Abstract
The late stages of 30S and 50S ribosomal subunits biogenesis have been studied in a wild-type (wt) strain of Escherichia coli (MC4100) subjected to a severe heat stress (45–46°C). The 32S and 45S ribosomal particles (precursors to 50S subunits) and 21S ribosomal particles (precursors to 30S subunits) accumulate under these conditions. They are authentic precursors, not degraded or dead-end particles. The 21S particles are shown, by way of a modified 3′5′ RACE procedure, to contain 16S rRNA unprocessed, or processed at its 5′ end, and not at the 3′ end. This implies that maturation of 16S rRNA is ordered and starts at its 5′-terminus, and that the 3′-terminus is trimmed at a later step. This observation is not limited to heat stress conditions, but it also can be verified in bacteria growing at a normal temperature (30°C), supporting the idea that this is the general pathway. Assembly defects at very high temperature are partially compensated by plasmid-driven overexpression of the DnaK/DnaJ chaperones. The ribosome assembly pattern in wt bacteria under a severe heat stress is therefore reminiscent of that observed at lower temperatures in E. coli mutants lacking the chaperones DnaK or DnaJ.
INTRODUCTION
The response of Escherichia coli to environmental stresses has been intensively studied: in general, the molecular components involved belong to the transcription machinery, i.e. the (p)ppGpp-dependent stringent response (1), the general stress response depending on the sigma factor σs (2), the envelope stress response controlled by the sigma factor σE (3) and the cytoplasmic heat-shock response controlled by the sigma factor σ32 (4). Beyond transcription, a special class of universal proteins, the molecular chaperones, fulfil both housekeeping and defence against stress function (5). One of the main players is the chaperone DnaK (HSP70) which retro-controls the heat-shock response in E.coli by sequestrating and inactivating the heat shock-specific transcription factor σ32(6). However, the HSP70 chaperone machine exerts a myriad of other functions (7), among which is the post-translational quality control of proteins, which involves the detection and either refolding or elimination, in co-operation with proteases, of proteins irreversibly unfolded (8). DnaK also is one of the extrinsic factors in ribosome assembly (9–11): it participates in the latter stages of ribosome biogenesis, because its inactivity (in a dnaK756-ts mutant) or absence (in a ΔdnaK null mutant) results in defective ribosome assembly and the accumulation at high temperatures of 21S ribosomal particles, which are precursors to mature 30S subunits, and of 32S and 45S ribosomal particles, which are precursors to mature 50S subunits (12,13).
A wild-type (wt) strain of E. coli K12 can grow up to 46°C (Figure 1 of refs. 14 and 15) in a minimal medium supplemented with methionine (16). We reasoned that a severe heat stress imposed to a wt strain of E. coli could mimic the pattern of ribosome assembly observed in a strain lacking DnaK (ΔdnaK) or DnaJ (ΔdnaJ), because the misfolded proteins would saturate the DnaK chaperone system, which is limiting (17). We show here that this is indeed what is observed: ribosomal particles, which accumulate under a severe heat stress, are genuine precursors to mature ribosomal subunits. We propose a model in which late stages of ribosome biogenesis are coupled to the protein quality control system through DnaK. We further demonstrate by use of a modification of the 3′5′ rapid amplification of complementary DNA ends (RACE) method that the maturation of the precursor 16S rRNA within the 21S precursor particles is ordered, and that it is initiated at its 5′ end. This conclusion appears to be true also at 30°C, and therefore not limited to heat stress conditions. The exact order of the processing events of the 16S rRNA in vivo has not been elucidated until now.
Figure 1.
(A–F) Sedimentation profiles of ribosomal subunits prepared from strain MC4100 labelled with [3H]-uridine for 1 h at 42°C (A), or 44°C (B), or 45°C (C) or 46°C (D), or from strain MC4100 pulse-labelled with [3H]-uridine for 1 min at 30°C (E), or from strain BB1553 (ΔdnaK) labelled with [3H]-uridine for 1 h at 37°C (F). In (E), pulse-labelling with [5-3H]-uridine (Amersham, TRK178) isotopically undiluted (28 Ci/mmol) for 1 min was abruptly terminated by the addition of NaN3 to give a final concentration of 10 mM and by pouring onto an equal volume of crushed ice. Sedimentation is from right to left. A260/ml, open circles. [3H] c.p.m. × 103, filled circles. (G) Bacterial growth from strain SR6618 [MC4100 Φ(groESL::lacZ)] was followed by measuring the A600/ml of the culture at 30°C. At A600/ml = 0.25, half of the culture was shifted to 45°C, and bacterial samples were withdrawn from both cultures at every 20 min for β-galactosidase assays. (H) β-Galactosidase specific activities were measured following standard procedures (19), and expressed per A600/ml of the culture at 30°C or at 45°C, i.e. in Miller units, as follows = 1000 × A420/ml / t × v × A600/ml, where A420/ml measures the concentration of the orthonitrophenol produced by hydrolysis of ONPG (ortho-nitrophenyl-β-galactoside), v the volume of culture used in the assay in ml, and t the time of hydrolysis in minutes. The difference between the β-galactosidase specific activities at 30°C and 45°C reflects the activity of the transcription factor σ32, and thus indirectly the decline of available DnaK, which if present would bind tightly to σ32 and inactivate it.
MATERIALS AND METHODS
Bacterial strains and growth medium
The bacterial strains used are all derived from E. coli K12 (Table 1). Ribosome assembly defects are generally exacerbated in an E. coli strain devoid of stringent control (relaxed phenotype) (18), and therefore MC4100 (dnaK + , relA1) was chosen for studying the ribosomal particles synthesized under a severe heat stress. Minimal medium A (19) supplemented with 0.4% glucose, 1 µg/ml thiamine, 0.2 % casamino acids and 0.1 mM L-tryptophan was used for labelling bacteria.
Table 1.
Bacterial and plasmid strains used
| Strain | Relevant genotype | Reference |
|---|---|---|
| E. coli | ||
| MC4100 | F-araD139 Δ (lacZYA-argF)U169 rpsL150, relA1,deoC1, ptsF25, rbsR flbB301 | 80, 81 |
| BB1553 | MC4100, ΔdnaK52 ::cmr (dnaK disrupted by a genetic determinant conferring resistance to chloramphenicol), sid B1 | 82 |
| SR6618 | MC4100 with a chromosomally integrated heat-shock promoterPgroES::lacZ fusion (RNA polymerase-σ32–transcribed). | 83 |
| Plasmids | ||
| pWKS30 | Low copy vector, ampR | 84 |
| pDM39 | pWKS30 dnaK+dnaJ+, ampR | 85 |
Labelling of bacteria and sedimentation analysis of ribosomal particles on sucrose gradients
Liquid cultures (50–200 ml) were inoculated with 0.005 vol of an overnight preculture grown in the same medium at 30°C. Incubation at 30°C in a shaking water-bath was continued for about 3 h, until the A600/ml reaches 0.25. For labelling at high temperatures, the culture was transferred to a second water-bath set up at the required temperature, and shaken for 1.5 h before adding the labelled uridine. The [5-3H]-uridine from Amersham (TRK178, 1 mCi/ml) of high specific activity (26 Ci/mmol) was isotopically diluted with non-radioactive uridine (Sigma, U-3750), and added to the culture at the final concentration of 3 µM and 1 µCi/ml. Incubation of bacteria was continued for 1 h. Bacteria were collected by centrifugation, divided into two batches (one for analysis of the ribosome assembly pattern, and the other one for 3′5′ RACE analysis). The bacterial pellets (usually 50–100 mg) were kept frozen at −20°C. Crude extracts were prepared in the TMNSH buffer (10 mM Tris–HCl pH 7.4, 10 mM magnesium acetate, 60 mM NH4Cl, 1 mM dithiothreitol) by grinding bacteria with alumina (Alcoa, A305) at 4°C, centrifuged to pellet the alumina, unbroken cells and cells debris, and then mixed if necessary with a small amount (5 A260 units) of non-radioactive wt E. coli ribosomes. Samples were adjusted to 400 mM NaCl, and immediately layered onto 35 ml 10–30% linear sucrose gradients prepared in 400 mM NaCl-containing TMNSH buffer, and then centrifuged at 3°C in a Beckman SW28 rotor for 17 h at 27 000 r.p.m. These ionic conditions result in complete ribosome dissociation (20). Each gradient was then collected in about 30 fractions by aspirating from the bottom of the gradient with a peristaltic pump. Optical density (A260/ml) was measured first, and then [3H]-radioactivity was measured by precipitating the whole fraction with 5% trichloracetic acid, collecting the acid-insoluble radioactivity on Millipore filters, and counting the [3H] c.p.m. by liquid scintillation.
Mapping of 5′ and 3′ extremities of p16S rRNA by 3′5′ RACE
5′3′ RACE experiments were performed on total RNA, phenol-extracted from whole cells, as described in ref. 21. We circularized 5 µg of total RNA with T4 RNA ligase (New England Biolabs, n° M0204) and subjected to RT-PCR across the 5′3′ junction, using the primer FA (5′ATCTGGATCCGATTCATGACTGGGGTGAAGTC 3′, with a 10-nt extension including a BamH1 site (underlined), and the primer RA (5′ATCTGAATTCGTTCGACTTGCATGTGTTAGGC 3′, with a 10-nt extension including an EcoR1 site (underlined), transcriptase reverse M-MuLV RT.RNase H− (Finnzymes, n° F-572), and Taq DNA polymerase (New England Biolabs, n° M0267). Polymerase chain reaction (PCR) was run on a Mastercycler personal apparatus by using the following program: initial denaturation (94°C × 2 min), then 30 cycles of [denaturation (94°C × 30 s), annealing (60°C × 30 s), extension (72°C × 1 min)], and finally 72°C × 10 min to finish the elongation. PCR products were separated by electrophoresis on 2.5% agarose gels, and were purified for sequencing, after preparative agarose gel electrophoresis, using the kit NucleoSpin Extract II (Macherey-Nagel). Sequencing was performed by MWG (Germany).
Thermo-denaturation of the RNA prior to the 3′5′ RACE
Five micrograms of RNA in a total volume of 20 µl containing the T4 RNA ligase buffer (New England Biolabs, n° B0204S) were incubated for 4 min at 75°C, and then rapidly frozen in a solid CO2 (dry ice)-ethanol bath for 1 min. To allow the samples to thaw slowly, they were then placed on ice for about 15 min, as described in ref. 22, and then 2 µl of T4 RNA ligase was added to start the 3′5′ RNA ligation.
Primer extension analysis
Primer RI (5′-ACGTGTTCACTCTTGAGACTTGG-3′) and RO (5′-GTTCGACTTGCATGTGTTAGGC-3′, the same as primer RA without the 10-nt extension including an EcoRI site at its 5′ end) were [32P]-labelled at their 5′ ends with T4 polynucleotide kinase and ATP-γ-[32P]. Either RI (Figure 5A) or RO (Figure 5B) was annealed to total RNA phenol-extracted as described in ref. 21 from MC4100 bacteria grown at 46°C. Mixtures were incubated for 4 min at 75°C, rapidly frozen in a solid CO2 (dry ice)-ethanol bath for 2 min, and transferred to ice. Primers were extended by AMV reverse transcriptase (Finnzymes, n° F-570L) in the presence of dATP, dCTP, dGTP, dTTP 5 mM each for 30 min at 45°C, and primer extension products were finally mixed with the loading buffer containing formamide, bromophenol blue and xylene cyanol (from the Thermo Sequenase Cycle Sequencing Kit, USB n°78500).
Figure 5.
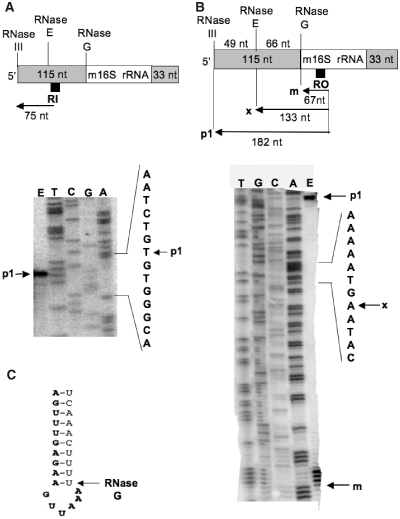
(A) Polyacrylamide gel electrophoresis of the cDNA product obtained by primer RI extension from total RNA isolated from MC4100 bacteria grown at 46°C. The upper part of the panel shows the position of annealing on the 16S rRNA of the [32P]-labelled primer RI (black square) used in this experiment, and the size (75 nt) of the expected cDNA product. The lower part of the panel shows the 8% polyacrylamide sequencing gel on which the primer extension product was loaded (lane E), together with a sequencing ladder (lanes T, C, G and A) generated by the use of the same primer. The arrow p1 indicates the position of migration of the [32P]-labelled cDNA in lane E. (B) Polyacrylamide gel electrophoresis of the cDNA products obtained by primer RO extension from total RNA isolated from MC4100 bacteria grown at 46°C. The upper part of the panel shows the position of annealing on the 16S rRNA of the [32P]-labelled primer RO (black square) used in this experiment, and the size of the expected cDNA products (182 nt and 67 nt) generated from p1 and m 16S rRNA, respectively; 133 nt is the predicted size of a cDNA product (x) generated from a precursor 16S rRNA species which would have been processed by the RNase E only (and not yet by the RNase G). The lower part of the panel shows the 6% polyacrylamide sequencing gel on which the primer extension products were loaded (lane E), together with a sequencing ladder (lanes T, C, G and A) generated by the use of the same primer. The arrows p1, x and m indicate the position of the [32P]-labelled cDNAs predicted (x) and observed (p1 and m) in lane E. (C) Reproduced from Figure 5 of ref. 33, shows the secondary structure proposed by these authors of the p1 16S rRNA near the processing site of the RNase G (nucleotides in bold belong to the m 16S rRNA).
In parallel, sequencing ladders were generated using the same sequencing kit, using either RI (Figure 5A) or RO (Figure 5B) as [32P]-labelled primers, and as a template a 275-bp DNA fragment containing 200 nt upstream of the 5′ end of the 16S rDNA sequence. This 275-bp DNA fragment was produced by PCR amplification of E. coli MC4100 chromosomal DNA with the primers PAF (5′-CCGCTGAGAAAAAGCGAAG-3′) and PAR (5′-GCCACTCGTCAGCAAAGAAG-3′). Primer extension products and the sequencing ladder generated by the use of the same primer were electrophorized on an 8% or 6% polyacrylamide/7 M urea sequencing gel. The gels were transfered to Whatman 3MM paper, vacuum-dried, and the [32P] radioactivity was visualized by a Typhoon PhosphoImager. The USB sequencing kit allows incorporation of 7-deaza-dG residues into the sequencing ladder products, whereas the primers extension products contain dG residues. This results in a slightly different electrophoretic mobility of a given oligonucleotide depending on the type of residues (7-deaza-dG vs dG) it harbours.
RESULTS
Under a severe heat stress, MC4100 bacteria accumulate ribosomal particles, which are genuine precursors to ribosomal subunits
Sedimentation pattern of ribosomal particles synthesized in bacteria under a severe heat stress
To observe ribosome assembly at different temperatures, MC4100 bacteria were grown at 30°C until they reached an A600/ml of 0.25, and then aliquots were shifted to 42°C, 44°C, 45°C or 46°C for 1.5 h (while the remaining of the culture continued at 30°C). The cultures were then labelled with [3H]-uridine for 1 h at the same temperature. Crude extracts were centrifuged on sucrose gradients in ionic conditions that promote complete dissociation of the 50S and 30S ribosomal subunits. The sedimentation pattern of ribosomal particles at 42°C (Figure 1A) was similar to that at 30°C (data not shown) and appeared perfectly normal. However at 44°C it became partially defective for the 50S subunit (accumulation of 45S particles, Figure 1B), at 45°C completely defective for both subunits (accumulation of 45S, 32S and 21S particles, Figure 1C), and even more pronounced when the temperature increased to 46°C (Figure 1D), as an inversion of the 45S/32S ratio was observed.
Measurement of the specific activities of β-galactosidase at 30°C and 45°C in the strain SR6618 (a derivative of MC4100 which carries a chromosomal copy of the gene lacZ under the control of the heat-shock promoter of the groESL operon), allowed us to evaluate the heat stress response of bacteria at 45°C (1.6-fold increase of the Miller units at 45°C versus 30°C, Figure 1G and H).
The accumulation of 45S, 32S and 21S particles at high temperatures was reminiscent of the situation observed either in the same strain pulse-labelled with [3H]-uridine at 30°C for 1 min (Figure 1E), or in a ΔdnaK strain labeled with [3H]-uridine at 37°C for 1 h (Figure 1F) (13). In both cases, the late steps of ribosome assembly were arrested, and 45S, 32S and 21S ribosomal ‘precursor’ particles accumulate. Further experiments were therefore designed to elucidate the nature of the ribosomal particles found in MC4100 at high temperature.
Ribosomal particles are neither degradation products nor dead-end particles, but they are authentic precursors to mature ribosomal subunits
30S and 50S ribosomal subunits are thermostable above 45°C both in vitro (23) and in vivo (24). To verify, however, that ribosomal particles found in MC4100 at 45°C are not heat-induced degradation products (25,26), ribosomes in strain MC4100 were labelled with [3H]-uridine during their synthesis at 30°C, and then bacteria were transferred to 46°C (or to 30°C as control) and incubated for 2 h in the same culture medium ‘without’ [3H]-uridine. Ribosomes extracted from such cells sedimented as 30S and 50S subunits, and no traces of abnormal ribosomal particles were found (Figure 2A). Therefore, the stability of ribosomes assembled at 30°C was not affected at 46°C. Regardless, heat-induced degradation products would have sedimented as a collection of heterogeneous particles, and they would not behave as homogeneous particles displaying the expected sedimentation coefficients (45S, 32S and 21S). The conditions of heat stress used here were therefore quite different from the thermal injury used by others (27), which lead to ribosome destruction.
Figure 2.
(A) Sedimentation profile of ribosomal subunits prepared from strain MC4100 labelled with [3H]-uridine at 30°C, and subsequently incubated for 2 h at 46°C in non-radioactive growth medium. (B) Sedimentation profile of ribosomal subunits prepared from strain MC4100 labelled with [3H]-uridine at 46°C, and subsequently incubated for 2 h at 30°C in growth medium containing 0.5 mM non-radioactive uridine and 500 µg/ml rifampicin. Sedimentation is from right to left. A260/ml, open circles. [3H] c.p.m. × 103, filled circles.
To determine whether these ribosomal particles were genuine precursors to mature ribosomes or dead-end particles, MC4100 bacteria were labelled with [3H]-uridine at 45°C (an aliquot of labelled bacteria was analysed as a control, and displayed the same sedimentation pattern as shown in Figure 1C), and then chased for 1.5 h at 30°C with a large excess of unlabelled uridine in the presence of 500 µg/ml rifampicin. The aim of adding rifampicin was to prevent any de novo rRNA synthesis. Under these conditions, 21S, 32S and 45S particles were converted almost completely to 30S and 50S ribosomal subunits, although a residual amount of 45S particles was still present (Figure 2B). This demonstrates that they were authentic precursors to mature subunits, and not dead-end particles. The transformation of the 45S particles into 50S subunits is the slowest step, as already known (28,29). The final demonstration that the 21S particles are true precursors to mature 30S subunits will be presented subsequently.
Processing of the precursor 16S rRNA accumulated under a severe heat stress is sequential, and it starts at its 5′ end
3′5′ RACE method with or without prior thermal RNA denaturation
Processing of 16S rRNA is initiated by an RNase III cleavage of the 30S rRNA primary transcript in the double-stranded processing stalk flanking the mature sequence (30). The resulting precursor 16S rRNA (p1, also called 17S rRNA) possesses 5′ and 3′ extensions (115 and 33 extra-nucleotides respectively, Figure 3A). Its 5′ end is matured by a combination of the endoribonuclease activities of RNase E and RNase G, while the 3′ end will be trimmed by an unknown RNase, to yield the mature m16S rRNA (31–33). Until now, the precise order of these cleavages was unknown.
Figure 3.
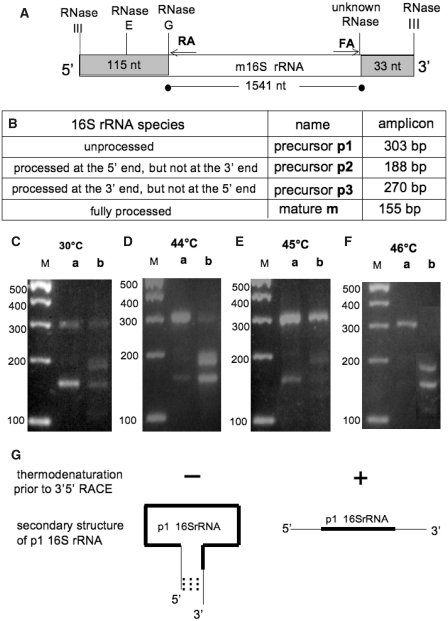
(A) Schematic processing of the p116S rRNA. The extra-sequences of 115 nt and 33 nt, flanking the m 16S rRNA at its 5′ and 3′ ends, respectively, are shown on a grey background. RA and FA are the primers used for 3′5′ RACE analysis. The site of annealing of RA to m 16S rRNA, and that of FA to the reverse complement of the m 16S rRNA, are indicated by arrows. Figure not drawn to scale. (B) Expected sizes in bp of the RT-PCR products (amplicons) obtained from the different species of 16S rRNA (p1, p2, p3 and m) by 3′5′ RACE. (C–F) Agarose gel electrophoresis of RT-PCR products obtained by 3′5′ RACE from total RNA isolated from MC4100 bacteria grown at 30°C (C), or 44°C (D), or 45°C (E) or 46°C (F). Each RNA sample was thermo-denatured (lanes b), or not (lanes a) prior to the 3′5′ ligation. The sizes (in bp) of the molecular weight markers are indicated to the left of each gel (M). (G) The thermodenaturation step dissociates the complementary sequences present at the 3′ and 5′ends of the p1 16S rRNA, and therefore offers to all the 16S rRNA species an equal chance to access to the T4 RNA ligase.
Here, we used the 5′3′ RACE technique to map the 5′ and 3′ ends of the 16S rRNA synthesized in MC4100 bacteria at the different temperatures. Briefly, aliquots of the [3H]-labelled bacteria, previously used to study the ribosome assembly patterns at the different temperatures (Figure 1), were phenol-extracted, and then the total RNA obtained was circularized by 3′5′ligation, and subjected to RT-PCR with primers designed to amplify across the 5′3′ junction of circularized 16S rRNA. With the 32-nt-long oligonucleotides RA and FA used as primers (Figure 3A), the sizes of the RT-PCR products (or amplicons) expected to be generated from the m16S rRNA and from the different possible precursor species (p1, p2 and p3) are indicated in the panel B of Figure 3.
The results in the lanes a of the panels (C–F) of Figure 3 show that:
A predominant PCR product of about 150 bp was obtained from bacteria grown at 30°C (panel C), revealing that most of the 16S rRNA was mature as expected. A weaker band migrating at about 300 bp was also seen.
A major PCR product of about 300 bp was detected in the cultures that were shifted for 1.5 h to 44°C, 45°C and 46°C (panels D, E and F respectively), revealing the presence of a major p1 16S rRNA species, as well as two other weak PCR products of 150 bp (generated from m16S rRNA) and about 200 bp (which could correspond in size to a p2 16rRNA, that is fully matured at its 5′ end but unprocessed at its 3′end). Sequencing of these RT-PCR products was necessary to confirm these data.
However, these results are in agreement with the detection of 21S particles at high temperatures (Figure 1), and they showed that heat stress had prevented the maturation of the 16S rRNA inside the 21S particles. They excluded definitively the possibility that these ribosomal particles were heat-induced degradation products instead of precursors.
The unprocessed p1 16S rRNA was the only species in which the 5′ and 3′ ends were annealed along nearly 20 bp, and this proximity will greatly favour the 3′5′ ligation by the T4 RNA ligase (Figure 3G). This was not the case for the other rRNAs, which could be under-represented in the 3′5′ RACE assay for this reason (titration of the T4 RNA ligase by the p1 16S rRNA). We therefore designed a step of thermo-denaturation of RNAs, prior to the 3′5′ ligation, to denature the 3′ and 5′ ends of the p1 16S rRNA (Figure 3G). The results obtained by this method are presented in the lanes b of the panels C, D, E and F of Figure 3. It can be seen in all cases (including 30°C, panel C) that the PCR product of about 200 bp, which appeared weakly without prior thermo-denaturation of the RNAs (lanes a), became more apparent, along with a concomitant decrease in the intensity of the PCR product of about 300 bp.
The thermo-denaturation of RNAs (targeting the p1 16S rRNA) was therefore helpful to detect other 16S rRNA species that could be barely seen without this step. In particular, a RT-PCR product of about 200 bp appeared distinctly, which could come from a p2 16S rRNA (matured at its 5′ end but not at its 3′ end). By contrast, no RT-PCR product of 270 bp was detected, which is characteristic of a p3 16S rRNA (matured at its 3′ end, but not at its 5′end). This suggests that the maturation of the p1 16S rRNA was sequential, starting from its 5′ end.
Sequencing of the amplicons
To confirm the previous data, the RT-PCR products of about 150 bp (lane a of Figure 3C), 300 bp (lane a of Figure 3D) and 200 bp (lane b of the Figure 3D) were purified by preparative agarose gel electrophoresis and sequenced. The results, presented in Figure 4, are the following:
The DNA sequence of the RT-PCR product of about 150 bp contained 155 nt as expected (Figure 4A), and it corresponded to the 3′5′ junction where the first nucleotide at the 5′ end (A in bold type) was ligated with the last nucleotide at the 3′ end (A in bold type) of m16S rRNA. The expected trimming of the precursor 16S rRNA therefore occured at 30°C.
The DNA sequence of the RT-PCR product of about 300 bp was expected to be precisely 155 + 115 + 33 = 303 nt, but surprisingly it was found to contain 305 nt (Figure 4B). The 115 extra-nucleotides derived from the 5′ end and the 33 extra-nucleotides derived from the 3′ end of the p1 16S rRNA were present as expected, but also two additional nucleotides (GA) at the 3′5′ junction. This unexpected observation, which will be studied and explained in the next section (3.2.3.), does not alter our main conclusion, that the RT-PCR product of 305 bp was derived from a p1 16S rRNA, which was unprocessed at both its 5′ and 3′ends.
The DNA sequence of the RT-PCR product of about 200 bp was precisely 188 nt as expected (Figure 4C). It corresponded to the 3′5′ junction (AA in bold type, underlined) of the first nucleotide at the 5′ end of m16S rRNA ligated with the last nucleotide at the 3′end of p1 16S rRNA, which revealed that the 5′ extra-sequence (115 nt) had been removed, whereas the 3′ extra-sequence (33 nt) was still present. In other words, it is derived from a typical p2 16S rRNA, trimmed at its 5′ end (by the RNases E and G), but not at its 3′ end.
Figure 4.
DNA sequence of the RT-PCR products of 155 bp (A), 305 bp (B) and 188 bp (C) obtained by 3′5′ RACE from 16S rRNA m, p1 and p2, respectively. The DNA sequences shown here are those found in the 16S ribosomal RNA. Sequences derived from the m16S rRNA are highlighted in a grey background. Arrows indicate that complete DNA sequences extend until to the sequence of oligonucleotide FA, and to the reverse complement of oligonucleotide RA. In each panel, the two nucleotides shown in bold and underlined correspond to the junction, generated by the RNA ligation, of the 5′ and 3′ ends of the 16S RNAs.
Our results support the conclusion that the trimming of 16S rRNA is ordered and starts at its 5′ end, because the only two types of precursor 16S rRNAs were p1 (unprocessed at both ends) and p2 (processed at its 5′ end, unprocessed at its 3′ end).
Primer extension analysis
The two additional nucleotides (GA) at the 3′5′ junction of the circularized p1 16S rRNA [see Figure 4B, and the paragraph (ii) of the preceding section] could come from either the 5′ end (therefore harbouring 117 instead of 115 extra-nucleotides) or the 3′ end (therefore harbouring 35 instead of 33 extra-nucleotides) of the p1 16S rRNA. Indeed the original article by Young and Steitz (Figure 4 of ref. 30) describes not only a site of cleavage by RNase III at 115 nt upstream of the 5′ end of m 16S rRNA, but in addition two alternative sites at 113 nt and 117 nt upstream of the 5′ end of m 16S rRNA.
The 5′ end of the p1 16S rRNA was therefore determined by primer extension analysis, with a [32P]-labelled primer RI, which was designed to generate a cDNA of either 75 nt or 77 nt depending upon the length (115 nt or 117 nt) of the extra-sequence present at the 5′ end of the p1 16S rRNA. Electrophoresis of this [32P]-labelled cDNA on a 8% polyacrylamide sequencing gel, together with a [32P]-labelled sequencing ladder (lanes T, C, G, A of Figure 5A) generated by the use of the same primer, demonstrated that this cDNA (arrow p1 in lane E of Figure 5A) had the same migration as an oligonucleotide present in lane T of Figure 5A, which revealed unambiguously that it possesses 75 nt, and not 77 nt. The slightly different electrophoretic mobility of this oligonucleotide in the lane E compared to that in lane T results from incorporation of dG residues into the cDNA, versus the 7-deaza-dG residues present in the same oligonucleotide of the sequencing ladder (‘Materials and Methods’ section). It follows that the two additional nucleotides (GA) at the 3′5′ junction of the circularized p1 16S rRNA were not derived from its 5′ end, but can only be derived from the 3′ terminus. Indeed a GA is present downstream of the annotated RNase III cleavage site located 33 nt downstream of the mature 3′ terminus (Figure 4 of ref. 30). We conclude that in our conditions the RNase III cleavage site is located 35 nt downstream of the 3′end of m 16S rRNA.
Primer extension analysis can also be used to check whether an intermediate processing step of the p1 16S rRNA by RNase E can be detected. If it is so, the [32P]-labelled primer RO (which has the same site of annealing as the primer RA) would generate, from total RNA extracted from MC4100 bacteria grown at 46°C, a cDNA of 133 nt (named x in Figure 5B), along with the cDNAs of 182 nt and 67 nt that were reverse transcribed from the p1 and m 16S rRNA, respectively. Electrophoresis of these [32P]-labelled primer extension products on a 6% polyacrylamide sequencing gel (lane E of Figure 5B), together with a [32P]-labelled sequencing ladder generated by the use of the same primer (lanes T, G, C, A of Figure 5B), did not reveal any cDNA corresponding to x. By contrast, the cDNA of 182 nt (p1) and 67 nt (m) were found in lane E at the expected positions. In the same lane E, four bands corresponding to four oligonucleotides adjacent in the lane T of Figure 5B were also unexpectedly found. Most likely, they are due to arrests of the reverse transcriptase elongating through a particular secondary structure (stem–loop) of the RNA template shown in Figure 5C, as proposed in ref. 33. In conclusion, no RNase E-dependent intermediate processing step of the 5′ end of p1 16S rRNA could be detected, a result in full agreement with the 3′5′ RACE data (Figure 3C–F). Probably the processing by RNase G takes place soon after that by RNase E, or takes place independently of a facultative cleavage by RNase E.
Ribosomal particles sedimenting at 21S contain precursor 16S rRNA that is either unprocessed (p1) or processed at its 5′ end only (p2)
In order to examine the status of their rRNAs, the 21S precursor particles, which were synthesized and [3H]-uridine labelled in bacteria grown at 45°C, were isolated by preparative sucrose gradient sedimentation, as shown in Figure 6A. An aliquot of these 21S particles was mixed with non-radioactive 50S and 30S subunits and rerun on a similar sucrose gradient. The result (Figure 6B) allowed us to estimate their degree of purity and to conclude, as they displayed the expected sedimentation coefficient, that they were not unstable or artefacts generated during the ultracentifugation step.
Figure 6.
(A) Preparative sucrose gradient sedimentation of ribosomal particles from strain MC4100 labelled with [3H]-uridine at 45°C. Fractions 18–21 were pooled, and 21S particles were concentrated by ultracentrifugation. (B) An aliquot of the 21S particles isolated from (A) was mixed with unlabelled 50S and 30S subunits from a wt strain and rerun on a new sucrose gradient under the same conditions. Sedimentation is from right to left. A260/ml, open circles. [3H] c.p.m. × 103, filled circles. (C) 16S rRNA phenol-extracted from the 21S particles isolated from (A) were subjected to 3′5′ RACE analysis, with (lane b) or without (lane a) a thermodenaturation step prior to the 3′5′ RNA ligation. The RT-PCR products of 305 bp and 188 bp were purified by preparative agarose gel electrophoresis (from lanes a and b, respectively) and sequenced. In lane c, an aliquot of the 16S rRNA was thermodenatured, but instead of rapid freezing leading to irreversible RNA denaturation (as shown in lane b), was then subjected to a slow cooling (a couple of hours at room temperature) leading to reversible RNA denaturation, prior to the 3′5′ RACE procedure: the RT-PCR products obtained under these conditions are similar to those obtained in the absence of any RNA denaturation step (lane a).
16S rRNA was phenol-extracted from these particles, and 5′3′ RACE analysis was performed as described before, with (lane b of Figure 6C) or without (lane a of Figure 6C) thermo-denaturation of the 16S rRNA prior to the 3′5′ ligation. The results of Figure 6C show that:
No 155 bp RT-PCR product (generated from m16S rRNA) was detected, which confirms the absence of contamination by 30S ribosomal subunits, as shown in Figure 6B.
Two RT-PCR products of about 300 bp and 200 bp were detected, with an inversion of their relative proportion depending on whether the thermo-denaturation step was used (lane b) or not (lane a) on the 16S rRNA prior to the 3′5′ RACE procedure. This result validates the importance of the thermo-denaturation step because it allowed the unambiguous detection of the 200-bp RT-PCR product, as described before and illustrated in Figure 3G. This RNA denaturation step was shown to be reversible by slow cooling at room temperature, as demonstrated in the lane c of Figure 6C.
Sequencing of these two amplicons, purified by preparative agarose gel electrophoresis, gave the same results as those presented in Figure 4B and C (RT-PCR products of 305 bp and 188 bp, respectively).
No 270-bp RT-PCR product was detected, which showed that no p3 16S rRNA (matured at its 3′ end, and not at its 5′ end) can be found. Therefore, under our conditions, the maturation of the 16S rRNA appeared to be sequential, and it starts at the 5′end. On the other hand, our 21S precursor particles are heterogeneous according to their 16S rRNA content because they contained either unprocessed (p1) or processed at its 5′ end (p2) 16S rRNA. It both cases, the 3′ end of the precursor 16S rRNA was not trimmed.
A plasmid-driven over-expression of DnaK/DnaJ assists to some extent ribosome assembly under severe heat stress
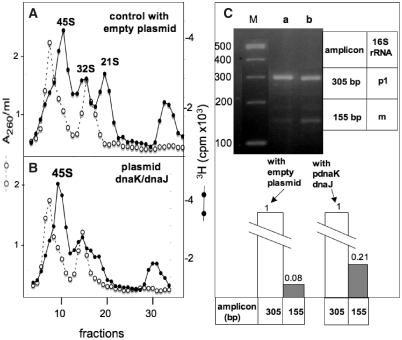
The resemblance of the ribosomal patterns observed in a wt dnaK strain (MC4100) at 45°C (Figure 1C) and in a ΔdnaK strain at 37°C (Figure 1F) suggests a common explanation, i.e. the lack in both cases of available DnaK (which is titrated by the thermo-denatured proteins in MC4100 under a severe heat stress, thus triggering the heat-shock response depicted in Figure 1H). A plasmid-driven over-expression of DnaK/DnaJ should logically compensate for the ribosome assembly defects in MC4100 at high temperatures, as already shown in the case of a ΔdnaK strain (13).
To prevent an excessive overproduction of DnaK/DnaJ, which is toxic for E. coli (34), MC4100 bacteria were transformed to ampicillin resistance with the low copy-number plasmid pDM39 (expressing the dnaK/dnaJ operon) or the corresponding empty vector pWSK30. Ribosomal particles synthesized and labelled with [3H]-uridine in each of these two transformants at 45°C were analysed by sucrose gradient sedimentation, and the data shown in Figure 7A and B indicate that the ribosome assembly defects were indeed less important in the strain over-expressing the dnaK/dnaJ operon (no 32S and significantly less 21S particles) than in the strain carrying the empty vector. However, the amount of particles sedimenting at 45S was roughly the same in both strains, indicating that either the final maturation from 45S to 50S subunits is the rate-limiting step or the most heat-sensitive, as already shown in the experiment of Figure 2B.
Figure 7.
(A and B) Sedimentation profiles of ribosomal subunits prepared from strains MC4100 pWKS30 (A) and MC4100 pDM39 (B) labelled with [3H]-uridine at 45°C. Sedimentation is from right to left. A260/ml, open circles. [3H] c.p.m. × 103, filled circles. (C) Total RNAs phenol-extracted from the same two bacterial batches were subjected to 3′5′ RACE analysis without prior thermodenaturation of RNA (lane a = MC4100 pWSK30, lane b = MC4100 pDM39). Relative DNA concentrations in the bands of the agarose gel were estimated using a Typhoon Trio phosphorImager and the ImageQuant software.
Total RNA was phenol-extracted from the same bacterial cultures and analysed by the 3′5′RACE method (without preliminary thermo-denaturation), and the results are shown in Figure 7C. RT-PCR products of 305 bp and 155 bp were detected in both cases, and their relative proportion (155 bp/305 bp) was higher (2.6-fold) when they were generated from RNA from the strain MC4100 pDM39 than from RNA in the control strain MC4100 pWKS30. This result, which agrees with the data in Figure 7A and B, indicated that a moderate over-expression of DnaK and DnaJ partially rescues the ribosome assembly defects at high temperature, and it supports our initial hypothesis concerning the role played by these chaperones in the process of ribosome biogenesis under heat stress conditions.
DISCUSSION
The central observation described in this work is that the late stages of ribosome assembly are arrested in a wt strain upon a severe heat stress (45–46°C). This result was not totally unexpected as we have already demonstrated that ribosome assembly is temperature-dependent and delayed in mutants lacking the chaperones DnaK or DnaJ (13). Thus, we predicted that a severe heat stress could cause a severe limitation in DnaK/DnaJ in a wt strain, because the misfolded proteins would titrate out all the free chaperones. As a consequence, the DnaK-dependent late stages in ribosome assembly would be hampered, similar to hampering of the DnaK-dependent feedback control of the heat-shock response in E. coli (4). The results presented in Figure 1 agree with this prediction. The uncontrolled σ32-dependent heat-shock response shown in Figure 1H argues for such a depletion of free DnaK. The moderate over-expression of DnaK, which partially rescues defects in ribosome assembly at high temperature (Figure 7), also supports this possibility.
It should be noticed that, in contrast to the late stages of ribosome assembly, rRNA transcription and the co-transcriptional early stages (leading to the 45S, 32S and 21S precursor particles) are not inhibited but on the contrary slightly increased following a temperature up-shift (35). The role of potential heat-shock promoters in all rRNA operons is however unclear (36,37). However, this observation argues against the possibility that impaired late ribosome assembly observed here is one among many other cellular processes that malfunction as a general and unfortunate consequence of stress.
How does the translation apparatus of E. coli adapt to higher temperatures? The effect of a shift-up of the bacterial growth temperature on the protein synthesis capacity has been studied for a long time (38–41). Several mechanisms play an essential role at high temperatures: (i) the post-translational protein quality control carried out by chaperones and proteases (8); (ii) the cytoplasmic σ32-mediated (4) and extracytoplasmic σE-mediated (42) heat stress responses, inducing a certain degree of thermotolerance (43,44); (iii) some specific HSPs involved in translation (45) in particular HSP15 (46) and YbeY (47–49). Two ribosomal protein genes, rpsL and rpmE, have σ32 promoters (50).
Our data emphasize a novel facet of the response of E. coli exposed to an elevated temperature, i.e. the arrest of late steps of ribosome biogenesis. True precursors to mature ribosomal subunits accumulate in these conditions: the benefit for E. coli and the physiological relevance of such a mechanism involving DnaK will be discussed at the end of this section. Nevertheless, data presented in this work provide a new tool for the isolation and further characterization of E. coli ribosomal precursor particles, and more generally for future structure-function relationship studies of the bacterial ribosome. These precursor particles can be reactivated in vivo (Figure 2B) and in vitro (data not shown), probably by a process of protein replacement that was recently described in the reactivation of stationary phase ribosomes (51).
Two unexpected conclusions of this work were reached through the 3′5′ RACE (Figures 3 and 4) and primer extension (Figure 5) analysis of the 16S rRNA synthesized in bacteria at high temperature:
First, the sites of cleavage by RNase III of the 30S rRNA primary transcript are located 115 nt upstream of the 5′ end of m16S rRNA, and 35 nt (not 33 nt) downstream of the 3′ end of m 16S rRNA, leading to the p1 16S rRNA (embedded into 21S precursor particles). Its 5′ end is matured by the endoribonuclease activities of RNase E and RNase G, together with the shortening (from 35 to 33 nt) of the extra-sequence at its 3′end, leading to the p2 16S rRNA (also embedded into 21S precursor particles). Interestingly, the p1 16S rRNA of the double mutant ΔybeY Δrnt was also extended by one or two nucleotides at the 3′ terminus, suggesting the involvement of RNase T in trimming the p1 16S RNA at the 3′ terminus prior to complete 3′ maturation (Figure 4D of ref. 49).
Second, this study describes for the first time the in vivo processing order of the 16S rRNA, which was currently unknown. We show here that the 16S rRNA inside the 21S precursor particles is a precursor, either unprocessed or partially processed at its 5′ end, but not processed at its 3′ end. The conclusion is that the processing of the p1 16S rRNA follows a defined order: the 5′ end is matured first by the RNases E and G. The trimming of the 3′ end performed by an unknown RNase takes place later, within 30S-like precursors (52) or in polysomes (53,54). This sequential maturation starting from the 5′ end fits well with the 5′ to 3′ transcriptionally coupled assembly of the 30S subunit in vivo (55). It also fits with the process of in vitro reconstitution of 30S subunits (56,57), which is initiated in the 5′ domain and terminates in the 3′ domain of 16S rRNA (58,59). We described a similar result when studying the processing of 16S rRNA in ΔdnaK or ΔdnaJ mutants at high temperatures (13). However it may be mentioned that: (a) in a double mutant lacking the RNases E and G, the lack of 5′ maturation considerably slows down but does not prevent the 3′ maturation of the 16S rRNA (33), and (b) the ordered transcription of rRNA domains from the 5′ to the 3′ end of both 16S and 23S rRNAs is surprisingly not essential for ribosome formation (60).
It remains to be determined whether the sequential processing of 16S rRNA starting from its 5′ end is the general pathway, or is due to the heat stress conditions used here. For example, the unknown RNase that maturates the 3′ end could be naturally thermosensitive or DnaK-dependent. This important point is currently under study, but the data shown in Figure 3C, lane b, i.e. a weak RT-PCR product of about 200 bp (most probably 188 bp) observed when starting from bacteria grown at 30°C, suggest that the ordered maturation of 16S rRNA described here is the common pathway at all temperatures.
The presence of the 5′ and 3′ extra-sequences in the p1 16S rRNA is important for in vivo ribosome biogenesis (this is why in vitro reconstitution that uses fully matured 16S rRNA as substrate mimics imperfectly in vivo assembly). The subsequent removal of these extra-sequences is essential, as the 5′ and 3′ ends of m16S rRNA are ∼80 Å apart within the 30S subunit. A recent report shows that the fidelity of translation is affected in a mutant that still carries an extra-sequence at the 5′end of its 16S rRNA (61).
Technical reasons prevented us from studying the maturation of the 23S rRNA in heat-stressed bacteria since: (i) an appropriate study would require the extraction of the 23S rRNA from each of the two 45S and 32S precursor particles, and therefore their initial resolution by sedimentation on preparative sucrose gradients. (ii) the extra-sequences in the precursor 23S rRNA are much shorter (3–7 nt at its 5′ end, and 7–9 nt at its 3′ end) than those in the p1 16S rRNA; therefore, the RT-PCR products generated by 3′5′ RACE will be poorly resolved by agarose gel electrophoresis. However, a recent study shows that the 23S rRNA maturation in E. coli is coordinated or coupled, i.e. 3′ and 5′ ends maturation proceeds concurrently at similar rates (62). This mode of processing is therefore completely different from that described here for the 16S rRNA. This is not surprising as, in contrast to the situation in the 30S subunit, the maturation of the 3′ and 5′ends of 23S rRNA is not necessary for the activity of the 50S subunit (63).
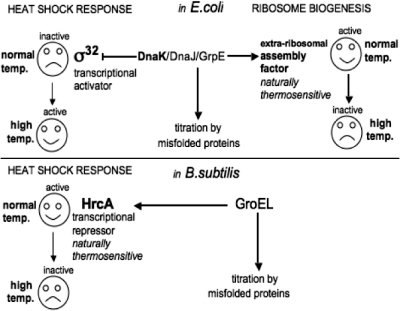
How does the DnaK chaperone machinery (DnaK/DnaJ/GrpE) (7) protect ribosome assembly at high temperatures? It has been claimed that there is a direct effect of DnaK on some ribosomal component(s) during in vitro reconstitution of 30S subunits (64,65). However, all the ribosomal components (proteins and rRNAs) are fully thermostable, even those extracted from a ΔdnaK null strain grown at 30°C (23). Therefore we favour the possibility that the target of DnaK, rather than some intrinsic ribosomal component, could be a naturally thermosensitive extra-ribosomal factor mediating the late stages of ribosome assembly. A large set of thermolabile proteins have been identified in E.coli (66). The benefit, or the physiological relevance, to the bacterial cell of such naturally thermosensitive components would be to establish, via DnaK, a regulatory link or coupling between the chaperone-mediated quality control of proteins and the late stages of ribosome biogenesis: upon severe heat shock, titration of DnaK by the heat-damaged proteins would leave the putative thermosensitive factor unprotected, retarding or halting late ribosome assembly, and preventing wasteful protein synthesis under conditions in which proteins tend to aggregate. Then, as the heat-damaged proteins are refolded or degraded, DnaK becomes available once again, and ribosome assembly can resume. Indeed, it is known that E. coli cells do maintain a reserve of ribosomal subunits, present but inactive, which can rapidly and efficiently promote the synthesis of new proteins during a nutritional shif-up (67). Such a DnaK-mediated mechanism would be reminiscent of the GroEL-mediated control of the heat- shock response in B. subtilis: the transcriptional repressor HrcA is protected by the chaperonin GroEL in normal conditions, but becomes inactive when GroEL is titrated by the unfolded proteins, resulting in the triggering of the heat-shock response (68). The similarity between this mechanism and our model of control by DnaK of the late stages of ribosome assembly is summarized in Figure 8.
Figure 8.
A model of the DnaK-controlled ribosome biogenesis in E. coli reminiscent of the HrcA-controlled heat-shock response in B. subtilis. The transcriptional repressor HrcA in B. subtilis (lower part) and the putative extra-ribosomal assembly factor involved in E. coli ribosome biogenesis (upper part) are both naturally thermosensitive and therefore dependent on GroEL and DnaK/DnaJ/GrpE, respectively, at high temperature. These chaperones, present in limiting amounts, are ‘titrated out’ by misfolded proteins under severe heat stress conditions, leaving HrcA and the putative factor unprotected, which turns ‘on’ the heat-shock response in B. subtilis, and turns ‘off’ the late stages of ribosome assembly in E. coli. Simultaneously, the E. coli transcription factor σ32, abandoned by DnaK, can freely recruit the RNA polymerase to the heat-shock promoters, thus turning on the heat-shock response.
Our model also predicts that stresses in protein synthesis other than heat will interfere with the late stages of ribosome assembly. Indeed antibiotics-treated E. coli cells manifest ribosome assembly defects (69), for example with chloramphenicol and erythromycin (70–73), neomycin and paromomycin (74) and kasugamycin (75).
Finally, chaperones cooperate in many pathways, including protein folding and degradation (8), and the control of the σ32-mediated heat-shock response in E. coli (76). It also is likely that the DnaK chaperone machine facilitates ribosome biogenesis within a network of chaperones, including GroEL/GroES (77) and some σ32-dependent HSPs other than DnaK (13). This also could be true for the trigger factor (TF) and HtpG, which associate with the ribosomal proteins S7 (78) and L2 (79), respectively. Ribosome assembly in a TF null mutant (Δtig) is currently under study.
FUNDING
The Centre National de la Recherche Scientifique (UPR 9073 CNRS) and the University Paris 7-Diderot. Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr Abdalla Al Refaii for preliminary experiments, Dr Eliane Hajnsdorf for help in the primer extension experiments, Dr Florent Busi for helpful discussions, Dr Günther Koraimann for the gift of the strain SR6618, and Dr Kyle Tanner for checking the English.
REFERENCES
- 1.Potrykus K, Cashel M. (p)ppGpp: still magical? Annu. Rev. Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 2.Hengge R. Proteolysis of sigmaS (RpoS) and the general stress response in Escherichia coli. Res. Microbiol. 2009;160:667–676. doi: 10.1016/j.resmic.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 3.Ades SE. Regulation by destruction: design of the sigma E envelope stress response. Curr. Opin. Microbiol. 2008;6:535–40. doi: 10.1016/j.mib.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Guisbert E, Yura T, Rhodius VA, Gross CA. Convergence of molecular, modeling, and systems approaches for an understanding of the E. coli heat shock response. Microbiol. Mol. Biol. Rev. 2008;72:545–554. doi: 10.1128/MMBR.00007-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alix JH. The work of chaperones. In: Nierhaus KH, Wilson DN, editors. Protein Synthesis and Ribosome Structure. Verlag, Weinheim: Wiley-VCH; 2004. pp. 529–562. [Google Scholar]
- 6.Rodriguez F, Arsene-Ploetze F, Rist W, Rüdiger S, Schneider-Mergener J, Mayer M, Bukau B. Molecular basis for regulation of the heat shock transcription factor sigma32 by the DnaK and DnaJ chaperones. Mol. Cell. 2008;32:347–358. doi: 10.1016/j.molcel.2008.09.016. [DOI] [PubMed] [Google Scholar]
- 7.Génevaux P, Georgopoulos C, Kelley WL. The Hsp70 chaperone machines of E. coli: a paradigm for the repartition of chaperone functions. Mol. Microbiol. 2007;66:840–857. doi: 10.1111/j.1365-2958.2007.05961.x. [DOI] [PubMed] [Google Scholar]
- 8.Liberek K, Lewandowska A, Zietkiewicz S. Chaperones in control of protein disaggregation. EMBO J. 2008;27:328–335. doi: 10.1038/sj.emboj.7601970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alix JH. Extrinsic factors in ribosome sssembly. In: Nierhaus KH, et al., editors. The Translational Apparatus: Structure, Function, Evolution. New York: Plenum; 1993. pp. 173–184. [Google Scholar]
- 10.Kaczanowska M, Ryden-Aulin M. Ribosome biogenesis and the translation process in E.coli. Microbiol. Mol. Biol. Rev. 2007;71:477–494. doi: 10.1128/MMBR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connolly K, Culver G. Deconstructing ribosome construction. Trends Biochem. Sci. 2009;34:256–263. doi: 10.1016/j.tibs.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El Hage A, Alix JH. Authentic precursors to ribosomal subunits accumulate in E. coli in the absence of functional DnaK chaperone. Mol. Microbiol. 2004;51:189–201. doi: 10.1046/j.1365-2958.2003.03813.x. [DOI] [PubMed] [Google Scholar]
- 13.Al Refaii A, Alix JH. Ribosome biogenesis is temperature-dependent and delayed in E. coli lacking the chaperones DnaK or DnaJ. Mol. Microbiol. 2009;71:748–762. doi: 10.1111/j.1365-2958.2008.06561.x. [DOI] [PubMed] [Google Scholar]
- 14.Ingraham JL, Marr AG. Effect of temperature, pressure, pH and osmotic stress on growth. In: Neidhardt FC, editor. Escherichia coli and Salmonella: Cellular and Molecular Biology. 2nd edn. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 1570–1578. [Google Scholar]
- 15.Rudolph B, Gebendorfer KM, Buchner J, Winter J. Evolution of Escherichia coli for growth at high temperatures. J. Biol. Chem. 2010;285:19029–19034. doi: 10.1074/jbc.M110.103374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mordukhova EA, Lee HS, Pan JG. Improved thermostability and acetic acid tolerance of Escherichia coli via directed evolution of homoserine o-succinyltransferase. Appl. Environ. Microbiol. 2008;24:7660–7668. doi: 10.1128/AEM.00654-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arsene F, Tomoyasu T, Bukau B. The heat shock response of E. coli. Int. J. Food Microbiol. 2000;55:3–9. doi: 10.1016/s0168-1605(00)00206-3. [DOI] [PubMed] [Google Scholar]
- 18.Alix JH, Guérin MF. Mutant DnaK chaperones cause ribosome assembly defects in E. coli. Proc. Natl Acad. Sci. USA. 1993;90:9725–9729. doi: 10.1073/pnas.90.20.9725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 20.Expert-Bezançon A, Guérin MF, Hayes DH, Legault L, Thibault J. Preparation of E. coli ribosomal subunits without loss of biological activity. Biochimie. 1974;56:77–89. doi: 10.1016/s0300-9084(74)80357-3. [DOI] [PubMed] [Google Scholar]
- 21.Redko Y, Bechhofer DH, Condon C. An unusual member of the RNAse III family of enzymes, catalyses 23S ribosomal RNA maturation in B. subtilis. Mol. Microbiol. 2008;68:1096–1106. doi: 10.1111/j.1365-2958.2008.06207.x. [DOI] [PubMed] [Google Scholar]
- 22.Uzan M, Favre R, Brody E. A nuclease that cuts specifically in the ribosome binding site of some T4 mRNAs. Proc. Natl Acad. Sci. USA. 1988;85:8895–8899. doi: 10.1073/pnas.85.23.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alix JH, Nierhaus KH. DnaK-facilitated ribosome assembly in E. coli revisited. RNA. 2003;9:787–793. doi: 10.1261/rna.5360203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tal M, Silberstein A, Moyner K. In vivo reassembly of 30S ribosomal subunits following their specific destruction by thermal shock. Biochim. Biophys. Acta. 1977;479:479–496. doi: 10.1016/0005-2787(77)90041-7. [DOI] [PubMed] [Google Scholar]
- 25.Dong H, Nilsson L, Kurland CG. Gratuitous overexpression of genes in Escherichia coli leads to growth inhibition and ribosome destruction. J. Bacteriol. 1995;177:1497–1504. doi: 10.1128/jb.177.6.1497-1504.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zundel MA, Basturea GN, Deutscher MP. Initiation of ribosome degradation during starvation in E. coli. RNA. 2009;15:977–983. doi: 10.1261/rna.1381309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomlins RI, Ordal ZJ. Precursor ribosomal ribonucleic acid and ribosome accumulation in vivo during the recovery of Salmonella typhimurium from thermal injury. J. Bacteriol. 1971;107:134–142. doi: 10.1128/jb.107.1.134-142.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varricchio F, Monier R. Ribosome patterns in Escherichia coli growing at various rates. J. Bacteriol. 1971;108:105–110. doi: 10.1128/jb.108.1.105-110.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michaels GA. Ribosome maturation of Escherichia coli growing at different growth rates. J. Bacteriol. 1972;110:889–894. doi: 10.1128/jb.110.3.889-894.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Young RA, Steitz JA. Complementary sequences 1700 nucleotides apart form a ribonuclease III cleavage site in E. coli ribosomal precursor RNA. Proc. Natl Acad. Sci. USA. 1978;75:3593–3597. doi: 10.1073/pnas.75.8.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srivastava AK, Schlessinger D. Mechanism and regulation of bacterial ribosomal RNA processing. Annu. Rev. Microbiol. 1990;44:105–129. doi: 10.1146/annurev.mi.44.100190.000541. [DOI] [PubMed] [Google Scholar]
- 32.Apirion D, Miczak A. RNA processing in prokaryotic cells. Bioessays. 1993;15:113–120. doi: 10.1002/bies.950150207. [DOI] [PubMed] [Google Scholar]
- 33.Li Z, Pandit S, Deutscher MP. RNAse G (CafA protein) and RNAse E are both required for the 5′ maturation of 16S ribosomal RNA. EMBO J. 1999;18:2878–2885. doi: 10.1093/emboj/18.10.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blum P, Ory J, Bauernfeind J, Krska J. Physiological consequences of DnaK and DnaJ overproduction in Escherichia coli. J. Bacteriol. 1992;174:7436–7444. doi: 10.1128/jb.174.22.7436-7444.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Condon C, Philips J, Fu ZY, Squires C, Squires CL. Comparison of the expression of the seven ribosomal RNA operons in Escherichia coli. EMBO. J. 1992;11:4175–4185. doi: 10.1002/j.1460-2075.1992.tb05511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newlands JT, Gaal T, Mecsas J, Gourse RL. Transcription of the Escherichia coli rrnB P1 promoter by the heat shock RNA polymerase (E sigma 32) in vitro. J. Bacteriol. 1993;175:661–668. doi: 10.1128/jb.175.3.661-668.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seoh HK, Weech M, Zhang N, Squires CL. rRNA antitermination functions with heat shock promoters. J. Bacteriol. 2003;185:6486–6489. doi: 10.1128/JB.185.21.6486-6489.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patterson D, Gillespie D. Effect of elevated temperatures on protein synthesis in Escherichia coli. J. Bacteriol. 1972;112:1177–1183. doi: 10.1128/jb.112.3.1177-1183.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamamori T, Ito K, Nakamura Y, Yura T. Transient regulation of protein synthesis in Escherichia coli upon shift-up of growth temperature. J. Bacteriol. 1978;134:1133–1140. doi: 10.1128/jb.134.3.1133-1140.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lemaux PG, Herendeen SL, Bloch PL, Neidhardt FC. Transient rates of synthesis of individual polypeptides in E. coli following temperature shifts. Cell. 1978;13:427–434. doi: 10.1016/0092-8674(78)90317-3. [DOI] [PubMed] [Google Scholar]
- 41.Farewell A, Neidhardt FC. Effect of temperature on in vivo protein synthetic capacity in Escherichia coli. J. Bacteriol. 1998;180:4704–4710. doi: 10.1128/jb.180.17.4704-4710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ades SE, Grigorova IL, Gross CA. Regulation of the alternative sigma factor sigma(E) during initiation, adaptation, and shutoff of the extracytoplasmic heat shock response in E.coli. J. Bacteriol. 2003;185:2512–2519. doi: 10.1128/JB.185.8.2512-2519.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shenhar Y, Rasouly A, Biran D, Ron EZ. Adaptation of Escherichia coli to elevated temperatures involves a change in stability of heat shock gene transcripts. Environ. Microbiol. 2009;11:2989–2997. doi: 10.1111/j.1462-2920.2009.01993.x. [DOI] [PubMed] [Google Scholar]
- 44.Guyot S, Pottier L, Ferret E, Gal L, Gervais P. Physiological responses of Escherichia coli exposed to different heat-stress kinetics. Arch. Microbiol. 2010;192:651–661. doi: 10.1007/s00203-010-0597-1. [DOI] [PubMed] [Google Scholar]
- 45.Rasouly A, Ron EZ. Interplay between the heat shock response and translation in Escherichia coli. Res. Microbiol. 2009;160:288–296. doi: 10.1016/j.resmic.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 46.Jiang L, Schaffitzel C, Bingel-Erlenmeyer R, Ban N, Korber P, Koning RI, de Geus DC, Plaisier JR, Abrahams JP. Recycling of aborted ribosomal 50S subunit-nascent chain-tRNA complexes by the heat shock protein Hsp15. J. Mol. Biol. 2009;386:1357–1367. doi: 10.1016/j.jmb.2008.10.079. [DOI] [PubMed] [Google Scholar]
- 47.Rasouly A, Schonbrun M, Shenhar Y, Ron EZ. YbeY, a heat shock protein involved in translation in Escherichia coli. J. Bacteriol. 2009;191:2649–2655. doi: 10.1128/JB.01663-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rasouly A, Davidovich C, Ron EZ. The heat shock protein YbeY is required for optimal activity of the 30S ribosomal subunit. J. Bacteriol. 2010;192:4592–4596. doi: 10.1128/JB.00448-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davies BW, Köhrer C, Jacob AI, Simmons LA, Zhu J, Aleman LM, Rajbhandary UL, Walker GC. Role of E.coli YbeY, a highly conserved protein, in rRNA processing. Mol. Microbiol. 2010;78:506–518. doi: 10.1111/j.1365-2958.2010.07351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wade JT, Roa DC, Grainger DC, Hurd D, Busby SJ, Struhl K, Nudler E. Extensive functional overlap between sigma factors in E. coli. Nat. Struct. Mol. Biol. 2006;13:806–81422. doi: 10.1038/nsmb1130. [DOI] [PubMed] [Google Scholar]
- 51.Pulk A, Liiv A, Peil L, Maiväli U, Nierhaus K, Remme J. Ribosome reactivation by replacement of damaged proteins. Mol. Microbiol. 2010;75:801–814. doi: 10.1111/j.1365-2958.2009.07002.x. [DOI] [PubMed] [Google Scholar]
- 52.Lindahl L. Intermediates and time kinetics of the in vivo assembly of Escherichia coli ribosomes. J. Mol. Biol. 1975;92:15–37. doi: 10.1016/0022-2836(75)90089-3. [DOI] [PubMed] [Google Scholar]
- 53.Mangiarotti G, Turco E, Ponzetto A, Altruda F. Precursor 16S RNA in active 30S ribosomes. Nature. 1974;247:147–148. doi: 10.1038/247147a0. [DOI] [PubMed] [Google Scholar]
- 54.Britton RA, Wen T, Schaefer L, Pellegrini O, Uicker WC, Mathy N, Tobin C, Daou R, Szyk J, Condon C. Maturation of the 5′ end of Bacillus subtilis 16S rRNA by the essential ribonuclease YkqC/RNase J1. Mol. Microbiol. 2007;63:127–138. doi: 10.1111/j.1365-2958.2006.05499.x. [DOI] [PubMed] [Google Scholar]
- 55.Cowling de Narvaez CC, Schaup HW. In vivo transcriptionally coupled assembly of E.coli ribosomal subunits. J. Mol. Biol. 1979;134:1–22. doi: 10.1016/0022-2836(79)90411-x. [DOI] [PubMed] [Google Scholar]
- 56.Bunner AE, Nord S, Wikström PM, Williamson JR. The effect of ribosome assembly cofactors on in vitro 30S subunit reconstitution. J. Mol. Biol. 2010 a;398:1–7. doi: 10.1016/j.jmb.2010.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bunner AE, Beck AH, Williamson JR. Kinetic cooperativity in Escherichia coli 30S ribosomal subunit reconstitution reveals additional complexity in the assembly landscape. Proc. Natl Acad. Sci. USA. 2010 b;107:5417–5422. doi: 10.1073/pnas.0912007107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Powers T, Daubresse G, Noller HF. Dynamics of in vitro assembly of 16 S rRNA into 30 S ribosomal subunits. J. Mol. Biol. 1993;232:362–374. doi: 10.1006/jmbi.1993.1396. [DOI] [PubMed] [Google Scholar]
- 59.Sykes MT, Williamson JR. A complex assembly landscape for the 30S ribosomal subunit. Annu. Rev. Biophys. 2009;38:197–215. doi: 10.1146/annurev.biophys.050708.133615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kitahara K, Suzuki T. The ordered transcription of RNA domains is not essential for ribosome biogenesis in Escherichia coli. Mol. Cell. 2009;34:760–766. doi: 10.1016/j.molcel.2009.05.014. [DOI] [PubMed] [Google Scholar]
- 61.Roy-Chaudhuri B, Kirthi N, Culver GM. Appropriate maturation and folding of 16S rRNA during 30S subunit biogenesis are critical for translational fidelity. Proc. Natl Acad. Sci. USA. 2010;107:4567–4572. doi: 10.1073/pnas.0912305107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gutgsell NS, Jain C. Coordinated regulation of 23S rRNA maturation in E. coli. J. Bacteriol. 2010;192:1405–1409. doi: 10.1128/JB.01314-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sirdeshmukh R, Schlessinger D. Why is processing of 23 S ribosomal RNA in Escherichia coli not obligate for its function? J. Mol. Biol. 1985;186:669–672. doi: 10.1016/0022-2836(85)90139-1. [DOI] [PubMed] [Google Scholar]
- 64.Maki JA, Schnobrich DJ, Culver GM. The DnaK chaperone system facilitates 30S ribosomal subunit assembly. Mol. Cell. 2002;10:129–138. doi: 10.1016/s1097-2765(02)00562-2. [DOI] [PubMed] [Google Scholar]
- 65.Maki JA, Southworth DR, Culver GM. Demonstration of the role of the DnaK chaperone system in assembly of 30S ribosomal subunits using a purified in vitro system. RNA. 2003;9:1418–1421. doi: 10.1261/rna.5139703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mogk A, Tomoyasu T, Goloubinoff P, Rudiger S, Roder D, Langen H, Bukau B. Identification of thermolabile E. coli proteins: prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 1999;18:6934–6949. doi: 10.1093/emboj/18.24.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Champney WS. Kinetics of ribosome synthesis during a nutritional shift-up in Escherischia coli K-12. Mol. Gen. Genet. 1977;152:259–266. doi: 10.1007/BF00693079. [DOI] [PubMed] [Google Scholar]
- 68.Reischl S, Wiegert T, Schumann W. Isolation and analysis of mutant alleles of the Bacillus subtilis HrcA repressor with reduced dependency on GroE function. J. Biol. Chem. 2002;277:32659–32667. doi: 10.1074/jbc.M201372200. [DOI] [PubMed] [Google Scholar]
- 69.Champney WS. The other target for ribosomal antibiotics: inhibition of bacterial ribosomal subunit formation. Infect. Disord. Drug Targets. 2006;6:377–390. doi: 10.2174/187152606779025842. [DOI] [PubMed] [Google Scholar]
- 70.Schlessinger D, Ono M, Nikolaev N, Silengo L. Accumulation of 30S preribosomal ribonucleic acid in an Escherichia coli mutant treated with chloramphenicol. Biochemistry. 1974;13:4268–4271. doi: 10.1021/bi00718a004. [DOI] [PubMed] [Google Scholar]
- 71.Sykes J, Metcalf E, Pickering JD. The nature of the proteins in ‘chloramphenicol particles' from E. coli A19 (Hfr rel met rns) J. Gen. Microbiol. 1977;98:1–16. doi: 10.1099/00221287-98-1-1. [DOI] [PubMed] [Google Scholar]
- 72.Siibak T, Peil L, Xiong L, Mankin A, Remme J, Tenson T. Erythromycin- and chloramphenicol-induced ribosomal assembly defects are secondary effects of protein synthesis inhibition. Antimicrob. Agents Chemother. 2009;53:563–571. doi: 10.1128/AAC.00870-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Siibak T, Remme J. Subribosomal particle analysis reveals the stages of bacterial ribosome assembly at which rRNA nucleotides are modified. RNA. 2010;16:2023–2032. doi: 10.1261/rna.2160010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Foster C, Champney WS. Characterization of a 30S ribosomal subunit assembly intermediate found in Escherichia coli cells growing with neomycin or paromomycin. Arch. Microbiol. 2008;189:441–449. doi: 10.1007/s00203-007-0334-6. [DOI] [PubMed] [Google Scholar]
- 75.Kaberdina AC, Szaflarski W, Nierhaus KH, Moll I. An unexpected type of ribosomes induced by kasugamycin: a look into ancestral times of protein synthesis? Mol. Cell. 2009;33:227–236. doi: 10.1016/j.molcel.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guisbert E, Herman C, Lu CZ, Gross CA. A chaperone network controls the heat shock response in E. coli. Genes Dev. 2004;18:2812–2821. doi: 10.1101/gad.1219204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.El Hage A, Sbai M, Alix JH. The chaperonin GroEL and other heat-shock proteins, besides DnaK, participate in ribosome biogenesis in Escherichia coli. Mol. Gen. Genet. 2001;264:796–808. doi: 10.1007/s004380000369. [DOI] [PubMed] [Google Scholar]
- 78.Martinez-Hackert E, Hendrickson WA. Promiscuous substrate recognition in folding and assembly activities of the trigger factor chaperone. Cell. 2009;138:923–934. doi: 10.1016/j.cell.2009.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Motojima-Miyazaki Y, Yoshida M, Motojima F. Ribosomal protein L2 associates with E. coli HtpG and activates its ATPase activity. Biochem. Biophys. Res. Commun. 2010;400:241–245. doi: 10.1016/j.bbrc.2010.08.047. [DOI] [PubMed] [Google Scholar]
- 80.Peters JE, Thate TE, Craig NL. Definition of the Escherichia coli MC4100 genome by use of a DNA array. J. Bacteriol. 2003;185:2017–2021. doi: 10.1128/JB.185.6.2017-2021.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ferenci T, Zhou Z, Betteridge T, Ren Y, Liu Y, Feng L, Reeves PR, Wang L. Genomic sequencing reveals regulatory mutations and recombinational events in the widely used MC4100 lineage of Escherichia coli K-12. J. Bacteriol. 2009;191:4025–4029. doi: 10.1128/JB.00118-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bukau B, Walker GC. Mutations altering heat shock specific subunit of RNA polymerase suppress major cellular defects of E. coli mutants lacking the DnaK chaperone. EMBO J. 1990;9:4027–4036. doi: 10.1002/j.1460-2075.1990.tb07624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wagner MA, Zahrl D, Rieser G, Koraimann G. Growth phase- and cell division-dependent activation and inactivation of the sigma32 regulon in Escherichia coli. J. Bacteriol. 2009;191:1695–1702. doi: 10.1128/JB.01536-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang RF, Kushner SR. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 85.Missiakas D, Georgopoulos C, Raina S. The Escherichia coli heat shock gene htpY: mutational analysis, cloning, sequencing, and transcriptional regulation. J. Bacteriol. 1993;175:2613–2624. doi: 10.1128/jb.175.9.2613-2624.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]