Abstract
Chlamydia trachomatis is an obligate intracellular bacterium that exhibits a unique biphasic developmental cycle that can be disrupted by growth in the presence of IFN-γ and β-lactams, giving rise to an abnormal growth state termed persistence. Here we have examined the expression of a family of non-coding RNAs (ncRNAs) that are differentially expressed during the developmental cycle and the induction of persistence and reactivation. ncRNAs were initially identified using an intergenic tiling microarray and were confirmed by northern blotting. ncRNAs were mapped, characterized and compared with the previously described chlamydial ncRNAs. The 5′- and 3′-ends of the ncRNAs were determined using an RNA circularization procedure. Promoter predictions indicated that all ncRNAs were expressed from σ66 promoters and eight ncRNAs contained non-templated 3′-poly-A or poly-AG additions. Expression of ncRNAs was studied by northern blotting during (i) the normal developmental cycle, (ii) IFN-γ-induced persistence and (iii) carbenicillin-induced persistence. Differential temporal expression during the developmental cycle was seen for all ncRNAs and distinct differences in expression were seen during IFN-γ and carbenicillin-induced persistence and reactivation. A heterologous co-expression system was used to demonstrate that one of the identified ncRNAs regulated the expression of FtsI by inducing degradation of ftsI mRNA.
INTRODUCTION
The pathogenic chlamydiae are obligate intracellular bacterial pathogens that cause important human diseases. Chlamydia trachomatis is the leading cause of preventable blindness in developing countries (1) and the most commonly reported sexually transmitted bacterial infection worldwide (2). Chlamydia trachomatis has been implicated in more serious disease sequelae that have been proposed to arise due to chronic inflammatory conditions associated with persistent infections [reviewed in (3)]. Chlamydia alternates between two morphological forms, the Elementary Body (EB) and the Reticulate Body (RB) (4). EBs are extra-cellular, metabolically inert forms, responsible for dissemination of infection by their ability to attach to and invade susceptible cells. Upon infection, EBs are internalized in membrane bound vacuoles termed inclusions. EBs differentiate into metabolically active forms, termed RBs and undergo repeated cycles of binary fission, leading to secondary differentiation back to EBs. The host cell then lyses or everts the inclusion (5), releasing EBs, that then infect neighboring cells [reviewed in (6)]. Under stressful growth conditions, imposed by immunological responses, antibiotics or nutrient deprivation, the developmental cycle is disrupted, resulting in the appearance of large, aberrant RBs [reviewed in (7)].
The normal developmental cycle and persistence in the presence of IFN-γ have been studied using DNA microarrays (8,9) and cell-biology techniques but the mechanisms for coordinated gene regulation and the changes associated with IFN-γ-induced persistence remain largely unknown (10). Temporal control of gene expression during the developmental cycle does not correlate with sigma factor expression (11) although one of the prototypical late genes, hctB (encoding one of the bacterial histone-like proteins) has been shown to be under control of the alternative sigma factor, σ28 (12). Interestingly, the other chlamydial histone-like protein, hctA, is expressed from a σ66 promoter (13) (σ66 is analogous to the major sigma factor σ70 in Escherichia coli), indicating that control of late gene expression may be more complex than an expression cascade directed by the use of alternative sigma factors (11). Niehus et al. (14) have shown that two representative mid-cycle genes are up-regulated under the influence of increased DNA supercoiling but the three representative late genes studied showed little or no response to changes in DNA supercoiling. The effect may be due to the increased G+C content of the promoter elements, specifically in the −10 and spacer regions. Grieshaber et al. (15) have shown that translation of hctA mRNA is controlled by the expression of a non-coding RNA [ncRNA, also termed small RNAs (sRNA)]. These findings were originally described in a co-expression study in E. coli in which expression of IhtA (Inhibitor of HctA) was able to rescue E. coli from the lethal effects of HctA expression (16). Subsequently these studies were extended and tested in C. trachomatis and it was determined that expression of IhtA translationally silenced the hctA mRNA but had no effect on the transcription of the hctA gene (15).
Identification of bacterial ncRNAs has increased dramatically with the advances in genomic technology and the appreciation of the importance of these non-coding molecules in regulatory pathways [reviewed in (17,18)]. Experimental approaches to identifying ncRNAs [reviewed in (19,20)] have been diverse and include (i) direct metabolic labeling of abundant ncRNAs and detection by gel electrophoresis, (ii) genetic screens for regulatory phenotypes that mapped to non-coding regions, (iii) cloning of size fractionated RNAs, (iv) bioinformatic identification of conserved intergenic regions, (v) co-purification with RNA-binding proteins, (vi) microarray analysis of intergenic regions and (vii) deep-sequencing of bacterial transcriptomes.
Albrecht et al. (21) have recently determined the transcriptomes of C. trachomatis L2b/UCH-1/proctititis EBs and RBs using a deep-sequencing approach. The sequence analyses were done on gradient-purified EBs and RBs at 24 h post infection (PI). Matched libraries were used to compare normal samples with samples that had been enriched for primary mRNA transcripts by removal of RNAs containing 5′-P groups (generally considered to be processed molecules) and the removal of 5′-PPP molecules (generally considered to be primary transcripts) by removal of the pyrophosphate group and ligation of a 5′-specific linker. They mapped 366 transcriptional start sites using this procedure. This analysis revealed fundamental differences between the transcriptomes of RBs and EBs. In addition, the analyses identified 16 putative ncRNAs in intergenic regions and 25 small anti-sense RNAs in ORFs.
We have used an Affymetrix Custom microarray that contains the intergenic regions of C. trachomatis serovar D to predict putative ncRNAs. Predicted ncRNAs were then verified using northern blotting and transcript mapping procedures. Expression of confirmed ncRNAs were shown to have developmentally regulated expression patterns, were subject to RNA modification and showed changes in expression when the organism were grown in persistence-inducing conditions in the presence of IFN-γ and carbenicillin. These regulatory RNAs may play a significant role in the control of transcription and translation of stage-specific genes and proteins in C. trachomatis and may contribute significantly to the temporal controls required for the differentiation processes associated with the developmental cycle.
MATERIALS AND METHODS
Chlamydia trachomatis growth and cell culture
Chlamydia trachomatis serovar D (strain UW-3/Cx) was grown in HeLa 229 cells cultivated at 37°C with 5% CO2 in high glucose-containing DMEM (Cellgro, Mediatech) supplemented with 10% heat-inactivated FBS. EBs were purified on density gradients of RenoCal-76 (Bracco Diagnostics, NJ, USA) as previously described (8). Monolayers were pre-treated with DEAE-Dextran (30 µg/ml) and infected at a multiplicity of infection (MOI) of 1. PI timing began immediately following the addition of infectious EBs to the monolayers. Growth in the presence of IFN-γ was done as described previously (10). IFN-γ (5 ng/ml) was used to induce persistence. The amount of IFN-γ used was determined based on a titration experiment for the minimum amount of IFN-γ that induce persistence and allowed for reactivation. Carbenicillin-persistence was induced by adding 2 µg/ml antibiotic to the media at the time of infection. Similar to the IFN-γ experiments, the amount of carbenicillin used was determined by a titration experiment that allowed for reactivation.
RNA purification
Total RNA was purified from infected HeLa 229 cultures (9 × 105 cells per well cultivated as monolayers in six well culture plates) at various times PI. At the designated times PI, the culture media was discarded and cells lysed in 1 ml per well lysis buffer containing Proteinase K (0.167 µg/µl) (MasterPureTM, Epicentre) and total nucleic acid was isolated. An aliquot of the lysate (300 µl) was treated with RNase A (0.0167 µg/µl) and used to purify DNA. The remainder of the lysate was treated with DNase I and used for total RNA preparation according to manufacturer’s instructions (MasterPure, Epicentre). Purified DNA was quantified by qPCR and used for RNA normalization to calculated C. trachomatis genome numbers as previously described (22). RNA for microarray analysis was further purified from residual contaminating DNA by treating with DNase I (0.3 U/1 µg RNA) at 37°C for 1 h (TurboDNAFree, Ambion, TX, USA). RNA was then precipitated using sodium acetate (0.1 volume) and ethanol (3 volumes) overnight at −20°C. RNA was then pelleted by centrifugation, washed with 80% ethanol and re-suspended in DPEC water (1 µg/µl). RNA (250 µg) was then enriched using the MICROBEnrichTM (Ambion) and MICROBExpressTM (Ambion) protocols, resulting in the removal of a significant portion of HeLa cell polyadenylated mRNA, 18S and 28S rRNAs in addition to bacterial 16S and 23S rRNA.
RNA was reverse transcribed, hybridized, washed, stained and scanned according to the Prokaryotic sample and array processing section in the GeneChip Expression Analysis Technical Manual (Affymetrix). In short, cDNA was generated from enriched RNA preparations using random primers (Invitrogen, CA, USA) and Superscript RT II (Invitrogen). Following cDNA generation, RNA was removed by adding NaOH (1 N) and incubating for 30 min at 65°C followed by neutralization in HCl (1 N). cDNA was then purified using MinElute columns (Qiagen). cDNA was then fragmented using DNase I (Amersham) using 0.6 U/1 μg of cDNA generated. Fragmented cDNA products were then end-labeled using GeneChip DNA labeling Reagent (Affymetrix) and Terminal Deoxynuceotidyl Transferase (Promega). Fragmented and labeled cDNA were analysed using 4–20% TBE gels (Invitrogen) using a gel shift assay using NeutrAvidin (NeutrAvidin, Pierce). The hybridization cocktail, consisting of labeled cDNA, control B2 oligo (Affymetrix), herring sperm DNA (Promega) and BSA (Invitrogen) was then hybridized to MPAUT-1 GeneChip and incubated for ∼ 16 h in the Affymetrix Hybridization Oven 640 (Affymetrix) at 45°C rotating at 60 r.p.m. Samples were then washed and stained on the Affymetrix Fluidics Station 450 (Affymetrix) using Streptavidin (Pierce Chemical), Anti-Streptavidin antibody (Vector Labratories) and R-Streptavidin Phycoerythrin (Molecular Probes) and using fluidics protocol ProKGE-WS2_450 in GCOS 1.1 (Affymetrix). After washing and staining, completed chips were scanned using the Genechip Scanner 3000 (Affymetrix). Data was scaled using GCOS 1.1 (Affymetrix) to a mean intensity of 1000 using the C. trachomatis ORFs as a scaling mask. The pivot file for the sample was then saved in GCOS as a text tab delimited file and the data was imported into GeneSpring 7.2 (Agilent) for data analysis.
RNA circularization, cDNA synthesis and cloning for 5′-/3′-end sequencing
The procedure for RNA 5′-/3′-end sequencing has been described previously (23) and was used with some modifications. A total of 8 µg of DNA-free total RNA prepared from 24 h PI cultures was treated with 10 U tobacco acid pyrophosphatase (Epicentre, WI, USA) for 30 min at 37°C to convert 5′-triphosphate groups of primary transcripts to 5′-monophosphates. Following organic extraction, RNA was treated overnight at 17°C with 40 U of T4 RNA ligase (New England Biolabs, MA, USA) to circularize transcripts. Following organic extraction and ethanol precipitation, 1.5 µg of self-ligated RNA was converted to cDNA using gene specific reverse primes and the Superscript III (200 U) reverse transcription kit (Invitrogen) in a 20 µl reaction. Incubation was carried out in 20 min intervals at 42, 50, 55 and 60°C. After heat inactivation of the reverse transcriptase for 5 min at 85°C, cDNA was treated with 1 U of RNase H (New England Biolabs) at 37°C for 20 min. A total of 1 µl of the reaction served as template in a subsequent standard 25 µl PCR reaction using 1× Advantage 2 Polymerase Mix [TITANIUM Taq DNA Polymerase, TaqStart Antibody (1.1 μg/μl)], 1× Advantage 2 SA PCR Buffer [Tris–HCl (pH 8.5), 1 mM, KCl 5 mM, MgCl2 0.2 mM] and 0.2 mM dNTP (Clontech, CA, USA) and primer pairs designed to amplify products representing successful self-ligated ncRNA transcripts. PCR products were separated by 10% PAGE TBE gel electrophoresis (BIORAD, CA, USA) and fragments of the expected size were excised, incubated overnight in 10 mM Tris–HCl buffer then re-amplified using the aforementioned PCR protocol. PCR products were separated by 3% agarose gel electrophoresis and fragments of the expected size were excised, purified and cloned into the pCR2.1-TOPO vector (Invitrogen). Positive colonies were picked, grown overnight in LB media with carbenicillin (50 µg/ml) and plasmid were prepared using MiniPlasmid Prep (Qiagen). Plasmids were sequenced using a standard M13 Forward primer.
NORTHERN BLOTTING
Northern blotting was performed using 10% TBE–PAGE–Urea gels for RNA electrophoresis in 1× TBE buffer. RNA was transferred by electro-blotting, using plate electrodes, for 1 h at 50 V, using 1× TBE buffer as transfer medium. Zeta-probe GT nylon membranes (BIORAD) were used for electro-blotting. Single-stranded biotin-labeled RNA Probes were prepared using a MAXIscript T7 Kit (Ambion). Target sequences were amplified by PCR using chlamydial DNA as a template and a minimal T7 promoter sequence was added 5′ of the anti-sense strand. The resultant PCR fragment was fractionated on agarose gels, extracted and used as a template for a larger scale PCR reaction. The PCR product was purified and was used as a DNA template for an in vitro transcription reaction using the MAXIscript T7 kit and biotin-16-UTP (Roche, IN, USA). The reaction was carried out for 1 h at 37°C, DNase treated for 15 min at 37°C, then the reaction was stopped by addition of 1 µl 0.5 M EDTA. Buffer components and excess NTPs were removed by gel filtration using NucAway Spin Columns (Ambion). Hybridization was carried out in UltraHyb (Ambion) medium overnight at 68°C. Blots were washed using NorthMax Low Stringency Buffer (Ambion) at room temperature (2 × 5 min) and with NorthMax High Stringency Buffer (Ambion) at 68°C (2 × 60 min). Northern blots were developed using alkaline phosphatase and streptavidin according to manufacturer’s protocols (BrightStarTM BioDetect, Ambion).
Quantitative RT–PCR
Primer/probe sets have been designed for C. trachomatis ompA, ftsI and CTIG270 using Primer Express software (Applied Biosystems). Standard curves were performed for each gene using purified chromosomal template DNA at concentrations ranging from 10 to 0.001 ng/ml (data not shown). Assays were performed (Universal PCR System, Applied Biosystems) using DNA preparations of a portion of the sample for RNA purifications that were used for northern blotting. Bacterial genome copy numbers were estimated by converting triplicate measurements of mean critical threshold (Ct) values to DNA concentrations (using Standard curves) and converting concentrations to copy numbers using the calculated molecular mass of the bacterial genome (7000 Sequence Detector, ABI Prism).
Functional analysis of CTIG270 in an E. coli surrogate system
The ftsI gene was fused in frame to a 5′-FLAG-tag sequence (N-DYKDDDDK-C) and cloned under the control of a plac promoter (from pCR2.1, Invitrogen) The FLAG-TAG was added using PCR with oligonucleotides containing the motif. Cloning was carried out in a promoterless pSMART-LCAmp (Lucigen, WI, USA). PCR was used to add an FseI site to the 3′-end and 5′-end of the promoter and the FLAG-tagged ftsI, respectively. Both amplicons were digested with FseI followed by ligation. The fusion product was then amplified by PCR and blunt end cloned into pSMART-LCAmp. CTIG270 was cloned under the control of an arabinose responsive promoter (pBAD) in pRANGER-BTB vector (Lucigen). Escherichia coli transformed with both plasmids was induced with IPTG and/or arabinose. Escherichia coli was harvested and analysed using northern blotting for both mRNA and ncRNA.
Escherichia coli transformed with both plasmids was grown to an OD600 of 0.6. The culture was split into three tubes. One tube was induced with arabinose (0.2%), the second tube was induced with IPTG (1 mM) and the third one was uninduced. All three tubes were incubated at 37°C on a shaker for 2.5 h. A sample was collected from each test condition and cells were centrifuged. Bacterial cells were used to prepare RNA (MasterPure, Epicentre) for northern blotting and protein for western blotting. Following sample collection each culture tube received the second inducer. The first tube was induced with IPTG (1 mM), the second tube was induced with arabinose (0.2%) and the third tube was induced with both IPTG (1 mM) and arabinose (0.2%). The culture tubes were incubated at 37°C on a shaker for 2.5 h and a second sample was collected from each tube as previously mentioned. Culture tubes were incubated at 37°C on a shaker for 2.5 h and the third sample was collected as previously mentioned. RNA and protein samples were resolved by gel electrophoreses and loading was normalized using OD600 measurements. Protein samples were resolved on a NuPAGE Bis–Tris gel and transferred onto a PVDF membrane according to manufactures instructions (Invitrogen). Western blotting was carried out using the ProteoQwest™ FLAG Chemiluminescent western blotting kit (SIGMA-ALDRICH, MO, USA). RNA samples used for CTIG270 detection were resolved on a 10% Urea PAGE gels, while RNA samples for ftsI detection were resolved on a 1% formaldehyde agarose gel (NorthernMaxTM, Ambion). RNA was then transferred to a Zeta probe GT Nylon membranes according to manufacturer’s instructions (BIORAD). Northern blotting was carried out using biotinylated RNA probes specific to CTIG270 and ftsI.
RESULTS AND DISCUSSION
Design, construction and testing of the chlamydial intergenic microarray
Using intergenic microarrays to determine the expression of ncRNAs represents a direct approach to screen for ncRNAs. We have designed and constructed a GeneChip Custom Affymetrix microarray to determine the expression levels of ORFs and untranslated elements in intergenic regions (IGRs) of trachomatis D (strain UW-3/CX) based on the genomic sequence (24). The intergenic regions of C. trachomatis D genome that are 50 bp or more are represented on the microarray as 25 nt probes tiled head to tail and covering the intergenic regions on both strands. The MICROBExpressTM protocol was chosen to enrich for bacterial mRNA and ncRNA based on its superior signal to noise ratio compared with other enrichment protocols (data not shown). Equivalent quantities of RNA were used and processed according to the Expression and Analysis Technical Manual, Prokaryotic Probe Processing (Affymetrix).
Criteria for identification of C. trachomatis ncRNAs
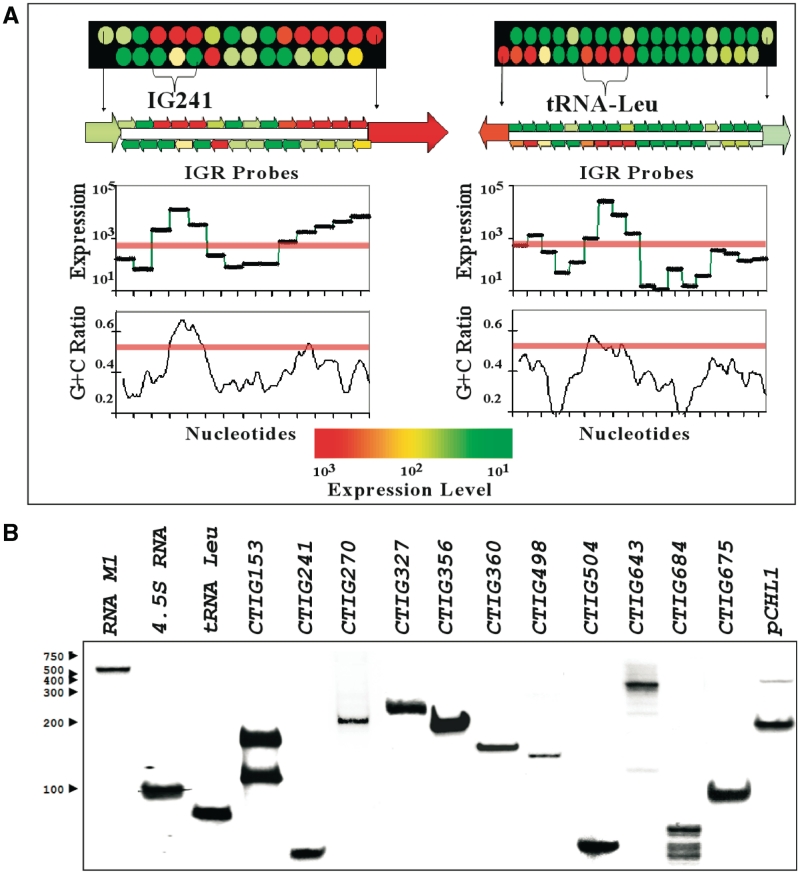
Potential ncRNAs were named using the IGR number corresponding to the downstream ORF using the CT numbers defined by Stephens et al. (24), e.g. an ncRNA located upstream of CT241 is referred to as CTIG241. Chlamydia trachomatis D RNA collected from 40 h PI samples was processed and used to characterize the expression of ncRNAs. Identification of potential ncRNAs was based on a number of criteria including: (i) sequential probes showing simultaneous expression (three or more), (ii) high levels of fluorescence (more than 1000), (iii) G+C ratio of the expressed region (elevated G+C ratios were noted in many of the original molecules that tested positive by northern blotting), (iv) presence of Rho-independent terminators at the presumed end of the ncRNA (not an absolute requirement) and (v) genomic conservation with other chlamydial species (not an absolute requirement). An example of this type of analysis is shown in Figure 1A in which the expression of a potential ncRNA (CTIG241) is compared with expression of tRNA-Leu-2. The IGRs and flanking ORFs are shown with corresponding expression levels and G+C content. Housekeeping ncRNAs were used as a validation tool for the microarray analyses as shown in Figure 1B. These data are shown in Table 1. We were able to detect all 37 tRNAs predicted in the genome of C. trachomatis D (including the tRNA-Leu-2 gene in Figure 1A). In addition, a number of highly conserved ncRNAs found in other bacterial genera were also detected. These included: (i) tmRNA, associated with the release of stalled ribosomes and the incorporation of a short peptide tag that targets the partial protein product for degradation (25), (ii) 4.5S RNA which forms part of a ribonucleoprotein complex that plays a role in protein secretion (26) and (iii) M1 RNA, which forms the catalytic subunit of RNase P, involved in 5′-tRNA processing (27). Chlamydia trachomatis D expressed 34 other intergenic sequences that met the criteria for detection of ncRNA by microarray analysis (Table 1). These included ncRNAs that could be classified as cis-acting anti-sense molecules in that the expressed transcript overlapped the 3′-end of a transcript from an annotated gene in the opposite direction [e.g. CTIG270 ncRNA and ftsI gene, CTIG153 ncRNA and CT152 (lolD)] and the previously described anti-sense transcripts from the cryptic plasmid pCHL1 (21,28). Intergenic, potential ncRNA were also found that fit into the category of trans-acting molecules, eight of which are shown in Figure 1B (CTIG241, 327, 356, 360, 498, 504, 643 and 684) and the remainder are listed in Table 1 (16 predicted ncRNAs were confirmed by northern blot analysis, as listed in Table 1). The latter category included the recently described trans-acting ncRNA from C. trachomatis, IhtA (15), which has been shown to translationally silence expression of the chlamydial histone HctA in a heterologous E. coli co-expression system. Sequences representing predicted ncRNAs reported in Table 1 were checked for putative ORFs using GLIMMER ver. 3.02 (29). Sequences were submitted to GLIMMER webserver (http://www.ncbi.nlm.nih.gov/genomes/MICROBES/glimmer_3.cgi). Sequences were also inspected by VectorNTI ver. 11 for start codons (ATG, GTG, TTG) that are in frame with a stop codon (TAA, TGA, TAG) that encoded ORFs >20 amino acids. Both methods showed that the only putative ORF might be located within the plasmid anti-sense transcript, a putative peptide of 57 amino acids with a TTG as its start codon and no identifiable Shine Delgarno upstream of the start codon. Based on these results, there is strong evidence that ncRNAs in Table 1 do not represent un-annotated ORFs.
Figure 1.
Identification of potential ncRNAs. (A) Criteria for identification involved the expression levels of a contiguous intergenic regions, G+C ratio and position with respect to flanking ORFs. Shown here is a comparison of the CTIG241 potential ncRNA and the tRNA-Leu. Threshold levels for inclusion are shown as pink bars across the tiled intergenic regions. Each of the circles in the upper panel indicate separate 25-mer probes for the upper and lower strands and their expression levels. The flanking ORFs are indicated as a single circle on one strand according to their orientation. (B) Northern blotting of potential ncRNAs from C. trachomatis at 40 h PI. Housekeeping small RNAs (RNase P, 4.5S RNA and tRNA-Leu-2) and several novel (CTIG153, 241, 270, 327, 356, 360, 498, 504, 643 and 684) and previously described IhtA (15) and plasmid anti-sense transcripts (28) RNAs are shown. Chlamydial ncRNAs were probed with strand-specific, biotin labeled probes using equivalent amounts of non-enriched RNAs.
Table 1.
Results of the late cycle intergenic microarray experiment showing all known ncRNAs [tRNA, tmRNA, 4.5SRNA, 5SRNA, M1RNA, ihtA(15) and pCHL1 plasmid anti-sense transcripts(21,28)] in addition to potential ncRNAs (NA, not attempted)
| Name | Approximate size | Classa | A.F.U. | Northern |
Deep sequencing (21) |
|
|---|---|---|---|---|---|---|
| Seq | Northern | |||||
| 37 tRNAs | 75–100 | tRNA | 25 927 | NA | √ | NA |
| tmRNA | 525 | PI | 24 598 | NA | √ | Positive |
| 4.5S RNA | 100 | PI | 15 917 | Positive | √ | Positive |
| 5S rRNA | 125 | Ribosomal | 32 001 | NA | √ | NA |
| M1 RNA | 400 | PI | 20 235 | Positive | √ | NA |
| ihtA | 150 | trans/3′-UTR | 9938 | Positive | √ | Positive |
| pCHL1 | 500/220 | Cis | NA | Positive | √ | Positive |
| CTIG153 | 150/100 | Cis | 884 | Positive | √ | Positive |
| CTIG241 | 75 | Trans | 6073 | Positive | √ | Positive |
| CTIG270 | 100 | Cis | 994 | Positive | √ | NA |
| CTIG327 | 250 | Trans | 1926 | Positive | − | NA |
| CTIG356 | 225 | trans/UTR | 7238 | Positive | √ | Positive |
| CTIG360 | 200 | trans/Riboswitch | 4533 | Positive | − | NA |
| CTIG498 | 100 | trans/3′-UTR | 3976 | Positive | − | NA |
| CTIG504 | 75 | trans/3′-UTR | 3959 | Positive | − | NA |
| CTIG582 | 200 | Trans | 2135 | Positive | − | NA |
| CTIG643 | 650 | Trans | 7279 | Positive | √ | NA |
| CTIG684 | 100 | Trans | 1420 | Positive | √ | Negative |
| CTIG805 | 200 | Trans | 5677 | Positive | √ | Negative |
| CTIG809 | 125 | Trans | 748 | Positive | − | NA |
| CTIG857 | 225 | Trans | 772 | Positive | − | NA |
| CTIG001 | 125 | Trans | 1396 | Negative | − | NA |
| CTIG059 | 50 | Trans | 1281 | Negative | − | NA |
| CTIG072 | 175 | Trans | 2780 | Negative | − | NA |
| CTIG181 | 75 | Trans | 4001 | Negative | − | NA |
| CTIG195 | 200 | Trans | 1852 | Negative | − | NA |
| CTIG237 | 325 | Trans | 6378 | Negative | − | NA |
| CTIG256 | 150 | Trans | 1941 | Negative | − | NA |
| CTIG268 | 175 | trans/5′-UTR | 5765 | Negative | − | NA |
| CTIG323 | 225 | Trans | 4479 | Negative | √ | NA |
| CTIG433 | 75 | Trans/5′-UTR | 2723 | Negative | − | NA |
| CTIG442 | 200 | Trans/3′UTR | 2339 | Negative | − | NA |
| CTIG444 | 125 | Cis | 3620 | Negative | − | NA |
| CTIG449 | 125 | Trans | 4248 | Negative | √ | NA |
| CTIG592 | 150 | Trans | 1335 | Negative | − | NA |
| CTIG660 | 125 | trans/3′-UTR | 14 347 | Negative | − | NA |
| CTIG663 | 250 | Trans | 2784 | Negative | − | NA |
| CTIG775 | 175 | Trans | 2877 | Negative | − | NA |
| CTIG813 | 225 | Trans | 1364 | Negative | − | NA |
| CTIG864 | 125 | Trans | 19 393 | Negative | − | NA |
Approximate sizes are based on the array (except for plasmid anti-sense transcript), the potential functional class, the average fluorescence of the probes representing the ncRNA.
aClass of ncRNA refers to its presumed mode of action. PI is protein interaction.
A.F.U., average fluorescence units.
Characterization of ncRNAs in C. trachomatis D
A subset of ncRNAs (10) was chosen for further analysis based on their expression levels at 24 and 48 h PI and expression differences during IFN-γ-induced persistence. These were compared with the previously described IhtA (15) and the pCHL1.2 plasmid anti-sense transcripts (21,28). These included cis-acting ncRNAs (CTIG153, 270 and pCHL1.2), potential trans-acting ncRNAs [CTIG241, 327, 356, 498, 504, 643, 675 (IhtA) and 684] and a potential riboswitch-like ncRNA (CTIG360)
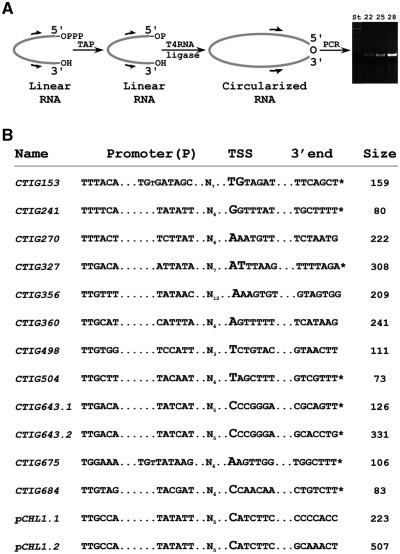
We determined the 5′- and 3′-ends of each molecule using an RNA circularization assay previously developed to assess the poly-A status of RNAs (23,30). A schematic representation of the procedure is shown in Figure 2A. Briefly, total RNA of a 24 h PI sample was treated with tobacco acid pyrophosphatase (to convert primary triphosphate to monophosphate groups) and subjected to end circularization by T4 RNA ligase. Guided by the intergenic microarray results we designed primers for cDNA synthesis of circularized RNA. After cDNA synthesis, junction fragments were amplified by PCR (Figure 3A), cloned and sequenced (10 clones per reaction). This analysis revealed the transcription start sites (TSS in Figure 2B) for all 12 ncRNAs, including those with more than one species (CTIG643.1/2 and pCHL1.1/2, each pair having the same 5′-end). The smaller fragment of CTIG153 could not be determined due to the constraints on primer design presented by the small size of the transcript. The 3′-end of each of the transcripts was also determined. The 3′-ends marked with an asterisk in Figure 3B were determined to have non-templated additions that were of variable sizes. Generally, specific transcripts had between 3–5 added nucleotides but the largest found was 27 nt. Although the additions were predominantly poly-A, several non-templated additions contained G residues.
Figure 2.
Determination of the 5′- and 3′-ends of chlamydial ncRNAs using an RNA circularization assay (23). (A) Schematic representation of the RNA circularization procedure, beginning with the removal of the 5′-pyrophosphate using tobacco acid phosphatase (TAP) followed by circularization using T4 RNA ligase. Primers were then designed to amplify the 5′/3′ junction. (B) The 5′- and 3′-ends of the ncRNAs determined in this study. The 5′-end is designated the TSS and the 3′-end and overall size of the ncRNAs is listed. ncRNAs that contained non-templated additions at the 3′-end (primarily poly-A additions of different lengths) are indicated by an asterisk. Promoter predictions were made by examination of the areas immediately upstream of the TSS. All of the predicted promoters were of the σ66 (major sigma factor) type and two had an extended −10 sequence.
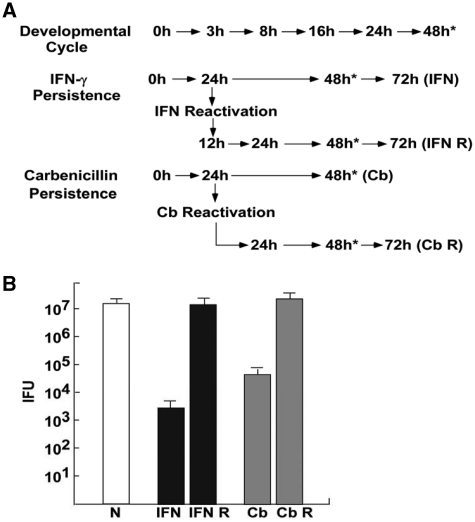
Figure 3.
The chlamydial developmental cycle and persistence induction and reactivation using IFN-γ and carbenicillin. (A) Samples were collected for northern blotting at the times indicated (IFN, IFN-γ persistence; IFN R, IFN-γ reactivation; Cb, carbenicllin persistence; Cb R, carbenicillin reactivation). (B) Titers of samples collected at 48 h PI for each of the experiments (indicated by the asterisks next to time points in Figure 4A). Both persistence induction conditions resulted in significant reductions in IFUs at 48 h PI which returned to near normal IFU numbers following reactivation.
It has been shown that prokaryotes possess two enzymes that can add nucleotides to the 3′-ends of RNA, namely Poly(A) polymerase I (PAP I) (31) and Polynucleotide phosphorylase (PNPase) (32) and that both enzymes were involved in modulating RNA stability (33,34). PAP I, an enzyme belonging to the nucleotidyl-transferase family, adds poly(A) extensions to the 3′-ends of mRNAs, as well as to tRNA and rRNA (35). PNPase not only synthesizes long, heteropolymeric tails in vivo, but also accounts for all of the observed residual polyadenylylation in PAP I deficient strains (36). Polyadenylation of RNAs by PAP I in E. coli plays a significant role in mRNA decay and general RNA quality control. It has been demonstrated that >90% of E. coli ORFs transcribed during exponential growth undergo some degree of polyadenylation by PAP I, either as full-length transcripts or decay intermediates (37). In prokaryotes, polyadenylated RNA molecules are usually degraded more efficiently than non-modified transcripts (38).
The TSSs determined by the RNA circularization procedure were used to predict potential promoters for the ncRNAs (Figure 2B). The promoters identified were all of the σ66 family (39) the major sigma factor in C. trachomatis) but functional characterization of the promoters was not performed and these remain predicted promoter elements at this point. Two of the ncRNA promoters have sequences characterized as encoding ‘extended’ −10 regions (40). Interestingly, one of these is CTIG675 (IhtA) as our determinations indicated that the TSS is located six bases downstream of the TSS previously determined by Grieshaber et al. (15). It is unclear at this point whether this finding is due to technical differences in the methods used or that IhtA may have more than one promoter, although Albrecht et al. (21) identified the same residue as we did for the TSS for IhtA.
Comparison of detection of ncRNAs by intergenic microarray to deep-sequencing
We have compared our intergenic microarray analysis with the recently published study by Albrecht et al. (21) using deep-sequencing. The primary differences between the studies are the strains used and the times at which samples were taken. We used C. trachomatis D (UW-3/Cx) at 40 h PI and they studied C. trachomatis L2b (UCH-1/proctitis) at 24 h PI. The relative strengths and weaknesses of the two techniques are reviewed in Sharma and Vogel (19). One particular difficulty that pertains to both procedures however is the ability to remove the vast excess of host structural RNAs to determine the bacterial transcriptome. In the Albrecht et al. (21) study, the organisms were gradient purified to reduce the host background. This may be somewhat problematic at earlier time points. However, our analyses were restricted to intergenic ncRNAs while the deep-sequencing approach allowed them to detect 25 putative anti-sense ncRNAs that lie within ORFs. We detected only three anti-sense ncRNAs that overlap ORFs based on the fact the anti-sense molecules extended beyond the ORF mRNAs on the opposite strand. Excluding housekeeping RNAs (i.e. tRNAs, tmRNA, 4.5S RNA, 5S RNA and M1 RNA) we found 34 candidate genes, of which 16 were confirmed by northern blotting. The Albrecht et al. (21) study found sequence reads for 11 intergenic ncRNAs we predicted. They confirmed nine of these by northern blotting. Four intergenic ncRNAs predicted by Albrecht et al. (21) were not found in our study. Nine ncRNAs were confirmed by northern blotting by both groups, as shown in Table 1. They detected two molecules by sequencing that were predicted by our criteria but which we could not confirm by northern blotting. The TSS analyses were very similar for molecules examined in both studies, i.e. IhtA and CTIG270 had identical TSS and CTIG684, CTIG241 and CTIG153 differed by 1 nt. The reasons for the differences in ncRNA detection are not clear at this time but may be related to the sampling time PI or the copy number of the predicted ncRNA transcripts.
Developmental expression of ncRNAs in C. trachomatis D
To determine the expression patterns of ncRNAs, we chose to examine (i) expression during the normal developmental cycle, (ii) expression during IFN-γ-induced persistence and reactivation and (iii) expression during carbenicillin-induced persistence and reactivation. We are primarily interested in the expression of ncRNAs during the development of IFN-γ-induced persistence and reactivation. Carbenicillin-induced persistence was included to determine changes that were specific to IFN-γ-induced persistence. DNA/RNA samples for the developmental cycle were taken at 0, 3, 8, 16, 24 and 48 h PI as described previously (8) and shown in Figure 3A. DNA/RNA was collected from IFN-γ-induced persistent samples at 24, 48, 72 h PI and 12, 24, 48, 72 h post reactivation (IFN and IFN R in Figure 3A). IFN-γ was used to induce persistence (5 ng/ml, as determined by a pilot titration experiment) and was added to the media 24 h prior to infection. Samples were reactivated at 24 h PI by changing media with IFN-γ-free media containing excess tryptophan (10) and samples were taken at the times shown in Figure 3A (IFN and IFN R). Carbenicillin-induced persistence was induced by the addition of antibiotic (2 µg/ml, as determined by a pilot titration experiment) at the time of infection. DNA/RNA was collected from carbenicillin-treated samples at 24 and 48 h PI and 24, 48 and 72 h post reactivation (Cb and Cb R in Figure 3A). As shown in Figure 3B, IFN-γ treatment resulted in a reduction of IFUs of approximately four logs at 48 h PI. Following reactivation for 48 h, the IFU values returned to ∼98% of control cultures, indicating that RBs were present but were in a non-dividing or persistent state. Similarly, carbenicillin treatment resulted in a distinct decrease in IFUs at 48 h PI, but following removal of the antibiotic (Cb reactivation), IFU numbers returned to control levels (Figure 3B).
Genome copy numbers for each of the samples was determined by qPCR as previously described (22) and the samples were then thoroughly DNased and RNA was then re-purified. Expression of ncRNAs was determined by northern blotting of RNA samples from equivalent numbers of bacteria (as determined by genomic normalization) for the developmental cycle and carbenicillin persistence. Samples used for IFN-γ persistence contained 20% of the genome numbers used for the other analyses due to the reduced chlamydial biomass associated with growth in the presence of IFN-γ. Samples have been grouped under each condition to allow for direct comparisons between normal and persistent growth conditions. The results in Figure 4 show the ncRNA expression patterns for potential ncRNAs that showed changes in expression during growth in the presence of IFN-γ (the genomic-locus of each ncRNA is also shown). The expression of multiple species of ncRNAs has been previously described for other bacteria (e.g. in E. coli for gadY, (41) and dsrA (42) and is believed to involve post-transcriptional processing that is dependent on the balance between protection by Hfq and degradation by RNase E. The functional significance has not been well-studied but, importantly Davis and Waldor (43) have shown that MicX of Vibro cholerae can be detected as both an active precursor and a processed form and that the smaller, processed molecule is significantly more stable and abundant. Interestingly, processing occurs in a Hfq and RNase E-dependent manner but the regulatory effects of MicX on its target mRNA are not dependent on Hfq. Chlamydia spp. do not contain obvious orthologs of Hfq or RNase E suggesting that regulation and processing may be very different in these organisms.
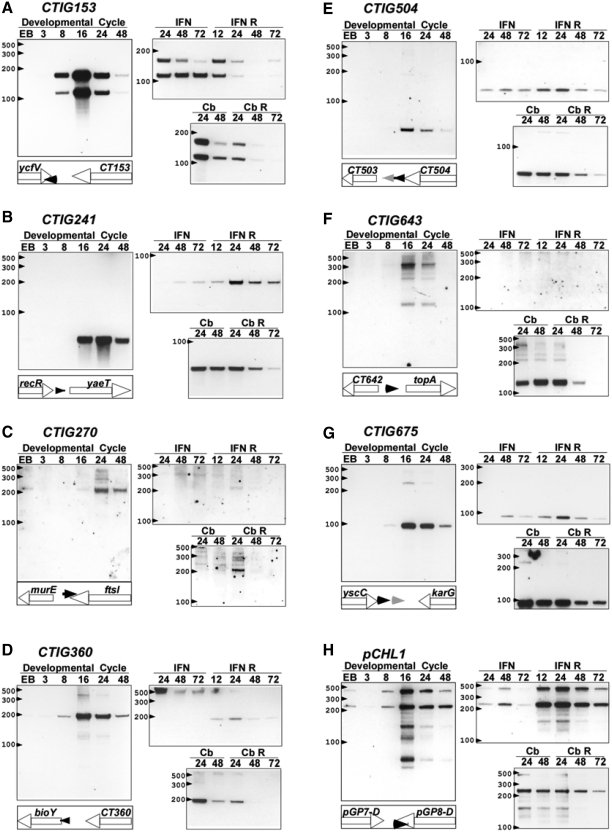
Figure 4.
Developmental expression of ncRNAs in C. trachomatis during the developmental cycle, IFN-γ-induced and carbenicillin-induced persistence and reactivation. Each panel (A–H) shows the expression pattern of the particular putative ncRNA under the conditions described in Figure 3. In addition the genomic position and orientation of the ncRNAs are shown in the schematic panel under the Developmental cycle panel (black triangles are putative ncRNAs and grey triangles are tRNAs).
Expression of ncRNAs during the normal developmental cycle and during IFN-γ-induced and carbenicillin-induced persistence
ncRNAs that showed significant changes in expression during IFN-γ-induced persistence are shown in Figure 4. In each case the results of northern blotting are shown for the complete developmental cycle and compared with expression patterns during the induction of persistence and reactivation. CTIG153 was expressed during the normal developmental cycle from ∼8 to 24 h PI and expression dropped dramatically by 48 h PI. This molecule was expressed as two species of ∼159 nt and 115 nt (Figure 4A). Only the longer species was amenable to RNA circularization analysis as mentioned previously. During IFN-γ treatment the larger species was found at reduced levels and by 72 h PI was nearly undetectable. Reactivation resulted in expression of both molecules in similar quantities and expression of both decreased significantly throughout the reactivation period. Similar finding were found during carbenicillin-induced persistence and reactivation, suggesting that similar processing events occurred during both procedures (Figure 4A). CTIG241 was expressed later in the developmental cycle (∼16–48 h PI) and was barely detectable during IFN-γ-induced persistence. Higher expression levels were found during the reactivation phase. This was distinct from the profile found during carbenicillin-induced persistence where strong expression was found under the influence of the antibiotic that waned during reactivation. The expression of CTIG270 was only detected late in the developmental cycle (24—48 h PI) and the 222 nt species was not detected during IFN-γ-induced persistence or reactivation. During carbenicillin-induced persistence the same species was only detected at 24 h post-reactivation.
The 241 nt CTIG360 molecule was detected from ∼8 to 48 h PI with a peak at 16 h PI (Figure 4D). This molecule overlaps the 5′-UTR of the bioY gene, a predicted ortholog of biotin transporters found in other bacterial species. Interestingly, the 241 nt molecule was not detected during IFN-γ-induced resistance but a higher molecular weight species (>500 nt) was clearly detectable. Following reactivation, the higher molecular weight species disappeared and the 241 nt species reappeared. This expression pattern was not found during carbenicillin-induced persistence. This pattern of expression is similar to that found with other genes or operons regulated by riboswitches but further studies are needed to confirm this. Expression of the very small CTIG504 was determined to be maximal at 16 h PI and expression decreased later in the developmental cycle. Expression was detected at all points during IFN-γ-induced persistence and reactivation while expression was seen during carbenicillin treatment but was reduced following reactivation. CTIG643 expression was detected at 16 and 24 h PI but was not detected during IFN-γ treatment. During carbenicillin treatment the lower molecular weight fragment, expressed at low levels during normal development, was expressed strongly during treatment but at declining levels during reactivation.
Expression of CTIG675 was strongest at 16 and 24 h PI and decreased at 48 h PI. This molecule was only weakly expressed during IFN-γ-induced persistence but was expressed at high levels during carbenicillin treatment. The plasmid anti-sense transcripts were first detected at 8 h PI and were expressed at high levels during the mid-cyle. Expression was barely detectable during IFN-γ treatment but the molecules were expressed at high levels during reactivation. Interestingly, only the lower molecular weight species was detected during carbenicillin treatment and reactivation. These results, although preliminary, argue that expression of this group of putative ncRNAs are developmentally regulated since identical RNA samples were loaded in each corresponding lane [e.g. compare the timing of expression of CTIG153 (Figure 4A) and CTIG241 (Figure 4B)].
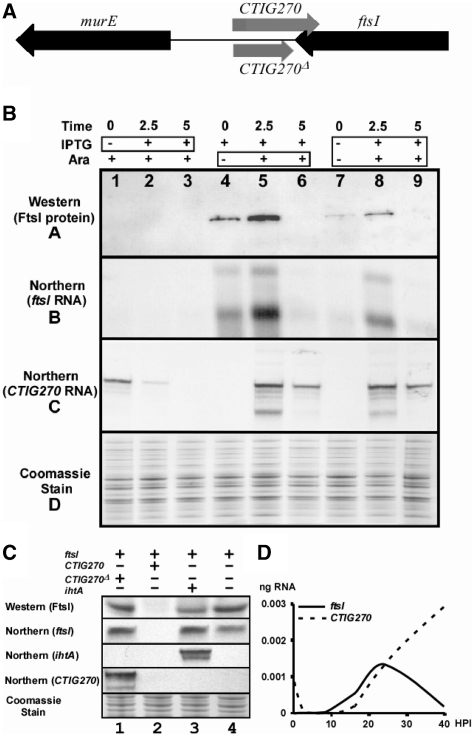
CTIG270 regulates FtsI expression in an E. coli heterologous system
FtsI has been shown to play an important role in peptidoglycan synthesis associated with bacterial cell division in other systems (44). The general block in cell division during IFN-γ-induced persistence (and most other forms of chlamydial persistence) led us to examine the effect of co-expression of these genes in an E. coli system. The anti-sense nature of CTIG270 and the ftsI gene are shown in Figure 5A. The ftsI gene was cloned into the pSMART LCAmp construct containing an in-frame FLAG-tag insert under the control of the lacZ promoter/operator system which expressed a FLAG-tag positive product of the predicted size, i.e. 75 kDa (Figure 5B). The ftsI construct was then co-transformed into the strain containing the inducible CTIG270 construct in a pRANGER-cat. Expression studies were done in the co-transformed strain to determine the effect of CTIG270 expression on FtsI translation (Figure 5). The analysis indicated that when CTIG270 was induced prior to induction of the ftsI target gene, no expression of the FtsI protein or mRNA was seen and that the CTIG270 transcript decreased over the induction period, a result that is consistent with digestion of both RNAs following interaction. Induction of the ftsI gene prior to co-induction of CTIG270 showed that FtsI protein and ftsI mRNA were made, followed by a transient increase at 2.5 h of arabinose induction [probably due to transient cross-induction of the lacZ promoter, as reported previously (45)] and complete disappearance of the protein and mRNA in the presence of the ncRNA for 5 h. Co-induction of both genes resulted in some expression of FtsI, ftsI mRNA and CTIG270 at 2.5 h, but at 5 h the ftsI mRNA (and therefore the FtsI protein) were not found. These results are consistent with a base-pairing interaction of the ncRNA with the target gene resulting in RNA degradation.
Figure 5.
Analysis of the co-expression of the chlamydial ncRNA CTIG270 and the chlamydial ftsI gene in E. coli. (A) A schematic showing the location of CTIG270 and flanking genes. (B) Results of co-expression assays. Samples were prepared using different co-induction protocols and samples were tested for the presence of the FtsI protein and mRNA and CTIG270. The western blot was developed with an α-Flag-tag antibody. The same samples were run on SDS-Page gels as loading controls and stained with Coomassie Blue. Northern blots for ftsI and CTIG270 were done from RNA preparations from the same samples. The co-induction analysis is shown at the top of the figure. The first three lanes show the results when stimulation with arabinose for 5 h is followed by the addition of IPTG for different times (boxed region) as indicated. In the second set of three lanes, IPTG induction for 5 h was followed by induction with arabinose (boxed region) for the times indicated. In the third set of three lanes induction of both the ncRNA and ftsI were stimulated with IPTG and arabinose (boxed region) for the times indicated. The results indicated that expression of CTIG270 results in the degradation of ftsI mRNA. (C) Co-expression controls in which the lanes shown are analogous to lane 9 in Figure 5A in that both plasmids are co-induced for the same length of time. A truncated form of CTIG270 (CTIG270Δ), which does not overlap the 3′-UTR of ftsI, is expressed in lane 1 but has no influence on the expression of FtsI. Expression of full-length CTIG270 however results in the degradation both ftsI mRNA and CTIG270 (Figure 5C, lane 2 and as shown in Figure 5A). Expression of an unrelated ncRNA (IhtA) that acts on hctA mRNA (15) also has very little effect on the expression of FtsI (lane 3). Expression of FtsI was not affected by the presence of the empty vector pRANGER BTB (lane 4). (D) Quantitative RT-PCR analysis of the expression profiles of ftsI and CTIG270 throughout the developmental cycle. Measurements were made in triplicate for each time point.
Control experiments are shown in Figure 5C, in which samples are co-induced as shown for lane 9 in Figure 5B. Expression of a truncated form of CTIG270, named CTIG270Δ is shown in lane 1. This ncRNA lacks the region of CTIG270 that overlaps the 3′-UTR of ftsI. Expression of this product does not result in the degradation of ftsI mRNA and expression of FtsI is apparent. Expression of full length CTIG270 (lane 2, Figure 5C) results in degradation of both ftsI mRNA and the CTIG270 ncRNA, as shown previously in Figure 5B. Expression of an unrelated ncRNA (ihtA) is shown in lane 3 and there is a very minimal effect on ftsI mRNA and FtsI protein expression. We believe the slightly lower amounts of FtsI protein may be due to the general cytotoxic effect of ihtA expression (15). Expression of FtsI protein is unaffected in the presence of the empty vector used to express ncRNAs (Figure 5C, lane 4).
The expression of ftsI and CTIG270 were measured during the chlamydial developmental cycle using qRT–PCR with Taqman primer probe sets. In each case the triplicate measurements were normalized to chlamydial genome copy numbers as described in the ‘Materials and Methods’ section. The results indicate that ftsI mRNA can first be detected ∼10–12 h PI and peaks at ∼24 h PI. After this point the levels of ftsI mRNA decrease to nearly undetectable levels by 40 h PI. The expression of CTIG270 follows ftsI expression by several hours and increases throughout the cycle. These data are consistent with the predicted degradation of ftsI mRNA by CTIG270 after ∼24 h PI. This does not imply that cell division stops at this point, as FtsI protein synthesized during the 10–24–h PI portion of the cycle should not be affected by CTIG270 expression. Instead, the regulatory process appears to allow for maximum FtsI expression during the period of rapid cell division by RBs.
CONCLUSIONS
Chlamydia trachomatis D/UW-3/Cx expresses numerous ncRNA that can be initially detected using an intergenic microarray and validated by northern blotting analyses. These include the subset of potential ncRNAs in this study which are potential cis-acting (anti-sense) and trans-acting ncRNAs in addition to a non-coding 5′-UTR of the bioY gene that structurally resembles a riboswitch. We have shown that these ncRNAs have distinct temporal expression patterns during the normal developmental cycle and several appear to have processing steps that result in the expression of molecules of different molecular weights. We have also shown that the expression of many ncRNAs is altered during growth conditions that induce persistent growth, particularly IFN-γ-induced persistence and carbenicillin-induced-persistence. These studies of ncRNAs in C. trachomatis provide a strong basis for mechanistic studies that may determine a role for ncRNA regulation in the development and persistence of C. trachomatis.
FUNDING
National Institutes of Health (AI 070693 to R.J.B.). Funding for open access charge: National Institutes of Health (R01 070693).
REFERENCES
- 1.Thylefors B, Negrel AD, Pararajasegaram R, Dadzie KY. Global data on blindness. Bull. World Health Organ. 1995;73:115–121. [PMC free article] [PubMed] [Google Scholar]
- 2.Gerbase AC, Rowley JT, Mertens TE. Global epidemiology of sexually transmitted diseases. Lancet. 1998;351(Suppl. 3):2–4. doi: 10.1016/s0140-6736(98)90001-0. [DOI] [PubMed] [Google Scholar]
- 3.Ward ME. In: Chlamydia: Intracellular Biology, Pathogenesis, and Immunity. Stephens RS, editor. Washington, D.C.: ASM Press; 1999. pp. 171–210. [Google Scholar]
- 4.Moulder JW. Interaction of chlamydiae and host cells in vitro. Microbiol. Rev. 1991;55:143–190. doi: 10.1128/mr.55.1.143-190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hybiske K, Stephens RS. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc. Natl Acad. Sci. USA. 2007;104:11430–11435. doi: 10.1073/pnas.0703218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abdelrahman YM, Belland RJ. The chlamydial developmental cycle. FEMS Microbiol. Rev. 2005;29:949–959. doi: 10.1016/j.femsre.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Hogan RJ, Mathews SA, Mukhopadhyay S, Summersgill JT, Timms P. Chlamydial persistence: beyond the biphasic paradigm. Infect. Immun. 2004;72:1843–1855. doi: 10.1128/IAI.72.4.1843-1855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belland RJ, Zhong G, Crane DD, Hogan D, Sturdevant D, Sharma J, Beatty WL, Caldwell HD. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc. Natl Acad. Sci. USA. 2003;100:8478–8483. doi: 10.1073/pnas.1331135100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicholson TL, Olinger L, Chong K, Schoolnik G, Stephens RS. Global stage-specific gene regulation during the developmental cycle of Chlamydia trachomatis. J. Bacteriol. 2003;185:3179–3189. doi: 10.1128/JB.185.10.3179-3189.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belland RJ, Nelson DE, Virok D, Crane DD, Hogan D, Sturdevant D, Beatty WL, Caldwell HD. Transcriptome analysis of chlamydial growth during IFN-gamma-mediated persistence and reactivation. Proc. Natl Acad. Sci. USA. 2003;100:15971–15976. doi: 10.1073/pnas.2535394100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Douglas AL, Hatch TP. Expression of the transcripts of the sigma factors and putative sigma factor regulators of Chlamydia trachomatis L2. Gene. 2000;247:209–214. doi: 10.1016/s0378-1119(00)00094-9. [DOI] [PubMed] [Google Scholar]
- 12.Yu HH, Tan M. Sigma28 RNA polymerase regulates hctB, a late developmental gene in Chlamydia. Mol. Microbiol. 2003;50:577–584. doi: 10.1046/j.1365-2958.2003.03708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fahr MJ, Douglas AL, Xia W, Hatch TP. Characterization of late gene promoters of Chlamydia trachomatis. J. Bacteriol. 1995;177:4252–4260. doi: 10.1128/jb.177.15.4252-4260.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niehus E, Cheng E, Tan M. DNA supercoiling-dependent gene regulation in Chlamydia. J. Bacteriol. 2008;190:6419–6427. doi: 10.1128/JB.00431-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grieshaber NA, Grieshaber SS, Fischer ER, Hackstadt T. A small RNA inhibits translation of the histone-like protein Hc1 in Chlamydia trachomatis. Mol. Microbiol. 2006;59:541–550. doi: 10.1111/j.1365-2958.2005.04949.x. [DOI] [PubMed] [Google Scholar]
- 16.Grieshaber NA, Sager JB, Dooley CA, Hayes SF, Hackstadt T. Regulation of the Chlamydia trachomatis histone H1-like protein Hc2 is IspE dependent and IhtA independent. J. Bacteriol. 2006;188:5289–5292. doi: 10.1128/JB.00526-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottesman S. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 2005;21:399–404. doi: 10.1016/j.tig.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 18.Storz G, Opdyke JA, Zhang A. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 2004;7:140–144. doi: 10.1016/j.mib.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 19.Sharma CM, Vogel J. Experimental approaches for the discovery and characterization of regulatory small RNA. Curr. Opin. Microbiol. 2009;12:536–546. doi: 10.1016/j.mib.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 20.Huttenhofer A, Vogel J. Experimental approaches to identify non-coding RNAs. Nucleic Acids Res. 2006;34:635–646. doi: 10.1093/nar/gkj469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albrecht M, Sharma CM, Reinhardt R, Vogel J, Rudel T. Deep sequencing-based discovery of the Chlamydia trachomatis transcriptome. Nucleic Acids Res. 2009;38:868–877. doi: 10.1093/nar/gkp1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ouellette SP, Hatch TP, AbdelRahman YM, Rose LA, Belland RJ, Byrne GI. Global transcriptional upregulation in the absence of increased translation in Chlamydia during IFNgamma-mediated host cell tryptophan starvation. Mol. Microbiol. 2006;62:1387–1401. doi: 10.1111/j.1365-2958.2006.05465.x. [DOI] [PubMed] [Google Scholar]
- 23.Vogel J, Hess WR. Complete 5′ and 3′ end maturation of group II intron-containing tRNA precursors. RNA. 2001;7:285–292. doi: 10.1017/s1355838201001960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 25.Withey JH, Friedman DI. A salvage pathway for protein synthesis: tmRNA and trans-translation. Annu. Rev. Microbiol. 2003;57:101. doi: 10.1146/annurev.micro.57.030502.090945. [DOI] [PubMed] [Google Scholar]
- 26.Poritz MA, Bernstein HD, Strub K, Zopf D, Wilhelm H, Walter P. An E. coli ribonucleoprotein containing 4.5S RNA resembles mammalian signal recognition particle. Science. 1990;250:1111–1117. doi: 10.1126/science.1701272. [DOI] [PubMed] [Google Scholar]
- 27.Kleineidam RG, Pitulle C, Sproat B, Krupp G. Efficient cleavage of pre-tRNAs by E. coli RNAse P RNA requires the 2′-hydroxyl of the ribose at the cleavage site. Nucleic Acids Res. 1993;21:1097–1101. doi: 10.1093/nar/21.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fahr MJ, Sriprakash KS, Hatch TP. Convergent and overlapping transcripts of the Chlamydia trachomatis 7.5-kb plasmid. Plasmid. 1992;28:247–257. doi: 10.1016/0147-619x(92)90056-g. [DOI] [PubMed] [Google Scholar]
- 29.Salzberg SL, Delcher AL, Kasif S, White O. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 1998;26:544–548. doi: 10.1093/nar/26.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Urban JH, Vogel J. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 2008;6:e64. doi: 10.1371/journal.pbio.0060064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paschal BM, McReynolds LA, Noren CJ, Nichols NM. RNA polymerases. Curr. Protoc. Mol. Biol. 2008 doi: 10.1002/0471142727.mb0308s84. Chapter 3, Unit 3 8. [DOI] [PubMed] [Google Scholar]
- 32.Grunberg-Manago M, Oritz PJ, Ochoa S. Enzymatic synthesis of nucleic acidlike polynucleotides. Science. 1955;122:907–910. doi: 10.1126/science.122.3176.907. [DOI] [PubMed] [Google Scholar]
- 33.Carpousis AJ, Vanzo NF, Raynal LC. mRNA degradation: a tale of poly(A) and multiprotein machines. Trends Genet. 1999;15:24–28. doi: 10.1016/s0168-9525(98)01627-8. [DOI] [PubMed] [Google Scholar]
- 34.Carpousis AJ, Khemici V, Ait-Bara S, Poljak L. Co-immunopurification of multiprotein complexes containing RNA-degrading enzymes. Methods Enzymol. 2008;447:65–82. doi: 10.1016/S0076-6879(08)02204-0. [DOI] [PubMed] [Google Scholar]
- 35.Li Z, Pandit S, Deutscher MP. Polyadenylation of stable RNA precursors in vivo. Proc. Natl Acad. Sci. USA. 1998;95:12158–12162. doi: 10.1073/pnas.95.21.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohanty BK, Kushner SR. Polynucleotide phosphorylase functions both as a 3′ right-arrow 5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc. Natl Acad. Sci. USA. 2000;97:11966–11971. doi: 10.1073/pnas.220295997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mohanty BK, Kushner SR. The majority of Escherichia coli mRNAs undergo post-transcriptional modification in exponentially growing cells. Nucleic Acids Res. 2006;34:5695–5704. doi: 10.1093/nar/gkl684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jasiecki J, Wegrzyn G. Growth-rate dependent RNA polyadenylation in Escherichia coli. EMBO Rep. 2003;4:172–177. doi: 10.1038/sj.embor.embor733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan M, Engel JN. Identification of sequences necessary for transcription in vitro from the Chlamydia trachomatis rRNA P1 promoter. J. Bacteriol. 1996;178:6975–6982. doi: 10.1128/jb.178.23.6975-6982.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitchell JE, Zheng D, Busby SJW, Minchin SD. Identification and analysis of ‘extended -10' promoters in Escherichia coli. Nucleic Acids Res. 2003;31:4689–4695. doi: 10.1093/nar/gkg694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Opdyke JA, Kang JG, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 2004;186:6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Repoila F, Gottesman S. Signal transduction cascade for regulation of RpoS: temperature regulation of DsrA. J. Bacteriol. 2001;183:4012–4023. doi: 10.1128/JB.183.13.4012-4023.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davis BM, Waldor MK. RNase E-dependent processing stabilizes MicX, a Vibrio cholerae sRNA. Mol. Microbiol. 2007;65:373–385. doi: 10.1111/j.1365-2958.2007.05796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Botta GA, Park JT. Evidence for involvement of penicillin-binding protein 3 in murein synthesis during septation but not during cell elongation. J. Bacteriol. 1981;145:333–340. doi: 10.1128/jb.145.1.333-340.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Narayanan N, Hsieh MY, Xu Y, Chou CP. Arabinose-induction of lac-derived promoter systems for penicillin acylase production in Escherichia coli. Biotechnol. Prog. 2006;22:617–625. doi: 10.1021/bp050367d. [DOI] [PubMed] [Google Scholar]