Abstract
Ribosome biogenesis requires multiple nuclease activities to process pre-rRNA transcripts into mature rRNA species and eliminate defective products of transcription and processing. We find that in mammalian cells, the 5′ exonuclease Xrn2 plays a major role in both maturation of rRNA and degradation of a variety of discarded pre-rRNA species. Precursors of 5.8S and 28S rRNAs containing 5′ extensions accumulate in mouse cells after siRNA-mediated knockdown of Xrn2, indicating similarity in the 5′-end maturation mechanisms between mammals and yeast. Strikingly, degradation of many aberrant pre-rRNA species, attributed mainly to 3′ exonucleases in yeast studies, occurs 5′ to 3′ in mammalian cells and is mediated by Xrn2. Furthermore, depletion of Xrn2 reveals pre-rRNAs derived by cleavage events that deviate from the main processing pathway. We propose that probing of pre-rRNA maturation intermediates by exonucleases serves the dual function of generating mature rRNAs and suppressing suboptimal processing paths during ribosome assembly.
INTRODUCTION
Ribosomes are RNA-based molecular machines for protein synthesis that are produced in a cell in a complex assembly pathway (1). The main structural components of the eukaryotic ribosome are 18S rRNA in the small subunit and 5.8S/25S rRNAs (5.8S/28S in higher eukaryotes) in the large subunit. These rRNAs are initially transcribed by RNA polymerase I (Pol I) as a single large precursor, which also contains spacer regions removed during pre-rRNA maturation. For example, the primary transcript in mammalian species, 47S pre-rRNA, contains external (5′ETS, 3′ETS) and internal (ITS1 and ITS2) transcribed spacers (Figure 1A, Supplementary Figure S1). The synthesis of ribosomal subunits involves several endonucleolytic cleavages within spacer regions followed by exonucleolytic trimming to form the 5′ and 3′ ends of the mature rRNAs (2,3). In Saccharomyces cerevisiae, 5′ exonucleases function in the maturation of the 5′ ends of 5.8S and 25S rRNAs (4,5). In addition, several 3′ exonucleases have been implicated in the generation of the 3′ end of 5.8S rRNA (6–11). Why these rRNA ends are generated through exonuclease trimming rather than precise cleavages is not exactly clear.
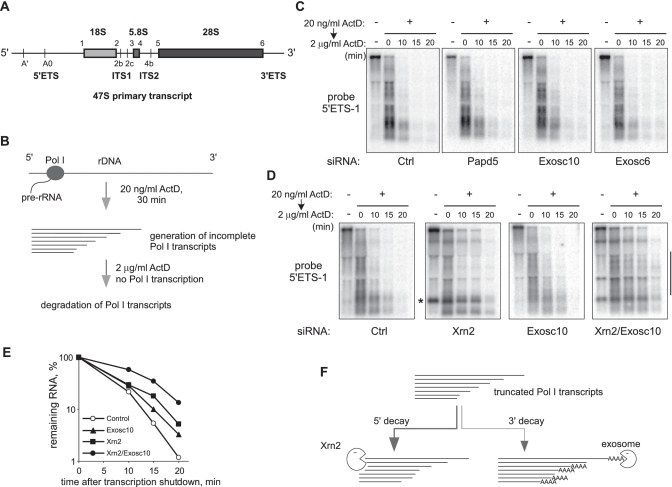
Figure 1.
Xrn2 contributes to the degradation of incomplete Pol I transcripts. (A) Structure of the full-length mouse Pol I transcript (47S pre-rRNA) showing major processing sites. (B) Incubation of cells with low doses (20 ng/ml) of ActD leads to abortive transcription by Pol I. Decay of the incomplete transcripts was assessed by analyzing RNA at different times points after blocking new transcription with 2 µg/ml ActD. (C and D) Northern hybridization to monitor decay of pre-rRNA transcripts in cells transfected with siRNAs targeting the indicated genes or control siRNA (Ctrl). Equal amounts of total RNA were loaded in each lane, the blots were hybridized with the oligonucleotide probe 5′ETS-1 complementary to nucleotides +346 to +375 in pre-rRNA [between the 5′ end of 47S and site A′, see (A)]. Asterisk indicates the 650-nt 5′-A′ fragment in Xrn2-depleted cells. (E) Levels of truncated Pol I transcripts were quantified by the phosphorimager analysis of smear intensity within the range indicated by a vertical bar in panel D and normalized to time point 0. (F) Degradation of abortive Pol I transcripts is bidirectional and involves both Xrn2 and the exosome.
The complex nature of ribosome assembly leads to inevitable errors in processing and folding of pre-rRNA. Surveillance mechanisms are crucial for eliminating defective precursors, thereby promoting structural and functional integrity of the final ribosomal particles (12). The exosome, a multisubunit protein complex containing 3′ exoribonuclease activities (6), plays an important role in rRNA quality control by initiating 3′ to 5′ degradation of defective pre-rRNAs in yeast (13,14). In many cases, it is still not well understood how the correct and defective rRNA precursors can be distinguished by the surveillance machinery. The TRAMP (Trf4/5p-Air1/2p-Mtr4p polyadenylation) complex is thought to provide an important regulatory role by tagging degradation substrates with short poly(A) tails that facilitate degradation by the exosome (15–17).
Ribosome biogenesis in mammals has been much less studied than in yeast, although many factors involved in this pathway are predicted to perform similar functions across eukaryotic species. Polyadenylated degradation intermediates have been identified among different RNA classes including rRNA in mammalian cells, suggesting conservation of the TRAMP/exosome surveillance function (18). We previously showed that aberrant Pol I transcripts in mouse cells can undergo polyadenylation, which is dependent on the mouse Trf4p homolog Papd5, and that degradation of these polyadenylated species involves the core exosome and an associated nuclear 3′ exonuclease, Exosc10 (PM/Scl-100) (19). The mammalian exosome and Exosc10 are present in the nucleolus (20,21), and are expected to be functionally analogous to their yeast counterparts with regard to pre-rRNA processing, although this remains to be experimentally demonstrated. The role of 5′ exonucleases in pre-rRNA maturation in mammalian species, to the best of our knowledge, has not been previously evaluated.
In this study, we report that both processing and decay of pre-rRNAs in mammalian cells rely heavily upon the 5′ to 3′ degradation carried out by the nuclear 5′ exonuclease Xrn2. A systematic evaluation of the mouse pre-rRNA maturation pathway revealed that multiple 5′ ends generated by endonucleolytic cleavages in pre-rRNA become targets for Xrn2. The mature 5′ ends of 5.8S and 28S rRNAs in mouse cells are processed by Xrn2 from longer 5′-extended precursors, demonstrating for the first time the conservation of an exonucleolytic trimming mechanism in their formation from yeast to mammals. Unexpectedly, abortive Pol I transcripts, discarded spacer fragments and aberrant precursors are all actively degraded from 5′ to 3′ and accumulate in cells upon Xrn2 depletion. Our data support a model in which proofreading of 5′ ends by Xrn2 is used repeatedly during pre-rRNA maturation as a mechanism to determine whether pre-rRNA intermediates are to be processed to mature forms or discarded.
MATERIALS AND METHODS
Cell culture, knockdowns and immunofluorescence
The mouse NIH 3T3-derived LAP3 cells were cultured as previously described (22). To downregulate expression of the desired genes, we introduced siGENOME SMARTpool siRNAs (Dharmacon) into cells using calcium phosphate transfection (19). Knockdown efficiency was monitored by RT-PCR using gene-specific primers and western blotting with antibodies against endogenous Xrn2 (Santa Cruz sc-99237). Immunofluorescence analysis was performed on paraformaldehyde-fixed cells (23) using antibodies against the HA tag (mouse 16B12, Covance, or rabbit C29F4, Cell Signaling), fibrillarin (MCA-38F3, EnCor Biotechnology), Nog1 (24) and Exosc10 (Sigma P4124). Cells were treated with ActD (Calbiochem) to inhibit Pol I transcription as previously described (19).
RNA analysis
Total cellular RNA was isolated from growing, subconfluent cells using Trizol (Invitrogen) and analyzed by northern hybridizations with 32P-labeled oligonucleotide probes (25). Primer extensions were performed according to a previous protocol (26), with the following modifications: a total of 2 pmol of a 32P-labeled primer was mixed with 0.5 µg RNA, annealed at 65°C for 5 min before adding the reaction mixture, incubated at 42°C for 5 min and then at 60°C for 40 min. Reactions were placed at 90°C for 5 min to inactive the enzyme, mixed with formamide loading dye and separated on a 6% sequencing gel. Oligonucleotide sequences and their locations relative to the mouse pre-rRNA sequence are listed in Supplementary Table S1.
RESULTS
Degradation of abortive pre-rRNA transcripts in mouse cells involves the 5′–3′ exoribonuclease Xrn2
In mouse cells, three rRNAs (18S, 5.8S and 28S) are transcribed by Pol I as a single 13.4-kb primary transcript, termed 47S pre-rRNA (Figure 1A). Inhibition of Pol I elongation with low doses of the anticancer agent actinomycin D (ActD) results in the generation of short heterogeneous transcripts, largely terminated within the 5′ETS region of pre-rRNA (27,28). Previously, we found that some of the abortive transcripts exhibited hallmark features of the polyadenylation-assisted 3′ degradation by the exosome, although the total levels of these RNAs were only weakly affected by knockdowns of components of the 3′ decay machinery (19). To better understand the mechanisms responsible for the decay of the abortive Pol I transcripts, we devised an assay in which we first treated cells for 30 min with a low dose (20 ng/ml) of ActD to generate these transcripts, and then shut down Pol I transcription completely with 2 µg/ml ActD to monitor decay rates (Figure 1B). We applied this assay to mouse cells in which various mammalian proteins predicted to act in RNA surveillance were knocked down using siRNAs, as was verified by reverse transcription (RT)-PCR (Supplementary Figure S2A). Northern hybridizations with a probe specific for the 5′ETS region of pre-rRNA revealed a smear of short RNAs accumulating in the presence of 20 ng/ml ActD, and these RNAs were largely destroyed within 20 min in cells that were transfected with a control nontargeting siRNA (Figure 1C, ‘Ctrl’). Consistent with our previous study (19), knockdowns of the putative poly(A) polymerase Papd5, the core exosome component Exosc6, or the exosome-associated nuclear 3′ exonuclease Exosc10/PM-Scl100 did not prevent the rapid decay of the abortive Pol I transcripts (Figure 1C). The possibility that 3′ decay might involve redundant nucleases also appeared unlikely since knocking down other putative mammalian 3′ exonucleases or simultaneously silencing Exosc10 and Exosc6 failed to prevent degradation of the abortive transcripts (Supplementary Figure S2B–E).
Surprisingly, the siRNA-mediated knockdown of the 5′ exonuclease Xrn2 dramatically altered the decay pattern of the abortive Pol I transcripts (Figure 1D, ‘Xrn2′). We reasoned that if a large fraction of the transcripts is rapidly degraded in the 5′ to 3′ direction by Xrn2, the contribution of the exosome-TRAMP pathway might become more apparent when Xrn2 function is inhibited. As shown in Figure 1D, simultaneously knocking down Xrn2 and Exosc10 further stabilized truncated Pol I transcripts, as compared with knockdown of either Xrn2 or Exosc10 alone. To estimate transcript decay rates, we measured hybridization signal within the range of the smear that was largely devoid of pre-rRNA cleavage products, which also accumulated upon Xrn2 depletion (such as marked by an asterisk, Figure 1D, see also below). This quantification confirmed the synergistic effect on the decay rate when both 5′ and 3′ exonucleases were depleted (Figure 1E). A similar effect on stability and an altered degradation pattern were also evident when the TRAMP component Papd5 was knocked down together with Xrn2 (Supplementary Figure S3). Thus, we conclude that truncated Pol I transcripts in mouse cells are degraded in both 5′ and 3′ directions, and that the 5′-end decay requires Xrn2 (Figure 1F).
Xrn2 is a nuclear protein and is required for the degradation of excised 5′ETS fragments
Xrn2 is a mammalian homolog (29) of the S. cerevisiae 5′ exoribonucleases Xrn1p/Kem1p and Rat1p, which are predominantly cytoplasmic and nuclear, respectively (30). Mammalian Xrn2 has been previously implicated in RNA Pol II transcription termination (31), intron degradation, pre-mRNA and microRNA metabolism (32), all of which are nuclear processes. Immunofluorescence analysis of HA-tagged Xrn2 in mouse cells confirmed that this protein is largely localized to the nucleus (Figure 2A). Xrn2 was also present in the nucleolus, as shown by costaining with several nucleolar proteins including fibrillarin, Nog1 and Exosc10, but was excluded from many areas brightly stained with a Hoechst dye, which likely represent condensed chromatin (Figure 2A). Given this localization pattern and the strong effect of Xrn2 depletion on the degradation of abortive Pol I transcripts, we decided to examine its functions in pre-rRNA metabolism in more detail.
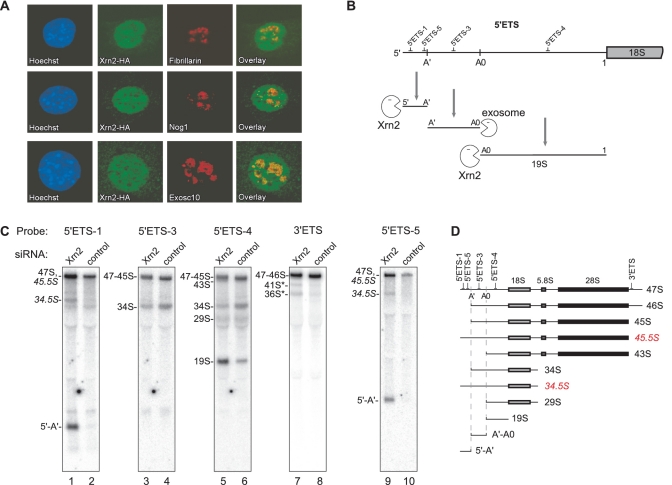
Figure 2.
Xrn2 is a nuclear protein that participates in the degradation of pre-rRNA spacer fragments and aberrant cleavage intermediates. (A) Indirect immunofluorescent staining of HA-tagged Xrn2 and endogenous nucleolar proteins. Nuclei were counterstained with Hoechst 33258. (B) Degradation of fragments released from 5′ETS by endonucleolytic cleavages at sites A′, A0 and 1 requires combined activities of Xrn2 and the exosome. The relative positions of hybridization probes are indicated at the top. (C) Northern hybridization analysis of pre-rRNA isolated from cells transfected with Xrn2-targeting or control siRNA. Membranes were hybridized with oligonucleotide probes to assess steady-state levels of various pre-rRNA species. (D) Structure of pre-rRNAs detected in hybridizations. Aberrant pre-rRNAs lacking A′ cleavage are highlighted in red.
The first processing step in mouse pre-rRNA occurs by endonucleolytic cleavage around position +650 relative to the transcription initiation site (33); however, no upstream fragment can be readily detected in cells. In hybridizations with probe 5′ETS-1, we noted accumulation of an RNA fragment prior to ActD treatment upon Xrn2 depletion (asterisk, Figure 1D). The size of this novel 5′ETS-derived RNA species (∼650 nt) suggested that it might represent the elusive 5′-A′ fragment. To confirm this, we hybridized total RNA from Xrn2-depleted and control cells with probes specific for different parts of the 5′ETS (Figure 2C, probe locations are indicated in Figure 2B). In addition to probe 5′ETS-1, the 650-nt species was detected with probe 5′ETS-5, complementary to the sequence immediately upstream of the A′ cleavage site (Figure 2C, lanes 1 and 9), but not with downstream probes 5′ETS-3 or 5′ETS-4 (Figure 2C, lanes 3–6). We conclude that under normal conditions, the 5′-A′ fragment formed by the A′ cleavage within 5′ETS is rapidly degraded by Xrn2.
We next asked whether Xrn2 is involved in the degradation of two other fragments excised during 5′ETS processing, A′-A0 and A0-1, the latter also called 19S (Figure 2B). Hybridizations with probe 5′ETS-3 did not reveal any detectable accumulation of the A′-A0 spacer fragment in Xrn2-depleted cells (Figure 2C, lane 3). By contrast, the 19S fragment accumulated after Xrn2 depletion (Figure 2C, lane 5), suggesting that its 5′ end, generated by processing at site A0, may provide another entry point for Xrn2. The A′-A0 fragment, which is not affected by Xrn2, was previously shown to depend on the exosome for 3′ degradation (26). Interestingly, the sequence immediately downstream from the A′ site stably associates with proteins after the A′ cleavage (34) and also serves as an anchor site for U3 snoRNA binding (35,36). The U3 snoRNA is the central component of the large early processing complexes formed on nascent pre-rRNAs (37–39). It is possible that proteins bound to the 5′ end generated by the A′ cleavage help to preserve the interaction between U3 snoRNA and pre-rRNA by shielding the 5′ end from an exonuclease attack.
Xrn2 depletion unmasks aberrant cleavage events in pre-rRNA maturation
Hybridization analysis revealed previously unobserved pre-rRNA species after Xrn2 depletion. One such RNA, designated 34.5S RNA, hybridized with probes 5′ETS-1 and 5′ETS-5 (Figure 2C, lanes 1 and 9), which do not detect the major 34S pre-rRNA. This pattern implies that unlike 34S, the 34.5S RNA is generated by an ITS1 cleavage occurring prior to the A′ cleavage in the 5′ETS. The latter is also known as the ‘primary’ cleavage in the mammalian pre-rRNA transcript, and it is thought to precede all other processing events (Supplementary Figure S1). Since the presence of 34.5S RNA suggested that the A′ cleavage might not be absolutely required for other cleavages to occur, we looked for additional pre-rRNAs extended into the 5′ region. For instance, cleavage at the 3′ end of the 28S rRNA sequence occurring prior to the A′ cleavage would be expected to generate 45.5S RNA instead of the normal 45S pre-rRNA (Figure 2D). 47S-45S precursors do not separate well on a gel due to their large size (∼13 kb), but can be distinguished through a series of hybridizations. After Xrn2 knockdown, there was a reproducible increase in the signal intensity of the 47S-45S band in hybridizations with probes 5′ETS-1 and 5′ETS-5 (Figure 2C, compare lanes 1 and 2, also 9 and 10). The lack of a corresponding signal increase with probes located 3′ relative to site A′, such as 5′ETS-3 or 5′ETS-4, indicated that this was not due to accumulating 46S or 45S pre-rRNAs (Figure 2C, lanes 3–6). Hybridization with probe 3′ETS next ruled out an increase in the primary 47S pre-rRNA (Figure 2C, lanes 7 and 8). We conclude that an aberrant, 5′-extended form of 45S indeed accumulates after Xrn2 depletion.
The observation that impaired Xrn2 function leads to accumulation of cleavage intermediates with an unremoved 5′-A′ segment challenges the long-standing view that the primary A′ cleavage is a prerequisite for mammalian pre-rRNA processing. Instead, our data suggest that cleavage at site A′ is simply a rapidly occurring event and when cleavages at other processing sites occur first, the resulting intermediates such as 45.5S and 34.5S are degraded from the 5′ end by Xrn2. By contrast, the 45S precursor, which undergoes further maturation (Supplementary Figure S1), is formed by the A′ cleavage that generates a 5′ end resistant to Xrn2 activity.
Xrn2 participates in processing within ITS1 and ITS2
We next investigated whether Xrn2 may be involved in later stages of pre-rRNA maturation. In S. cerevisiae, impairment of the major 5′ exonucleases Rat1p and Xrn1p leads to accumulation of 5′-extended forms of precursors to 25S (5) and 5.8S rRNA (4), indicating a role of exonucleolytic trimming in their 5′-end maturation, but whether the same mechanism is used in the corresponding processing steps in mammals has remained unknown. The precise location of ITS cleavages in mammals has not been established either, although early mapping efforts indicated two possible cleavage sites within the ITS1 in mammalian pre-rRNA (2b and 2c, see Figure 1A) and a provisional cleavage site 4b in the ITS2 (2). Analysis of mouse pre-rRNA with probes hybridizing within the ITS1 (Figure 3A, lanes 1–6) revealed a complex pattern of intermediates and ITS1-derived fragments consistent with the separation of the 18S and 5.8S/28S rRNA precursors by alternate endonucleolytic cleavages in ITS1 (Figure 3B).
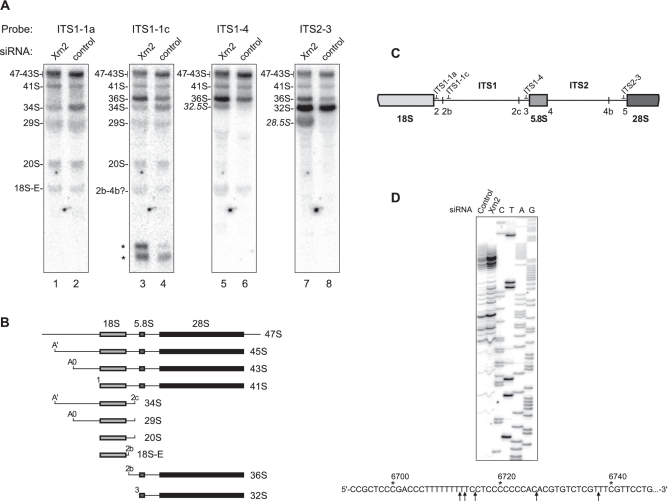
Figure 3.
Xrn2 is involved in processing of the ITS1 and ITS2. (A) Northern hybridizations to assess steady-state levels of pre-rRNAs derived by cleavages in the ITS regions. RNA was isolated from cells transfected with Xrn2-targeting or control siRNA and hybridized with the indicated oligonucleotide probes. 5′-extended precursors are indicated by italics. Asterisks, short ITS1-derived fragments. (B) Structure of the major precursors analyzed in (A). (C) Location of probes relative to cleavage sites in the ITS regions. (D) Primer extension analysis of the ITS1 region upstream from the 5.8S sequence. RNA extracted from the control or Xrn2 siRNA-transfected cells was reverse transcribed using primer pEx-5.8S-19 and products were separated next to sequencing reactions performed on a cloned ITS1 sequence with the same primer. Arrows on the pre-rRNA sequence indicate positions of major stops detected in Xrn2-depleted cells.
To study the effect of Xrn2 depletion on the formation of the 5′ end of 5.8S rRNA, we performed hybridization with probe ITS1-4, which is located immediately upstream from site 3 (Figure 3C), which is the normal 5′ end of the 32S pre-rRNA (Figure 3B). This hybridization showed that Xrn2 knockdown caused accumulation of a novel intermediate that comigrated with 32S pre-rRNA, and was designated 32.5S (Figure 3A, lane 5). The lack of a similar accumulation with probes located upstream of site 2c (Figure 3A, lanes 1 and 3; Figure 2C, lanes 3 and 5) shows that 32.5S is not a 3′-extended form of 34S. To examine the 5′ end of 32.5S, we performed primer extension with an oligonucleotide annealing at the junction of the ITS1 and 5.8S rRNA sequences. This revealed a number of RT stops, located 139–164 nt upstream of the 5.8S rRNA sequence, which increased in intensity upon depletion of Xrn2 (Figure 3D), indicating that 32.5S RNA represents a mixture of 5′-extended 32S pre-rRNA forms. We were unable to analyze the substrate-product relationship between 32.5S and 32S pre-rRNA due to the exceedingly long half-life of the 32S pre-rRNA in mouse cells [1–2 h, as shown by a previous metabolic labeling analysis (40)]. However, considering the analogous role of the yeast exonucleases Rat1p and Xrn1p in 5′-end trimming the 27SA3 pre-rRNA (4), which is equivalent to the mammalian 32.5S pre-rRNA, these data strongly suggest that the ITS1-derived extensions are removed by Xrn2 to generate 32S pre-rRNA.
Hybridization with probe ITS1-1c, located between the two major sites 2b and 2c (Figure 3C), indicates additional functions for Xrn2 in ITS1 processing. An increase in the 36S pre-RNA and reduced levels of the 34S pre-rRNA in Xrn2-depleted cells (Figure 3A, lanes 3 and 4) indicate a preferential use of site 2b over 2c in separating the 18S and 5.8S/28S processing branches. A similar change in the 34S/36S ratio was observed after functional inhibition of pre-60S subunit assembly factors in previous studies (40), although the reason for this effect is not yet clear. Accumulation of short ITS1-derived products (asterisks, Figure 3A, lanes 3 and 4) upon Xrn2 depletion suggests that this nuclease may also be involved in the degradation of discarded ITS1 fragments.
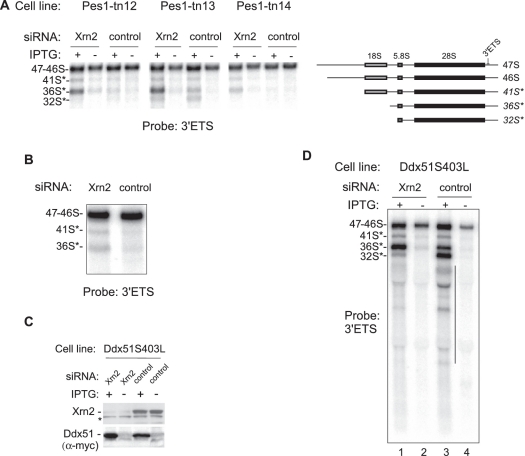
Another novel pre-rRNA observed after Xrn2 knockdown had a diffuse appearance on the blot as it comigrated with the abundant mature 28S rRNA (Figure 3A, lane 7). Because this pre-rRNA hybridized with probe ITS2-3, which is immediately upstream of the 28S sequence (Figure 3C), we designated this precursor 28.5S pre-rRNA, consistent with our nomenclature for other 5′-extended species. We reasoned that if the 28.5S pre-rRNA is generated from 32S pre-rRNA by cleavage within ITS2, then inhibition of ITS2 processing should reduce 28.5S levels. To test this, we used the previously described dominant-negative mutants of the assembly factor Pes1 (40). This protein, together with Bop1 and Wdr12, is part of the evolutionarily conserved Nop7 complex (Nop7–Erb1–Ytm1 in yeast), mutations in which can interfere with ITS2 processing and prevent the formation of the large subunit rRNAs (41–43). We transfected three cell lines in which different Pes1 mutants were expressed under the control of an IPTG-regulated promoter (40) with either control or Xrn2-targeting siRNAs (Figure 4A) and analyzed pre-rRNAs by hybridizations. As expected, inhibition of ITS2 processing reduced the levels of the 28.5S pre-rRNA after Xrn2 knockdown (Figure 4B).
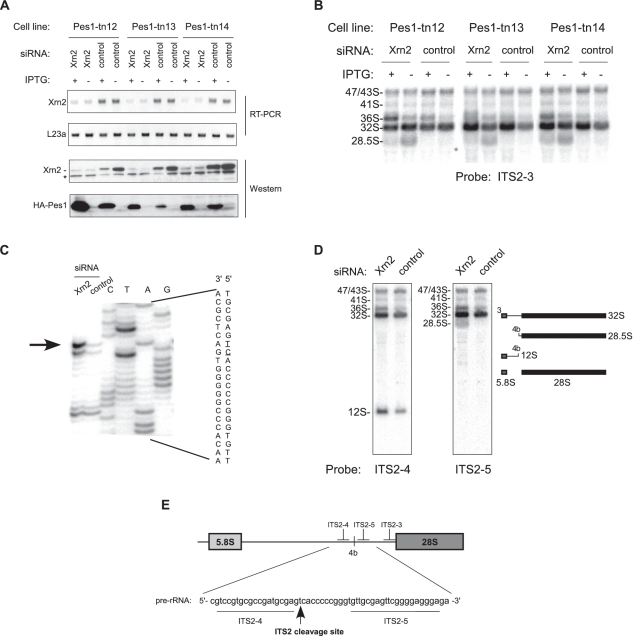
Figure 4.
5′-end maturation of 28S rRNA requires exonucleolytic trimming by Xrn2. (A) IPTG-inducible HA-Pes1 mutant cell lines (40) were transfected with either control or Xrn2-targeting siRNA. The efficiency of Xrn2 depletion was assessed by RT-PCR and western blotting. Ribosomal protein L23a mRNA was used a standard for RT-PCR. Induction of Pes1 mutants was verified with antibodies against the HA tag. Asterisk, a cross-reacting protein. (B) Northern analysis of pre-rRNA from the same cells shows accumulation of 28.5S pre-rRNA after downregulation of Xrn2. Inhibition of ITS2 processing by induction of Pes1 mutants reduces 28.5S formation. (C) Primer extension analysis with primer ITS2-pEx1 to map the main cleavage site in the ITS2. The nucleotide sequence shown with the 5′ end at the top is colinear with pre-rRNA; the cleavage site is indicated by an arrow and underlined in the sequence. (D) Hybridizations with probes ITS2-4 and ITS2-5 to confirm the correct identification of the main ITS2 cleavage site; relative probe positions are shown in (E).
To examine the role of Xrn2 in ITS2 processing in more detail, we next mapped the 5′ end of the 28.5S pre-rRNA by primer extension analysis. Xrn2 depletion led to a significantly increased RT stop at position −281 in ITS2 relative to the mature 5′ end of 28S (Figure 4C). Hybridizations of pre-rRNA with probes flanking this site showed that the 28.5S pre-rRNA was detected with probe ITS2-5 but not ITS2-4, while the upstream product of the ITS2 cleavage 12S pre-rRNA hybridized only with ITS2-4 (Figure 4D), confirming that the stop identified in primer extension corresponds to the major ITS2 cleavage site in mouse pre-rRNA (Figure 4E). These data are also in agreement with the early S1 nuclease mapping experiments that placed the 3′ end of the mouse 12S pre-rRNA ∼295 nt apart from the 5′ end of the 28S sequence (44).
Based on these data, we conclude that the 5′ end of the 28S rRNA in mammals is generated by a mechanism similar to that in yeast, in both cases starting with an endonucleolytic cleavage in the ITS2, followed by an exonucleolytic trimming of the 5′ extension, which in mammals is carried out by Xrn2. Moreover, Xrn2 performs a similar role in the formation of 5.8S precursors by trimming their 5′ ends after an initial cleavage in the ITS1.
Xrn2 mediates decay of defective pre-rRNAs with unremoved 3′ETS
Accumulation of multiple 5′-extended pre-rRNA species after Xrn2 depletion shows that surveillance of 5′ ends during mammalian ribosome biogenesis is widespread and can be adopted for a variety of tasks, including the generation of mature ends and decay of aberrant pre-rRNAs. However, because removing 5′ extensions from aberrant 5′-extended species makes them look like regular precursors, it is not easy to determine whether Xrn2 continues degradation past the mature 5′ ends. To address this problem, we took advantage of the observation that in addition to stalling pre-60S ribosome maturation, Pes1 mutants inhibit removal of the 3′ETS sequence from precursors (40). The presence of unremoved 3′ETS extensions thus allows the pre-rRNAs in stalled particles to be distinguished experimentally from normal intermediates. To assess the contribution of Xrn2 to degradation of the defective pre-rRNAs in Pes1 mutant cells, we hybridized RNA isolated from three different mutant lines (Figure 4A) with a probe specific for the 3′ETS region. As shown in Figure 5A, knockdown of Xrn2 in cells that were induced to express Pes1 mutants substantially increased the amount of the aberrant species 36S*, and to some extent 41S*, detected with the 3′ETS-specific probe. This result suggested that degradation of these aberrant 3′-extended pre-rRNAs could involve Xrn2. A small but reproducible increase in 3′-extended species was also observed when Xrn2 was knocked down in nontransfected cells (Figure 5B), presumably because a lack of timely 3′ETS processing can also occur at a low frequency under normal conditions.
Figure 5.
Downregulation of Xrn2 leads to accumulation of aberrant 3′-extended pre-rRNA species. (A) Accumulation of 3′ETS-extended ‘star’ species after Pes1-mutant induction in Xrn2-depleted cells. Hybridization analysis was performed with the same RNA as in Figure 4B. (B) Xrn2 depletion leads to increased levels of aberrant 3′-extended pre-rRNAs in normal cells. A magnified view of the top portion of the 3′ETS panel in Figure 2C is shown. (C) siRNA-mediated Xrn2 depletion in cells that express a dominant-negative mutant of Ddx51 inducible with IPTG. Western blotting was used to verify knockdown of Xrn2 and the induction of the mutant Myc-tagged Ddx51S403L; asterisk, a cross-reacting protein band. (D) Northern hybridization to detect 3′-extended pre-rRNA species. Hybridization of the decay products (vertical line) with a 3′ETS probe indicates heterogeneity of their 5′ ends.
To corroborate these results, we next analyzed cells that inducibly expressed a dominant-negative form of Ddx51, a putative mammalian helicase that functions in the displacement of U8 snoRNA from pre-rRNA (23). Expression of a catalytically inactive Ddx51 in cells was previously shown to inhibit the removal of the 3′ ETS from 28S rRNA precursors (23). Hybridization analysis of pre-rRNA isolated from cells after the induction of the myc-tagged Ddx51S403L mutant protein (Figure 5C) revealed 3′-extended precursors, such as 41S*, 36S* and 32S*, several smaller fragments and a smear of degradation products (Figure 5D, lane 3), consistent with 5′ decay of the aberrant 3′-extended species. Downregulation of Xrn2 using siRNA resulted in the accumulation of the 36S*, and at the same time reduced the amounts of 32S* and shorter decay products (Figure 5D, compare lanes 3 and 1). These data indicate that Xrn2 participates in the degradation of defective 3′-extended pre-rRNAs, with the 5′ terminus of 36S* likely being the main initial entry point. The bands present in the smear of the 3′ETS-containing decay products suggest that the degradation of these large pre-rRNAs is gradual, possibly reflecting disassembly of portions of a preribosomal particle. Together, these findings indicate that the role of Xrn2 in mammalian ribosome assembly includes the degradation of defective pre-rRNAs formed during pre-60S ribosome maturation.
DISCUSSION
The data presented here show that in mammalian cells, proofreading of the 5′ RNA ends by the exoribonuclease Xrn2 directs both the formation of mature 5′ ends in rRNAs and degradation of defective or discarded pre-rRNA species (Figure 6). Substrates of Xrn2 include normal precursors, excised fragments of transcribed spacers and aberrant pre-rRNA species such as the abortive Pol I transcripts or improperly processed intermediates. Given this wide range of targets, Xrn2 most probably acts by attacking any 5′ ends formed during pre-rRNA processing, but this activity can be utilized for a variety of purposes. For example, the timely formation of higher-order structures on RNA and/or binding of proteins would be expected to create ‘road blocks’ for Xrn2 and allow the mature ends to form. The absence or a delayed formation of such structural elements (or perhaps their reversal in misassembled complexes) would lead to substrate degradation. An attractive feature of this model is that a variety of normal and defective substrates could be distinguished by the same exonuclease without necessitating specific adaptors to recognize the aberrant forms.
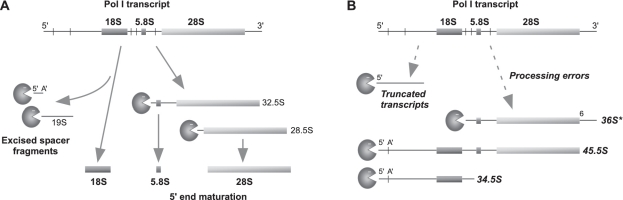
Figure 6.
Processing and quality control by Xrn2 in mammalian pre-rRNA processing. During pre-rRNA maturation, a variety of 5′ ends become targets of Xrn2. (A) In normal processing, Xrn2 trims 5′ ends in 32.5S and 28.5S pre-rRNA and degrades processing byproducts such as the excised spacer fragments generated during 18S rRNA formation. (B) Errors in transcription and processing of pre-rRNA are corrected through degradation of aberrant species by Xrn2. Truncated Pol I transcripts and abnormally cleaved pre-rRNAs (as illustrated by 34.5S, 45.5S and 36S*) are attacked and presumably targeted for degradation by Xrn2.
A novel role for mammalian Xrn2 in the nucleolus
Early biochemical studies demonstrated the presence of a processive 5′-3′ exonuclease in mammalian nucleoli (45). Immunofluorescence analysis shows that Xrn2 is present throughout the nucleus in mouse cells, including the nucleolus (Figure 2A). Proteomic studies of purified nucleoli have also identified human XRN2 as a nucleolar component [(46), http://www.lamondlab.com/NOPdb3.0/]. Consistent with the nucleolar localization, our results demonstrate that Xrn2 is an active player in pre-rRNA processing and turnover in mouse cells. The important role of Xrn2 in mammalian ribosome biogenesis is evident from the strong accumulation of a number of pre-rRNA-derived species in Xrn2 depletion experiments, even though the average efficiency achieved in our siRNA-mediated knockdowns was only 70–80%, as estimated from western analysis (Figure 4A and data not shown). This is also significant as redundancy in nucleases is typically observed in RNA decay pathways (47) and we expect that additional 5′ exonucleases may supplement Xrn2 activities in mammalian cells. A mammalian homolog of the S. cerevisiae Rrp17p, which was recently reported to function as a 5′ exonuclease during ribosome biosynthesis (48), might be one such candidate.
Besides constructing mature 5′ ends of rRNA, Xrn2 might also facilitate pre-rRNA maturation in indirect ways. A recent study (49) reported that AtXRN2, an Arabidopsis thaliana homolog of Xrn2, shortens the 5′ETS in this organism to expose the primary cleavage site for endonucleolytic processing by the U3 snoRNA-containing complex. The accumulation of a 5′-extended form of the mouse 45S pre-rRNAs upon Xrn2 depletion (45.5S RNA, Figure 2C) implies a partial inhibition of the A′ cleavage, although accumulation of the 5′-A′ fragment in the same cells (Figure 2C, lanes 1 and 9) argues that a prior Xrn2 trimming is not required for the A′ cleavage in mammals to occur. An alternative, but not mutually exclusive, possibility is that by acting in turnover of discarded 5′ETS fragments, mammalian Xrn2 promotes recycling of proteins bound to excised spacers and this in turn stimulates processing of nascent pre-rRNAs. Incorporation of Xrn2 into preribosomes could be also important for the proper assembly of these complexes in addition to providing an exoribonuclease activity, and this will need to be addressed in the future by analyzing Xrn2 mutants defective in catalytic function.
Role of the 5′- and 3′-end decay pathways in pre-rRNA surveillance
The homologs of Xrn2 in S. cerevisiae Rat1p and Xrn1p have been previously implicated in the termination of Pol I transcription (50,51), trimming 5′ ends during pre-rRNA maturation (4,5,50) and degradation of several excised pre-rRNA spacer fragments (52), whereas the quality-control functions in ribosome biogenesis have been attributed mainly to the 3′ to 5′ decay by the exosome. In yeast strains defective in the export of preribosomes from the nucleus, pre-rRNA is polyadenylated by the TRAMP complex and degraded by the exosome (14). A variety of aberrant pre-rRNA molecules and spacer fragments also accumulated in exosome and TRAMP mutant strains (6,13,15,53). Recent studies, however, suggest that 5′-end surveillance is also active in yeast ribosome biogenesis. For instance, depletion of Rat1p increases the amount of polyadenylated pre-rRNAs that accumulate in an rrp6Δ strain and reveals their 5′ heterogeneity (54).
Our results show that in mammalian ribosome synthesis, the 5′ degradation performed by Xrn2 is actively involved in removing diverse types of aberrant processing intermediates. The 5′ degradation by Xrn2 also plays a prominent role in the decay of truncated Pol I transcripts after ActD treatment (Figure 1D–F). The 3′ decay mediated by the Trf4p homolog Papd5 and the exosome (19) might primarily operate in this case on the transcripts that cannot be completely degraded by Xrn2, based on the observations that depletion of Papd5 and exosome components preferentially stabilized longer transcripts, in which bound proteins and/or secondary structures are more likely to restrict Xrn2 progression (Wang,M and Pestov,D.G., unpublished data).
Analysis of pre-rRNA degradation in this work and previous studies (19,26) provides strong evidence that a complete removal of all processing byproducts in mammalian cells requires the combined activities of 5′ and 3′ exonucleases. Bidirectional degradation could presumably serve to improve the overall efficiency of turnover of discarded fragments. Alternatively, degradation initiated from either a 5′ or 3′ end may be required simply because the opposite end is not accessible. This is likely the case for fragments excised during 5′ETS processing, which accumulate only after either Exosc10 depletion (26) or Xrn2 depletion (Figure 2B). Xrn2 belongs to the eukaryotic 5PX family of exonucleases (55) that exhibit processive activity toward 5′-monophosphate-ended RNA substrates and can be blocked by a 5′ triphosphate or secondary structures in RNA (56,57). Another level of regulation could be provided by activating factors such as Rai1p, which in yeast was shown to improve the ability of Rat1p to degrade regions with secondary structures and also act as a pyrophosphohydrolase on 5′ ends of RNA (58). Determining the individual structural features that control exonuclease access and the extent of digestion on different pre-rRNAs will be an important next step toward understanding how the 3′ and 5′ exonuclease activities are coordinated during ribosome biogenesis.
Differences and similarities in yeast and mammalian pre-rRNA processing
Most of the accumulated knowledge on eukaryotic ribosome assembly derives from studies in S. cerevisiae, and many pre-rRNA processing events in higher organisms remain incompletely defined. The A′ cleavage, absent in yeast, and also known as the ‘primary cleavage’ in mammals, is one of the two early processing steps within the 5′ETS (26) that require the U3 snoRNA (37). Our results indicate that other cleavages in mouse pre-rRNA can occasionally occur prior to A′, but the resulting aberrant precursors are rapidly destroyed by Xrn2. These data raise the interesting possibility that the A′ cleavage could provide an early checkpoint in pre-rRNA maturation by coupling the correct docking of a U3 snoRNA complex to the formation of a new stable 5′ end protected from exonucleolytic degradation.
In this study, we also for the first time demonstrate a mechanistic similarity between mammalian cells and budding yeast in processing the 5′ ends of 5.8S and 28S rRNAs. First, by analogy to previous yeast studies (4), we found that Xrn2 is involved in generating 5′ ends of the 5.8S rRNA precursors after initial ITS1 cleavages in mammals (Figure 3). In addition, the final step of 28S formation in mammals involves 5′-end trimming, which is performed by Xrn2 (Figure 4), and this parallels the mechanism of the 25S 5′-end formation in yeast (5). Surprisingly, Xrn2 depletion led to an accumulation of previously unobserved small fragments derived from ITS1 (asterisks, Figure 3A, lane 3), indicating the existence of several separate cleavage sites within mouse ITS1. This shows that mammalian processing in this spacer, which separates the small and large ribosomal subunit rRNA sequences, is more complex than previously thought, and will require further investigation.
Why does pre-rRNA processing need exonucleases?
The data reported in this work reinforce the idea that many cleavages in pre-rRNA do not follow a strict order, in agreement with a variety of previous observations made in both higher and lower eukaryotic organisms (26,35,59,60). One unexpected conclusion from our study is that the observed order in the mammalian pathway derives in part from Xrn2 attacking 5′ ends of aberrantly cleaved precursors. Knockdown of Xrn2 reveals pre-rRNA species that are normally present at very low levels in cells, such as the 5′-extended 34.5S and 45.5S RNAs, resulting from a missing cleavage in 5′ETS, and aberrant species with unremoved 3′ETS extensions due to lack of timely processing at site 6 (Figure 6B). These data support the view that the assembly of eukaryotic ribosomes can proceed through more than one path, being perhaps similar in this aspect to the assembly of bacterial ribosomes (61). Furthermore, our results imply that inspection of newly formed ends during pre-rRNA maturation by exonucleases has a role in directing assembly along the optimal route. Increasing efficiency in this system through the elimination of intermediates with abnormal cleavage patterns may indeed be the reason why so many endonucleolytic cleavages in pre-rRNA are coupled to subsequent exonucleolytic proofreading.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Institutes of Health (GM074091 to D.P.). Funding for open access charge: The National Institutes of Health (GM074091).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to John Woolford and Natalia Shcherbik for critical reading and helpful comments on the article.
REFERENCES
- 1.Kressler D, Hurt E, Baßler J. Driving ribosome assembly. Biochim. Biophys. Acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 2.Eichler DC, Craig N. Processing of eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 1994;49:197–239. doi: 10.1016/s0079-6603(08)60051-3. [DOI] [PubMed] [Google Scholar]
- 3.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 4.Henry Y, Wood H, Morrissey JP, Petfalski E, Kearsey S, Tollervey D. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 1994;13:2452–2463. doi: 10.1002/j.1460-2075.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geerlings TH, Vos JC, Raué HA. The final step in the formation of 25S rRNA in Saccharomyces cerevisiae is performed by 5′–>3′ exonucleases. RNA. 2000;6:1698–1703. doi: 10.1017/s1355838200001540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′–>5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 7.Briggs MW, Burkard KT, Butler JS. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J. Biol. Chem. 1998;273:13255–13263. doi: 10.1074/jbc.273.21.13255. [DOI] [PubMed] [Google Scholar]
- 8.van Hoof A, Lennertz P, Parker R. Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP and RNase P RNAs in yeast. EMBO J. 2000;19:1357–1365. doi: 10.1093/emboj/19.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faber AW, Van Dijk M, Raué HA, Vos JC. Ngl2p is a Ccr4p-like RNA nuclease essential for the final step in 3′-end processing of 5.8S rRNA in Saccharomyces cerevisiae. RNA. 2002;8:1095–1101. doi: 10.1017/s1355838202021027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ansel KM, Pastor WA, Rath N, Lapan AD, Glasmacher E, Wolf C, Smith LC, Papadopoulou N, Lamperti ED, Tahiliani M, et al. Mouse Eri1 interacts with the ribosome and catalyzes 5.8S rRNA processing. Nat. Struct. Mol. Biol. 2008;15:523–530. doi: 10.1038/nsmb.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gabel HW, Ruvkun G. The exonuclease ERI-1 has a conserved dual role in 5.8S rRNA processing and RNAi. Nat. Struct. Mol. Biol. 2008;15:531–533. doi: 10.1038/nsmb.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lafontaine DLJ. A `garbage can' for ribosomes: how eukaryotes degrade their ribosomes. Trends Biochem. Sci. 2010;35:267–277. doi: 10.1016/j.tibs.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 13.Allmang C, Mitchell P, Petfalski E, Tollervey D. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 2000;28:1684–1691. doi: 10.1093/nar/28.8.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dez C, Houseley J, Tollervey D. Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae. EMBO J. 2006;25:1534–1546. doi: 10.1038/sj.emboj.7601035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 16.Vanácová S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wyers F, Rougemaille M, Badis G, Rousselle J, Dufour M, Boulay J, Régnault B, Devaux F, Namane A, Séraphin B, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 18.Slomovic S, Fremder E, Staals RHG, Pruijn GJM, Schuster G. Addition of poly(A) and poly(A)-rich tails during RNA degradation in the cytoplasm of human cells. Proc. Natl Acad. Sci. USA. 2010;107:7407–7412. doi: 10.1073/pnas.0910621107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shcherbik N, Wang M, Lapik YR, Srivastava L, Pestov DG. Polyadenylation and degradation of incomplete RNA polymerase I transcripts in mammalian cells. EMBO Rep. 2010;11:106–111. doi: 10.1038/embor.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brouwer R, Allmang C, Raijmakers R, van Aarssen Y, Egberts WV, Petfalski E, van Venrooij WJ, Tollervey D, Pruijn GJ. Three novel components of the human exosome. J. Biol. Chem. 2001;276:6177–6184. doi: 10.1074/jbc.M007603200. [DOI] [PubMed] [Google Scholar]
- 21.Lejeune F, Li X, Maquat LE. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell. 2003;12:675–687. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- 22.Strezoska Z, Pestov DG, Lau LF. Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5. 8S RRNA processing and 60S ribosome biogenesis. Mol. Cell. Biol. 2000;20:5516–5528. doi: 10.1128/mcb.20.15.5516-5528.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srivastava L, Lapik YR, Wang M, Pestov DG. Mammalian DEAD box protein Ddx51 acts in 3′ end maturation of 28S rRNA by promoting the release of U8 snoRNA. Mol. Cell. Biol. 2010;30:2947–2956. doi: 10.1128/MCB.00226-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lapik YR, Misra JM, Lau LF, Pestov DG. Restricting conformational flexibility of the switch II region creates a dominant-inhibitory phenotype in Obg GTPase Nog1. Mol. Cell. Biol. 2007;27:7735–7744. doi: 10.1128/MCB.01161-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pestov DG, Lapik YR, Lau LF. Assays for ribosomal RNA processing and ribosome assembly. Curr. Protoc. Cell Biol. 2008 doi: 10.1002/0471143030.cb2211s39. Chapter 22, Unit 22.11. [DOI] [PubMed] [Google Scholar]
- 26.Kent T, Lapik YR, Pestov DG. The 5′ external transcribed spacer in mouse ribosomal RNA contains two cleavage sites. RNA. 2009;15:14–20. doi: 10.1261/rna.1384709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fetherston J, Werner E, Patterson R. Processing of the external transcribed spacer of murine rRNA and site of action of actinomycin D. Nucleic Acids Res. 1984;12:7187–7198. doi: 10.1093/nar/12.18.7187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hadjiolova KV, Hadjiolov AA, Bachellerie JP. Actinomycin D stimulates the transcription of rRNA minigenes transfected into mouse cells. Implications for the in vivo hypersensitivity of rRNA gene transcription. Eur. J. Biochem. 1995;228:605–615. doi: 10.1111/j.1432-1033.1995.0605m.x. [DOI] [PubMed] [Google Scholar]
- 29.Shobuike T, Sugano S, Yamashita T, Ikeda H. Characterization of cDNA encoding mouse homolog of fission yeast dhp1+ gene: structural and functional conservation. Nucleic Acids Res. 1995;23:357–361. doi: 10.1093/nar/23.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson AW. Rat1p and Xrn1p are functionally interchangeable exoribonucleases that are restricted to and required in the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 1997;17:6122–6130. doi: 10.1128/mcb.17.10.6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.West S, Gromak N, Proudfoot NJ. Human 5′ –> 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- 32.Morlando M, Ballarino M, Gromak N, Pagano F, Bozzoni I, Proudfoot NJ. Primary microRNA transcripts are processed co-transcriptionally. Nat. Struct. Mol. Biol. 2008;15:902–909. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kass S, Craig N, Sollner-Webb B. Primary processing of mammalian rRNA involves two adjacent cleavages and is not species specific. Mol. Cell. Biol. 1987;7:2891–2898. doi: 10.1128/mcb.7.8.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kass S, Sollner-Webb B. The first pre-rRNA-processing event occurs in a large complex: analysis by gel retardation, sedimentation, and UV cross-linking. Mol. Cell. Biol. 1990;10:4920–4931. doi: 10.1128/mcb.10.9.4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hartshorne T, Toyofuku W. Two 5′-ETS regions implicated in interactions with U3 snoRNA are required for small subunit rRNA maturation in Trypanosoma brucei. Nucleic Acids Res. 1999;27:3300–3309. doi: 10.1093/nar/27.16.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borovjagin AV, Gerbi SA. The spacing between functional Cis-elements of U3 snoRNA is critical for rRNA processing. J. Mol. Biol. 2000;300:57–74. doi: 10.1006/jmbi.2000.3798. [DOI] [PubMed] [Google Scholar]
- 37.Kass S, Tyc K, Steitz JA, Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990;60:897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- 38.Dragon F, Gallagher JEG, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grandi P, Rybin V, Bassler J, Petfalski E, Strauss D, Marzioch M, Schäfer T, Kuster B, Tschochner H, Tollervey D, et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002;10:105–115. doi: 10.1016/s1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]
- 40.Lapik YR, Fernandes CJ, Lau LF, Pestov DG. Physical and functional interaction between Pes1 and Bop1 in mammalian ribosome biogenesis. Mol. Cell. 2004;15:17–29. doi: 10.1016/j.molcel.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 41.Strezoska Z, Pestov DG, Lau LF. Functional inactivation of the mouse nucleolar protein Bop1 inhibits multiple steps in pre-rRNA processing and blocks cell cycle progression. J. Biol. Chem. 2002;277:29617–29625. doi: 10.1074/jbc.M204381200. [DOI] [PubMed] [Google Scholar]
- 42.Grimm T, Hölzel M, Rohrmoser M, Harasim T, Malamoussi A, Gruber-Eber A, Kremmer E, Eick D. Dominant-negative Pes1 mutants inhibit ribosomal RNA processing and cell proliferation via incorporation into the PeBoW-complex. Nucleic Acids Res. 2006;34:3030–3043. doi: 10.1093/nar/gkl378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang L, Sahasranaman A, Jakovljevic J, Schleifman E, Woolford JLJ. Interactions among Ytm1, Erb1, and Nop7 required for assembly of the Nop7-subcomplex in yeast preribosomes. Mol. Biol. Cell. 2008;19:2844–2856. doi: 10.1091/mbc.E07-12-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bowman LH, Goldman WE, Goldberg GI, Hebert MB, Schlessinger D. Location of the initial cleavage sites in mouse pre-rRNA. Mol. Cell. Biol. 1983;3:1501–1510. doi: 10.1128/mcb.3.8.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lasater LS, Eichler DC. Isolation and properties of a single-strand 5′–3′ exoribonuclease from Ehrlich ascites tumor cell nucleoli. Biochemistry. 1984;23:4367–4373. doi: 10.1021/bi00314a019. [DOI] [PubMed] [Google Scholar]
- 46.Ahmad Y, Boisvert F, Gregor P, Cobley A, Lamond AI. NOPdb: Nucleolar Proteome Database–2008 update. Nucleic Acids Res. 2009;37:D181–D184. doi: 10.1093/nar/gkn804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 48.Oeffinger M, Zenklusen D, Ferguson A, Wei KE, El Hage A, Tollervey D, Chait BT, Singer RH, Rout MP. Rrp17p is a eukaryotic exonuclease required for 5′ end processing of Pre-60S ribosomal RNA. Mol. Cell. 2009;36:768–781. doi: 10.1016/j.molcel.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zakrzewska-Placzek M, Souret FF, Sobczyk GJ, Green PJ, Kufel J. Arabidopsis thaliana XRN2 is required for primary cleavage in the pre-ribosomal RNA. Nucleic Acids Res. 2010;38:4487–4502. doi: 10.1093/nar/gkq172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.El Hage A, Koper M, Kufel J, Tollervey D. Efficient termination of transcription by RNA polymerase I requires the 5′ exonuclease Rat1 in yeast. Genes Dev. 2008;22:1069–1081. doi: 10.1101/gad.463708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kawauchi J, Mischo H, Braglia P, Rondon A, Proudfoot NJ. Budding yeast RNA polymerases I and II employ parallel mechanisms of transcriptional termination. Genes Dev. 2008;22:1082–1092. doi: 10.1101/gad.463408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petfalski E, Dandekar T, Henry Y, Tollervey D. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol. Cell. Biol. 1998;18:1181–1189. doi: 10.1128/mcb.18.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zanchin NI, Goldfarb DS. The exosome subunit Rrp43p is required for the efficient maturation of 5.8S, 18S and 25S rRNA. Nucleic Acids Res. 1999;27:1283–1288. doi: 10.1093/nar/27.5.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fang F, Phillips S, Butler JS. Rat1p and Rai1p function with the nuclear exosome in the processing and degradation of rRNA precursors. RNA. 2005;11:1571–1578. doi: 10.1261/rna.2900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zuo Y, Deutscher MP. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stevens A, Poole TL. 5′-exonuclease-2 of Saccharomyces cerevisiae. Purification and features of ribonuclease activity with comparison to 5′-exonuclease-1. J. Biol. Chem. 1995;270:16063–16069. doi: 10.1074/jbc.270.27.16063. [DOI] [PubMed] [Google Scholar]
- 57.Poole TL, Stevens A. Structural modifications of RNA influence the 5′ exoribonucleolytic hydrolysis by XRN1 and HKE1 of Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 1997;235:799–805. doi: 10.1006/bbrc.1997.6877. [DOI] [PubMed] [Google Scholar]
- 58.Xiang S, Cooper-Morgan A, Jiao X, Kiledjian M, Manley JL, Tong L. Structure and function of the 5′–>3′ exoribonuclease Rat1 and its activating partner Rai1. Nature. 2009;458:784–788. doi: 10.1038/nature07731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bowman LH, Rabin B, Schlessinger D. Multiple ribosomal RNA cleavage pathways in mammalian cells. Nucleic Acids Res. 1981;9:4951–4966. doi: 10.1093/nar/9.19.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kos M, Tollervey D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell. 2010;37:809–820. doi: 10.1016/j.molcel.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Talkington MWT, Siuzdak G, Williamson JR. An assembly landscape for the 30S ribosomal subunit. Nature. 2005;438:628–632. doi: 10.1038/nature04261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.