Abstract
Telomere maintenance and DNA repair are crucial processes that protect the genome against instability. RTEL1, an essential iron–sulfur cluster-containing helicase, is a dominant factor that controls telomere length in mice and is required for telomere integrity. In addition, RTEL1 promotes synthesis-dependent strand annealing to direct DNA double-strand breaks into non-crossover outcomes during mitotic repair and in meiosis. Here, we review the role of RTEL1 in telomere maintenance and homologous recombination and discuss models linking RTEL1’s enzymatic activity to its function in telomere maintenance and DNA repair.
INTRODUCTION
Telomeres are protective structures at the end of chromosomes, the maintenance of which is essential for genomic stability (1). In vertebrates, telomeres are composed of repetitive TTAGGG DNA (2) and associated proteins, which form a core complex known as the Shelterin complex (3). Telomeres protect chromosomes by distinguishing chromosomal ends from DNA double-strand breaks (DSBs), a function that is essential in avoiding chromosome end-to-end fusions and inappropriate recombination events (4).
Telomeres can form a protective lariat-like structure, referred to as the telomeric-loop, or T-loop (5). T-loops are created through strand invasion by the 3′ single-stranded overhang of telomeric DNA into duplex telomere repeats. This strand invasion displaces the identical sequence strand of the duplex telomeric DNA and so forms a displacement-loop (D-loop) at the base of the T-loop. The D-loop is also an intermediate in the DNA repair pathway via homologous recombination (HR) (6). This repair pathway is the main method for repairing DSBs when sister chromatid templates are available and is also required for meiotic recombination. How the T-loop is resolved during replication or how the invaded strand in the D-loop structure is displaced to promote repair to a non-crossover (NCO) outcome is still largely unknown.
In addition to the T-loop configuration, the guanine (G)-rich nature of the telomere may also pose a challenge for telomere maintenance. In vitro, single-stranded G-rich telomeric sequences are capable of forming stable structures called G-quadruplex (or G4) DNA (7). In vivo, G4 DNA might form at telomeres during replication, repair and transcription. The resolution of T-loops and telomeric G4 DNA could be important for telomere maintenance, and therefore, genomic stability.
Rtel1 (for Regulator of Telomere Length 1) is an essential helicase that plays a crucial role in telomere maintenance and DNA repair (8,9). Rtel1 was originally discovered as the dominant factor in setting telomere length in mice (9). In the absence of Rtel1, telomeres are not maintained and chromosome fusions are observed. In addition, RTEL1 was found to be a key protein in the repair of DSBs (8,10). It disrupts D-loops in vitro and promotes synthesis-dependent strand annealing (SDSA) in vivo. Notably, RTEL1 activity is not limited to mitotic cells, as RTEL-1 is required to regulate meiotic recombination in Caenorhabditis elegans. Here, we review the emerging role of RTEL1 at telomeres and in DNA repair and introduce a model linking the anti-recombinase activity of RTEL1 to its functions in telomere maintenance and DNA repair.
RTEL1 AND TELOMERE LENGTH REGULATION IN THE MOUSE
Telomeres shorten with each round of DNA replication (11). Typical and sporadic losses of telomeric DNA are compensated for by the enzyme telomerase. In yeast (12), maize (13) and Arabidopsis (14), the telomere length set point seems to be determined by multiple genes. In mice, telomere length is controlled by genetic (9) as well as epigenetic factors (15). Rtel1 was identified as a dominant genetic factor setting telomere length in mice (9,16). Most laboratory mice including Mus musculus have long telomeres, with lengths between 25 and 150 kb (17), but a related mouse species, Mus spretus, has telomeres between 5 and 15 kb (18), similar to the telomere length in human cells (19–21). Hodes and co-workers (16) found that telomeres of Mus spretus-derived chromosomes in the offspring of crosses between M. spretus with M. musculus were significantly longer than in the M. spretus parent, suggesting that a dominant genetic mechanism was elongating M. spretus telomeres during development. Genotype mapping pointed to a locus on distal chromosome 2 containing a dominant factor(s) determining telomere length setting in mice. This factor was shown to be Rtel1 based on the finding that Rtel1 expression from the M. musculus parent was required to elongate the telomeres of M. spretus-derived chromosomes (9). Mice lacking Rtel1 die around Day 10–11 with defects in multiple organs. The average telomere length of Rtel1-deficient embryonic stem cells is around 68% of that in wild-type cells (9). It is unknown how Rtel1 determines this telomere length equilibrium and how the difference between M. musculus and M. spretus Rtel1 determines a long or short telomere phenotype in these mice. However, differences between M. musculus and M. spretus Rtel1 are found in the promoter region, in the last four exons of the gene, and in mRNA splice variants (9). In addition, it is unknown if RTEL1 also determines telomere length in humans. However, no association was found between telomere length and single-nucleotide polymorphisms (SNPs) in RTEL1 (22).
RTEL1 AND OTHER FeS CLUSTER-CONTAINING HELICASES
RTEL1 belongs within the DEAH subfamily of the Superfamily 2 (SF2) helicases and is classified as a RAD3-related helicase with 5′ to 3′ directionality (23,24). An iron–sulfur (FeS) domain classifies RTEL1 within a very small subclass of FeS cluster-containing DNA helicases (25). Xeroderma pigmentosum group D (XPD) is the founding member of this subclass, which also contains ChlR1 and FANCJ, in addition to RTEL1 (26). Mutations in XPD, FANCJ and ChlR1 are responsible for the genetic disorders xeroderma pigmentosum (XP) (27), Fanconi anemia (FA) (28–31) and Warsaw breakage syndrome (32). Thus far, heritable mutations in RTEL1 have not been linked to specific human genetic syndromes. However, two independent studies identified intronic SNPs in RTEL1 associated with glioma susceptibility (33,34). Furthermore, RTEL1 is located in a gene-rich cluster (20q13.3) that is amplified in several human cancers (35–38). It is unclear if RTEL1 is directly implicated in these malignancies, since this cluster also contains other tumor susceptibility genes (38–40). The key characteristic of the XPD family of helicases is the conservation of four cysteine residues, which bind iron ions to form an FeS cluster (25). Removal of the archaeal XPD FeS domain abolishes its helicase activity and can destabilize its tertiary structure (25,41). The crystal structures of archaeal XPDs are consistent with a role of the FeS domain in separating the two strands of the DNA duplex (41–43). This is supported by the findings that the FeS cluster-containing domain of archaeal XPD recognizes the single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA) junction and that the DNA duplex is opened near the FeS domain (44). Based on the sequence similarity of the helicase core domain, it is likely that other FeS cluster-containing helicases, including RTEL1, also recognize the ssDNA–dsDNA junction. Disruption of the FeS cluster in RAD3 (yeast XPD) causes defective excision repair resulting in ultraviolet (UV) sensitivity in vivo (25). Clinically relevant mutations giving rise to syndromes in XPD and FANCJ patients cause the destabilization of the FeS cluster in archaeal XPD (25,41,42).
The severity of FA, XP and Warsaw breakage syndrome emphasize the important roles of the helicases containing FeS clusters in the maintenance of genome stability. Changes in the availability of iron and in the biogenesis of FeS clusters will likely influence RTEL1, XPD, FANCJ and ChlR1 protein levels. Interference with the packaging of FeS clusters into proteins was recently found to contribute to genomic instability (45). It is likely that compromised levels of FeS containing helicases are responsible for this phenotype.
In addition to its helicase motifs and FeS domain, a conserved eight amino acid PIP [proliferating cell nuclear antigen (PCNA)-interacting protein (46)] box is present at the C-terminus of the RTEL1 protein. PCNA is a highly conserved eukaryotic protein that functions in DNA replication as a sliding clamp encircling the DNA and acts as a cofactor for DNA polymerases. Currently, the RTEL1–PCNA interaction is speculative, but the RTEL1 PIP box could be necessary to properly orient the protein relative to DNA.
RTEL1 ANTAGONIZES HOMOLOGOUS RECOMBINATION IN VITRO
Although RTEL1 was originally predicted to be a helicase involved in recombination and repair, a biochemical function had remained elusive. Recent studies have now shown that RTEL1 antagonizes homologous recombination events upon DSB formation (8,10). After a DSB is sensed, the two duplex ends are resected to give 3′ ssDNA that is subsequently bound by Replication Protein A (RPA). RPA is then displaced by RAD51 recombinase to form a nucleoprotein filament, which searches for and invades the intact homologous dsDNA template (47), resulting in the formation of a D-loop structure (48). The invaded 3′ end is extended by a DNA synthesis reaction, copying the information on the homologous DNA template. Subsequently, the strand is displaced and annealed to the other processed DNA end, and the repair reaction is completed by additional DNA synthesis and ligation. A DSB repaired via this SDSA pathway yields a NCO repair product (49,50). Alternatively, if the other processed DNA strand is also captured in the D-loop, a double Holliday junction intermediate will be formed. Resolution of a double Holliday junction can give rise to crossover (CO) repair products (51).
Human RTEL1 possesses biochemical properties consistent with a role in antagonizing HR by promoting SDSA (8,10,52). In vitro, purified RTEL1 inhibits the formation of D-loops in an ATP-dependent manner (Figure 1) (8). In addition, RTEL1 was found to actively reverse HR by disrupting preformed D-loops in the presence of calcium (8), which activates and stabilizes the nucleoprotein filament, making it capable of performing the strand exchange reaction (53). In contrast to RTEL1, BLM, the helicase defective in human Bloom’s syndrome, and FANCJ are unable to dissociate preformed D-loops in the presence of calcium (8,54,55), but can catalyze the disruption of a RAD51 nucleoprotein filament in an unstable state (54,55). RTEL1 has no detectable disruption activity toward single-stranded RAD51 nucleoprotein filaments (8), indicating a unique anti-recombinase mechanism of RTEL1. Thus, the biochemical activity of RTEL1 is distinct from other known anti-recombinases, including yeast Srs2 and mammalian FBH1, BLM, FANCJ and RECQL5, all of which can disrupt single-stranded RAD51 nucleoprotein filaments (54–59). In the presence of RPA, RTEL1 was shown to preferentially disrupt a 3′ ssDNA invasion D-loop with a 5′ overhang but had almost no activity on a D-loop with a 5′ ssDNA invasion and a 3′ overhang, or a substrate with no overhang (10). The biochemical activity of RTEL1 is perhaps most similar to the yeast helicase Mph1, which is also able to disrupt D-loops; however, Mph1 was shown to have similar affinity for D-loops with a 5′ ssDNA invasion and 3′ overhang, with a 3′ ssDNA invasion and 5′ overhang and with no overhang (60). While Mph1 promotes NCO repair of mitotic DSBs (60), it is thus far unclear whether Mph1 might function to regulate meiotic recombination. The fact that RTEL1 can unwind a 3′ end invaded D-loop supports the hypothesis that RTEL1 stimulates SDSA-mediated DSB repair, during which such enzymatic activity is required to disassemble the D-loop intermediate and thereby promote NCO repair products (52). Detailed analysis of the phenotypes of C. elegans rtel-1 mutants and human Rtel1-deficient cells is consistent with the hypothesis that the biochemical activity of RTEL1 in disrupting D-loops is responsible for both mitotic repair and regulating meiotic recombination.
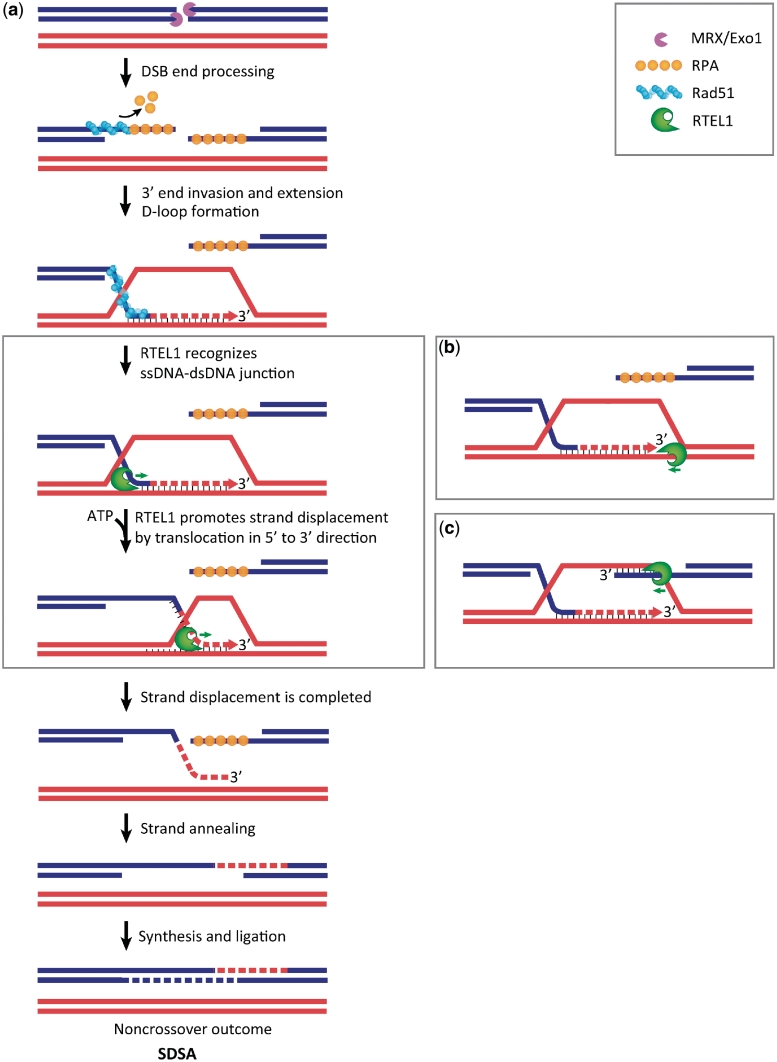
Figure 1.
Model for RTEL1 promoting synthesis-dependent strand annealing (SDSA). (a) After a DSB is sensed, the two duplex ends are resected to give 3′-ssDNA that is subsequently bound by RPA (orange). RPA is replaced by RAD51 recombinase (blue) to form a nucleoprotein filament that searches and invades the homologous dsDNA, forming a D-loop. The invaded strand serves as a primer for DNA synthesis, which copies the information on the homologous DNA strand. RTEL1 (green) promotes SDSA by the displacement of a 3′-end invaded D-loop. Although the mechanism by which RTEL1 promotes strand displacement is unknown, RTEL1 is proposed to recognize the ssDNA–dsDNA junction via its FeS domain, and act as a 5′- to 3′-helicase. The displaced strand can now anneal to the other processed DNA end and the repair reaction will be completed by DNA synthesis and ligation, resulting in a non-crossover outcome. Note that RTEL1 may displace invading strands from both of the processed DNA ends, however, only one invading strand is shown for clarity. Alternative models of RTEL1 action include: (b) RTEL1 translocation in a 5′- to 3′-direction along template strand, as opposed to the invading strand. (c) Although it is thought that SDSA occurs earlier than double Holliday junction formation, it is also possible, though less likely, that RTEL1 may act on the other processed DNA end. This activity might prevent the other processed DNA end from becoming part of the repair reaction, known as second end capture, and so prevent the formation of a double Holliday junction, which could be resolved as a crossover product.
TELOMERE LOSS IN Rtel1-DEFICIENT CELLS
Chromosome ends with low or undetectable telomere repeats are abundant in Rtel1 null ES cells (9), suggesting significant telomere loss. One possible explanation for this telomere phenotype might be that Rtel1 is required to open the D-loop structure within the T-loop (Figure 2a). Importantly, the telomeric D-loop resembles the preferred RTEL1 substrate: a 3′ ssDNA invaded D-loop with a 5′ overhang. Potentially, telomeric D-loops could be inappropriately processed by the HR pathway since this structure resembles a strand-invasion intermediate in HR. This was observed in mammalian cells expressing a truncated form of TRF2 (telomere repeat-binding factor 2) in which HR dependent large T-loop-sized circular telomeric DNAs were generated (61). In the absence of RTEL1, T-loops might be less efficiently resolved. Failing to open the T-loop for replication and/or transcription could lead to large telomeric deletions causing the telomere phenotype seen in Rtel1-deficient cells. In addition, Rtel1 could be required to prevent the 3′ single-stranded telomere end from invading the telomeres of other chromosomes (Figure 2b). This activity might prevent telomere recombination events, chromosome entanglements and subsequent breakage when attached chromosomes attempt to segregate during mitosis. Two other helicases important for telomere maintenance are the human RecQ family members Werner syndrome protein (WRN) and BLM. Both helicases interact biochemically and functionally with TRF1, TRF2, and POT1 (protection of telomeres 1) (62–65). In vitro, WRN and BLM have been implicated in unwinding the telomeric D-loop (63,64,66). Future experiments will determine if RTEL1 similarly unwinds telomeric D-loop structures and if this activity is required in vivo.
Figure 2.
Model for RTEL1 anti-recombinase function in telomere maintenance. (a) A telomere end is protected from being recognized as a DSB by folding back onto itself in a structure known as the T-loop. At the base of the T-loop, the naturally occurring 3′-ssDNA overhang invades the double-stranded repetitive telomere DNA forming a D-loop. The similarity of D-loop intermediates during DNA repair and telomere maintenance have led to the proposal that RTEL1 (green) may reverse T-loops. (b) D-loops can also form at telomeres by invasion of the 3′-ssDNA telomere overhang into sister or non-sister telomeres, leading to telomere strand-exchange and chromosome entanglements. RTEL1 may reverse inappropriate T-loop formation and thereby prevent telomere strand exchange.
An alternative explanation for the short telomere phenotype of Rtel1 null cells could be that RTEL1, which shares sequence similarity with the helicase FANCJ, may also be required to resolve G-quadruplex structures (67,68). G4 DNA could naturally be present at the telomere or arise during lagging strand replication of G-rich telomeric DNA, as was proposed (9). In addition, hybrid G4 DNA/RNA structures might form following transcription of telomeric DNA (69–71). FANCJ patient cells present genomic deletions corresponding to a G4 DNA signature motif (67) and are hypersensitive to telomestatin (68), a compound which specifically binds G4 DNA (72,73). In dog-1 (C. elegans FANCJ) mutants, large G-tract deletions throughout the genome and mild telomeric instability was observed (74–76). By contrast, in rtel-1 mutants genomic G-tract deletions were not detected and a telomere phenotype was not reported (8,74). It is interesting to note that C. elegans rtel-1 dog-1 double mutant animals display a synthetic lethal phenotype in which sterility results from replication-associated problems in the germline prior to meiosis [(8); Youds and Boulton, unpublished data]. Thus, some overlapping function for rtel-1 and dog-1 during replication is likely, at least in C. elegans. Experiments to conditionally knock out both Rtel1 and FancJ might reveal partially overlapping roles with regard to G4 DNA maintenance, particularly at telomeres. In addition to FANCJ, the 3′ to 5′ helicases WRN and BLM are able to unwind G4 DNA in vitro (77–79). Table 1 compares the existing data on RTEL1 and FANCJ.
Table 1.
Summary of the relationship of RTEL1 and FANCJ
| dog-1/FancJ/FANCJ | rtel-1/Rtel1/RTEL1 | |
|---|---|---|
| Best BLAST hit | rtel-1/Rtel1/RTEL1 | dog-1/FancJ/FANCJ |
| Domains of interest in human protein | Helicase domains including DEXDc, HELICc2, DEAD_2 and DinG, BRCA1-interaction site, Fe-S motif | Helicase domains including DEXDc2, DEAD_2, HELICx2 and DinG, PIP box, Fe-S motif |
| Synthetic lethal in C. elegans when combined with mutation in | rtel-1 | dog-1, mus-81, him-6/BLM, rcq-5/RECQ5 |
| DNA damage sensitivity? | Yes: ICL-inducing agents | Yes: ICL-inducing agents, camptothecin |
| Telomere phenotype? | dog-1: mild instability | Rtel1/RTEL1: dramatic loss |
| Elevated recombination? | None identified | Yes |
| Activity on in vitro biochemical substrate | G4 DNA, forked duplex DNA, D-loop | D-loop recombination intermediate |
| Activity with respect to recombination | Inhibits RAD51 strand exchange | Disassembles D-loop intermediate |
| Human patients identified? | Yes: Fanconi anemia subgroup J | None identified |
A recent study revealed that mammalian telomeres are difficult regions to replicate and resemble fragile sites (80,81). In the absence of Trf1, the replication fork has a greater tendency to stall when encountering telomeric DNA. It was proposed that Trf1 might act by recruiting or activating helicases like Rtel1 and Blm to telomeres (80). Rtel1-deficient ES cells and single Rtel1 and Blm knockdown mouse embryonic fibroblasts (MEFs) both show a mild fragile-telomere phenotype, which in MEFs is epistatic to the deletion of Trf1 (80). These results indicate that Rtel1 and Trf1 act in the same pathway to suppress telomere fragility. Additional functional studies of the biochemical and cellular activities of RTEL1, FANCJ, WRN and BLM may reveal more specific and redundant roles of these proteins in the regulation of the telomeric D-loop, G-quadruplexes, and other biological structures.
RTEL1 REGULATES MITOTIC AND MEIOTIC RECOMBINATION IN VIVO
Besides the requirement of Rtel1 for telomere maintenance, a role in DNA repair was envisioned upon studying differentiating Rtel1-deficient ES cells (9). Interestingly, within hours of the induction of differentiation, Rtel1-deficient ES cells showed a variety of chromosomal abnormalities, including broken chromosomes and chromatid gaps, pointing to a more general role of Rtel1 in maintaining genomic stability. Subsequently, a role for RTEL-1/RTEL1 in regulating HR was elucidated from work in C. elegans and human cell lines (8).
In the budding yeast Saccharomyces cerevisiae, the DNA helicase Srs2 is an anti-recombinase that regulates HR (56,57). Yeast SRS2 is synthetic lethal with SGS1, the sole S. cerevisiae RecQ helicase that bears homology to five human RecQ helicases, particularly to human BLM (82). This synthetic lethality is due to an accumulation of toxic recombination intermediates (82). No sequence homologs of SRS2 exist in higher organisms, but the SGS1 homolog in C. elegans is him-6/BLM (83). Therefore, based on the synthetic lethality of SRS2 and SGS1 in yeast, a screen to find a candidate for a C. elegans gene with an Srs2-like function was conducted by searching for genes that showed synthetic lethality when knocked-down in a him-6/BLM mutant strain. rtel-1 was identified as being synthetic lethal with him-6/BLM, making it a candidate for the SRS2 analog in C. elegans. Furthermore, rtel-1 showed synthetic lethal phenotypes when combined with mutations in mus-81, dog-1 and rcq-5 (8). The synthetic lethality in these strains correlated with an accumulation of RAD-51 foci in the germline, a marker of persistent HR intermediates. Furthermore, rtel-1 mutation causes synthetic sick phenotypes when combined with deletions in either rfs-1 or helq-1, two genes that encode proteins functioning late in HR to disassemble RAD-51 from double-stranded DNA filaments (84).
Caenorhabditis elegans rtel-1 and dog-1 mutants and human cells deficient for RTEL1 and FANCJ are sensitive to DNA interstrand crosslinks (ICLs) (8,28–31,75). Based on epistasis experiments in C. elegans, dog-1 acts in the fcd-2/FANCD2 pathway, while rtel-1 appears to act in a separate pathway for ICL repair. rtel-1 mutants are also sensitive to camptothecin (8). Surprisingly, C. elegans rtel-1 mutants and RTEL1 depleted cells are not sensitive to ionizing radiation (IR). However, it is possible that in the absence of RTEL1, all DSBs are efficiently repaired with a CO outcome. In accordance with a role of RTEL1 in antagonizing recombination, recombination frequencies were increased in RTEL1-depleted HeLa cells (8).
In addition to mitotic DNA repair, RTEL1 is also required to promote NCO repair during meiosis (10). Meiotic DSBs are not randomly distributed along chromosomes, but tend to occur in specific regions (85). Through homologous recombination, DSBs can be repaired by either a CO or a NCO event (86). The number of DSBs created exceeds the number of final CO events, in some organisms by more than ten times (87,88). How specific DSBs are selected to become COs is unknown, but the mechanism behind this ensures that each pair of homologs gets at least one CO (known as the obligate CO). In addition, a mechanism called CO interference regulates the distribution of COs along the chromosome in such a way that COs tend to occur further apart from each other than expected by chance (89). When the number of meiotic DSBs is reduced, the number of COs is maintained at the expense of NCOs; this is called CO homeostasis (90).
Recently, C. elegans RTEL-1 was shown to enforce both meiotic CO interference and homeostasis (10). Wild-type C. elegans only ever have a single CO per chromosome (91,92). rtel-1 mutants showed up to three COs per chromosome that were randomly distributed, suggesting a lack of CO interference. Furthermore, the CO increase was not due to increased formation of meiotic DSBs (10), distinguishing the mechanism of RTEL-1 function from that of condensin complex components (92,93). When DSBs were generated using IR, wild-type animals showed a small increase in COs, while rtel-1 mutants displayed a large, dose-dependent increase in COs, supporting a function for RTEL-1 in maintaining CO homeostasis. In yeast, it was shown that NCOs and COs appear with different kinetics during meiosis, suggesting that they are regulated by different mechanisms (49,94). In addition, NCOs are primarily repaired by SDSA in yeast meiosis (94). Thus, it is likely that NCOs occur via meiotic SDSA and that the D-loop disassembly ability of RTEL-1 is required for this pathway. Interestingly, the increase in the number of ZHP-3 foci, a protein restricted to recombination foci (95), was relatively small compared with the increase in CO events in rtel-1 mutants (10). Furthermore, after rtel-1 mutants were treated with IR, the number of COs increased, but ZHP-3 foci did not. These results indicate that two classes of meiotic recombination events are elevated in rtel-1 mutants: (i) ZHP-3-associated, obligate-type COs (small increase) and (ii) COs produced by recombination events not associated with ZHP-3 (predominant). Like in S. cerevisiae (86,96), this second class of COs requires MUS-81 and is likely interference-independent (10). This finding indicates that meiotic HR intermediates not reversed by RTEL-1 during SDSA will be MUS-81 substrates resulting in COs, which is consistent with the embryonic lethality of rtel-1 mus-81 double mutants (8).
Mets and Meyer (92) have shown that the average number of DSBs per C. elegans chromosome in wild-type meiosis is 2.1, while just one of these is resolved into a CO. How RTEL-1 is recruited and regulated in a way that just half of the DSBs induced during wild-type meiosis result in a CO is an intriguing question. Designated COs may be protected from RTEL-1 activity by certain proteins; alternatively, RTEL-1 activity may be carefully regulated such that RTEL-1 only becomes active after the obligate COs have been completed.
It will be of interest to determine if RTEL1 is also required for CO interference and homeostasis in mammals. Human RTEL1 could have similar functions in meiosis since it was shown to disassemble D-loop recombination intermediates in vitro (8,10). In mice, the highest Rtel1 expression is detected in spermatogonia and meiotic spermatocytes within the testes (9). Although full knockout of Rtel1 function is lethal, the requirement of mouse Rtel1 in meiosis could potentially be studied if a (pre)meiotic-Cre transgenic line were available, since mice with conditional knock-out alleles have been established (97).
CONCLUDING REMARKS
RTEL1 is an essential helicase, a dominant factor in setting telomere length in mice and is required for telomere and genome maintenance. Furthermore, RTEL1 is a key protein in mitotic and meiotic DSB repair and promotes NCO repair outcomes through SDSA. As it is currently unknown how RTEL1 is recruited and regulated during DSB repair, in both mitotic and meiotic cells, determining its post-translational modifications and interacting proteins may shed light on these questions. Live-cell imaging of fluorescent-tagged RTEL1 and co-localization studies with other DNA repair proteins will enhance our understanding of the biological function and dynamics of RTEL1.
The data available on RTEL1 so far do not exclude the possibility that RTEL1 is preferentially recruited to, or has higher enzymatic activity at G-rich regions in the genome. In humans, 25 618 meiotic recombination hot spots have been identified (98). Interestingly, potential G4 DNA-forming sequences were found to be significantly enriched within recombination hot spots (99). Also, G4 DNA was predicted to be formed within a 50-bp window around all of the top seven previously reported short recombination enriched sequences (98,99). It is possible that RTEL1 is recruited to meiotic DSBs by G4 DNA structures at recombination hot spots to prevent COs from occurring. Possible recruitment to G4 DNA could be direct or indirect via binding to other G4 DNA-binding proteins. In yeast, Mre11 and the meiosis-specific protein Hop1 bind G4 DNA, and Hop1 itself promotes G4 DNA formation (100,101); thus, homologs and binding partners of these proteins are potential recruiters of RTEL1 to G4 DNA.
Finally, given that RTEL1 has already been shown to play multiple roles in maintaining genome stability, both at telomeres and more generally at sites of mitotic DNA damage, as well as regulating meiotic recombination, RTEL1 could be classified as having a tumor suppressive function. Certainly, Rtel1-deficient cells will undergo uncontrolled HR and that may result in telomere loss and/or gain and chromosomal rearrangements and translocations, all of which are characteristic of cancer cells. Conversely, upregulated RTEL1 function might prevent HR when it is needed as a legitimate means for repair. RTEL1 mutation has already been associated with risk of glioma (33,34), and RTEL1 was shown to be overexpressed in gastrointestinal tract tumors (35). A challenge ahead will be to determine whether RTEL1 dysfunction plays a causative role in these and other cancers.
FUNDING
The Canadian Institutes of Health Research (MOP38075, GMH79042 to P.M.L.); Terry Fox Foundation (018006 to P.M.L.); Cancer Research UK (to S.J.B.). S.J.B. is a Royal Society Wolfson Research Merit Award holder. Funding for open access charge: Canadian Cancer Society Research Institute and the Terry Fox Foundation.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Ester Falconer for helpful comments and critically reading this article.
REFERENCES
- 1.Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 2.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl Acad. Sci. USA. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 4.de Lange T. How telomeres solve the end-protection problem. Science. 2009;326:948–952. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 6.de Lange T. T-loops and the origin of telomeres. Nat. Rev. Mol. Cell. Biol. 2004;5:323–329. doi: 10.1038/nrm1359. [DOI] [PubMed] [Google Scholar]
- 7.Sen D, Gilbert W. Guanine quartet structures. Methods Enzymol. 1992;211:191–199. doi: 10.1016/0076-6879(92)11012-8. [DOI] [PubMed] [Google Scholar]
- 8.Barber LJ, Youds JL, Ward JD, McIlwraith MJ, O'Neil NJ, Petalcorin MI, Martin JS, Collis SJ, Cantor SB, Auclair M, et al. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135:261–271. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding H, Schertzer M, Wu X, Gertsenstein M, Selig S, Kammori M, Pourvali R, Poon S, Vulto I, Chavez E, et al. Regulation of murine telomere length by Rtel: an essential gene encoding a helicase-like protein. Cell. 2004;117:873–886. doi: 10.1016/j.cell.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 10.Youds JL, Mets DG, McIlwraith MJ, Martin JS, Ward JD, ONeil NJ, Rose AM, West SC, Meyer BJ, Boulton SJ. RTEL-1 enforces meiotic crossover interference and homeostasis. Science. 2010;327:1254–1258. doi: 10.1126/science.1183112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lansdorp PM. Major cutbacks at chromosome ends. Trends Biochem. Sci. 2005;30:388–395. doi: 10.1016/j.tibs.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Walmsley RM, Petes TD. Genetic control of chromosome length in yeast. Proc. Natl Acad. Sci. USA. 1985;82:506–510. doi: 10.1073/pnas.82.2.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burr B, Burr FA, Matz EC, Romero-Severson J. Pinning down loose ends: mapping telomeres and factors affecting their length. Plant Cell. 1992;4:953–960. doi: 10.1105/tpc.4.8.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shakirov EV, Shippen DE. Length regulation and dynamics of individual telomere tracts in wild-type Arabidopsis. Plant Cell. 2004;16:1959–1967. doi: 10.1105/tpc.104.023093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiang YJ, Calado RT, Hathcock KS, Lansdorp PM, Young NS, Hodes RJ. Telomere length is inherited with resetting of the telomere set-point. Proc. Natl Acad. Sci. USA. 2010;107:10148–10153. doi: 10.1073/pnas.0913125107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu L, Hathcock KS, Hande P, Lansdorp PM, Seldin MF, Hodes RJ. Telomere length regulation in mice is linked to a novel chromosome locus. Proc. Natl Acad. Sci. USA. 1998;95:8648–8653. doi: 10.1073/pnas.95.15.8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kipling D, Cooke HJ. Hypervariable ultra-long telomeres in mice. Nature. 1990;347:400–402. doi: 10.1038/347400a0. [DOI] [PubMed] [Google Scholar]
- 18.Starling JA, Maule J, Hastie ND, Allshire RC. Extensive telomere repeat arrays in mouse are hypervariable. Nucleic Acids Res. 1990;18:6881–6888. doi: 10.1093/nar/18.23.6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cross SH, Allshire RC, McKay SJ, McGill NI, Cooke HJ. Cloning of human telomeres by complementation in yeast. Nature. 1989;338:771–774. doi: 10.1038/338771a0. [DOI] [PubMed] [Google Scholar]
- 20.Allshire RC, Gosden JR, Cross SH, Cranston G, Rout D, Sugawara N, Szostak JW, Fantes PA, Hastie ND. Telomeric repeat from T. thermophila cross hybridizes with human telomeres. Nature. 1988;332:656–659. doi: 10.1038/332656a0. [DOI] [PubMed] [Google Scholar]
- 21.de Lange T, Shiue L, Myers RM, Cox DR, Naylor SL, Killery AM, Varmus HE. Structure and variability of human chromosome ends. Mol. Cell Biol. 1990;10:518–527. doi: 10.1128/mcb.10.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mirabello L, Yu K, Kraft P, De Vivo I, Hunter DJ, Prescott J, Wong JY, Chatterjee N, Hayes RB, Savage SA. The association of telomere length and genetic variation in telomere biology genes. Hum. Mutat. 2010;31:1050–1058. doi: 10.1002/humu.21314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudolf J, Makrantoni V, Ingledew WJ, Stark MJ, White MF. The DNA repair helicases XPD and FancJ have essential iron–sulfur domains. Mol. Cell. 2006;23:801–808. doi: 10.1016/j.molcel.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 26.White MF. Structure, function and evolution of the XPD family of iron–sulfur-containing 5′–>3′ DNA helicases. Biochem. Soc. Trans. 2009;37:547–551. doi: 10.1042/BST0370547. [DOI] [PubMed] [Google Scholar]
- 27.Andressoo JO, Hoeijmakers JH. Transcription-coupled repair and premature ageing. Mutat. Res. 2005;577:179–194. doi: 10.1016/j.mrfmmm.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 28.Bridge WL, Vandenberg CJ, Franklin RJ, Hiom K. The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nat. Genet. 2005;37:953–957. doi: 10.1038/ng1627. [DOI] [PubMed] [Google Scholar]
- 29.Levitus M, Waisfisz Q, Godthelp BC, de Vries Y, Hussain S, Wiegant WW, Elghalbzouri-Maghrani E, Steltenpool J, Rooimans MA, Pals G, et al. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat. Genet. 2005;37:934–935. doi: 10.1038/ng1625. [DOI] [PubMed] [Google Scholar]
- 30.Levran O, Attwooll C, Henry RT, Milton KL, Neveling K, Rio P, Batish SD, Kalb R, Velleuer E, Barral S, et al. The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat. Genet. 2005;37:931–933. doi: 10.1038/ng1624. [DOI] [PubMed] [Google Scholar]
- 31.Litman R, Peng M, Jin Z, Zhang F, Zhang J, Powell S, Andreassen PR, Cantor SB. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell. 2005;8:255–265. doi: 10.1016/j.ccr.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 32.van der Lelij P, Chrzanowska KH, Godthelp BC, Rooimans MA, Oostra AB, Stumm M, Zdzienicka MZ, Joenje H, de Winter JP. Warsaw breakage syndrome, a cohesinopathy associated with mutations in the XPD helicase family member DDX11/ChlR1. Am. J. Hum. Genet. 2010;86:262–266. doi: 10.1016/j.ajhg.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, Simon M, Marie Y, Boisselier B, Delattre JY, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat. Genet. 2009;41:899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wrensch M, Jenkins RB, Chang JS, Yeh RF, Xiao Y, Decker PA, Ballman KV, Berger M, Buckner JC, Chang S, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat. Genet. 2009;41:905–908. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bai C, Connolly B, Metzker ML, Hilliard CA, Liu X, Sandig V, Soderman A, Galloway SM, Liu Q, Austin CP, et al. Overexpression of M68/DcR3 in human gastrointestinal tract tumors independent of gene amplification and its location in a four-gene cluster. Proc. Natl Acad. Sci. USA. 2000;97:1230–1235. doi: 10.1073/pnas.97.3.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muleris M, Almeida A, Gerbault-Seureau M, Malfoy B, Dutrillaux B. Identification of amplified DNA sequences in breast cancer and their organization within homogeneously staining regions. Genes Chromosomes Cancer. 1995;14:155–163. doi: 10.1002/gcc.2870140302. [DOI] [PubMed] [Google Scholar]
- 37.Shinomiya T, Mori T, Ariyama Y, Sakabe T, Fukuda Y, Murakami Y, Nakamura Y, Inazawa J. Comparative genomic hybridization of squamous cell carcinoma of the esophagus: the possible involvement of the DPI gene in the 13q34 amplicon. Genes Chromosomes Cancer. 1999;24:337–344. [PubMed] [Google Scholar]
- 38.Pitti RM, Marsters SA, Lawrence DA, Roy M, Kischkel FC, Dowd P, Huang A, Donahue CJ, Sherwood SW, Baldwin DT, et al. Genomic amplification of a decoy receptor for Fas ligand in lung and colon cancer. Nature. 1998;396:699–703. doi: 10.1038/25387. [DOI] [PubMed] [Google Scholar]
- 39.Ozon S, Byk T, Sobel A. SCLIP: a novel SCG10-like protein of the stathmin family expressed in the nervous system. J. Neurochem. 1998;70:2386–2396. doi: 10.1046/j.1471-4159.1998.70062386.x. [DOI] [PubMed] [Google Scholar]
- 40.Schurmann A, Massmann S, Joost HG. ARP is a plasma membrane-associated Ras-related GTPase with remote similarity to the family of ADP-ribosylation factors. J. Biol. Chem. 1995;270:30657–30663. doi: 10.1074/jbc.270.51.30657. [DOI] [PubMed] [Google Scholar]
- 41.Fan L, Fuss JO, Cheng QJ, Arvai AS, Hammel M, Roberts VA, Cooper PK, Tainer JA. XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations. Cell. 2008;133:789–800. doi: 10.1016/j.cell.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu H, Rudolf J, Johnson KA, McMahon SA, Oke M, Carter L, McRobbie AM, Brown SE, Naismith JH, White MF. Structure of the DNA repair helicase XPD. Cell. 2008;133:801–812. doi: 10.1016/j.cell.2008.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolski SC, Kuper J, Hanzelmann P, Truglio JJ, Croteau DL, Van Houten B, Kisker C. Crystal structure of the FeS cluster-containing nucleotide excision repair helicase XPD. PLoS Biol. 2008;6:e149. doi: 10.1371/journal.pbio.0060149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pugh RA, Honda M, Leesley H, Thomas A, Lin Y, Nilges MJ, Cann IK, Spies M. The iron-containing domain is essential in Rad3 helicases for coupling of ATP hydrolysis to DNA translocation and for targeting the helicase to the single-stranded DNA-double-stranded DNA junction. J. Biol. Chem. 2008;283:1732–1743. doi: 10.1074/jbc.M707064200. [DOI] [PubMed] [Google Scholar]
- 45.Veatch JR, McMurray MA, Nelson ZW, Gottschling DE. Mitochondrial dysfunction leads to nuclear genome instability via an iron–sulfur cluster defect. Cell. 2009;137:1247–1258. doi: 10.1016/j.cell.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warbrick E. PCNA binding through a conserved motif. Bioessays. 1998;20:195–199. doi: 10.1002/(SICI)1521-1878(199803)20:3<195::AID-BIES2>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 47.Sung P, Krejci L, Van Komen S, Sehorn MG. Rad51 recombinase and recombination mediators. J. Biol. Chem. 2003;278:42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- 48.Kasamatsu H, Robberson DL, Vinograd J. A novel closed-circular mitochondrial DNA with properties of a replicating intermediate. Proc. Natl Acad. Sci. USA. 1971;68:2252–2257. doi: 10.1073/pnas.68.9.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allers T, Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106:47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- 50.Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, Smith GR. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127:1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwacha A, Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995;83:783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- 52.Adelman CA, Boulton SJ. Metabolism of postsynaptic recombination intermediates. FEBS Lett. 2010;584:3709–3716. doi: 10.1016/j.febslet.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 53.Bugreev DV, Mazin AV. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc. Natl Acad. Sci. USA. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes Dev. 2007;21:3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sommers JA, Rawtani N, Gupta R, Bugreev DV, Mazin AV, Cantor SB, Brosh RM., Jr FANCJ uses its motor ATPase to destabilize protein–DNA complexes, unwind triplexes, and inhibit RAD51 strand exchange. J. Biol. Chem. 2009;284:7505–7517. doi: 10.1074/jbc.M809019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- 57.Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- 58.Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fugger K, Mistrik M, Danielsen JR, Dinant C, Falck J, Bartek J, Lukas J, Mailand N. Human Fbh1 helicase contributes to genome maintenance via pro- and anti-recombinase activities. J. Cell. Biol. 2009;186:655–663. doi: 10.1083/jcb.200812138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prakash R, Satory D, Dray E, Papusha A, Scheller J, Kramer W, Krejci L, Klein H, Haber JE, Sung P, et al. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 2009;23:67–79. doi: 10.1101/gad.1737809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 62.Lillard-Wetherell K, Machwe A, Langland GT, Combs KA, Behbehani GK, Schonberg SA, German J, Turchi JJ, Orren DK, Groden J. Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2. Hum. Mol. Genet. 2004;13:1919–1932. doi: 10.1093/hmg/ddh193. [DOI] [PubMed] [Google Scholar]
- 63.Opresko PL, Mason PA, Podell ER, Lei M, Hickson ID, Cech TR, Bohr VA. POT1 stimulates RecQ helicases WRN and BLM to unwind telomeric DNA substrates. J. Biol. Chem. 2005;280:32069–32080. doi: 10.1074/jbc.M505211200. [DOI] [PubMed] [Google Scholar]
- 64.Opresko PL, Otterlei M, Graakjaer J, Bruheim P, Dawut L, Kolvraa S, May A, Seidman MM, Bohr VA. The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol. Cell. 2004;14:763–774. doi: 10.1016/j.molcel.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 65.Opresko PL, von Kobbe C, Laine JP, Harrigan J, Hickson ID, Bohr VA. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem. 2002;277:41110–41119. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- 66.Ghosh A, Rossi ML, Aulds J, Croteau D, Bohr VA. Telomeric D-loops containing 8-oxo-2′-deoxyguanosine are preferred substrates for Werner and Bloom syndrome helicases and are bound by POT1. J. Biol. Chem. 2009;284:31074–31084. doi: 10.1074/jbc.M109.027532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.London TB, Barber LJ, Mosedale G, Kelly GP, Balasubramanian S, Hickson ID, Boulton SJ, Hiom K. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J. Biol. Chem. 2008;283:36132–36139. doi: 10.1074/jbc.M808152200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu Y, Shin-ya K, Brosh RM., Jr FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol. Cell. Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Luke B, Lingner J. TERRA: telomeric repeat-containing RNA. EMBO J. 2009;28:2503–2510. doi: 10.1038/emboj.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu Y, Suzuki Y, Ito K, Komiyama M. Telomeric repeat-containing RNA structure in living cells. Proc. Natl Acad. Sci. USA. 2010;107:14579–14584. doi: 10.1073/pnas.1001177107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu Y, Suzuki Y, Komiyama M. Click chemistry for the identification of G-quadruplex structures: discovery of a DNA–RNA G-quadruplex. Angew. Chem. Int. Ed. Engl. 2009;48:3281–3284. doi: 10.1002/anie.200806306. [DOI] [PubMed] [Google Scholar]
- 72.Kim MY, Vankayalapati H, Shin-Ya K, Wierzba K, Hurley LH. Telomestatin, a potent telomerase inhibitor that interacts quite specifically with the human telomeric intramolecular g-quadruplex. J. Am. Chem. Soc. 2002;124:2098–2099. doi: 10.1021/ja017308q. [DOI] [PubMed] [Google Scholar]
- 73.Shin-ya K, Wierzba K, Matsuo K, Ohtani T, Yamada Y, Furihata K, Hayakawa Y, Seto H. Telomestatin, a novel telomerase inhibitor from Streptomyces anulatus. J. Am. Chem. Soc. 2001;123:1262–1263. doi: 10.1021/ja005780q. [DOI] [PubMed] [Google Scholar]
- 74.Cheung I, Schertzer M, Rose A, Lansdorp PM. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat. Genet. 2002;31:405–409. doi: 10.1038/ng928. [DOI] [PubMed] [Google Scholar]
- 75.Youds JL, Barber LJ, Ward JD, Collis SJ, O'Neil NJ, Boulton SJ, Rose AM. DOG-1 is the Caenorhabditis elegans BRIP1/FANCJ homologue and functions in interstrand cross-link repair. Mol. Cell. Biol. 2008;28:1470–1479. doi: 10.1128/MCB.01641-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao Y, Tarailo-Graovac M, O'Neil NJ, Rose AM. Spectrum of mutational events in the absence of DOG-1/FANCJ in Caenorhabditis elegans. DNA Repair. 2008;7:1846–1854. doi: 10.1016/j.dnarep.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 77.Mohaghegh P, Karow JK, Brosh RM, Jr, Bohr VA, Hickson ID. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fry M, Loeb LA. Human werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J. Biol. Chem. 1999;274:12797–12802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- 79.Sun H, Karow JK, Hickson ID, Maizels N. The Bloom's syndrome helicase unwinds G4 DNA. J. Biol. Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 80.Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Durkin SG, Glover TW. Chromosome fragile sites. Annu. Rev. Genet. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 82.Gangloff S, Soustelle C, Fabre F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet. 2000;25:192–194. doi: 10.1038/76055. [DOI] [PubMed] [Google Scholar]
- 83.Wicky C, Alpi A, Passannante M, Rose A, Gartner A, Muller F. Multiple genetic pathways involving the Caenorhabditis elegans Bloom's syndrome genes him-6, rad-51, and top-3 are needed to maintain genome stability in the germ line. Mol. Cell. Biol. 2004;24:5016–5027. doi: 10.1128/MCB.24.11.5016-5027.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ward JD, Muzzini DM, Petalcorin MI, Martinez-Perez E, Martin JS, Plevani P, Cassata G, Marini F, Boulton SJ. Overlapping mechanisms promote postsynaptic RAD-51 filament disassembly during meiotic double-strand break repair. Mol. Cell. 2010;37:259–272. doi: 10.1016/j.molcel.2009.12.026. [DOI] [PubMed] [Google Scholar]
- 85.Handel MA, Schimenti JC. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat. Rev. Genet. 2010;11:124–136. doi: 10.1038/nrg2723. [DOI] [PubMed] [Google Scholar]
- 86.Bishop DK, Zickler D. Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell. 2004;117:9–15. doi: 10.1016/s0092-8674(04)00297-1. [DOI] [PubMed] [Google Scholar]
- 87.Anderson LK, Reeves A, Webb LM, Ashley T. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics. 1999;151:1569–1579. doi: 10.1093/genetics/151.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Moens PB, Chen DJ, Shen Z, Kolas N, Tarsounas M, Heng HH, Spyropoulos B. Rad51 immunocytology in rat and mouse spermatocytes and oocytes. Chromosoma. 1997;106:207–215. doi: 10.1007/s004120050241. [DOI] [PubMed] [Google Scholar]
- 89.Jones GH. The control of chiasma distribution. Symp. Soc. Exp. Biol. 1984;38:293–320. [PubMed] [Google Scholar]
- 90.Martini E, Diaz RL, Hunter N, Keeney S. Crossover homeostasis in yeast meiosis. Cell. 2006;126:285–295. doi: 10.1016/j.cell.2006.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hillers KJ, Villeneuve AM. Chromosome-wide control of meiotic crossing over in C. elegans. Curr. Biol. 2003;13:1641–1647. doi: 10.1016/j.cub.2003.08.026. [DOI] [PubMed] [Google Scholar]
- 92.Mets DG, Meyer BJ. Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell. 2009;139:73–86. doi: 10.1016/j.cell.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tsai CJ, Mets DG, Albrecht MR, Nix P, Chan A, Meyer BJ. Meiotic crossover number and distribution are regulated by a dosage compensation protein that resembles a condensin subunit. Genes Dev. 2008;22:194–211. doi: 10.1101/gad.1618508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McMahill MS, Sham CW, Bishop DK. Synthesis-dependent strand annealing in meiosis. PLoS Biol. 2007;5:e299. doi: 10.1371/journal.pbio.0050299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bhalla N, Wynne DJ, Jantsch V, Dernburg AF. ZHP-3 acts at crossovers to couple meiotic recombination with synaptonemal complex disassembly and bivalent formation in C. elegans. PLoS Genet. 2008;4:e1000235. doi: 10.1371/journal.pgen.1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hollingsworth NM, Brill SJ. The Mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev. 2004;18:117–125. doi: 10.1101/gad.1165904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wu X, Sandhu S, Ding H. Establishment of conditional knockout alleles for the gene encoding the regulator of telomere length (RTEL) Genesis. 2007;45:788–792. doi: 10.1002/dvg.20359. [DOI] [PubMed] [Google Scholar]
- 98.Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 99.Mani P, Yadav VK, Das SK, Chowdhury S. Genome-wide analyses of recombination prone regions predict role of DNA structural motif in recombination. PLoS ONE. 2009;4:e4399. doi: 10.1371/journal.pone.0004399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ghosal G, Muniyappa K. Saccharomyces cerevisiae Mre11 is a high-affinity G4 DNA-binding protein and a G-rich DNA-specific endonuclease: implications for replication of telomeric DNA. Nucleic Acids Res. 2005;33:4692–4703. doi: 10.1093/nar/gki777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Muniyappa K, Anuradha S, Byers B. Yeast meiosis-specific protein Hop1 binds to G4 DNA and promotes its formation. Mol. Cell. Biol. 2000;20:1361–1369. doi: 10.1128/mcb.20.4.1361-1369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]