight against 50 mM Tris, pH 8.0, 1 mM DTT, 5% glycerol.
In order to inactivate the proofreading activity of human Pol ε, residues 275–277 (DIE) in the highly conserved ExoI motif were changed by site-directed mutagenesis to AIA and confirmed by sequencing.
Human Pol ε lacking the GST tag was prepared as described above, with the following modifications. The human Pol ε was coexpressed in autoinduction medium (52) with pRK603, which allows coexpression of TEV protease, at 25°C until the culture was saturated. Peak fractions from the HisTrap column were pooled, dialyzed into 50 mM HEPES, pH 7.5, 1 mM DTT, 5% glycerol and bound to SP sepharose. Bound protein was eluted with a 0–1 M NaCl gradient. Peak fractions were pooled, dialyzed into 50 mM Tris, pH 7.5, 1 mM DTT, 5% glycerol, 100 mM NaCl and bound to Q Sepharose. Bound protein was eluted with a 100 mM to 1 M NaCl gradient. Peak fractions were pooled, concentrated and passed through a pre-equilibrated Superdex-200 sizing column. Fractions containing the purified 140 kDa protein were pooled, dialyzed into 50 mM Tris, pH 8.0, 1 mM DTT, 5% glycerol and aliquots were frozen and stored at −80°C.
Primer extension assays
An 18-mer DNA oligo, 5′-CCTCTTCGCTATTACGCC-3′, was charged with 32P at its 5′-end by incubating with T4 polynucleotide kinase (Invitrogen) and γ-32P-labeled ATP (Perkin Elmer) for 30 min at 37°C. Unincorporated 32P was separated by passage over an illustra MicroSpin G-25 column (GE Healthcare) and the purified radiolabeled primer was then annealed to a complementary 45mer DNA oligonucleotide carrying the sequence, 5′-TTGCAGCACATCCCCCTTTCGCCAGCTGGCGTAATAGCGAAGAGG, at a final concentration of 2 µM. Reaction conditions were 50 mM Tris pH 7.4, 8 mM MgCl2, 1 mM DTT, 10% glycerol, 200 µM dNTPs, 100 nM DNA primer-template. Reactions were started by the addition of the indicated amounts of enzyme and incubated at 37°C for various timepoints. Reactions were stopped by the addition of an equal volume of 95% formamide, denatured for 5 min at 95°C, chilled on ice and then run on denaturing polyacrylamide gels. The gels were then dried and exposed to a phosphorimaging screen and scanned using a Typhoon Trio+ Imager (GE Healthcare). dNTP incorporation was quantitated using ImageQuant5.2 software(GE Healthcare). Exonuclease activity was assessed by withholding dNTPs from the reaction.
lacZ Mutant frequency determination
The lacZ forward mutation assay was performed essentially as described (54). Briefly, double-stranded M13mp2 DNA containing a 407-nt ssDNA gap was used as a substrate in reactions containing 0.15 nM DNA, 50 mM Tris–Cl, pH 7.4, 8 mM MgCl2, 2 mM DTT, 100 µg/ml BSA, 10% glycerol, 250 µM dNTPs and 1.5 nM Pol ε. After incubation at 37°C, the DNA was checked for complete gap-filling by 0.8% agarose gel electrophoresis at 60 V for 16 h. The filled gap product was then transfected into E. coli cells, which were used to determine the frequency of light blue and colorless plaques that occurred as a result of mutations arising during DNA synthesis. In this assay, accurate DNA synthesis yields dark blue plaques. LacZ mutant frequencies were calculated from combining at least two independent experiments unless otherwise indicated. DNA from mutant plaques was subsequently purified and the lacZ gene sequenced. Error rates were calculated as described (54).
Rate constant determinations by steady state kinetics
Misinsertion
Reaction mixtures consisted of 50 mM Tris pH 7.4, 8 mM MgCl2, 1 mM DTT, 10% glycerol, 100 nM DNA primer-template, 1 nM enzyme and variable amounts of either dATP or dTTP. Reactions were conducted under empirically determined single-hit conditions, such that dNTP incorporation was linear with time and did not exceed 20% primer utilization. Km and Vmax were calculated using non-linear regression fit to the Michaelis–Menten kinetics (GraphPad) and kcat, Fins and Fext were calculated as described (5). The oligonucleotides used as substrates were 5′-CCTCTTCGCTATTACGCC-3′ for the primer and 5′-TTGCAGCACATCCCCCTTTCGCCAGCTGGCGTAATAGCGAAGAGG-3′ for the template.
Extension from a terminal mispair
The oligonucleotides used were 5′-CCTCTTCGCTATTACGCCA-3′ for the primer containing a correctly paired primer terminus, 5′-CCTCTTCGCTATTACGCCT-3′ for the primer containing a mispaired primer terminus and 5′-TTGCAGCACATCCCCCTTTCGCCAGCTGGCGTAATAGCGAAGAGG-3′ for the template. Reaction mixtures were as described above, except that dGTP was used.
RESULTS
Expression and purification of recombinant human Pol ε
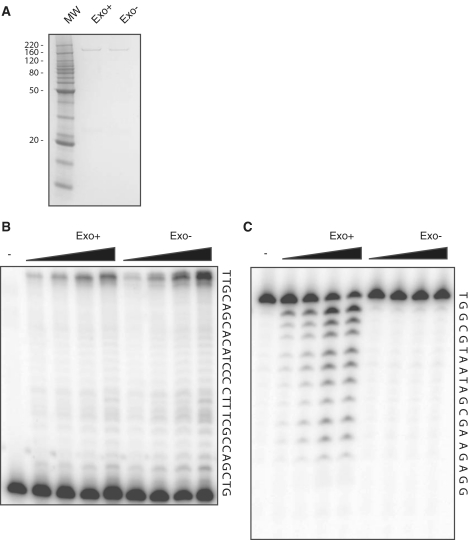
The purification of the endogenous 4-subunit DNA polymerase ε from both yeast and human cells is labor intensive (43,55). Johansson and colleagues circumvented this problem by expressing in yeast a 152 kDa fragment of the yeast enzyme. This fragment contains all six conserved polymerase and exonuclease motifs (56) and is similar in size to catalytically active fragments observed in earlier purifications of endogenous Pol ε (43). We undertook a similar approach to express and purify a fragment corresponding to the N-terminal 140 kDa of human Pol ε (Pol ε-N140), which also contains all six conserved polymerase and exonuclease domains. We have also taken advantage of a tandem affinity tag to facilitate purification. A GST tag was fused in frame to the N-terminus of Pol ε, while a 6x His tag was added to the C-terminus, resulting in a protein with a predicted molecular weight of 168 kDa. This allowed for a convenient two-step purification, first by passage over a Ni2+ column and elution with imidazole, followed by passage over a glutathione sepharose column and elution with glutathione. We recovered a highly purified 170 kDa form of the polymerase (Figure 1A) and verified its identity by mass spectrometry.
Figure 1.
Purification and polymerization and exonuclease assays of human Pol ε. (A) Purified Exo+and Exo− human Pol ε was loaded onto a 10% SDS–polyacrylamide gel and stained with Coomassie. Molecular weight marker (MW) is shown with selected molecular weights indicated in kDa. (B) Image of DNA synthesis reaction products resolved on a 16% denaturing polyacrylamide gel. Lanes 2–5 are 1, 2, 5 and 10 min at 37°C with 1 nM Exo+Pol ε. Lanes 6–9 are 1, 2, 5 and 10 min at 37°C with 1 nM Exo− Pol ε. Lane 1 is a control reaction with no enzyme. (C) Image of 3′–5′ exonuclease reaction products resolved on a 16% denaturing polyacrylamide gel. Lanes 2–5 are 1.4, 2.8, 5.6 and 8.6 nM Exo+ Pol ε. Lanes 6–9 are 1.4, 2.8, 5.6 and 8.6 nM Exo− Pol ε. Reactions were performed as described in ‘Materials and Methods’ section. Template DNA sequences are shown to the side of each substrate.
We next wanted to ensure that the purified form of Pol ε-N140 was active for DNA polymerase activity. We measured the ability of the catalytic fragment to incorporate dNTPs into a radiolabeled 18mer oligonucleotide primer annealed to a 45-mer DNA template in the presence of Mg2+. Both the exo+ and exo− forms of human Pol ε were able to catalyze nucleotide addition to the end of the primer (Figure 1B). Full-length product was observed even in the shortest time points measured under conditions in which the enzyme was limiting, indicating that the human Pol ε is a processive enzyme. Additionally, the amount of primer extended by the exo− enzyme was within 2-fold that of the exo+ enzyme, consistent with observations made with the yeast enzyme (56).
The high fidelity of yeast Pol ε is due in part to its proofreading exonuclease activity. Human Pol ε contains all three conserved exonuclease motifs and endogenous Pol ε purified from HeLa cells is exonuclease-proficient. We tested whether our human Pol ε-N140 was able to catalyze exonucleolytic degradation. The enzyme was incubated with a primer-template substrate containing a primer-terminal mispair under the same reaction conditions as the primer extension reactions but lacking dNTPs. The human Pol ε was proficient for robust 3′–5′ exonuclease activity (Figure 1C).
In order to ensure that the exonuclease-inactivation mutation truly inactivated proofreading activity, we carried out identical reactions using the enzyme with the putative catalytic exonuclease aspartate residues mutated to alanines. The putative exonuclease-inactive human Pol ε enzyme showed no exonuclease activity even upon extended incubation with excess enzyme (Figure 1C), demonstrating that the exo- Pol ε mutant is exonuclease-deficient.
Fidelity of human DNA polymerase ε
To determine the in vitro replication fidelity of human Pol ε, we used the lacZ forward mutation assay. In this assay, a DNA polymerase synthesizes across a 407-nt single strand DNA gap contained within a 7.2 kb double strand circular M13mp2 DNA. It should be noted that this is the identical substrate used to determine the replication fidelities of yeast Pols ε and δ as well as human Pol δ (57–59). The assay allows the observation of all 12 possible base–base mismatches, as well as insertions and deletions in a large number of sequence contexts, from which individual error rates can be calculated (54). Additionally, more complex mutations like large deletions between direct repeats can be observed. Each human Pol ε-N140 preparation was observed by agarose gel electrophoresis to completely fill in the single strand gap (data not shown).
Exonuclease-proficient human Pol ε-N140 gave an overall lacZ mutant frequency of 6.5 × 10−4 (Table 1), which is essentially the background of the assay [6.4 × 10−4 ref. (60)]. This is similar to the only previously reported value of the fidelity of mammalian Pol ε, 7.1 × 10−4, (61), which was of the endogenous enzyme purified from calf thymus that is proficient for exonucleolytic proofreading. Most, if not all, errors in our collection were likely due to previously characterized base damage occurring during substrate preparation and thus were not sequenced.
Table 1.
lacZ mutant frequencies for wild-type and exo− human Pol ε
| hPol ε Exo+ | hPol ε Exo− | |
|---|---|---|
| Total number of plaques | 27 792 | 26 089 |
| Total number of mutant plaques | 18 | 119 |
| lacZ mutant frequency (×10−4) | 6.5 | 46 |
| Number of mutants sequenced | nd | 119 |
| Total number of mutations | nd | 123 |
nd = not determined.
Exonuclease-deficient human Pol ε-N140 gave an overall lacZ mutant frequency ≥7.1-fold higher than the exonuclease-proficient enzyme (46 × 10−4, Table 1). This increase indicates that the proofreading contributed significantly to overall human Pol ε fidelity, similar to what was seen with the yeast Pol ε (59). These data came from two independent experiments using two different gapped substrate preparations. Since the data from both sets were similar to each other and statistically significant, the data from both experiments were subsequently combined into one set.
While no high resolution structure of the catalytic subunit from any Pol ε enzyme currently exists, the crystal structures of a growing number of related B family DNA polymerases have been reported (62–64). In each, the N-terminus is not in direct contact with the polymerase active site, indicating that the GST tag is unlikely to affect DNA synthesis or fidelity. However, the N-terminal domain of yeast Pol δ does contact the last downstream unpaired template nucleotide (63), indicating the possibility of altering fidelity indirectly. We directly tested whether the N-terminal GST tag affected fidelity by measuring lacZ mutation frequencies in both its presence and its absence. We used the TEV cleavage sequence present in the construct and the AcTEV protease to remove the GST tag from the recombinant protein. There was no difference in lacZ mutant frequency between two independent experiments where the GST tag was undisturbed (40 × 10−4 and 56 × 10−4) and one experiment after the GST tag was cleaved (45 × 10−4), such that data from all three experiments were combined. The close agreement in fidelity between the two constructs strongly suggests that the GST tag does not interfere with human Pol ε replication fidelity.
Error rates for different classes of mutations
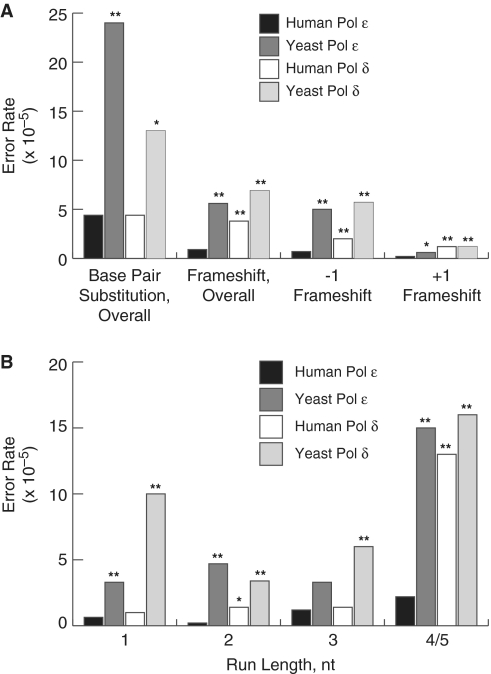
In order to determine both the nature and location of errors made by human Pol ε, we sequenced the lacZα-complementation sequence from a collection of independent mutant clones. The most common errors observed were base pair substitutions, though single-base deletions and insertions were detected for each construct and were spread throughout the entire detectable sequence. Additionally, only 3.4% (4 out of 119) of the clones contained more than one mutation in the lacZ sequence, which is too few to be informative regarding clustered mutations. Based on the observed mutant frequency, the relative amounts of each error made and the known number of detectable sites for each error we were able to calculate the rates at which exonuclease-deficient human Pol ε made each class of error, as well as the rates for specific mispairs (Figure 2A).
Figure 2.
Base substitution and frameshift error rates for human and yeast Pol ε and human and yeast Pol δ. (A) Error rates for base pair substitutions, overall frameshifts, −1 and +1 frameshifts. Error rates were calculated as described (54). Error rates are shown for human Pol ε (black bars, this study), yeast Pol ε [dark gray bars, (59)], human Pol δ [white bars, (58)] and yeast Pol δ [light gray bars, (57)]. P-values are shown above each comparison to a human Pol ε error rate where the difference was found to be significant. The (asterisk and double asterisk) symbols indicate P-values where P ≤ 0.05 and 0.001, respectively. (B) Single nucleotide deletion dependence on homonucleotide run length. Error rates were calculated for −1 deletions in non-iterated nucleotides (1) or runs of the indicated number nucleotides (2, 3, 4/5). The legend and source of data are as in (A).
Base pair substitutions
Human Pol ε was highly accurate for overall base–base mispairs (4.4 × 10−5, Figure 2A). This is 5.5-fold more accurate than the average base pair substitution error rate reported for yeast Pol ε (59). While significantly more accurate for base substitutions than the yeast enzyme, the proportion of base–base mismatches to all other errors remained similar between the two orthologs under similar reaction conditions (compare 72 versus 76%).
The most common base substitution errors observed with the exonuclease-deficient human Pol ε were T→A transversions (4.3 × 10−5, Figure 3). These errors result from misinsertion of an incoming dTMP opposite a template T and are the second most common base pair substitution made by the yeast Pol ε [24 × 10−5, ref. (59)].
Figure 3.
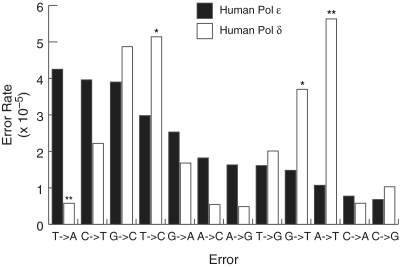
Fidelity of individual base pair substitutions for human Pol ε and human Pol δ. Error rates for each of the 12 possible mispairs for the exonuclease-deficient forms of human Pol ε (black bars, this study) and human Pol δ [white bars, (58)] were calculated as described (54). P-values are shown above each comparison to a human Pol ε error rate where the difference was found to be significant. The (asterisk and double asterisk) symbols indicate P-values where P ≤ 0.05 and 0.005, respectively.
The second most commonly observed errors with exo− human Pol ε were C→T transitions (4.0 × 10−5, Figure 3). Misinsertion of dAMP opposite a template C followed by polymerase extension results in C→T transitions. This also appears to be an error common to Pol ε as it is the most frequent base–base mispair made by the yeast Pol ε (59). However, C→T is also considered a background error in the lacZ forward mutation assay. They are thought to arise from DNA synthesis past cytosines that were deaminated in the DNA template during substrate preparation (54). C→T transitions have now been seen with multiple DNA Pol ε preparations from different organisms and using a number of independently produced substrate preparations, so it is likely that a high number of C•dA errors is a unique error signature of Pol ε.
Exo− human Pol ε generated A→T transversions at a relatively low rate (1.1 × 10−5, Figure 3). This discrimination against A•dA mispairs by the human enzyme distinguishes it from yeast Pol ε as well as both yeast and human Pol δ. Yeast Pol ε makes A→T transversions at an 11-fold higher rate than that observed for the human enzyme. Yeast and human Pol δ both make A→T transversions at a relatively high rate (12 × 10−5 and 5.2 × 10−5, respectively).
Single-base deletions
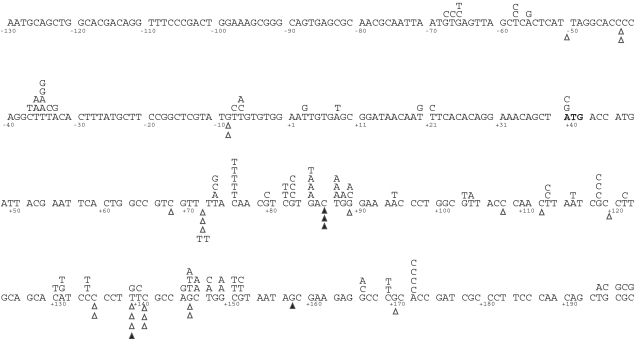
Human Pol ε was highly accurate for deletion of a single nucleotide (0.71 × 10−5, Figure 2A). The deletions that did occur were distributed evenly across the spectrum, with no hotspots identified (Figure 4). Remarkably, this enzyme was nearly as accurate for 4–5 homonucleotide runs as it was for shorter runs or non-iterated nucleotides (Figure 2B, black bars) (65).
Figure 4.
Spectrum of base substitution and single-base frameshift mutations made by exonuclease-deficient human Pol ε in the lacZ gene. Base substitutions are shown above the sequence, while deletions (open triangles) and insertions (closed triangles) are shown below. The numbering is relative to the +1 at the transcription start site.
Human Pol ε also generated a unique spectrum of single-base deletions in that it was highly accurate for frameshifts involving a template A. No deletions of A nucleotides were observed either in non-iterated adenine nucleotides or in A runs of any length, resulting in an error rate of ≤0.14 × 10−5 for adenine deletions. Taken together with the base pair substitution data, this observation suggests that human Pol ε may be particularly proficient at discriminating between correctly and incorrectly paired bases at template adenines.
Single-base insertions
With an observed error rate of 0.16 × 10−5 (Figure 2A), human Pol ε was highly accurate for insertion errors. While four out of five insertion errors were at sites flanked on either side by different nucleotides, the sample size is too small to determine the extent to which human Pol ε is able generate insertion errors through mechanisms different than the classical Streisinger slippage. The small sample size also complicates analysis of the yeast Pol ε, which is also accurate for insertion errors (59). Regardless, both yeast Pol ε and human Pol ε differ remarkably from yeast and human Pol δ, for which almost all insertion errors either occur in existing homonucleotide runs or expand an existing nucleotide and can be accounted for by slippage intermediates.
Steady state kinetic analysis
Results from the lacZ forward mutation assay showed that T→A transversions were the most common types of base substitution error made by exonuclease-deficient human Pol ε (Figure 3). The increased mispair efficiency could be due to increased misinsertion efficiency (Fins) of dTMP opposite template T, increased extension efficiency (Fext) from a pre-existing primer terminal T•T mispair or a combination of the two. We tested at which steps human Pol ε was more efficient by using steady-state kinetics to measure Km and kcat, which were calculated as described (66,67). The misinsertion and extension efficiencies, Fins and Fext, are defined as (kcat/Km)wrong /(kcat/Km)right where wrong and right refer to the incorrectly and correctly paired nucleotides, respectively.
The catalytic efficiency for misinsertion decreased dramatically for human Pol ε relative to that for correct insertion (Compare 0.55 to 69, Table 2). This was mainly due to a 36-fold increase in the Km for the incorrect dTTP relative to correct dATP, as the kcat decreased only 3.5-fold for the incorrect nucleotide. The larger degree of change in Km relative to kcat is consistent with observations from other B family DNA polymerases (68,69).
Table 2.
Steady-state rate constants for human DNA Pol ε
| Primer terminus | Incoming dNTP | Km (µM) | kcat (min−1) | kcat/Km | Fins |
|---|---|---|---|---|---|
| Correct insertion versus misinsertion | |||||
| 5′ –T- | dATP | 0.055 ± 0.025 | 3.8 ± 0.25 | 69 | 1 |
| 3′ –A–T– | |||||
| 5′ –T– | dTTP | 2.0 ± 1.4 | 1.1 ± 0.053 | 0.55 | 8.0 × 10−3 |
| 3′ –A–T– | |||||
| Extension from matched versus mismatched primer termini | Fext | ||||
| 5′ –T– | dGTP | 0.028 ± 0.018 | 1.0 ± 0.087 | 36 | 1 |
| 3′ –A–C– | |||||
| 5′ –T– | dGTP | 0.087 ± 0.069 | 1.2 ± 0.010 | 14 | 0.39 |
| 3′ –T–C– | |||||
The Fext for human Pol ε for a preexisting T•T mispair at the primer terminus was 49-fold higher than the Fins observed for misinsertion (compare 3.9 × 10−1 to 8.0 × 10−3, Table 2). The difference between Fins and Fext is again largely explained by differences in Km. While the Km values for the next correct nucleotide were similar for the two experiments (compare 0.055 versus 0.028 µM for insertion and extension, respectively, Table 2), the Km for insertion of the correct nucleotide extending from a preexisting primer terminal T•T mispair only increased 3.1-fold (0.087 versus 0.028 µM) while the kcat remained similar to that for a correctly paired A•T primer terminus. Thus, while human Pol ε was slower to insert the incorrectly paired dT opposite template T, it readily extended once T•T mispair was formed.
DISCUSSION
Our in vitro studies strongly suggest that human Pol ε, which contains an intrinsic 3′–5′ proofreading exonuclease activity, is a highly accurate DNA polymerase. Even when its exonuclease activity is inactivated and Pol ε is unable to correct its own replication errors, its intrinsic fidelity remains high. This means that the human enzyme has an intrinsically strong base selection function. With respect to specific types of errors, human Pol ε is faithful for base–base mismatches, though it retains a unique error specificity. A major contribution to the high degree of accuracy observed for human Pol ε comes from its extremely low error rate for introducing frameshift errors, particularly in the context of long repeated sequences. In addition, both exonuclease-proficient and -deficient human Pol ε enzymes are processive.
The results presented here allow comparison to be made between the replication fidelities of the yeast and human homologues of both Pol ε and Pol δ. About 75% of the errors made by Pol ε (from either humans or yeast) are base pair substitutions. In contrast, such substitutions constitute ∼43% of the errors made by human and yeast Pol δ. However, the average base pair substitution error rates observed for both human Pols ε and δ were essentially identical [4.4 × 10−5, Figure 2A, this study versus 4.4 × 10−5, (58)] when using the same DNA substrate. A direct comparison of human Pol ε and human Pol δ error rates for specific mispairs shows that even though the two enzymes make base pair substitutions at the same average rate overall, they have different error specificities for individual mispairs (Figure 3). The most striking difference in base pair substitution specificity is seen for T→A and A→T transversions. Both yeast and human Pol ε make T→A transversions at a relatively high rate. This is in direct contrast with Pol δ, for which T→A transversions are among the least commonly made errors [Figure 3 and ref. (57)]. The high rates of T→A transversions observed for both human and yeast Pol ε are unique among B family polymerases. T→A transversions are among the rarest mispairs seen with phage RB69 gp43 and eukaryotic Pol α and Pol ζ (58,70–72). The differences between the enzymes might be indicative of differences in geometry of the polymerase active site in the different taxa and Pol B subfamilies. In fact, each of the three polymerase motifs shared by all B family polymerases contains residues that are conserved within Pol ε homologs but diverged in other B family polymerases. This feature might be reflected in reduced discrimination, which may be biologically relevant since T•T mispairs are among the least well corrected by the mismatch repair system (73).
Since human Pol ε has a relatively high T→A error rate, we compared steady-state rate constants for insertion versus misinsertion and for extension from correct versus incorrect primer termini. Human Pol ε strongly discriminated at the insertion step (Fins for dTTP, Table 2), with the small differences in turnover seen for the correct versus incorrect nucleotide similar to the same values measured for human Pol δ (68). This high discrimination at the insertion step is in contrast to the poor discrimination seen with extension from an existing mispair. Human Pol ε efficiently extends from an existing T•T mispair at the primer terminus with similar efficiency to a correctly paired A•T at the primer terminus (Fext, Table 2). These observations suggest that human Pol ε arrives at an increased propensity for T→A transversions primarily through mispair extension. The mechanism responsible for the relative lack of A→T transversions for the human enzyme relative to the yeast enzyme remains an open and interesting question.
While yeast Pols ε and δ delete single nucleotides overall at rates similar to each other, human Pol ε deletes single nucleotides at a 2.8-fold lower rate than human Pol δ. It was noted for yeast Pol ε that while deletions of non-iterated nucleotides occurred at the same rate as deletions from runs of 2 nt or 3 nt, deletions from four to five homonucleotide runs were more frequent, similar to patterns observed for yeast Pol α and RB69 DNA Pol (59). Yeast and human Pol δ demonstrate a similar pattern with the exception that yeast Pol δ deletes non-iterated nucleotides at a relatively high rate. Human Pol ε is similar too, in that the error rates for non-iterated, 2- and 3-nt runs were similar to each other (Figure2B, black bars). However, human Pol ε is much more accurate for homonucleotide runs of four or five bases, the longest runs present in the target sequence. As this pattern is different from other B family DNA polymerases, it raises the possibility that the human Pol ε active site has unique features that result in higher fidelity when in contact with extended runs of the same nucleotide.
One important question is why the human enzyme is significantly more accurate than its yeast homolog. Several factors might contribute to the high fidelity. First, if the yeast model of dividing the labor at the replication fork is applicable to human cells and Pol ε is responsible for leading strand replication, then it would also be responsible for synthesizing many more base pairs in the human genome, which is 260 times larger than that of yeast. Increased accuracy would help reduce errors made during genome duplication prior to the involvement of subsequent repair pathways. Second, if the role of Pol ε in human cells has assumed different priorities from its yeast homolog, then the differing fidelity constraints might have selected for a more accurate enzyme in humans. Another possibility, raised previously by immunohistochemistry experiments, is the impact of heterochromatin replication (17). Heterochromatin contains large numbers of repetitive sequence elements. It is much more abundant in human cells, where it makes up ∼20% of the genome, than in yeast, where it is largely restricted to the mating type loci, rDNA and subtelomeric regions (74). Replicating heterochromatin would likely require the presence of a polymerase that is highly faithful for replicating repetitive sequences, a task for which human Pol ε, with its high accuracy for homonucleotide runs, would be well suited.
Another important role for the specificity of Pol ε replication fidelity is in suppression of particular types of tumors (51). The results of Albertson et al. (51) make clear that mammalian Pols δ and ε, or at least their exonuclease activities, differ significantly from those of their yeast homologues. In yeast the effects on mutation rate of inactivating proofreading are consistently stronger for Pol δ mutants than for Pol ε mutants (75,76). Mouse is the interesting exception, as Pol ε exo− mutants have higher mutant frequencies in each tissue examined. Mutation rates in Pol ε exo− MEFs are equal to or higher than those derived from Pol δ exo− mice (51). A sequenced collection of spontaneous Hprt1 mutants from Pol ε exo−/exo− MEFs revealed predominantly base substitution mutations with no frameshift errors. At this level, the in vivo mouse error spectrum is similar to what we observed with human Pol ε in vitro. However, we should point out that interpretations are being made with caution, since any replication errors made by the proofreading-deficient polymerases in the mouse system in vivo are presumably subject to mismatch error correction. Additionally, the in vitro replication fidelity of mouse Pol ε is unknown.
The unique error signature of exonuclease-deficient human Pol ε can now hopefully be used in combination with in vivo data to determine the precise roles of Pol ε during replication in humans. Additionally, it should now be possible to investigate the effects that various active site residues have on DNA synthesis fidelity and what roles these residues might have in preventing mutagenesis, microsatellite instability and cancer in human cells.
FUNDING
National Institutes of Health (ES016780 and RR020152); Tulane University, start-up funds. Funding for open access charge: National Institutes of Health (4R00ES016780).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank Dr Jim Karam and Dr Art Lustig for thoughtful comments on the article. We would also like to thank Dr Robert Petrovich and Lori Edwards of the Protein Expression Core Facility at NIEHS for expert technical assistance and advice. The authors also thank Dr Chau-Wen Chou at the LSUHSC Proteomics Core Facility, New Orleans, for mass spectrometry help.
REFERENCES
- 1.Kunkel TA, Burgers PM. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 2008;18:521–527. doi: 10.1016/j.tcb.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waga S, Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature. 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- 3.Shcherbakova PV, Pavlov YI. 3′–>5′ exonucleases of DNA polymerases epsilon and delta correct base analog induced DNA replication errors on opposite DNA strands in Saccharomyces cerevisiae. Genetics. 1996;142:717–726. doi: 10.1093/genetics/142.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karthikeyan R, Vonarx EJ, Straffon AF, Simon M, Faye G, Kunz BA. Evidence from mutational specificity studies that yeast DNA polymerases delta and epsilon replicate different DNA strands at an intracellular replication fork. J. Mol. Biol. 2000;299:405–419. doi: 10.1006/jmbi.2000.3744. [DOI] [PubMed] [Google Scholar]
- 5.Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol. Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masumoto H, Sugino A, Araki H. Dpb11 controls the association between DNA polymerases alpha and epsilon and the autonomously replicating sequence region of budding yeast. Mol. Cell Biol. 2000;20:2809–2817. doi: 10.1128/mcb.20.8.2809-2817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muramatsu S, Hirai K, Tak YS, Kamimura Y, Araki H. CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol (epsilon}, and GINS in budding yeast. Genes Dev. 2010;24:602–612. doi: 10.1101/gad.1883410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 10.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl Acad. Sci. USA. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Northam MR, Robinson HA, Kochenova OV, Shcherbakova PV. Participation of DNA polymerase zeta in replication of undamaged DNA in Saccharomyces cerevisiae. Genetics. 2010;184:27–42. doi: 10.1534/genetics.109.107482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morrison A, Bell JB, Kunkel TA, Sugino A. Eukaryotic DNA polymerase amino acid sequence required for 3′—5′ exonuclease activity. Proc Natl Acad. Sci. USA. 1991;88:9473–9477. doi: 10.1073/pnas.88.21.9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simon M, Giot L, Faye G. The 3′ to 5′ exonuclease activity located in the DNA polymerase delta subunit of Saccharomyces cerevisiae is required for accurate replication. EMBO J. 1991;10:2165–2170. doi: 10.1002/j.1460-2075.1991.tb07751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dua R, Levy DL, Campbell JL. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae pol epsilon and its unexpected ability to support growth in the absence of the DNA polymerase domain. J. Biol. Chem. 1999;274:22283–22288. doi: 10.1074/jbc.274.32.22283. [DOI] [PubMed] [Google Scholar]
- 15.Kesti T, Flick K, Keranen S, Syvaoja JE, Wittenberg C. DNA polymerase epsilon catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol. Cell. 1999;3:679–685. doi: 10.1016/s1097-2765(00)80361-5. [DOI] [PubMed] [Google Scholar]
- 16.Pavlov YI, Shcherbakova PV. DNA polymerases at the eukaryotic fork-20 years later. Mutat. Res. 2010;685:45–53. doi: 10.1016/j.mrfmmm.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuss J, Linn S. Human DNA polymerase epsilon colocalizes with proliferating cell nuclear antigen and DNA replication late, but not early, in S phase. J. Biol. Chem. 2002;277:8658–8666. doi: 10.1074/jbc.M110615200. [DOI] [PubMed] [Google Scholar]
- 18.Nishida C, Reinhard P, Linn S. DNA repair synthesis in human fibroblasts requires DNA polymerase delta. J. Biol. Chem. 1988;263:501–510. [PubMed] [Google Scholar]
- 19.Syvaoja J, Linn S. Characterization of a large form of DNA polymerase delta from HeLa cells that is insensitive to proliferating cell nuclear antigen. J. Biol. Chem. 1989;264:2489–2497. [PubMed] [Google Scholar]
- 20.Syvaoja J, Suomensaari S, Nishida C, Goldsmith JS, Chui GS, Jain S, Linn S. DNA polymerases alpha, delta, and epsilon: three distinct enzymes from HeLa cells. Proc. Natl Acad. Sci. USA. 1990;87:6664–6668. doi: 10.1073/pnas.87.17.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aboussekhra A, Biggerstaff M, Shivji MK, Vilpo JA, Moncollin V, Podust VN, Protic M, Hubscher U, Egly JM, Wood RD. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80:859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- 22.Mozzherin DJ, Fisher PA. Human DNA polymerase epsilon: enzymologic mechanism and gap-filling synthesis. Biochemistry. 1996;35:3572–3577. doi: 10.1021/bi952142p. [DOI] [PubMed] [Google Scholar]
- 23.Shivji MK, Podust VN, Hubscher U, Wood RD. Nucleotide excision repair DNA synthesis by DNA polymerase epsilon in the presence of PCNA, RFC, and RPA. Biochemistry. 1995;34:5011–5017. doi: 10.1021/bi00015a012. [DOI] [PubMed] [Google Scholar]
- 24.Wu X, Guo D, Yuan F, Wang Z. Accessibility of DNA polymerases to repair synthesis during nucleotide excision repair in yeast cell-free extracts. Nucleic Acids Res. 2001;29:3123–3130. doi: 10.1093/nar/29.14.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogi T, Limsirichaikul S, Overmeer RM, Volker M, Takenaka K, Cloney R, Nakazawa Y, Niimi A, Miki Y, Jaspers NG, et al. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol. Cell. 2010;37:714–727. doi: 10.1016/j.molcel.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 26.Sobol RW, Horton JK, Kuhn R, Gu H, Singhal RK, Prasad R, Rajewsky K, Wilson SH. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 27.Parlanti E, Locatelli G, Maga G, Dogliotti E. Human base excision repair complex is physically associated to DNA replication and cell cycle regulatory proteins. Nucleic Acids Res. 2007;35:1569–1577. doi: 10.1093/nar/gkl1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navas TA, Sanchez Y, Elledge SJ. RAD9 and DNA polymerase epsilon form parallel sensory branches for transducing the DNA damage checkpoint signal in Saccharomyces cerevisiae. Genes Dev. 1996;10:2632–2643. doi: 10.1101/gad.10.20.2632. [DOI] [PubMed] [Google Scholar]
- 29.Navas TA, Zhou Z, Elledge SJ. DNA polymerase epsilon links the DNA replication machinery to the S phase checkpoint. Cell. 1995;80:29–39. doi: 10.1016/0092-8674(95)90448-4. [DOI] [PubMed] [Google Scholar]
- 30.Lou H, Komata M, Katou Y, Guan Z, Reis CC, Budd M, Shirahige K, Campbell JL. Mrc1 and DNA polymerase epsilon function together in linking DNA replication and the S phase checkpoint. Mol. Cell. 2008;32:106–117. doi: 10.1016/j.molcel.2008.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osborn AJ, Elledge SJ. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 2003;17:1755–1767. doi: 10.1101/gad.1098303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calzada A, Hodgson B, Kanemaki M, Bueno A, Labib K. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev. 2005;19:1905–1919. doi: 10.1101/gad.337205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee J, Gold DA, Shevchenko A, Shevchenko A, Dunphy WG. Roles of replication fork-interacting and Chk1-activating domains from Claspin in a DNA replication checkpoint response. Mol. Biol. Cell. 2005;16:5269–5282. doi: 10.1091/mbc.E05-07-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ehrenhofer-Murray AE, Kamakaka RT, Rine J. A role for the replication proteins PCNA, RF-C, polymerase epsilon and Cdc45 in transcriptional silencing in Saccharomyces cerevisiae. Genetics. 1999;153:1171–1182. doi: 10.1093/genetics/153.3.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iida T, Araki H. Noncompetitive counteractions of DNA polymerase epsilon and ISW2/yCHRAC for epigenetic inheritance of telomere position effect in Saccharomyces cerevisiae. Mol. Cell Biol. 2004;24:217–227. doi: 10.1128/MCB.24.1.217-227.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith JS, Caputo E, Boeke JD. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol. Cell Biol. 1999;19:3184–3197. doi: 10.1128/mcb.19.4.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kukimoto I, Elderkin S, Grimaldi M, Oelgeschlager T, Varga-Weisz PD. The histone-fold protein complex CHRAC-15/17 enhances nucleosome sliding and assembly mediated by ACF. Mol. Cell. 2004;13:265–277. doi: 10.1016/s1097-2765(03)00523-9. [DOI] [PubMed] [Google Scholar]
- 38.Poot RA, Dellaire G, Hulsmann BB, Grimaldi MA, Corona DF, Becker PB, Bickmore WA, Varga-Weisz PD. HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. EMBO J. 2000;19:3377–3387. doi: 10.1093/emboj/19.13.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tackett AJ, Dilworth DJ, Davey MJ, O’Donnell M, Aitchison JD, Rout MP, Chait BT. Proteomic and genomic characterization of chromatin complexes at a boundary. J. Cell Biol. 2005;169:35–47. doi: 10.1083/jcb.200502104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kesti T, Frantti H, Syvaoja JE. Molecular cloning of the cDNA for the catalytic subunit of human DNA polymerase epsilon. J. Biol. Chem. 1993;268:10238–10245. [PubMed] [Google Scholar]
- 41.Li Y, Asahara H, Patel VS, Zhou S, Linn S. Purification, cDNA cloning, and gene mapping of the small subunit of human DNA polymerase epsilon. J. Biol. Chem. 1997;272:32337–32344. doi: 10.1074/jbc.272.51.32337. [DOI] [PubMed] [Google Scholar]
- 42.Crute JJ, Wahl AF, Bambara RA. Purification and characterization of two new high molecular weight forms of DNA polymerase delta. Biochemistry. 1986;25:26–36. doi: 10.1021/bi00349a005. [DOI] [PubMed] [Google Scholar]
- 43.Hamatake RK, Hasegawa H, Clark AB, Bebenek K, Kunkel TA, Sugino A. Purification and characterization of DNA polymerase II from the yeast Saccharomyces cerevisiae. Identification of the catalytic core and a possible holoenzyme form of the enzyme. J. Biol. Chem. 1990;265:4072–4083. [PubMed] [Google Scholar]
- 44.Uitto L, Halleen J, Hentunen T, Hoyhtya M, Syvaoja JE. Structural relationship between DNA polymerases epsilon and epsilon* and their occurrence in eukaryotic cells. Nucleic Acids Res. 1995;23:244–247. doi: 10.1093/nar/23.2.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu W, Linn S. Proteolysis of the human DNA polymerase epsilon catalytic subunit by caspase-3 and calpain specifically during apoptosis. Nucleic Acids Res. 2000;28:4180–4188. doi: 10.1093/nar/28.21.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dua R, Edwards S, Levy DL, Campbell JL. Subunit interactions within the Saccharomyces cerevisiae DNA polymerase epsilon (pol epsilon) complex. Demonstration of a dimeric pol epsilon. J. Biol. Chem. 2000;275:28816–28825. doi: 10.1074/jbc.M002376200. [DOI] [PubMed] [Google Scholar]
- 47.Li Y, Pursell ZF, Linn S. Identification and cloning of two histone fold motif-containing subunits of HeLa DNA polymerase epsilon. J. Biol. Chem. 2000;275:23247–23252. doi: 10.1074/jbc.M002548200. [DOI] [PubMed] [Google Scholar]
- 48.Asturias FJ, Cheung IK, Sabouri N, Chilkova O, Wepplo D, Johansson E. Structure of Saccharomyces cerevisiae DNA polymerase epsilon by cryo-electron microscopy. Nat. Struct. Mol. Biol. 2006;13:35–43. doi: 10.1038/nsmb1040. [DOI] [PubMed] [Google Scholar]
- 49.Chui G, Linn S. Further characterization of HeLa DNA polymerase epsilon. J. Biol. Chem. 1995;270:7799–7808. doi: 10.1074/jbc.270.14.7799. [DOI] [PubMed] [Google Scholar]
- 50.Chilkova O, Stenlund P, Isoz I, Stith CM, Grabowski P, Lundstrom EB, Burgers PM, Johansson E. The eukaryotic leading and lagging strand DNA polymerases are loaded onto primer-ends via separate mechanisms but have comparable processivity in the presence of PCNA. Nucleic Acids Res. 2007;35:6588–6597. doi: 10.1093/nar/gkm741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Albertson TM, Ogawa M, Bugni JM, Hays LE, Chen Y, Wang Y, Treuting PM, Heddle JA, Goldsby RE, Preston BD. DNA polymerase epsilon and delta proofreading suppress discrete mutator and cancer phenotypes in mice. Proc. Natl Acad. Sci. USA. 2009;106:17101–17104. doi: 10.1073/pnas.0907147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 53.Magnusdottir A, Johansson I, Dahlgren LG, Nordlund P, Berglund H. Enabling IMAC purification of low abundance recombinant proteins from E. coli lysates. Nat. Methods. 2009;6:477–478. doi: 10.1038/nmeth0709-477. [DOI] [PubMed] [Google Scholar]
- 54.Bebenek K, Kunkel TA. Analyzing fidelity of DNA polymerases. Methods Enzymol. 1995;262:217–232. doi: 10.1016/0076-6879(95)62020-6. [DOI] [PubMed] [Google Scholar]
- 55.Chui GS, Linn S. Purification of mammalian polymerases: DNA polymerase epsilon. Methods Enzymol. 1995;262:93–98. doi: 10.1016/0076-6879(95)62012-5. [DOI] [PubMed] [Google Scholar]
- 56.Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Regulation of B family DNA polymerase fidelity by a conserved active site residue: characterization of M644W, M644L and M644F mutants of yeast DNA polymerase epsilon. Nucleic Acids Res. 2007;35:3076–3086. doi: 10.1093/nar/gkm132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fortune JM, Pavlov YI, Welch CM, Johansson E, Burgers PM, Kunkel TA. Saccharomyces cerevisiae DNA polymerase delta: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J. Biol. Chem. 2005;280:29980–29987. doi: 10.1074/jbc.M505236200. [DOI] [PubMed] [Google Scholar]
- 58.Schmitt MW, Matsumoto Y, Loeb LA. High fidelity and lesion bypass capability of human DNA polymerase delta. Biochimie. 2009;91:1163–1172. doi: 10.1016/j.biochi.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shcherbakova PV, Pavlov YI, Chilkova O, Rogozin IB, Johansson E, Kunkel TA. Unique error signature of the four-subunit yeast DNA polymerase epsilon. J. Biol. Chem. 2003;278:43770–43780. doi: 10.1074/jbc.M306893200. [DOI] [PubMed] [Google Scholar]
- 60.Kunkel TA. The mutational specificity of DNA polymerase-beta during in vitro DNA synthesis. Production of frameshift, base substitution, and deletion mutations. J. Biol. Chem. 1985;260:5787–5796. [PubMed] [Google Scholar]
- 61.Thomas DC, Roberts JD, Sabatino RD, Myers TW, Tan CK, Downey KM, So AG, Bambara RA, Kunkel TA. Fidelity of mammalian DNA replication and replicative DNA polymerases. Biochemistry. 1991;30:11751–11759. doi: 10.1021/bi00115a003. [DOI] [PubMed] [Google Scholar]
- 62.Berman AJ, Kamtekar S, Goodman JL, Lazaro JM, de Vega M, Blanco L, Salas M, Steitz TA. Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases. EMBO J. 2007;26:3494–3505. doi: 10.1038/sj.emboj.7601780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase delta. Nat. Struct. Mol. Biol. 2009;16:979–986. doi: 10.1038/nsmb.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang J, Sattar AK, Wang CC, Karam JD, Konigsberg WH, Steitz TA. Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69. Cell. 1997;89:1087–1099. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- 65.Kesti T, McDonald WH, Yates JR, 3rd, Wittenberg C. Cell cycle-dependent phosphorylation of the DNA polymerase epsilon subunit, Dpb2, by the Cdc28 cyclin-dependent protein kinase. J. Biol. Chem. 2004;279:14245–14255. doi: 10.1074/jbc.M313289200. [DOI] [PubMed] [Google Scholar]
- 66.Boosalis MS, Petruska J, Goodman MF. DNA polymerase insertion fidelity. Gel assay for site-specific kinetics. J. Biol. Chem. 1987;262:14689–14696. [PubMed] [Google Scholar]
- 67.Petruska J, Goodman MF, Boosalis MS, Sowers LC, Cheong C, Tinoco I., Jr Comparison between DNA melting thermodynamics and DNA polymerase fidelity. Proc. Natl Acad. Sci. USA. 1988;85:6252–6256. doi: 10.1073/pnas.85.17.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Choi JY, Chowdhury G, Zang H, Angel KC, Vu CC, Peterson LA, Guengerich FP. Translesion synthesis across O6-alkylguanine DNA adducts by recombinant human DNA polymerases. J. Biol. Chem. 2006;281:38244–38256. doi: 10.1074/jbc.M608369200. [DOI] [PubMed] [Google Scholar]
- 69.Mendelman LV, Petruska J, Goodman MF. Base mispair extension kinetics. Comparison of DNA polymerase alpha and reverse transcriptase. J. Biol. Chem. 1990;265:2338–2346. [PubMed] [Google Scholar]
- 70.Bebenek A, Carver GT, Dressman HK, Kadyrov FA, Haseman JK, Petrov V, Konigsberg WH, Karam JD, Drake JW. Dissecting the fidelity of bacteriophage RB69 DNA polymerase: site-specific modulation of fidelity by polymerase accessory proteins. Genetics. 2002;162:1003–1018. doi: 10.1093/genetics/162.3.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Niimi A, Limsirichaikul S, Yoshida S, Iwai S, Masutani C, Hanaoka F, Kool ET, Nishiyama Y, Suzuki M. Palm mutants in DNA polymerases alpha and eta alter DNA replication fidelity and translesion activity. Mol. Cell Biol. 2004;24:2734–2746. doi: 10.1128/MCB.24.7.2734-2746.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhong X, Garg P, Stith CM, Nick McElhinny SA, Kissling GE, Burgers PM, Kunkel TA. The fidelity of DNA synthesis by yeast DNA polymerase zeta alone and with accessory proteins. Nucleic Acids Res. 2006;34:4731–4742. doi: 10.1093/nar/gkl465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brown TC, Jiricny J. Different base/base mispairs are corrected with different efficiencies and specificities in monkey kidney cells. Cell. 1988;54:705–711. doi: 10.1016/s0092-8674(88)80015-1. [DOI] [PubMed] [Google Scholar]
- 74.Grewal SI, Jia S. Heterochromatin revisited. Nature reviews. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 75.Morrison A, Sugino A. The 3′–>5′ exonucleases of both DNA polymerases delta and epsilon participate in correcting errors of DNA replication in Saccharomyces cerevisiae. Mol. Gen. Genet. 1994;242:289–296. doi: 10.1007/BF00280418. [DOI] [PubMed] [Google Scholar]
- 76.Tran HT, Gordenin DA, Resnick MA. The 3′–>5′ exonucleases of DNA polymerases delta and epsilon and the 5′–>3′ exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae. Mol. Cell Biol. 1999;19:2000–2007. doi: 10.1128/mcb.19.3.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]