Abstract
The Epstein–Barr virus (EBV) is an oncogenic human Herpes virus found in ∼15% of diffuse large B-cell lymphoma (DLBCL). EBV encodes miRNAs and induces changes in the cellular miRNA profile of infected cells. MiRNAs are small, non-coding RNAs of ∼19–26 nt which suppress protein synthesis by inducing translational arrest or mRNA degradation. Here, we report a comprehensive miRNA-profiling study and show that hsa-miR-424, -223, -199a-3p, -199a-5p, -27b, -378, -26b, -23a, -23b were upregulated and hsa-miR-155, -20b, -221, -151-3p, -222, -29b/c, -106a were downregulated more than 2-fold due to EBV-infection of DLBCL. All known EBV miRNAs with the exception of the BHRF1 cluster as well as EBV-miR-BART15 and -20 were present. A computational analysis indicated potential targets such as c-MYB, LATS2, c-SKI and SIAH1. We show that c-MYB is targeted by miR-155 and miR-424, that the tumor suppressor SIAH1 is targeted by miR-424, and that c-SKI is potentially regulated by miR-155. Downregulation of SIAH1 protein in DLBCL was demonstrated by immunohistochemistry. The inhibition of SIAH1 is in line with the notion that EBV impedes various pro-apoptotic pathways during tumorigenesis. The down-modulation of the oncogenic c-MYB protein, although counter-intuitive, might be explained by its tight regulation in developmental processes.
INTRODUCTION
The Epstein–Barr virus (EBV) asymptomatically infects >95% of the adult population world wide, but sporadically induces infectious mononucleosis. In the vast majority of cases, the virus establishes a lifelong latent infection without further complications. Under certain circumstances, however, the virus has oncogenic potential in vivo which is reflected by its ability to growth-transform B-lymphocytes in vitro (1). It is associated with not only tumors of B-cell origin such as Burkitt’s and Hodgkin’s lymphoma, but also carcinomas of the stomach and the oropharyngeal cavity (nasopharyngeal carcinoma). EBV is present in virtually all cases of peripheral NK- and T-cell lymphoma and can be detected in ∼15% of diffuse large B-cell lymphoma (DLBCL) [for a recent review, see (2)]. In tumor cells, EBV expresses a limited set of so-called latent proteins as well as the highly transcribed non-protein encoding EBER RNAs. EBV encodes a set of 25 miRNAs that are also found in EBV-positive tumors (3–5). Furthermore, an EBV-encoded small nucleolar RNA (snoRNA) was previously discovered (6). MiRNAs are short, 19–26 nt RNAs with partial homology to sequences in their target mRNAs. Loading of miRNA to the 3′-untranslated region (3′UTR) of its target mRNA by RNA-induced-silencing-complex (RISC) results in either translational repression or mRNA degradation ultimately leading to reduced protein synthesis [for review, see (7–10)]. In addition to its own non-coding RNAs, EBV induces cellular non-coding RNAs (ncRNAs) such as vault RNA (11) and cellular miRNAs (12). Conversely, EBV infection represses several cellular miRNAs (5). The aim of this study was to establish the cellular and viral miRNA profile of DLBCL and to identify and analyze potential targets for the deregulated miRNAs.
MATERIAL AND METHODS
Patients tissue and immunohistochemistry
Freshly snap frozen B-cell lymphoma tissue samples used for miRNA expression analysis and qRT-PCR were retrieved from the files of the Institute for Surgical Pathology, University Hospital, Zürich, Switzerland. Diagnosis was done on formalin fixed, paraffin embedded tissue of the corresponding patient according to the WHO-classification system by two pathologists (TR and MT). The study was approved by the local Ethical Committee. Immunohistochemical staining was conducted on paraffin-embedded sections by an automated staining system (Discovery Ventana Medical System, Tucson, AZ) using Ventana ultraViewTM Universal DAB Detection Kit with the commercially available antibody anti-SIAH1 (Novus Biologicals, catalog number: H00006477-M01A, Clone: 2F2, dilution 1:100). Antigen-retrieval was performed with a TRIS-based buffer for 90 min at 98°C.
Cell lines and constructs
HEK 293-T, U2932 and their EBV positive derivatives where cultivated at 37°C, 5% CO2 in RPMI 1640 GlutaMax™ medium supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, USA) and 1x Antibiotics/Antimycotics (Sigma, St. Louis, USA). Additionnaly, EBV-GFP virus infected U2932 cells were maintained under selective pressure of G418 (200 mg/ml, Invitrogen). The dual luciferase reporter plasmid pMIR-RL has been described elsewhere (5). 3′UTR fragments of c-MYB (accession number: NM_005375; SIAH1 (accession number: NM_003031), c-SKI (Accession number: NM_003036.3) and LATS2 (Accession number: NM_014572) mRNA were cloned via PCR amplification from testis cDNA and ligated into the corresponding SacI and SpeI sites of pMIR-RL. PCR products of genomic DNA containing hsa-miR-424 precursor sequences flanked by ∼100 bp were cloned into eukaryotic expression vector pSG5 (Stratagene, La Jolla, USA) using EcoRI and BglII restriction sites. Respective miRNA expression was confirmed separately by northern blotting. MiR-expression vector pSG5-miR-155 was described previously (13).
Small RNA cloning
Small RNA cloning was carried out by Vertis Biotechnology (Weihenstephan, Germany) and has been described earlier (5). The small RNA fraction from each four available lymphoma and normal tissues was isolated using miRVana-microRNA isolation kit (Ambion, Austin, USA). RNA was run on 12.5% polycrylamide gel and stained with SYBRgreenII. Fraction from 15 to 40 nt was re-eluted and dissolved in water. Further on, small RNAs were poly(A)-tailed using poly(A) polymerase, and an adaptor was ligated to the 5′ phosphate of the miRNAs: (5′ end adaptor, 3′ nucleotides: 5′-GCCTCCCTCGCGCCATCAGCTNNNNGACCTTGGCTGTCACTCA-3′). NNNN represents a ‘barcode’ sequence. Next, first-strand-cDNA synthesis was performed using an oligo(dT)- linker primer and M-MLV-RNaseH reverse transcriptase (3′ end oligo dT linker primer; 61 bases: 5′-GCCTTGCCAGCCCGCTCAGACGAGACATCGCCCCG C(T)25-30). The resulting cDNAs were PCR amplified in 22 cycles using the high-fidelity Phusion polymerase (Finnzymes, Espoo, Finland). The 120–135-bp amplification products were confirmed by polyacrylamide gel electrophoresis (PAGE) analysis. Both cDNAs pools were mixed in equal amounts and subjected to gel fractionation. The 120–135-bp fraction was electroeluted from 6% PAA-gels. After isolation with Nucleospin Extract II (Macherey and Nagel, Bethlehem, USA), cDNA pools were dissolved in 5 mM Tris–HCl (pH 8.5) with a concentration of 10 ng/ml and used in single-molecule sequencing. Massively parallel sequencing was performed by 454 Life Sciences (Branford, USA) using the Genome Sequencer 20 system.
Sequence annotation
The adapter sequences and poly(A) tails were removed from the sequences. The remaining sequences of length 15 bases or longer were subjected to later analysis. Due to the disturbing signal from poly(A) tails in 454 sequencing, the sequences were analyzed semi-manually, distinguishing the false ‘A’ signals in the sequences. The set of sequences were first compared with the mature sequences of known miRNAs from miRBase v12.0. The sequences that are not perfectly matching may be artifacts of the cloning procedure or a result of non-templated modification of mature miRNAs and were annotated according to the best blast hit to the database. The rest of the sequences were again compared with the hairpin sequences of known miRNAs from miRBase v12.0, and the star sequences of known miRNAs that have not been annotated in the database were identified. Remaining reads were blasted against non-coding RNA databases including rRNA, tRNA, snoRNA, etc., retrieved from Genbank (http://www.ncbi.nlm.nih.gov/Genbank/), snoRNABase (http://www-snorna.biotoul.fr/) and fRNAdb (http://www.ncrna.org/frnadb/blast). The annotated sequences were discarded from the sequence set. The non-coding RNA sequences from EBV were identified by the blast against EBV genome retrieved from Genbank (Acc. No.: AJ507799). Remaining sequences were subjected to miRDeep (14) algorithm for searching of novel miRNAs.
Differential expression detection
Relative expression level changes was calculated by counting the numbers of respective miRNA reads normalized to the total number of annotated miRNAs in percent. Ratio of resulting abundance in library (or cloning frequency) was calculated between normal tissue control (tonsil) and respective lymphoma entity giving relative expression level change. Potentially interesting miRNA candidates where chosen according to the criteria of a 2-fold expression level change and >50 reads within one of the examined library. After extracting all known miRNA and other non-coding RNA sequences from the two libraries we applied mirDeep algorithm (14) to the remaining reads. The EBV genome sequence (Accession number: AJ507799) was included to a modified miRDeep search.
In silico target prediction
Selected miRNA candidates where checked by to up to five different available target prediction algorithms (PicTar 4.1 (http://pictar.bio.nyu.edu/cgi-bin/PicTar_vertebrate.cgi), mirbase (http://microrna.sanger.ac.uk/targets/v5/), TargetScan (http://www.targetscan.org/), miRTar (http://mirtar.mbc.nctu.edu.tw/) and miRNA viewer (http://cbio.mskcc.org/cgi-bin/miRNAviewer/miRNAviewer.pl). For each miRNA candidate, very stringent conditions where set in search for overlapping target predictions (genes were only considered as possible targets if one less than the given algorithms resulted in common predictions). To further narrow the amount of potential target genes the resulting list was aligned to their known relevance to lymphomagenesis by a PubMed search.
Reverse transcription and semi-quantitative RT–PCR
Tissue samples where homogenized under RNAse-free conditions and total RNA was isolated by standard TRIzol® reagent (Invitrogen, Carlsbad, USA) according to the manufacture’s recommendations. DNAseI (Invitrogen, Carlsbad, USA) treated RNA was poly-Adenylated using Ambion’s (Austin, USA) poly(A)-tailing Kit followed by chloroform-phenol purification. Reverse transcription reaction was carried out by application of SuperScript™ III First strand synthesis system (Invitrogen, Carlsbad, USA) using a poly(T) adapter primer (5′-GCGAGCACAGAATTAATACGACTCACTATAGG(T)12VN*-3′). Semi-quantitative RT–PCR was done as described previously (15) on Light-Cycler 1.5 System using LightCycler® FastStart DNAMasterPLUS SYBR Green I Kit (Roche Applied Science, Mannheim, Germany). The primers used are: Reverse primer (5′-GAGCACAGAATTAATACGAC-3′) miR-specific forward primers (miR-424: 5′-CAGCAGCAATTCATGTTTTGAA-3′; miR-155: 5′-TTAATGCTAATCG TGATAGGGGTAA-3′). Human 5.8s rRNA was used for internal normalization (primer: 5′-CTACGCCTGTCTGAGCGTCGCTT-3′). Following programs were established for quantitative PCR: (i) initial denaturation 10 min 95°, denaturation 10 s, annealing 63°C 5 s, amplification 72°C 5 s for miR-155; (ii) initial denaturation 5 min 95°C, denaturation 10 s, annealing 66°C 5 s, amplification 72°C 5 s for 5.8 s rRNA; (iii) initial denaturation 5 min 95°C, denaturation 10 s, annealing from 68°C to 61°C (touch-down step starting with cycle 10, decrease of 1°/cycle) 5 s, amplification 72°C 5 s for miR-424. Cycle number was adjusted individually for each miRNA according to their expression level. Relative expression level changes were calculated according to 2(−ΔΔcT)-method as described previously (16).
Transfection and luciferase assay
Plasmid transfections for luciferase assays were performed with 165 ng pMIR-RL, 333 ng pSG5 per 1 × 105 cells in a 24-well plate using Fugene®HD reagent (Roche Applied Science, Mannheim, Germany) as described by the manufacturer’s instructions. Luciferase activity was measured 48-h post-transfection using dual-glo luciferase reporter system as described by the manufacturer’s instructions (Promega, San Luis Obispo, USA). Luciferase activity was measured on Wallac Victor2 (Perkin-Elmer, Waltham, USA) multiplate reader in LUIMITRAC™ 96-well plates (Greiner Bio-one, Frickenhausen, Germany).
Northern blot
Total RNA was extracted from cells using TriZol Reagent (Invitrogen, Carlsbad) following the vendor’s recommendations. Total RNA measuring 25 μg was routinely electrophoresed through 12% urea-polyacrylamide gel and then electroblot-transferred to nylon membrane Hybond XL (Amersham) for 1 h at 2 mA/cm2. The membrane was EDC chemically cross-linked as described (17). Blots were hybridized with radioactive-labeled antisense probe overnight and then washed twice for 15 min with 5 × SSC, 1% SDS and twice for 15 min with 1 × SSC, 1% SDS. As radioactive probes, we used RNA probes labeled with miRVana Probe construction kit (Ambion) according to the manufacturer’s instructions. The following antisense probes were used: miR-424: 5′-UUC AAA ACA UGA AUU GCU GCU G-3′, miR-155: 5′-CCC CUA UCA CGA UUA GCA UUA A-3′.
MicroRNA blocking assay and western blot
MicroRNA blocking was done by transfecting 0.75 nMol per 106 cells miR-155 or miR-424 inhibitors or respective miRNA mimics (Ambion). Used cells were U2932 and EBV infected subclones transfected with lipid based siPORT™ NeoFX™ Transfection Agent (Ambion) according to manufactures recommendation. Anti-miR™ miRNA Inhibitors—Negative Control #1 (Ambion) served as mock transfectant. Transfection efficacy was assessed by using a FAM-labeled Oligonucleotide (Ambion). After transfection procedure cells were cultivated for 3 days, counted and protein extracts were created by adding 5× sample buffer (Promega). Protein levels were normalized according to the counted cell number. Western blot was performed as previously (18). In brief, after electrophoresis proteins were transferred onto Nitrocellulose membranes, blocked in 2.5% PBS-milk powder. Anti-c-MYB Antibody (Upstate, mouse clone 1-1) was diluted 1:500, anti-SIAH1 1:250 (Novus Biological, mouse clone 2F2) and anti-β-catenin (Cell signaling) 1:1000 in 1% PBS-milk powder an incubated for 1 h at room temperature. Rabbit anti-β-actin (Cell signaling; 1:1000 in 0.5% PBS/BSA) detection served as a loading control. After three times each 10 min washing in 0.1% PBS/Tween secondary anti-mouse peroxidase-antibody (Biorad) was diluted 1:1000 in 2.5% PBS-milk powder or peroxidase-anti rabbit IgG (Sigma-Aldrich) 1:20 000 and incubated for 45 min at room temperature. After additional washing steps proteins were detected using ECL chemiluminescence kit (GE Healthcare) and visualized on X-ray films.
Immunohistochemistry
Immunohistochemical staining was conducted on paraffin-embedded sections by an automated staining system (Discovery Ventana Medical System, Tucson, AZ) using Ventana ultraViewTM Universal DAB Detection Kit with the commercially available antibody anti-SIAH1 (Novus Biologicals, catalog number: H00006477-M01A, Clone: 2F2, dilution 1:100). Antigen-retrieval was performed with a TRIS-based buffer for 90 min at 98°C.
Statistical analysis
Densitometric quantification for northern blot was done by using ImageQuant software (Molecular Dynamics, GE-Healthcare, USA). Statistical analysis of the Luciferase-Assay and quantitative RT–PCR was performed with GraphPad Prism 4.0 (Graph Pad software, La Jolla, CA, USA).
RESULTS
Small RNA library annotation
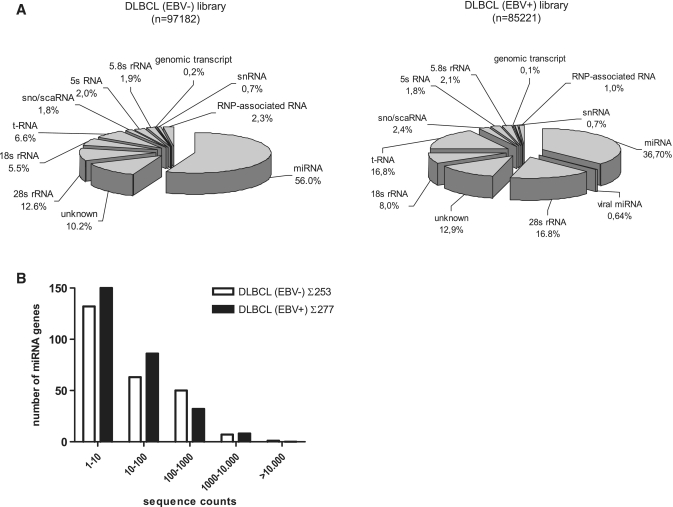
To generate miRNA cDNA libraries, tissue samples of four EBV-positive and four EBV-negative DLBCLs were pooled. The presence or absence of EBV in the tissues was determined by EBER in situ hybridization. All of the EBV-positive DLBCLs exhibited an EBV latency type I (EBER+/EBNA1+/LMP1-/EBNA2-/BZLF1-). Characteristics of the used tissues are shown in Supplementary Table S1. Library annotation resulting from 454 sequencing was done semi-manually as described before (5). In the library of EBV-negative DLBCL, ∼56% of a total of 97 182 reads represented known miRNA sequences, while 37% of a total of 85 221 reads were miRNA derived in the EBV-positive DLBCL library. Figure 1A shows the distribution of miRNA and other small ncRNA sequences in the two libraries as indicated. The number of unidentified sequences ranged between 10–13%. We also monitored the sequence count distribution of all expressed miRNAs in the two tumor entities. Sequence counts covered a range of five orders of magnitude (Figure 1B). The majority (>50%) of cellular miRNA species where present at very low level (1–10) reads, while only 11–19% were abundantly expressed (100–1000 reads). Of note, miR-16 accounted for 19.6% of all reads in the EBV-negative and for 14.7% of all reads in the EBV-positive DLBCL.
Figure 1.
Overview of small RNA and miRNA gene expression in primary B-cell lymphoma tissues obtained by deep sequencing of miRNA cDNA libraries. (A) Proportions of various classes of small RNAs detected by sequencing of EBV-negative DLBCL (total reads: 97 182) and EBV-positive DLBCL (total reads: 85 221) libraries as indicated. ScaRNA, small cajal body RNA; snRNA, small nuclear RNA; snoRNA, small nucleolar RNA; rRNA, ribosomal RNA; tRNA, transfer RNA; RNP-associated, ribonucleo-protein complex derived RNAs; unknown, derived from unannotated/intergenic regions or sequencing artefacts. (B) Distribution of miRNA genes expressed according to their sequence counts in EBV-positive and -negative DLBCL. Shown are the numbers of unique miRNA genes plotted as a function of their expression levels as defined by a given range of sequence counts in the respective libraries of small RNAs.
The advantage of miRNA cloning and sequencing is the potential identification of so far unknown miRNAs. We found six different candidate sequences matching the human genome and fitting mirDeep algorithm miRNA prediction criteria (14). Of these, five sequences were described in the Sanger Database while this analysis was under way (data not shown). A sixth sequence (ACAUACUUCUUUAUAUGCCCAU) homologous to hsa-miR-1-2* was found four times but could not be verified by northern blotting in the uninfected DLBCL line U2932, or the EBV-infected derivatives thereof. This potential miRNA was not pursued further.
Analysis of EBV-encoded miRNA expression
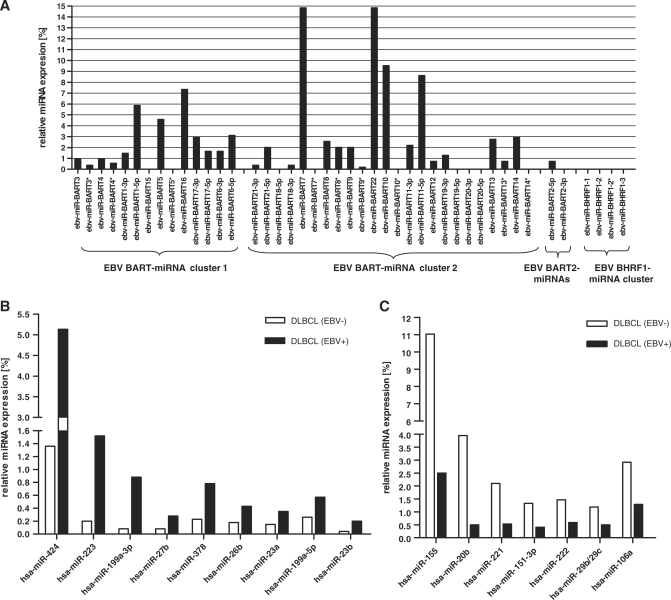
In the library derived from EBV-positive DLBCL, virus-encoded miRNAs represented 1.7% of all identified miRNA reads (Figure 1A). We found the strongest expression for EBV-miR-BART7 and -BART22 (14.9% each), -BART10 (9.6%), -BART11-5p (8.6%) and -BART16 (7.3%), where 100% is considered as the total number of EBV miRNA reads in the library. The DLBCL samples contained the previously described EBV-encoded miR-BART21 and -BART22 (5). The distribution of the various EBV-miRNAs is graphically shown in Figure 2A and listed in Supplementary Table S2. In the EBV-positive tumors, we found all known EBV-encoded miRNAs with the exception of those encoded by the BHRF1 cluster and also no reads from EBV-miR-BART15 and -BART20. This is in line with our previous data for EBV-positive NPC (5) and a report about viral miRNA expression in EBV-infected gastric carcinoma (19) where the BHRF1-derived miRNAs were also not present.
Figure 2.
miRNA expression in primary DLBCL tissue detected by 454 deep sequencing. (A) Distribution of EBV-encoded miRNAs in EBV-positive DLBCL. The total count of EBV-derived miRNAs was set to 100%. No reads were obtained for ebv-miR-BHRF1-1, -2, -3 and ebv-miR-BART-15 and -20. (B) Human cellular miRNAs miR-223, miR-199a-3p, miR-424, miR-27b, miR-378 and miR-26b are upregulated in EBV-positive versus -negative tissues. (C) Human cellular miRNAs miR-20b, miR-155, miR-221, miR-222, miR-151-3p, miR-29b/c and miR-106b are downregulated in EBV positive tissues. The criteria for miRNA selection were a 2-fold change and ≥50 reads (∼0.05%) abundance in one of the analyzed libraries. MiRNA expression levels are shown as percentages of the total miRNA reads in the libraries.
MicroRNA expression pattern in EBV-positive versus -negative DLBCL
We then compared the cellular miRNA expression levels in the EBV-positive versus -negative DLBCLs. We set an abundance threshold of >50 reads (corresponding to ∼0.05% of all reads) in one of the examined libraries and an expression level change of ≥2-fold. This resulted in a distinct miRNA profile characteristic for EBV-positive DLBCLs. Nine miRNAs (hsa-miR-424, -223, -199a-3p, -199a-5p, -27b, -378, -26b, -23a, -23b) were found to be upregulated and seven miRNAs (hsa-miR-155, -20b, -221, -151-3p, -222, -29b/c, -106a) were downregulated as shown in Figure 2B and 2C, respectively. Some of the deregulated miRNAs had already been linked to cancer (Table 1). Clustered miRNAs (miR-27b/miR-23b; miR-221/miR-222; miR-20b/miR-106b) exhibited similar expression changes suggesting not only co-transcription, but also co-processing events. The results of the sequence analysis are summarized in Supplementary Table S3.
Table 1.
Overview of miRNA candidates misregulated in EBV positive relative to EBV negative counterpart DLBCL library and their known relation to cancer
| microRNA species | Fold change | Known relation to cancer | Refererence | Name of journal | Issue |
|---|---|---|---|---|---|
| hsa-miR-223 | 4.48 up | miR-223, miR-26b, miR-23b, miR-23a are significantly upregulated in kidney and bladder cancers | Gottardo et al. 2007 | Urologic Oncology | 25(5): 387–392 |
| E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in AML | Pulikkan et al. 2010 | Blood | 4;115(9): 1768–1778 | ||
| hsa-miR-199a/b-3p | 3.81up | Expression is correlated with worse overall and event free survival in AML | Garzon et al. 2008 | Blood | 111(6): 3183–3189 |
| hsa-miR-424 | 3.78 up | PU.1 activates the transcription of miR-424 and is involved in stimulating monocyte differentiation | Rosa et al. 2007 | Proceedings of the National Academy of Sciences of the United States of America | 104(50): 19849–19854 |
| Acts pro-differentiative and modulates cell cycle regulators in AML | Forrest et al. 2010 | Leukemia | 24(2): 460–466 | ||
| hsa-miR-27b | 3.56 up | Expression of miR-27b contributes to in vitro angiogenesis | Kuehbacher 2008 | Trends in Pharmacological Science | 29(1): 12–15 |
| hsa-miR-378 | 3.43 up | MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression | Lee et al., 2007 | Proceedings of the National Academy of Sciences of the United States of America | 104(51): 20350–20355 |
| hsa-miR-26b | 2.35 up | Reduced expression in hepatocellular carcinoma | Ji et al. 2009 | New England Journal of Medicine | 361(15): 1437–1447 |
| hsa-miR-23a | 2.27 up | Upregulated in EBV+ lymphoblastoid cell lines | Mrazek et al. 2008 | Nucleic Acids Research | 35(10): e73 |
| hsa-miR-199a-5p | 2.17 up | ||||
| hsa-miR-23b | 2.11 up | MicroRNA-23b mediates urokinase and c-met downmodulation and a decreased migration of human hepatocellular carcinoma cells | Salvi et al. 2009 | FEBS Journal | 276(11): 2966–2982 |
| Suppressed by c-MYC | Gao et al. 2009 | Nature | 458(72,3,9): 762–765 | ||
| hsa-miR-20b | 7.91 down | Oncogenic potential of the miR-106-363 cluster and has implication in human T-cell leukemia | Landais et al. 2007 | Cancer Research | 67(12): 5699–5707 |
| hsa-miR-155 | 4.41 down | Oncogene, upregulated in EBV+ Lymphoblastoid cell lines | Mrazek et al. 2007 | Nucleic Acids Research | 35(10): e73 |
| Costinean et al. 2006 | Proceedings of the National Academy of Sciences of the United States of America | 103(18): 7024–7029 | |||
| hsa-miR-221 | 3.99 down | miR-221 were more highly expressed in ABC-type than GCB-type cell lines; over-expressed in de novo DLBCLtransformed DLBCL | Lawrie et al. 2007 | International Journal of Cancer | 121(5): 1156–1161 |
| hsa-miR-151-3p | 3.26 down | ||||
| hsa-miR-222 | 2.49 down | See miR-221 | |||
| hsa-miR-29b/29c | 2.37 down | mir-29 regulates Mcl-1 (anti-apoptotic BCL-2 member) protein expression and apoptosis | Mott et al. 2007 | Oncogene | 26(42): 6133–6140 |
MicroRNA candidate expression validation by RT–PCR
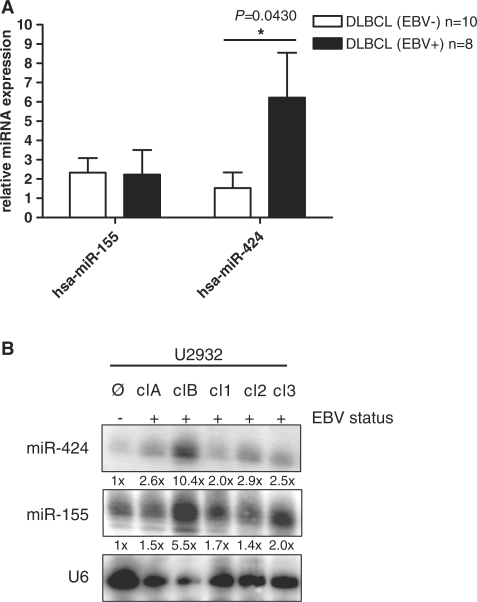
It was previously shown that miR-155 is strongly upregulated in human B-cell lymphomas (20) and that miR-155 has the properties of an oncogene (21). We had found that miR-155 is also strongly upregulated in NPC as compared to normal control tissue (5); in addition, EBV-infection of BL cell lines resulted in dramatically increased amounts of this miRNA (12). In contrast, here we found a relative decrease of miR-155 expression in the EBV-positive as compared to the EBV-negative DLBCLs (3.5% versus 15.3% representation, respectively; see Figure 2C and Supplementary Table S3. To analyze this in more detail, we extracted RNA from snap-frozen tumor tissues (10 lymphomas from EBV-negative and eight from EBV-positive tissues) and measured miRNA levels by qRT-PCR in comparison to RNA extracted from non-transformed tonsillar tissue. The value obtained from this latter tissue was set to 1. As shown in Figure 3A, the relative levels of miR-155 in the EBV-positive and -negative DLBCL tissue were approximately the same by this method, but were still much higher than in the tonsil control tissue.
Figure 3.
miR-155 and -424 expression validation in primary DLBCL and DLBCL cell lines. (A) MiR-155 and miR-424 expression validation by quantitative RT–PCR. Total RNA was extracted from three tonsil tissues, and each 10 EBV positive and eight negative DLBCL (including tissues previously used for small RNA-library generation), were used for qRT-PCR analysis. Bars indicate relative expression levels compared to normal control. *P-vlaue < 0.05, student’s t-test, paired. (B) miR-155 and -424 expression validation by northern blot in DLBCL cell lines. Total RNA of U2932 cell line and EBV-positive clones (A, B, 1, 2 and 3) was detected using miRNA-specific probes and quantified by densitometric analysis. Given numbers display relative expression levels compared to loading control U6 snRNA.
In addition to miR-155 which is a known oncogene, we focused our further experiments on miR-424 which was found in a feed-back circuit with PU.1 (22), a key transcription factor employed by EBNA2-mediated transformation of B-cells (23). We also compared miR-424 levels by qRT-PCR analysis because miR-424 showed the strongest relative expression increase in EBV-positive DLBCL versus EBV-negative DLBCL (0.94% versus 0.25%, respectively). As depicted in Figure 3A, we found an ∼5-fold increase (P = 0.0430) in miR-424 in the EBV-positive cases of DLBCL. In contrast to miR-155, which was elevated in the tumor versus non-tumor tissue, we did not observe a statistically significant increase in miR-424 levels in EBV-negative DLBCL as compared to the tonsil tissue but a strong upregulation in the EBV-positive versus -negative cases.
The relative up- or down-regulation of various miRNAs was further analyzed by northern blotting using the U6 snRNA as a standard. We confirmed the upregulation of miRNAs miR-424, -223, -199a-3p, -27b, -26b and -23a, and the downregulation of miR-155, -221 and -222 while miR-20b and miR-106 (downregulated by sequencing) were found upregulated by northern blotting. The quantification of the northern blots is shown in Supplementary Table S4; the northern blots are shown in Supplementary Figure S1A–C. We assume that the different methods applied were responsible for these discrepancies. However, the up- or downregulation was confirmed in the majority of miRNAs analyzed.
The levels of miR-155 and miR-424 were then analyzed in the EBV-negative U2932 DLBCL cell line and five EBV-convertants thereof. As shown in Figure 3B, EBV-infection increased the amounts of the two miRNAs to a varying degree. The clones all expressed detectable amounts of LMP1; clones clA and cl1 which contain no or only minute amounts of EBNA2 did also contain elevated levels of both miR-155 and miR-424. We had found in other experiments that the expression of EBNA2 alone, neither in U2932 DLBCL cells (P. Trivedi, unpublished data) nor in DG75 cells (F. Grässer, unpublished data), induced these microRNAs. We then looked at qRT-PCR results in 11 EBER-positive DLBCLs. Of these, five were LMP1-positive (latency II or III). Out of these LMP1-positive lymphomas three patients were immuncompromised. Interestingly, we observed a 4-fold increase of miR-155 and a 7-fold, but statistically not significant increase of miR-424 expression in the LMP1-positive lymphomas compared to the LMP1-negative ones. There was a trend towards the upregulation of miR-155 (P = 0.0639) when we compared LMP1-negative lymphomas with LMP1-positive DLBCLs (Supplementary Figure S2). Taken together, our data indicate an upregulation of miR-155 in DLBCL cell lines and in primary lymphoma tissue, all displaying EBV latency II or III, suggesting a role for LMP1.
In silico miRNA target prediction
We used PicTar, MirTar and TargetScan miRNA target prediction algorithms to select a potential set of genes that might be regulated by miR-155 and miR-424. The data were combined to obtain overlapping results in order to achieve a high-significance level. We initially obtained a list of 126 genes for miR-155 and of 27 genes for miR-424. The predictions were further narrowed down by the known relation of the targets to EBV biology and/or lymphomagenesis. We finally arrived at a list of eight potential miRNA target genes for miR-155 as shown in Supplementary Figure S3. The mRNA of the AID (activation-induced cytidine deaminase) gene was already shown by others to be a target for miR-155 (24,25). The same approach for miR-424 narrowed the targets down to five candidates. C-MYB was predicted to be a target of both miR-424 and miR-155.
Deregulated miR-424 inhibits SIAH1, miR-155/miR-424 inhibit c-MYB and miR-155 targets the c-SKI-3′UTR
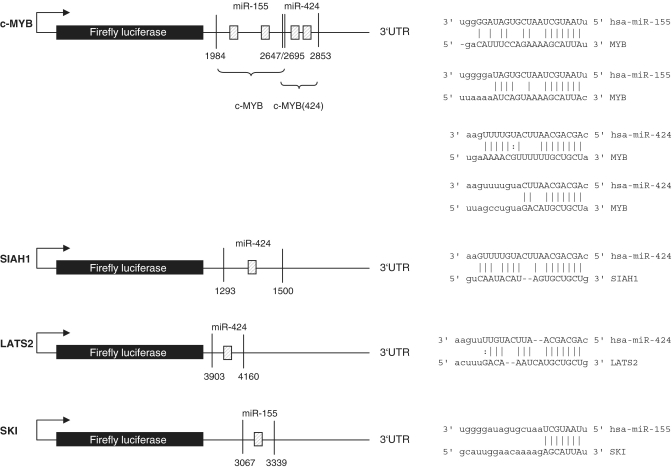
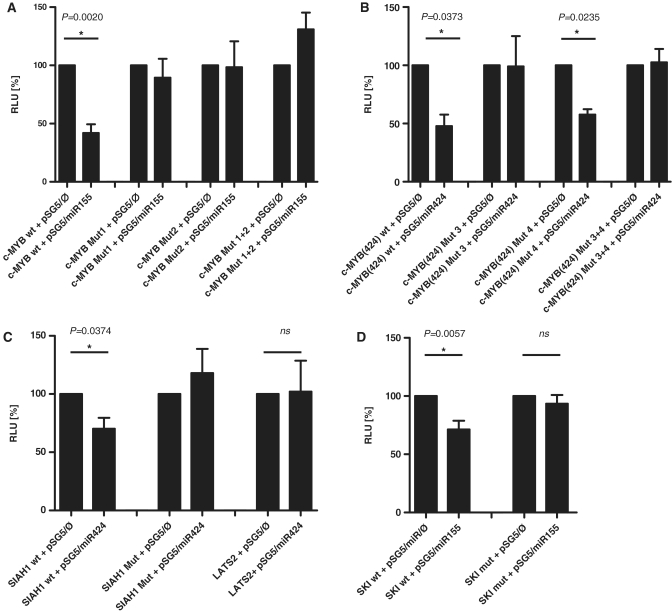
We chose to analyze the effects of miR-424 and miR-155 on the 3′UTRs of c-MYB, SIAH1, c-SKI and LATS2. The predicted binding sites for these are shown in Figure 4. Two binding sites each for miR-424 and miR-155 were predicted for the c-MYB 3′UTR which was therefore likely to be a target that deserved to be analyzed. SIAH1, c-SKI and LATS2 had one potential binding site. We wanted to analyze SIAH1 because a potential link to β-catenin regulation by LMP1 via SIAH1 was suggested previously (26). The 3′UTRs of the four genes were cloned into a luciferase reporter (5) construct and co-transfected into HEK 293-T cells with the appropriate miRNA expression vector. The expression of miR-424 from the vector pSG5 was verified by northern blotting as shown in Supplementary Figure S4. The expression of miR-155 from pSG5-miR-155 was described previously (13). We observed a significant decrease (P = 0.002) of luciferase activity for c-MYB with miR-155 by ∼60% (Figure 5A). We then mutated the two predicted binding sites for miR-155 either individually or in combination. Surprisingly, the construct lost its responsiveness when the miR-155 binding sites were mutated individually or in combination (Figure 5A). This result suggested that the two candidate miR-155-binding sites are functional and that they apparently cooperate. This issue was not pursued further.
Figure 4.
Target predictions for selected mRNA targets. Schematic overview of the luciferase constructs and putative bindings sites of miR-155 and -424 to their predicted mRNA targets c-MYB, SIAH1 and LATS2. The locations of the potential binding sites are indicated by shaded boxes and by the sequence alignments depicted on the right side.
Figure 5.
Luciferase reporter assays. Firefly luciferase reporters fused to the c-MYB, SIAH1, c-SKI and LATS2-3′UTRs were cotransfected with miR-155 and -424 into HEK 293-T cells in the indicated combinations. (A) MiR-155 targets c-MYB. The proximal binding site is denoted Mut1, the distal binding site is denoted Mut2, and the mutation of both binding sites is denoted Mut1+2. (B) MiR-424 targets c-MYB. The distal binding site for is denoted Mut3, the proximal site is denoted Mut4, and the mutation of both sites is called Mut 3+4. (C) miR-424 targets SIAH1 but not LATS2. The single miR-424-binding site in the SIAH1 3′UTR was mutated and denoted SIAH1 Mut. The predicted binding site for miR-424 in the LATS2-3′-UTR was not mutated as the wt-3′UTR was not responsive to miR-424. (D) miR-155 targets c-SKI. The single binding site for miR-155 was mutated and denoted SKI Mut. The values represent at least three experiments carried out in triplicate. ns: not significant.
We next assayed the c-MYB-luciferase construct which contained the two miR-424 candidate binding sites and found a reduction of ∼60% (P = 0.0373) when miR-424 was co-expressed; here, the mutation of the potential distal binding site (denoted 2853 in Figure 4) lost the responsiveness to miR-424 (P = 0.0235), while mutation of the potential proximal binding site (denoted 2695 in Figure 4) had no effect indicating that only the distal candidate binding site was functional (Figure 5B).
In a further set of experiments, miR-424 reduced the activity of the SIAH1-3′UTR reporter by ∼30% which is statistically significant (P = 0.0374), and the mutation of the predicted binding site resulted in a loss of responsiveness to miR-424 whilst the LATS2-3′UTR was not influenced by miR-424 (Figure 5C). Control transfections with irrelevant miRNAs did also have no effect on the activity of the analyzed 3′UTR constructs (data not shown). Finally, the 3′UTR of c-SKI, when assayed with miR-155 showed a clear downregulation (P = 0.0057). After the mutation of the candidate binding site, the c-SKI-3′UTR lost its responsiveness demonstrating that the target site is functional. This is shown in Figure 5D.
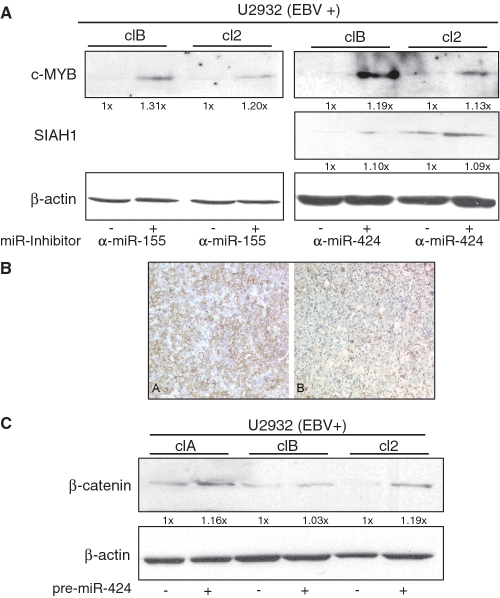
Blocking of miR-155 and miR-424 expression leads to higher protein level of c-MYB and SIAH1 in vivo
Next, we measured c-MYB and SIAH1 protein levels in the presence or absence of miR-155 and miR-424 inhibitor (Figure 6A). Transfection efficiency was 86–88% (Supplementary Figure S5). Endogenous c-MYB protein level was increased when miR-155 was inhibited in all the clones of the EBV-positive U2932 DLBCL line compared to scrambled control oligonucleotide. Likewise, antisense to miR-424 increased c-MYB and SIAH1 protein levels in these cells (Figure 6A). Downregulation of miR-155 and miR-424 was demonstrated by northern blotting (Supplementary Figure S6). In summary, c-MYB is negatively regulated by both miR-155 and -424, while SIAH1 is down-modulated by miR-424 and c-SKI is potentially downregulated by miR-155.
Figure 6.
(A) Induction of c-MYB and SIAH1 protein levels by antisense inhibition of miR-155 and -424. Lysates of the EBV-positive U2932 cell clones transfected with indicated antisense inhibitors to miR-155, -424 or a scrambled oligonucleotide as a negative control were analyzed by western blotting using anti-c-MYB or anti-SIAH1 antibody. The protein input was determined by staining for β-actin. (B) Immunhistochemical staining for SIAH1: strong membranous positivity in a high percentage of tumor cells in an EBV- (A, left) DLBCL versus low expression in an EBV+ DLBCL (B, right). (C) Overexpression of miR-424 results in increased levels of β-catenin. The EBV-positive subclones clA, clB and cl2 of the DLBCL line U2932 or were treated with control oligonucleotide or with pre-miR-424 for the expression of miR-424. The levels of β-catenin and β-actin (as input control) were determined by western-blot analysis.
Impact of EBV-altered miR-424 on the β-catenin/wnt signaling pathway
SIAH1 is an E3-ubiquitin ligase that, besides multiple other functions, is involved in regulating the β-catenin pathway and is assumed to function as a tumor suppressor (27). In a previous report, it was shown that the EBV-encoded oncogene LMP1 upregulates β-catenin through inhibition of SIAH1 expression (26). Moreover, a recent report demonstrated that SIAH1 downregulates β-catenin by direct ubiquitinylation (28). To gain further insight into the functional relevance of miR-424 over-expression in the EBV-positive DLBCL, we assayed the levels of β-catenin upon inhibition of SIAH1. As shown in Figure 6A, inhibition of miR-424 clearly leads to increased levels of SIAH1 demonstrating a direct link between this miRNA and SIAH1. This protein blot confirms the experiments that showed a downregulation of the SIAH1-3′UTR by miR-424 (Figure 5C). This observation is confirmed in primary lymphoma tissue where SIAH1 protein expression demonstrated by immunohistochemistry appears to be downregulated in EBV+ versus EBV- DLBCLs (Figure 6B). Conversely, we can clearly show an upregulation of β-catenin after ectopic expression of miR-424 in two out of three different EBV+ subclones of the U2932 cell line (Figure 6C). So far, a deregulation of β-catenin has not been shown directly in DLBCL, however, overexpression of β-catenin has been found in cutaneous B-cell lymphomas (29).
DISCUSSION
This is the second report aimed at elucidating the miRNA profile of tumors infected by EBV using a deep-sequencing approach. In a first report, we had compared the miRNA levels in endemic nasopharyngeal carcinoma (NPC) to non-infected, healthy control tissue (5). In NPC, we detected two novel EBV microRNAs designated ebv-miR-BART21 and -BART22 that were both present in DLBCL samples analyzed here pointing at a role for these miRNAs in EBV-driven tumor transformation. Furthermore, we did not detect ebv-mir-BART15 and -BART20 as observed in NPC where we found no or only one read for -BART15 and -BART20 (5). Of the EBV miRNAs, the strongest expression was observed for ebv-miR-BART22, -BART10 and -BART16. In contrast to NPC samples where the EBV-encoded miRNAs accounted for 5–19% of the total, the EBV miRNAs represented only 1.7% of the total miRNA counts in DLBCL. It is therefore unclear whether EBV-miRNAs expressed at low or very low levels carry out a function in the transformed cell.
MiRNAs from the BHRF1 cluster were reported to repress the chemokine CXCL-11 in DLBCLs of immunocompromised (HIV+) patients with an EBV latency type III (30). In contrast, the lymphomas analyzed in our miRNA expression study were derived from immunocompetent patients. In accordance with the patients’ immune status, the EBV-positive DLBCLs did not express LMP or EBNA2 corresponding to an EBV latency type I. Accordingly, miRNAs derived from the BHRF1-cluster were not found as the types I and II latency do not appear to support the expression of these transcripts. This finding matches the situation described for NPC (5,31), gastric carcinoma (19) and peripheral T-cell lymphoma (32). The absence of these miRNAs was also reported for an EBV-positive type I Burkitt’s lymphoma (BL) cell line (3), but so far, no data were available for primary, EBV-positive DLBCL of immunocompetent, HIV-negative patients. The BHRF1 miRNAs might thus play only a role in the induction of the B-cell lymphoma in HIV-infected individuals.
EBV infection has a profound impact on cellular miRNA expression. Both absolute levels of miRNAs appear to be downregulated due to viral infection (33) as well as relative levels of cellular miRNAs are also strongly deregulated by EBV infection (5,19,31,18,34). Nevertheless, this global downregulation of miRNAs occurred in in vitro generated lymphoblastoid cell lines and did not undergo a selection driven by the host immune system during tumor formation. Recently published data including our own had shown that miR-155 is upregulated by EBV-infection (12,34,35). MiR-155, encoded by the BIC gene, has oncogenic potential (21). In primary cases of BL, however, no BIC mRNA was found regardless of the EBV status (36) while the same group reported BIC upregulation in Hodgkin-lymphoma and DLBCL (37). By deep-sequence analysis, we found a relative downregulation of miR-155 in the EBV-positive DLBCL versus EBV-negative DLBCL. In contrast, the qRT-PCR showed an upregulation in both, EBV+ and EBV- DLBCL as compared to reactive tonsil tissue. To analyze the possible contribution of LMP1, we compared by qRT-PCR the levels of miR-155 and -424 in EBV-positive DLBCLs that were, by immunohistochemistry, either LMP1-positve or LMP1-negative. There was a trend towards upregulation in LMP1-positive samples, but no statistically significant difference (Supplementary Figure S2). The patient’s statistics are shown in Supplementary Table S5. The northern blot analysis of EBV-infected versus uninfected DLBCL U2932 lines showed a 1.4–5.5-fold upregulation of miR-155 in the EBV-infected cell line (which has a different latency state than tumors) in accordance with previous data for EBV-infected BL cell lines (12). It is thus possible that either the differences in the miR-155 levels were due to differences in the tissue samples employed or methodological biases may account for the observed variations. However, we have recently shown by deep sequencing and by qRT-PCR analysis that the sequence count obtained by deep sequencing of small cDNA libraries yields highly reproducible results (38). The expression of miR-29b was lower in the EBV-positive than the -negative cases. This miRNA was previously shown to be upregulated by LMP1 (39); however, the lower level of miR-29b in the EBV-positive cases analyzed in this report might be due to the fact that our samples exhibited a type I latency (LMP1-negative).
While all other ncRNA species stayed more or less constant (Figure 1A and B), the proportion of miRNAs and tRNAs changed drastically between EBV negative and EBV positive DLBCL. In EBV-positive DLBCL, we identified ∼40% less miRNAs reads compared to the other small RNA library (Figure 1A, right panel). This suggests that EBV might have a modulatory influence on miRNA transcription or processing pathways in vivo, consistent with an earlier publication (33). In contrast, the relative amount of tRNA increased roughly 2-fold in EBV-negative to 5-fold in EBV positive DLBCL, possibly reflecting a so far unknown advantageous mechanism of EBV to protein metabolism or an enhanced Pol III activity (40).
MiR-424 was the miRNA that was most strongly upregulated by EBV-infection in primary DLBCL tissue and also the cell lines studied here, both by sequence analysis as well as by qRT-PCR. In addition, there was a significant upregulation in the lymphoma samples as compared to non-transformed tonsil tissue. The function of miR-424 might vary depending on the cellular context of its expression/repression. Enhanced levels of miR-424 of 5.5-fold have been reported for colon cancer (41), while it was downregulated 2.8-fold in chronic lymphatic leukemia (CLL) (42). In addition, miR-424 is stronger expressed in T-ALL as compared to B-ALL and may serve as a marker to distinguish these two entities (43). MiR-424 also appears to be involved in cellular differentiation (44,45). Interestingly, miR-424 is induced by the transcription factor PU.1 during monocyte/macrophage differentiation. PU.1 in turn is induced by EBV-infection and plays a role in EBNA2-mediated gene expression (23).
Here, we demonstrate that miR-424 downregulates the protein level of the ubiquitin ligase SIAH1. MiR-424 in turn is strongly upregulated by EBV-infection in DLBCL samples, in EBV-infected DLBCL lines and also when the primary lymphoma tissues were compared to tonsil tissue by qRT-PCR. SIAH1 was found to act as a tumor suppressor (27,46), probably via induction of apoptosis (47) by interference with the JNK-pathway (29,48). Furthermore, it was shown that β-catenin levels in Burkitt’s lymphoma lines were upregulated via LMP1-induced down-modulation of SIAH1 (26). Because DLBCL usually do not express the viral LMP1 oncogene, the upregulation of β-catenin via miR-424-induced reduction of SIAH1 offers a framework to explain the function of deregulated miR-424 in this tumor entity. SIAH1 is a down-stream effector of p53, and it is conceivable that induction of miR-424 prevents p53-mediated apoptosis in the virus-infected cell. However, miR-424 is also strongly induced in EBV-negative DLBCL as compared to primary tonsil tissue (Figure 3A) indicating that miR-424 plays a role in the induction or maintenance of the DLBCL phenotype in the absence of the virus. SIAH1 is a negative regulator of β-catenin protein expression and we could indeed demonstrate that downregulation of SIAH1 correlates with increased β-catenin levels.
We demonstrate that c-MYB is a target of both miR-155 and miR-424. C-MYB has oncogenic potential (49–51) and its expression is tightly regulated during B-cell development (52). Various miRNAs, i.e. miR-34a (53), miR-15a (54) or miR-150 (55) regulate c-MYB protein expression. C-MYB and miR-15a regulate each other’s expression via a feed-back loop (56) in accordance with a prediction that c-MYB, among other transcription factors, regulates miRNA gene expression (57). In our analysis, miR-15a represented 4.67% of all reads in the EBV-negative DLBCL and 4.28% of all reads in the EBV-positive DLBCLs indicating that expression of c-MYB is also tightly regulated in DLBCL. The seed sequence for miR-424 and miR-15a is the same; thus, the increase of miR-424 might add an additional layer of repression. The level of miR-424 rose from 0.98% in uninfected to 1.92% of the total miRNA count in EBV-infected DLBCL. It is unclear whether the additional expression of both miR-155 and -424 increases the repression of c-MYB because we obtained no experimental evidence of a cooperative effect of miR-155 and miR-424 when the two miRNAs were co-expressed with the c-MYB 3′UTR reporter (data not shown). In addition, miR-424 and miR-15a might compete for the binding site(s). We have recently reported that the myo6-3′UTR is a target for both miR-145 and -143 (38). There, we also found no cooperation of the two miRNAs when tested together on the myo6-3′UTR. So far, we have no explanation for the observation that already the mutation of each of the individual binding sites on the c-MYB-3′UTR results in ablated responsiveness to miR-155. Our data imply that the two binding sites cooperate with each other. Clarification of this problem will require additional experiments beyond the scope of this article.
The effect of miR-424 upregulation and the concomitant downregulation of SIAH1 is inline with the role for SIAH1 as a tumor suppressor, while the seemingly counter-intuitive decrease in c-MYB (and possibly c-SKI) expression might best be explained by its tightly regulated expression during B-cell development. The effect of miR-155 on the 3′-UTR of the c-SKI proto-oncogene could be demonstrated by luciferase assay and by mutation of the potential binding site but not by upregulation of c-SKI protein levels after inhibition of miR-155. Upregulation of miR-155 in EBV-positive DLBCL was recently shown to inhibit TGF-β1 via inhibition of SmaD5 by miR-155 (58). These authors pointed out that it was unclear how Smad3, which is also inhibited in DLBCL was regulated in these cells as they did not find a direct effect of miR-155 on the SmaD3 3′-UTR. Downregulation of c-SKI might reduce its inhibitory effect on SmaD3 which in turn activates the growth stimulation by TGF- β1. Both c-SKI and c-MYB are usually considered to be proto-oncogenes involved in transcriptional activation (59,60) while their function in development and differentiation is poorly understood. It is possible that both c-MYB and c-SKI are involved in differentiation, i.e. towards a promyelocytic phenotype (61). Their expression might be incompatible with their expression in the DLBCL setting explaining their downregulation by miR-155. In summary, we show that EBV infection has a profound influence on the cellular miRNA profile in DLBCL.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Krebshilfe (grant 107166 to G.M. and F.G.); German Bundesministerium für Bildung und Forschung (grant NGFN to S.B. and #01GS0801/4 to F.G.); Max-Planck-Society (to Meister lab, partial); German Bundesministerium für Bildung und Forschung (grant NGFN-Plus #01GS0801/5 to G.M.); Bavarian Ministry for Education and Science (BayGene to G.M.); MIUR, AIRC, Progetto strategico (ISS9ACF/1) and FISM (2007/R/17) (to A.F. and P.T.). Funding for open access charge: Grant 107166 from the Deutsche Krebshilfe (to G.M. and F.G.) Grant NGFN-Plus #01GS0801/4 from the German Ministry of Education (to F.G.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Weihong Qi from Functional Genomics Center Zurich (FGCZ) for performing the modified miRDeep analysis and Ruth Nord for expert technical assistance.
REFERENCES
- 1.Rickinson AB, Kieff E. In: Fields Virology. Knipe DM, Howley PM, editors. Vol. 2. Philadelphia: Lippincott-Raven; 2007. pp. 2655–2700. [Google Scholar]
- 2.Delecluse HJ, Feederle R, O’Sullivan B, Taniere P. Epstein Barr virus-associated tumours: an update for the attention of the working pathologist. J. Clin. Pathol. 2007;60:1358–1364. doi: 10.1136/jcp.2006.044586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA. 2006;12:733–750. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu JY, Pfuhl T, Motsch N, Barth S, Nicholls J, Grässer F, Meister G. Identification of novel Epstein-Barr virus microRNA genes from nasopharyngeal carcinomas. J. Virol. 2009;83:3333–3341. doi: 10.1128/JVI.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hutzinger R, Feederle R, Mrazek J, Schiefermeier N, Balwierz PJ, Zavolan M, Polacek N, Delecluse HJ, Huttenhofer A. Expression and processing of a small nucleolar RNA from the Epstein-Barr virus genome. PLoS Pathog. 2009;5:e1000547. doi: 10.1371/journal.ppat.1000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meister G. miRNAs get an early start on translational silencing. Cell. 2007;131:25–28. doi: 10.1016/j.cell.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 8.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- 10.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nandy C, Mrazek J, Stoiber H, Grässer FA, Huttenhofer A, Polacek N. Epstein-barr virus-induced expression of a novel human vault RNA. J. Mol. Biol. 2009;388:776–784. doi: 10.1016/j.jmb.2009.03.031. [DOI] [PubMed] [Google Scholar]
- 12.Mrazek J, Kreutmayer SB, Grässer FA, Polacek N, Huttenhofer A. Subtractive hybridization identifies novel differentially expressed ncRNA species in EBV-infected human B cells. Nucleic Acids Res. 2007;35:e73. doi: 10.1093/nar/gkm244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barth S, Pfuhl T, Mamiani A, Ehses C, Roemer K, Kremmer E, Jaker C, Hock J, Meister G, Grässer FA. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 2008;36:666–675. doi: 10.1093/nar/gkm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 15.Shi L, Cheng Z, Zhang J, Li R, Zhao P, Fu Z, You Y. hsa-mir-181a and hsa-mir-181b function as tumor suppressors in human glioma cells. Brain Res. 2008;1236:185–193. doi: 10.1016/j.brainres.2008.07.085. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Pall GS, Hamilton AJ. Improved northern blot method for enhanced detection of small RNA. Nat. Prot. 2008;3:1077–1084. doi: 10.1038/nprot.2008.67. [DOI] [PubMed] [Google Scholar]
- 18.Motsch N, Pfuhl T, Mrazek J, Barth S, Grässer FA. Epstein-Barr Virus-encoded latent membrane protein 1 (LMP1) induces the expression of the cellular microRNA miR-146a. RNA Biol. 2007;4:131–137. doi: 10.4161/rna.4.3.5206. [DOI] [PubMed] [Google Scholar]
- 19.Kim do N, Chae HS, Oh ST, Kang JH, Park CH, Park WS, Takada K, Lee JM, Lee WK, Lee SK. Expression of viral microRNAs in Epstein-Barr virus-associated gastric carcinoma. J. Virol. 2007;81:1033–1036. doi: 10.1128/JVI.02271-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl Acad. Sci. USA. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc. Natl Acad. Sci. USA. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosa A, Ballarino M, Sorrentino A, Sthandier O, De Angelis FG, Marchioni M, Masella B, Guarini A, Fatica A, Peschle C, et al. The interplay between the master transcription factor PU.1 and miR-424 regulates human monocyte/macrophage differentiation. Proc. Natl Acad. Sci. USA. 2007;104:19849–19854. doi: 10.1073/pnas.0706963104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laux G, Adam B, Strobl LJ, Moreau Gachelin F. The Spi-1/PU.1 and Spi-B ets family transcription factors and the recombination signal binding protein RBP-J kappa interact with an Epstein-Barr virus nuclear antigen 2 responsive cis-element. EMBO J. 1994;13:5624–5632. doi: 10.1002/j.1460-2075.1994.tb06900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dorsett Y, McBride KM, Jankovic M, Gazumyan A, Thai TH, Robbiani DF, Di Virgilio M, San-Martin BR, Heidkamp G, Schwickert TA, et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teng G, Hakimpour P, Landgraf P, Rice A, Tuschl T, Casellas R, Papavasiliou FN. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621–629. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jang KL, Shackelford J, Seo SY, Pagano JS. Up-regulation of beta-catenin by a viral oncogene correlates with inhibition of the seven in absentia homolog 1 in B lymphoma cells. Proc. Natl Acad. Sci. USA. 2005;102:18431–18436. doi: 10.1073/pnas.0504054102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roperch JP, Lethrone F, Prieur S, Piouffre L, Israeli D, Tuynder M, Nemani M, Pasturaud P, Gendron MC, Dausset J, et al. SIAH-1 promotes apoptosis and tumor suppression through a network involving the regulation of protein folding, unfolding, and trafficking: identification of common effectors with p53 and p21(Waf1) Proc. Natl Acad. Sci. USA. 1999;96:8070–8073. doi: 10.1073/pnas.96.14.8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dimitrova YN, Li J, Lee YT, Rios-Esteves J, Friedman DB, Choi HJ, Weis WI, Wang CY, Chazin WJ. Direct ubiquitination of {beta}-catenin by siah-1 and regulation by the exchange factor TBL1. J. Biol. Chem. 2010;285:13507–13516. doi: 10.1074/jbc.M109.049411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellei B, Pacchiarotti A, Perez M, Faraggiana T. Frequent beta-catenin overexpression without exon 3 mutation in cutaneous lymphomas. Mod. Pathol. 2004;17:1275–1281. doi: 10.1038/modpathol.3800181. [DOI] [PubMed] [Google Scholar]
- 30.Xia T, O’Hara A, Araujo I, Barreto J, Carvalho E, Sapucaia JB, Ramos JC, Luz E, Pedroso C, Manrique M, et al. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 2008;68:1436–1442. doi: 10.1158/0008-5472.CAN-07-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cosmopoulos K, Pegtel M, Hawkins J, Moffett H, Novina C, Middeldorp J, Thorley-Lawson DA. Comprehensive profiling of Epstein-Barr virus microRNAs in nasopharyngeal carcinoma. J. Virol. 2009;83:2357–2367. doi: 10.1128/JVI.02104-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jun SM, Hong YS, Seo JS, Ko YH, Yang CW, Lee SK. Viral microRNA profile in Epstein-Barr virus-associated peripheral T cell lymphoma. Br. J. Haematol. 2008;140:320–323. doi: 10.1111/j.1365-2141.2008.07186.x. [DOI] [PubMed] [Google Scholar]
- 33.Godshalk SE, Bhaduri-McIntosh S, Slack FJ. Epstein-Barr virus-mediated dysregulation of human microRNA expression. Cell cycle. 2008;7:3595–3600. doi: 10.4161/cc.7.22.7120. [DOI] [PubMed] [Google Scholar]
- 34.Cameron JE, Fewell C, Yin Q, McBride J, Wang X, Lin Z, Flemington EK. Epstein-Barr virus growth/latency III program alters cellular microRNA expression. Virology. 2008;382:257–266. doi: 10.1016/j.virol.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gatto G, Rossi A, Rossi D, Kroening S, Bonatti S, Mallardo M. Epstein-Barr virus latent membrane protein 1 trans-activates miR-155 transcription through the NF-kappaB pathway. Nucleic Acids Res. 2008;36:6608–6619. doi: 10.1093/nar/gkn666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kluiver J, Haralambieva E, de Jong D, Blokzijl T, Jacobs S, Kroesen BJ, Poppema S, van den Berg A. Lack of BIC and microRNA miR-155 expression in primary cases of Burkitt lymphoma. Genes, Chromosomes Cancer. 2006;45:147–153. doi: 10.1002/gcc.20273. [DOI] [PubMed] [Google Scholar]
- 37.Kluiver J, Poppema S, de Jong D, Blokzijl T, Harms G, Jacobs S, Kroesen BJ, van den Berg A. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J. Pathol. 2005;207:243–249. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- 38.Szczyrba J, Loprich E, Wach S, Jung V, Unteregger G, Barth S, Grobholz R, Wieland W, Stohr R, Hartmann A, et al. The MicroRNA Profile of Prostate Carcinoma Obtained by Deep Sequencing. Mol. Cancer Res. 2010;8:529–538. doi: 10.1158/1541-7786.MCR-09-0443. [DOI] [PubMed] [Google Scholar]
- 39.Anastasiadou E, Boccellato F, Vincenti S, Rosato P, Bozzoni I, Frati L, Faggioni A, Presutti C, Trivedi P. Epstein-Barr virus encoded LMP1 downregulates TCL1 oncogene through miR-29b. Oncogene. 2010;29:1316–1328. doi: 10.1038/onc.2009.439. [DOI] [PubMed] [Google Scholar]
- 40.Felton-Edkins ZA, Kondrashov A, Karali D, Fairley JA, Dawson CW, Arrand JR, Young LS, White RJ. Epstein-Barr virus induces cellular transcription factors to allow active expression of EBER genes by RNA polymerase III. J. Biol. Chem. 2006;281:33871–33880. doi: 10.1074/jbc.M600468200. [DOI] [PubMed] [Google Scholar]
- 41.Wang YX, Zhang XY, Zhang BF, Yang CQ, Chen XM, Gao HJ. Initial study of microRNA expression profiles of colonic cancer without lymph node metastasis. J. Dig. Dis. 2010;11:50–54. doi: 10.1111/j.1751-2980.2009.00413.x. [DOI] [PubMed] [Google Scholar]
- 42.Pallasch CP, Patz M, Park YJ, Hagist S, Eggle D, Claus R, Debey-Pascher S, Schulz A, Frenzel LP, Claasen J, et al. miRNA deregulation by epigenetic silencing disrupts suppression of the oncogene PLAG1 in chronic lymphocytic leukemia. Blood. 2009;114:3255–3264. doi: 10.1182/blood-2009-06-229898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fulci V, Colombo T, Chiaretti S, Messina M, Citarella F, Tavolaro S, Guarini A, Foa R, Macino G. Characterization of B- and T-lineage acute lymphoblastic leukemia by integrated analysis of MicroRNA and mRNA expression profiles. Genes, Chromosomes Cancer. 2009;48:, 1069–1082. doi: 10.1002/gcc.20709. [DOI] [PubMed] [Google Scholar]
- 44.Forrest AR, Kanamori-Katayama M, Tomaru Y, Lassmann T, Ninomiya N, Takahashi Y, de Hoon MJ, Kubosaki A, Kaiho A, Suzuki M, et al. Induction of microRNAs, mir-155, mir-222, mir-424 and mir-503, promotes monocytic differentiation through combinatorial regulation. Leukemia. 24:460–466. doi: 10.1038/leu.2009.246. [DOI] [PubMed] [Google Scholar]
- 45.Schmeier S, MacPherson CR, Essack M, Kaur M, Schaefer U, Suzuki H, Hayashizaki Y, Bajic VB. Deciphering the transcriptional circuitry of microRNA genes expressed during human monocytic differentiation. BMC Genom. 2009;10:595. doi: 10.1186/1471-2164-10-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsuzawa S, Takayama S, Froesch BA, Zapata JM, Reed JC. p53-inducible human homologue of Drosophila seven in absentia (Siah) inhibits cell growth: suppression by BAG-1. EMBO J. 1998;17:2736–2747. doi: 10.1093/emboj/17.10.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Medhioub M, Vaury C, Hamelin R, Thomas G. Lack of somatic mutation in the coding sequence of SIAH1 in tumors hemizygous for this candidate tumor suppressor gene. Int. J. Cancer. 2000;87:794–797. [PubMed] [Google Scholar]
- 48.Xu Z, Sproul A, Wang W, Kukekov N, Greene LA. Siah1 interacts with the scaffold protein POSH to promote JNK activation and apoptosis. J. Biol. Chem. 2006;281:303–312. doi: 10.1074/jbc.M509060200. [DOI] [PubMed] [Google Scholar]
- 49.Clappier E, Cuccuini W, Kalota A, Crinquette A, Cayuela JM, Dik WA, Langerak AW, Montpellier B, Nadel B, Walrafen P, et al. The C-MYB locus is involved in chromosomal translocation and genomic duplications in human T-cell acute leukemia (T-ALL), the translocation defining a new T-ALL subtype in very young children. Blood. 2007;110:1251–1261. doi: 10.1182/blood-2006-12-064683. [DOI] [PubMed] [Google Scholar]
- 50.Lahortiga I, De Keersmaecker K, Van Vlierberghe P, Graux C, Cauwelier B, Lambert F, Mentens N, Beverloo HB, Pieters R, Speleman F, et al. Duplication of the MYB oncogene in T cell acute lymphoblastic leukemia. Nat. Genet. 2007;39:593–595. doi: 10.1038/ng2025. [DOI] [PubMed] [Google Scholar]
- 51.Persson M, Andren Y, Mark J, Horlings HM, Persson F, Stenman G. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc. Natl Acad. Sci. USA. 2009;106:18740–18744. doi: 10.1073/pnas.0909114106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bender TP, Thompson CB, Kuehl WM. Differential expression of c-myb mRNA in murine B lymphomas by a block to transcription elongation. Science. 1987;237:1473–1476. doi: 10.1126/science.3498214. [DOI] [PubMed] [Google Scholar]
- 53.Navarro F, Gutman D, Meire E, Caceres M, Rigoutsos I, Bentwich Z, Lieberman J. miR-34a contributes to megakaryocytic differentiation of K562 cells independently of p53. Blood. 2009;114:2181–2192. doi: 10.1182/blood-2009-02-205062. [DOI] [PubMed] [Google Scholar]
- 54.Chung EY, Dews M, Cozma D, Yu D, Wentzel EA, Chang TC, Schelter JM, Cleary MA, Mendell JT, Thomas-Tikhonenko A. c-MYB oncoprotein is an essential target of the dleu2 tumor suppressor microRNA cluster. Cancer Biol. Ther. 2008;7:1758–1764. doi: 10.4161/cbt.7.11.6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-MYB. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 56.Zhao H, Kalota A, Jin S, Gewirtz AM. The c-myb proto-oncogene and microRNA-15a comprise an active autoregulatory feedback loop in human hematopoietic cells. Blood. 2009;113:505–516. doi: 10.1182/blood-2008-01-136218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee J, Li Z, Brower-Sinning R, John B. Regulatory circuit of human microRNA biogenesis. PLoS Comp. Biol. 2007;3:e67. doi: 10.1371/journal.pcbi.0030067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rai D, Kim SW, McKeller MR, Dahia PL, Aguiar RC. Targeting of SMAD5 links microRNA-155 to the TGF-beta pathway and lymphomagenesis. Proc. Natl Acad. Sci. USA. 107:3111–3116. doi: 10.1073/pnas.0910667107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol. Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 60.Sakhinia E, Faranghpour M, Liu Yin JA, Brady G, Hoyland JA, Byers RJ. Routine expression profiling of microarray gene signatures in acute leukaemia by real-time PCR of human bone marrow. Br. J. Haematol. 2005;130:233–248. doi: 10.1111/j.1365-2141.2005.05594.x. [DOI] [PubMed] [Google Scholar]
- 61.Grässer FA, Graf T, Lipsick JS. Protein truncation is required for the activation of the c-myb proto-oncogene. Mol. Cell. Biol. 1991;11:3987–3996. doi: 10.1128/mcb.11.8.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.